Abstract
Hepatitis C virus (HCV) causes acute and chronic liver disease in humans, including chronic hepatitis, cirrhosis, and hepatocellular carcinoma. The polyprotein encoded in the HCV genome is co- and posttranslationally processed by host and viral peptidases, generating the structural proteins Core, E1, E2 and p7, and five non-structural proteins. The two envelope proteins E1 and E2 are heavily glycosylated. Studying the glycan moieties attached to the envelope E2 glycoprotein is important because the N-linked glycans on E2 envelope protein are involved in the interaction with some human neutralizing antibodies, and may also have a direct or indirect effect on protein folding. In the present study we report the mass spectrometric characterization of the glycan moieties attached to the E2 glycoprotein. The mass spectrometric analysis clearly identified the nature, composition and microheterogeneity of the sugars attached to the E2 glycopeptides. All 11 sites of glycosylation on E2 protein were characterized, and the majority of these sites proved to be occupied by high mannose glycans. However, complex type oligosaccharides, which have not been previously identified, were exclusively observed at two N-linked sites and their identity and heterogeneity were determined.
Introduction
HCV infects over 170 million people worldwide. Infection develops into chronic hepatitis, which is one of the most prevalent causes of liver cirrhosis and represents the most frequent indication for liver transplantation. HCV is a small, enveloped positive-strand RNA virus belonging to the Flaviviridae family [1]. The genome of HCV is ∼ 9.5 kb encoding for a single polyprotein between 3010 and 3033 amino acids in length. A combination of host and viral peptidases are involved in processing the polyprotein, which results in at least nine different proteins. The HCV polyprotein is synthesized on the endoplasmic reticulum (ER) associated ribosomes and is processed co- and posttranslationally generating the structural proteins Core, E1, E2, and p7, and five nonstructural proteins [2]. The two envelope proteins E1 and E2 are heavily N-glycosylated and they are believed to be type 1 transmembrane protein with N terminal ectodomains and C terminal hydrophobic anchors [3]. Together, they are expected to form the viral envelope [4]. During their synthesis, the ectodomains of HCV glycoproteins are targeted to the ER lumen where they are modified by N-linked glycosylation. This suggests that HCV glycoprotein complexes contain a retention signal for localization in an intracellular compartment [5]. These glycoproteins interact to form a noncovalent heterodimeric complex that accumulates in ER-like structures. In principle, ER localization of a protein can be the consequence of actual retention in this organelle or of retrieval from the Golgi [6, 7].
Glycans have been shown to be essential for proper functioning of a protein, and, therefore, may play a significant biological role including locating a protein within the cell, protection of the protein against proteolytic attack, induction and maintenance of the spatial conformation in a biologically active form, facilitation of the extracellular secretion as well as direction and modulation of the immune response [8]. A consensus sequence for N-glycosylation has been reported, Asn-Xaa-Ser/Thr/Cys, in which Xaa may be any amino acid except Pro [9, 10]. One glycosylation site on a protein may have multiple glycan structures (microheterogeneity), and one protein may have different structures at different sites (macroheterogeneity). Structural heterogeneity is an important characteristic of oligosaccharides and significantly complicates the structural analysis of glycoproteins.
Previous studies have shown that the HCV envelope proteins are highly modified by N-linked glycans [11-14]. It was reported that E1 has up to 6 glycosylation sites in comparison with E2, which has 11 potential glycosylation sites [15]. A global sequence analysis of the potential glycosylation sites in E2 indicated that nine of the 11 sites are strongly conserved. The two remaining sites N94 (N476)♣ and N158 (N540) showed a level of conservation of 75% and 89%, respectively (♣the numbers correspond to the amino acid sequence of HCV E2 protein, amino acids 1- 333 (383–715) from strain HCV-1a. The positions of the residue are indicated corresponding to the positions in the HCV polyprotein of reference strain H (GenBank access number AF009606)).
Expression of HCV glycoprotein E2 followed by total or partial deglycosylation indicates that a large number of the glycosylation sites are occupied [15]. Previous studies indicated that some of the N-linked glycans on E2 protein mediate their recognition by human neutralizing antibodies [16]. Furthermore, N-linked glycans are known to play a role in protein folding, and this effect can be either direct or indirect [8, 17]. The presence of a large polar sugar moiety is thought to affect the folding at least locally, by orienting polypeptide segments towards the surface of protein domains.
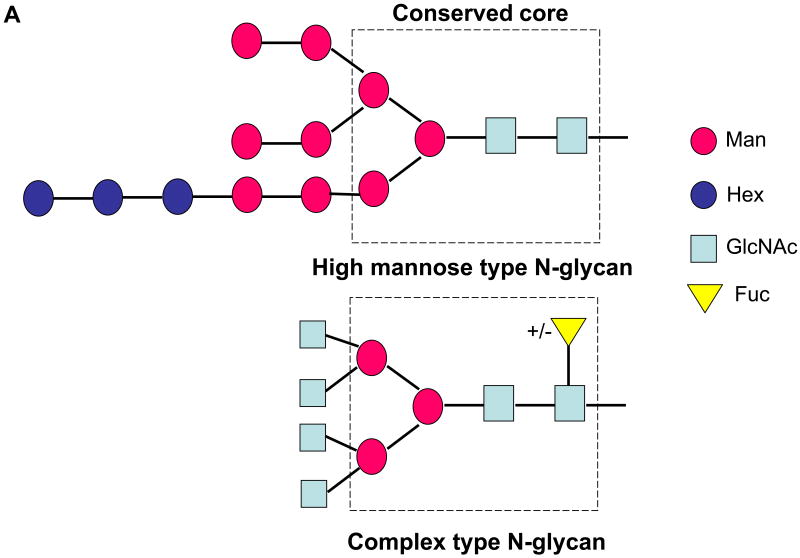
The major types of N-glycans attached to glycoproteins are: (i) high mannose, consisting primarily of mannose (Man) with a maximum number of nine mannoses possible unless not fully processed, (ii) complex type glycans, mainly composed of N-acetylglucosamine (GlcNAc) and galactose (Gal) with or without sialic acid, where a fucose (Fuc) may be added to the first GlcNAc in the core (Figure 1A); and (iii) hybrid type glycans which are composed of mannose, GlcNAc-Gal and with or without sialic acid. There have been several reports about the glycan types attached to the E2 protein. Most of them reported that the glycans are only high mannose type oligosaccharides [5]. It is believed that the lack of complex type glycosylation on the E2 protein excludes the step involving the transit through the medial Golgi and it has been confirmed by immunofluorescence that HCV glycoprotein heterodimer is located in the ER or an ER-like compartment [3, 18]. The notation ManX, where X ranges from 4 to 9 in the case of the observed tryptic and chymotryptic glycopeptides, indicates that X mannose residues are attached to the chitobiose core (GlcNAc β(1-4) GlcNAc). The glycans associated with the HCV glycoprotein heterodimer have been previously characterized by HPLC and three species have been observed: Man9, 8 and 7-GlcNAc2.
Figure 1.
(A) Schematic representation of a precursor glycan (top) and a complex type (bottom) glycan. The dotted square indicates the trimannosyl chitobiose core (Man3GlcNAc2) found in all mature N-linked glycan types. (B) Amino acid sequence of E2, starting from amino acid 1 to 333 (383–715) from strain HCV-1a. The positions of the residues are indicated corresponding to the positions in the HCV polyprotein of reference strain H (GenBank access number AF009606) [38]. The 11 sites of glycosylation are shown in bold and italics.
Whereas unmodified proteins can often be studied by X-ray crystallography or nuclear magnetic resonance spectroscopy, these methods may not provide reliable structural information about the oligosaccharide moieties of glycoproteins. Recently, mass spectrometry (MS) using matrix assisted laser desorption/ ionization (MALDI) and/or electrospray ionization (ESI) [19-21] have become increasingly used in glycobiology [22-24], where no other structural technique can match MS for the range of structural problems that can be addressed, the complexity of samples that can be analyzed successfully, and the quantity of structural information that can be obtained from sub-nanomolar amounts of material. The main advantages of MS are its sensitivity, accuracy, speed and applicability to mixtures. The masses of glycans on a protein can be determined either as free oligosaccharides, after their release from the protein and derivatization, or while still attached to the peptide backbone [25]. This mass information is indicative of the sugar composition and, can often be used to predict sequences based on prior knowledge of biosynthetic pathways. Sequencing of glycoproteins by MS requires characteristic fragment ions (Hex+, m/z 163.1 and HexNAc+, m/z 204.1) to be present in the mass spectra or tandem MS (MS/MS) data [26]. Hence, data from MS analyses of chemical and enzymatic degradations are usually required to supplement molecular and fragment ion information to define structural features such as sugar type, branching and stereochemistry. Complete structural characterization of a glycan however, also requires definition of branching, linkages, configurations, and the identification of similar sugar isomers. Glycosylation analysis is recognized as one of the challenges in mass spectrometry and therefore, to achieve this goal, liquid chromatography and mass spectrometry (LC-MS) methods are invaluable. The combination of LC for the separation and MS/MS for the detection and structural analysis of glycans and glycopeptides provides detailed information at high sensitivity [27]. The most convenient approach for the determination of the composition of the oligosaccharides present in a given protein preparation regardless of their position on the protein backbone(s) requires their release by enzymes (peptide N-glycosidases F and A, endo-glycosidases) or by chemical elimination procedures [28]. Typically on-line LC coupled to ESI MS followed by MS/MS, are widely applied to the analysis of oligosaccharide derivatives [26, 28, 29]. Another approach for the study of glycoproteins is the combination of on-line (capillary electrophoresis) CE-ESI MS as it provides high resolution, sensitivity and relatively short times of analysis [30, 31]. Identification of specific glycosylation sites requires a lengthy process often involving trypsin digestion to produce glycopeptides, separation of the glycopeptides by high performance liquid chromatography (HPLC), and characterization of each fraction by MS with or without enzymatic release, to determine the oligosaccharide and peptide structures of each fraction. Because full scan mass spectra do not always yield unequivocal structural information, tandem mass spectrometry is often necessary to identify unique fragments that correspond to the oligosaccharide moieties [32]. In addition, computer software such as GlycoMod [33], can be used to easily and quickly determine the possible N-glycan composition corresponding to experimentally determined masses of trypsin generated glycopeptides.
