Abstract
We report the crystal structure of Thermus aquaticus DNA polymerase I in complex with an inhibitory Fab, TP7, directed against the native enzyme. Some of the residues present in a helical conformation in the native enzyme have adopted a γ turn conformation in the complex. Taken together, structural information that describes alteration of helical structure and solution studies that demonstrate the ability of TP7 to inhibit 100% of the polymerase activity of the enzyme suggest that the change in conformation is probably caused by trapping of an intermediate in the helix-coil dynamics of this helix by the Fab. Antibodies directed against modified helices in proteins have long been anticipated. The present structure provides direct crystallographic evidence. The Fab binds within the DNA binding cleft of the polymerase domain, interacting with several residues that are used by the enzyme in binding the primer:template complex. This result unequivocally corroborates inferences drawn from binding experiments and modeling calculations that the inhibitory activity of this Fab is directly attributable to its interference with DNA binding by the polymerase domain of the enzyme. The combination of interactions made by the Fab residues in both the polymerase and the vestigial editing nuclease domain of the enzyme reveal the structural basis of its preference for binding to DNA polymerases of the Thermus species. The orientation of the structure-specific nuclease domain with respect to the polymerase domain is significantly different from that seen in other structures of this polymerase. This reorientation does not appear to be antibody-induced and implies remarkably high relative mobility between these two domains.
TP7 is an antibody that has been used as a thermolabile inhibitor in hot-start variations of PCR (1) by using Thermus aquaticus DNA polymerase I (TaqP) as the enzymatic component. Binding and inhibition studies (2) and a recent computational modeling study (3) have shown that inhibitory activity of TP7 and its Fab (TP7) is probably caused by its interaction with TaqP residues necessary for binding the primer:template complex. We present structural evidence for this hypothesis in a crystal structure of TaqP complexed with TP7. The structure is also of additional significance in demonstrating the mechanism of polymerase inhibition by entities that are not substrate analogs. The structure of TaqP including the polymerase (pol) and the structure specific nuclease (nuc) domains (4), that of the pol domain alone (5), and that of TaqP complexed with a primer:template complex (6) all have been reported. The structure of the native enzyme in a different crystal form (7) also has been determined (H.M.K.M., unpublished work). Structure of unliganded TP7, which recognizes and binds to native TaqP or the pol domain but does not recognize any peptides derived from the pol domain is, likewise, available (2, 3). In the structure of the complex, there is considerable distortion of several residues of TaqP from their native helical conformation. These conformationally perturbed residues are part of the assembled antigenic epitope that interacts with the antibody paratope. These observations strongly suggest that TP7 is directed against an altered native form of TaqP. Structural and binding studies have suggested that some antibodies could be directed against altered native forms of an injected antigen (11, 31–34). In particular, work on Themiste zostericola myohemerythrin has shown that mAbs induced by both protein and pertinent peptide react much more strongly with unwound forms of helix C in that protein and has shown how an antipeptide antibody can recognize an altered native form of a protein (9, 12). However, this report presents the direct observation of a complex of a protein antigen with an Fab directed against and recognizing an altered helical structure. Moreover, although large, segmental motion in DNA polymerases has been documented, especially as an accompaniment to substrate binding (4–6), the unwinding of almost two turns of an α helix observed in this structure is unprecedented.
MATERIALS AND METHODS
Crystals were obtained by using TaqP (30) purified by crystallization (7) and TP7 purified as reported (2). Crystals of TaqP, washed in 20 mM Tris⋅HCl (pH 7.4) and 47% saturated ammonium sulfate, were dissolved in and extensively dialyzed against ammonium sulfate free buffer. The protein then was concentrated to 0.5 mg/ml, was mixed with a stoichiometric equivalent of TP7 Fab, and was left standing over 2–3 days at 4°C. The complex was concentrated to 5 mg/ml, and crystals were grown from 22% PEG3350 solutions in 200 mM Tris⋅HCl (pH 7.4) and 0.1% NaN3 at 20°C by using vapor diffusion in hanging drops. The drops were made up by mixing equal volumes of protein and well solutions. A total of 414,722 observations were made on seven crystals (average size = 0.3 × 0.4 × 0.4 mm) on a Siemens X1,000 multiwire area detector (Siemens, Madison, WI) at room temperature and was reduced to 81,111 unique reflections by using xds (13). Space group was determined to be P1 with a = 76.6 Å, b = 89.1 Å, c = 89.3 Å, α = 100.7°, β = 115.3°, and γ = 95.3° with one complex molecule in the cell yielding a solvent content of 69%. Merging statistics are presented in Table 1.
Table 1.
