Abstract
Through a convergence of functional genomic and proteomic studies, we identify Bora as a previously unknown cell cycle protein that interacts with the Plk1 kinase and the SCF–β-TrCP ubiquitin ligase. We show that the Bora protein peaks in G2 and is degraded by proteasomes in mitosis. Proteolysis of Bora requires the Plk1 kinase activity and is mediated by SCF–β-TrCP. Plk1 phosphorylates a conserved DSGxxT degron in Bora and promotes its interaction with β-TrCP. Mutations in this degron stabilize Bora. Expression of a nondegradable Bora variant prolongs the metaphase and delays anaphase onset, indicating a physiological requirement of Bora degradation. Interestingly, the activity of Bora is also required for normal mitotic progression, as knockdown of Bora activates the spindle checkpoint and delays sister chromatid segregation. Mechanistically, Bora regulates spindle stability and microtubule polymerization and promotes tension across sister kinetochores during mitosis. We conclude that tight regulation of the Bora protein by its synthesis and degradation is critical for cell cycle progression.
Introduction
Cell cycle progression is controlled by regulated expression of cell cycle proteins, by their posttranslational modifications, and by their temporally and spatially regulated proteolysis (Murray, 2004). Among these regulatory mechanisms, protein expression at specific cell cycle stages plays an important role. A comprehensive genomic and proteomic study in yeast indicates that genes periodically expressed in the cell cycle control the assembly and the activity of regulatory complexes important for cell cycle progression (de Lichtenberg et al., 2005). Furthermore, periodically expressed proteins are also more frequently targeted by other regulatory mechanisms, such as proteolysis and phosphorylation, indicating that periodically expressed genes represent a key group of cell cycle regulators (de Lichtenberg et al., 2005). Human genome contains 566 genes transcriptionally induced in G2 or in mitosis, half of which encode functionally uncharacterized novel proteins (Whitfield et al., 2002). To understand the mechanism of mitotic regulation in human cells, we have undertaken a genomic approach to investigate the function of genes transcriptionally induced in G2/M. We initially focused on novel genes with the best induction profile in G2/M in a siRNA screen using quantitative assays for their functions in mitotic entry and progression. This has led to the identification of several important regulators of mitosis and cytokinesis (Zhao and Fang, 2005; Wong and Fang, 2006;Zhao et al., 2006; Seki and Fang, 2007). In this paper, we report our characterization of a G2-induced gene, FLJ22624, that is required for normal mitotic progression.
A distant homologue of FLJ22624, identified in Drosophila melanogaster as dBora, is required for asymmetrical division of sensory cells (Hutterer et al., 2006). dBora interacts genetically with the Aurora A pathway in vivo, and the recombinant dBora protein modestly activates the Aurora A kinase activity in vitro. The Aurora A–dBora pathway controls the asymmetrical localization of cell fate determinants and orientation of the mitotic spindle and, therefore, mediates asymmetrical cell division.
In an independent proteomic study on the function and regulation of the mitotic polo-like kinase (Plk) 1 using mass spectrometry, we rediscovered FLJ22624/Bora as a Plk1-interacting protein. Plk1 controls multiple transitions in mitosis and cytokinesis, and one of its key biochemical functions is to phosphorylate its substrates and to target them for degradation by the SCF–β-TrCP ubiquitin ligase in the cell cycle (Barr et al., 2004). Indeed, we identified subunits of the SCF–β-TrCP ligase as Bora-interacting proteins by mass spectrometry, suggesting an active mechanism for Bora turnover in the cell cycle.
The SCF–β-TrCP ligase consists of the core enzyme Skp1-Cul1 and an F-box protein, β-TrCP (Cardozo and Pagano, 2004; Ang and Wade Harper, 2005; Petroski and Deshaies, 2005). β-TrCP directly interacts with substrates and acts as an adaptor protein to bridge substrates to the ligase, thereby targeting them for destruction. The timing of substrate degradation, to a large extent, is not controlled by the activity of the SCF ligase but instead is determined by the posttranslational modifications of substrates in the cell cycle. The majority of the β-TrCP substrates contains a DSGxxS degron, and β-TrCP recognizes this degron when both Ser are phosphorylated, although other noncanonical degrons have also been reported (Jin et al., 2003; Watanabe et al., 2004, 2005; Kanemori et al., 2005). Thus, the SCF–β-TrCP ligase is a key enzyme that acts together with cell cycle kinases to control the rapid and irreversible proteolysis of cell cycle proteins and to mediate unidirectional transitions in the cell cycle. Although there are two paralogues of the F-box protein β-TrCP (β-TrCP1 and β-TrCP2) in human genome, they have identical biochemical activity and act similarly (Cardozo and Pagano, 2004; Ang and Wade Harper, 2005; Petroski and Deshaies, 2005).
In this paper, we report the function and regulation of Bora in mitosis. We show that the Bora protein accumulates in G2 and is degraded in a Plk1- and β-TrCP–dependent manner in mitosis. Both the accumulation and the degradation of Bora control mitotic progression. During mitosis, the Bora protein regulates the spindle stability and microtubule polymerization and is required for efficient chromosome congression, full interkinetochore tension, and timely anaphase onset. In contrast, expression of a nondegradable Bora delayed anaphase onset, indicating a functional importance of Bora degradation. We conclude that Bora is a novel cell cycle regulator essential for mitosis.
Results
The levels of the Bora protein fluctuate in the cell cycle
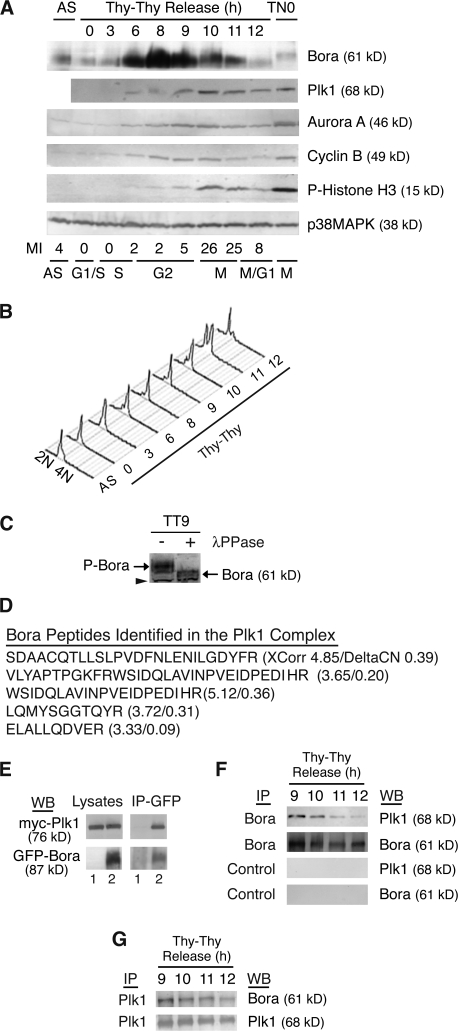
Bora is induced transcriptionally in G2 (http://genome-www.stanford.edu/cgi-bin/Human-CellCycle/Hela/graph/mwhitSearch.pl?gene=IMAGE:824753; Whitfield et al., 2002). We examined the levels of the Bora protein across the cell cycle and compared it to key cell cycle regulators. HeLa S3 cells were arrested at the G1/S boundary by a double thymidine treatment (Thy–Thy) and then released into fresh media (Fang et al., 1998b). The cell cycle profile of released cells was analyzed by FACS (Fig. 1, A and B). The mitotic time-points were also determined by levels of phosphorylated histone H3 (Fig. 1 A). The levels of Bora were low at the G1/S boundary but increased in late S, peaked in G2 (8–9 h after release), and then gradually decreased in mitosis (Fig. 1 A). The Bora protein accumulated earlier than other mitotic regulators, such as Aurora A and cyclin B, and its level decreased at a time when these mitotic regulators peaked (10 h after release; Fig. 1 A). Consistent with this, the levels of Bora were low in unsynchronized cells (AS) and in late prometaphase cells shaken-off from the thymidine-nocodazole–arrested culture (TN0), even though a low level of Bora was still detectable in prometaphase cells (Fig. 1 A). Bora is phosphorylated in G2 and in mitosis, as treatment of G2/M lysates with λ-phosphatase reduced the mobility of Bora (Fig. 1 C). We concluded that Bora is a cell cycle–regulated protein.
Figure 1.
Bora is a cell cycle protein interacting with Plk1. (A and B) The levels of Bora fluctuate in the cell cycle. HeLa S3 cells were synchronized at the G1/S boundary by a double thymidine arrest (Thy–Thy), released into fresh media, and harvested at the indicated times. Protein levels were analyzed by Western blotting (A) and cell cycle profile was assayed by FACS with a propidium iodine staining and with an anti-MPM2 antibody staining (A and B). The MPM2 antibody recognizes mitotic phosphoproteins and was used here to determine mitotic index. p38MAPK served as a loading control. AS, unsynchronized cells; TN0, prometaphase cells synchronized by a thymidine-nocodazole arrest. (C) Phosphorylation of Bora in the cell cycle. G2 cells were collected at 9 h (TT9) after release from a double thymidine arrest and cell lysates were incubated with or without λ-phosphatase (λ-PPase). In a Western blot analysis, the phosphorylation state of Bora was determined by its mobility in SDS-PAGE. Arrowhead points to a cross reacting band, which also serves as a loading control. P-Bora, phospho-Bora. (D) Identification of Bora as a Plk1-interacting protein. Listed are peptides of Bora identified by mass spectrometry in the GFP-S-Plk1 complexes together with their XCorr and DeltaCN scores. (E) Ectopically expressed Bora interacts with Plk1. Myc-Plk1 was cotransfected with GFP (lane 1) or GFP-Bora (Lane 2) into HeLa cells and cells were harvested at 48 h after transfection. Cell lysates and the anti-GFP immunoprecipitates (IP) were assayed by Western blotting (WB). (F and G) Endogenous Bora interacts with Plk1 during Bora degradation. HeLa S3 cells were synchronized as in A. Endogenous Bora (F) or Plk1 (G) was immunoprecipitated with respective antibodies and associated proteins were analyzed by Western blotting. Control IP in F corresponded to immunoprecipitation with nonspecific IgG.