In the present study, the mass spectrometric characterization of the glycopeptides from HCV E2 envelope glycoprotein, using nano-liquid chromatography-tandem mass spectrometry (nanoLC/MS/MS) on a Q-Tof hybrid mass spectrometer is reported. The mass spectrometric analyses allowed the identification of the composition of the N-linked oligosaccharides attached to the E2 protein. All 11 sites of glycosylation on E2 were characterized, and the majority of these sites proved to be only of high mannose type. However, complex type oligosaccharides, for which there has been little evidence [34], were exclusively observed at two N-linked sites, and their identity and heterogeneity were determined. This information should prove valuable in the structural modeling of the E2 glycoprotein either entirely in silico or based on eventual crystal structure of the deglycosylated protein.
Experimental
Materials
HCV E2 envelope glycoprotein (recombinant) was purchased from Austral Biologicals (San Ramon, CA). Urea, dithiothreitol, iodoacetamide, 96% formic acid, ammonium bicarbonate and ethanol were purchased from Sigma-Aldrich (St. Louis, MO). Sequencing grade-modified trypsin was obtained from Promega (Madison, WI). Chymotrypsin was purchased from Roche Diagnostics Corporation (Indianapolis, IN). Acetonitrile was purchased from Caledon Laboratories, Ltd. (Georgetown, Ontario). Purified water (17.8 MΩ) was obtained from an in-house Hydro Picopure 2 system. All chemicals were used without further purification unless otherwise specified.
HCV E2 sample preparation
HCV E2 (10 μg) expressed in Chinese hamster ovary (CHO) cells was diluted in 75 μl of buffer containing 5 M Urea and 100 mM Tris (pH 7.8) (hydroxymethyl aminomethane) and 100 mM dithiothreitol was added. The protein was denaturated for 1.5 h at 65°C. Carboxymethylation was performed by adding 100 mM iodacetamide followed by incubation in the dark for 1 h at room temperature. The reaction was quenched by adding 2.5 μl dithiothreitol (100 mM) and incubated for 15 min at room temperature. Prior to digestion, the protein sample was purified, using a Hewlett Packard 1100 HPLC system (Wilmington, DE) equipped with a Vydak C4 column (4.6 mm i.d., 5 μm particles) (Grace Vydac, CA), a diode array detector, and a fraction collector. Gradient elution was carried out with two solvents: A: water, 0.1% formic acid and B: acetonitrile 90%, water 10%, 0.1% formic acid. A linear gradient at 1ml/min of 5% B–60% B over 50 min was used to purify the proteins. Fractions were collected, lyophilized, and redissolved in 50 μl 60% acetonitrile, 0.1% formic acid just prior to MALDI- time of flight (TOF) MS analysis.
In solution digestion of HCV E2
The HPLC fractions found by mass spectrometry to contain the E2 glycoprotein were combined and lyophilized. The protein was redissolved in 50 μl 50 mM ammonium bicarbonate buffer pH 7.5, and trypsin was added using an enzyme: substrate ratio of 1:50, and digested overnight at 37°C. A subsequent digestion was performed in which chymotrypsin was added at an enzyme: substrate ratio of 1:50. The reaction was allowed to proceed overnight at 25°C. After digestion the reaction mixtures were lyophilized and were redissolved in 0.1% formic acid just prior the nanoLC/MS/MS analysis.
Mass spectrometry
MALDI-TOF mass spectra were acquired on a Voyager-DE STR mass spectrometer (Applied Biosystems, Framingham, MA), equipped with a nitrogen laser (λ=337 nm). The measurements were performed in the linear positive ion mode. Samples (0.5 μl) were spotted onto a 100 sample stainless steel MALDI plate and mixed on target with 0.5 μl of a saturated solution of α-cyanohydroxycinnamic acid in 45:45:10 (v:v:v) water:ethanol:formic acid. The mass spectra were calibrated externally using a mixture of standard peptides with a mass accuracy of greater than 0.01%. Data were acquired in linear mode using an accelerating voltage of 23000 volts, a grid percentage of 80%, and a delay time of 300 nanoseconds.
nanoLC/MS/MS analyses were performed on a Waters Q-Tof Premier mass spectrometer equipped with a nanoAcquity UPLC system and a NanoLockspray source (Waters, Milford, MA). Separations were performed using a 3 μm nanoAquity Atlantis column dC18 100 μm × 100 mm (Waters) at a flow rate of 300 nL/min. A nanoAquity trapping column 5 μm C18 180 μm × 200 mm (Waters) was positioned in-line with the analytical column. Trapping of a 2 μl aliquot of the digested sample was performed for 3 min at 5 μl/min flow rate. Peptides were eluted using a linear gradient from 98% A (water/ 0.1% formic acid (v/v)) and 2% B (acetonitrile/ 0.1% formic acid (v/v)) to 95% B over 60 minutes. Mass spectrometer settings for MS analyses were as follows: capillary voltage of 3.5 kV, cone voltage of 30 V, collision energy of 8.0 V, and scan range of 200-2000 Da. The tandem mass spectra were obtained in data dependant acquisition mode, and a fixed range of collision energies between 30 V and 40 V were applied in order to ensure good fragmentation of the glycopeptides. Peptides were identified by mass and charge measurement with in silico digestion using the BioLynx Protein/ Peptide Editor, feature of MassLynx V4.0 (Micromass, UK). For the unambiguous determination of glycosylated peptides, the GlycoMod program, available on the internet as part of the ExPASY suite of proteomics tools at http://www.expasy.ch/tools/glycomod, was utilized [33].
Structural images
To illustrate the structure of HCV E2 an existing homology model made by Yagnik et al. [35] was used, based on Tick-borne encephalitis virus envelope glycoprotein E virus (TBEV). The crystal structure of TBEV (1SVB) [36] was used to build the E2 homology model, and the RasMol program for molecular visualization, was utilized (http://www.umass.edu/microbio/rasmol/[37]).
Results and Discussion
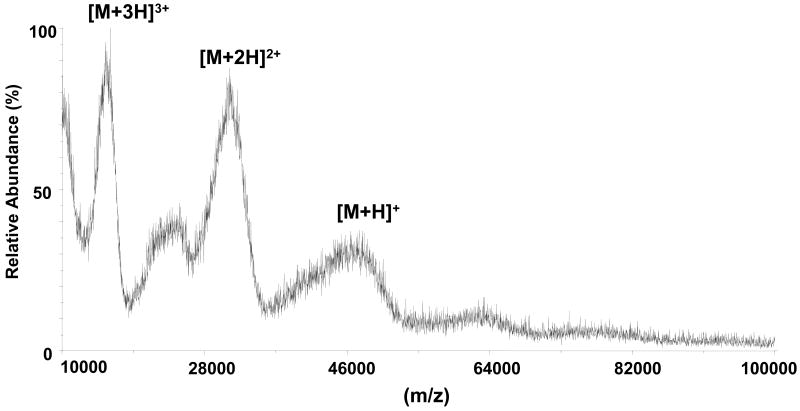
The E2 glycoprotein has been reported to contain primarily high mannose glycans [15, 16]. In order to determine specific glycan structures on E2, the intact native protein was first analyzed by MALDI-TOF MS. The amino acid sequence of the glycoprotein E2 is shown in Figure 1B, with the numbering starting from Ala1 to Lys333 (which corresponds to the numbering Ala383 to Lys715 within the entire HCV polyprotein of reference strain H GenBank access number AF009606) [38]. The 11 consensus glycosylation sites on E2, highlighted in Figure 1B, have been shown to be primarily occupied with high mannose glycans [15]. As shown in Figure 2, the MALDI-TOF mass spectrum of the intact glycosylated protein contains broad ions which correspond in mass to singly, doubly and triply charged molecular ions. E2 has a theoretical molecular weight of 36.5 kDa, but the observed mass of the singly charged ion in MALDI-TOF mass spectrum was approximately 50 kDa, indicating a large amount of N-linked glycosylation with extensive heterogeneity, which hampers the ability to determine the exact molecular weight of the intact protein.
Figure 2.
MALDI-TOF mass spectrum (mass range 10,000–100,000 Da) of HCV E2 showing singly, doubly, and triply charged molecular ions. The broadness of the peaks is due to of the extensive glycosylation of E2.
As alternatives to MALDI-TOF MS, methods such as LC-MS can be applied for the study of glycoproteins [23] by providing detailed information at high sensitivity. Glycopeptides can be identified using LC–MS of the enzymatic digestion mixture in the MS mode, based on characteristic ions arising from in-source decay which can be monitored by generating extracted ion chromatograms of these ions, for example, m/z 204.1 (protonated HexNAc) and m/z 366.1 (protonated HexNAc1Hex1) [39-42]. A search of the MS/MS data can be performed for these characteristic fragment ions [43]. Another approach for the detection of the glycopeptides in LC-MS analyses is through parent ion detection (PID) [40]. We performed an initial experiment in which the tryptic and chymotryptic digests were analyzed on the Q-Tof using PID. Identification of glycosylated peptides was accomplished using the parent ion detection mode for specific sugar oxonium ions (Hex+, m/z 163.1 and HexNAc+, m/z 204.1). A mass spectrum at low collision energy (5V) followed by one at high collision energy (30V) was acquired for each peptide. The presence of specific sugar fragment ions in the spectrum at high collision energy was diagnostic for the presence of glycopeptides (data not shown).
