Abstract
Background
Little is known about the molecular mechanism which regulates how the whole cranium is shaped. Mouse models currently available for genetic research include several hundreds of unique inbred strains and genetically engineered mutants.
By cross comparing their genomic structures, we can elucidate the cause of any differences in the phenotype between two strains. The craniometry of subspecies, or closely related species, of mice provide a good systemic model to study the relationship between genetic variance and cranial shape evolution. The lack of a quantified framework for comparing and analyzing mouse cranial shape has been a problem. For this reason, we performed quantitative analysis of cranial shape morphology between several mouse strains.
Results
This article reports on a craniometric assay of seven mouse strains: four inbred strains (C57BL/6J, BALB/cA, C3H/HeJ, and CBA/JNCr) from Mus musculus domesticus (M. m. domesticus); one closed colony strain (ICR) from M. m. domesticus; one inbred strain (MSM/Ms) from Mus musculus molossinus; and, Mus spretus as a strain from a species other than M. m. domesticus. We performed linear measurements and geometric morphometrics. Geometric morphometrics revealed that the cranial characteristics of each strains were clearly distinguishable. We obtained mean scores for each species using the tpsRelw Program and plotted them.
Conclusion
Geometric morphometrics proved to be useful for identifying and classifying variations in form, and it revealed that M. spretus has a slender cranium when compared with our other strains. The mean cranial shape of C3H or CBA was more similar to MSM/Ms, which is derived from M. m. molossinus, than to either C57BL/6J, BALB, or ICR which are derived from M. m. domesticus. Future work in this field will aid in elucidating the mechanism of whole cranial shape regulation.
Background
Over 450 inbred mouse strains have been described and developed up until now, providing plentiful phenotypes and genomic backgrounds for genetic studies. Most inbred laboratory strains are known to have originated from a limited founder population of Mus musculus musculus and M. m. domesticus housed within a small number of research facilities and laboratories [1]. Most of these strains have been bred for over 150 generations; they are isogenic and homogeneous for over 98.6% of their genomes [2]. In the past few years, there have been several articles on the phylogenetic relationships among these inbred strains based on the available genome data. Wade et al reported on the fine structure of variation in the mouse genomes using SNPs [3]. They suggested these genomes to be mosaics, most being derived from M. m. domesticus and a minority from other subspecies. Recently, Sakai et al used the information from more than 1200 microsatellite loci to refine the fine phylogenetic relationships among inbred strains [1].
Here, we studied craniometric relationships among mouse strains. The mouse cranium has many significant features. Firstly, like all vertebrate crania, it is a complex structure comprising and protecting many important organs: brain; jaws; and, sensory organs, such as the eyes, the ears, and the nose. It is therefore appropriate and logical that the morphological diversity of the cranium should reflect phylogenetic and functional aspects of the adaptive evolution of each species after speciation. Secondly, the morphology of hard tissues, such as cranial bones, is easily measured and compared. Thirdly, the cranium is constructed from multiple bony components: the developmental process of the formation of the bones, and the sutures between them, has already been described in detail [4-6]. Fourthly, it is already known that mutations at many loci affect craniometric characteristics [7]. Considering the above, it would seem reasonable to assume that the craniometry of subspecies, or of closely related species, of mice provides a good systemic model for studying the relationship between cranial evolution and genetic variance.
The main purpose of this paper is to find whether the craniometric relationship and the phylogenic pattern, which is obtained from genomic data, among our subject species, correspond or not. To solve this question, the first thing that was necessary to be done was to select an appropriate method to quantify the cranial morphology of each sample. Next, we had to examine whether utilizing this method we could identify the quantitative differences among those species.
Questions concerning the extent and nature of morphological diversity, among closely related species and subspecies, are best addressed within a morphospace [8], and geometric morphometrics is a powerful tool for providing a morphospace for studying this diversity. Traditional multivariate methods are known to have weaknesses [9,10]. Relative warp analysis, which is a technique within the family of geometric morphometrics, offers a useful way to quantify shape variation independently of variation in the three parameters: size; translation; and, orientation. Algorithms for calculating relative warps are given by Bookstein [10] and Rohlf [11]. In this approach, landmark coordinates for all the specimens are superimposed using Procrustes methods [12], such that the fit between all the specimens is as close as possible. A reference specimen is computed and established as the mean of the fitted landmark coordinates. Relative warp analysis can be performed using the tpsRelw program by F. J. Rohlf [13]. We used geometric morphometrics in a craniometric assay of several mouse strains, subspecies and species: it was revealed that the cranial characteristics of each strain, subspecies or species were clearly distinguishable.
