Abstract
Fructose-1,6-(bis)phosphate aldolase is a ubiquitous enzyme that catalyzes the reversible aldol cleavage of fructose-1,6-(bis)phosphate and fructose 1-phosphate to dihydroxyacetone phosphate and either glyceral-dehyde-3-phosphate or glyceraldehyde, respectively. Vertebrate aldolases exist as three isozymes with different tissue distributions and kinetics: aldolase A (muscle and red blood cell), aldolase B (liver, kidney, and small intestine), and aldolase C (brain and neuronal tissue). The structures of human aldolases A and B are known and herein we report the first structure of the human aldolase C, solved by X-ray crystallography at 3.0 Å resolution. Structural differences between the isozymes were expected to account for isozyme-specific activity. However, the structures of isozymes A, B, and C are the same in their overall fold and active site structure. The subtle changes observed in active site residues Arg42, Lys146, and Arg303 are insufficient to completely account for the tissue-specific isozymic differences. Consequently, the structural analysis has been extended to the isozyme-specific residues (ISRs), those residues conserved among paralogs. A complete analysis of the ISRs in the context of this structure demonstrates that in several cases an amino acid residue that is conserved among aldolase C orthologs prevents an interaction that occurs in paralogs. In addition, the structure confirms the clustering of ISRs into discrete patches on the surface and reveals the existence in aldolase C of a patch of electronegative residues localized near the C terminus. Together, these structural changes highlight the differences required for the tissue and kinetic specificity among aldolase isozymes.
Keywords: isozyme specificity, structural enzymology, protein–protein interactions, isozyme-specific residues, structure/function
Fructose-1,6-(bis)phosphate aldolases are ubiquitous enzymes that catalyze the reversible cleavage of fructose-1,6-(bis)phosphate (Fru-1,6-P2) and fructose 1-phosphate (Fru-1-P) to dihydroxy-acetone phosphate (DHAP) and either glyceraldehyde-3-phosphate (G3P) or glyceraldehyde, respectively. The mechanisms of these aldolases occur by two distinct chemical paths (Rutter 1964). Class I aldolases of animals and higher plants use covalent catalysis through a Schiff-base intermediate with ketose sugar substrates. Class II aldolases of most bacteria and fungi require a divalent metal cation as a cofactor. Among the class I enzymes found in mammals, there are three tissue-specific isozymes of aldolase that have similar molecular masses and catalytic mechanisms: aldolase A (expressed primarily in muscle and red blood cells), aldolase B (expressed primarily in liver, kidney, and small intestine), and aldolase C (expressed primarily in brain, smooth muscle, and neuronal tissue) (Lebherz and Rutter 1969).
The aldolase isozymes are similar in sequence (Rottmann et al. 1987) with 66% identity between human A and B, 68% identity between B and C, and 78% identity between A and C. All isozymes have strictly conserved residues in the active site consisting of Asp33, Arg42, Lys107, Lys146, Glu187, Ser271, Arg303, and Lys229 (which forms the Schiff-base intermediate) (Choi et al. 2001). They are immunochemically distinct and have different catalytic properties (Penhoet et al. 1969), such that the ratio of the specificity constants (kcat/Km) for the two hexose substrates of aldolase C is between those of A and B. Aldolases A and B have well-defined roles in glycolysis and gluconeogenesis, but the physiological role of aldolase C remains elusive.
Aldolase C is found in actively proliferating and cancer cells (Schapira et al. 1970; Skala et al. 1987) and has been identified as zebrin in fetal rodent brain (Ahn et al. 1994). Its gene expression is enhanced by nerve-growth-factor-induced B factor (NGF1-B), which is a transcription factor of the thyroid/steroid/retinoid nuclear receptor gene family. NGF1-B expression in rat brain embryos (Buono et al. 1997) parallels that of rat aldolase C (Makeh et al. 1994). Together, these results suggest a neurodevelopmental role for aldolase C (Ahn et al. 1994), but biochemical data are unable to identify its physiological role. Thus, its structure may offer insight into its role in cell growth, vertebrate development, and/or other metabolic processes.
X-ray crystallographic structures of rabbit muscle aldolase A (Sygusch et al. 1987), human muscle aldolase A (Gamblin et al. 1991), and human liver aldolase B (Dalby et al. 2001) demonstrate that these vertebrate enzymes are structurally similar. All possess a homotetrameric αβ-barrel structure with the active site of each monomer located at the center of the αβ-barrel. It should be noted that the last 20 amino acids of the C terminus is visualized in only a few of the published aldolase structures and is observed in different conformations even within an isozyme (Blom and Sygusch 1997; Dalby et al. 2001). One of these conformations shows the C terminus forming an arm-like structure attached by a highly flexible hinge that wraps around the surface sometimes contacting the active site (Gamblin et al. 1991; Blom and Sygusch 1997). While the ablation of the C terminus produces an essentially inactive enzyme (Rose et al. 1965; Berthiaume et al. 1993), the coupling mechanism between the catalytic cycle and the conformational change of the C terminus remains unclear.
Aldolase not only plays a key role in glycolysis, but also binds to macromolecules unrelated to glycolysis such as F-actin (Wang et al. 1996), vacuolar H+-ATPase (Lu et al. 2001), GLUT4 (Kao et al. 1999), cell surface adhesins (Jewett and Sibley 2003), S100A1 (Zimmer and Van Eldik 1986), thrombospondin-related anonymous protein (TRAP) (Buscaglia et al. 2003), brain dynein light chain LC8 (Navarro-Lérida et al. 2004), RNA (Kiri and Goldspink 2002), and tubulin (Karkhoff-Schweizer and Knull 1987). Thus, aldolase may be used for other functions in the cell, a concept that has been described as “moonlighting” (Jeffery 1999; Copley 2003). These interactions may be specific to each isozyme. For example, studies have shown that aldolases associate with target proteins in a tissue-specific manner (Kusakabe et al. 1997). Aldolase B binds to the liver cytoskeleton with higher affinity than to muscle or brain cytoskeleton (Kusakabe et al. 1997). Aldolase C binds less tightly to the cytoskeleton compared with the other two isozymes and this has been attributed to its more acidic pI (Kusakabe et al. 1997).