Bora interacts with Plk1
In an independent project, we investigated the function and regulation of Plk1 by purifying Plk1 complexes from mitotic cells stably expressing a GFP- and S-peptide–tagged Plk1 transgene. Proteomic analysis of associated proteins by mass spectrometry using liquid chromatography tandem mass spectrometry revealed that Bora is a Plk1-interacting protein (Fig. 1 D). This interaction was confirmed in a transient transfection experiment by coimmunoprecipitation of myc-Plk1 with GFP-Bora but not with GFP (Fig. 1 E).
As Plk1 interacts with and promotes the degradation of several cell cycle regulators (Barr et al., 2004), we investigated the interaction of Bora and Plk1 during down-regulation of Bora. HeLa S3 cells were synchronized to late G2 and early mitosis by a release from a double thymidine arrest for 9–10 h (TT9 and 10). Plk1 and Bora mutually coimmunoprecipitated with each other in G2 and in mitosis (TT9 and 10; Fig. 1, F and G). As cells exit from mitosis (TT12), the extent of Plk1-Bora association was substantially reduced, partly because of the decreased abundance of Bora. This interaction is specific, as control IgG precipitated neither Plk1 nor Bora (Fig. 1 F), and the anti-Bora antibody did not immunoprecipitate other mitotic kinases such as Aurora A (not depicted). We concluded that Bora interacts with Plk1 at late G2 and in mitosis.
Bora is degraded by proteasomes in a Plk1-dependent manner
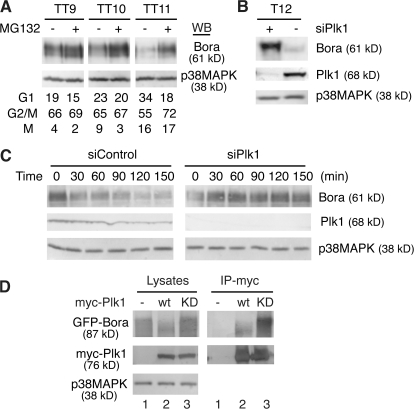
Next, we investigated whether the down-regulation of Bora is mediated by proteasome-dependent proteolysis. HeLa S3 cells were synchronized by a double thymidine arrest and then released into fresh media (Fang et al., 1998b). At 7, 8, and 9 h after release (corresponding to early to late G2), the proteasome inhibitor MG132 was added to the culture media and cells were harvested 2 h later. Inhibition of proteasomes stabilized Bora, especially at late G2 and mitosis (Fig. 2 A, TT10 and 11). This stabilization was not caused by a change in the cell cycle profile of proteasome-treated cells as assayed by FACS. We concluded that Bora is degraded by proteasomes.
Figure 2.
Bora is degraded in a Plk1- and proteasome-dependent manner. (A) Proteasome-dependent degradation of Bora. HeLa S3 cells were synchronized with a double thymidine arrest and then released into fresh media. At 7, 8, and 9 h after release, the proteasome inhibitor MG132 or control DMSO was added and cells were collected 2 h later (TT9, 10, and 11, respectively). Protein levels were analyzed by Western blotting and cell cycle profile was determined by FACS. (B) Plk1 controls the Bora protein level. HeLa cells were transfected with a siRNA targeted to Plk1 (siPlk1) or a control siRNA, synchronized with thymidine for 18 h, and then released. At 10 h after release, 1 μM taxol was added and mitotic cells harvested 2 h later (T12). Protein levels were determined by Western blotting. (C) Plk1 controls Bora half-life. HeLa cells were synchronized by a double thymidine treatment and transfected with a control siRNA (siControl) or siPlk1 during the second thymidine arrest. At 8 h after release from the second thymidine arrest, 100 μg/ml cyclohexamine was added and cells were harvested at 0, 30, 60, 90, 120, and 150 min later. Half-life of Bora was determined by Western blotting. (D) Plk1 kinase activity is required for Bora degradation. GFP-Bora was cotransfected with a control vector (lane 1), myc-Plk1 (lane 2), or myc-Plk1-K82R (lane 3; KD, kinase dead) and cells were harvested at 48 h after transfection. Total cell lysates and the anti-myc immunoprecipitates were analyzed by Western blotting.
Degradation of Bora requires Plk1, as knockdown of Plk1 by a specific siRNA or inhibition of Plk1 by the small molecule inhibitor BI 2536 promoted accumulation of Bora in mitosis (Fig. 2 B and not depicted). Knockdown of Plk1 also increased the half-life of Bora (Fig. 2 C), although this effect could be indirect, as depletion of Plk1 also delays the entry into mitosis (Hansen et al., 2004), a cell cycle phase in which Bora is degraded. Furthermore, the kinase activity of Plk1 is required for the degradation of Bora, as cotransfection of the kinase-active myc-Plk1 with GFP-Bora destabilized GFP-Bora (Fig. 2 D, left, lanes 2 vs. 1), whereas cotransfection of the kinase-dead myc-Plk1-K82R mutant stabilized GFP-Bora (Fig. 2 D, left, compare lanes 1–3). In contrast, GFP-Bora coimmunoprecipitated with myc-Plk1 independent of its kinase activity (Fig. 2 D, right), indicating that binding of Bora to Plk1 is not sufficient for its degradation and that Plk1-mediated phosphorylation is required.
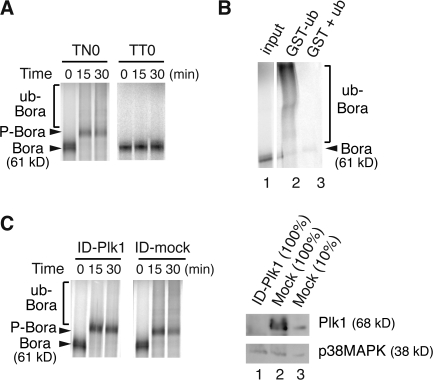
Next, we reconstituted the ubiquitination of Bora in extracts of synchronized HeLa S3 cells in vitro. When incubated, in the presence of recombinant ubiquitin, ATP, and MG132, with extracts (TN0) of prometaphase cells arrested by a thymidine-nocodazole treatment, 35S-labeled Bora migrated as a smear above the phosphorylated Bora (P-Bora; Fig. 3 A, left). This slow migrating smear corresponded to ubiquitinated Bora, as GST-ubiquitin conjugates purified from the TN0 extracts by glutathione beads contained the slow migrating 35S-Bora-GST-ub conjugates (Fig. 3 B, lane 2). Furthermore, ubiquitination of Bora in TN0 extracts requires Plk1, as immunodepletion of Plk1 from extracts (>90% depletion) substantially reduced the degree of Bora ubiquitination (Fig. 3 C). Ubiquitination of Bora is cell cycle regulated, as incubation of 35S-Bora with extracts (TT0) of G1 cells arrested by a double thymidine treatment generated neither phosphorylation nor ubiquitination of Bora (Fig. 3 A, right). Thus, Bora is ubiquitinated in mitotic extracts in a Plk1-dependent manner.
Figure 3.
Ubiquitination of Bora requires Plk1. (A) In vitro ubiquitination assay. In vitro–synthesized 35S-Bora was incubated, in the presence of MG132, with extracts of HeLa S3 cells arrested by a thymidine-nocodazole treatment (TN0) or by a double thymidine treatment (TT0). Samples were taken at the indicated times and assayed by SDS-PAGE. P-Bora, phosphorylated Bora. (B) In vitro ubiquitination assay described in A was performed in TN0 extracts for 60 min in the presence of GST-ubiquitin (lane 2) or GST plus nontagged ubiquitin (lane 3) and the ubiquitin conjugates were purified by Glutathione beads and assayed by SDS-PAGE. Lane 1, 10% input of 35S-Bora. White line indicates that intervening lanes have been spliced out. (C) TN0 extracts were either mock depleted or depleted by an anti-Plk1 antibody at 4°C for 1 h. Ubiquitination assay in the depleted extracts was performed as described in A and the depletion efficiency was determined by Western blotting. ID, immunodepletion. In lane 3, 1/10 of the extracts (relative to lanes 1 and 2) was loaded to determine the depletion efficiency.
Bora is ubiquitinated in a β-TrCP1–dependent manner
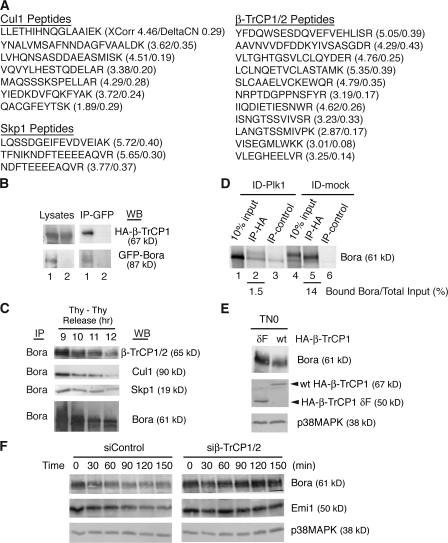
To investigate the mechanism of Bora degradation, we purified the Bora complex from HeLa S3 cells stably expressing a GFP and S-peptide–tagged Bora transgene. Proteomic analysis of associated proteins by mass spectrometry revealed three subunits of the SCF–β-TrCP ligase (Cul1, Skp1, and β-TrCP1/2) as Bora-interacting proteins (Fig. 4 A). Interaction between β-TrCP1 and Bora was confirmed in a transient transfection experiment by coimmunoprecipitation of HA-β-TrCP1 with GFP-Bora but not with GFP (Fig. 4 B). Furthermore, endogenous SCF–β-TrCP ligase coprecipitated with the anti-Bora antibody beads but not with the nonspecific IgG beads (Fig. 4 C and not depicted).