Results
Linear Distance Measurements
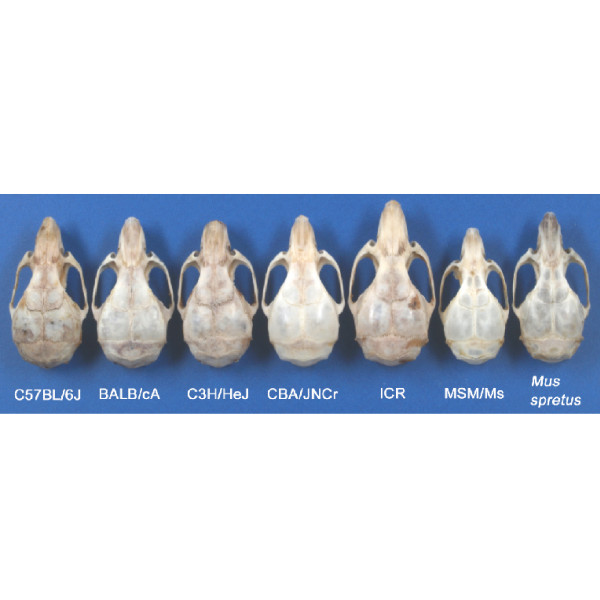
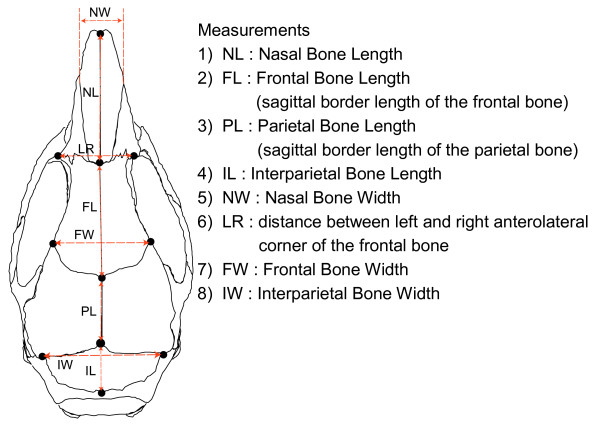
Figure 1 shows an example of a cranium from each strain (female, 12 weeks old). The crania of the different strains differ from each other in their general appearance. We first performed a preliminary morphometric study using conventional linear distance measurements. We placed 11 landmarks on the cranium, based upon which we made eight measurements (Fig. 2). These landmarks were located on the boundaries between the bones, of different parts of the cranium, for measuring the dimensions of these bones. Table 1 shows the results of linear distance measurements for each sample. The standard deviations of the measurements indicate that intraspecies variations in the specimens are sufficiently small.
Figure 1.
Cranial Shape Variations in Seven Mouse Strains (Female, 12 Weeks Old).
Figure 2.
Linear Measurements of the Cranium.
Table 1.
Summary of the Linear Measurements Among Mouse Strains.
| No. | Strain | Sex | distance (mm) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| NL | FL | PL | IL | NW | LR | FW | IW | |||
| 1 | C57BL/6J | ♂ | 7.7 | 7.7 | 4 | 3.6 | 2.7 | 5 | 5.3 | 8.4 |
| 2 | 7.6 | 7.6 | 4 | 3.5 | 2.6 | 5.1 | 5.5 | 8.3 | ||
| 3 | 7.4 | 7.8 | 4 | 3.5 | 2.6 | 5 | 5.3 | 8.1 | ||
| 4 | 7.8 | 7.8 | 4.1 | 3.5 | 2.7 | 5.2 | 5.4 | 8.1 | ||
| 5 | 7.8 | 7.8 | 3.7 | 3.5 | 2.6 | 5.1 | 5.4 | 8 | ||
| 6 | 7.6 | 7.8 | 4 | 3.7 | 2.7 | 5.1 | 5.3 | 8.4 | ||
| 7 | ♀ | 7.6 | 7.8 | 3.8 | 3.7 | 2.6 | 5.1 | 5.3 | 7.8 | |
| 8 | 7.1 | 7.8 | 3.7 | 3.6 | 2.5 | 5.2 | 5.5 | 7.8 | ||
| 9 | 7.7 | 7.7 | 3.7 | 3.5 | 2.6 | 5.1 | 5.2 | 8 | ||
| Mean | 7.589 | 7.756 | 3.889 | 3.6 | 2.622 | 5.1 | 5.356 | 8.1 | ||
| SD | 0.22 | 0.073 | 0.162 | 0.1 | 0.067 | 0.071 | 0.101 | 0.229 | ||
| 10 | BALB/cA | ♂ | 8 | 7.2 | 4.8 | 3.1 | 2.6 | 5 | 5.1 | 9 |
| 11 | 8.3 | 7.1 | 5.1 | 3.1 | 2.5 | 5.1 | 5 | 9 | ||
| 12 | 8.1 | 7.3 | 5.1 | 2.9 | 2.7 | 5 | 5.2 | 9 | ||
| 13 | ♀ | 7.1 | 6.3 | 4.7 | 2.9 | 2.5 | 4.8 | 5.5 | 8.6 | |
| 14 | 7.8 | 6.8 | 4.6 | 3.0 | 2.5 | 5 | 5.1 | 8.9 | ||
| 15 | 8.2 | 6.6 | 4.6 | 3.0 | 2.5 | 4.8 | 5.1 | 8.7 | ||
| Mean | 7.917 | 6.883 | 4.817 | 3.0 | 2.55 | 4.95 | 5.167 | 8.867 | ||
| SD | 0.436 | 0.387 | 0.232 | 0.1 | 0.084 | 0.122 | 0.175 | 0.175 | ||
| 16 | C3H/HeJ | ♂ | 7.5 | 7 | 4.9 | 3.3 | 2.5 | 5 | 4.6 | 8.8 |
| 17 | 7.3 | 7 | 4.8 | 3.4 | 2.5 | 5 | 4.8 | 8.8 | ||
| 18 | 7.3 | 7.1 | 4.8 | 3.4 | 2.6 | 4.9 | 4.7 | 8.8 | ||
| 19 | ♀ | 7.5 | 7.1 | 4.8 | 3.4 | 2.5 | 4.9 | 5 | 8.9 | |
| 20 | 7.2 | 6.6 | 4.6 | 3.4 | 2.7 | 4.9 | 5.1 | 8.7 | ||
| 21 | 7.7 | 7 | 4.6 | 3.4 | 2.6 | 4.9 | 5 | 8.7 | ||
