OXYLIPINS AS SIGNALING MOLECULES IN DIVERSE LIFE FORMS
Oxylipins are biologically active signaling molecules derived from oxygenated polyunsaturated fatty acids and are found ubiquitously in most living organisms. In mammals, the eicosanoids, which include prostaglandins, are one of the best-studied groups of biologically important oxylipins. In addition to their essential roles in numerous other physiological functions, eicosanoids function as signaling molecules in vertebrate and invertebrate animals and in eukaryotic microbes (Stanley, 2006).
The discovery of prostaglandins and related biologically active substances was recognized by the award of a Nobel Prize in Physiology or Medicine in 1982. Shortly after this, the pioneering work published in Plant Physiology by Vick and Zimmerman (1984) provided one of the first insights into the biosynthesis of jasmonate (JA), an oxylipin signaling molecule in plants. Indeed, of the various oxylipins synthesized enzymatically through the oxylipin (also known as octadecanoid) pathway, the plant hormones JAs (e.g. jasmonic acid and its methyl ester, MeJA) are often considered to be the structural and, in some cases, functional analogues of prostaglandins in animals. JAs are potent regulators of genes involved in cell growth and biotic and abiotic stress responses. Over the last decade or so, the JA signaling pathway has been studied extensively in dicot plant species, such as Arabidopsis (Arabidopsis thaliana), tomato (Solanum lycopersicum), and tobacco (Nicotiana tabacum), and to a somewhat limited extent in monocot plants, such as barley (Hordeum vulgare) and rice (Oryza sativa). Although much remains to be learnt, both forward and reverse genetic studies, particularly in Arabidopsis, have greatly expanded our understanding of the potential roles of JAs in plants.
The biosynthesis of JAs has recently been reviewed (Wasternack, 2007), and a general overview is shown in Figure 1. Following synthesis, JAs are perceived by as yet unknown receptor proteins, and this presumably activates a signal transduction pathway that culminates in the transcriptional activation or repression of a large number of JA-responsive genes. JAs inhibit root elongation, and this property has been extensively exploited for the identification of JA signaling mutants. One of the first JA signaling mutants identified was the Arabidopsis coronatine insensitive1 (coi1) mutant. Root elongation of coi1 mutant seedlings showed reduced sensitivity to JAs but also to coronatine (COR), a functional JA homolog and toxin produced by the bacterial pathogen Pseudomonas syringae (Feys et al., 1994). The coi1 mutant displays defects in many, if not all, JA-dependent functions, such as fertility, secondary metabolite biosynthesis, pest and pathogen resistance, and wound responses. The COI1 locus encodes an F-box protein, and because F-box proteins are integral parts of multi-protein complexes involved in protein ubiquitination, it was speculated that COI1 is required for removal of repressors of the JA signaling pathway (Xie et al., 1998). However, until very recently, the nature of the COI1-targeted repressors has remained elusive. Similarly, two other JA signaling loci, JAR1 (JASMONATE RESISTANT1) and JIN1 (JASMONATE INSENSITIVE1), were identified from analyses of the Arabidopsis jar1 and jin1 mutants, which also show reduced sensitivity to exogenous JAs. JAR1 encodes a JA amino acid synthetase involved in conjugating jasmonic acid to Ile (Staswick and Tiryaki, 2004). JIN1 (also known as MYC2) encodes a basic helix-loop-helix-type transcription factor involved in the transcriptional regulation of JA-responsive gene expression (Lorenzo et al., 2004). Despite extensive characterizations of individual mutants, the exact nature of the functional relationships among these three major players (i.e. COI1, JAR1, and JIN1/MYC2) of JA signaling has long been enigmatic. Importantly, the recent cloning of the JAI3 (JASMONATE INSENSITIVE3) locus (Chini et al., 2007) has filled a significant gap in our overall understanding of the JA signaling pathway by mechanistically linking the functions of COI1, JAR1, and JIN1, as well as revealing the nature of the long-sought repressors of JA signaling.
