Abstract
Background
The Euthyneura are considered to be the most successful and diverse group of Gastropoda. Phylogenetically, they are riven with controversy. Previous morphology-based phylogenetic studies have been greatly hampered by rampant parallelism in morphological characters or by incomplete taxon sampling. Based on sequences of nuclear 18S rRNA and 28S rRNA as well as mitochondrial 16S rRNA and COI DNA from 56 taxa, we reconstructed the phylogeny of Euthyneura utilising Maximum Likelihood and Bayesian inference methods. The evolution of colonization of freshwater and terrestrial habitats by pulmonate Euthyneura, considered crucial in the evolution of this group of Gastropoda, is reconstructed with Bayesian approaches.
Results
We found several well supported clades within Euthyneura, however, we could not confirm the traditional classification, since Pulmonata are paraphyletic and Opistobranchia are either polyphyletic or paraphyletic with several clades clearly distinguishable. Sacoglossa appear separately from the rest of the Opisthobranchia as sister taxon to basal Pulmonata. Within Pulmonata, Basommatophora are paraphyletic and Hygrophila and Eupulmonata form monophyletic clades. Pyramidelloidea are placed within Euthyneura rendering the Euthyneura paraphyletic.
Conclusion
Based on the current phylogeny, it can be proposed for the first time that invasion of freshwater by Pulmonata is a unique evolutionary event and has taken place directly from the marine environment via an aquatic pathway. The origin of colonisation of terrestrial habitats is seeded in marginal zones and has probably occurred via estuaries or semi-terrestrial habitats such as mangroves.
Background
Within the phylum Mollusca, Gastropoda represent the largest and most diverse group in terms of species numbers, niche selection and life history strategies. They have been traditionally classified into three main subclasses: Prosobranchia, Opisthobranchia and Pulmonata [1-3]. Within Gastropoda, Opisthobranchia and Pulmonata have been united as Euthyneura and have since Spengel [4] been contrasted to the Streptoneura (= Prosobranchia). The latter, however, are commonly accepted as being paraphyletic [5].
Although a plethora of morphological and anatomical data on Gastropoda have accumulated over the last centuries, it was not until the 1980s that the development of cladistic methodology allowed for analysing these data in a phylogenetic framework e. g. [5-9]. Nevertheless, euthyneuran gastropods have often been neglected in these studies.
Several new studies based on morphological data have indicated that Euthyneura is a taxon clearly distinct from the remaining Gastropoda, belonging to a larger monophyletic group, the Heterobranchia. The latter also include the paraphyletic Heterostropha [10]. Euthyneura are characterised by several autapomorphies [11]. However, difficulties with respect to establishing a natural system of Euthyneura are well known [12] and can be attributed to the large number of homoplasies and convergent evolution of character traits.
Recent phylogenetic studies have focussed on several subgroups of Euthyneura, producing partly conflicting results.
Pulmonata have been both analysed morphologically [6,13] and on the basis of molecular data [14,15]. They have mostly been recovered monophyletic [6,8,11,13,16]. However, phylogenetic relationships of subgroups within Pulmonata have not been conclusively resolved yet. [11,17].
Opisthobranchia are often rendered paraphyletic in phylogenetic analyses regardless of morphological or molecular systematic approaches [11,18-22]. Moreover, positions of enigmatic taxa within the phylogenetic system of Opisthobranchia, such as the Sacoglossa, Acochlidiacea, Umbraculoidea or Acteonoidea also remain unresolved so far [21,22].
To date, no sound phylogenetic hypothesis for the Euthyneura exists. Recent morphological analyses by Dayrat and Tillier [11] yielded very poor resolution within Euthyneura and demonstrated the need to explore new datasets in order to critically analyse the phylogeny of this controversial group of gastropods. Upcoming molecular systematic studies have mostly utilised single genetic markers comprising partial sequences only [17] or have not included all major lineages particularly respective to Pulmonata [19]. These studies only provided limited new insights into the phylogeny of Euthyneura. However, a sound phylogenetic hypothesis of a taxon is the prerequisite to reconstruct evolutionary changes in the group of interest.
To this end, we present an extensive phylogenetic analysis of the relationships of Euthyneura by using a multi-gene dataset including nuclear and mitochondrial genes. For the first time a broad taxon sampling of all major euthyneuran subgroups is considered. Based on the phylogenetic hypotheses proposed here, we discuss evolutionary trends within Euthyneura. In particular, we propose new hypotheses how invasion of freshwater and terrestrial habitats as major evolutionary events of the Gastropoda, has occurred.
Results
Sequence alignment and statistical tests
In total we aligned sequences of 56 taxa (Table 1) for the large taxon set and 34 taxa for the reduced taxon set. Due to ambiguous homologisation of certain nucleotide positions in the alignments we excluded parts of high variability which were mainly due to inserts in certain taxa from the alignments. The lengths of the obtained alignments (after removal of ambiguous nucleotide positions) for the different datasets (complete taxon number versus reduced taxon number for combined analyses) are shown in Table 2.
Table 1.
Information on taxon sampling. Taxon names, localities, accession numbers provided (taxonomic classification in suprafamilial categories follows Bouchet et al. [38]); sequences generated in current study are marked with an asterisk; -: missing sequences
| Taxon | Family | Locality | 18S | 16S | COI | 28S |
|---|---|---|---|---|---|---|
| PULMONATA | ||||||
| BASOMMATOPHORA | ||||||
| SIPHONARIOIDEA | ||||||
| Siphonaria alternata | Siphonariidae | Bermuda | AY427523 | EF489299* | - | AY427488 |
| Siphonaria concinna | Siphonariidae | South Africa | EF489334* | EF489300* | EF489378 | EF489353* |
| Siphonaria capensis | Siphonariidae | South Africa | EF489335* | EF489301* | EF489379* | EF489354* |
| Siphonaria serrata | Siphonariidae | South Africa | EF489336* | EF489302* | EF489380* | - |
| AMPHIBOLOIDEA | ||||||
| Salinator cf. fragilis | Amphibolidae | Australia, NT | - | EF489303* | EF489381* | EF489355* |
| Phallomedusa solida (formerly Salinator solida) | Amphibolidae | Genbank | DQ093440 | DQ093484 | DQ093528 | DQ279991 |
| Amphibola crenata | Amphibolidae | New Zealand, Wellington | EF489337* | EF489304* | - | EF489356* |
| HYGROPHILA | ||||||
| ACROLOXOIDEA | ||||||
| Acroloxus lacustris | Acroloxidae | Germany | AY282592 | EF489311* | AY282581 | EF489364* |
| PLANORBOIDEA | ||||||
| Ancylus fluviatilis | Planorbidae | Germany | AY282593 | EF489312* | AY282582 | EF489365* |
| Bulinus tropicus | Bulinidae | Zimbabwe | AY282594 | EF489313* | AY282583 | EF489366* |
| Planorbis planorbis | Planorbidae | Germany | EF012192 | EF489315* | EF012175 | EF489369* |
| Physella acuta | Physidae | France, Atlantic | AY282600 | AY651241 | AY282589 | EF489368* |
| LYMNAEOIDEA | ||||||
| Lymnaea stagnalis | Lymnaeidae | Germany | EF489345* | EF489314* | EF489390* | EF489367* |