| Mean | 7.417 | 6.967 | 4.75 | 3.3 | 2.567 | 4.933 | 4.867 | 8.783 | ||
| SD | 0.183 | 0.186 | 0.122 | 0.0 | 0.082 | 0.052 | 0.197 | 0.075 | ||
| 22 | CBA/JNCrj | ♂ | 7.2 | 7.2 | 4.6 | 3.7 | 2.3 | 5.1 | 5.2 | 8.6 |
| 23 | 7.5 | 7.7 | 4.5 | 3.7 | 2.3 | 5.1 | 5.1 | 8.6 | ||
| 24 | 7.5 | 7.4 | 4.5 | 3.7 | 2.3 | 5.1 | 5 | 8.6 | ||
| 25 | ♀ | 7.6 | 7.5 | 4.3 | 3.9 | 2.4 | 5.1 | 5.3 | 8.7 | |
| 26 | 7.6 | 7.5 | 4.3 | 3.8 | 2.4 | 5 | 5.1 | 8.6 | ||
| 27 | 7.7 | 7.5 | 4.3 | 3.8 | 2.4 | 5 | 5.1 | 8.8 | ||
| Mean | 7.517 | 7.467 | 4.417 | 3.8 | 2.35 | 5.067 | 5.133 | 8.65 | ||
| SD | 0.172 | 0.163 | 0.133 | 0.1 | 0.055 | 0.052 | 0.103 | 0.084 | ||
| 28 | ICR | ♂ | 8 | 7.4 | 4.4 | 3.2 | 2.9 | 5.2 | 5.1 | 8.6 |
| 29 | 7.8 | 7.4 | 4.4 | 3.2 | 2.8 | 5.4 | 5 | 8.6 | ||
| 30 | 8.1 | 7.4 | 4.4 | 3.2 | 2.7 | 5.4 | 5 | 8.6 | ||
| 31 | ♀ | 8.8 | 7.4 | 4.4 | 3.4 | 2.9 | 5.4 | 5 | 8.6 | |
| 32 | 8.8 | 7.2 | 4.5 | 3.5 | 2.7 | 5.4 | 4.8 | 8.5 | ||
| 33 | 8.4 | 7.4 | 4.5 | 3.2 | 2.9 | 5.5 | 4.8 | 8.5 | ||
| Mean | 8.317 | 7.367 | 4.433 | 3.3 | 2.817 | 5.383 | 4.95 | 8.567 | ||
| SD | 0.422 | 0.082 | 0.052 | 0.1 | 0.098 | 0.098 | 0.122 | 0.052 | ||
| 34 | MSM/Ms | ♂ | 6.8 | 5.4 | 4.4 | 3.0 | 2.1 | 4.4 | 4.1 | 7.2 |
| 35 | 6.9 | 5.4 | 4.4 | 3.1 | 2.2 | 4.3 | 4.1 | 7.2 | ||
| 36 | 6.7 | 5.8 | 4.6 | 3.0 | 2.2 | 4.3 | 4.1 | 7.2 | ||
| 37 | 6.7 | 5.8 | 4.5 | 3.1 | 2.1 | 4.3 | 4.1 | 7.2 | ||
| 38 | ♀ | 6.7 | 5.2 | 4.5 | 3.1 | 2.1 | 4.3 | 4 | 7.2 | |
| 39 | 6.7 | 5.7 | 4.4 | 3.2 | 2.1 | 4.4 | 3.9 | 7.2 | ||
| 40 | 6.8 | 5.7 | 4.3 | 3.2 | 2.2 | 4.4 | 3.8 | 7.2 | ||
| 41 | 6.8 | 5.8 | 4.6 | 3.1 | 2.2 | 4.5 | 3.9 | 7.2 | ||
| Mean | 6.763 | 5.6 | 4.463 | 3.1 | 2.15 | 4.363 | 4 | 7.2 | ||
| SD | 0.074 | 0.233 | 0.106 | 0.08 | 0.053 | 0.074 | 0.12 | 9E-16 | ||
| 42 | M. spretus | ♂ | 8 | 6 | 4.7 | 3.4 | 2.3 | 4.2 | 4.7 | 7.2 |
| 43 | 8 | 6.1 | 4.9 | 3.3 | 2.1 | 4.6 | 4.3 | 7.2 | ||
| 44 | 7.9 | 6.1 | 4.7 | 3.3 | 2.2 | 4.6 | 4.6 | 7.2 | ||
| 45 | 8.4 | 6 | 4.6 | 3.4 | 2.2 | 4.7 | 4.4 | 7.2 | ||
| 46 | ♀ | 8 | 6.1 | 4.8 | 3.4 | 2 | 5 | 4.3 | 7.2 | |
| 47 | 7.8 | 5.8 | 4.8 | 3.3 | 2.4 | 4.8 | 4.2 | 7.2 | ||
| 48 | 8.2 | 5.9 | 4.8 | 3.3 | 2.1 | 5 | 4.3 | 7.2 | ||
| 49 | 8.3 | 5.9 | 4.8 | 3.3 | 2.1 | 4.8 | 4.4 | 7.3 | ||
| Mean | 8.075 | 5.988 | 4.763 | 3.3 | 2.175 | 4.713 | 4.4 | 7.213 | ||
| SD | 0.205 | 0.113 | 0.092 | 0.0 | 0.128 | 0.259 | 0.169 | 0.035 | ||
NL, Nasal Bone Length; FL, Frontal Bone Length (sagittal border length of frontal bone); PL, Parietal Bone Length (sagittal border length of parietal bone); IL, Interparietal Bone Length; NW, Nasal Bone Width; LR, distance between left and right anterolateral corner of the frontal bone; FW, Frontal Bone Width; IW, Interparietal Bone Width.