Figure 4.
Bora is ubiquitinated in a β-TrCP1–dependent manner. (A) Identification of the SCF–β-TrCP subunits as Bora-interacting proteins. Listed are peptides of Cul1, Skp1, and β-TrCP1/2 identified by mass spectrometry in the GFP–S-Bora complex together with their XCorr and DeltaCN scores. (B) Bora associates with β-TrCP1 in vivo. HA-β-TrCP1 was cotransfected into HeLa cells with either GFP-Bora (lane 1) or GFP (lane 2) and cells were harvested at 48 h after transfection. Cell lysates and the anti-GFP immunoprecipitates were assayed by Western blotting. (C) Endogenous Bora and SCF–β-TrCP interact during the degradation of Bora. HeLa S3 cells were synchronized as described in Fig. 1 A. Endogenous Bora was immunoprecipitated and associated proteins were analyzed by Western blotting. (D) Bora interacts with β-TrCP1 in a Plk1-dependent manner. TN0 extracts were either mock depleted or depleted of Plk1 as described in Fig. 3 C. In vitro–synthesized 35S-Bora was first incubated with depleted extracts and then with nonlabeled HA-β-TrCP1 that had been immunoprecipitated by the anti-HA antibody/protein A beads. Proteins associated with HA-β-TrCP1 beads were assayed by SDS-PAGE. IP-control corresponds to a control immunoprecipitation of HA-β-TrCP1 with nonspecific IgG. Input, 10% of Bora after incubation with the TN0 extracts. Numbers below lanes 2 and 5 represent β-TrCP1-bound Bora relative to their respective total inputs. (E) β-TrCP1 controls the levels of Bora in vivo. HeLa cells were transfected with HA-β-TrCP1 or HA-β-TrCP1-δF and then synchronized by a thymidine-nocodazole arrest. Mitotic cells were shaken off at 42 h after transfection and protein levels in mitotic cells were determined by Western blotting. (F) β-TrCP1 controls Bora half-life. HeLa cells were synchronized by a double thymidine treatment and transfected with a control siRNA (siControl) or a previously characterized siRNA targeting both β-TrCP1 and 2 (si β-TrCP1/2; Mailand et al., 2006) during the second thymidine arrest. At 8 h after release from the second thymidine arrest, cyclohexamine was added and cells harvested 0, 30, 60, 90, 120, and 150 min later. Half-life of Bora was determined by Western blotting.
The interaction between Bora and SCF–β-TrCP ligase requires the Plk1 activity. In the presence of TN0 extracts, in vitro–translated 35S-Bora efficiently coprecipitated with HA-β-TrCP1 (Fig. 4 D, compare lanes 5 and 6). This interaction depends on Plk1 in the TN0 extracts, as immunodepletion of Plk1 abolished the complex formation (Fig. 4 D, lane 2).
The stability of endogenous Bora is controlled by β-TrCP. Expression of a dominant-negative mutant (β-TrCP1-δF) of β-TrCP1 that lacks the F box (aa 30–179 deleted; Hansen et al., 2004) stabilized the endogenous Bora in mitosis (Fig. 4 E), which is consistent with the ability of β-TrCP1-δF to stabilize other substrates of the SCF–β-TrCP ligase such as Emi1 (Hansen et al., 2004). Furthermore, knockdown of β-TrCP using a previously characterized siRNA (Mailand et al., 2006) increased the half-lives of both Bora and Emi1 (Fig. 4 F). Thus, Bora levels are under the control of the β-TrCP1 pathway in vivo.
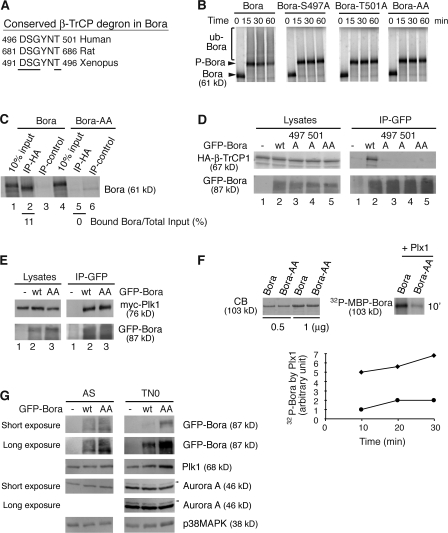
A DSGxxT degron is required for Bora degradation
Next, we determined the molecular recognition of Bora by β-TrCP. β-TrCP recognizes in substrates a consensus motif, DSGxxS, in which both serine residues are phosphorylated (Cardozo and Pagano, 2004; Ang and Wade Harper, 2005; Petroski and Deshaies, 2005). Although Bora does not contain such a motif, it does have the 496-DSGYNT-501 sequence that is conserved among human, rat, and Xenopus laevis (Fig. 5 A). Thus, we investigated the role of this sequence in Bora degradation by mutating S497 and T501 to A, either singly or doubly. In the in vitro ubiquitination assay performed in TN0 extracts, mutations of S497, T501, or both blocked ubiquitination of Bora (Fig. 5 B), indicating that these two residues are critical for its degradation. Consistent with this, the 35S-Bora-AA mutant failed to interact with HA-β-TrCP1 both in the in vitro binding assay and in the cotransfection experiment in vivo (Fig. 5, C and D). Thus, S497 and T501 are directly recognized by β-TrCP. In contrast, these two residues are dispensable for the interaction between Bora and Plk1, as GFP-Bora-AA efficiently coprecipitated the myc-Plk1 in cotransfection experiments (Fig. 5 E). This experiment further indicated that mutations of S497 and T501 do not globally alter the folding of the mutant proteins.
Figure 5.
The DSGxxT degron is required for Bora degradation. (A) The conserved β-TrCP degron in Bora from different species. Residues important for recognition by β-TrCP are underlined. (B) The β-TrCP degron is required for ubiquitination of Bora. In vitro ubiquitination assays were performed in TN0 extracts as described in Fig. 3 A. Bora-AA has both S497 and T501 mutated to A. (C) In vitro binding of HA-β-TrCP1 to Bora and Bora-AA were performed in nondepleted TN0 extracts, as described in Fig. 4 D. Numbers below lanes 2 and 5 represent β-TrCP1–bound Bora relative to their respective total inputs. (D and E) The β-TrCP degron is required for the association of Bora with β-TrCP1 but not for the association of Bora with Plk1 in vivo. HA-β-TrCP1 (D) or myc-Plk1 (E) was cotransfected into HeLa cells with either GFP (lane 1) or GFP-Bora (wild type or mutants) and cells were harvested 48 h after transfection. Cell lysates and the anti-GFP immunoprecipitates were assayed by Western blotting. (F) Plk1 phosphorylates the Bora degron. Recombinant MBP-Bora and MBP-Bora-AA were phosphorylated by purified recombinant Plx1 (X. laevis homologue of Plk1) in the presence of radioactive ATP. The kinetics of Plx1-mediated phosphorylation was quantified and plotted. The amounts of MBP-Bora and MBP-Bora-AA were determined by Coomassie blue staining (CB). ♦, Bora; •, Bora-AA. (G) The DSGxxT degron is recognized in vivo. HeLa cells were transfected with GFP, GFP-Bora, or GFP-Bora-AA and harvested as asynchronous culture (AS) at 34 h after transfection or as prometaphase cells (TN0) at 48 h after transfection after a thymidine-nocodazole arrest. Total cell lysates were analyzed by Western blotting. Arrowheads point to the active and phosphorylated Aurora A (Hirota et al., 2003).
The DSGYNT degron was directly phosphorylated by Plk1, as the Bora-AA mutant was phosphorylated much less efficiently by recombinant Plx1, the X. laevis homologue of Plk1 (Fig. 5 F). Furthermore, this degron controls the levels of the Bora protein in vivo. HeLa cells were transfected with either GFP-Bora or GFP-Bora-AA, and the levels of expressed proteins were determined in asynchronous cells and in prometaphase cells arrested by a thymidine-nocodazole treatment (Fig. 5 G). The level of GFP-Bora-AA was only marginally higher than that of GFP-Bora in asynchronous cells, the majority of which were in G1, whereas GFP-Bora-AA was significantly more abundant than GFP-Bora in prometaphase cells, a cell cycle stage with active Plk1. Interestingly, ectopic expression of the nondegradable Bora variant also increased the level of Plk1 in prometaphase cells, whereas expression and phosphorylation of Aurora A were not affected (Fig. 5 G).
Degradation of Bora is required for normal mitotic progression
To analyze the physiological requirement of Bora degradation, we constructed HeLa cell lines stably expressing GFP-Bora and GFP-Bora-AA resistant to knockdown (GFP-rBora and GFP-rBora-AA). Multiple clones were analyzed and data from representative clones are presented in this section. To avoid complication of mitotic defects arising from knockdown of Bora (see Figs. 7 and 8), we analyzed the effect of rBora and rBora-AA expression on mitosis without depletion of endogenous Bora.