| CHILINOIDEA | ||||||
| Chilina sp. 1 | Chilinidae | Chile | EF489338* | EF489305* | EF489382* | EF489357* |
| Chilina sp. 2 | Chilinidae | Chile | - | EF489306* | EF489383* | EF489358* |
| Latia neritoides | Latiidae | New Zealand, Waikato | EF489339* | EF489307* | EF489384* | EF489359* |
| EUPULMONATA | ||||||
| TRIMUSCULOIDEA | ||||||
| Trimusculus afra | Trimusculidae | Senegal, Gorée | EF489343* | EF489309* | EF489388* | - |
| ELLOBIOIDEA | ||||||
| Myosotella myosotis | Ellobiidae | Croatia | EF489340* | AY345053 | EF489385* | EF489360* |
| Ophicardelus costellaris | Ellobiidae | New Zealand, Wellington | EF489342* | - | EF489387* | EF489362* |
| Ophicardelus ornatus | Ellobiidae | Genbank | DQ0934442 | DQ093486 | DQ093486 | DQ279994 |
| Carychium minimum | Carychiidae | Germany | EF489341* | EF489308* | EF489386* | EF489361* |
| OTINOIDEA | ||||||
| Otina ovata | Otinidae | France | EF489344* | EF489310* | EF489389* | EF489363* |
| SYSTELLOMMATOPHORA | ||||||
| ONCHIDIOIDEA | ||||||
| Onchidium verruculatum | Onchidiidae | Australia, QLD | AY427522 | EF489316* | EF489391* | AY427487 |
| Onchidella floridana | Onchidiidae | Bermuda | AY427521 | EF489317* | EF489392* | AY427486 |
| STYLOMMATOPHORA | ||||||
| CLAUSILIOIDEA | ||||||
| Albinaria sp. | Clausiliidae | Genbank | AY546382 | AY546342 | AY546262 | - |
| HELICOIDEA | ||||||
| Arianta arbustorum | Helicidae | Genbank | AY546383 | AY546343 | AY546263 | AY014136 |
| ARIONOIDEA | ||||||
| Arion silvaticus | Arionidae | Genbank | AY145365 | AY947380 | AY987918 | AY145392 |
| ENOIDEA | ||||||
| Ena montana | Enidae | Genbank | AY546396 | AY546356 | AY546276 | - |
| ATHORACOPHOROIDEA | ||||||
| Athoracophorus bitentaculatus | Athoracophoridae | AF047198 | - | AY150090 | AY014018 | |
| OPISTHOBRANCHIA | ||||||
| APLYSIOMORPHA | ||||||
| AKEROIDEA | ||||||
| Akera bullata | Akeridae | Genbank | AY427502 | AF156127 | AF156143 | AY427466 |
| APLYSIOIDEA | ||||||
| Aplysia californica | Aplysiidae | Genbank | AY039804 | AF192295 | AF077759 | AY026366 |
| THECOSOMATA | ||||||
| CAVOLINIOIDEA | ||||||
| Hyalocylis striata | Cavoliniidae | Genbank | DQ237966 | - | DQ237999 | DQ237985 |
| Cavolinia uncinnata | Cavoliniidae | Genbank | DQ237964 | - | DQ237997 | DQ237983 |
| GYMNOSOMATA | ||||||
| CLIONOIDEA | ||||||
| Spongiobranchaea australis | Pneumodermatidae | Genbank | DQ237969 | - | DQ238002 | DQ237988 |
| Pneumoderma cf. atlantica | Pneumodermatidae | Genbank | DQ237970 | - | DQ238003 | DQ237989 |
| SACOGLOSSA | ||||||
| CYLINDROBULLOIDEA | ||||||
| Cylindrobulla beauii | Cylindrobullidae | USA, Florida | EF489347* | EF489321* | - | EF489371* |
| PLACOBRANCHIDOIDEA | ||||||
| Elysia viridis | Placobranchidae | Genbank | AY427499 | AJ223398 | DQ237994 | AY427462 |
| UMBRACULOIDEA | ||||||
| Umbraculum umbraculum | Umbraculidae | Australia, NSW | AY165753 | EF489322* | DQ256200 | AY427457 |
| CEPHALASPIDEA | ||||||
| HAMINOEOIDEA | ||||||
| Haminoea hydatis | Haminoeidae | France, Atlantic | AY427504 | EF489323* | DQ238004 | AY427468 |
| DIAPHANOIDEA | ||||||
| Diaphana sp. | Diaphanidae | Norway, Kattegat | - | EF489325* | EF489394* | EF489373* |
| Toledonia globosa | Diaphanidae | Scotia Arc, Atlantic | EF489350* | EF489327* | EF489395* | EF489375* |
| PHILINOIDEA | ||||||
| Cylichna gelida | Cylichnidae | Scotia Arc, Atlantic | EF489349* | EF489326* | - | EF489374* |
| Scaphander lignarius | Cylichnidae | Spain Mediterranean Sea | EF489348* | EF489324* | - | EF489372* |
| ACOCHLIDIACEA | ||||||
| HEDYLOPSOIDEA | ||||||
| Unela glandulifera | Parhedylidae | Croatia | AY427517 | EF489328* | - | AY427482 |
| Pontohedyle milaschevitchi | Parhedylidae | Italy | AY427519 | EF489329* | - | AY427484 |
| Hedylopsis spiculifera | Hedylopsidae | Italy | AY427520 | - | - | AY427485 |
| NUDIPLEURA | ||||||
| PLEUROBRANCHOIDEA | ||||||
| Tomthompsonia antarctica | Pleurobranchidae | Antarctica | AY427492 | EF489330* | DQ237992 | AY427452 |
| Pleurobranchus peroni | Pleurobranchidae | Australia, NSW | AY427494 | EF489331* | DQ237993 | AY427455 |
| BATHYDORIDOIDEA | ||||||
| Bathydoris clavigera | Bathydorididae | Genbank | AY165754 | AF249222 | AF249808 | AY427444 |
| TRITONIOIDEA | ||||||
| Dendronotus dalli | Dendronotoidae | Genbank | AY165757 | AF249252 | AF249800 | AY427450 |
| LOWER HETEROBRANCHIA | ||||||
| Orbitestella sp. | Orbitestellidae | New Zealand, North Island | EF489352* | EF489333* | EF489397* | EF489377* |
| ACTEONOIDEA | ||||||
| Rictaxis punctocaelatus | Acteonidae | USA, California | EF489346* | EF489318* | EF489393* | EF489370* |
| Pupa solidula | Acteonidae | Australia, QLD | AY427516 | EF489319* | DQ238006 | AY427481 |
| Hydatina physis | Aplustridae | Australia, NSW | AY427515 | EF489320* | - | AY427480 |
| PYRAMIDELLOIDEA | ||||||
| Turbonilla sp. | Pyramidellidae | New Zealand, North Island | EF489351* | EF489332* | EF489396* | EF489376* |
| CAENOGASTROPODA | ||||||
| Littorina littorea | Littorinidae | Genbank | X91970 | DQ093481 | AY345020 | AJ488672 |
Table 2.
Information on sequence alignments of the different datasets and models of sequence evolution for Bayesian analyses
| Gene region and taxon set | Number of taxa | Length of alignment (after removal of ambiguous positions) | Excluded nucleotide positions | Model of sequence evolution |
|---|---|---|---|---|
| 18S rRNA (large taxon set) | 53 | 1843 | 228–302 | TRN+I+G |
| 773–1044 | α = 0.3539 | |||
| 1500–1523 | pinv = 0.3977 | |||
| 1777–1990 | ||||
| 18S rRNA (reduced taxon set) | 34 | 1826 | 228–302 | TRN+I+G |
| 766–1037 | α = 0.3202 | |||
| 1490–1513 | pinv = 0.4220 | |||
| 1750–1977 | ||||
| 28S rRNA (large taxon set) | 52 | 1123 | 540–565 | GTR+I+G |
| 656–712 | α = 0.5337 | |||
| 1076–1125 | pinv = 0.2449 | |||
| 28S rRNA (reduced taxon set) | 34 | 1099 | 543–562 | GTR+I+G |
| 675–726 | α = 0.3905 | |||
| 1052–1074 | pinv = 0.0889 | |||
| 16S rRNA (large taxon set) | 49 | 452 | 318–450 | HKY+I+G |
| 486–587 | α = 0.6450 | |||
| 664–681 | pinv = 0.2421 | |||
| 16S rRNA (reduced taxon set) | 34 | 452 | 318–450 | K81uf+I+G |
| 486–587 | α = 0.5878 | |||
| 664–681 | pinv = 0.2173 | |||
| CO1 (large taxon set) | 47 | 597 | GTR+I+G | |
| α = 0.4170 | ||||
| pinv = 0.2722 | ||||
| CO1 (reduced taxon set) | 34 | 597 | GTR+I+G | |
| α = 0.3975 | ||||
| pinv = 0.3077 | ||||
| CO1 (without 3rd codon position) (large taxon set) | 47 | 400 | 3rd codon positions | TIM+I+G |
| α = 0.3496 | ||||
| pinv = 0.3879 | ||||
| CO1 (without 3rd codon position) (reduced taxon set) | 34 | 400 | 3rd codon positions | TIM+I+G |
| α = 0.3435 | ||||
| pinv = 0.4759 | ||||
Performance of the chi-square-test in PAUP yielded a homogeneous base composition in the 28S-alignments (P = 0.999). The 18S- (P = 0.001) and 16S- alignments (P = 0.007) showed heterogeneous base composition mainly due to the sequences of Nudipleura (Bathydoris clavigera, Dendronotus dalli, Pleurobranchus peroni, Tomthompsonia antarctica) (for 18S) and Orbitestellidae (for 16S). The COI-alignment showed heterogeneous base composition when using all three codon positions (P = 0.000). However, we identified substitution saturation in the third codon position (Iss 0.830 >Iss·c 0.771 for large taxon set; Iss 0.753 >Iss·c 0.737 for reduced taxon set) and subsequently removed third codon positions from the alignments. The P-value of the chi-square test then changed to 1.00 indicating homogeneous base composition for all taxa.