Data measurement and refinement
| Resolution, Å | Reflections* | Rmerge† | 〈I/σ(I)〉 | Rcryst‡ | Rfree§ |
|---|---|---|---|---|---|
| 37.0–4.2 | 95.2 | 0.047 | 12.3 | 0.185 | 0.247 |
| 4.2–3.3 | 90.3 | 0.050 | 11.4 | 0.169 | 0.243 |
| 3.3–2.9 | 90.2 | 0.066 | 12.8 | 0.170 | 0.237 |
| 2.9–2.7 | 90.7 | 0.065 | 10.2 | 0.187 | 0.257 |
| 2.7–2.5 | 87.5 | 0.083 | 9.7 | 0.218 | 0.279 |
| 2.5–2.3 | 75.9 | 0.122 | 8.0 | 0.250 | 0.309 |
| All | 82.0 | 0.074 | 8.2 | 0.189 | 0.253 |
% measured with I > 2σ(I).
∑ (I − 〈I〉)/∑I.
∑ |Fo − Fc|/∑Fo.
For 5% of observed reflexions (4,080) chosen at random over 8–2.3 Å.
The structure was determined by molecular replacement, with data between 8 and 4 Å that had I > 2σI, by using AMORE (14) as implemented in CCP4 (15). Search models were derived from the native structure (4) for TaqP and from the structure of unliganded TP7 (3) for the Fab. Rotational solutions were obtained separately for pol and nuc domains of TaqP and for TP7. The highest rotation peak for TP7 was chosen arbitrarily to fix the origin as it had the largest signal-to-noise ratio with a correlation coefficient of 15.0 (next highest = 8.9), and relative translation vectors for the other two parts were obtained independently. Correlation coefficient for pol was 19.7 (next highest = 11.4), and the R-value was 44.2% (next lowest = 46.4). Corresponding parameters for nuc were 18.8 (12.6) and 44.7 (48.3). Internal consistency was verified by keeping pol, nuc, TP7 + pol, TP7 + nuc, or pol + nuc fixed in turn and finding self-consistent relative translation vectors for other components of the complex. In every case, the top peak was the correct solution. The maximum rms deviation (rmsd) from respective initial translation solutions was 2.4 Å. Translation results were confirmed by using the T2 and phased translation function calculations in CCP4. The molecular replacement solution was verified independently in xplor (16).
Refinement with xplor, using the Engh and Huber parameter set (43), used reflections from 8 to 2.3 Å with I > 2σI; the magnitude of “wa” was set by the check procedure. All model building used o (17). Cross validation with free R-values (18) used 5% (4,080) of reflections chosen randomly over 8–2.3 Å. Patterson correlation refinement (19), as well as rigid body refinement with the elbow angle for Fab varied from 130° to 180°, indicate that there was a substantial change in this angle relative to unliganded TP7. Using a TP7 model with an elbow angle of 146° (compared with 154° for unliganded TP7) gave the lowest R-value of 0.420. This model for TP7, along with the enzyme model from the initial translation function solution, was refined as a collection of six segmented rigid bodies (variable and constant domains of heavy and light chains of TP7, nuc, and pol domains for TaqP), resulting in an R-value of 0.365 for data between 8 and 3 Å. Refinement, using the slow cooling simulated annealing protocol (16), and model building into segment deleted 2fo-fc, 3fo-fc, and fo-fc maps with sigmaa-weighted (20) coefficients against the 2.3-Å data set resulted in a crystallographic R-value of 0.263 and a free R-value of 0.326. Residues in the TaqP-Fab interface were built into Bhat omit maps (21) as were residues 280–310 in TaqP, connecting the pol and nuc domains. Restrained B factor refinement, a limited search for solvent molecules (around the pol domain and Fab), followed by positional refinement at 2.3 Å resulted in the final crystallographic R-value of 0.189 and free R-value of 0.253. Crystallographic and free R-values as a function of resolution are in Table 1.