The identities of the glycans attached at each N-glycosylation site were determined from the LC-MS analyses of the tryptic and chymotryptic digests of reduced and alkylated E2 protein. From the LC/MS/MS analysis of the tryptic digest, 4 of 11 consensus N-linked sites were unambiguously determined. The primary sequence of E2 contains only a few potential trypsin cleavage sites, therefore, longer proteolytic fragments, containing multiple glycosylation sites were formed. Consequently, these peptides were not observed in the LC-MS analysis of the tryptic digest. In order to elucidate the glycosylation pattern at the remaining sites, chymotrypsin was used to generate shorter proteolytic fragments, which would contain a single glycosylation position within each peptide. Glycopeptides with the same backbone structure containing different glycan moieties (site microheterogeneity) show a specific pattern in ESI-MS [26]. Those glycopeptides ions forming a charge envelope are separated by an absolute mass difference of 162 amu (hexose), in the case of high mannose/hybrid type oligosaccharides. In addition, collision activated dissociation (CAD) spectra of glycopeptides contain carbohydrate marker ions, such as at m/z 163 (Hex+), 204 (HexNAc+), or 366 (HexHexNAc+), which simplify their identification if these masses are extracted from the MS/MS total ion current. Different batches of E2 glycoprotein were used in these experiments and slight differences were noticed in the results, especially in the relative abundance of some glycoforms compared to others, as well as in the number of attached mannose residues. However, similar glycosylation patterns of the protein were observed in all experiments.
The N-linked glycosylation sites in E2 glycoprotein contain a broad variety of high mannose glycans, ranging from the minimal core structure (Man3) to, at most, 9 hexose residues attached to the trimannosyl chitobiose moiety (Hex3Man9). It should be noted that the relative abundances correspond to the abundance ratios observed in the raw data, and that differences in sensitivity and ionization efficiency were not considered. The relative abundance of the glycoforms contained at one N-linked site was determined from the deconvoluted mass spectrum over the chromatographic range containing the charge envelops of the ions for the corresponding glycopeptides. The peptides determined for each glycosylation site and the corresponding glycan populations are presented in Tables 1 and 2.
Table 1.
Glycosylation sites and amino acid sequences of high mannose glycopeptides observed in LC-MS/MS analysis of a tryptic digest of reduced and alkylated E2.
| Sitea | Tryptic peptide | Glycopeptide mass | Glycan typec | Relative abundanced | ||
|---|---|---|---|---|---|---|
| Theoretical | Observed | Mass accuracies
(ppm) |
||||
|
N66
(N448) |
65FNSSGCPER73b | 2106.798 | 2106.763 | 16 | Man 4 | 0.61 |
| 2268.851 | 2268.800 | 22 | Man 5 | 0.31 | ||
| 2430.904 | 2430.855 | 20 | Man 6 | 1 | ||
| 2592.956 | 2592.910 | 17 | Man 7 | 0.57 | ||
| 2755.008 | 2754.980 | 10 | Man 8 | 0.54 | ||
| 2917.060 | 2917.015 | 15 | Man 9 | 0.42 | ||
| 3079.112 | 3079.045 | 21 | Hex1Man9 | 0.38 | ||
| 3241.164 | 3241.115 | 15 | Hex2Man9 | 0.20 | ||
| 3403.216 | 3403.181 | 10 | Hex3Man9 | 0.15 | ||
|
N194
(N576) |
181VCGAPPCVIGGVGNNTLLCPTDCFRK206b | 4401.872 | 4401.825 | 11 | Man 7 | 0.6 |
| 4563.925 | 4563.867 | 13 | Man 8 | 1 | ||
| 4725.978 | 4725.930 | 10 | Man 9 | 0.44 | ||
|
N241
(N623) |
233LWHYPCTINYTIFK246b | 3071.428 | 3071.460 | 10 | Man 5 | 0.54 |
| 3233.482 | 3233.523 | 13 | Man 6 | 1 | ||
| 3395.435 | 3395.487 | 15 | Man 7 | 0.65 | ||
| 3557.487 | 3557.509 | 6 | Man 8 | 0.38 | ||
| 3719.539 | 3719.503 | 10 | Man 9 | 0.52 | ||
| 3881.591 | 3881.632 | 11 | Hex1Man9 | 0.44 | ||
| 4043.644 | 4043.664 | 5 | Hex2Man9 | 0.70 | ||
| 4205.696 | 4205.722 | 7 | Hex3Man9 | 0.47 | ||
|
N263
(N645) |
258LEAACNWTRGERCDLEDR275b | 3676.469 | 3676.497 | 8 | Man 7 | 0.25 |
| 3838.521 | 3838.589 | 17 | Man 8 | 0.30 | ||
| 4000.573 | 4000.600 | 7 | Man 9 | 0.70 | ||
| 4162.625 | 4162.650 | 6 | Hex1Man9 | 1 | ||
| 4324.677 | 4324.748 | 16 | Hex2Man9 | 0.45 | ||
| 4486.729 | 4486.758 | 8 | Hex3Man9 | 0.65 | ||
The numbers in parenthesis represent the positions of the N glycosylation sites which correspond to the positions in the HCV polyprotein of reference strain H (GenBank access number AF009606).
Represents a carboxymethylated cysteine residue.
The notation ManX indicates that X mannose residues are attached to the chitobiose core of each N-linked high mannose glycan.
The relative abundance was determined from deconvoluted mass spectrum over the mass range containing the glycopeptide ions corresponding to glycopeptides with the same amino acid sequence.
Table 2.
Glycosylation sites and amino acid sequences of high mannose glycopeptides observed in LC-MS/MS analysis of a chymotryptic digest of reduced and alkylated E2.
| Sitea | Chymotryptic peptide | Glycopeptide mass | Glycan typec | Relative abundanced | ||
|---|---|---|---|---|---|---|
| Theoretical | Observed | Mass accuracy
(ppm) |
||||
|
N35
(N417) |
32INTNGSW38 | 1682.678 | 1682.676 | 1 | Man 3 | 0.5 |
| 1844.731 | 1845.732 | 1 | Man 4 | 0.95 | ||
| 2006.784 | 2006.774 | 4 | Man 5 | 0.82 | ||
| 2168.837 | 2168.816 | 9 | Man 6 | 1 | ||
| 2330.889 | 2330.873 | 7 | Man 7 | 0.37 | ||
| 2492.942 | 2492.954 | 5 | Man 8 | 0.25 | ||
| 2654.995 | 2655.016 | 8 | Man 9 | 0.25 | ||
|
N66
(N448) |
66NSSGCPERL74 | 2015.799 | 2015.830 | 15 | Man 4 | 0.85 |
| 2177.852 | 2177.829 | 10 | Man 5 | 0.5 | ||
| 2339.905 | 2339.921 | 7 | Man 6 | 1 | ||
| 2501.957 | 2501.953 | 2 | Man 7 | 0.37 | ||
| 2664.010 | 2664.007 | 1 | Man 8 | 0.36 | ||
| 2826.063 | 2826.069 | 2 | Man 9 | 0.12 | ||
|
N94
(N476) |
88GPISYANGSGLDERPYCW105b | 3743.481 | 3743.457 | 6 | Man 8 | 0.17 |
| 3905.534 | 3905.519 | 4 | Man 9 | 0.50 | ||
| 4067.587 | 4067.628 | 10 | Hex1Man 9 | 1 | ||
| 4229.640 | 4229.704 | 15 | Hex2Man 9 | 0.25 | ||
| 4391.692 | 4391.766 | 17 | Hex3Man 9 | 0.22 | ||
| 0.18 | ||||||
|
N150
(N532) |
148GANDTDVFVL157 | 2428.137 | 2428.156 | 8 | Man 6 | 1 |
| 2590.031 | 2590.055 | 9 | Man 7 | 0.45 | ||
| 2752.084 | 2752.131 | 17 | Man 8 | 0.85 | ||
| 2914.137 | 2914.143 | 2 | Man 9 | 0.42 | ||
| 3076.190 | 3076.207 | 6 | Hex1Man 9 | 0.54 | ||
| 3238.243 | 3238.293 | 15 | Hex2Man 9 | 0.52 | ||
|
N158
(N540) |
156VLNNTRPPLGNWF168 | 2743.222 | 2743.241 | 7 | Man 5 | 0.39 |
| 2905.275 | 2905.262 | 4 | Man 6 | 1 | ||
| 3067.327 | 3067.361 | 11 | Man 7 | 0.15 | ||
| 3229.380 | 3229.405 | 8 | Man 8 | 0.10 | ||
| 3391.433 | 3391.451 | 5 | Man 9 | 0.07 | ||
|
N174
(N556) |
173MNSTGF178 | 1547.581 | 1547.580 | 1 | Man3 | 0.1 |
| 1709.634 | 1709.645 | 6 | Man4 | 0.4 | ||
| 1871.687 | 1871.690 | 2 | Man 5 | 0.62 | ||
| 2033.740 | 2033.732 | 4 | Man 6 | 1 | ||
| 2195.792 | 2195.799 | 3 | Man 7 | 0.30 | ||
| 2357.845 | 2357.844 | 1 | Man 8 | 0.31 | ||
| 2519.898 | 2519.901 | 1 | Man 9 | 0.27 | ||
|
N241
(N623) |
235HYPCTINY242 | 2225.855 | 2225.835 | 9 | Man 5 | 0.09 |
| 2387.908 | 2387.883 | 10 | Man 6 | 1 | ||
| 2549.950 | 2549.960 | 4 | Man 7 | 0.28 | ||
| 2712.013 | 2711.960 | 20 | Man 8 | 0.19 | ||
| 2874.066 | 2874.069 | 1 | Man 9 | 0.13 | ||
|
N263
(N645) |
251VGGVEHRLEAACNW264 | 2756.148 | 2756.120 | 10 | Man5 | 0.1 |
| 2918.201 | 2918.175 | 9 | Man 6 | 1 | ||
| 3080.253 | 3080.230 | 7 | Man 7 | 0.31 | ||
| 3242.306 | 3242.282 | 7 | Man 8 | 0.33 | ||
| 3404.359 | 3404.325 | 10 | Man 9 | 0.5 | ||
The numbers in parenthesis represent the positions of the N glycosylation sites which correspond to the positions in the HCV polyprotein of reference strain H (GenBank access number AF009606).
Represents a carboxymethylated cysteine residue.
The notation ManX indicates that X mannose residues are attached to the chitobiose core of each N-linked high mannose glycan.
The relative abundance was determined from the deconvoluted mass spectrum over the mass range containing the glycopeptide ions corresponding to glycopeptides with the same amino acid sequence.