In order to understand the cellular properties of aldolase isozymes, it is important to correlate the structure and function of these proteins. The key distinctions among the isozymes likely reside in the minimal set of isozyme-specific residues (ISRs), which are residues strictly conserved among each paralog. From a sequence alignment of 21 vertebrate aldolase isozymes, the ISRs for each isozyme have been identified (11 A ISRs, 12 B ISRs, and 4 C ISRs). When these ISRs are mapped onto their corresponding tertiary structures, they were found to cluster on the surface in two distinct patches, the distal surface patch (DSP) and the terminal surface patch (TSP) (Pezza et al. 2003). At the time of that study, a structure for aldolase C was unknown and herein the ISRs are mapped onto the aldolase C structure.
Since nature has already honed the minimal number of residues that must be retained for isozyme function and overall fold, isozymes are excellent tools for assigning function to structure. As a first step toward understanding the cellular function of aldolase at the molecular level, the structures of the three aldolase isozymes, A, B, and C, are required. Subsequent structural comparisons may expose isozyme-specific differences that confer distinct cellular functions. Herein, we report the cloning, expression, and structure determination of human aldolase C and compare the structure with those of the other human aldolase isozymes.
Results
Overall structure and activity
To determine if the kinetic constants of recombinant human aldolase C are comparable with those of the enzyme isolated from mammalian tissue, we determined the steady-state kinetic constants for the substrates Fru-1,6-P2 and Fru-1-P. The kcat toward Fru-1,6-P2 was 5.2 ± 0.2 sec−1 and the Km was 10.7 ± 0.5 μM. The kcat toward the substrate Fru-1-P was 2.8 ± 0.3 sec−1 and the Km was 16,000 ± 2000 μM. The Km values of cloned aldolase C agree with previously reported values of recombinant human C (Kusakabe et al. 1994). The kcat for Fru-1,6-P2 of aldolase C is intermediate to those of aldolases A and B, at 12.1 sec−1 and 0.67 sec−1, respectively (Penhoet and Rutter 1971), while the Km of aldolase C is closer to that of A rather than B (at 10 μM and 0.8 μM, respectively [Pezza et al. 2003]).
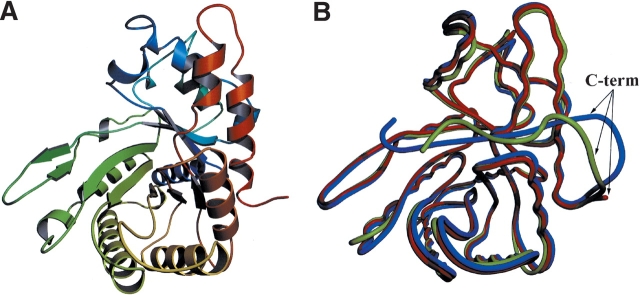
The structure of human aldolase C was solved to 3.0 Å resolution by X-ray crystallography (Fig. 1A ▶). Phases were determined by molecular replacement using the aldolase A-DHAP complex model from rabbit muscle (Choi et al. 2001; PDB accession code 1J4E) and the final structure refined to an R-work = 0.225 and R-free = 0.261. The structure comprises residues 2–343 as the C-terminal 20 amino acids were disordered in this structure and could not be accurately modeled (Fig. 1A ▶). The disorder of the C terminus is consistent with the aldolase A and B structures (Sygusch et al. 1987; Choi et al. 1999, 2001; Dalby et al. 1999, 2001). The tertiary and quaternary structures of human aldolase C are essentially identical to that of the unliganded structures of human muscle aldolase A (Gamblin et al. 1991; PDB accession code 1ALD) and liganded human liver aldolase B (Dalby et al. 2001; PDB accession code 1QO5) with rms deviations of 0.62 Å and 0.71 Å, respectively, between main-chain Cα atoms (Fig. 1B ▶).
Figure 1.
The structure of aldolase C monomer (A) depicted as a ribbon diagram ramped from blue to red from the N terminus to the C terminus and (B) depicted as a coil overlaid (in red) with the structures of aldolase A (blue; PDB accession code 1ALD) and aldolase B (green; PDB accession code 1QO5).
Active site
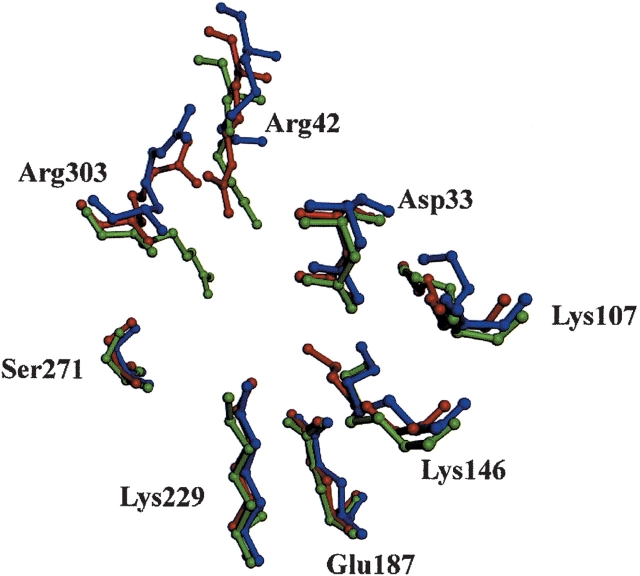
The positions of active-site residues (Asp33, Lys107, Glu187, Ser271, and Lys229) remain essentially unchanged among the unliganded structures of human aldolase A, B, and C (PDB accession codes 1ALD and 1QO5 for aldolases A and B, respectively; Figs. 2 ▶, 3 ▶). (Unless otherwise stated, the aldolase C structure will be compared with the unliganded aldolase A structure 1ALD and the ammonium sulfate bound aldolase B structure. Note that the structure of unliganded aldolase B is unknown.) The residues Arg42, Lys146, and Arg303 exhibit subtle changes that form new interactions or conformations. Of these, Arg42 demonstrates only a minor change in its guanidinium side chain conformation among the isozymes.
Figure 2.