RESULTS & DISCUSSION
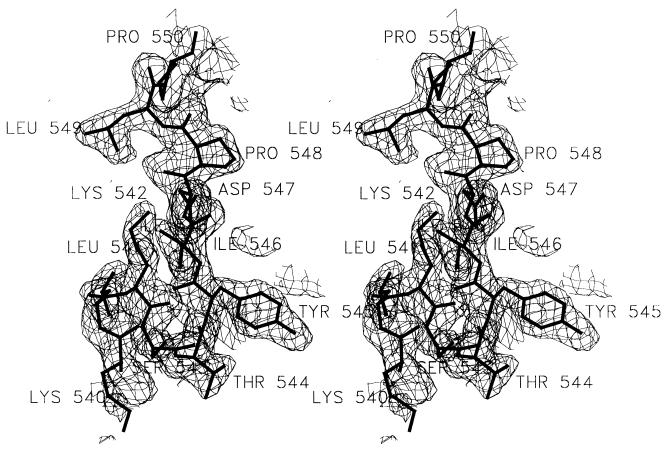
A portion of the electron density map, with the refined model of a part of helix I superimposed, is shown in Fig. 1. Electron density in maps was not clear enough to place residues 792–796 of TaqP as well as atoms beyond Cβ for residues 483–489. Residues 1–4 in the heavy chain of TP7 also could not be positioned unambiguously; density for atoms beyond Cβ for K38 in the light chain was likewise not present. The current model includes 1,246 of 1,255 protein residues and 263 solvent molecules modeled as water oxygens. Only one water molecule is close to the binding interface, mediating an interaction between Y103-OH in the heavy chain and N583 O in TaqP. N583 is a ligand to DNA in a reported TaqP-DNA complex (6). Analysis using procheck (22) indicates that conformational parameters are within limits expected for a structure refined at 2.3 Å with 2.2% of residues in disallowed areas of the Ramachandran (44) map. rmsd is 0.01 Å for bond lengths, 1.6° for bond angles, 1.7° for improper angles, and 4.7 Å2 for B factors between bonded atoms.
Figure 1.
Electron density map with refined model superimposed. Shown is a stereo representation of a 2Fo − Fc map, contoured at 1.1 σ. Phases were generated by removing residues 520–560 in TaqP and refining the structure for 40 cycles with tight geometric restraints. Final refined model for residues 540–550 in TaqP is superimposed. All residues are labeled. The figure was made by using setor (47).
Structure of the Complex.
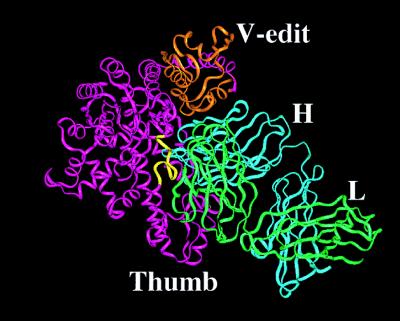
The overall structure of the complex reveals a TP7 Fab conformation close to its unliganded form. Larger but highly localized changes are evident in TaqP. Superposition (45) of the pol domains from the native (4) and complexed structures (441 common Cα atoms) shows that the backbones of the two in regions far from contact with the Fab are very similar with an rmsd of 0.8 Å. Folding of the nuc domain is also very similar (rmsd of 0.6 Å for 187 Cα atoms) to that reported (4), although its orientation relative to the pol domain is remarkably different (Fig. 2a). A 168° rotation of the nuc domain about an axis passing through the centroids of the vestigial editing nuclease (v-edit) domain and the thumb subdomain, with a concomitant unwinding and reorientation of several residues in the connector region (residues 280–310), places the nuc domain in contact with some of the helices in the fingers subdomain. This new orientation of the domain brings the nuc active site to within 38 Å of that of pol and implies that the connector region is highly flexible allowing, the nuc and pol domains substantial range in relative disposition. However, this alternate orientation of the nuc domain in TaqP, like that in the other reported structure of the enzyme (4), does not provide an obvious structural basis for the current model for primer excision and nick translation by TaqP. The geometric relationships postulated for the DNA-structure-specific nature of substrate recognition by the nuc domain are not satisfied by either of the two structures (ref. 23 and V. Lyamichev, personal communication).
Figure 2.
Changes in TaqP on complex formation. (a) Superposition of the Cα drawing (gray ribbon) of native TaqP (4) with that (pink) of the enzyme in the complex. Polymerase (pol) and structure-specific exonuclease (nuc) in native structure are labeled. New orientation of the nuc domain shown is highlighted in blue. Zones of residues showing a mainchain rmsd >2.0 Å in the complex are labeled 1 through 5. Residues 527 to 552 in helix I are highlighted in red. (b) Expanded view of changes in Helix I. A Cα ribbon drawing of helix I in native TaqP (gray) is superimposed over that in this complex (red). The figures were made with grasp (48).
The cause of the rearrangement of the nuc domain is, at present, obscure. It is possible that reorientation of the nuc domain in the complex has been caused by the Fab binding because many close contacts between residues 260–290 of the nuc domain and residues 1L-26L of the Fab result when the former is placed in the same relative orientation to the pol domain as in the native structure (4). It is also equally likely that the reorientation is spontaneous. Radius of gyration measurements imply a different, more compact configuration for TaqP (4), and, in the structure of TaqP determined in a different crystal form (ref. 7 and H.M.K.M., unpublished results), although folding of the pol domain is almost identical (Cα rmsd of 0.7 Å) to native, the nuc domain is present in the same orientation as it is in this complex in the absence of any ligands.