To corroborate our interpretation, the experimental glycopeptide masses were submitted to GlycoMod, a software tool used for determination of glycosylation compositions from mass spectrometric data [33]. This program compares the experimental mass of a glycopeptide to a list of precompiled masses of possible monosaccharide compositions, taking into account the possible peptides containing the N-X-S/T/C motif. For example, the observed m/z of 1011.90 (corresponding to a quadruply charged ion), was assigned to the tryptic peptide 233–246, corresponding to the mass of 4043.66 Da, containing a carboxymethylated cysteine residue, with an attached high mannose glycan of the composition Hex2Man9GlcNAc2. Multiple monosaccharide compositions, however, were proposed as possible matches within a mass tolerance of 0.1 Da, in addition to the assigned Hex2Man9GlcNAc2 structure, which has been previously proposed by Duvet et al [3]. In order to verify the glycan structure, the MS/MS data of the [M+4H]4+ ion of m/z 1011.90 was acquired. The fragment ions observed by MS/MS confirmed the assigned glycan composition Hex2Man9GlcNAc2 and the identity of the tryptic peptide 233–246 containing the glycosylation site N241 (data not shown). The number of hexose residues larger than 9 indicates incomplete processing of the precursor glycan. As mass spectrometry using conventional CAD can not distinguish between the isobaric monosaccharides mannose and glucose, the structures containing more than 9 mannose residues are indicated as Hex1-3Man9 (Tables 1 and 2). In the chymotryptic digest, the same site N241 was observed containing only Man9 residues in total, and in both digests the most abundant ion was the one corresponding to Man6 structure (Table 1 and 2). As explained above, the differences in the number of mannose residues is caused by the slight differences between the E2 glycoprotein batches that were used. The tryptic glycopeptide 65–73, containing the glycosylation site N66 (N448), indicates the presence of high mannose species, with a maximum of nine mannose residue attached to the trimannosyl chitobiose core (Hex3Man9) and with Man6 as the most abundant glycan. In the chymotryptic digest, the peptide 66-74 containing the N66 site of glycosylation showed a maximum of Man9 attached to the site, with Man6 being also the most abundant ion.
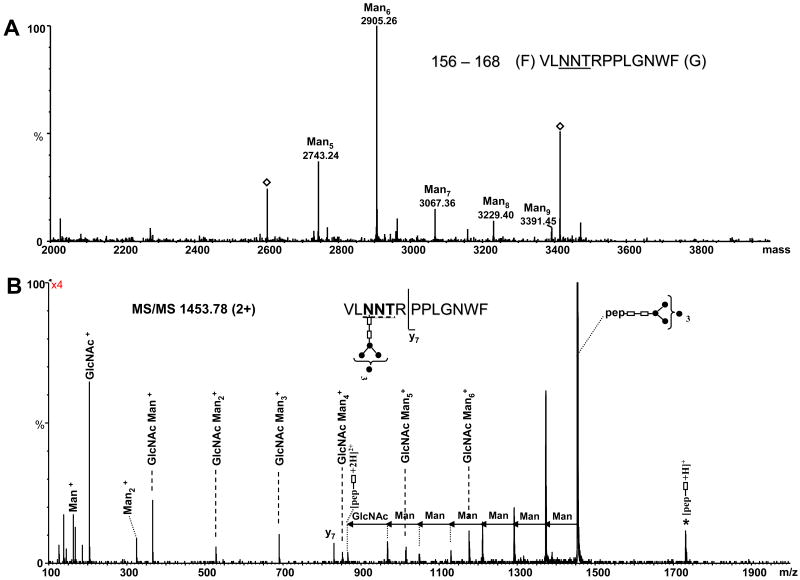
The MS and MS/MS spectra of the chymotryptic peptide 156–168 corresponding to the glycosylation site N158 (N540) are presented in Figure 3. The high mannose species observed in the deconvoluted mass spectrum (Figure 3A) were assigned to glycopeptide 158–168 carrying five to nine mannose units attached to the GlcNAc2 moiety (see Table 2). The notation ManX indicates a high mannose glycan with X the number of mannose residues attached to the chitobiose moiety (GlcNAc- GlcNAc). Among these, Man6 is the most abundant, followed by Man5 and Man7. The identity of the glycopeptide containing the Man6 glycan, corresponding to the deconvoluted mass 2905.26 Da, was determined from the CAD data of the doubly charged precursor of m/z 1453.78 (Figure 3B). The precursor ion represents the base peak in the MS/MS and the observed fragment ions originate from the successive neutral loss of hexoses from the non-reducing end of the oligosaccharide. The glycan was fragmented down to one GlcNAc moiety attached to the peptide backbone, which was observed as both doubly (m/z 866.28) and singly charged fragment ions (m/z 1731.56) in the spectrum. Another fragmentation pathway of the precursor led to formation of singly protonated sugar ions, which are characteristic of high mannose glycans. A single backbone fragment ion observed at m/z 830.42 (y7), which resulted from the cleavage of the amide bond between Arg161 and Pro162, confirmed the identity of the peptide as 156–168.
Figure 3.
(A) Deconvoluted mass spectrum over the mass range 2000–4000, showing the glycopeptides with the amino acid sequence 156–168 and high mannose glycans with composition from Man5 to Man9. The masses indicated by an empty diamond could not be identified. (B) MS/MS of the precursor ion m/z of 1453.78 (2+) corresponding to the chymotryptic peptide 156–168 containing the Man6 glycan at position N158 (N540). The glycopeptide carrying the glycan is indicated at the top (right) and the consensus sequence is underlined. The mass differences of 81 and 101.5 amu between doubly charged fragment ions indicate loss of mannose and N-acetyl glucosamine, respectively. For a better illustration, the tandem mass spectrum was multiplied by four. Filled circles represent Man residues; empty rectangles represent GlcNAc. The MS/MS spectra are presented in raw format, because in this manner they are illustrative for the different fragmentation pathways observed for the described glycopeptides. The number in the bracket indicates the number of Man residues attached to the trimannosyl chitobiose core. The star denotes singly charged glycosylated ions fragment ions derived from the precursor by fragmentation of the glycan and charge reduction.
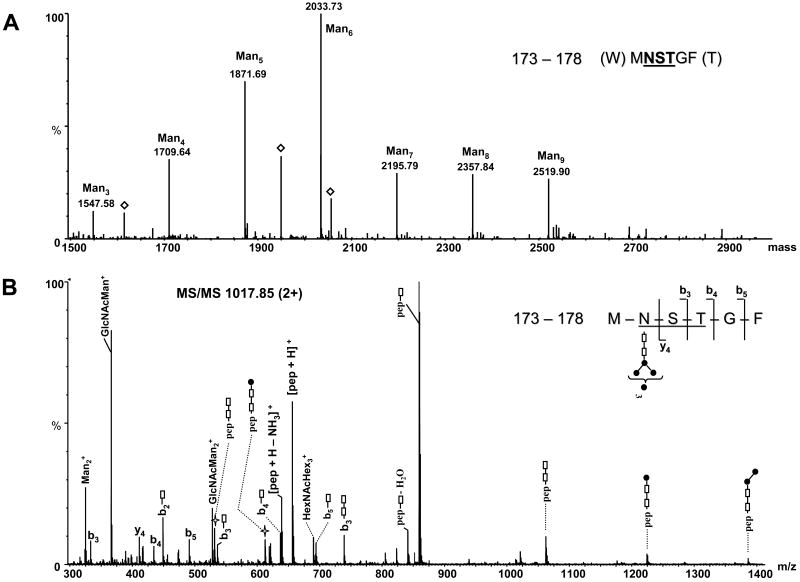
The microheterogeneity of glycosylation site N174 (N556) observed in glycopeptide 173–178 is consistent with a high mannose type glycan (Man3-9) and is presented in Figure 4A. The masses in the deconvoluted spectrum were assigned to glycopeptide 173–178 containing glycans with a length ranging from Man3 to Man9. The Man5 and Man6 are the most abundant populations, followed by Man4, Man7, Man8 and Man9, which are almost equally represented. The tandem mass spectrum of the doubly charged ion of m/z 1017.85, corresponding to glycopeptide 173–178 which contains a Man6 glycan is presented in Figure 4B. This precursor ion also fragmented to the intact peptide backbone (m/z 859.66) with a GlcNAc moiety attached at site N174. Interestingly, the protonated peptide ion of m/z 656.66 resulted from the complete loss of the oligosaccharide, by glycosidic bond cleavage between the sugar and the Asn side chain. Unlike the other MS/MS data, a large number of specific backbone cleavages were observed (b3, b4, b5 and y4) and the peptide backbone could be almost completely assigned based on this spectrum (Figure 4B). In addition, backbone fragments still containing one or two GlcNAc units were abundant. This fragmentation pattern and the data discussed above suggest that the presence of backbone fragmentation may be dependent on the amino acid sequence and length of the peptide containing the sugar, on the position of the glycosylation site within the peptide backbone, or on a combination of these factors.
Figure 4.
ESI-MS of chymotryptic peptide 173–178: (A) Deconvoluted mass spectrum over the mass range 1500–3000, showing the glycopeptides with the amino acid sequence 173–178 containing high mannose glycans with composition from Man3 to Man9. The empty diamonds represent unrelated ions. (B) MS/MS of the precursor ion m/z 1017.85 (2+) corresponding to the chymotryptic peptide 173–178 containing the Man6 glycan at the glycosylation site N174 (N556). The glycopeptide and the structure of glycan are indicated at the top (right). The presence of the backbone b and y ions confirms the peptide sequence. Filled circles represent Man residues; empty rectangles represent GlcNAc. The number in the bracket represents maximum of Man residues attached to the trimannosyl chitobiose core. The majority of the observed ions are singly charged and the empty star denotes doubly charged ions.
For the tryptic peptide 181–206 containing the glycosylation site N194 (N576), a maximum of Man9 residues were observed, with Man8 being the most abundant ion. In the chymotryptic digest the peptide 191-204 containing the same site of glycosylation was seen having the same number of mannoses residues attached to this N-linked site (data not shown).