The active site of aldolase C (red) with residues depicted as ball-and-stick, overlaid with the active site residues of aldolase A (blue; PDB accession code 1ALD) and aldolase B (green; PDB accession code 1QO5).
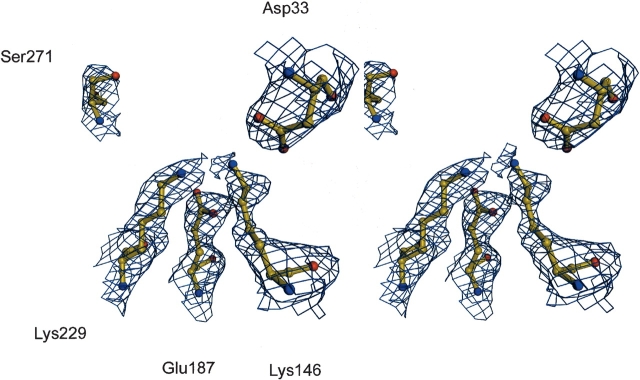
Figure 3.
Stereo view of the active site residues of the aldolase C structure. The residues are shown as ball-and-stick models. The 2Fo-Fc electron density map contoured at 1σ is depicted as blue cages.
Lys146, a residue required for carbon–carbon bond cleavage (Morris and Tolan 1994; Choi et al. 2001), is closer to the catalytic Lys229 with a distance of 3.0 Å compared with the longer distance in aldolase A of 5.3 Å and B of 6.3 Å (distances were measured between the ɛ-amino groups of Lys146 and Lys229). All distances are accurate to within ±0.44 Å, as calculated from a Luzzatti plot. The distances between Lys146 and Lys229 in all liganded structures of aldolase A are >6.0Å. Thus, the smallest distance between Lys229 and Lys146 exists in the unliganded structure of C.
Of the active-site residues, Arg303 shows the largest displacement of the side chain position relative to that in aldolases A and B. The distance between Arg303 and Lys229 in aldolase C is 9.5 Å (measured from guanidine of Arg to ɛ-amino group of Lys), which is significantly different from the equivalent distances in isozymes A and B of 8.4 Å and 10.6 Å, respectively. As previously observed, a comparison of all conformations of Arg303 in liganded and unliganded structures of aldolase A shows that arginine’s flexible side chain can adopt multiple conformations either proximal or distal to the catalytic Lys229 (the distal position is adopted in the presence of bound substrate) (Choi et al. 1999). The aldolase C structure shows that the Arg303 conformation is neither distal to the active site nucleophile as seen in aldolase A (1ALD), nor proximal to the active site nucleophile as seen in B structures, but lies somewhere in between (Fig. 2 ▶). Note that in liganded structures of aldolase A (6ALD, 1J4E, 1QO5), Arg303 takes on the same conformation as seen in B. Thus, the conformation of Arg303 in aldolase C is unique compared with unliganded and liganded aldolase structures.
Isozyme-specific residues
Of the four aldolase C ISRs (Pezza et al. 2003), Arg314, Thr324, and Glu332 are visible in this structure. The other ISR, Gly350, is located in the disordered C-terminal region and therefore its position could not be determined. The aldolase C ISRs are colocalized in the C-terminal region of the protein and Arg314, Thr324, and Glu332 form unique interactions in aldolase C. Arg314 forms hydrogen bonds with Asn311 (measured from guanidine group of Arg to amide carbonyl of Asn, 2.8 Å). This interaction occurs exclusively in aldolase C since residue 314 in A and B is usually replaced by glycine. Residue 311 is interesting since it is an ISR in aldolases A and B (Lys and Ala, respectively). Thus, an interaction occurring in an aldolase C ISR (Arg314–Asn311) cannot occur in the other two isozymes.
The aldolase C ISR, Glu332, makes a salt bridge with Lys71 (group of Glu to ɛ-amino group of Lys 2.7 Å). In human aldolase A, such an interaction is unlikely since residues 332 and 71 are the hydrophobic residues Leu and Pro, respectively, and are located 6.6 Å apart. In human aldolase B, Met332 and Gln71 are too distant to make an interaction (5.3 Å). Thus, the Glu332–Lys71 salt bridge is another example of an interaction that occurs only in the aldolase C isozyme. Finally, the side chain hydroxyl of ISR Thr324 forms hydrogen bonds with the carbonyl oxygens of Val61 and Ala320.
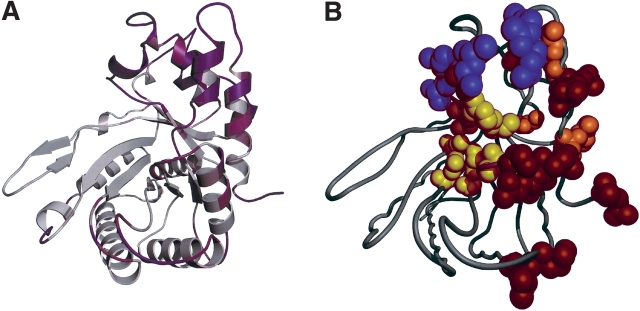
Mobile areas of the aldolase C surface were found by temperature factor analysis (Fig. 4A ▶), and the locations of the regions with elevated B-factors were compared with those of the surface ISR patches (Fig. 4B ▶). The average temperature factor for the aldolase C structure is 35.7 Å2, while that for the ISRs in aldolase C is 49.5 Å2. Mapping of the temperature factors on the aldolase C structure shows that although there are some other areas of the structure with relatively high temperature factors, they comprise solely loops on the outside of the protein, whereas the TSP and DSP comprise mostly helices (Fig. 4B ▶).
Figure 4.
Diagram of the aldolase C monomer (A) depicted as a ribbon diagram with the temperature factors mapped onto the structure ramped in color from low B-factor (white) to high B-factor (magenta) and (B) depicted as a coil with ISR (TSP in purple, DSP in orange), negative patch (red; see Fig. 5 ▶), and active site (yellow) residues shown as space-filling models. Orientations of A and B are as in Figure 1 ▶.