Morphospace Axes
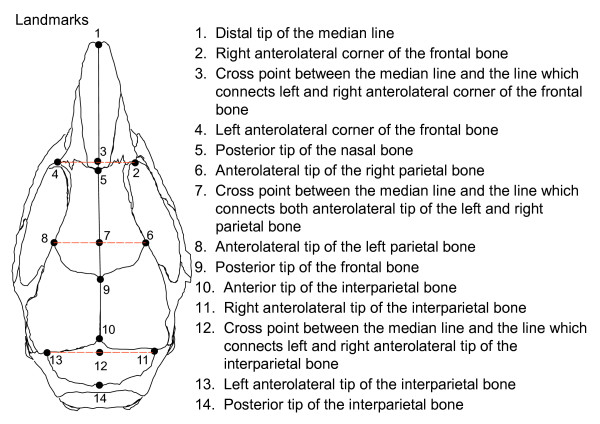
It is not easy to integrate the results of linear measurements analyses into a comparative morphological study systematically. We therefore performed morphological comparisons, with shape variables, using geometric morphometric methods [10]. We photographed all specimens in dorsal view, using a Keyence Digital Microscope; thusly, 14 biologically homologous cranial landmarks were digitized (Fig. 3).
Figure 3.
Landmarks Used to Describe the Cranial Shape in Geometric Morphometric Analyses.
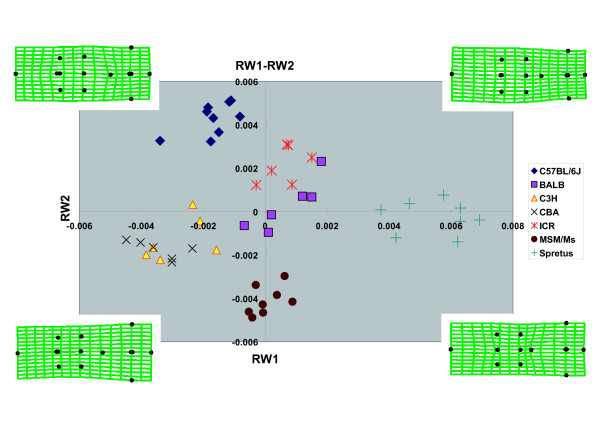
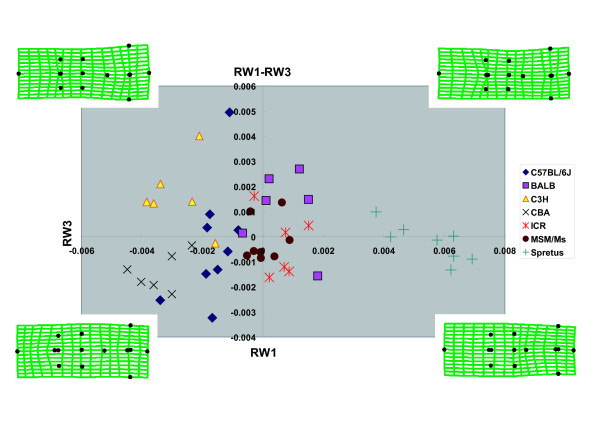
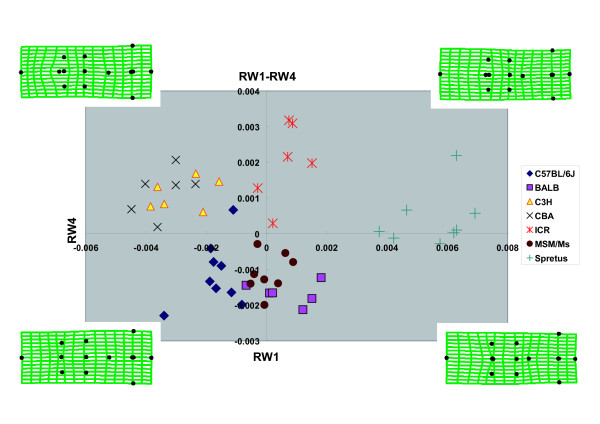
Four relative warp axes (RW1-4) together accounted for 75.1% of the total shape variation in the data. The major axes of shape variation in the crania of the seven strains can be seen in the plot (Fig. 4) of all specimens on RW1 and RW2 which accounted for 30.6% and 28.0%, respectively, of all variation in the samples. Significant interspecific clustering was observed in this morphospace. RW1 accounted for 30.6% of the variance and described the length of the frontal bone and the sphenoidal angle of the parietal bone (Fig. 4). M. spretus was characterized by relatively short frontal bones and a short sphenoidal angle of the parietal bone; in contrast, CBA possessed relatively long frontal bones and a long sphenoidal angle of the parietal bone. RW2 accounted for 28.0% of the variance and described the width of the frontal bone (Fig. 4). C57BL/6J had relatively wide frontal bones in contrast to the narrow frontal bones of MSM. RW3 accounted for 9.1% of the variance and described the length of the nasal bone (Fig. 5). BALB/cA was characterized by relatively long nasal bones when compared with CBA/JNCr. The axes RW1 and RW3 together discriminated C3H (positive value) and CBA (negative value). RW4 accounted for 7.4% of the variance and described the width of the region around the nasal border of the frontal bone (Fig. 6). This width was wider in ICR, and narrower in MSM. The axes RW1 and RW4 together discriminated ICR (large positive value) and BALB (large negative value).