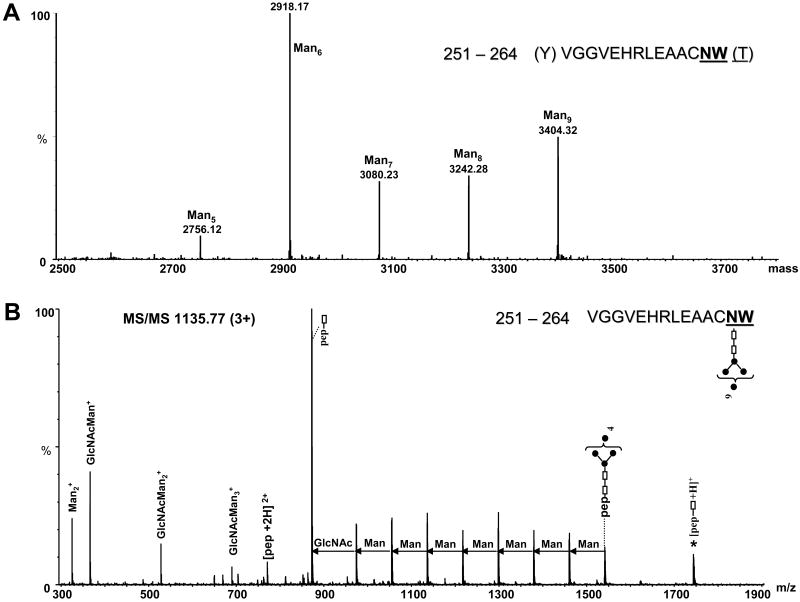
The deconvoluted mass spectrum of the region of the chromatogram where glycoforms of the peptide 251–264 (N645) appeared is presented in Figure 5A. These data indicate the presence of high mannose glycans with a minimum Man5 and maximum Man9 composition. Man6 is the most abundant among these species. The tandem mass spectrum of the triply charged ion of m/z 1135.77 (Figure 5B) is consistent with the proposed glycan composition (Man9) for the peptide 251–264. The major fragmentation pathway arises from successive cleavage of the monosaccharides from the non-reducing end of the glycan. The composition of the glycan was deduced from the mass difference between these fragments. The doubly charged molecular peptide ion of m/z 770.83 is less abundant, while the ion of m/z 872.37, assigned to the peptide 251–264 with a single GlcNAc moiety attached at N263, represents the most abundant ion (Figure 5B). This N-linked site showed slight differences in the number of attached mannoses, where up to Hex3Man9 were identified for N263 in the tryptic peptide 258-275, compared to Man9 that were observed in the chymotryptic peptide 251-264. This high number of hexoses suggests incomplete processing of the precursor glycan. As discussed above, these results are due to the differences in the protein batches that were used (Table 1 and 2).
Figure 5.
(A) Deconvoluted mass spectrum over the mass range 2500–4000, showing the glycopeptides with the amino acid sequence 251–264 containing high mannose glycans with composition from Man5 to Man9. (B) MS/MS of the precursor ion m/z 1135.77 (3+) corresponding to chymotryptic glycopeptide 251–264 containing the Man9 glycan at position N263 (N645). The amino acid sequence of the glycopeptide and the structure of the glycan are indicated at the top (right). The successive mass difference of 81 amu between doubly charged fragment ions is indicative for a high mannose glycan. Filled circles represent Man residues; empty rectangles represent GlcNAc. The number in the bracket represents maximum of Man residues attached to the glycosylation site. The star denotes singly charged ions formed by fragmentation of the glycan and charge reduction of the precursor ion.
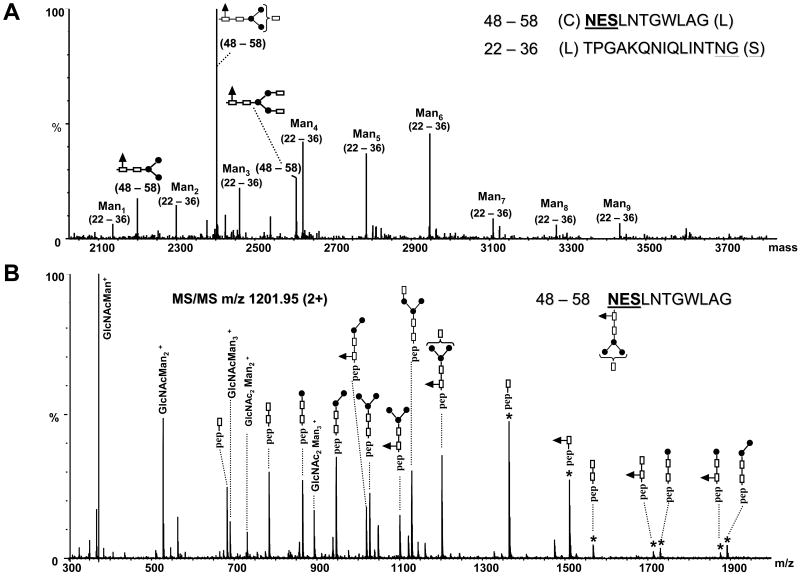
The deconvoluted mass spectrum of the region of the chromatogram where glycopeptides 48–58 and 22–36, and their glycoforms containing the N-linked sites N48 and N35 respectively elute, is shown in Figure 6A. For the site N35, a maximum of Man9 residues was observed, with Man4, Man5 and Man6 being the most abundant species. The deconvoluted masses were assigned to each peptide containing different glycan structures. The mass spectrum was deconvoluted over the entire range containing the charge envelops of multiply charged ions, for the corresponding glycopeptides. The identity of these species was confirmed by MS/MS of the corresponding multiply charged ions (data not shown). Small amounts of the species Man1 and Man2 were observed in the spectrum, indicating that, to some extent, the glycopeptides underwent in-source decomposition. For the peptide 48–58, complex type glycans were exclusively observed at site N48 (N430). In the deconvoluted mass spectrum (Figure 6A), ions for these glycoforms with masses at m/z 2198.90, 2401.98 and 2605.08 were observed and they are separated by a mass interval of 203 amu. This mass corresponds to the sugar N-acetylglucosamine, therefore indicating the presence of zero, one or two terminal GlcNAc residues. This was assigned as N-acetylglucosamine because this monosaccharide is expected to elongate the Man3 core structure in complex N-linked glycans. The identity of the peptide observed with this sugar moiety and the composition of the glycan were determined from the MS/MS of the doubly charged precursor of m/z 1201.95 (Figure 6B). Singly charged fragment ions separated by 146 amu clearly indicate the presence of fucose (Fuc) attached at the first GlcNAc residue of the Man3GlcNAc2 core. The singly charged ion of m/z 1510.68 corresponds in mass to amino acids 48–58, plus a GlcNAcFuc rest. The ion of m/z 1364.68 corresponds in mass to residues 48–58 with a single GlcNAc. Two series of fragment ions were observed in the MS/MS spectrum (Figure 6B): one series is composed of doubly charged fragment ions that result from the successive neutral loss of monosaccharides from the non-reducing end. The other series of ions are formed by charge reduction of the precursor ion. These data are consistent with the glycan structure GlcNAcFuc – Man3GlcNAc2 at the site N48. The deconvoluted masses of m/z 2198.90 and 2605.08 correspond in mass to residues 48–58 plus the complex type glycans Fuc – Man3GlcNAc2 and GlcNAc2Fuc – Man3GlcNAc2, respectively (see Figure 6A and Table 3). Interestingly, these glycopeptides could result from anomalous chymotrypsin activity, which cleaved after glycine residues in both cases instead of residues specific for this enzyme. However, in the absence of backbone fragmentation, the observed mass of the peptide by itself cannot rule out the possibility that cleavage at other residues occurred; the peptide QHKFNSSGCP, bearing the site N66 (N448) and with the cysteine alkylated, has a mass (1160.50) similar to peptide 48–58 (1160.55), and, thus, could also represent a plausible candidate. Cleavage after proline, however, is very uncommon for all serine proteases, irrespective of their specificity, and this plus the observed mass error argues against (but does not disprove) assignment of the peptide as QHKFNSSGCP. There is also the possibility that a peptide modification or protein mutation occurred, as E2 is a viral recombinant protein, and that, instead of cleavage after glycine, this ion may arise from an unidentified amino acid sequence that contains multiple mutations/modifications. No evidence, however, for the presence of mutations was observed in the rest of our data. Therefore, the most likely origin of this ion is that it is due to a peptide formed by a non-specific cleavage occurring after glycine, as chymotrypsin is less specific than trypsin. Because complex types N-glycans have a well defined sugar composition, this enables the identification of the peptide containing this type of glycan from the MS/MS data (i.e. the fragment containing a single GlcNAc moiety). The backbone fragment with the amino acid sequence 48–58 was the only peptide that matched the mass of these glycopeptide fragment ions. Usually chymotrypsin cleaves the protein backbone after aromatic amino acids at higher rates than after other amino acids, but under different circumstances (glycan type, conformation of the protein) it also may cleave other amino acids [44]. These data show that the presence of the glycan moieties attached to E2 can alter the specificity of chymotrypsin. The formation of glycopeptides from non-specific proteolytic cleavages complicates the assignment of the N-linked sites and of the corresponding glycan structures using MS data alone.
Figure 6.
(A) Deconvoluted mass spectrum over the mass range 2000–4000, illustrating the microheterogeneity of the glycopeptides 48–58 and 22–36. The consensus sequences for N-glycosylation are underlined. For the clarity of the figure, the complex type glycans determined for the peptide 48–58 are schematically represented. (B) MS/MS of the precursor ion of m/z 1201.95 (2+) corresponding to the anomalous chymotryptic glycopeptide 48–58 containing the indicated complex type glycan (at the top, right) attached to the N48 (N430) site. Filled circles represent Man residues, empty rectangles represent GlcNAc, and the filled triangles represent a Fuc residue. The rectangle in the bracket indicates the additional presence of a GlcNAc residue. The star denotes singly charged glycopeptide fragment ions resulted from the fragmentation of the glycan and charge reduction of the precursor.
Table 3.