Additionally, we observed substitution saturation in the 16S-alignments (Iss 1.071 >Iss·c 0.784 for large taxon set; Iss 1.02 >Iss·c 0.750 for reduced taxon set).
The relative rate test revealed that evolutionary rates are different in the investigated taxa and genetic markers. This is especially true for the two ribosomal genes 18S rRNA and 28S rRNA, where the Nudipleura show extremely high Z-Scores (e. g. 18S: Dendronotus dalli vs Orbitestella sp. = 12.19; 28S: Dendronotus dalli vs Turbonilla sp. = 5.34).
Investigation of differences in incongruence length between the four different sequence partitions and two taxon sets revealed that combination of the partitions improves phylogenetic signal with a p value of 0.01 (10.000 replicates).
Phylogenetic analyses
We performed different analyses which provided mostly congruent results regarding phylogenetic relationships of subgroups within Euthyneura.
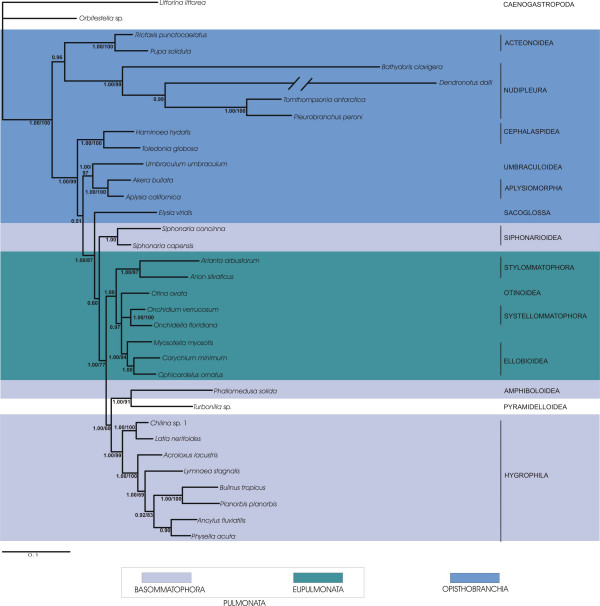
Our analyses of concatenated sequences of 18S rRNA, 28S rRNA, 16S rRNA and COI of the large taxon set comprised 56 taxa representing all of the major taxa of Euthyneura (Fig. 1) and four non-euthyneuran taxa. The reduced dataset included 34 taxa with Trimusculoidea, Acochlidiacea, Thecosomata and Gymnosomata not represented (Figs. 2 and 3). Although 16S-sequences and 3rd codon positions of the COI sequences exhibited varying degrees of substitution saturation we used all sequences in our combined analyses of the large taxon set (Fig. 1) and the reduced taxon set (Fig. 3) to avoid the loss of phylogenetic signal at lower taxonomic levels (e. g. relationships within superfamilies). In a previous study, Thollesson [18] demonstrated that 16S rRNA sequences provide useful information in recontruction of phylogenetic relationships within Euthyneura. However, to accommodate substitution saturation and to test its influence on the phylogeny inferred, we additionally performed combined analyses of the reduced dataset excluding 16S-sequences and 3rd codon positions of COI sequences (Fig. 2).
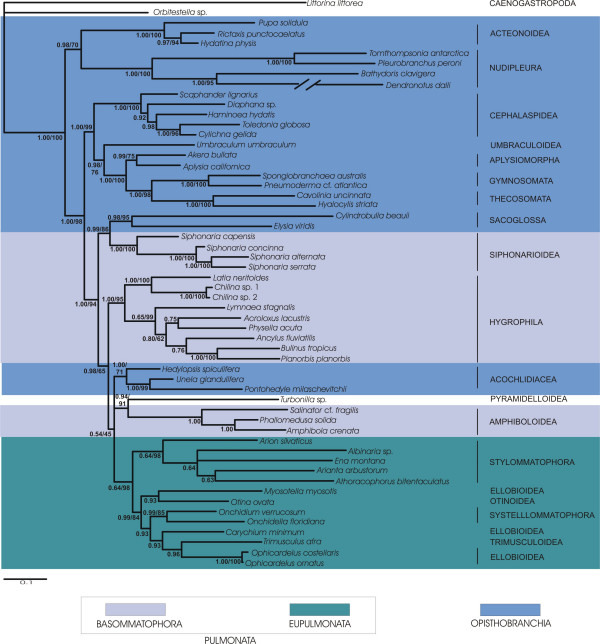
Figure 1.
Bayesian inference phylogram of euthyneuran relationships (large taxon set). Concatenated sequences of 18S rRNA, 28S rRNA, 16S rRNA and COI DNA of all taxa studied were used. 50% majority rule consensus tree. Posterior probabilities and bootstrap support of Maximum Likelihood analysis provided at the branches. Taxonomic classification follows Bouchet et al. [38]. Euthyneuran taxa are marked by colour frames.
Figure 2.
Bayesian inference phylogram of euthyneuran relationships (reduced taxon set). Sequences of 18S, 28S and CO1 (without 3rd codon positions) were used. 50% majority rule consensus tree. Posterior probabilities and bootstrap support of Maximum Likelihood analysis provided at the branches. Taxonomic classification follows Bouchet et al [38]. Euthyneuran taxa are marked by colour frames.
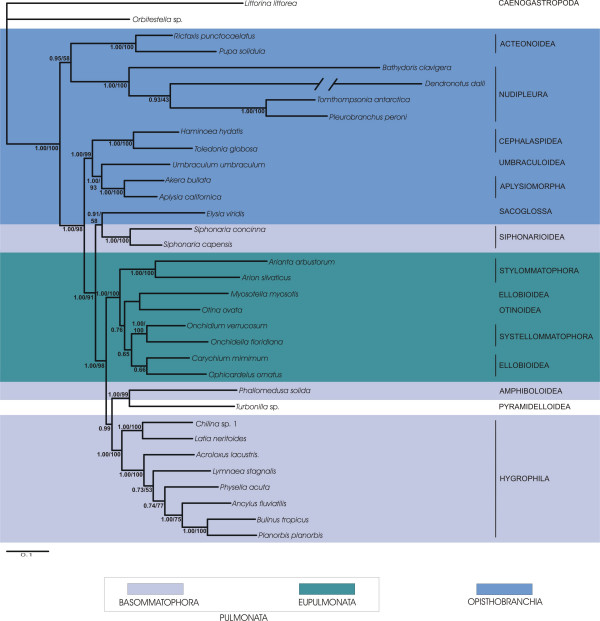
Figure 3.
Bayesian inference phylogram of euthyneuran relationships (reduced taxon set). We used concatenated sequences of 18S rRNA, 28S rRNA, 16S rRNA and COI DNA. 50% majority rule consensus tree. Posterior probabilities and bootstrap support of Maximum Likelihood analysis provided at the branches. Taxonomic classification follows Bouchet et al. [38]. Euthyneuran taxa are marked by colour frames.
The first striking result of this study is the inclusion of the taxon Turbonilla sp. (Pyramidelloidea) within Euthyneura as sister taxon to the Amphiboloidea (Figs. 1, 2 and 3). Within the Euthyneura Opisthobranchia are polyphyletic (Figs. 1 and 3) or paraphyletic (Fig. 2). The Opisthobranchia comprise several distinct clades. The most basal offshoot within Euthyneura is a clade composed of Acteonoidea and Nudipleura, both forming a sister group relationship. However, the internal branches leading towards Acteonoidea and Nudipleura are relatively long. Additionally, terminal branches of Bathydoris clavigera and especially Dendronotus dalli are extremely long indicating high substitution rates in these taxa. Therefore, this result should be considered with caution.