Structural Changes on Complex Formation.
Distinctly visible changes in both antigen and Fab accompany complex formation. The biggest change in the Fab at the domain level is a decrease in elbow angle from 154° in the unliganded Fab to 146° in the complex. Changes in elbow angle of this and larger magnitude have been observed in other cases (8, 9). There is currently no evidence that such changes necessarily are correlated with ligand binding (8–10). There is little change in the orientation of VH relative to VL domain compared with unliganded TP7. The complementarity-determining region (CDR) loops in the L chain and H1 and H2 are close to canonical conformations they have in unliganded TP7 (3). rmsd for 59 frame-work Cα atoms in the VL domain between unliganded and liganded TP7, after superposition [using residues 1–21, 36–44, 62–88 and 99–110, numbered according to Kabat (24)], is 0.8 Å. rmsd for 65 Cα atoms in the VH domain (using residues 5–28, 36–48, 68–87 and 107–110) is 1.1 Å. Changes in the CDRs of the Fab, using the above superposition, are at the low end of those observed in other Fab-antigen complexes (8). rmsd for all atoms of CDRs from their positions in unliganded TP7 is 1.6 Å, with Y96H and Y99H showing the largest mainchain and sidechain deviations of 2.7 and 4.6 Å respectively. There is also a large (3.2 Å, mainchain) movement of framework residue Thr 87H.
Much larger changes are seen in regions of TaqP contacted by the TP7 Fab. There are several segments of TaqP mainchain that show >2-Å rmsds between the native and complexed forms of the enzyme (Fig. 2 a and b). These segments are distributed among the v-edit, thumb, and palm subdomains of TaqP; residues 386–408 in the v-edit domain (zone 1, Fig. 2a); residues 418–514 (zone 2) and 520–555 (zone 3) in the thumb subdomain; residues 580–590 (zone 4) in the palm subdomain; and residues 714–736 (zone 5) in the fingers subdomain. The largest mainchain rmsd (7.5 Å) is shown by residue Tyr 545 in helix I of the thumb. All zones except the third are close to their native conformations (Fig. 2a) but are displaced relative to the centroid of the pol domain. By using Cα atoms of residues 316–820 to calculate the mass-weighted centroid of the native pol domain, differences in rms distances of Cα atoms of each of the zones between complex and native are 0.2, 1.3, 0.6, −0.5, and −0.3 Å, respectively. Except for the second zone, all are relatively small. Larger movement of zone 2 appears to be triggered by changes in zone 3, especially those in residues 527–552 that form the large helix I in native TaqP (4).
In the complex, residues 540–542 and 543–545 form two successive γ turns (25). A result of this unwinding is an increase in inter-residue distances in this region of TaqP. Overall length of helix I (Cα distance) has increased from 36.7 Å in native to 39.2 Å in the complex. There are even larger changes at the carboxyl terminus of helix I (for example, residue 544 to residue 551, which is 12 Å in native and is 18 Å in the complex), which are attenuated at the ends of the helix by looping out of residues 540–543. Movement of zone 2 away from the centroid appears, in part, to be an adjustment of the structure to accommodate this increase in length. B factors for residues in helix I are not significantly higher than those for other parts of the structure in either of two native pol domain structures reported (4, 5) or in this complex.
Interactions Stabilizing the Complex.
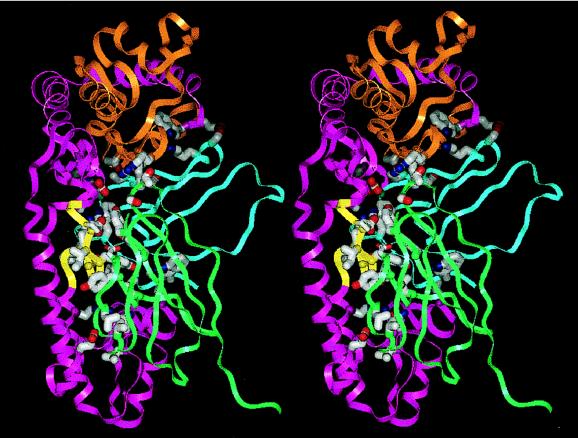
Fig. 3 shows an overall view of the complex, and Fig. 4 shows a detailed view of the TaqP-TP7 interface. As observed in other Fab-antigen complexes, a mix of favorable (27) van der Waals contacts (Table 2 and Fig. 4) and hydrogen bonds between the CDRs of the Fab and TaqP stabilize the complex. Residues contacted in TaqP are drawn from the thumb (mainly helix I), fingers, and the v-edit subdomains. The Fab interface with helix I is predominantly hydrophobic whereas that with the v-edit domain is rich in hydrogen bonds; there are no salt links. A total of 17 residues from five CDRs and 4 frame-work residues of TP7 make 43 favorable van der Waals contacts and an additional 28 hydrogen bonds with 27 residues in TaqP (Table 2). The CDRs are not involved equally, with 13 residues of CDRs H3, H2, and L2 providing a total of 32 van der Waals contacts and 22 hydrogen bonds whereas CDR H1 provides none. The complex buries 1,705 Å2 of solvent accessible area (46), using a probe radius of 1.6 Å, compared with separated components in the same conformation (Table 2), with TP7 contributing 814 Å2 and TaqP contributing 891 Å2.