Figure 4.
Scatter Plot of Scores on Relative Warps 1 and 2. Deformation grids indicate general shapes of each quadrant.
Figure 5.
Scatter Plot of Scores on Relative Warps 1 and 3. Deformation grids indicate general shapes of each quadrant.
Figure 6.
Scatter Plot of Scores on Relative Warps 1 and 4. Deformation grids indicate general shapes of each quadrant.
Correlation Between Relative Warp and Centroid Size
Shape and size are correlated in many cases. So we performed analysis to find the precise correlation between size and shape. Analyses of cranial size used the standard measure of geometric scale, which is centroid size (CS), defined as the square root of the squared distance between each landmark and the centroid of the landmark configurations summed over all landmarks. Then we performed a univariate regression analysis between each of the first 4 relative warps and CS (Table 2).
Table 2.
Summary of the Regression Analysis of the First 4 RW Scores on the Centroid Size.
| R | F | Probability | |
|---|---|---|---|
| RW1 | 0.315374 | 5.190951 | 0.027293 |
| RW2 | 0.576152 | 23.35412 | 1.48E-05 |
| RW3 | 0.017629 | 0.014612 | 0.904302 |
| RW4 | 0.388199 | 8.339589 | 0.005847 |
RW, relative warp; R, correlation coefficient
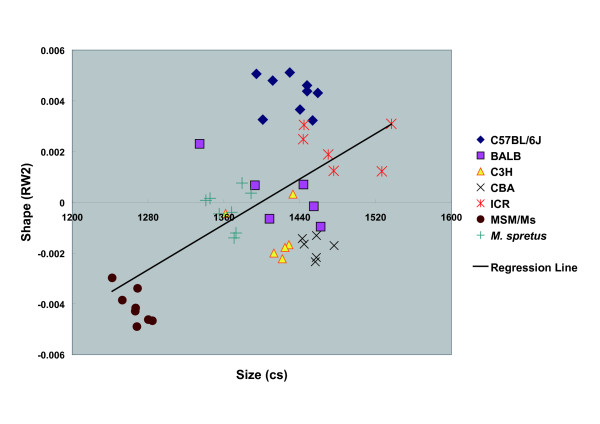
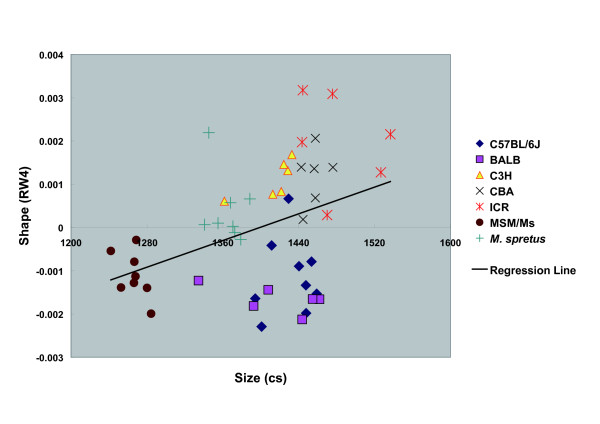
RW2 produced a significant regression on CS (P = 1.48E-05, R = 0.576152) (Fig. 7), indicating that the RW2 axis largely describes shape variation associated with size. RW4 produced a weak regression on CS (P = 0.005847, R = 0.388199) (Fig. 8). Regressions of RWs 1 or 3 on CS were not significant (P > 0.05). In terms of size, MSM/Ms animals are smaller than are the others, and they have relatively small RW2 values (Fig. 7).
Figure 7.
Regression of Relative Warp 2 Scores on Centroid Size. Scatterplot summarizing relations between the size (centroid size [CS]) and shape (relative warp 2 [RW]) variation. Regression (n = 49) of RW2 scores on CS (F = 23.35412; P = 1.48E-05; R2 = 0.331951).
Figure 8.
Regression of Relative Warp 4 Scores on Centroid Size. Scatterplot summarizing relations between the size (centroid size [CS]) and shape (relative warp 4 [RW]) variation. Regression (n = 49) of RW4 scores on CS (F = 8.339589; P = 0.005847; R2 = 0.150698).