The second opisthobranch clade comprises the Cephalaspidea as sister to the Umbraculoidea plus Aplysiomorpha (Figs. 2 and 3). When Thecosomata and Gymnosomata are included (Fig. 1), they represent the immediate sister taxon to the Aplysiomorpha. The latter three taxa are sister to Umbraculoidea. The Sacoglossa are recovered closely related to the Pulmonata as sister to the Siphonariidae (Figs. 1 and 3). However, support for this sistergroup relationship is non existent in the analyses of the reduced dataset. Nevertheless, inclusion of Sacoglossa within Pulmonata is well supported in all analyses, rendering the Pulmonata paraphyletic. Inclusion of the enigmatic Acochlidiacea in the large taxon placed them within Pulmonata (Fig. 1).
Within Pulmonata we recovered monophyly for Eupulmonata. Whereas the Bayesian statistical support for a monophyletic Eupulmonata is rather low in the analysis of the large taxon set (Fig. 1) possibly due to missing data in these taxa (especially in Stylommatophora, see Table 1), the combined analyses of the reduced taxon set showed a high posterior probability (Fig. 2) and additionally high bootstrap support for a monophyletic Eupulmonata (Fig. 3). Within Eupulmonata, Onchidioidea are a well supported monophyletic clade whereas the Bayesian posterior probability for a monophyletic Stylommatophora is low in the analysis of the large taxon set probably due to the reasons mentioned above. Ellobioidea are poorly supported and their monophyly cannot be recovered unequivocally. Basommatophora are paraphyletic. Within Basommatophora, Siphonariidae and Hygrophila are well supported monophyletic taxa.
Two clades can be distinguished within Hygrophila. The first clade consists of Chilina spp. and Latia neritoides whereas the second clade is comprised of higher limnic Basommatophora. The relationships in this latter clade differ in the different analyses. Within higher limnic Basommatophora only a close relationship of Bulinus tropicus and Planorbis planorbis is consistent and well supported in all analyses.
Reconstruction of character evolution
Based on the phylogenetic hypothesis for Euthyneura deduced with a Bayesian analysis of the reduced data set we reconstructed the ancestral character states and character evolution relating to habitat types in euthyneuran subgroups.
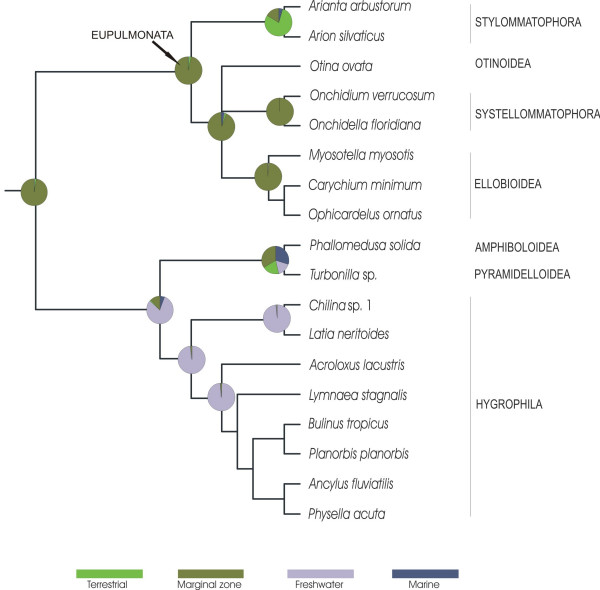
We calculated posterior probabilities for each of the four habitat types for certain ancestral nodes within Pulmonata (Fig. 4). It can be seen from this analysis that colonisation of freshwater habitats occurred only once within Pulmonata by Hygrophila. The ancestor of Hygrophila probably already occurred in a freshwater habitat. Colonisation of terrestrial habitats in Eupulmonata has its seeds in marginal zones such as estuaries or mangroves. The ancestor of Eupulmonata, Amphiboloidea plus Pyramidelloidea and Hygrophila probably already lived in a marginal habitat as indicated by the high proportion of posterior probabilities for this habitat type at the respective node (Fig. 4). Otina ovata within Eupulmonata and Turbonilla sp. both inhabit marine environments.
Figure 4.
Ancestral character state reconstruction of habitat types in Eupulmonata and Hygrophila. We used Bayesian inference methods and mapped them onto part of Figure 2. Pie charts symbolise the relative proportions of posterior probablilites for each of the four character states relating to habitat types.
Discussion
Euthyneura
With the current study we present a comprehensive molecular phylogenetic analysis of Euthyneura including representatives of all major subgroups. We used different approaches to infer a phylogenetic hypothesis for this taxon of Gastropoda. These approaches yielded for the most part constant results respective to the deeper nodes in the tree and indicated only minor differences at generic or familial level.
The unification of Opisthobranchia and Pulmonata in the group Euthyneura has been widely accepted since its original definition by Spengel [4]. Consequently, the monophyly of Euthyneura has mostly been accepted in recent systematic and phylogenetic investigations [5,9,11-13,15,23,24]. Although the defining character of Euthyneura, euthyneury, is considered to be a result of multiple convergence [16,17], several autapomorphic characters supporting monophyly have been proposed for Euthyneura [11,16,17].
Our results contradict a monophyletic Euthyneura, since Pyramidelloidea are included within euthyneurans as sister group to the Amphiboloidea. This is in congruence with Grande et al. [19] who found Pyramidelloidea deeply nested within Pulmonata in their molecular phylogenetic analysis. Although not having included Pyramidelloidea in his molecular phylogenetic analysis of Euthyneura, Thollesson [18] proposed a possible synapomorphy of Pyramidelloidea and Euthyneura in the presence of a gap in the helix G16 of the 16S rRNA gene, thus supporting inclusion of Pyramidelloidea within Euthyneura as proposed here. Studies considering additional pyramidelloid and other basal heterobranch taxa currently undertaken in the group of the senior author will hopefully shed more light on the position of the enigmatic Pyramidelloidea within Heterobranchia (Euthyneura plus Heterostropha sensu Ponder and Waren [10]). Additionally, new character complexes, such as secondary structures of ribosomal genes, could provide further valuable phylogenetic information for the taxa in question, as has been demonstrated recently by Lydeard et al. [25].
Opisthobranchia
Although the separation between Opisthobranchia and Pulmonata is rather distinct because of e. g. characters of the nervous system, it has been difficult to propose sound autapomorphies for a monophyletic Opisthobranchia [12]. Although the subgroups of Opisthobranchia can be clearly distinguished and appear to be monophyletic [21,26-29], monophyly of the Opisthobranchia altogether could not be shown by most recent phylogenetic analyses, regardless of whether they used morphological or molecular data [18,19,21,22,30]. The Opisthobranchia appear to be paraphyletic (cp. e. g. [7,18]) rather than a natural grouping. Our data confirm this latter assumption by rendering the Opisthobranchia either polyphyletic or paraphyletic (depending on the analysis). Filtering our MCMC trees of the Bayesian analysis of the reduced dataset (including all genes) under the constraint of a monophyletic Opisthobranchia revealed that out of 100.000 trees not one single tree supports a monophyletic Opisthobranchia. Application of the Approximately Unbiased test [31] to both reduced datasets also rejected a monophyletic Opisthobranchia at the 5% significance level. In contrast, we can distinguish several clades within Opisthobranchia clearly distinct from each other gradually leading to the pulmonate level of organisation.
One such clade comprises the Nudipleura and Acteonoidea which in our trees appears at the base of the Euthyneura. Acteonoidea have traditionally been regarded as the most primitive opisthobranchs or have even been excluded from Opisthobranchia [22,27,32] whereas Nudipleura are derived [21,22]. However, this unexpected grouping of both taxa has recently continuously been revealed by molecular systematic studies. [20,21,33]. Nevertheless, statistical support in our analyses is rather low and good synapomorphies for this sister group relationship are still warranted. Moreover, we observed deviant base composition and rate heterogeneity in Nudipleura which also could attribute to the basal position of this taxon and possibly artificially groups them with Acteonoidea.