Figure 3.
Overall View of the complex. Cα ribbon drawings of the pol domain (magenta), with the V-edit subdomain colored orange, and light (green) and heavy chains (cyan) are shown. Residues 540–550 in the helix are colored yellow. The figure was made with grasp (48).
Figure 4.
Stereo view of the TaqP-TP7 interface. Parts of Cα ribbons of the pol domain and TP7, color coded as in Fig. 3, are shown. Side-chain atoms of all residues listed in Table 2 are shown as stick drawings. Carbon, nitrogen, and oxygen atoms of the sidechains are colored gray, blue, and red respectively. The figure was made with grasp (48).
Table 2.
Interactions in the complex and buried surface area
| CDR178 | Residues contacted in TaqP† | Number of van der Waals contacts (Number of H-bonds) | Buried surface area, Å2‡ |
|---|---|---|---|
| L1 (Y32 Y34) | Y545 P548 D547 | 5 (1) | 41 |
| L2 (D50 L54 G56) | E471 R536 I546 | 3 (3) | 83 |
| L3 (S91 S92) | R313 K314 | 2 (2) | 64 |
| H1 | None | 0 (0) | 9 |
| H2 (S53 T55 T56 D57 K63 S64) | S383 T385 T386 E388 E397 W398 A568 A570 R728 | 10 (15) | 184 |
| H3 (G98 Y99 W100 Y101) | S515 K542 S543 Y545 N580 | 15 (4) | 128 |
| FRAME (L46L Y49L V58L W105H) | P481 L484 N485 K511 R512 T544 Y545 | 8 (3) | 305 |
| All | 43 (28) | 814 |
Evidence obtained from solution state studies (28) on binding to and inhibition of TaqP by TP7 has indicated strongly that the Fab binds to regions of TaqP involved in DNA binding. A minimally biased modeling study reported recently (3) also has suggested that the most likely regions of TaqP contacted by TP7 CDRs are regions corresponding to helix I in the thumb subdomain and several other residues in the palm subdomain and the v-edit domain. TP7 in the complex binds within the DNA binding cleft of TaqP, recognizing an extremely complex epitope assembled from the thumb, fingers, and the v-edit regions of the enzyme (Table 2 and Fig. 4). Of the 24 residues in TaqP contacted by TP7 in the complex, 11 were identified as potential contacts in the docking study. The modeling study used a rigid body docking procedure (29) and was therefore unable to anticipate the large changes resulting from complex formation.
It is not clear whether there is any functional significance to the change in conformation of helix I associated with its binding to DNA. We note, however, that several residues from this region provide ligands to DNA, and potential flexibility here might be advantageous in the process of recognition and stable complex formation although intermediate states may not necessarily be visible in the final enzyme–DNA complex determined crystallographically. Structural and functional studies on DNA polymerases (26) have demonstrated that parts of these enzymes are mobilized by the stimulus of DNA binding. But no significant changes have been observed in the regions corresponding to the residues (540–548) that have the largest change in conformation in the present complex, although these residues provide some ligands for DNA binding. Changes that accompany DNA binding (6) are in a section of residues (480–515) that are closer to the N terminus of helix I (nearer the tip of the thumb) than to its carboxyl terminus.
Factors Responsible for Change in Conformation.
It is evident that TaqP in the complex with TP7 is present in a conformation that is different from that adopted by the enzyme alone. It is also clear that there is comparatively little change in the conformation of TP7 from its unliganded form (3). In light of these two observations, and the fact that no additives capable of inducing conformational changes were used in crystallization, the changed conformation of residues in helix I in the complex can be attributed to a limited number of factors. One idea is that the antibody was elicited in vivo against an altered form of the native enzyme. In this model, features particular to the way the antigen is presented to the immune system, such as the adjuvant or some unanticipated reaction in the immunized mouse, lead to the selection of antibodies that recognize a subtly altered form of the enzyme. From this point of view, reactivity with native enzyme is essentially cross-reactivity, and structural accommodation on interaction with the antibody is the adoption by the enzyme of a conformation that otherwise would be disfavored. From extensive work on mAbs against T. zostericola myohemerythrin it has been suggested that a substantial fraction of antibodies are directed against such altered native forms of proteins (11, 32–34).