Glycosylation sites and amino acid sequences of complex-type glycopeptides observed in LC-MS/MS analysis of a chymotryptic digest of reduced and alkylated E2.
| Sitea | Chymotryptic peptide | Glycopeptide mass | Glycan typec | Relative abundanced | ||
|---|---|---|---|---|---|---|
| Theoretical | Observed | Mass accuracy
(ppm) |
||||
|
N41
(N423) |
39HINSTAL45 | 1849.794 | 1849.786 | 4 | Man3-GlcNAc | 0.28 |
| 1995.852 | 1995.831 | 10 | Man3-GlcNAc-Fuc | 1 | ||
| 2132.873 | 2132.860 | 6 | Man 6 | 0.80 | ||
| 1970.820 | 1970.811 | 4 | Man 5 | 0.88 | ||
| 1808.767 | 1808.769 | 1 | Man 4 | 0.20 | ||
| 1646.714 | 1647.711 | 2 | Man 3 | 0.10 | ||
|
N48
(N430) |
48NESLNTGWLAG58b | 2198.921 | 2198.902 | 8 | Man3-Fuc | 0.18 |
| 2402.001 | 2401.984 | 7 | Man3-GlcNAc-Fuc | 1 | ||
| 2605.080 | 2605.071 | 4 | Man3-GlcNAc2-Fuc | 0.29 | ||
The numbers in parenthesis represent the positions of the N glycosylation sites which correspond to the positions in the HCV polyprotein of reference strain H (GenBank access number AF009606).
Anomalous chymotryptic cleavage sites.
The notation ManX indicates that X mannose residues are attached to the chitobiose core of each N-linked high mannose glycan. The rest of the notations indicate complex type glycans.
The relative abundance was determined from the deconvoluted mass spectrum over the mass range containing the glycopeptide ions corresponding to glycopeptides with the same amino acid sequence.
In addition to N48, complex type glycans were determined for the site N41 (N423), observed in peptide 39–45 (Table 3). The sugar composition of the complex type glycans was determined from the MS/MS data of the glycopeptides observed as doubly charged ions of m/z 926.38 (1849.78 Da) as: GlcNAc – Man3GlcNAc2 and m/z 999.39 (1995.83) as GlcNAcFuc – Man3GlcNAc2, respectively (data not shown). High mannose type oligosaccharides ranging from Man3 to Man6 were also observed at the site N41 (peptide 39–45, Table 3) and the corresponding glycopeptides eluted slightly later than those carrying the complex type glycans. Interestingly, for both sites containing complex type glycans, the Man3GlcNAc-Fuc species is the most abundant ion. Regarding the sugar composition, we may say that these are not mature complex type glycans. This is the first time when complex type glycans were positively identified on the E2 glycoprotein. Although it has not been reported that these complex glycans transit through the medial Golgi, it is possible that they are not sufficiently long enough to develop into mature structures, and thereby are immediately translocated into the ER compartment.
Tables 1, 2 and 3 summarize the glycosylation sites identified after tryptic and chymotryptic digestions, indicating the position of glycosylation and the observed glycopeptides, as well as the glycopeptide neutral masses. Using GlycoMod software the E2 glycopeptides resulted from both chymotrypsin as well as trypsin digests, with a mass accuracy of 0.1 Da were identified. Multiple monosaccharide compositions were proposed as possible matches in some instances, and the correct glycan structure was determined from MS/MS analyses. The data identification, however, was unreliable when nonspecific chymotryptic cleavages occurred, so that manual interpretation of the MS/MS data was mandatory in order to ascertain the correct amino acid sequence of the glycopeptides and the composition of the attached glycans. As presented in Table 2, short glycopeptides were observed (e.g. 32-38 or 173-178) eluting from the C18 column. From our experiments it does not appear that short glycopeptides were lost during the trapping process, however this fact can not be absolutely excluded.
The envelope proteins play a major role in a virus life cycle. Envelope proteins are known to be involved in viral entry into the cell by binding to a receptor present on the host cell and inducing fusion between the viral envelope and the membrane of the host cell [16]. The E2 protein is heavily glycosylated with 11 potential sites of glycosylation. Nine of these 11 sites are highly conserved, suggesting that the glycosylation may play an essential role in some biological functions or conformation of the glycoprotein [15]. In the early secretory pathway, the glycans play a role in protein folding and in certain sorting events [17]. It is known that during glycosylation of a protein, a precursor oligosaccharide composed of GlcNAc, Man and glucose (Glc), with the composition Glc3Man9GlcNAc2 is transferred to nascent proteins in the ER in a co-translational event [17]. The diversity of these N-linked oligosaccharide structures on mature glycoproteins arises from major modification of this precursor structure, which occurs posttranslationally. While still in the ER, the glucose residues are quickly removed from the oligosaccharides of most glycoproteins. This process continues in the Glogi apparatus, and, thus, glucose is not observed on the mature glycoprotein [3]. From our data, peptides with levels of mannosylation higher than Man9 were detected, indicating incomplete processing. Incomplete processing of glycans might be a function of protein processing while still in the ER compartment, with a high level of mannosylation being present in HCV in its natural setting. Incomplete processing in which the hexose residues in excess of the expected Man9 maximum are glucoses could also be explained by the folding of the protein, which might alter the accessibility of these glycans to the processing by glucosidases and mannosidases. The hypothesis that steric hindrance interferes with glycan processing is consistent with our hypothesis that the glycans play a significant role in stabilization of protein tertiary structure. In the low energy CID experiments involved for the structural characterization of E2 glycopeptides one can not differentiate between isobaric mannose and glucose structures, consequently the glycans observed as having a number of hexoses higher than Man9 are depicted as Hex1-3Man9.
To accurately characterize a glycoprotein, mass spectrometry has proven to have a tremendous ability to identify the type of glycans as well as the sites of glycosylation on a protein. For example, we have used a combination of MALDI-TOF and LC followed by nanoLC/MS/MS in order to characterize the glycan structures attached to the human immunodeficiency virus (HIV) gp120 glycoprotein, and high mannose glycans and hybrid glycans were found attached to the protein [32, 45]. Although LC-MS analysis of released glycans may provide a detailed picture of the structure of the glycans derived from a protein or any complex protein mixture, information on the original attachment sites of the glycans and the underlying proteins is lost. This critical information can either be obtained by LC-MS analysis of the remaining peptides after glycan release based on Asn to Asp conversion but cannot connect specific glycans to specific sites or, by the direct analysis of glycopeptides, which provides the connection between glycan type and location.
In the present paper the MS/MS spectra of high mannose oligosaccharides were readily differentiated from those of hybrid/complex type and provided immediate information about the glycans on E2. Large glycopeptides with a high sugar to peptide ratio are expected to be highly sensitive to in-source fragmentation by glycosidic bond cleavage, and, therefore, need special analysis conditions. Furthermore, efficient formation of multiply charged ions of glycopeptides is crucial for their detection within the mass range of the instrument as well as for high resolution of the isotopic pattern. The major collision induced dissociation fragmentation pathway of high mannose glycan containing peptides can be characterized by the successive loss of the sugar moieties from the non-reducing end of the glycan, thereby generating a series of ions containing the peptide backbone and the remaining sugars attached at the reducing end [26]. In addition to the losses from the non-reducing end, tandem mass spectra of complex type glycans contain at least one additional series of ions that result from the initial loss of the α(1 – 6) linked fucose from the first GlcNAc residue of the trimannosyl chitobiose core. Although they are not usually observed in the CAD spectra of protonated glycopeptides, ions due to backbone cleavages along the amino acid chain, commonly with complete or partial loss of the glycan chain, have also been observed in the MS/MS spectra. These data allow for the identification of the peptide which contain the glycans, as well as the precise location of the glycan. The observation of amino acid backbone cleavages may depend on several factors: (i) the amino acid sequence; (ii) the number of amino acid residues contained in a glycopeptide; (iii) the position of the sugar chain within the glycopeptide or, (iv) a combination of these factors.
To successfully investigate the glycans and their role in the structure and function of the E2 protein, a structure of the glycoprotein or, at a minimum, a working model of the glycoprotein is required. To date there is no crystal structure available for the E2 protein. Therefore, the homology model of E2 was based on the Tick-borne encephalitis virus envelope glycoprotein E virus (TBEV). Because of that, it would be reasonable to assume that its physical relationship to the viral membrane would also be similar. Previous studies showed that Flavivirus envelope glycoprotein E from TBEV shows functional similarity to E2 [36] and these proteins are similar from the point of view of the parameters in these fold recognition structures [35]. Moreover, the organization of E2 into multiple antigenic domains has similarities to the large envelope glycoprotein E on TBEV and also with the envelope protein E1 from Semliki Forest virus, an alphavirus [46] having similar structural and functional properties [47]. Thus, in order to map the location of the glycans on a model structure, the TBEV structure (1SVB) was selected as a good candidate for a homology model of E2 [36]. These viruses undergo structural rearrangements at low pH environments, which hypothetically lead to the exposure of initially, buried hydrophobic residues, and this process is believed to be part of an endocytosis entry pathway [47]. Therefore, a published homology model for the E2 protein based on the TBEV structure was employed as a working model [35].
The location of the glycans on the E2 was thus mapped to the homology model and is presented in Figure 7. The two complex- type glycans that were newly identified by mass spectrometry are represented in green. The N41 (N423) site previously described as buried in this model [35] rather than surface exposed, is located in a region rich in β-sheet structural elements. By comparison to N41, N48 (N430) is mainly surface exposed, and located on the opposite site of the molecule, being nearly parallel to the location of N41. Based on a hydrophobicity plot, Yagnik et al. predicted that the region between amino acids 35-55 (418-438) (thus encompassing the two complex type glycans) is hydrophobic and mainly surrounded by β-sheet structures [35]. The distance between the two glycosylation sites in this model is 19Å. This region of the protein 35-55 has not been previously characterized to have a specific biological function, but considering the high β-sheet content, one might speculate that it is involved in the folding process of the E2 protein, maybe implicating the complex type glycans that are located in its vicinity, which may have an impact on the 3-D structure and folding of the protein. It has been previously reported that modification of the complex type glycans by sialylation could affect the potential biological activity of the virus, possibly by reducing the infectivity of the primate lentivirus [48]. Furthermore, in the case of gp120 it was reported that the removal of the fucose associated with sialylated glycans changed the protein conformation, most likely by exposing epitopes that were previously buried [49]. It has been demonstrated that the N-linked glycans of the HIV envelope glycoprotein limit its immunogenicity and restrict binding of certain antibodies to their epitopes on the virion surface [50].