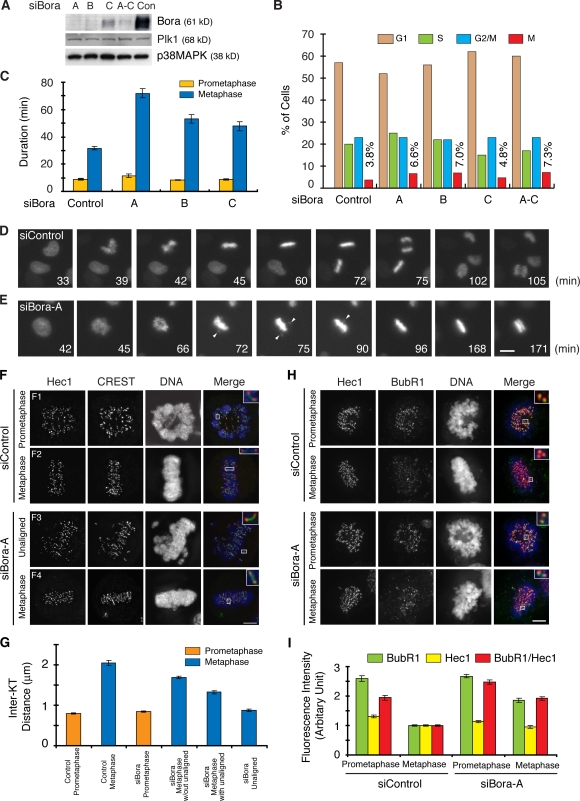
Figure 7.
Knockdown of Bora delays anaphase onset. (A and B) Analysis of Bora knockdown efficiency. HeLa cells were either control transfected or transfected with siRNAs targeting three regions of the Bora gene (siBora-A, -B, and -C and a mixture of all three) and collected at 52 h after transfection. Protein levels were determined by Western blotting (A) and cell cycle profile was determined by FACS (n > 20,000 cells; B). (C–E) Kinetics of mitotic progression in control and Bora-depleted cells was analyzed by time-lapse microscopy in HeLa cells stably expressing GFP-histone H2B. Cell images were captured every 3 min and the durations of prometaphase and metaphase were quantified by counting more than 50 mitotic cells in each transfection (C). Still frames from movies of representative cells are shown in D for control transfection and in E for Bora knockdown. Arrowheads point to unaligned chromosomes. (F–I) Analysis of tension across sister kinetochores (F and G) and the spindle checkpoint signals (H and I). Shown in F and H are maximum projections from deconvolved z stacks of representative control or Bora-depleted HeLa cells stained for Hec1 (red), CREST (green; F) or BubR1 (green; H), and DNA (blue). Insets show a single z slice of the boxed regions. Interkinetochore (Inter-KT) distance was quantified (n > 45 kinetochore pairs for each category) and plotted (G). Hec1 and BubR1 (n = 60 kinetochores) signals were quantified in 10 control and Bora-depleted cells (I). The BubR1 kinetochore signal normalized with the Hec1 intensity of the same kinetochores was also plotted. For metaphase cells, only chromosomes aligned at the metaphase plate were quantified in I. The siBora metaphase without unaligned category in G represents chromosomes in metaphase cells (F4) that contain no unaligned chromosomes. The siBora metaphase with unaligned category represents chromosomes on the metaphase plate in knockdown cells (F3) that contain unaligned chromosomes. The unaligned chromosomes in these cells (F3) were quantified in a separate category (siBora unaligned). Error bars show standard error. Bar: (D and E) 10 μm; (F and H) 5 μm.
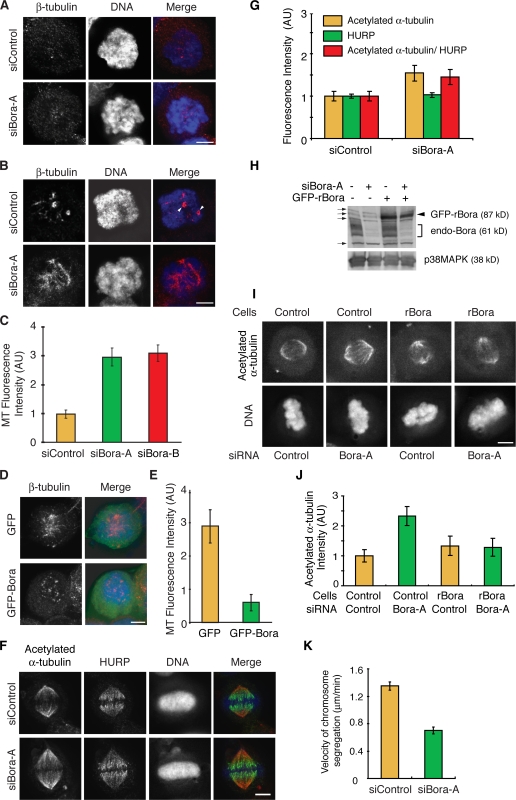
Figure 8.
Bora controls spindle stability and microtubule growth in mitosis. (A–E) Shown in A, B, and D are maximum projections from deconvolved z stacks of representative HeLa cells stained for β-tubulin (red) and DNA (blue). Cells were transfected with siRNA (A–C) or with GFP/GFP-Bora (D and E). Transfected cells were treated with 5 μg/ml nocodazole for 10 min at 37°C, washed, released into fresh media, and fixed at 0 min (A) and 6 min (B–E) after release. Centrosomes are marked by arrowheads in B. Images from representative cells were acquired under constant exposure. The amounts of microtubules repolymerized at 6 min after release were quantified from 10 mitotic cells in each transfection and plotted in C and E. (F and G) Shown in F are maximum projections from deconvolved z stacks of representative control or Bora-depleted HeLa cells stained for acetylated α-tubulin (red), HURP (green), and DNA (blue). Images from representative cells were acquired under constant exposure. Mean HURP and acetylated α-tubulin immunofluorescence intensities on metaphase spindles was quantified (n = 20 half-spindles from 10 cells; G). Acetylated α-tubulin signal normalized against the HURP intensity of the same cell was also plotted. (H–J) Control and GFP-rBora–expressing HeLa cells were transfected with a control siRNA or with siBora-A and the knockdown efficiency was determined by Western blotting (H). Long exposure of Western blot uncovered cross reacting bands (H, arrows), one of which comigrated with GFP-rBora. The levels of acetylated α-tubulin were assayed by immunofluorescence staining (I), quantified, and plotted (J). (K) Velocity of sister chromatid segregation at anaphase A was measured in control and Bora knockdown cells from time-lapse movies shown in Fig. 7 (D and E; n = 32 anaphase cells for each quantification). AU, arbitrary units. Error bars show standard error. Bars, 5 μm.
rBora and rBora-AA-c1 (clone 1) cells expressed transgenes at levels between one and twofold of the endogenous Bora, whereas rBora-AA-c2 (clone 2) cells expressed the transgene at a level ∼16 times that of the endogenous protein (Fig. 6 A). Importantly, rBora was degraded in prometaphase cells (TN0) but rBora-AA was stable (Fig. 6 B), which is consistent with the requirement of the DSGYNT degron for Bora degradation.
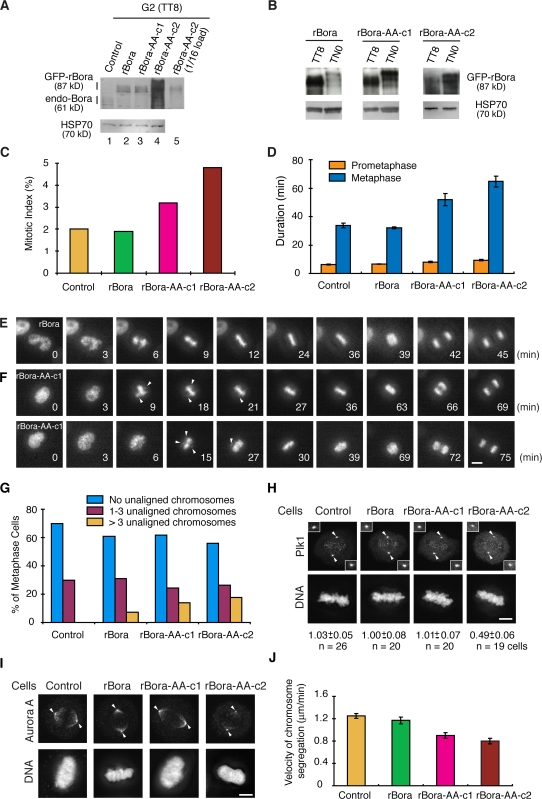
Figure 6.
Expression of nondegradable Bora accumulates cells in mitosis and delays anaphase onset. (A–C) Control, GFP-rBora, GFP-rBora c1, and GFP-rBora c2 cells were collected at G2 (TT8) or in mitosis (TN0), and the levels of GFP fusion proteins and endogenous (endo) Bora were analyzed by Western blotting (A and B). Mitotic index of unsynchronous cells were determined by FACS (n > 20,000 cells) and plotted (C). (D–F) Kinetics of mitotic progression in control, GFP-rBora, GFP-rBora-c1, and GFP-rBora-c2 cells was analyzed by time-lapse microscopy upon transient transfection of GFP-histone H2B. The levels of GFP fusion proteins in stable cell lines were far below that of GFP-H2B. Cell images were captured every 3 min and the durations of prometaphase and metaphase were quantified by counting more than 40 mitotic cells in each transfection (D). Still frames from movies of representative cells were shown in E for GFP-rBora and in F for GFP-rBora-AA-c1. Arrowheads point to unaligned chromosomes. (G) Control, GFP-rBora, GFP-rBora-c1, and GFP-rBora-c2 cells were stained for β-tubulin, and DNA and distribution of metaphase cells (n > 70 metaphase cells) were quantified. (H and I) Localization of Plk1 (H) and Aurora A (I) were shown in maximum projections from deconvolved z stacks of representative metaphase cells stained for DNA and Plk1 (H) or Aurora A (I). Images in each channel were acquired under constant exposure and processed identically for different cell lines. Arrowheads point to centrosome regions. Insets in H show centrosomal Plk1 signals, and the numbers below images indicate the relative centrosomal Plk1 signal intensity. (J) Velocity of sister chromatid segregation at anaphase A was measured from the time-lapse movies shown in D–F (n > 42 anaphase cells for each quantification). Error bars show standard error. Bars: (E and F) 10 μm; (H and I) 5 μm.