A second and related model is induced fit. It is conceivable that TP7 initially cross-reacts with native TaqP and coerces the enzyme residues into their conformation seen in the complex in a multistep process. In this model, the antibody is selected in vivo for reactivity with native enzyme, and it so happens that the selection is favored even though structural adaptation by the enzyme is required for high-affinity binding from an initially lower affinity complex. The property of induced fit or antibody-induced conformational change in the antigen has been suggested for some neutralizing antibodies against polio virus (42) and for several antibodies against Escherichia coli β-galactosidase (36). At present, there is no evidence from solution state studies (2, 28) for a multistep interaction in the present case. Nevertheless, if antibody-induced distortion is a step in complex formation, the marginal conformational stability (see below) of residues 540–545 in helix I would facilitate it.
A third model is that the native enzyme continually is cycling among several similarly probable conformers and that the antibody selectively stabilizes one and perturbs the equilibrium distribution of species toward the stabilized conformer. Computational and experimental studies have shown that proteins are flexible entities that constantly undergo several kinds of motion in solution, including helix–coil transitions (35). In this view, TP7 might be directed against a form of TaqP in which helix I occupies just one of the ensemble of nonhelical conformations dynamically available to the helix in solution. Ability to selectively bind one conformer in thermodynamic equilibrium with others with consequent shift of equilibrium in favor of the recognized conformer has been proposed for some antibodies against β-galactosidase (36). Catalytic antibodies directed against transition state analogues (37), structures of some of which are known (38–40), are capable of behaving in precisely this fashion.
Analysis of sequence in helix I (527–552) in terms of helix propagation propensity values of Scheraga (41) indicates that residues 540–545 might be the least stable members. By using a five-residue sliding window and tabulated propensity values (35), residues 540–545 have average propensity values that are 5–10% smaller than other residues. Residues 548 and 550 are prolines that are known to severely compromise helix stability, suggesting that helix I, as a hydrogen bonded entity, terminates at residue 547. As a consequence, residues 544 and 545, two of four terminal residues at the carboxyl end of the helix, make fewer backbone hydrogen bonds than residues in the body of the helix. Thus, extrahelical interactions being comparable, the combination of above factors predisposes this portion of helix I toward unwinding. Because only the end result is visualized in this crystal structure, it is not possible to choose definitively between the several models proposed on the basis of currently available data.
Mode of Inhibition and Basis of Preference for Thermus Polymerases.
Solution state experiments (2, 28), besides indicating strongly that TP7 binds to DNA recognition and binding regions of the enzyme, also have provided evidence that, although the Fab recognizes other polymerases from Thermus species, it does not recognize those from other, more distantly related species tested. Our docking study (3) provided a probable structural interpretation of both of these activities of TP7. The structure of the complex provides definitive structural evidence for conclusions derived from those studies. Residues 485, 539, 543, 568, 575, 577, 580, 583, 745, and 750 of TaqP make stabilizing interactions with the template strand of DNA in a reported TaqP-DNA complex (6). Of these, the Fab in the present complex makes either van der Waals contacts or hydrogen bonds with residues 485, 543, 568, 580, and 583. Similarly, TP7 makes stabilizing interactions with residues 515, 516, 536, and 540, which contact the primer strand of DNA. It thus seems clear that the ability of TP7 to compete with primer:template complex in binding to TaqP has its origins in its recognition of and binding to TaqP residues involved in interacting with DNA. Its inhibitory capability is obviously a consequence of its abolition of primer:template complex binding to the enzyme.
TP7 also interacts with some residues in the 383–397 region in the v-edit domain of TaqP (Table 2, Fig. 4). Taken together, the interactions with the pol and v-edit domains provide a structural rationale for the inability of TP7 to recognize other bacterial polymerases such as E. coli DNA polymerase I. The sequences of TaqP and E. coli DNA polymerase I in the v-edit region are completely different; the sequences in the respective pol domains in the region of interaction are also significantly different, although the changes are largely chemically conservative (3). These differences are undoubtedly the reason for nonrecognition of E. coli protein by TP7.