Figure 7.
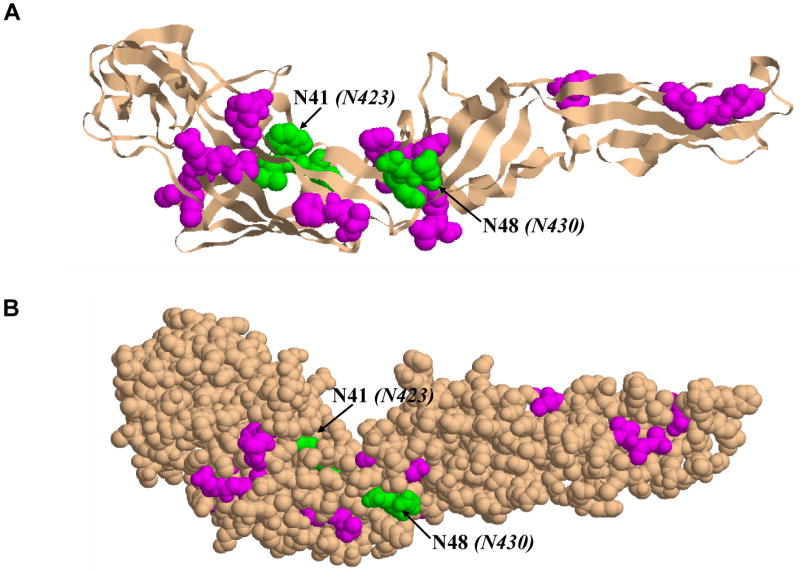
HCV E2 homology model based on the Tick Borne Encephalitis Virus envelope glycoprotein E (TBEV) structure (PDB id: 1SVB): (A) Cartoon representation of E2 glycoprotein showing the location of the glycans attached to the protein. The nine sites of glycosylation found to have attached only high mannose type sugars are highlighted in magenta, and the two sites found to have complex type glycans attached are shown in green. (B) Another view of the E2 glycoprotein illustrated in space filling representation. The same colors as described above were used to indicate the location of the glycans on E2.
Viral envelope proteins usually contain N-linked glycans which can play a major role in their folding, entry functions or in modulating the immune response [50, 51]. Previous studies indicated that mutation of some glycosylation sites in the HCV envelope glycoproteins can reduce or abolish HCV pseudoparticles (HCVpp) infectivity without affecting incorporation of the glycoproteins into the particles [16], possibly by changes in the local conformation of the antibody recognition sites. Moreover, E2 N-linked glycans at position N41 (N423) and N66 (N448) were reported as high mannose and have been shown to be essential for the entry functions of HCV envelope glycoproteins [16]. Interestingly, N41 (N423) is one of the glycan sites that was identified here as containing both complex type glycans as well as high mannose type glycans, indicating the microheterogeneity of this specific site. Based on these observations, one might predict that the presence of this glycan at the N41 (N423) site is essential for proper folding of the protein as well as for antibody recognition on E2. In addition, a recent study showed that the loss of glycosylation at N41 (N423) site leads to noncovalent heterodimer formation as well as CD81 binding, indicating that the removal of a large sugar moiety leads to better exposure of the CD81 binding site [52]. Furthermore, they observed reduction in HCVpp infectivity upon glycan removal. Glycosylations at sites N35 (N417), N94 (N476), N150 (N532) and N263 (N645) have also been shown to modulate HCVpp entry. Furthermore, N174 (N556) and N241 (N623) were indicated to have a direct effect on protein folding [16]. The presence of a large, polar oligosaccharide is indeed known to affect protein folding by orienting polypeptide segments toward the surfaces of the protein domains [17, 53]. Moreover, to establish the natural properties of the virus, lectin-binding assays were performed by Sato et al. in order to characterize the glycan moiety on the surface of HCV particles recovered from sera of infected patients [34]. This study suggested that the envelope glycoproteins E1 and E2 of HCV might contain complex type glycans, and their results also indicated that the N-linked glycans are present on the surface of native virions of HCV [34]. These authors also postulated that the selectivity of HCV and hepatitis B virus (HBV) in binding different lectins is related to the nature of the carbohydrate structures on the virion surface. Using different types of lectins and based on their binding efficiency, it was concluded that these viruses would contain complex type sugar chains and that the sugar moieties present on HCV virions are very similar to those for HBV. However, no further evidence for the presence and location of complex type glycans on E2 was presented. Although no possible biological function of these complex type glycans was reported, we hypothesize that there may be a correlation between the types of N-linked glycans and the structure and function of the E2 protein, but this hypothesis remains to be investigated in a future study.
Conclusions
The mass spectrometric characterization of the glycopeptides from HCV E2 envelope glycoprotein using nanoLC/MS/MS is reported. Using tandem mass spectrometry the glycan moieties associated with all 11 consensus sites of glycosylation on E2 were clearly characterized. These mass spectrometric results are consistent with previously reported high mannose type N-glycans. More importantly, in addition to high mannose glycans, complex type oligosaccharides were identified at two N-linked sites and their identity was established from MS/MS data. These complex type glycans were observed at positions N41 (N423) and N48 (N430). These data show the unambiguous mass spectrometric characterization of the glycans attached to the E2 protein. To summarize, it is our hope that these results may lead to a better understanding of the hepatitis C virus envelope E2 protein processing, and, finally, may help in the elucidation of the structure of the protein.
Acknowledgments
This research was supported in part by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences. The authors want to thank Dr. Leesa Deterding and Dr. Alina Zamfir for the critical review of the manuscript as well as for informative discussions throughout the project.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rehermann B, Nascimbeni M. Immunology of hepatitis B virus and hepatitis C virus infection. Nat Rev Immunol. 2005;5(3):215–29. doi: 10.1038/nri1573. [DOI] [PubMed] [Google Scholar]
- 2.Randall G, Rice CM. Hepatitis C virus cell culture replication systems: their potential use for the development of antiviral therapies. Curr Opin Infect Dis. 2001;14(6):743–7. doi: 10.1097/00001432-200112000-00013. [DOI] [PubMed] [Google Scholar]
- 3.Duvet S, Cocquerel L, Pillez A, Cacan R, Verbert A, Moradpour D, Wychowski C, Dubuisson J. Hepatitis C virus glycoprotein complex localization in the endoplasmic reticulum involves a determinant for retention and not retrieval. J Biol Chem. 1998;273(48):32088–95. doi: 10.1074/jbc.273.48.32088. [DOI] [PubMed] [Google Scholar]
- 4.Keck ZY, Sung VM, Perkins S, Rowe J, Paul S, Liang TJ, Lai MM, Foung SK. Human monoclonal antibody to hepatitis C virus E1 glycoprotein that blocks virus attachment and viral infectivity. J Virol. 2004;78(13):7257–63. doi: 10.1128/JVI.78.13.7257-7263.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deleersnyder V, Pillez A, Wychowski C, Blight K, Xu J, Hahn YS, Rice CM, Dubuisson J. Formation of native hepatitis C virus glycoprotein complexes. J Virol. 1997;71(1):697–704. doi: 10.1128/jvi.71.1.697-704.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Munro S. An investigation of the role of transmembrane domains in Golgi protein retention. Embo J. 1995;14(19):4695–704. doi: 10.1002/j.1460-2075.1995.tb00151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nilsson T, Hoe MH, Slusarewicz P, Rabouille C, Watson R, Hunte F, Watzele G, Berger EG, Warren G. Kin recognition between medial Golgi enzymes in HeLa cells. Embo J. 1994;13(3):562–74. doi: 10.1002/j.1460-2075.1994.tb06294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wormald MR, Dwek RA. Glycoproteins: glycan presentation and protein-fold stability. Structure. 1999;7(7):R155–60. doi: 10.1016/s0969-2126(99)80095-1. [DOI] [PubMed] [Google Scholar]
- 9.Gavel Y, von Heijne G. Sequence differences between glycosylated and non-glycosylated Asn-X-Thr/Ser acceptor sites: implications for protein engineering. Protein Eng. 1990;3(5):433–42. doi: 10.1093/protein/3.5.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kornfeld R, Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu Rev Biochem. 1985;54:631–64. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 11.Dubuisson J, Hsu HH, Cheung RC, Greenberg HB, Russell DG, Rice CM. Formation and intracellular localization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia and Sindbis viruses. J Virol. 1994;68(10):6147–60. doi: 10.1128/jvi.68.10.6147-6160.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grakoui A, Wychowski C, Lin C, Feinstone SM, Rice CM. Expression and identification of hepatitis C virus polyprotein cleavage products. J Virol. 1993;67(3):1385–95. doi: 10.1128/jvi.67.3.1385-1395.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ralston R, Thudium K, Berger K, Kuo C, Gervase B, Hall J, Selby M, Kuo G, Houghton M, Choo QL. Characterization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia viruses. J Virol. 1993;67(11):6753–61. doi: 10.1128/jvi.67.11.6753-6761.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spaete RR, Alexander D, Rugroden ME, Choo QL, Berger K, Crawford K, Kuo C, Leng S, Lee C, Ralston R, et al. Characterization of the hepatitis C virus E2/NS1 gene product expressed in mammalian cells. Virology. 1992;188(2):819–30. doi: 10.1016/0042-6822(92)90537-y. [DOI] [PubMed] [Google Scholar]
- 15.Goffard A, Dubuisson J. Glycosylation of hepatitis C virus envelope proteins. Biochimie. 2003;85(34):295–301. doi: 10.1016/s0300-9084(03)00004-x. [DOI] [PubMed] [Google Scholar]