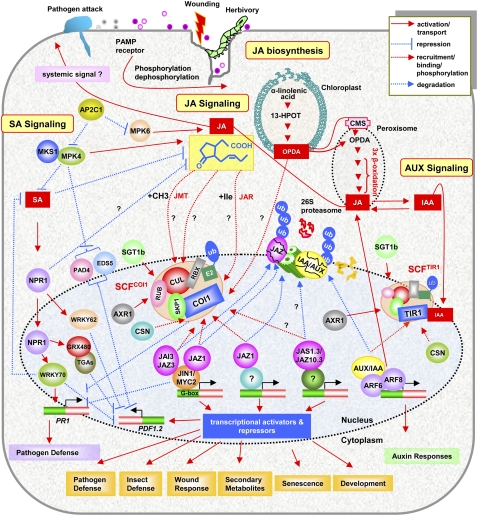
Figure 1.
An integrated view of JA biosynthesis and signaling, including signaling interactions between JA and SA and JA and auxin (IAA) in Arabidopsis. Biotic and abiotic stresses, such as pathogen and insect attack and wounding, generate signals/elicitors that activate a phosphorylation cascade that regulates JA biosynthesis and signaling. Briefly, JAs are derived from α-linolenic acid liberated from membrane phospholipids by the action of phospholipase A. α-Linolenic acid is first converted to 13-hydroperoxy linolenic acid (13-HPOT) and then to 12-OPDA in the chloroplasts in a series of reactions catalyzed by 13-lipoxygenase, allene oxide synthase, and allene oxide cyclase, respectively. 12-OPDA is then transported to peroxisomes either passively or actively by the ABC (ATP-binding cassette) transporter COMATOSA (CMS). 12-OPDA is subsequently reduced by 12-OXOPHYTODIENOATE REDUCTASE3 to 3-oxo-2-(2′-pentenyl)-cyclopentane-1-octanoic acid, which then undergoes three cycles of β-oxidation in the peroxisomes to produce jasmonic acid. Jasmonic acid is further modified in the cytosol to produce various jasmonic acid derivatives. For instance, jasmonic acid is converted to volatile oxylipin MeJA by a JA methyl transferase or conjugated into several amino acids by an amino acid synthetase encoded by the JAR1 gene. JAZ proteins act as negative regulators of the transcriptional regulator JIN1/MYC2, and their JA- and SCFCOI1-dependent degradation liberates JIN1/MYC2 from repression. JIN1/MYC2, by possibly binding to the conserved G-box element found in the promoters, coordinates a transcriptional cascade that involves other transcriptional activators and repressors from AP2/ERF, WRKY, and MYBs to modulate distinct JA-dependent functions. JA may also be transported to distal tissue that has not been directly challenged to activate systemic gene expression. The cross talk between JA and SA and JA and auxin signaling occurs at multiple points. As explained in the text, NPR1, MPK4, WRKY70, SCFCOI1, and JIN1/MYC2 are some of the major players involved in these interactions. Similarly, the JA-auxin cross talk is modulated by CSN, SGT1b, AXR1, and ARFs. Please note that for SA and auxin signaling, only those components known to interact with JA signaling are shown. See text for further details and abbreviations.
Our aim in this Update article is to briefly review these recent findings that have added fresh insights into our understanding of how JA signals are transmitted within the cell. Our particular focus will be on the roles of a recently discovered class of repressors whose destruction through a COI1-mediated ubiquitination pathway is required for the transcriptional activation of the JA-dependent gene expression. In addition, the emerging roles of the transcriptional regulator JIN1/MYC2 that acts immediately downstream from these repressors in coordinating a transcriptional cascade will be briefly reviewed. Finally, positive and negative feedback loops regulating JA biosynthesis and signaling and some recent examples of interactions between JA and other hormonal signaling pathways will be considered. Readers particularly interested in JA biosynthesis should refer to other recent reviews on this topic (Browse, 2005; Wasternack, 2007).
ACTIVATION OF JA SIGNALING BY REPRESSOR REMOVAL
Many plant processes are controlled by repressors of downstream transcriptional networks, and the degradation of these repressors under external stimuli and by plant hormones provides a rapid regulatory trigger system. The involvement of protein degradation pathways in JA signaling became apparent after the identification of the COI1 gene encoding an F-box protein with Leu repeats (Xie et al., 1998). Indeed, COI1 or SCFCOI1 is an integral part of a highly conserved multi-protein complex called the SCF E3 ubiquitin ligase complex. The SCF complex is found in all eukaryotes and consists of a Skp1 (S-phase kinase-associated protein)-related protein, a cullin, a RING-box protein, and an F-box protein. The SCF complex is involved in marking proteins with ubiquitin tags to facilitate their degradation by the 26S proteasome (for review, see Stone and Callis, 2007). The F-box protein component (e.g. SCFCOI1) is known to be responsible for the specificity of the SCF complexes to particular targets. However, as stated above, repressor proteins potentially targeted by SCFCOI1 have been unknown. Recently, three laboratories have simultaneously converged on a family of genes that fulfils this role.