Acknowledgments
We are grateful to Drs. Sherin Abdel-Meguid, Sudhakar Babu, David Davies, and Ian Wilson for previewing an earlier version of the manuscript and for many helpful suggestions. Thanks also to Carolyn Hinchman and Michelle Shanahan for preparing the TP7 mAb. We thank Frederick Biomedical SuperComputing Center for computer time and Karol Miaskiewicz for help with running our programs. K.M. thanks the American Cancer Society for grant support. Structural biology at Fels is supported by a grant from the Lucille P. Markey Foundation. R.M. thanks Xcyte Therapies for support.
ABBREVIATIONS
- pol
polymerase domain including the vestigial editing nuclease domain
- nuc
structure specific nuclease domain
- v-edit
vestigial editing nuclease
- TaqP
Thermus aquaticus DNA polymerase I
- CDR
complementarity-determining region
- rmsd
rms deviation
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, Biology Department, Brookhaven National Laboratory, Upton, NY 11973 (PDB ID code 1bgx).
References
- 1. Sharkey D J, Scalice E R, Christy K G, Atwood S M, Daiss J L. Bio/Technology. 1994;12:506–509. doi: 10.1038/nbt0594-506. [DOI] [PubMed] [Google Scholar]
- 2.Scalice E R, Sharkey D J, Daiss J L. J Immunol Methods. 1995;183:15–26. doi: 10.1016/0022-1759(95)00019-7. [DOI] [PubMed] [Google Scholar]
- 3.Murali R, Helmer-Citterich M, Sharkey D J, Scalice E R, Daiss J L, Sullivan M A, Krishna Murthy H M. Protein Eng. 1998;11:79–86. doi: 10.1093/protein/11.2.79. [DOI] [PubMed] [Google Scholar]
- 4.Kim Y, Eom S H, Wang J, Lee D, Suh S, Steitz T A. Nature (London) 1995;376:612–616. doi: 10.1038/376612a0. [DOI] [PubMed] [Google Scholar]
- 5.Korolev S, Nayal M, Barnes W, Cera E, Waksman G. Proc Natl Acad Sci USA. 1995;92:9264–9268. doi: 10.1073/pnas.92.20.9264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eom S H, Wang J, Steitz T A. Nature (London) 1996;382:278–281. doi: 10.1038/382278a0. [DOI] [PubMed] [Google Scholar]
- 7.Urs U K, Sharkey D J, Peat T J, Hendrickson W A, Krishna Murthy H M. Proteins Struct Funct Genet. 1995;23:111–114. doi: 10.1002/prot.340230112. [DOI] [PubMed] [Google Scholar]
- 8.Wilson I A, Stanfield R L. Curr Opin Struct Biol. 1994;4:857–867. doi: 10.1016/0959-440x(94)90267-4. [DOI] [PubMed] [Google Scholar]
- 9.Wilson I A, Stanfield R L. Curr Opin Struct Biol. 1993;3:113–118. [Google Scholar]
- 10.Davies D R, Cohen G H. Proc Natl Acad Sci USA. 1996;93:7–12. doi: 10.1073/pnas.93.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tainer J A, Deal C D, Geysen H M, Roberts V A, Getzoff E D. Int Rev Immunol. 1991;7:165–188. doi: 10.3109/08830189109061773. [DOI] [PubMed] [Google Scholar]
- 12.Stanfield R L, Fieser T M, Lerner R A, Wilson I A. Science. 1990;248:712–719. doi: 10.1126/science.2333521. [DOI] [PubMed] [Google Scholar]
- 13.Kabsch W. J Appl Crystallogr. 1988;21:916–924. [Google Scholar]
- 14.Navaza J. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 15.CCP4, The CCP4 Suite: Programs for Protein Crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 16.Brünger A T. xplorManual, Version 3.0. New Haven, CT: Yale Univ. Press; 1990. [Google Scholar]
- 17.Jones T A, Zou J-Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 18.Brunger A T. Nature (London) 1992;355:472–474. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 19.Brunger A T. Acta Crystallogr A. 1990;46:46–57. doi: 10.1107/s0108767390002355. [DOI] [PubMed] [Google Scholar]
- 20.Read R J. Acta Crystallogr A. 1986;42:140–149. [Google Scholar]
- 21.Bhat T N. J Appl Crystallogr. 1988;21:279–281. [Google Scholar]
- 22.Laskowski R A, MacArthur M W, Moss D S, Thornton J M. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 23.Lyamichev V, Brow M A D, Dahlberg J E. Science. 1993;260:778–783. doi: 10.1126/science.7683443. [DOI] [PubMed] [Google Scholar]
- 24.Kabat, E. A., Wu, T. T., Reid-Miller, M., Perry, H. M. & Gottesman, K. S. Sequences of Proteins of Immunological Interest (U.S. Department of Health and Human Services, Washington, DC).