Expression of rBora-AA increased mitotic index (Fig. 6 C), although the distribution of cells in G1, S, and G2 were not altered (not depicted). Time-lapse analysis of mitotic progression upon transfection of GFP-Histone H2B indicated that the duration of prometaphase (from nuclear envelope breakdown to the initial formation of the metaphase plate) was not altered, but the duration of metaphase (from initial metaphase to anaphase onset) was lengthened by >60% in rBora-AA–expressing cells (Fig. 6, D–F). The higher the levels of Bora-AA expression, the longer the metaphase (compare c1 and 2 clones). Furthermore, rBora-AA–expressing metaphase cells frequently had more than three chromosomes unaligned outside of the metaphase plate for extended time, although these chromosomes eventually incorporated into the metaphase plate before anaphase onset (Fig. 6, F and G). Similarly, rBora-expressing cells, but not the GFP-expressing control cells, also had a small percentage of metaphase cells with greater than three unaligned chromosomes (Fig. 6 G), probably because of the increased level of Bora protein in these cells. We conclude that degradation of Bora is required for timely mitotic progression through metaphase.
Bora functions in the Aurora A–Plk1 pathway (unpublished data). Thus, we analyzed the localization of Plk1 and Aurora A during mitosis. Expression of Bora, either the wild-type or the nondegradable variant, at its physiological level did not alter the localization of either kinase within the detection sensitivity of our assays (Fig. 6, H and I). However, expression of rBora-AA at a level 16 times that of the endogenous protein greatly diminished Plk1 signals on centrosomes and Aurora A signals on spindle pole microtubules in rBora-AA-c2 cells (Fig. 6, H and I). Quantitative analysis indicated a 50% reduction in the centrosomal Plk1 signals in rBora-AA-c2 metaphase cells compared with rBora metaphase cells. Changes in Plk1 and Aurora A function are likely to contribute to mitotic defects observed in cells expressing nondegradable Bora.
As the majority of endogenous Bora was degraded before metaphase, we expected an effect of nondegradable Bora on chromosome movement in anaphase. Expression of rBora did not affect the rate of anaphase chromosome segregation (Fig. 6 J), as rBora was degraded before anaphase onset (Fig. 6 B). However, expression of rBora-AA reduced the velocity of chromosome segregation by 25–35% at anaphase A (Fig. 6 J), although the fidelity of sister chromatid segregation was not affected in our assay (not depicted). Thus, timely degradation of Bora is required for efficient anaphase.
Bora controls interkinetochore tension and is required for timely anaphase onset
We investigated the function of Bora in the cell cycle. Bora did not associate with any specific cellular structure across the cell cycle, based on our anti-Bora antibody staining and our analysis of GFP-Bora stable cell lines (unpublished data). Thus, Bora likely acts as a soluble factor. Efficient knockdown of Bora by three independent siRNAs (Fig. 7 A) increased the mitotic index, even though the distribution of cells in G1, S, and G2 phase was not altered (Fig. 7 B). Time-lapse analysis of HeLa cells stably expressing GFP-Histone H2B indicated that knockdown of Bora did not substantially affect the progression through prometaphase but significantly prolonged the length of metaphase (Fig. 7, C–E). Furthermore, metaphase cells depleted of Bora frequently had unaligned chromosomes outside of the metaphase plate for a prolonged time (Fig. 7 E). Even though these unaligned chromosomes eventually congressed to the metaphase plate, cells still stayed at metaphase without unaligned chromosomes for an extended time which was followed by sister chromatid segregation.
Anaphase onset requires the establishment of interkinetochore tension generated by the pulling force derived from dynamic turnover of attached microtubules. A prolonged metaphase suggested a lack of tension across sister kinetochores, which can be measured by interkinetochore distance (Maiato et al., 2004). In control cells, the presence of the pulling force increased the interkinetochore distance in control cells from prometaphase (0.80 ± 0.02 μm) to metaphase (2.05 ± 0.07 μm; Fig. 7, F and G). However, the interkinetochore distance of chromosomes aligned at the metaphase plate in Bora-depleted cells was substantially shorter than that in control metaphase cells (Fig. 7 G). Thus, depletion of Bora reduced the interkinetochore tension in metaphase cells.
Microtubule attachment to kinetochores and tension across sister kinetochores are monitored by the spindle checkpoint proteins Mad2 and BubR1, respectively (Chan et al., 1999; Skoufias et al., 2001). We observed a twofold increase in kinetochore BubR1 signals for chromosomes aligned at the metaphase plate in Bora knockdown cells compared with control metaphase cells (Fig. 7, H and I), confirming a defect in interkinetochore tension. In contrast, kinetochore Mad2 signals were comparable between Bora-depleted and control metaphase cells, indicating that kinetochores from chromosomes at the metaphase plate are attached to microtubules in Bora-depleted cells (unpublished data). Thus, knockdown of Bora activates the tension-sensitive spindle checkpoint and delays anaphase onset.
Bora controls spindle dynamics in mitosis
As interkinetochore tension is generated by dynamic polymerization and depolymerization of kinetochore microtubules in metaphase cells (Maiato et al., 2004), we directly measured the microtubule growth in Bora knockdown cells. siRNA-transfected mitotic cells were treated with nocodazole to completely depolymerize spindle microtubules and then washed into fresh media to allow microtubules to repolymerize. Although knockdown of Bora did not affect the depolymerization of spindle microtubules (t = 0 min; Fig. 8 A), mitotic cells depleted of Bora repolymerized microtubules with a substantially faster kinetics (Fig. 8, B and C). 6 min after release from nocodazole treatment, mitotic cells depleted of Bora had three times more microtubules polymerized compared with control mitotic cells (Fig. 8 C). Conversely, ectopic expression of Bora reduced the rate of microtubule repolymerization (Fig. 8, D and E).
An independent assay for spindle turnover is the level of acetylated α-tubulin, a marker for stabilized microtubules (Piperno et al., 1987; de Pennart et al., 1988). The mean immunofluorescence intensity of acetylated α-tubulin was increased by over 50% in the metaphase spindle of Bora knockdown cells compared with that of control cells (Fig. 8, F and G). This change in microtubule stability specifically resulted from Bora knockdown, as expression of a Bora transgene resistant to the Bora siRNA rescued the acetylated α-tubulin phenotype in Bora knockdown cells (Fig. 8, H–J). Changes in the spindle dynamics are expected to affect the rate of chromosome segregation at anaphase. Indeed, the velocity of anaphase chromosome movement was reduced by 40% in Bora knockdown cells (Fig. 8 K). Thus, Bora regulates the spindle stability and microtubule polymerization, which, in turn, controls the interkinetochore tension at metaphase and the rate of chromosome segregation at anaphase.
Discussion
Through functional analyses of G2-induced genes and the Plk1 proteome, we identified Bora as a novel cell cycle regulator and report here its function and regulation in mitosis. We found that mitotic Plk1 phosphorylates Bora on its conserved DSGxxT degron, which promotes its interaction with and degradation by the SCF–β-TrCP ligase. Cell cycle–regulated degradation of Bora is crucial for normal mitotic progression, and expression of a nondegradable variant of Bora resulted in prolonged mitosis and a delay in anaphase onset. Interestingly, even though the majority of Bora is degraded before metaphase, the activity of Bora is also important for mitosis. Bora controls spindle stability and regulates interkinetochore tension and anaphase chromosome movement. We conclude that controlled synthesis and proteolysis of Bora regulate mitotic progression. As Bora has been linked to the Aurora A and Plk1 pathways in the cell cycle (Hutterer et al., 2006; unpublished data), it is likely that the function of Bora in mitosis is mediated through its regulation of these two kinases.
Function of Bora in the cell cycle
Bora is induced in G2 and degraded in mitosis, indicating that Bora functions from late G2 to early mitosis. We showed here that Bora is required for normal progression through mitosis, as knockdown of Bora increased the mitotic index. Although spindle assembly and passage through prometaphase are normal overall in Bora knockdown cells, metaphase cells tend to have unaligned chromosomes that persist for an extended time. Even when all the chromosomes are aligned at the metaphase plate, cells depleted of Bora still stay longer at metaphase. Mechanistically, depletion of Bora results in a reduction in the interkinetochore tension and activation of the tension-sensitive spindle checkpoint, leading to a cell cycle delay/arrest (Fig. 7).
A reduction in the interkinetochore tension in Bora-depleted cells, in theory, could result from a change in the kinetochore or centrosome structure, a change in the spindle dynamics, or an altered interaction between astral microtubules and the cell cortex (Maiato et al., 2004). We found that knockdown of Bora did not affect the centrosome or the spindle pole structure during mitosis, as determined by immunofluorescence staining of ninein, centrin, centroilin, pericentrin, and γ-tubulin (unpublished data). Similarly, mitotic cells depleted of Bora seem to have functional kinetochores, as determined by attachment of microtubules to kinetochores and by the presence of an active spindle checkpoint (Fig. 7). Interestingly, spindle microtubules in Bora-depleted cells repolymerized threefold faster than control cells after treatment with nocodazole, whereas ectopic expression of Bora at high levels substantially reduced the rate of microtubule growth (Fig. 8). Consistent with these observations, Bora-depleted cells have a more stable mitotic spindle, as indicated by higher levels of the acetylated α-tubulin in the spindle (Fig. 8). Thus, Bora controls the stability and growth of spindle microtubules to promote a full interkinetochore tension. As proper microtubule dynamics control the anaphase chromosome segregation (Ganem et al., 2005), Bora also plays a role in anaphase chromosome movement.