A second well supported opisthobranch clade in the current analysis of the large taxon set comprises the Cephalaspidea, Umbraculoidea and Aplysiomorpha plus Thecosomata and Gymnosomata (Pteropoda). This close grouping of Cephalaspidea, Aplysiomorpha and Pteropoda has been suggested lately by several studies [17,33]. The position of Umbraculoidea within this clade, although well supported in our current analyses, should be considered with caution since only one umbraculoid taxon has been included. Moreover, no morphological synapomorphies are known to support a close relationship of Umbraculoidea and Aplysiomorpha/Pteropoda. More umbraculoid taxa must be considered in future studies to clarify the phylogenetic position of this taxon.
The phylogenetic position of Sacoglossa within Opisthobranchia/Euthyneura has been a matter of debate [20-22]. Although morphologically well defined as a monophylum [26,27], different analyses assign Sacoglossa equivocally to different clades within the Euthyneura. Our data suggest a close affinity to primitive Pulmonata. However, Sacoglossa are definitely a taxon that needs to be paid more attention to in future phylogenetic studies.
The same holds true for the Acochlidiacea, an enigmatic taxon inhabiting interstitial marine habitats and even freshwater systems, a feature unique within Opisthobranchia. Due to their very small size (often less than 1mm in length) hardly anything is known about their morphology and life history. Recent morphological investigations utilising modern computer generated 3-D reconstructions [34,35] have shed some light on the anatomy of these enigmatic gastropods, however, more studies are warranted. The phylogenetic position of the Acochlidiacea is still unclarified. Vonnemann et al. [21] found them as a basal taxon within Opisthobranchia, while our results imply a position within Pulmonata.
Pulmonata
The monophyly of Pulmonata is widely accepted on the basis of morphological characters [6,7,11,13,16]. In contrast, our data suggest paraphyly of Pulmonata due to the inclusion of Sacoglossa, Pyramidelloidea and possibly Acochlidiacea. If our phylogenetic hypothesis is correct, one must postulate that pulmonate autapomorphies (such as acquisition of a pneumostome and pulmonary vessels as well as the presence of a procerebrum and dorsal bodies) have secondarily been reduced in Sacoglossa, Acochlidiacea and Pyramidelloidea.
Paraphyly or even polyphyly of Pulmonata was recovered by molecular systematic studies [14,19,36].
Within Pulmonata we can distinguish two monophyletic clades: Eupulmonata and Hygrophila. The unification of Siphonarioidea and Amphiboloidea in the Thalassophila based on morphological data [13] could not be confirmed by our data.
The Eupulmonata comprise a group of marine, semi-terrestrial and truly terrestrial gastropods showing a high diversity with regards to species number and life history characters. Monophyly of this taxon is strongly supported by Wade and Mordan [15] and the present results, however no morphological apomorphy is known to date [9,10]. Relationships within Eupulmonata have been disputed in the past. Monophyly of Stylommatophora is undoubted [11,13-15,17,37] and can be supported by several autapomorphies [11,13]. Our results also show a monophyletic Stylommatophora. Haszprunar and Huber [16] proposed a close relationship of Ellobiidae, Trimusculidae and Stylommatophora on the basis of apomorphic characters of the nervous system, whereas Dayrat and Tillier [11,12] consider Stylommatophora and Systellommatophora (Onchidioidea and Veronicelloidea) to be closely related. Neither of these two hypotheses are confirmed by our findings since we find a close relationship between ellobioid taxa, Otinoidea, Onchidioidea and Trimusculoidea. The phylogenetic status of the Ellobioidea cannot be conclusively clarified from our data since they appear to be paraphyletic in the two analyses utilising all sequence data. Exclusion of 16S-sequences and 3rd codon positions of COI-sequences, however, recovers them monophyletic.
The Basommatophora sensu Nordsieck [13] and Bouchet et al. [38] are paraphyletic in our analyses. No topology in our set of 100.000 MCMC trees of the Bayesian analysis of the reduced dataset supports a monophyletic Basommatophora. Moreover, testing an alternative tree topology with a constrained monophyletic Basommatophora with the Approximately Unbiased test [31] rejected monophyly of Basommatophora at the significance level of 5%. This is not surprising since the monophyly of Basommatophora has already been disputed by Hubendick [39]. Within Basommatophora the Hygrophila are a well supported and defined group of pulmonates. Our results support earlier hypotheses of a common origin of these freshwater taxa [12,13,17,40]. For the first time, however, crucial taxa like Latia are included in a molecular study. Within Hygrophila two clades can be distinguished; on the one hand the Chilinoidea (sensu Boss [3], including Chilinidae and Latiidae), and on the other the higher limnic Basommatophora. Chilina and Latia are sister-species, a relationship that has already been mentioned earlier [13,39,41]. Higher limnic Basommatophora can be divided into four well distinguishable families [39] while interfamilial relationships of these taxa vary in the current analyses thus rendering them beyond the scope of this study. These relationships are discussed in other publications [42,43].
Former taxonomic classifications of Pulmonata united a group of primitive taxa (Ellobiidae, Otinidae, Amphibolidae, Siphonariidae, Chilinidae and Latiidae) in the Archaeopulmonata [44]. Other authors followed this classification [6,3,45]. However, our results clearly indicate a paraphyletic nature of these taxa and therefore we propose the disuse of the name Archaeopulmonata.
Colonisation of freshwater and terrestrial habitats
One key step in the evolution of Euthyneura was the invasion of freshwater and terrestrial habitats by Pulmonata resulting in a multitude of taxa worldwide adapted to these habitats. It is undisputed that the first pulmonate gastropods were marine organisms [12,46]. Marine pulmonates can be found in truly marine submersed environments (such as Williamia) and, more frequently, in upper littoral zones (Siphonaria, Myosotella, Trimusculus). Certain taxa even occur in estuaries or mangroves (Onchidiidae, certain Ellobiidae, Amphibolidae). Terrestrial pulmonates inhabit a wide spectrum of habitats, ranging from caves to lowlands to mountains and from boreal to tropical ecological zones. The same holds true for freshwater taxa, which occur in different types of limnic environments worldwide.
Our reconstruction of character evolution relating to habitat type (Fig. 4) shows that within Pulmonata the freshwater habitat has only been conquered once by the Hygrophila. Calculation of posterior probabilities for the different habitat types at specific nodes indicates that the ancestor of Hygrophila probably already lived in a freshwater habitat. Acknowledging that there are extremely few onchidioidean species reported to live in brackish or freshwater it would not change the picture that the major radiation into freshwater happened only once leading to the Hygrophila. The same is true for the few acochlidiaceans that are known from freshwater habitats. Freshwater members of both taxa were not included in our data set, for the time being, we would also hypothesize a similar evolutionary pathway excluding a terrestrial step.
In non-pulmonates, the situation is different. 'Prosobranchs', such as the superfamily Cerithoidea, comprise predominantly marine taxa. Nevertheless, freshwater was invaded independently in several cerithoid lineages [47]. Even the evolution of life history traits such as viviparity in the freshwater invaders was correlated with this evolutionary step [48].
The ancestor of Eupulmonata and Hygrophila, in contrast, appears to have lived in a marginal zone like supralittoral zones, estuaries or mangroves. Therefore we conclude, that colonisation of freshwater in Pulmonata occurred via an aquatic pathway directly from the marine realm and not via a terrestrial step.
The terrestrial habitats have most probably also been invaded via marginal zones, as can be seen from the Bayesian reconstruction of ancestral character states in Eupulmonata. Involvement of freshwater systems in colonisation of terrestrial habitats is less likely, since the freshwater taxa (Hygrophila) are clearly separated from Eupulmonata and the posterior probability for a freshwater habitat is basically non-existant at the base of the Eupulmonata.
Terrestriality on the contrary is thought to have evolved much more than 10 times independently (discussed in Barker [49]). Barker [49] already emphasized that recent ellobiid terrestriality was clearly derived from marine littoral ancestors and not secondarily from terrestrial ancestors.
Invasion of marine habitats by Otina ovata within Eupulmonata and Turbonilla sp. obviously is the result of a secondary colonisation originating in marginal zones.
Conclusion
Euthyneura are considered the crown group of Gastropoda. Within this taxon Gastropoda have reached their peak in species richness and ecological diversity. This obvious evolutionary success can probably be attributed to several factors. Marine Opisthobranchia, e. g., have evolved several clades specialised on less utilised food resources such as sponges or cnidarians. A key innovation in the evolution of Pulmonata was the colonization of freshwater and terrestrial habitats.