The cloning of mutated genes in JA-insensitive mutants has so far provided vital information about the signaling events involved in this pathway. In contrast to the recessive coi1, jar1, and jin1 mutations, the relatively less-studied jai3 mutation confers a dominant JA insensitivity phenotype (Chini et al., 2007). To identify the molecular nature of this mutation, Chini et al. (2007) have sequenced the chromosomal region around the genetically mapped location of jai3 for possible mutations. This exercise identified a point mutation in a gene of unknown function. This mutation is predicted to cause the aberrant splicing of this gene, presumably leading to the production of a protein truncated at the C terminus. As expected, transgenic expression of the jai3 mutant protein in wild-type plants, but not the wild-type JAI3, produces the jai3 mutant phenotype, confirming that the jai3 mutation was indeed responsible for the dominant JA insensitivity phenotype. The JAI3 protein contains a ZIM (zinc finger protein expressed in inflorescence meristem) domain (Shikata et al., 2003). Because JAI3 was both an early JA-responsive gene and required for JA sensitivity, it was renamed as JASMONATE ZIM DOMAIN3 (JAZ3; Chini et al., 2007).
Thines et al. (2007), on the other hand, have used a reverse genetic approach to identify the possible functions of the early JA-inducible genes in Arabidopsis stamens. An earlier study had found that, as early as 30 min after JA treatment, several genes encoding individual members of the JAZ protein family showed strong induction in the stamens of the JA-deficient 12-oxophytodienoate reductase3 mutant (Mandaokar et al., 2006). Thines et al. (2007) studied the functions of these genes by overexpressing and knocking out the expression of individual JAZ genes. Disappointingly, none of the lines studied displayed any discernible JA-dependent phenotype, possibly due to functional redundancy. However, when a truncated version of JAZ1 called JAZ1Δ3 was transgenically expressed in Arabidopsis, a coi1-like phenotype characterized by male sterility, JA insensitivity, and increased resistance to infection by the bacterial pathogen P. syringae pv. tomato was observed. JAZ1Δ3 transgenic lines also show increased susceptibility to the herbivorous insect Spodoptera exigua (Chung et al., 2008). These transgene-conferred phenotypes were suggestive of a role for JAZ1 in JA signaling. The additional work by both Chini et al. (2007) and Thines et al. (2007) has led to the conclusion that JAZ1 and JAI3/JAZ3 are indeed the long-sought repressors of the JA signaling pathway, and their SCFCOI1-dependent ubiquitination is required for the activation of JA-responsive gene expression. Indeed, the JA-dependent degradation of JAZs could be inhibited by a specific inhibitor of the 26S proteasome activity or in the coi1 mutant background. Furthermore, results from yeast two-hybrid studies showed that SCFCOI1 interacts with the JAI3/JAZ3 C-terminal region, the same domain that is truncated in the jai3 mutant. Another interesting finding was that the interaction between SCFCOI1 and JAZ1 or JAI3/JAZ3 was promoted by JA-Ile in a highly specific manner but not by jasmonic acid, MeJA, COR, or the JA precursor 12-oxo-phytodionic acid (12-OPDA; Chini et al., 2007; Thines et al., 2007). This finding has obvious implications about the identity of the biologically active signal in this signaling pathway (see below).
How does JA- and SCFCOI1-dependent degradation of JAZ repressors transcriptionally regulate the JA signaling pathway? JAZ proteins do not contain any DNA-binding domain. This is an indication that they may interact with other proteins to regulate gene expression (see also Vanholme et al., 2007 for additional discussion). Interestingly, both JAZ1 and JAI3/JAZ3 each interact with JIN1/MYC2, a transcriptional regulator in JA signaling (see also below), in yeast two-hybrid assays (Chini et al., 2007). This finding suggests that, in the absence of a JA signal, JAZ1 and JAI3/JAZ3 repress JIN1/MYC2. This repression most likely occurs in the nucleus, as both JAZ repressors and JIN1/MYC2 are found in the nucleus during normal growth and development (Lorenzo et al., 2004; Chini et al., 2007; Thines et al., 2007). Upon sensing of the JA signals, JAZ repressors are recruited to the SCF E3 complex for ubiquitination and subsequent degradation by the proteasome. The removal of these repressors then paves the way for JIN1/MYC2 to regulate JA-dependent gene expression (Fig. 1).