How does Bora control spindle stability? It has been reported previously that Bora enhances the kinase activity of Aurora A in the presence of PP1 (Hutterer et al., 2006), a protein phosphatase associated with Aurora A in mitosis (Barr and Gergely, 2007). In addition, we found that Bora not only binds to Plk1, but also regulates its kinase activity (unpublished data). As both Plk1 and Aurora A have been reported to promote the assembly and function of mitotic spindle (Sumara et al., 2004; van Vugt et al., 2004; Barr and Gergely, 2007; Lenart et al., 2007), Bora is likely to regulate spindle dynamics through the Plk1 and Aurora A pathways in mitosis.
Even though depletion of Bora generated defects at metaphase and anaphase, the execution point of Bora may not necessarily have to be at these cell cycle stages, as it is possible that the lack of Bora protein in depleted G2 or early mitotic cells alters the cell physiology in such a way that results in spindle defects at later mitotic stages. Alternatively, it is possible that a subpopulation of Bora is resistant to proteolysis at metaphase (Figs. 1 A and 6 B) and that it is this population of Bora that carries out its mitotic function.
Degradation of Bora by the SCF–β-TrCP1 pathway in a Plk1-dependent manner
The following evidences indicate that Bora is degraded by the SCF–β-TrCP pathway and that the Plk1 kinase is essential for this degradation. First, the levels of the Bora protein vary in the cell cycle, peaking at G2 before mitotic entry (Fig. 1). Down-regulation of Bora in early mitosis requires the proteasome activity (Fig. 2). Second, Bora interacts with Plk1 and the kinase activity of Plk1 is required for the degradation of Bora (Figs. 1–3 ). In fact, Plk1 directly phosphorylates the conserved DSGxxT degron in Bora (Fig. 5). Third, β-TrCP1 directly interacts with Bora through its DSGxxT phosphodegron in a Plk1-dependent manner and this interaction is required for degradation of Bora in vivo (Figs. 4–6 ).
Bora is degraded after Plk1 activation in early mitosis but before metaphase, which is consistent with the fluctuation of the Bora levels in the cell cycle and with our proteasome inhibitor experiments (Figs. 1, 2, and 6). Thus, Bora joins a group of cell cycle regulators, such as Wee1, Emi1, and Claspin, whose degradation is mediated by SCF–β-TrCP and controlled by Plk1. Plk1 interacts with a subset of its substrates through polo-box domain (PBD), which requires a prior priming phosphorylation. In the case of mitotic substrates, such as Wee1 and Emi1, the priming kinase usually is Cdk1 (Barr et al., 2004; Hansen et al., 2004; Moshe et al., 2004; Watanabe et al., 2004, 2005). Other cell cycle–regulated kinases also function as priming kinases (Neef et al., 2007). For example, Claspin is phosphorylated by ATR upon DNA replication stress and prime phosphorylated Claspin is then phosphorylated and inactivated by Plk1, leading to the termination of the replication checkpoint signaling (Yoo et al., 2004; Bennett and Clarke, 2006; Mailand et al., 2006; Mamely et al., 2006; Peschiaroli et al., 2006). We found that Plk1 directly phosphorylates the DSGxxT degron in Bora (Fig. 5). It is likely that the trigger for mitotic degradation of Bora is the activation of Plk1, although we cannot formally exclude the possibility that priming phosphorylation of Bora by another kinase enhances subsequent phosphorylation of its degron by Plk1.
Physiological role of Bora proteolysis
The importance of Bora degradation is underscored by the fact that expression of a nondegradable Bora-AA variant at the physiological protein level slowed down metaphase progression and delayed anaphase onset (Fig. 6). Furthermore, at high levels of expression, Bora-AA, but not Bora, increased the level of mitotic Plk1 (Fig. 5) and affected the cellular localization of mitotic Plk1 and Aurora A (Fig. 6). This increase in Plk1 level is likely mediated through a stabilization of the active Plk1 in the Bora-AA–Plk1 complex, suggesting the existence of a mechanism for active turnover of Plk1 in normal mitotic cells. Similarly, a reduction in Plk1 signals on mitotic centrosomes in Bora-AA-c2 cells likely results from a sequestration of Plk1 in the noncentrosomal Bora-AA–Plk1 complex, which may prevent Plk1 from association with prime phosphorylated centrosomal substrates through its PBD, an interaction required for targeting Plk1 to centrosomes (Barr et al., 2004). Consistent with this, Bora directly interacts with PBD in Plk1 (unpublished data).
Expression of rBora-AA at high levels also reduced the Aurora A signals on spindle poles without affecting its protein level or its phosphorylation state (Figs. 5 and 6). It has been reported that in the presence of PP1, Bora enhances the kinase activity of Aurora A (Hutterer et al., 2006), which may explain the effect of rBora-AA on the localization of Aurora A. Alternatively, as Plk1 is required for targeting Aurora A to centrosomes (De Luca et al., 2006), effect of Bora-AA on Aurora A may be mediated through Plk1.
We speculate that a failure to degrade endogenous Bora in mitosis could allow this protein to accumulate in the cell cycle and that high levels of Bora accumulated after multiple rounds of cell divisions would eventually interfere with Plk1 and Aurora A function in normal cells. At the cellular level, expression of Bora at high levels also reduces microtubule growth during mitosis, possibly as a result of a perturbation of Aurora A and/or Plk1 (Fig. 8). Thus, degradation of Bora in mitosis provides a mechanism for cells to regulate the level of Plk1 and the localization of Plk1 and Aurora A and to control the assembly and function of the mitotic spindle.
It is interesting to note that the Bora gene is located in a region of chromosome 13 (13q21-q22) that harbors a putative breast cancer susceptibility gene (Rozenblum et al., 2002). The same region of the chromosome has also been implicated as a common site for somatic deletions in a variety of malignant tumors. Elucidation of the function and regulation of Bora will aid our understanding of its potential role in tumorigenesis in the future.
Materials and methods
Plasmids, recombinant proteins, and antibodies
Human Bora cDNA was subcloned from plasmid BC056876 (Open Biosystems) into pCS2-FA and pCS2-eGFP-FA. Point mutations were generated using the QuickChange Site-Directed Mutagenesis kit (Stratagene). MBP-tagged full-length Bora and His-tagged Bora 1–312 aa and 355–559 aa were expressed in Escherichia coli and purified. His-tagged Bora 1–312 aa and 355–559 aa were used for antiserum production and for antibody purification. Active recombinant Plx1 was expressed in Sf9 cells for 44 h and then treated with 250 nM okadaic acid for 4 h before harvesting (Kumagai and Dunphy, 1996). The following antibodies were obtained from commercial sources: anti-phosphohistone H3 antibody (Cell Signaling Technology); mouse anti–β-tubulin (Sigma-Aldrich); and anti–cyclin B1, anti-Plk1, anti-Aurora A, anti-HA, anti-myc, and anti-p38MAPK (Santa Cruz Biotechnology, Inc.).
Cell culture and transfection
HeLa and HeLa S3 cells were cultured in DME containing 10% FBS (Invitrogen) and antibiotics. Cells were synchronized at the G1/S boundary by a double thymidine treatment (TT0; 18-h thymidine arrest and 9-h release followed by 18-h thymidine arrest; Fang et al., 1998b). Alternatively, cells were synchronized at prometaphase by a thymidine-nocodazole arrest (TN0; 18-h thymidine arrest and 5-h release followed by 14-h nocodazole arrest; Fang et al., 1998a,b).
siRNAs were synthesized by Dharmacon, Inc. and transfected using DharmaFect 1 (Dharmacon, Inc.). siRNAs targeting Bora were 5′-CTATGAGACTTCAGATGTA-3′, 5′-TAACTAGTCCTTCGCCTAT-3′, and 5′-TATCGGACCTCAGTGATTC-3′. siRNA against Plk1 (Hansen et al., 2004) and β-TrCP1/2 (Mailand et al., 2006) were 5′-AGATTGTGCCTAAGTCTCT-3′ and 5′-AAGUGGAAUUUGUGGAACAUC-3′, respectively. The control siRNA was 5′-CGTACGCGGAATACTTCGATT-3′. All three siRNAs against Bora efficiently reduced the expression of Bora and gave a similar phenotype, confirming the specificity of knockdown. siRNA targeting Bora has been described previously (Hutterer et al., 2006), but the reported siRNA resulted in a <50% reduction in the Bora protein. A previous study reported that depletion of Bora in U2OS cells led to multipolar spindles (Hutterer et al., 2006). However, knockdown of Bora with reported siRNA or with our three specific siRNAs in HeLa cells and in U2OS cells did not result in multipolar spindles under our transfection conditions. Thus, Bora is not required for spindle organization during mitosis in human cells.
DNA transfection was performed using Effectene (QIAGEN) or Lipofectamine 2000 (Invitrogen). Transfected cells were harvested between 36 and 48 h after transfection and analyzed by immunoprecipitation and Western blotting.
Bora gene resistant to siBora-A (rBora) was constructed by silent mutations in coding sequence. GFP-rBora, GFP-rBora-AA, and control GFP were subcloned into the pBabe vector and subsequently transfected into Phoenix-Ampho packaging cells. The viral supernatants were used to infect HeLa cells and stable clones isolated and characterized after selection for 2 wk with 0.4 μg/ml puromycin.