Electrostatic potential surface
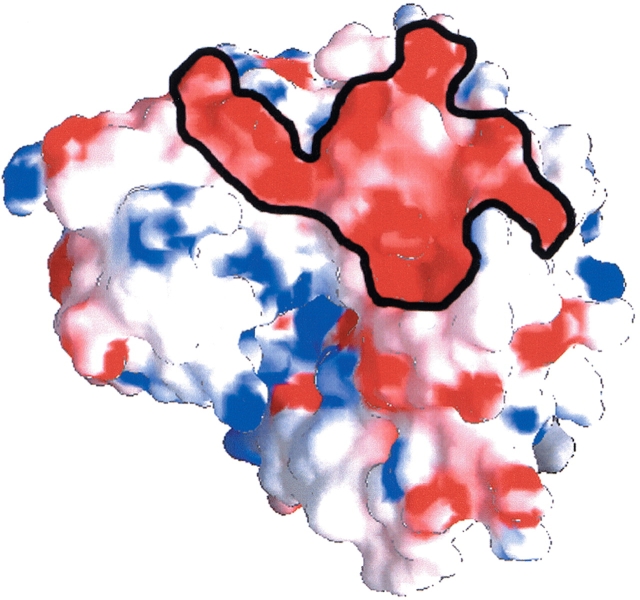
Since the calculated isoelectric points (pIs) of the isozymes differ significantly, the electrostatic potential surface was calculated to analyze the charge distribution. pIs for human aldolases A, B, and C were calculated using the pI determination program of the Biology Workbench (http://workbench.sdsc.edu) (Subramaniam 1998). Aldolase C has a calculated pI of 6.4, lower than that of aldolases A and B, with pI values of 8.1 and 7.8, respectively. These values are consistent with the electrophoretic mobilities and empirically determined pIs of the aldolase isozymes (Susor et al. 1975; Kusakabe et al. 1997). This difference in the pI is reflected in the dramatically different electrostatic potential surface of the C enzyme (Fig. 5 ▶). Overall, negative charge was distributed across the surface of the protein and in particular there is a cluster of negative charges that comprises a “negative” patch (Fig. 5 ▶).
Figure 5.
The electrostatic surface potential of aldolase C mapped onto the space-filling model with white (neutral), blue (positive, 15kτ), and red (negative, −15kτ). The negative surface patch outlined by a black line (Pro230, Pro240, Thr244, Glu246, Glu247, Gln 274, Glu275, Glu277, Glu278, Glu325, Glu326, Arg330, Tyr342, Glu343, and Gly344) includes negatively charged residues but also encompasses an area containing residues of other charge. The electrostatic potential surface of aldolase C was calculated and rendered using the program GRASP (Nicholls et al. 1991).
The residues that define the negative patch include glutamates 246, 247, 275, 277, 278, 325, 326, and 343. Although ISRs are localized on the surface of the enzyme (Pezza et al. 2003), none are located in this “negative” patch (Fig. 4B ▶). This patch is located adjacent to the TSP, which comprises ISRs in the vicinity of the acidic C-terminal region (Kusakabe et al. 1997). (The calculated pI of the C-terminal region containing residues 344–363 is 4.1.) It should be noted that the negative patch does not coincide with the intersubunit interfaces of the aldolase tetramer.
Discussion
Overall structure and activity
Overall, the protein architecture of the three human aldolase isozymes, A, B, and C, is retained. The constellation of enzyme active site residues is fully preserved in aldolase C, highlighting the fact that (1) there is little plasticity in the ability of alternate arrangements of amino acids to carry out the aldolase reaction effectively, and (2) kinetic differences among isozymes of aldolase are not affected solely at the level of the active site but are the result of subtle changes in protein dynamics or electrostatics distant from the active site. We hypothesize that small changes in the active site are insufficient to account for the tissue-specific isozymic differences.
Active site and the role of the C terminus
The positions of active-site residues remain mostly unchanged among the unliganded structures of human aldolase A, B, and C (PDB accession codes 1ALD and 1QO5 for A and B, respectively) (Figs. 2 ▶, 3 ▶). Those residues exhibiting differences include Lys146, Arg42, and Arg303, which show subtle changes in conformation (vide supra). These residues have mechanistic or substrate binding roles and thus changes in these positions may alter the kinetic properties of the isozymes. For example, two of these residues, Arg303 and Arg42, are involved in one of the two alternate C6-binding sites (Choi et al. 1999). The third residue, Lys146, is in a position to perturb the pKa values of important residues in catalysis, such as the Schiffbase Lys229 or general acid/base catalyst Asp33 toward neutrality (Allen 1998; Choi et al. 2001), and the results herein further support this hypothesis. That is, the subtle changes in position of Lys146 may alter the reactivity of these essential residues.
The active site residue that exhibits the greatest flexibility is Arg303, as seen in several previous structures (Choi et al. 2001). In unliganded aldolase A structures, the side chain of this moiety is proximal to the active site Lys229, while in liganded structures, it is distal to this nucleophile. Unexpectedly, in aldolase C, Arg303 adopts a conformation that is neither proximal nor distal to the catalytic lysine, but lies somewhere between the conformations observed in liganded and unliganded structures of other aldolases. It is tempting to conclude that the orientation of Arg303 offers a simple correlation with isozyme kinetic differences. Additional structural information, specifically a structure of aldolase B crystallized in the absence of ammonium sulfate, is needed to paint a complete picture of Arg303’s conformations in the absence of bound ligand.
The inherent flexibility of the Arg303 side chain observed herein is consistent with its proposed role as a “trigger” residue. The C terminus has been invoked in models coupling the opening and closing of this portion of the protein over the active site with substrate binding and product release. This role was first suggested by the structure of aldolase A observed at the end of the catalytic cycle (Blom and Sygusch 1997). In that structure, Arg303 forms a salt bridge with Glu354 of the C terminus, resulting in an ordered structure of the C terminus and an active site that is partially occluded. Whereas when aldolase A was trapped in the middle or the beginning of the catalytic cycle, Arg303 forms a salt bridge with a substrate phosphate (Choi et al. 1999, 2001) and thus is unavailable for binding to the C terminus. It may be significant that position 354 is strictly conserved as Glu in aldolase A, whereas it is a Gln in aldolases B and C, defining it as an aldolase A ISR.