Cluster Analyses
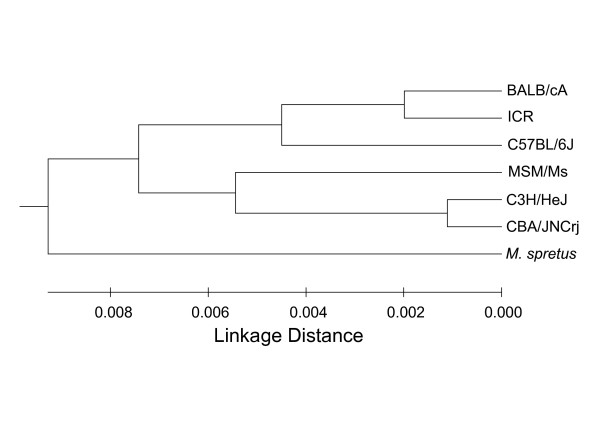
We obtained mean scores for each species using the tpsRelw Program and plotted them in RW1 and RW2. Based on the mean distance between species, we generated a phylogenic tree using the UPGMA Method (Fig. 9). This tree reflects morphological similarity between species; however, only a part of this pedigree matches the tree that was created from the genomic sequence (for example C3H and CBA), whereas some parts of this pedigree did not match the genomic sequence tree.
Figure 9.
Cluster Analysis of the Mean Cranial Shape of Each Species. Cluster analysis based on the distance between mean cranial shape of each species on the scatter plot for relative warps 1 and 2.
Discussion
The current classical laboratory strains were derived (in unequal percentages) from three parental subspecies: Mus musculus domesticus; M. m. musculus; and, M. m. castaneus. M. m. domesticus is common in Western Europe, Africa, and the Near East [14]. The range of M. m. musculus spans from Eastern Europe to Japan, across Russia and northern China. M. m. castaneus is found from Sri Lanka to South-East Asia, including the Indo-Malayan archipelago. None of these subspecies was completely isolated genetically, and in the common regions there is evidence of genetic exchange ranging from limited introgression to complete blending. Such exchanges have occurred between M. m. musculus and M. m. domesticus in Europe, and between M. m. musculus and M. m. castaneus in China and Japan. In Japan, M. m. musculus and M. m. castaneus have hybridized extensively and have given rise to a unique population referred to as Mus musculus molossinus [15,16]. M. spretus is the well-known western Mediterranean short-tailed mouse, a different species of the genus Mus.
In this study, we performed geometric morphometrics, a method developed by Fink and Zelditch (1995)[17]. There are three main points with respect to these results to take note of. Firstly, the cranial morphology of each strain is clearly distinguishable by geometric morphometrics. RW1 and RW2 discriminated among C57BL/6J, MSM/Ms, and M. spretus, although C3H overlapped with CBA and BALB overlapped with ICR. RW1 and RW3 discriminated between C3H and CBA, and RW1 and RW4 distinguished ICR from BALB. Secondly, according to our morphometric data, the mean cranial shapes of C3H and CBA were morphologically close to each other, at least with respect to RW1 and RW2 which represent the major axes of shape variation in the crania of the seven strains we studied. Strains C3H and CBA are considered genealogically closely related [18] and a small genetic distance between them has been demonstrated by SNP analysis [19]. On the other hand, M. spretus, a different species from the other six strains, showed a quite different mean cranial shape. Thirdly, our data also suggests that genetically close strains do not always possess morphologically similar crania; for example, the mean cranial shape of C3H and CBA were more similar to MSM/Ms, which is derived from M. m. molossinus [20], than to either C57BL/6J, BALB, or ICR which are paradoxically genetically closer strains. There are several possible explanations for this inconsistency. The first is that the genomic regions, which specifically control cranial morphology, are well conserved compared with the overall low homology of genomes between two genetically distant but morphologically similar species: in this case, only small regions are synapomorphic. The second possibility is that some epigenetic factors affect the morphology. The third possibility is that the resemblance of cranial morphology is caused by homoplasy: in this case, functional factors predominate over phylogenetic factors. The fourth, unknown environmental effects can also account for the results.
There have been craniometric studies in mice or other animals by several groups. Hallgrímsson et al (2004) reported that A/WySnJ mice had different craniofacial shapes compared with C57BL/6J mice [21]. Klingenberg et al (2004) compared the mandibles between the strains LG/J and SM/J utilizing geometric morphometrics [22]. In this study, we analyzed seven strains, and our results are applicable to comparative morphological studies of mice. Mutations in cis-regulatory sequences have been implicated as being the predominant source of variation in morphological evolution. Fondon and Garner (2004) reported that variation in repeats, in coding regions, is associated with morphology [23]. In the case of cichlid fish, some loci are significantly associated with jaw shape [24]. It is possible that some of our results from geometric morphometrics could be explained by association with some particular genomic structure. It is also possible that the epigenetic factors or functional factors could account for the results. For example, epigenetic interactions were reported by Hallgrímsson et al (2007)[25], and functional effects were analyzed by Zelditch et al (2004, 2006)[26,27], or Willmore et al (2006)[28]. Recently, there have been many reports describing the detailed expression patterns, or the functional studies, of many genes during cranial development. Many examples of gene mutations that affect cranial morphology have been accumulating [7]; however, the developmental process of the cranium is so complicated that many questions remain unanswered.