Proteomics
Tandem purification of the Plk1 and Bora complexes.
We established HeLa S3 cell lines stably expressing the GFP- and S-tagged Plk1 or Bora at levels comparable to their respective endogenous proteins. Tagged Plk1 and Bora complexes were purified through two affinity steps (Cheeseman et al., 2004). Purified complexes were eluted in urea and proteolytically digested with trypsin as previously reported (Washburn et al., 2001).
Liquid chromatography tandem mass spectrometry.
A 100-μm i.d. capillary with a 5-μm pulled tip was packed with 10 cm of 5-μm Aqua C18 material (Phenomenex). The desalting column was then equilibrated for 30 min with Buffer A (5% acetonitrile and 0.1% formic acid) and the protein digest was pressure loaded onto it. The column was placed inline with an 1100 quaternary HPLC (Agilent) and analyzed by using a one-step separation which consisted of a 120-min gradient from 0% to 100% Buffer B (80% acetonitrile and 0.1% formic acid). As peptides eluted from the microcapillary column, they were electrosprayed directly onto an LTQ mass spectrometer (Thermo Fisher Scientific) with the application of a distal 2.4-kV spray voltage. A cycle of one full-scan mass spectrum (400–1,400 m/z) was followed by three data-dependent tandem MS spectra at 35% normalized collision energy. Application of MS scan functions and HPLC solvent gradients were controlled by the XCalibur datasystem (Thermo Fisher Scientific).
Analysis of tandem MS.
Tandem MS spectra were analyzed by using the following software analysis protocol. Poor-quality spectra were removed from the dataset by using an automated spectral quality assessment algorithm (Bern et al., 2004). Tandem MS spectra remaining after filtering were searched with the SEQUEST algorithm (Eng et al., 1994) against a Human Database concatenated to a decoy database in which the sequence for each entry in the original database was reversed (Peng et al., 2003). No enzyme specificity was considered for any search. SEQUEST results were assembled and filtered by using the DTASelect program (version 2.0; Tabb et al., 2002). DTASelect 2.0 uses a linear discriminant analysis to dynamically set XCorr and DeltaCN thresholds for the entire dataset to achieve a user-specified false-positive rate (5% in this analysis). The false-positive rates are estimated by the program from the number and quality of spectral matches to the decoy database.
In vitro ubiquitination and binding assays
HeLa S3 cells were synchronized at prometaphase by a thymidine-nocodazole treatment (TN0), collected, and then rinsed twice in ice-cold phosphate-buffered saline and once in hypotonic lysis buffer (20 mM Hepes-KOH, pH 7.6, 5 mM KCl, 1.5 mM MgCl2, 1 mM DTT, a protease inhibitor cocktail [Complete; Roche], 0.5 μM microcystin, and an energy mix). The cell pellet was then resuspended in 1 vol hypotonic lysis buffer, swollen on ice for 30 min, and then lysed by freezing in liquid nitrogen and thawing at 37°C. Cell lysates were passed through a 21-gauge needle 20 times and centrifuged at the maximum speed in a microfuge at 4°C. The clarified supernatant was aliquoted, frozen in liquid nitrogen, and stored at −80°C (Stewart and Fang, 2005).
For ubiquitination assays, extracts were supplemented with 1.25 mg/ml ubiquitin, 2 μM ubiquitin-aldehyde, 10 μM MG132, and an energy mix (Stewart and Fang, 2005). 9 μl of the extracts were then added to 1 μl 35S-labeled substrates synthesized by in vitro translation (Promega). Reactions were proceeded at 30°C for the indicated times and the extent of ubiquitination was analyzed by SDS-PAGE.
HA-β-TrCP1 and Bora and its mutants were synthesized by in vitro transcription and translation (TNT) systems in reticulocyte lysates (Promega). 5 μl of the in vitro–translated 35S-Bora was incubated with 10 μl TN0 extracts for 20 min and then with nonlabeled HA-β-TrCP1 that had been immunoprecipitated by the anti-HA antibody/protein A beads. The immunoprecipitated protein beads were washed five times in the XB buffer (10 mM Hepes-KOH, pH 7.8, 100 mM KCl, 1 mM MgCl2, 0.1 mM CaCl2, and 50 mM sucrose), containing 500 mM KCl and 0.5% NP-40, and two times in the XB buffer.
Immunoprecipitation and immunofluorescence
Antibodies against Bora, Plk1, or GFP were coupled to Affi-Prep Protein A beads (Bio-Rad Laboratories) at a concentration of 0.3 mg/ml. HeLa or HeLa S3 cell pellets were lysed in the NP-40 lysis buffer (50 mM Tris-HCl, pH 8.0, 140 mM NaCl, 1% NP-40, 10% glycerol, 0.1 mM EDTA, 1 mM DTT, 0.5 μM microcystin, and 10 μg/ml each of leupeptin, pepstatin, and chymostatin). Lysates were centrifuged and incubated with Protein A beads coupled with preimmune rabbit IgG at 4°C for 1 h. The precleared lysates were then incubated with anti-Bora, Plk1, or GFP antibodies/Protein A beads at 4°C overnight. Beads were recovered by centrifugation, washed five times with the lysis buffer in the presence of 500 mM NaCl and twice with the lysis buffer, analyzed by SDS-PAGE, and immunoblotted with appropriate antibodies.
HeLa cells were cultured on poly-Lysine–coated glass coverslips overnight. The coverslips were fixed with methanol at −20°C or with 4% paraformaldehyde in PBS at the room temperature. Alternatively, cells were extracted with PHEMT (60 mM Pipes, 25 mM Hepes, pH 6.9, 10 mM EGTA, 4 mM MgCl2, and 0.5% Triton X-100) and then fixed with 1.6% PFA. After fixation, cells were permeabilized and blocked with PBS-BT (PBS, 0.1% Triton X-100, and 3% BSA) at the room temperature for 30 min. Coverslips were subsequently incubated in primary and secondary antibodies diluted in PBS-BT. Images were acquired with Openlab 5.2 (Improvision) under a microscope (Axiovert 200M; Carl Zeiss, Inc.) using a 100× oil immersion lens. Deconvolved images were obtained using AutoDeblur and AutoVisualizer (versions 9.1; AutoQuant Imaging).
For time-lapse microscopy, HeLa/GFP-H2B cells or HeLa cells stably expressing rBora were cultured in Leibovitz's L-15 medium (Invitrogen) supplemented with 10% FBS (Invitrogen) and 2 mM l-glutamine (Invitrogen). Cells were placed in a 37°C heated microscope chamber and observed on a microscope (Axiovert 200M) with a 20× lens. Images were acquired in the GFP channel every 3 min for 5 h with Openlab 5.2 software (Improvision). For experiments in Fig. 6 (D–F), HeLa cells stably expressing rBora were transiently transfected with GFP-H2B and imaged 40 h after transfection.
Acknowledgments
We are grateful to members of the Fang laboratory for discussions.
This work was supported by a Burroughs-Wellcome Career Award in Biomedical Research (G. Fang) and grants from National Institutes of Health (GM062852 to G. Fang and HL079442 and RR11823-10 to J.R. Yates).
Abbreviations used in this paper: PBD, polo-box domain; Plk, polo-like kinase.