A comparison of the local structure near the C terminus of aldolase C with that of A and B uncovers the fact that a salt bridge between the barrel portion of the protein and the C terminus, which occurs in aldolase C (between Glu332 and Lys71), is prevented by the replacement of 332 with Leu in aldolase A and Met in aldolase B. This is one among many examples cited herein showing that an amino acid that is conserved among orthologs prevents an interaction that occurs in paralogs. The importance of this and other interactions to kinetics and protein binding will be tested in the future via site-directed mutagenesis.
Isozyme-specific residues
Previously, ISRs have been shown to be concentrated on the surface of the aldolase tetramer (Pezza et al. 2003). The ISRs form patches that do not coincide with the enzyme oligomeric interfaces or crystal contacts. Three of the four aldolase C ISRs are visualized in this structure and have been shown to form unique interactions absent in aldolases A and B. Certain regions of the protein surface may be responsible for differential isozymic activities by providing alternate binding surfaces for docking of the C terminus of aldolase itself or of different protein partners. Alternatively, if a residue is in close proximity to an ISR, then it may be affecting kinetics from a distance and allow communication between the active site and the surface or another location in the protein. One reason for the surface localization of so many ISRs may be their role in cellular functions aside from, or related to, their direct role in catalysis. Previous work has demonstrated the participation of specific residues as structural determinants of ligand specificity and catalytic properties. In the case of NO synthase, the structural basis for isozyme specificity is embedded in sequence differences outside the active site as well as conformational flexibility in the active site. This flexibility is also determined by ISRs outside the active site (Fedorov et al. 2004). In the case of enolase, the sequences that differ between the three enolase isozymes were mapped onto the three-dimensional structure of the enzyme. Three areas with relatively high densities of isozyme-specific substitutions were found and proposed to be likely sites of contact with other macromolecules (Lebioda and Stec 1991).
A correlation between elevated temperature factors derived from X-ray crystal structures and sites of protein–protein interactions has been observed in studies of proteases and their inhibitors (Hubbard et al. 1991). Examination of the temperature factors from aldolase structures shows that the most disordered or mobile regions are the same among all aldolase isozymes and colocalize in the area of the ISRs (Pezza et al. 2003). This includes the essential C terminus of the protein, which is highly disordered in most previously solved aldolase structures (and now aldolase C). Aldolase C completes the picture, showing the correlation between areas of flexibility in the isozymes and the ISR patches, which we hypothesize are sites of protein–protein interactions.
The electrostatic surface of aldolase C is much more negatively charged than that of aldolases A or B. We have noted a region of high negative charge close to the TSP which may act (1) along with the TSP as a binding site for protein partners that prefer an acidic binding sequence or (2) as a unique (docking) site for one of the conformations of the mobile C terminus. Complete understanding of the differential cellular activities of aldolase isozymes awaits the determination of the substrate complexes of aldolases B and C and the elucidation of the regulation and structure of aldolase complexes with target proteins.
Materials and methods
Materials
Restriction endonucleases were purchased from New England Biolabs. CM-Sepharose Fast Flow was from Amersham/Pharmacia Inc. All other materials were reagent grade or better.
Expression plasmid construction
The aldolase C open reading frame was amplified from the aldolase C expressed sequence tag (EST) (accession no. F07971) using oligodeoxyribonucleotides 5′-CCC CAT ATG CCC CAC TCA TAC CCA GCC CTT 3′ (forward) and 5′-GGG AAG CTT AGTAGG CAT GGT TGG CAA TGT AG 3′ (reverse). The product was digested with restriction endonucleases Nde I and Hind III and ligated into pET21a (Novagen). The complete open reading frame was amplified again using oligodeoxyribonucleotides NCO1: 5′-ACGTACGTCCATGGCACCTCACTCGTACCCAGCCCTT-3′ (forward) and PET.RC: 5′-TCAGCGGTGGCAGCAGCCAAC-3′ (reverse) and subsequently cloned into the high-copy expression plasmid pPB1 (Beernink and Tolan 1992) using restriction endonucleases Nco I and Hind III (the boldface nucleotides in oligodeoxyribonucleotide NCO1 refer to the engineered Nco I site). DNA sequence analysis revealed three mutations, encoding substitutions L209X, S275N, and I358V, which were from the EST and/or introduced during subsequent amplifications. These were repaired via PCR-assisted site-directed mutagenesis (Ho et al. 1989). The following oligodeoxyribonucleotides were used to repair L209X: 1150U: 5′-AGAAGGTCTTGGCTGCAGTGTACA AGGCCCTGAGTG-3′ (forward) and 1150L: 5′GCCTTGTACACTGCAGCCAAGACCTTCTC-3′ (reverse). The following primers were used to repair S275N: 950U: 5′-TGTCTGGCGGCCAGAGCGAAGAAGAGGCATCATTC-3′ (forward) and 950L: 5′-TTCTTCGCTCTGGCCGCCAGACAGGAAGG-3′ (reverse). The following flanking primers were used to generate the full-length product and to repair I358V: geneU: 5′-AACGACCGAGC GCAGCGAGTCAGTGAGC-3′ (forward) and HINDL: 5′-GAC CCCAAGCTTAGTAGGCATGGTTGGCAATGTAGAGTGAC-3′ (reverse). Underlined nucleotides refer to the codon that was changed. The boldface nucleotides in oligodeoxyribonucleotides 1150U and 1150L were changes that created a Pst I site. The boldface nucleotides in oligodeoxyribonucleotides 950U and 950L were changes that created an Eae I site and those in HINDL created a Hind III site. The final product was confirmed to encode human aldolase C by DNA sequence determination.
Protein expression, purification, characterization, and crystallization
Aldolase C was purified as previously described (Morris and Tolan 1993) using affinity elution from CM-Sepharose Fast Flow with the following modifications. The extract was dialyzed against 14 mM MES, 6 mM glycine, 6 mM KOH, and 1 mM dithiothreitol (pH 6.2). The column was washed with 44 mM MOPS, 11 mM glycine, 11 mM KOH, and 1 mM dithiothreitol (pH 7.0) and elution took place with Fru-1,6-P2 in 25 mM TAPS, 13 mM glycine, 13 mM KOH, and 1 mM dithiothreitol (pH 8.2). Aldolase C was purified to ≥95% purity as determined by SDS-PAGE. Protein was precipitated with 70% saturated ammonium sulfate and stored at 4°C until ready to use. Activity of recombinant aldolase C toward the substrates Fru-1,6-P2 and Fru-1-P was measured as described previously (Morris and Tolan 1993) using a coupled assay (Racker 1952).