Another member of the JAZ family, JASMONATE ASSOCIATED1 (JAS1), also appears to be involved in JA signaling (Yan et al., 2007). This gene was first identified by a microarray screen for JA-regulated transcripts. The JAS1.3 locus is an alternatively transcribed gene, producing two different isoforms in Arabidopsis. The overexpression and RNAi-mediated silencing of the shorter isoform of JAS1, designated as JAS1.3/JAZ10.3, made the plants less and more sensitive to growth repression by MeJA. In contrast, the overexpression of the longer isoform of this gene did not produce any JA-dependent growth phenotype. JAS1.3/JAZ10.3 overexpressing plants also showed reduced growth inhibition after wounding (Yan et al., 2007), another JA-dependent phenotype. The molecular mechanism(s) of JAS1.3/JAZ10.3-mediated growth phenotype is currently far from clear. However, these findings collectively suggest that the different members of the JAZ protein family have essential roles in multiple JA-dependent functions.
MULTIPLICITY OF JA SIGNALS AND RECEPTORS
The finding that JA-Ile, a jasmonic acid-Ile conjugate, but not jasmonic acid itself, MeJA, COR, or the JA precursor 12-OPDA promotes the interaction between SCFCOI1-JAZ complexes in yeast two-hybrid assays (Thines et al., 2007) raises new questions regarding the exact nature of the JA signal(s) perceived by putative JA receptors. This finding might imply that jasmonic acid may not be the signal directly responsible for the activation of the JA signaling pathway, but possibly it undergoes further modifications to be converted to a biologically active signal. In contrast to this, in other hormone signaling pathways (e.g. auxin), conjugation of amino acids to plant hormones is often used as a versatile mechanism for rapid reduction of the hormone levels and, consequently, down-regulation of the signaling pathway (Woodward and Bartel, 2005). When required, hormone-amino acid conjugates are rapidly hydrolyzed to release the active hormone and this activates the signaling pathway. As mentioned above, the product of the JAR1 locus conjugates Ile to jasmonic acid. However, the jar1 mutant does not display all the defects observed in the coi1 mutant, suggesting that JA-Ile may not be the only signal responsible for the activation of the JA signaling pathway.
Interestingly, although JA-Ile produced by JAR1 promotes the interaction between JAZ and SCFCOI1, a recent report found that wound-induced expression JAZ and SCFCOI1-dependent genes in the jar1 mutant was similar to that in wild-type plants (Chung et al., 2008). This and other relatively subtle JA defects observed in the jar1 mutant could be due to the fact that wound-induced JA-Ile levels in this mutant are still relatively high (approximately 10%–25% of wild-type levels; Chung et al., 2008). This indicates that JAR1 is perhaps not the only enzyme conjugating JA to Ile in Arabidopsis. In addition to Ile, the JAR family of related GH3 enzymes has the potential to conjugate JA to other amino acids such as Trp, Val, and Leu as recently found in tobacco (Wang et al., 2008). In addition, both JA-Ile and 12-OPDA induce distinct but overlapping patterns of gene expression as jasmonic acid and MeJA (Taki et al., 2005; Wang et al., 2008). The 12-OPDA-induced gene expression also appears to be independent from SCFCOI1 in Arabidopsis (Taki et al., 2005). JA is converted to MeJA by a JA methyl transferase (Fig. 1), while at least in tobacco, exogenously applied MeJA is hydrolyzed by a MeJA esterase to produce jasmonic acid (Wu et al., 2008). Silencing the expression of the NaMJE gene encoding this MeJA-cleaving esterase in transgenic tobacco inhibits MeJA- but not jasmonic acid-induced insect resistance (Wu et al., 2008). It is not known whether the conversion of MeJA to jasmonic acid plays any role in transducing airborne signals emitted by the nearby plants through JA signaling. However, it is likely that there might be multiple JA-derived signals affecting the stability of different SCFCOI1-JAZ complexes. The presence of different JA-derived signals could be used as a means to regulate endogenous hormone levels but also help to rapidly respond to different endogenous and exogenous cues.