References
- Ang, X.L., and J. Wade Harper. 2005. SCF-mediated protein degradation and cell cycle control. Oncogene. 24:2860–2870. [DOI] [PubMed] [Google Scholar]
- Barr, A.R., and F. Gergely. 2007. Aurora-A: the maker and breaker of spindle poles. J. Cell Sci. 120:2987–2996. [DOI] [PubMed] [Google Scholar]
- Barr, F.A., H.H. Sillje, and E.A. Nigg. 2004. Polo-like kinases and the orchestration of cell division. Nat. Rev. Mol. Cell Biol. 5:429–440. [DOI] [PubMed] [Google Scholar]
- Bennett, L.N., and P.R. Clarke. 2006. Regulation of Claspin degradation by the ubiquitin-proteosome pathway during the cell cycle and in response to ATR-dependent checkpoint activation. FEBS Lett. 580:4176–4181. [DOI] [PubMed] [Google Scholar]
- Bern, M., D. Goldberg, W.H. McDonald, and J.R. Yates III. 2004. Automatic quality assessment of peptide tandem mass spectra. Bioinformatics. 20:I49–I54. [DOI] [PubMed] [Google Scholar]
- Cardozo, T., and M. Pagano. 2004. The SCF ubiquitin ligase: insights into a molecular machine. Nat. Rev. Mol. Cell Biol. 5:739–751. [DOI] [PubMed] [Google Scholar]
- Chan, G.K., S.A. Jablonski, V. Sudakin, J.C. Hittle, and T.J. Yen. 1999. Human BUBR1 is a mitotic checkpoint kinase that monitors CENP-E functions at kinetochores and binds the cyclosome/APC. J. Cell Biol. 146:941–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman, I.M., S. Niessen, S. Anderson, F. Hyndman, J.R. Yates III, K. Oegema, and A. Desai. 2004. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 18:2255–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lichtenberg, U., L.J. Jensen, S. Brunak, and P. Bork. 2005. Dynamic complex formation during the yeast cell cycle. Science. 307:724–727. [DOI] [PubMed] [Google Scholar]
- De Luca, M., P. Lavia, and G. Guarguaglini. 2006. A functional interplay between Aurora-A, Plk1 and TPX2 at spindle poles: Plk1 controls centrosomal localization of Aurora-A and TPX2 spindle association. Cell Cycle. 5:296–303. [DOI] [PubMed] [Google Scholar]
- de Pennart, H., E. Houliston, and B. Maro. 1988. Post-translational modifications of tubulin and the dynamics of microtubules in mouse oocytes and zygotes. Biol. Cell. 64:375–378. [DOI] [PubMed] [Google Scholar]
- Eng, J.K., A.L. McCormack, and J.R. Yates III. 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5:976–989. [DOI] [PubMed] [Google Scholar]
- Fang, G., H. Yu, and M.W. Kirschner. 1998. a. The checkpoint protein MAD2 and the mitotic regulator CDC20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes Dev. 12:1871–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, G., H. Yu, and M.W. Kirschner. 1998. b. Direct binding of CDC20 protein family members activates the anaphase-promoting complex in mitosis and G1. Mol. Cell. 2:163–171. [DOI] [PubMed] [Google Scholar]
- Ganem, N.J., K. Upton, and D.A. Compton. 2005. Efficient mitosis in human cells lacking poleward microtubule flux. Curr. Biol. 15:1827–1832. [DOI] [PubMed] [Google Scholar]
- Hansen, D.V., A.V. Loktev, K.H. Ban, and P.K. Jackson. 2004. Plk1 regulates activation of the anaphase promoting complex by phosphorylating and triggering SCFbetaTrCP-dependent destruction of the APC Inhibitor Emi1. Mol. Biol. Cell. 15:5623–5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota, T., N. Kunitoku, T. Sasayama, T. Marumoto, D. Zhang, M. Nitta, K. Hatakeyama, and H. Saya. 2003. Aurora-A and an interacting activator, the LIM protein Ajuba, are required for mitotic commitment in human cells. Cell. 114:585–598. [DOI] [PubMed] [Google Scholar]
- Hutterer, A., D. Berdnik, F. Wirtz-Peitz, M. Zigman, A. Schleiffer, and J.A. Knoblich. 2006. Mitotic activation of the kinase Aurora-A requires its binding partner Bora. Dev. Cell. 11:147–157. [DOI] [PubMed] [Google Scholar]
- Jin, J., T. Shirogane, L. Xu, G. Nalepa, J. Qin, S.J. Elledge, and J.W. Harper. 2003. SCFbeta-TRCP links Chk1 signaling to degradation of the Cdc25A protein phosphatase. Genes Dev. 17:3062–3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanemori, Y., K. Uto, and N. Sagata. 2005. Beta-TrCP recognizes a previously undescribed nonphosphorylated destruction motif in Cdc25A and Cdc25B phosphatases. Proc. Natl. Acad. Sci. USA. 102:6279–6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai, A., and W.G. Dunphy. 1996. Purification and molecular cloning of Plx1, a Cdc25-regulatory kinase from Xenopus egg extracts. Science. 273:1377–1380. [DOI] [PubMed] [Google Scholar]
- Lenart, P., M. Petronczki, M. Steegmaier, B. Di Fiore, J.J. Lipp, M. Hoffmann, W.J. Rettig, N. Kraut, and J.M. Peters. 2007. The small-molecule inhibitor BI 2536 reveals novel insights into mitotic roles of polo-like kinase 1. Curr. Biol. 17:304–315. [DOI] [PubMed] [Google Scholar]
- Maiato, H., J. DeLuca, E.D. Salmon, and W.C. Earnshaw. 2004. The dynamic kinetochore-microtubule interface. J. Cell Sci. 117:5461–5477. [DOI] [PubMed] [Google Scholar]
- Mailand, N., S. Bekker-Jensen, J. Bartek, and J. Lukas. 2006. Destruction of Claspin by SCFbetaTrCP restrains Chk1 activation and facilitates recovery from genotoxic stress. Mol. Cell. 23:307–318. [DOI] [PubMed] [Google Scholar]
- Mamely, I., M.A. van Vugt, V.A. Smits, J.I. Semple, B. Lemmens, A. Perrakis, R.H. Medema, and R. Freire. 2006. Polo-like kinase-1 controls proteasome-dependent degradation of Claspin during checkpoint recovery. Curr. Biol. 16:1950–1955. [DOI] [PubMed] [Google Scholar]
- Moshe, Y., J. Boulaire, M. Pagano, and A. Hershko. 2004. Role of Polo-like kinase in the degradation of early mitotic inhibitor 1, a regulator of the anaphase promoting complex/cyclosome. Proc. Natl. Acad. Sci. USA. 101:7937–7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray, A.W. 2004. Recycling the cell cycle: cyclins revisited. Cell. 116:221–234. [DOI] [PubMed] [Google Scholar]
- Neef, R., U. Gruneberg, R. Kopajtich, X. Li, E.A. Nigg, H. Sillje, and F.A. Barr. 2007. Choice of Plk1 docking partners during mitosis and cytokinesis is controlled by the activation state of Cdk1. Nat. Cell Biol. 9:436–444. [DOI] [PubMed] [Google Scholar]
- Peng, J., J.E. Elias, C.C. Thoreen, L.J. Licklider, and S.P. Gygi. 2003. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J. Proteome Res. 2:43–50. [DOI] [PubMed] [Google Scholar]
- Peschiaroli, A., N.V. Dorrello, D. Guardavaccaro, M. Venere, T. Halazonetis, N.E. Sherman, and M. Pagano. 2006. SCFbetaTrCP-mediated degradation of Claspin regulates recovery from the DNA replication checkpoint response. Mol. Cell. 23:319–329. [DOI] [PubMed] [Google Scholar]
- Petroski, M.D., and R.J. Deshaies. 2005. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6:9–20. [DOI] [PubMed] [Google Scholar]
- Piperno, G., M. LeDizet, and X.J. Chang. 1987. Microtubules containing acetylated α-tubulin in mammalian cells in culture. J. Cell Biol. 104:289–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenblum, E., P. Vahteristo, T. Sandberg, J.T. Bergthorsson, K. Syrjakoski, D. Weaver, K. Haraldsson, H.K. Johannsdottir, P. Vehmanen, S. Nigam, et al. 2002. A genomic map of a 6-Mb region at 13q21-q22 implicated in cancer development: identification and characterization of candidate genes. Hum. Genet. 110:111–121. [DOI] [PubMed] [Google Scholar]
- Seki, A., and G. Fang. 2007. CKAP2 is a spindle-associated protein degraded by APC/C-CDH1 during mitotic exit. J. Biol. Chem. 282:15103–15113. [DOI] [PubMed] [Google Scholar]
- Skoufias, D.A., P.R. Andreassen, F.B. Lacroix, L. Wilson, and R.L. Margolis. 2001. Mammalian mad2 and bub1/bubR1 recognize distinct spindle-attachment and kinetochore-tension checkpoints. Proc. Natl. Acad. Sci. USA. 98:4492–4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, S., and G. Fang. 2005. Anaphase-promoting complex/cyclosome controls the stability of TPX2 during mitotic exit. Mol. Cell. Biol. 25:10516–10527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumara, I., J.F. Gimenez-Abian, D. Gerlich, T. Hirota, C. Kraft, C. de la Torre, J. Ellenberg, and J.M. Peters. 2004. Roles of polo-like kinase 1 in the assembly of functional mitotic spindles. Curr. Biol. 14:1712–1722. [DOI] [PubMed] [Google Scholar]
- Tabb, D.L., W.H. McDonald, and J.R. Yates III. 2002. DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 1:21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vugt, M.A., B.C. van de Weerdt, G. Vader, H. Janssen, J. Calafat, R. Klompmaker, R.M. Wolthuis, and R.H. Medema. 2004. Polo-like kinase-1 is required for bipolar spindle formation but is dispensable for anaphase promoting complex/Cdc20 activation and initiation of cytokinesis. J. Biol. Chem. 279:36841–36854. [DOI] [PubMed] [Google Scholar]
- Washburn, M.P., D. Wolters, and J.R. Yates III. 2001. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19:242–247. [DOI] [PubMed] [Google Scholar]
- Watanabe, N., H. Arai, Y. Nishihara, M. Taniguchi, T. Hunter, and H. Osada. 2004. M-phase kinases induce phospho-dependent ubiquitination of somatic Wee1 by SCFbeta-TrCP. Proc. Natl. Acad. Sci. USA. 101:4419–4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, N., H. Arai, J. Iwasaki, M. Shiina, K. Ogata, T. Hunter, and H. Osada. 2005. Cyclin-dependent kinase (CDK) phosphorylation destabilizes somatic Wee1 via multiple pathways. Proc. Natl. Acad. Sci. USA. 102:11663–11668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield, M.L., G. Sherlock, A.J. Saldanha, J.I. Murray, C.A. Ball, K.E. Alexander, J.C. Matese, C.M. Perou, M.M. Hurt, P.O. Brown, and D. Botstein. 2002. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol. Biol. Cell. 13:1977–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, J., and G. Fang. 2006. HURP controls spindle dynamics to promote proper interkinetochore tension and efficient kinetochore capture. J. Cell Biol. 173:879–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo, H.Y., A. Kumagai, A. Shevchenko, and W.G. Dunphy. 2004. Adaptation of a DNA replication checkpoint response depends upon inactivation of Claspin by the Polo-like kinase. Cell. 117:575–588. [DOI] [PubMed] [Google Scholar]
- Zhao, W.M., and G. Fang. 2005. MgcRacGAP controls the assembly of the contractile ring and the initiation of cytokinesis. Proc. Natl. Acad. Sci. USA. 102:13158–13163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, W.M., A. Seki, and G. Fang. 2006. Cep55, a microtubule-bundling protein, associates with centralspindlin to control the midbody integrity and cell abscission during cytokinesis. Mol. Biol. Cell. 17:3881–3896. [DOI] [PMC free article] [PubMed] [Google Scholar]