Before crystallization, the ammonium sulfate slurry was exchanged with crystallization buffer (1 mM HEPES at pH 7.0) using an UltraFree centrifugal concentrator (Millipore) with a 10-kDa MW cutoff membrane. A final protein concentration of 6–15 mg/mL was used for crystallization by the hanging drop vapor diffusion method. Initial conditions were identified from the Hampton Research crystal screens I and II and were further optimized by screening around the initial conditions and using the Hampton Research additive screen. The final crystallization conditions consisted of equal volumes of protein and reservoir solution (25% PEG 8000, 0.2 M calcium acetate, 0.1 M Tris at pH 8.0, 4% glucose), which were equilibrated against 0.5 mL of reservoir at 18°C. Plate or rod-like crystals appeared within 2 wk. Crystals were dehydrated (Heras et al. 2003; Kuo et al. 2003) via vapor diffusion by equilibrating overnight over a solution of 25% PEG 8K, 0.2 M calcium acetate, 0.1 M Tris (pH 8.0), and 4% glucose and then immersed briefly in reservoir solution containing 20% glycerol as a cryoprotectant. Crystals were immediately frozen directly in a stream of N2 gas cooled by liquid nitrogen.
Data collection
Data were collected at −180°C on a Rigaku RU-300 X-ray generator equipped with Osmic Confocal Max-Flux multilayer optics and a RAXIS-IV++ area detector. A crystal-to-detector distance of 160 mm and 0.5° oscillations were used. The programs DENZO and SCALEPACK (Otwinowski and Minor 1997) were used to index and reduce the data, respectively. The space group is P1 with unit cell dimensions of a = 83.9 Å, b = 122.0 Å, c = 129.5 Å, α = 108.0°, β = 108.6°, γ = 97.4°. A complete data set to 3.0 Å was obtained from a single crystal. Assuming a Matthews coefficient of 2.3 Å3 /Da (Matthews 1968), 12 aldolase monomers (three tetramers) were calculated to be in the asymmetric unit. Crystals of aldolase C did not diffract to significantly higher resolution at synchrotron light sources, indicating that crystal quality limited resolution.
X-ray diffraction and crystal structure analysis
Phase information was obtained by molecular replacement with the program MOLREP (Vagin and Teplyakov 1997) using an initial model created by Swiss Modeller (Peitsch 1996) from the aldolase C sequence and the monomeric structure of rabbit muscle aldolase A (sequence identity of 82.9%; PDB accession code 1J4E). The monomeric model was transformed into a tetramer based on the tetrameric structure of aldolase A. Using a high-resolution cutoff of 6 Å, the solution from Molrep for three tetramers had an R-factor of 55.2%. This solution was pursued since there were reasonable contacts between tetramers with no overlap with the symmetry-related molecules. The initial model was refined in the program CNS (Brünger et al. 1998). After a single round of rigid-body refinement, torsional annealing at 6000K, and minimization, R-free was <0.45. For the calculation of R-free during refinement, 10% of all reflections were excluded. Noncrystallographic symmetry (NCS) matrices were determined between monomers. After including strict NCS parameters in refinement, R-free decreased significantly to 0.36. Each subsequent round of refinement included positional, slow-cooling, and B-group refinement, followed by manual rebuilding using the molecular graphics program O (Jones et al. 1991). The structure was refined to 3.0 Å resolution with a crystallographic R-factor of 0.225 and a free R-factor of 0.261. The structure is well defined with an average B-factor of 35.9 Å2 for main chain and side chain atoms. The Ramachandran plot showed that 88% of the residues were in the “favored” regions defined by PROCHECK (Morris et al. 1992), with the remaining 12% of the residues falling in the “allowed” regions. Final refinement and model statistics are tabulated in Table 1. Ribbon diagrams and ball-and-stick figures were rendered with the programs Molscript (Kraulis 1991) and Povray (Persistance of Vision Ray Tracer, Version 3.1). It should be noted that the numbering of aldolase A, B, and C residues is identical in the structures deposited in the Protein Data Bank.
Table 1.
Data collection and refinement statistics
| Data collection | |
| Resolution (Å) | 100–3.0 |
| No. of reflections | 901,035 |
| No. of unique reflections | 89,550 |
| Completeness (overall/outer shell) (%) | 97.2 (95.5) |
| Rsym (overall/outer shell) | 0.097 (0.34) |
| I/σ(overall/outer shell) | 5.3 (1.5) |
| Refinement statistics | |
| Protein atoms (nonhydrogen) | 2624 |
| Resolution (Å) | 3.0 |
| Rcryst | 0.255 |
| Rfree | 0.261 |
| Mean B-factor (Å2) | 35.9 |
| Rms deviation | |
| Bonds (Å) | 0.007 |
| Angles (deg) | 1.40 |
| Dihedrals (deg) | 21.95 |
| Impropers (deg) | 0.99 |
The coordinates of the refined structure have been deposited in the Protein Data Bank, accession code 1XFB.
Acknowledgments
This work was supported by NIH grant GM60616 (to D.R.T. and K.N.A.).
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.04915904.
References
- Ahn, A.H., Dziennis, S., Hawkes, R., and Herrup, K. 1994. The cloning of zebrin II reveals its identity with aldolase C. Development 120 2081–2090. [DOI] [PubMed] [Google Scholar]
- Allen, K.N. 1998. Reactions of enzyme-derived enamines. In Comprehensive biological catalysis (ed. M. Sinnott), pp. 135–172. Academic Press, New York.