TRANSCRIPTIONAL COORDINATION OF JA SIGNALING
As mentioned above, JAI3/JAZ3 most likely suppresses the transcription factor JIN1/MYC2, which, acting early on in the signaling pathway, can either positively or negatively modulate diverse JA-dependent functions. In particular, the JA-dependent expression of pathogen and insect defense genes is differentially regulated by JIN1/MYC2. In the jin1/myc2 mutant, JA-dependent induction of wound and insect response genes was significantly attenuated, and, as a result, jin1/myc2 mutant plants showed increased susceptibility to an insect pest (Dombrecht et al., 2007). In contrast, the JA-dependent induction of pathogen defense genes was heightened in the jin1/myc2 mutant, which showed increased resistance to bacterial and fungal pathogens (Anderson et al., 2004; Lorenzo et al., 2004). Accumulating evidence indicates that plants are able to coordinate their responses depending on the type of attack so that the metabolic cost of plant defense can be minimized. Indeed, although both herbivory and pathogen attack activate JA signaling, the defense genes that are activated are functionally specialized against insect pests or pathogens, respectively (De Vos et al., 2005). JIN1/MYC2 and similar other molecular switches might perhaps be required to fine-tune plant defense against different biological threats.
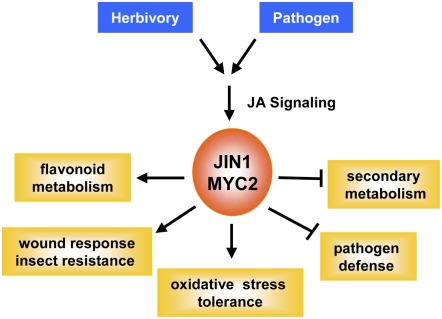
Recent research has also showed that, in addition to pathogen and insect defense, JIN1/MYC2 differentially regulates other JA-dependent functions in Arabidopsis. For instance, in addition to insect resistance, JIN1/MYC2 positively regulates JA-mediated oxidative stress tolerance and flavonoid metabolism. In contrast, JA-dependent pathogen defense and the biosynthesis of secondary metabolites (e.g. biosynthesis of indole glucosinolates) are negatively regulated by JIN1/MYC2 (Dombrecht et al., 2007; Fig. 2). It was proposed that these regulatory controls are mediated by JIN1/MYC2 by coordinating a transcriptional cascade involving a number of other transcription factors (AP2/ERFs, MYBs, zinc fingers, and WRKYs), each with demonstrated roles in regulating downstream gene expression (Dombrecht et al., 2007).
Figure 2.
JIN1/MYC2 differentially regulates different JA-dependent phenotypes. Arrows and blunt arrows indicate positive and negative regulation, respectively. See text for details.
JIN1/MYC2 does not have any obvious roles in fertility, although this trait is regulated by SCFCOI1. Fertility might be controlled by other SCFCOI1-regulated transcription factors. Recent analysis of the T-DNA insertion mutants of the two JA-responsive MYB transcription factor genes, MYB21 and MYB24, indicated their involvement in fertility (Mandaokar et al., 2006), although whether these MYB transcription factors interact with SCFCOI1 and/or JAZ repressors is currently unknown.
NEGATIVE AND POSITIVE REGULATORY FEEDBACK LOOPS IN JA SIGNALING
The relatively broad effects of hormone signaling pathways on multiple plant physiological processes demand that signaling pathways are tightly and coordinately regulated, preferably at multiple points. So far, both negative and positive feedback regulatory loops that regulate JA biosynthesis and signaling have been identified. First, JA biosynthesis genes are activated by JAs, suggesting that JAs positively regulate their own biosynthesis through a positive feedback loop. The recent identification of the Arabidopsis FATTY ACID OXYGENATION UPREGULATED2 gene that encodes a Ca2+-permeant nonselective cation channel suggested that cation fluxes are an important part of this positive feedback loop (Bonaventure et al., 2007).