Conclusion
Geometric morphometrics revealed that the cranial characteristics of each species and subspecies were clearly distinguishable. M. spretus, a different species from the other 6 strains, has an extremely slender cranium compared with the other strains. Hallgrímsson et al argued that A/WySnJ mice exhibit altered facial morphology which results from a reduction in the growth of the maxillary process during formation of the face, and that this is relevant to evolutionary changes in facial prognathism in nonhuman primates and in human evolution [21]. M. spretus has a long maxillary and is the opposite to A/WySnJ mice. M. spretus diverged from M. musculus about 3 million years ago and developed into a different mouse species. Our data also suggests that genetically close strains do not always possess morphologically similar crania; the mean cranial shape of C3H and CBA were more similar to MSM/Ms, which is derived from M. m. molossinus [20], than to either C57BL/6J, BALB, or ICR which are paradoxically genetically closer.
Methods
Specimens
The mice used in this study were all 12 weeks old. C57BL/6J, BALB/cA, C3H/HeJ, and ICR mice were purchased from Clea Japan Inc. (Meguro Ward, Tokyo, Japan). CBA/JNCr mice were purchased from Charles River Laboratories Japan Inc. (Kohoku Ward, Yokohama, Japan). MSM/Ms mice and Mus spretus were obtained from the Riken BioResource Center (Tsukuba, Ibaraki, Japan). After euthanasia, the mice were skinned and the crania were removed and cleaned. All animal experiments were carried out with the approval of the Ethical Committee at the Center for Animal Resources and Development, Kumamoto University (D-18-090, A-19-154).
Linear Distance Measurement
We measured distances between 11 landmarks directly using digital calipers (Fig. 2). Statistical analyses were performed using the ystat 2000 Program.
Morphometrics
Crania were photographed in dorsal view using a Keyence Digital Microscope and 14 landmarks were digitized (Fig. 3). Relative warp analysis was performed using the tpsRelw Program.
Regression Analysis Between Size and Shape
The calculation of centroid size (CS) was done in the CoordGen6f Program, part of the Integrated Morphometrics Programs (IMP), produced in Matlab6 [29]. Univariate regression analysis was done in Microsoft Excel.
Cluster Analysis
Cluster analyses (unweighted pair-group average [UPGA]) were performed for interspecies distance between their mean shapes (acquired with the tpsRelw Program) on a scatter plot of relative warp (RW) 1 versus RW2, and a dendrogram was constructed.
Authors' contributions
MK is responsible for executing all work involved in this paper. KY conceived of this study and supervised the data analyses. Both authors read and approved the final manuscript.
Contributor Information
Minoru Kawakami, Email: mkawa@gpo.kumamoto-u.ac.jp.
Ken-ichi Yamamura, Email: yamamura@gpo.kumamoto-u.ac.jp.
Acknowledgements
We wish to thank Toshihiko Shiroishi of the Mammalian Genetics Laboratory, National Institute of Genetics, for permission to use the Keyence Digital Microscope and for helpful suggestions. We are grateful to the Riken BioResource Center for providing MSM/Ms and M. spretus. Our special thanks are due to Naomi Nakagata for his assistance in collecting the materials. We also thank Nobuki Nakanishi for comments on the manuscript. This work was supported in part by a Grant-in-Aid from the Ministry of Education, Science, Culture and Sports of Japan and a grant from the Osaka Foundation of Promotion of Clinical Immunology. No other monies were received from any public or private agency or corporation.
References
- Sakai T, Kikkawa Y, Miura I, Inoue T, Moriwaki K, Shiroishi T, Satta Y, Takahata N, Yonekawa H. Origins of mouse inbred strains deduced from whole-genome scanning by polymorphic microsatellite loci. Mamm Genome. 2005;16(1):11–19. doi: 10.1007/s00344-004-3013-9. [DOI] [PubMed] [Google Scholar]
- Barton NH, Keightley PD. Understanding quantitative genetic variation. Nat Rev Genet. 2002;3(1):11–21. doi: 10.1038/nrg700. [DOI] [PubMed] [Google Scholar]
- Wade CM, Kulbokas EJ, Kirby AW, Zody MC, Mullikin JC, Lander ES, Lindblad-Toh K, Daly MJ. The mosaic structure of variation in the laboratory mouse genome. Nature. 2002;420(6915):574–578. doi: 10.1038/nature01252. [DOI] [PubMed] [Google Scholar]
- Eswarakumar VP, Monsonego-Ornan E, Pines M, Antonopoulou I, Morriss-Kay GM, Lonai P. The IIIc alternative of Fgfr2 is a positive regulator of bone formation. Development. 2002;129(16):3783–3793. doi: 10.1242/dev.129.16.3783. [DOI] [PubMed] [Google Scholar]
- Iseki S, Wilkie AO, Morriss-Kay GM. Fgfr1 and Fgfr2 have distinct differentiation- and proliferation-related roles in the developing mouse skull vault. Development. 1999;126(24):5611–5620. doi: 10.1242/dev.126.24.5611. [DOI] [PubMed] [Google Scholar]
- Rice DP, Rice R, Thesleff I. Molecular mechanisms in calvarial bone and suture development, and their relation to craniosynostosis. Eur J Orthod. 2003;25(2):139–148. doi: 10.1093/ejo/25.2.139. [DOI] [PubMed] [Google Scholar]