- Beernink, P.T. and Tolan, D.R. 1992. Construction of a high-copy “ATG vector” for expression in Escherichia coli. Protein Expr. Purif. 3 332–336. [DOI] [PubMed] [Google Scholar]
- Berthiaume, L., Tolan, D.R., and Sygusch, J. 1993. Differential usage of the carboxyl-terminal region among aldolase isozymes. J. Biol. Chem. 268 10826–10835. [PubMed] [Google Scholar]
- Blom, N. and Sygusch, J. 1997. Product binding and role of the C-terminal region in class I D-fructose 1,6-bisphosphate aldolase. Nat. Struct. Biol. 4 36–39. [DOI] [PubMed] [Google Scholar]
- Brünger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. 1998. Crystallography & NMR system: A new software suite for macro-molecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54 905–921. [DOI] [PubMed] [Google Scholar]
- Buono, P., Conciliis, L.D., Izzo, P., and Salvatore, F. 1997. The transcription of the human fructose-bisphosphate aldolase C gene is activated by nerve-growth-factor-induced B factor in human neuroblastoma cells. Biochem. J. 323 245–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscaglia, C.A., Coppens, I., Hol, W.G., and Nussenzweig, V. 2003. Sites of interaction between aldolase and thrombospondin-related anonymous protein in plasmodium. Mol. Biol. Cell 14 4947–4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, K.H., Mazurkie, A.S., Morris, A.J., Utheza, D., Tolan, D.R., and Allen, K.N. 1999. Structure of a fructose-1,6-bis(phosphate) aldolase liganded to its natural substrate in a cleavage-defective mutant at 2.3 Å. Biochemistry 38 12655–12664. [DOI] [PubMed] [Google Scholar]
- Choi, K.H., Shi, J., Hopkins, C.E., Tolan, D.R., and Allen, K.N. 2001. Snapshots of catalysis: The structure of fructose-1,6-(bis)phosphate aldolase covalently bound to the substrate dihydroxyacetone phosphate. Biochemistry 40 13868–13875. [DOI] [PubMed] [Google Scholar]
- Copley, S.D. 2003. Enzymes with extra talents: Moonlighting functions and catalytic promiscuity. Curr. Opin. Chem. Biol. 7 265–272. [DOI] [PubMed] [Google Scholar]
- Dalby, A., Dauter, Z., and Littlechild, J.A. 1999. Crystal structure of human muscle aldolase complexed with fructose 1,6- bisphosphate: Mechanistic implications. Protein Sci. 8 291–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalby, A.R., Tolan, D.R., and Littlechild, J.A. 2001. The structure of human liver fructose-1,6-bisphosphate aldolase. Acta Crystallogr. D Biol. Crystallogr. 57 1526–1533. [DOI] [PubMed] [Google Scholar]
- Fedorov, R., Vasan, R., Ghosh, D.K., and Schlichting, I. 2004. Structures of nitric oxide synthase isoforms complexed with the inhibitor AR-R17477 suggest a rational basis for specificity and inhibitor design. Proc. Natl. Acad. Sci. 101 5892–5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamblin, S.J., Davies, G.J., Grimes, J.M., Jackson, R.M., Littlechild, J.A., and Watson, H.C. 1991. Activity and specificity of human aldolases. J. Mol. Biol. 219 573–576. [DOI] [PubMed] [Google Scholar]
- Heras, B., Edeling, M.A., Byriel, K.A., Jones, A., Raina, S., and Martin, J.L. 2003. Dehydration converts DsbG crystal diffraction from low to high resolution. Structure (Camb.) 11 139–145. [DOI] [PubMed] [Google Scholar]
- Ho, S., Hunt, H., Horton, R., Pullen, J., and Pease, L. 1989. Site-directed mutagenesis by overlapping extension using the polymerase chain reaction. Gene 77 51–59. [DOI] [PubMed] [Google Scholar]
- Hubbard, S.J., Campbell, S.F., and Thornton, J.M. 1991. Molecular recognition. Conformational analysis of limited proteolytic sites and serine proteinase protein inhibitors. J. Mol. Biol. 220 507–530. [DOI] [PubMed] [Google Scholar]
- Jeffery, C.J. 1999. Moonlighting proteins. Trends Biochem. Sci. 24 8–11. [DOI] [PubMed] [Google Scholar]
- Jewett, T.J. and Sibley, L.D. 2003. Aldolase forms a bridge between cell surface adhesins and the actin cytoskeleton in apicomplexan parasites. Mol. Cell 11 885–894. [DOI] [PubMed] [Google Scholar]
- Jones, T.A., Zou, J.Y., Cowan, S.W., and Kjeldgaard, M. 1991. Improved methods for building protein models in electron density maps and the location of errors in these methods. Acta Crystallogr. A 47 110–119. [DOI] [PubMed] [Google Scholar]
- Kao, A.W., Noda, Y., Johnson, J.H., Pessin, J.E., and Saltiel, A.R. 1999. Aldolase mediates the association of F-actin with the insulin-responsive glucose transporter GLUT4. J. Biol. Chem. 274 17742–17747. [DOI] [PubMed] [Google Scholar]
- Karkhoff-Schweizer, R. and Knull, H.R. 1987. Demonstration of tubulinglycolytic enzyme interactions using a novel electrophoretic approach. Biochem. Biophys. Res. Commun. 146 827–831. [DOI] [PubMed] [Google Scholar]
- Kiri, A. and Goldspink, G. 2002. RNA-protein interactions of the 3′ untranslated regions of myosin heavy chain transcripts. J. Muscle Res. Cell Motil. 23 119–129. [DOI] [PubMed] [Google Scholar]
- Kraulis, P.J. 1991. MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24 946–950. [Google Scholar]
- Kuo, A., Bowler, M.W., Zimmer, J., Antcliff, J.F., and Doyle, D.A. 2003. Increasing the diffraction limit and internal order of a membrane protein crystal by dehydration. J. Struct. Biol. 141 97–102. [DOI] [PubMed] [Google Scholar]