Previously published phylogenies [11,17,19] did not allow an exact inference of possible pathways respective to pulmonate distribution. Up to now, it was not clear whether the invasion of freshwater happened directly from the marine environment or via a terrestrial step. Based on our phylogenetic hypothesis of Euthyneura and especially Pulmonata we are now able to trace an evolutionary scenario regarding colonisation of different habitats by these Gastropoda.
Our study on the phylogeny of Euthyneura has clearly shown, that traditional classification of these Gastropoda needs to be reconsidered. We were able to present well supported clades and reconstructed the evolutionary history of these clades regarding invasion of habitats and occupation of ecological niches. This should serve as the basis for future discussions on the evolution of this successful group of gastropod molluscs.
Methods
Taxon sampling and specimen collection
A total of 53 representative taxa of most major extant lineages of Euthyneura were included in the current phylogenetic study. Additionally, we included two basal heterobranch species (Turbonilla sp. and Orbitestella sp.) as well as the caenogastropod Littorina littorea. Specimens were collected from different locations worldwide (for details see Table 1). They were collected from the field by hand, snorkelling or scuba diving, preserved in 80–100% ethanol and stored cooled until further processing.
For some taxa (Stylommatophora, Amphibolidae, Ellobiidae, Opisthobranchia and Littorinidae) we obtained published sequences from Genbank). Accession numbers of all sequences used in the analyses are listed in Table 1.
DNA extraction, PCR and sequencing
Genomic DNA was extracted from muscle tissue or entire animals using the DNeasy Tissue Kit (Qiagen, Hilden, Germany) or the protocol given in [43]. Voucher depositions are listed in Additional file 1. We amplified fragments of four genes, including 18S rDNA, 28S rDNA, 16S rDNA and COI. For details of primers used and PCR protocols see Additional file 2.
After the amplification products were purified from an agarose gel using the QIAquick Gel Extraction Kit from Qiagen (Hilden, Germany). The DNA was subjected to cycle sequencing using the ABI Prism Big Dye terminator kit (Perking-Elmer, Norwalk, CT, USA) or the CEQ DTCS Quick Start Kit (Krefeld, Germany). DNA sequences were obtained with an ABI 377 automated DNA sequencer at the Scientific Research Lab, Frankfurt/Main and by using a CEQ 2000 Beckmann Coulter sequencer. All fragments were sequenced in both directions using the same primer sequences as used for the PCR. For 18S and 28S several internal primers were used (Additional file 3).
Sequence alignment
Sequence alignments were performed with CLUSTAL W implemented in the software package BioEdit Version 7.0.5 [50]. Default parameters were used but subsequent manual correction was necessary and was performed with BioEdit. Table 2 provides an overview on the number of taxa and lengths of the sequence alignments used in the analyses. Several base positions have been excluded from the alignments of 18S rRNA, 28S rRNA and 16S rRNA (for details see Table 2) prior to reconstructing phylogeny due to inserts in certain taxa or due to high variability within the alignments (especially in the 28S- and 16S-fragment).
Statistical tests
We investigated base compositions by means of the software PAUP 4.0b10 [51]. To test the degree of substitution saturation we used the test of Xia et al. [52], as implemented in the software package DAMBE [53]. A relative rate test was performed with the software k2WuLi [54] in order to test for rate heterogeneity in the sequences. In order to examine whether there are significant differences in incongruence length between the four datasets, a homogeneity-partition test implemented in PAUP 4.0b10 [51] was performed. Alternative tree topologies were tested for the reduced taxon sets using the Approximately Unbiased (AU) test [31]. The likelihood at each nucleotide position was calculated for each alternative topology (constrained monophyletic Opisthobranchia, constrained monophyletic Basommatophora) as well as the topology under scrutiny using PAUP 4.0b10 [51]. Likelihoods were used to calculate p-values using CONSEL version 0.1 [55].
Phylogenetic analyses
Prior to phylogenetic analyses, we determined the optimal model of sequence evolution for each of the four partitions and each of the taxon sets using Modeltest 3.7 [56] based on the Akaike information criterion (for details of the models determined refer to Table 2).
We applied several different analyses to our data. First, we used a concatenated alignment of all four partitions including all taxa for which sequences were available (large taxon sets). Missing sequences for certain taxa in certain partitions were coded as missing data in the respective alignments.
Secondly, we also followed a more conservative approach by analysing a combined dataset of all four partitions with only those taxa for which sequences of all partitions were available (reduced taxon set, 34 taxa). For all analyses Bayesian inference methods were applied with Mr Bayes 3_03b [57], using separate models of evolution for each of the four partions. The tree space was explored using four chains of a Markov Chain Monte Carlo algorithm (one cold, three heated) for 1,000000 generations. Likelihoods of the trees started at around -67470 and quickly converged upon a stable value of about -54600 after approximately 7,000 generations. Thus we computed a 50% majority rule consensus tree with the first 1,000 trees (= 10,000 generations) ignored as burn-in. Posterior probabilities were calculated for each node in the tree. We consider a BPP value of 0.95 and higher to give good support to a node.
Additional Maximum likelihood analyses were conducted for all data sets using RAxML-VI-HPC version 4.0.0 [58]). This program allows analyzing large combined data sets and uses a GTR model based approach with internal estimation of all free model parameters for all data partitions. Bootstrapping with 100 replicates was performed.
Trees were rooted with the caenogastropod Littorina littorea.
Reconstruction of character evolution
We reconstructed character evolution regarding habitat types with a Bayesian approach as implemented in the BayesTraits software package [59]. The BayesMultiState option allows to automatically find posterior distribution of models of evolution using the reversible-jump MCMC method. Ancestral character states were reconstructed for selected nodes in the phylogeny resulting in respective probabilities of all character states for that node. Data on habitat types colonised by the investigated taxa were taken from the literature. Information on coding for individual taxa can be taken from Additional file 4. Rate deviation was set to 8.0. A hyperprior approach was used with exponential prior seeded from a uniform on the intervall 0 to 30. Thus, acceptance rates between 20 and 30 % were achieved, which are in the preferred range.
Authors' contributions
AKK, CA and BS designed the study. AKK and CA collected most of the material. AKK analysed the data for phylogenetic inferences and drafted the manuscript. AKK and CA performed the ancestral character state reconstruction and CA was additionally involved in the final preparation of the manuscript. AD and KK provided the gene sequences and also provided input to the manuscript. BS provided background information. All authors have read and approved the final manuscript version.
Supplementary Material
Information on voucher depositions for specimens. The table provides information on locality of voucher depositions of specimens (tissue, shells, DNA) of samples investigated.
Information on PCR conditions. The table provides information on fragments amplified, PCR primers used, and PCR protocols.
Information on internal sequencing primers. The table provides the nucleotide sequences for the internal sequencing primers of the 18S rRNA gene and the 28S rRNA fragment.
Coding scheme for ancestral habitat reconstruction. Information is provided of coding of habitat types (marine, freshwater, terrestrial habitat and marginal zones) of the investigated species.
Contributor Information
Annette Klussmann-Kolb, Email: Klussmann-Kolb@bio.uni-frankfurt.de.
Angela Dinapoli, Email: a.dinapoli@bio.uni-frankfurt.de.
Kerstin Kuhn, Email: k.kuhn@bio.uni-frankfurt.de.
Bruno Streit, Email: streit@bio.uni-frankfurt.de.
Christian Albrecht, Email: Christian.Albrecht@allzool.bio.uni-giessen.de.
Acknowledgements
We are indebted to many colleagues who helped in collecting material from all over the world: Lydia Bousset (Thiverval-Grignon, FRA), the late David Brown (London, UK), Benoit Dayrat (Merced, USA), Andrzej Falniowski (Krakow, POL), Jeff Goddard (Santa Barbara, USA), Martin Haase (Greifwald, GER), Marc Hasenbank (Wellington, NZ), Alan Hodgson (Grahamstown, SA), Aslak Jørgensen (Charlottenlund, DK), Michael Schrödl and Enrico Schwabe (Munich, GER), Sid Staubach (Frankfurt, GER), Magda Szarowska (Krakow, POL), Richard Willan (Darwin, AUS), Tim Wollesen (Frankfurt, GER).