JA also rapidly activates the transcription of genes encoding JAZ repressors (Chini et al., 2007) while facilitating, as stated above, their destruction at the protein level. This destruction and subsequent resynthesis of JAZ repressors during JA signaling would reset the signaling pathway, avoiding a run-away response. Another level of control in JA signaling is exerted at the JIN1/MYC2 level. JIN1/MYC2 controls transcriptional activation of the JAZ-encoding genes (Chini et al., 2007). MYC2 expression itself is both positively and negatively regulated by JAs. The negative regulation of JIN1/MYC2 during JA signaling is proposed to occur through the mitogen-activated protein kinase pathways regulated by MKK3 and MPK6 (Takahashi et al., 2007). In addition, JIN1/MYC2 can negatively regulate its own expression, possibly by binding to the conserved G-box found in its own promoter (Dombrecht et al., 2007). Additional control points in JA signaling probably exist. In particular, complex cross communication between JA and other hormonal signaling pathways might help fine-tune JA biosynthesis and signaling (see below).
The roles of protein phosphorylation/dephosphorylation pathways in negative and positive regulation of JA biosynthesis and signaling are just emerging. Importantly, protein phosphorylation often precedes the ubiquitination process, which, as discussed above, is critical for the activation of the JA signaling pathway. Although whether Arabidopsis JAZ repressors are phosphorylated before being ubiquitinated is not known, a recent report indicated that PPS3, the potato homolog of JAI3/JAZ3, is phosphorylated by StMPK1, which shows close sequence similarity to Arabidopsis MPK6 (Katou et al., 2005).
In addition to phosphorylation, protein dephosphorylation pathways modulate JA levels. For instance, in response to wounding, the PP2C-type phosphatase AP2C1 negatively regulates mitogen-activated protein kinase signaling pathways as deduced from the analysis of the Arabidopsis ap2c1 mutant, which contains increased levels of wound-induced JAs and displays enhanced resistance to a phytophagous mite (Schweighofer et al., 2007).
JA SIGNALING AND HORMONAL CROSS TALK
It is becoming evident that plant hormone signaling pathways extensively interact during plant growth and development as well as during adaptation to biotic and abiotic stresses. This hormonal cross talk is indeed intriguingly complex and often dose-, species-, tissue-, and inducer-specific. The JA signaling pathway is no exception to this. Over the years, many components that are shared between JA and various other plant hormone signaling pathways have been identified. Cross talk is mostly inferred from the observation that genetic ablation of the individual shared components (or nodes) compromises both pathways or if one hormone brings about physiological changes mainly by promoting the synthesis or action of another hormone.
JA-SALICYLIC ACID INTERACTIONS
The mutually antagonistic interactions between salicylic acid (SA) and JA pathways first became evident from the analysis of SA- and JA-marker gene expression in SA and JA signaling mutants in Arabidopsis. Indeed, mutations that disrupt JA signaling (e.g. coi1) lead to the enhanced basal and inducible expression of the SA marker gene PR1, while mutations that disrupt SA signaling (e.g. npr1) lead to the concomitant increases in the basal or induced levels of the JA marker gene PDF1.2. Interestingly, exogenous SA promotes the JA-dependent induction of the defense gene PDF1.2 when applied at low concentrations. However, at higher SA concentrations, the induction of PDF1.2 by JA is reduced, leading to the proposal that the interaction between these two pathways might be dose dependent (Mur et al., 2006). Plants treated with SA or inoculated with virulent strains of P. syringae pv. tomato show compromised resistance to Alternaria brassicicola, a necrotrophic pathogen sensitive to JA-dependent defenses, possibly due to suppression of JA-dependent defenses known to be effective against necrotrophic pathogens (Spoel et al., 2007). Interestingly, however, inoculations with an avirulent strain of P. syringae that trigger hypersensitive response do not compromise A. brassicicola resistance, suggesting that cross talk between SA and JA signaling is also specific to pathogen strain (Spoel et al., 2007).
The antagonistic interaction between SA and JA signaling is at least partly mediated by NONINDUCIBLE PR1 (NPR1), a master regulator of SA signaling, but also responds to oxidative events (Spoel et al., 2003, 2007). Interestingly, the SA-JA antagonism and the involvement of NPR1 is reminiscent to the inhibition of prostaglandin biosynthesis by aspirin, which acts by trans-acetylation of cylooxygenases and by inhibiting the transcription from cyclooxygenase-encoding genes involved in prostaglandin synthesis through the function of transcriptional repressor IKβ, an NPR1 homolog in animals. In tobacco, the role of NPR1 in regulating JA-SA cross talk also appears to be different from that in Arabidopsis. In insect-attacked tobacco, NPR1 down-regulates SA biosynthesis, and this leads to the up-regulation of JA biosynthesis and signaling that is required for defense against insect attack (Rayapuram and Baldwin, 2007). Therefore, the mechanism of antagonistic interaction between SA and JA pathways seems to vary between different species.