- Kusakabe, T., Motoki, K., and Hori, K. 1994. Human aldolase C: Characterization of the recombinant enzyme expressed in Escherichia coli. J. Biochem. (Tokyo) 115 1172–1177. [DOI] [PubMed] [Google Scholar]
- ———. 1997. Mode of interactions of human aldolase isozymes with cytoskeletons. Arch. Biochem. Biophys. 344 184–193. [DOI] [PubMed] [Google Scholar]
- Lebherz, H.G. and Rutter, W.J. 1969. Distribution of fructose diphosphate aldolase variants in biological systems. Biochemistry 8 109–121. [DOI] [PubMed] [Google Scholar]
- Lebioda, L. and Stec, B. 1991. Mapping of isozymic differences in enolase. Int. J. Biol. Macromol. 13 97–100. [DOI] [PubMed] [Google Scholar]
- Lu, M., Holliday, L.S., Zhang, L., Dunn Jr., W.A., and Gluck, S.L. 2001. Interaction between aldolase and vacuolar H+-ATPase: Evidence for direct coupling of glycolysis to the ATP-hydrolyzing proton pump. J. Biol. Chem. 276 30407–30413. [DOI] [PubMed] [Google Scholar]
- Makeh, I., Thomas, M., Hardelin, J.-P., Briand, P., Kahn, A., and Skala, H. 1994. Analysis of a brain specific isozyme: Expression and chromatin structure of the rat aldolase C gene and transgenes. J. Biol. Chem. 269 4194–4200. [PubMed] [Google Scholar]
- Matthews, B.W. 1968. Solvent content of protein crystals. J. Mol. Biol. 33 491–497. [DOI] [PubMed] [Google Scholar]
- Morris, A.J. and Tolan, D.R. 1993. Site-directed mutagenesis identifies aspartate 33 as a previously unidentified critical residue in the catalytic mechanism of rabbit aldolase A. J. Biol. Chem. 268 1095–1100. [PubMed] [Google Scholar]
- ———. 1994. Lysine-146 of rabbit muscle aldolase is essential for cleavage and condensation of the C3–C4 bond of fructose 1,6-bis(phosphate). Biochemistry 33 12291–12297. [DOI] [PubMed] [Google Scholar]
- Morris, A.L., MacArthur, M.W., Hutchinson, E.G., and Thornton, J.M. 1992. Stereochemical quality of protein structure coordinates. Proteins 12 345–364. [DOI] [PubMed] [Google Scholar]
- Nicholls, A., Sharp, K., and Honig, B. 1991. Protein folding and association: Insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11 281–296. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. and Minor, W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276 307–326. [DOI] [PubMed] [Google Scholar]
- Peitsch, M.C. 1996. ProMod and Swiss-Model: Internet-based tools for automated comparative protein modelling. Biochem. Soc. Trans. 24 274–279. [DOI] [PubMed] [Google Scholar]
- Penhoet, E.E. and Rutter, W.J. 1971. Catalytic and immunochemical properties of homomeric and heteromeric combinations of aldolase subunits. J. Biol. Chem. 246 318–323. [PubMed] [Google Scholar]
- Penhoet, E.E., Kochman, M., and Rutter, W.J. 1969. Molecular and catalytic properties of aldolase C. Biochemistry 8 4396–4402. [DOI] [PubMed] [Google Scholar]
- Pezza, J.A., Choi, K.H., Berardini, T.Z., Beernink, P.T., Allen, K.N., and Tolan, D.R. 2003. Spatial clustering of isozyme-specific residues reveals unlikely determinants of isozyme specificity in fructose-1,6-bisphosphate aldolase. J. Biol. Chem. 278 17307–17313. [DOI] [PubMed] [Google Scholar]
- Racker, E. 1952. Enzymatic synthesis and breakdown of desoxyribose phosphate. J. Biol. Chem. 196 347–365. [PubMed] [Google Scholar]
- Rose, I.A., O’Connell, E.L., and Mehler, A.H. 1965. Mechanism of the aldolase reaction. J. Biol. Chem. 240 1758–1765. [PubMed] [Google Scholar]
- Rottmann, W.H., Deselms, K.R., Niclas, J., Camerato, T., Holman, P.S., Green, C.J., and Tolan, D.R. 1987. The complete amino acid sequence of the human aldolase C isozyme derived from genomic clones. Biochimie 69 137–145. [DOI] [PubMed] [Google Scholar]
- Rutter, W.J. 1964. Evolution of aldolase. Fed. Proc. 23 1248–1257. [PubMed] [Google Scholar]
- Schapira, F., Reuber, M.D., and Hatzfeld, A. 1970. Resurgence of two fetal-type of aldolases (A and C) in some fast-growing hepatomas. Biochem. Biophys. Res. Commun. 40 321–327. [DOI] [PubMed] [Google Scholar]
- Skala, H., Vibert, M., Lamas, E., Maire, P., Schweighoffer, F., and Kahn, A. 1987. Molecular cloning and expression of rat aldolase C messenger RNA during development and hepatocarcinogenesis. Eur. J. Biochem. 163 513–518. [DOI] [PubMed] [Google Scholar]
- Subramaniam, S. 1998. The biology workbench—A seamless database and analysis environment for the biologist. Proteins 32 1–2. [PubMed] [Google Scholar]
- Susor, W.A., Penhoet, E., and Rutter, W.J. 1975. Fructose-diphosphate aldolase, pyruvate kinase, and pyridine nucleotide-linked activities after electrophoresis. Methods Enzymol. 41 66–73. [DOI] [PubMed] [Google Scholar]
- Sygusch, J., Beaudry, D., and Allaire, M. 1987. Molecular architecture of rabbit skeletal muscle aldolase at 2.7-Å resolution. Proc. Natl. Acad. Sci. 84 7846–7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin, A. and Teplyakov, A. 1997. MOLREP: An automated program for molecular replacement. J. Appl. Cryst. 30 1022–1025. [Google Scholar]
- Wang, J., Morris, A.J., Tolan, D.R., and Pagliaro, L. 1996. The molecular nature of the F-actin binding activity of aldolase revealed with site-directed mutants. J. Biol. Chem. 271 6861–6865. [PubMed] [Google Scholar]
- Zimmer, D.B. and Van Eldik, L.J. 1986. Identification of a molecular target for the calcium-modulated protein S100. Fructose-1,6-bisphosphate aldolase. J. Biol. Chem. 261 11424–11428. [PubMed] [Google Scholar]