- 16.Goffard A, Callens N, Bartosch B, Wychowski C, Cosset FL, Montpellier C, Dubuisson J. Role of N-linked glycans in the functions of hepatitis C virus envelope glycoproteins. J Virol. 2005;79(13):8400–9. doi: 10.1128/JVI.79.13.8400-8409.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Imperiali B, O'Connor SE. Effect of N-linked glycosylation on glycopeptide and glycoprotein structure. Curr Opin Chem Biol. 1999;3(6):643–9. doi: 10.1016/s1367-5931(99)00021-6. [DOI] [PubMed] [Google Scholar]
- 18.Op De Beeck A, Cocquerel L, Dubuisson J. Biogenesis of hepatitis C virus envelope glycoproteins. J Gen Virol. 2001;82(Pt 11):2589–95. doi: 10.1099/0022-1317-82-11-2589. [DOI] [PubMed] [Google Scholar]
- 19.Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Electrospray ionization for mass spectrometry of large biomolecules. Science. 1989;246(4926):64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 20.Smith RD, Loo JA, Edmonds CG, Barinaga CJ, Udseth HR. New developments in biochemical mass spectrometry: electrospray ionization. Anal Chem. 1990;62(9):882–99. doi: 10.1021/ac00208a002. [DOI] [PubMed] [Google Scholar]
- 21.Hillenkamp F, Karas M, Beavis RC, Chait BT. Matrix-assisted laser desorption/ionization mass spectrometry of biopolymers. Anal Chem. 1991;63(24):1193A–1203A. doi: 10.1021/ac00024a002. [DOI] [PubMed] [Google Scholar]
- 22.Harvey DJ. Matrix-assisted laser desorption/ionization mass spectrometry of carbohydrates. Mass Spectrom Rev. 1999;18(6):349–450. doi: 10.1002/(SICI)1098-2787(1999)18:6<349::AID-MAS1>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 23.Bunkenborg J, Pilch BJ, Podtelejnikov AV, Wisniewski JR. Screening for N-glycosylated proteins by liquid chromatography mass spectrometry. Proteomics. 2004;4(2):454–65. doi: 10.1002/pmic.200300556. [DOI] [PubMed] [Google Scholar]
- 24.Dell A, Morris HR. Glycoprotein structure determination by mass spectrometry. Science. 2001;291(5512):2351–6. doi: 10.1126/science.1058890. [DOI] [PubMed] [Google Scholar]
- 25.Balen B, Krsnik-Rasol M, Zamfir AD, Milosevic J, Vakhrushev SY, Peter-Katalinic J. Glycoproteomic survey of Mammillaria gracillis tissues grown in vitro. J Proteome Res. 2006;5(7):1658–66. doi: 10.1021/pr0600327. [DOI] [PubMed] [Google Scholar]
- 26.Harvey DJ. Collision-induced fragmentation of underivatized N-linked carbohydrates ionized by electrospray. J Mass Spectrom. 2000;35(10):1178–90. doi: 10.1002/1096-9888(200010)35:10<1178::AID-JMS46>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 27.Jiang H, Desaire H, Butnev VY, Bousfield GR. Glycoprotein profiling by electrospray mass spectrometry. J Am Soc Mass Spectrom. 2004;15(5):750–8. doi: 10.1016/j.jasms.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 28.Mechref Y, Novotny MV. Structural investigations of glycoconjugates at high sensitivity. Chem Rev. 2002;102(2):321–69. doi: 10.1021/cr0103017. [DOI] [PubMed] [Google Scholar]
- 29.Zaia J. Mass spectrometry of oligosaccharides. Mass Spectrom Rev. 2004;23(3):161–227. doi: 10.1002/mas.10073. [DOI] [PubMed] [Google Scholar]
- 30.Zamfir AD. Recent advances in sheathless interfacing of capillary electrophoresis and electrospray ionization mass spectrometry. J Chromatogr A. 2007;1159:2–13. doi: 10.1016/j.chroma.2007.03.115. [DOI] [PubMed] [Google Scholar]
- 31.Zamfir AD, Bindila L, Lion N, Allen M, Girault HH, Peter-Katalinic J. Chip electrospray mass spectrometry for carbohydrate analysis. Electrophoresis. 2005;26(19):3650–73. doi: 10.1002/elps.200500101. [DOI] [PubMed] [Google Scholar]
- 32.Zhu X, Borchers C, Bienstock RJ, Tomer KB. Mass spectrometric characterization of the glycosylation pattern of HIV-gp120 expressed in CHO cells. Biochemistry. 2000;39(37):11194–204. doi: 10.1021/bi000432m. [DOI] [PubMed] [Google Scholar]
- 33.Cooper CA, Gasteiger E, Packer NH. GlycoMod--a software tool for determining glycosylation compositions from mass spectrometric data. Proteomics. 2001;1(2):340–9. doi: 10.1002/1615-9861(200102)1:2<340::AID-PROT340>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 34.Sato K, Okamoto H, Aihara S, Hoshi Y, Tanaka T, Mishiro S. Demonstration of sugar moiety on the surface of hepatitis C virions recovered from the circulation of infected humans. Virology. 1993;196(1):354–7. doi: 10.1006/viro.1993.1488. [DOI] [PubMed] [Google Scholar]
- 35.Yagnik AT, Lahm A, Meola A, Roccasecca RM, Ercole BB, Nicosia A, Tramontano A. A model for the hepatitis C virus envelope glycoprotein E2. Proteins. 2000;40(3):355–66. doi: 10.1002/1097-0134(20000815)40:3<355::aid-prot20>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 36.Rey FA, Heinz FX, Mandl C, Kunz C, Harrison SC. The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature. 1995;375(6529):291–8. doi: 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- 37.Sayle RA, Milner-White EJ. RASMOL: biomolecular graphics for all. Trends Biochem Sci. 1995;20(9):374. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
- 38.Kuiken C, Combet C, Bukh J, Shin IT, Deleage G, Mizokami M, Richardson R, Sablon E, Yusim K, Pawlotsky JM, Simmonds P. A comprehensive system for consistent numbering of HCV sequences, proteins and epitopes. Hepatology. 2006;44(5):1355–61. doi: 10.1002/hep.21377. [DOI] [PubMed] [Google Scholar]
- 39.Itoh S, Kawasaki N, Ohta M, Hyuga M, Hyuga S, Hayakawa T. Simultaneous microanalysis of N-linked oligosaccharides in a glycoprotein using microbore graphitized carbon column liquid chromatography-mass spectrometry. J Chromatogr A. 2002;968(12):89–100. doi: 10.1016/s0021-9673(02)00951-2. [DOI] [PubMed] [Google Scholar]
- 40.Ritchie MA, Gill AC, Deery MJ, Lilley K. Precursor ion scanning for detection and structural characterization of heterogeneous glycopeptide mixtures. J Am Soc Mass Spectrom. 2002;13(9):1065–77. doi: 10.1016/S1044-0305(02)00421-X. [DOI] [PubMed] [Google Scholar]
- 41.Sullivan B, Addona TA, Carr SA. Selective detection of glycopeptides on ion trap mass spectrometers. Anal Chem. 2004;76(11):3112–8. doi: 10.1021/ac035427d. [DOI] [PubMed] [Google Scholar]
- 42.Wang F, Nakouzi A, Angeletti RH, Casadevall A. Site-specific characterization of the N-linked oligosaccharides of a murine immunoglobulin M by high-performance liquid chromatography/electrospray mass spectrometry. Anal Biochem. 2003;314(2):266–80. doi: 10.1016/s0003-2697(02)00693-0. [DOI] [PubMed] [Google Scholar]
- 43.Wuhrer M, Deelder AM, Hokke CH. Protein glycosylation analysis by liquid chromatography-mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2005;825(2):124–33. doi: 10.1016/j.jchromb.2005.01.030. [DOI] [PubMed] [Google Scholar]
- 44.Neil GL, Niemann C, Hein GE. Structural specificity of alpha-chymotrypsin: polypeptide substrates. Nature. 1966;210(5039):903–7. doi: 10.1038/210903a0. [DOI] [PubMed] [Google Scholar]
- 45.Cutalo JM, Deterding LJ, Tomer KB. Characterization of glycopeptides from HIV-I(SF2) gp120 by liquid chromatography mass spectrometry. J Am Soc Mass Spectrom. 2004;15(11):1545–55. doi: 10.1016/j.jasms.2004.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lescar J, Roussel A, Wien MW, Navaza J, Fuller SD, Wengler G, Wengler G, Rey FA. The Fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell. 2001;105(1):137–48. doi: 10.1016/s0092-8674(01)00303-8. [DOI] [PubMed] [Google Scholar]
- 47.McMinn PC. The molecular basis of virulence of the encephalitogenic flaviviruses. J Gen Virol. 1997;78:2711–22. doi: 10.1099/0022-1317-78-11-2711. [DOI] [PubMed] [Google Scholar]
- 48.Hu H, Shioda T, Moriya C, Xin X, Hasan MK, Miyake K, Shimada T, Nagai Y. Infectivities of human and other primate lentiviruses are activated by desialylation of the virion surface. J Virol. 1996;70(11):7462–70. doi: 10.1128/jvi.70.11.7462-7470.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bolmstedt A, Biller M, Hansen JE, Moore JP, Olofsson S. Demonstration of peripheral fucose units in N-linked glycans of human immunodeficiency virus type 1 gp 120: effects on glycoprotein conformation. Arch Virol. 1997;142(12):2465–81. doi: 10.1007/s007050050255. [DOI] [PubMed] [Google Scholar]
- 50.Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. Antibody neutralization and escape by HIV-1. Nature. 2003;422(6929):307–12. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 51.Hebert DN, Zhang JX, Chen W, Foellmer B, Helenius A. The number and location of glycans on influenza hemagglutinin determine folding and association with calnexin and calreticulin. J Cell Biol. 1997;139(3):613–23. doi: 10.1083/jcb.139.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Owsianka AM, Timms JM, Tarr AW, Brown RJ, Hickling TP, Szwejk A, Bienkowska-Szewczyk K, Thomson BJ, Patel AH, Ball JK. Identification of conserved residues in the E2 envelope glycoprotein of the hepatitis C virus that are critical for CD81 binding. J Virol. 2006;80(17):8695–704. doi: 10.1128/JVI.00271-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Imperiali B, Shannon KL. Differences between Asn-Xaa-Thr-containing peptides: a comparison of solution conformation and substrate behavior with oligosaccharyltransferase. Biochemistry. 1991;30(18):4374–80. doi: 10.1021/bi00232a002. [DOI] [PubMed] [Google Scholar]