- Richman JM, Lee SH. About face: signals and genes controlling jaw patterning and identity in vertebrates. Bioessays. 2003;25(6):554–568. doi: 10.1002/bies.10288. [DOI] [PubMed] [Google Scholar]
- Foote M. The evolution of morphological diversity. Annu Rev Ecol Syst. 1997;28:129–152. doi: 10.1146/annurev.ecolsys.28.1.129. [DOI] [Google Scholar]
- Marcus L. Proceedings of the Michigan morphometrics workshop Museum of Zoology special publication. Ann Arbor University of Michigan; 1988. Traditional morphometrics; pp. 77–122. [Google Scholar]
- Bookstein FL. Morphometric tools for landmark data: geometry and biology. Cambridge, UK: Cambridge University Press; 1991. [Google Scholar]
- Rohlf FJ. Contributions to morphometrics. Madrid CSIC; 1993. Relative warp analysis and an example of its application to mosquito wings; pp. 132–159. [Google Scholar]
- Rohlf FJ, Slice D. Extensions of the Procrustes method for the optimal superimpositon of landmarks. Syst Zool. 1990;39:40–59. doi: 10.2307/2992207. [DOI] [Google Scholar]
- Morphometrics at SUNY Stony Brook. http://life.bio.sunysb.edu/morph/
- Auffray JC, Vanlerberghe F, Britton-Davidian J. The house mouse progression in Eurasia: a palaeontological and archaeozoological approach. Biological Journal – Linnean Society London. 1990;41:13–25. doi: 10.1111/j.1095-8312.1990.tb00818.x. [DOI] [Google Scholar]
- Yonekawa H, Moriwaki K, Gotoh O, Miyashita N, Matsushima Y, Shi LM, Cho WS, Zhen XL, Tagashira Y. Hybrid origin of Japanese mice "Mus musculus molossinus": evidence from restriction analysis of mitochondrial DNA. Mol Biol Evol. 1988;5(1):63–78. doi: 10.1093/oxfordjournals.molbev.a040476. [DOI] [PubMed] [Google Scholar]
- Guénet JL, Bonhomme F. Wild mice: an ever-increasing contribution to a popular mammalian model. Trends Genet. 2003;19(1):24–31. doi: 10.1016/S0168-9525(02)00007-0. [DOI] [PubMed] [Google Scholar]
- Fink WL, Zelditch ML. Phylogenetic analysis of ontogenetic shape transformations: A reassessment of the piranha genus Pygocentrus (Teleostei) Systematic Biology. 1995;44:343–360. doi: 10.2307/2413597. [DOI] [Google Scholar]
- Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the Mouse Embryo A Laboratory Manual Second Edition. Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- Pletcher MT, McClurg P, Batalov S, Su AI, Barnes SW, Lagler E, Korstanje R, Wang X, Nusskern D, Bogue MA. et al. Use of a dense single nucleotide polymorphism map for in silico mapping in the mouse. PLoS Biol. 2004;2(12):e393 . doi: 10.1371/journal.pbio.0020393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikkawa Y, Miura I, Takahama S, Wakana S, Yamazaki Y, Moriwaki K, Shiroishi T, Yonekawa H. Microsatellite database for MSM/Ms and JF1/Ms, molossinus-derived inbred strains. Mamm Genome. 2001;12(9):750–752. doi: 10.1007/s003350030008. [DOI] [PubMed] [Google Scholar]
- Hallgrímsson B, Dorval CJ, Zelditch ML, German RZ. Craniofacial variability and morphological integration in mice susceptible to cleft lip and palate. J Anat. 2004;205(6):501–517. doi: 10.1111/j.0021-8782.2004.00356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingenberg CP, Leamy LJ, Cheverud JM. Integration and modularity of quantitative trait locus effects on geometric shape in the mouse mandible. Genetics. 2004;166(4):1909–1921. doi: 10.1534/genetics.166.4.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fondon JW, Garner HR. Molecular origins of rapid and continuous morphological evolution. Proc Natl Acad Sci USA. 2004;101(52):18058–18063. doi: 10.1073/pnas.0408118101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson RC, Streelman JT, Kocher TD. Directional selection has shaped the oral jaws of Lake Malawi cichlid fishes. Proc Natl Acad Sci USA. 2003;100(9):5252–5257. doi: 10.1073/pnas.0930235100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallgrimsson B, Lieberman DE, Liu W, Ford-Hutchinson AF, Jirik FR. Epigenetic interactions and the structure of phenotypic variation in the cranium. Evolution & development. 2007;9(1):76–91. doi: 10.1111/j.1525-142X.2006.00139.x. [DOI] [PubMed] [Google Scholar]
- Zelditch ML, Lundrigan BL, Garland T Jr. Developmental regulation of skull morphology. I. Ontogenetic dynamics of variance. Evolution & development. 2004;6(3):194–206. doi: 10.1111/j.1525-142X.2004.04025.x. [DOI] [PubMed] [Google Scholar]
- Zelditch ML, Mezey J, Sheets HD, Lundrigan BL, Garland T Jr. Developmental regulation of skull morphology II: ontogenetic dynamics of covariance. Evolution & development. 2006;8(1):46–60. doi: 10.1111/j.1525-142X.2006.05074.x. [DOI] [PubMed] [Google Scholar]
- Willmore KE, Leamy L, Hallgrimsson B. Effects of developmental and functional interactions on mouse cranial variability through late ontogeny. Evolution & development. 2006;8(6):550–567. doi: 10.1111/j.1525-142X.2006.00127.x. [DOI] [PubMed] [Google Scholar]
- Mathworks. MATLAB. The Mathworks, Natick, MA; 2000. [Google Scholar]