DNA samples and few sequences of Opisthobranchia were provided by Verena Vonnemann (Hamburg, GER). Claudia Nesselhauf (Frankfurt, GER) helped in sequencing some of the material.
Heiko Schmidt (Vienna) was helpful in applying the AU test.
We are thankful to Benoit Dayrat and three anonymous referees for valuable comments.
This project was funded by the German Science Foundation (KL 1303/1-3, KL 1303/4-1) and by the Hessian Ministry of Science and Culture to AKK and by the Freunde und Förderer of Frankfurt University to BS.
References
- Thiele J. Handbuch der systematischen Weichtierkunde. II. Subclassis Opisthobranchia. Jena: Gustav Fischer Verlag; 1931. [Google Scholar]
- Fretter V, Graham A. British Prosobranch Molluscs. London: Ray Society; 1962. [Google Scholar]
- Boss KJ. In: Synopsis and classification of living organisms. Parker SP, editor. New York: McGraw Hill Book Company; 1982. Mollusca; pp. 945–1166. [Google Scholar]
- Spengel JW. Die Geruchsorgane und das Nervensystem der Mollusken. Z Wiss Zool. 1881;35:333–383. [Google Scholar]
- Ponder WF, Lindberg D. Towards a phylogeny of gastropod molluscs: an analysis using morphological characters. Zool J Linn Soc. 1997;119:83–265. doi: 10.1006/zjls.1996.0066. [DOI] [Google Scholar]
- Tillier S. Relationships of gymnomorph gastropods (Mollusca: Gastropoda) Zool J Linn Soc. 1984;82:345–362. [Google Scholar]
- Haszprunar G. The Heterobranchia – a new concept of the phylogeny of the higher Gastropoda. Z Zool Syst Evolutionsforsch. 1985;23:15–37. [Google Scholar]
- Haszprunar G. On the origin and evolution of major gastropod groups, with special reference to the streptoneura. J Moll Stud. 1988;54:367–441. doi: 10.1093/mollus/54.4.367. [DOI] [Google Scholar]
- Salvini-Plawen L, Steiner G. In: Origin and evolutionary radiation of the Mollusca. Taylor J, editor. Oxford: Oxford University Press; 1996. Synapomorphies and plesiomorphies in higher classification of Mollusca; pp. 29–51. [Google Scholar]
- Ponder WF, Waren A. Classification of the Caenogastropoda and Heterostropha – a list of the family-group names and higher taxa. Malacol Rev. 1988;4(Suppl 4):288–328. [Google Scholar]
- Dayrat B, Tillier S. Evolutionary relationships of euthyneuran gastropods (Mollusca): a cladistic re-evaluation of morphological characters. Zool J Linn Soc. 2002;135:403–470. doi: 10.1046/j.1096-3642.2002.00018.x. [DOI] [Google Scholar]
- Dayrat B, Tillier S. In: Molecular Systematics and phyleogeography of molluscs. Lydeard C, Lindberg D, editor. Washington: Smithsonian Books; 2003. Goals and limits of phylogenetics – The Euthyneuran Gastropods; pp. 161–184. [Google Scholar]
- Nordsieck H. Phylogeny and system of Pulmonata. Arch Moll. 1992;121:31–52. [Google Scholar]
- Tillier S, Masselot M, Tillier A. In: Origin and Evolutionary Radiation of the Mollusca. Taylor J, editor. Oxford: Oxford University Press; 1996. Phylogenetic relationships of the pulomante gastropods from rRNA sequences, and tempo and age of the stylommatophoran radiation; pp. 267–284. [Google Scholar]
- Wade CM, Mordan PB. Evolution within the gastropod molluscs; using the ribosomal RNA gene-cluster as an indicator of phylogenetic relationships. J Moll Stud. 2000;66:565–570. doi: 10.1093/mollus/66.4.565. [DOI] [Google Scholar]
- Haszprunar G, Huber G. On the central nervous system of Smeagolidae and Rhodopidae, two families questionably allied to the Gymnomorpha (Gastropoda, Euthyneura) J Zool. 1990;220:185–199. [Google Scholar]
- Dayrat B, Tillier A, Lecointre G, Tillier S. New clades of euthyneuran Gastropods (Mollusca) from 28S rRNA sequences. Mol Phyl Evol. 2001;19:225–235. doi: 10.1006/mpev.2001.0926. [DOI] [PubMed] [Google Scholar]
- Thollesson M. Phylogenetic analysis of Euthyneura (Gastropoda) by means of the 16S rRNA gene: use of a 'fast'gene for 'higher-level'phylogenies. Proc R Soc Lond B. 1999;266:75–83. doi: 10.1098/rspb.1999.0606. [DOI] [Google Scholar]
- Grande C, Templado J, Cervera JL, Zardoya R. Molecular Phylogeny of Euthyneura (Mollusca:Gastropoda) Mol Biol Evol. 2004;21:303–313. doi: 10.1093/molbev/msh016. [DOI] [PubMed] [Google Scholar]
- Grande C, Templado J, Cervera JL, Zardoya R. Phylogenetic relationships among Opisthobranchia (Mollusca: Gastropoda) based on mitochondrial cox 1, trnV, and rrnL genes. Mol Phyl Evol. 2004;33:378–388. doi: 10.1016/j.ympev.2004.06.008. [DOI] [PubMed] [Google Scholar]
- Vonnemann V, Schrödl M, Klussmann-Kolb A, Wägele H. Reconstruction of the phylogeny of the Opisthobranchia (Mollusca, Gastropoda) by means of 18S and 28S rRNA gene sequences. J Moll Stud. 2005;71:113–125. doi: 10.1093/mollus/eyi014. [DOI] [Google Scholar]
- Wägele H, Klussmann-Kolb A. Opisthobranchia (Mollusca, Gastropoda) - more than just slimy slugs. Shell reduction and its implications on defence and foraging. Front Zool. 2005;2(1):3. doi: 10.1186/1742-9994-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillier S, Masselot M, Guerdoux J, Tillier A. Monophyly of major gastropod taxa tested from partial 28S rRNA sequences, with emphasis on Euthyneura and Hot-Vent Limpets Peltosporoidea. Nautilus. 1994. pp. 122–140.