- 25.Hutchinson E G, Thornton J M. Protein Sci. 1996;5:212–220. doi: 10.1002/pro.5560050204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joyce C M, Steitz T A. Annu Rev Biochem. 1994;63:777–822. doi: 10.1146/annurev.bi.63.070194.004021. [DOI] [PubMed] [Google Scholar]
- 27.Sheriff S, Hendrickson W A, Smith J L. J Mol Biol. 1987;197:273–296. doi: 10.1016/0022-2836(87)90124-0. [DOI] [PubMed] [Google Scholar]
- 28.Daiss J L, Scalice E R, Sharkey D J. J Immunol Methods. 1995;172:147–163. doi: 10.1016/0022-1759(94)90102-3. [DOI] [PubMed] [Google Scholar]
- 29.Ausiello G, Cesareni G, Helmer-Citterich M. Proteins Struct Funct Genet. 1997;28:556–567. [PubMed] [Google Scholar]
- 30.Lawyer F C, Stoffel S, Saiji R K, Myambo K, Drummond R, Gelfand D H. J Biol Chem. 1989;264:6427–6437. [PubMed] [Google Scholar]
- 31.Laver W G, Air G M, Webster R G, Smith-Gill S J. Cell. 1990;61:553–556. doi: 10.1016/0092-8674(90)90464-p. [DOI] [PubMed] [Google Scholar]
- 32.Geysen H M, Tainer J A, Rodda S J, Mason T J, Alexander H, Getzoff E D, Lerner R A. Science. 1987;235:1184–1190. doi: 10.1126/science.3823878. [DOI] [PubMed] [Google Scholar]
- 33.Getzoff E D, Geysen H M, Rodda S J, Alexander H, Tainer J A, Lerner R A. Science. 1987;235:1191–1196. doi: 10.1126/science.3823879. [DOI] [PubMed] [Google Scholar]
- 34.Fieser T M, Tainer J A, Geysen H M, Houghten R A, Lerner R A. Proc Natl Acad Sci USA. 1987;84:8568–8572. doi: 10.1073/pnas.84.23.8568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Creighton T E. Proteins: Structures and Molecular Properties. New York: Freeman; 1987. [Google Scholar]
- 36.Celada F, Strom R. Biopolymers. 1983;22:465–473. doi: 10.1002/bip.360220159. [DOI] [PubMed] [Google Scholar]
- 37.Lerner R A, Benkovic S J, Schultz P G. Science. 1991;252:659–667. doi: 10.1126/science.2024118. [DOI] [PubMed] [Google Scholar]
- 38.Haynes M R, Stura E A, Hilvert D, Wilson I A. Science. 1994;263:646–652. doi: 10.1126/science.8303271. [DOI] [PubMed] [Google Scholar]
- 39.Zhou G W, Guo J, Huang W, Fletterick R J, Scanlan T S. Science. 1994;265:1059–1064. doi: 10.1126/science.8066444. [DOI] [PubMed] [Google Scholar]
- 40.Golinelli-Pimpeneau A, Gigat B, Bizehard T, Navaza J, Saludjian P, Zemet R, Tawfik D S, Eshbar Z, Green B S, Knosow M. Structure. 1994;2:175–183. doi: 10.1016/s0969-2126(00)00019-8. [DOI] [PubMed] [Google Scholar]
- 41.Scheraga H A. Pure Appl Chem. 1978;50:315–324. doi: 10.1351/pac196715030503. [DOI] [PubMed] [Google Scholar]
- 42.Wien M W, Filman D J, Stura E A, Guillot S, Delpeyroux F, Crainic R, Hogle J M. Nat Struct Biol. 1995;2:232–243. doi: 10.1038/nsb0395-232. [DOI] [PubMed] [Google Scholar]
- 43.Engh R A, Huber R. Acta Crystallogr A. 1991;47:392–397. [Google Scholar]
- 44.Ramachandran G N, Sasisekharan V. Adv Protein Chem. 1968;23:283–437. doi: 10.1016/s0065-3233(08)60402-7. [DOI] [PubMed] [Google Scholar]
- 45.Kleywegt G J, Jones T A. Structure. 1995;3:540–545. doi: 10.1016/s0969-2126(01)00187-3. [DOI] [PubMed] [Google Scholar]
- 46.Connolly M L. Science. 1983;221:707–713. doi: 10.1126/science.6879170. [DOI] [PubMed] [Google Scholar]
- 47.Evans S V. J Mol Graph. 1993;11:134–138. doi: 10.1016/0263-7855(93)87009-t. [DOI] [PubMed] [Google Scholar]
- 48.Nicholls A, Sharp K A, Honig B. Proteins Struct Funct Genet. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]