- Yoon SH, Kim W. Phylogeny of some gastropod molluscs derived from 18S rRNA sequences with emphasis on Euthyneura. Nautilus. 2000;114:84–92. [Google Scholar]
- Lydeard C, Holznagel WE, Ueshima R, Kurabayashi A. Systematic implications of extreme loss or reduction of mitochondrial LSU rRNA helical-loop structure in gastropods. Malacologia. 2002;44:349–352. [Google Scholar]
- Jensen KR. Phylogenetic systematics and classification of the Sacoglossa (Mollusca, Gastropoda, Opisthobranchia) Phil Trans R Soc Lond B. 1996;351:91–122. doi: 10.1098/rstb.1996.0006. [DOI] [Google Scholar]
- Mikkelsen PM. The evolutionary relationships of Cephalaspidea s. l. (Gastropoda: Opisthobranchia): a phylogenetic analysis. Malacologia. 1996;37:375–442. [Google Scholar]
- Wägele H, Willan R. Phylogeny of the Nudibranchia. Zool J Linn Soc. 2000;130:83–181. doi: 10.1006/zjls.1999.0214. [DOI] [Google Scholar]
- Medina M, Walsh PJ. Molecular systematics of the order Anaspidea based on mitochondrial DNA sequences (12S, 16S, and COI) Mol Phyl Evol. 2000;15:41–58. doi: 10.1006/mpev.1999.0736. [DOI] [PubMed] [Google Scholar]
- Wägele H, Vonnemann V, Wägele W. In: Molecular Systematics and Phylogeography of Mollusks. Lydeard C, Lindberg DR, editor. Washington: Smithsonian Books; 2003. Toward a phylogeny of the Opisthobranchia; pp. 185–228. [Google Scholar]
- Shimodaira H. An Approximately Unbiased Test of Phylogenetic Tree Selection. Syst Biol. 2002;51:492–508. doi: 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
- Mikkelsen PM. Shelled opisthobranchs. Adv Mar Biol. 2002;42:67–136. doi: 10.1016/s0065-2881(02)42013-5. [DOI] [PubMed] [Google Scholar]
- Klussmann-Kolb A, Dinapoli A. Systematic position of the pelagic Thecosomata and Gymnosomata within Opisthobranchia (Mollusca, Gastropoda) – revival of the Pteropoda. J Zool Syst Evol Res. 2006;44:118–129. doi: 10.1111/j.1439-0469.2006.00351.x. [DOI] [Google Scholar]
- Sommerfeldt N, Schrödl M. Microanatomy of Hedylopsis ballantinei, a new interstitial acochlidian gastropod from the Red Sea, and its significance for phylogeny. J Moll Stud. 2005;71:153–165. doi: 10.1093/mollus/eyi018. [DOI] [Google Scholar]
- Neusser TP, Heß M, Haszprunar G, Schrödl M. Computer-based three-dimensional reconstruction of the anatomy of Microhedyle remanei (Marcus, 1953), an interstitial acochlidian gastropod from Bermuda. J Morph. 2006;267:231–247. doi: 10.1002/jmor.10398. [DOI] [PubMed] [Google Scholar]
- Knudsen B, Kohn AB, Nahir B, MacFadden CS, Moroz LL. Complete DNA sequence of the mitochondrial genome of the sea-slug, Aplysia californica : Conservation of the gene order in Euthyneura. Mol Phyl Evol. 2006;38:459–469. doi: 10.1016/j.ympev.2005.08.017. [DOI] [PubMed] [Google Scholar]
- Wade CM, Mordan PB, Naggs F. Evolutionary relationships among the pulmonate land snails and slugs (Pulmonata, Stylommatophora) Biol J Linn Soc. 2006;87:593–610. doi: 10.1111/j.1095-8312.2006.00596.x. [DOI] [Google Scholar]
- Bouchet P, Rocroi P, Fryda J, Hausdorf B, Ponder W, Valdes A, Waren A. Classification and Nomenclator of Gastropod Families. Malacologia. 2005;47:1–397. [Google Scholar]
- Hubendick B. In: Pulmonates. Systematics, evolution and ecology. Fretter V, Peake J, editor. Vol. 24. New York, London, San Francisco: Academic Press; 1978. Systematics and comparative morphology of the Basommatophora; pp. 1–47. [Google Scholar]
- Salvini-Plawen L. Origin, phylogeny and classification of the phylum Mollusca. Iberus. 1990;9:1–33. [Google Scholar]
- Vaught KC. A classification of the living Mollusca. Melbourne, F.L : American Malacologists, Inc; 1989. [Google Scholar]
- Albrecht C, Wilke T, Kuhn K, Streit B. Convergent evolution of shell shape in freshwater limpets: the African genus Burnupia. Zool J Linn Soc. 2004;140:577–588. doi: 10.1111/j.1096-3642.2003.00108.x. [DOI] [Google Scholar]
- Albrecht C, Kuhn K, Streit B. A molecular phylogeny of Planorboidea (Gastropoda: Pulmonata): insights from enhanced taxon sampling. Zool Scr. 2007;36:27–39. doi: 10.1111/j.1463-6409.2006.00258.x. [DOI] [Google Scholar]
- Morton JE. The evolution of the Ellobiidae with a discussion on the origin of the Pulmonata. Proc Zool Soc Lond. 1955;125:127–168. [Google Scholar]
- Harbeck K. Die Evolution der Archaeopulmonata. Zoo Verh Leiden. 1996;305:3–133. [Google Scholar]
- Hubendick B. Phylogenie und Tiergeographie der Siphonariidae. Zur Kenntnis der Phylogenie in der Ordnung Basommatophora und des Ursprungs der Pulmonatengruppe. Zool Bid Upp. 1945;24:1–216. [Google Scholar]
- Lydeard C, Holznagel WE, Glaubrecht M, Ponder WF. Molecular phylogeny and evidence for multiple origins of freshwater gastropods of the circum-global, diverse superfamily Cerithioidea (Mollusca: Caenogastropoda) Mol Phyl Evol. 2002;22:399–406. doi: 10.1006/mpev.2001.1072. [DOI] [PubMed] [Google Scholar]
- Köhler F, von Rintelen T, Meyer A, Glaubrecht M. Multiple origin of viviparity in Southeast Asian gastropods (Cerithioidea: Pachychilidae) and its evolutionary implications. Evolution. 2004;58:2215–2226. doi: 10.1111/j.0014-3820.2004.tb01599.x. [DOI] [PubMed] [Google Scholar]
- Barker GM. In: The biology of terrestrial Mollusca. Barker GM, editor. Oxford: CABI Publishing; 2001. Gastropods on Land: Phylogeny, Diversity and Adaptive Morphology. I; pp. 1–146. [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequences alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Swofford DL. PAUP. Phylogenetic analysis using parsimony (and other methods). Version 4_10b. Sunderland, Mass: Sinauer Associates. 2002.
- Xia X, Xie Z, Salemi M, Chen L, Wang Y. An index of substitution saturation and its application. Mol Phyl Evol. 2003;26:1–7. doi: 10.1016/S1055-7903(02)00326-3. [DOI] [PubMed] [Google Scholar]
- Xia X, Xie Z. DAMBE: data analysis in molecular biology and evolution. J Hered. 2001;92:371–373. doi: 10.1093/jhered/92.4.371. [DOI] [PubMed] [Google Scholar]
- Wu CI, Li WH. Evidence for higher rates of nucleotides substitution in rodents than in man. Proc Nat Acad Sci. 1985;82:1741–1745. doi: 10.1073/pnas.82.6.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics. 2001;17:1246–1247. doi: 10.1093/bioinformatics/17.12.1246. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MrBAYES; Bayesian inference for phylogeny. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Stammatakis A. RaxML-VI-HPC: Maximum Likelihood-based Phylogenetic Analyses with Thousands of Taxa and Mixed Models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Pagel M, Meade A. Bayesian analysis of correlated evolution of discrete characters by reversible-jump Markov chain Monte Carlo. Am Nat. 2006;147:808–825. doi: 10.1086/503444. [DOI] [PubMed] [Google Scholar]
- Wollscheid E, Wägele H. Initial results on the molecular phylogeny of the Nudibranchia (Gastropoda, Opisthobranchia) based on 18S rDNA data. Mol Phyl Evol. 1999;13:215–226. doi: 10.1006/mpev.1999.0664. [DOI] [PubMed] [Google Scholar]
- Winnepenninckx B, Backeljau T, De Wachter R. Small ribosomal subunit RNA and the phylogeny of Mollusca. Nautilus. 1994. pp. 98–110.
- Bargues MD, Mas-Coma S. Phylogenetic analysis of lymnaeid snails based on 18S rDNA sequences. Mol Biol Evol. 1997;14:569–577. doi: 10.1093/oxfordjournals.molbev.a025794. [DOI] [PubMed] [Google Scholar]
- Simon S, Frati F, Beckenbach A, Crespi B, Li H, Flook P. Evolution, weighting and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann Ent Soc Am. 1994;87:651–701. [Google Scholar]
- Schwenk K, Sand A, Boersma M, Brehm M, Mader E, Offerhaus D, Spaak P. Genetic markers, genealogies and biogeographic patterns in the Cladocera. Aqu Ecol. 1998;32:37–51. doi: 10.1023/A:1009939901198. [DOI] [Google Scholar]
- Folmer O, Black M, Heah W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome C oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3:294–299. [PubMed] [Google Scholar]
- Stothard JR, Rollinson D. Partial DNA sequences from the mitochondrial cytochrome oxidase subunit I (COI) gene can differentiate the intermediate snail hosts Bulinus globosus und B. nasutus (Gastropoda: Planorbidae) J Nat Hist. 1997;31:727–737. doi: 10.1080/00222939700770361. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Information on voucher depositions for specimens. The table provides information on locality of voucher depositions of specimens (tissue, shells, DNA) of samples investigated.
Information on PCR conditions. The table provides information on fragments amplified, PCR primers used, and PCR protocols.
Information on internal sequencing primers. The table provides the nucleotide sequences for the internal sequencing primers of the 18S rRNA gene and the 28S rRNA fragment.
Coding scheme for ancestral habitat reconstruction. Information is provided of coding of habitat types (marine, freshwater, terrestrial habitat and marginal zones) of the investigated species.