Acting downstream from NPR1, WRKY70 is a versatile transcription factor with roles in multiple signaling pathways and physiological processes. WRKY70 regulates the antagonistic interactions between SA and JA pathways (Fig. 1). Overexpression of WRKY70 leads to the constitutive expression of the SA-responsive PR genes and increased resistance to SA-sensitive pathogens but reduces resistance to JA-sensitive pathogens. In contrast, suppression of WRKY70 leads to increased expression from JA-responsive genes and increased resistance to a pathogen sensitive to JA-dependent defenses (Li et al., 2004a). WRKY70 is also implicated in suppressing SA levels when SA levels are particularly high (Wang et al., 2006). Another WRKY transcription factor that negatively regulates JA signaling in an NPR1-dependent manner is WRKY62 (Mao et al., 2007; Fig. 1). The hierarchical relationship between WRKY70 and WRKY62 is unknown.
The Arabidopsis mpk4 mutant exhibits constitutively active SA-dependent defense responses (e.g. increased SA levels, constitutive expression of PR1, and increased resistance to P. syringae) in the absence of pathogen attack. In contrast, the JA-dependent induction of the PDF1.2 gene was abolished in the mpk4 mutant (Petersen et al., 2000). Therefore, MPK4 is proposed to be a positive regulator of JA-dependent responses while a negative regulator of SA biosynthesis and signaling. As expected, overexpression of MKS1, a MPK4 substrate, also regulates SA signaling through interaction with WRKY transcription factors. However, no effect of MKS1 overexpression on JA signaling was found (Andreasson et al., 2005). Therefore, it is unknown whether MPK4 phosphorylates any JA signaling component during its positive regulation of JA signaling or if the reduced JA-dependent responses found in the mpk4 mutant is simply due to antagonistic effects of the enhanced SA biosynthesis and signaling. SA-JA interaction in Arabidopsis is also regulated by an SA-inducible glutaredoxin, GRX480, which interacts with the TGA-type transcription factors involved in the regulation of SA-inducible PR genes and suppresses the JA-responsive expression of PDF1.2 (Ndamukong et al., 2007). More recently, the involvement of ESR/ESP (epithiospecifying senescence regulator) and WRKY53 (Miao and Zentgraf, 2007) in SA-JA interaction has also been reported (Fig. 1).
In addition to COI1, the transcriptional regulator JIN1/MYC2 also has a role in antagonizing SA signaling in plants during infection by P. syringae. Increased PR1 expression and resistance is found in P. syringae-infected jin1/myc2 plants that show increased resistance to this pathogen (Laurie-Berry et al., 2006). It is not clear, however, whether JIN1/MYC2 directly represses PR1 or whether the increased PR1 expression observed in the jin1/myc2 mutant is due to indirect effects of the compromised JA signaling on SA signaling and subsequent responses.
JA-ETHYLENE INTERACTIONS
The interaction between JA and ethylene signaling is rather complex, and both synergistic and antagonistic interactions have been reported, depending on the stress conditions examined. Adding to this complexity, the role of ethylene in biotrophic pathogen-plant interactions could be different than that in necrotrophic pathogen-plant interactions (Broekaert et al., 2006). JA and ethylene synergistically induce a subset of defense genes following pathogen inoculation in Arabidopsis. For instance, induction of PDF1.2 by A. brassicicola requires both JA and ethylene signaling pathways (Penninckx et al., 1998). The cellulose synthase gene CeSA3/CEV1 controls a point of convergence between these two pathways as a negative regulator of both pathways, as deduced from the analysis of the cev1 mutant that displays constitutively active JA and ethylene responses (Ellis et al., 2002). The ETHYLENE RESPONSE FACTOR1 transcription factor also functions at the crossroad of JA and ethylene signaling as a positive regulator of both pathways (Lorenzo et al., 2003).
In contrast to these synergistic interactions, JA and ethylene signaling pathways act in a mutually antagonistic fashion in modulating ozone-induced cell death. Most, if not all, JA signaling and biosynthetic mutants show increased ozone sensitivity. In contrast to the effect of JA signaling, the ethylene signaling pathway promotes ozone-induced spread of lesion development (for review, see Overmyer et al., 2003).
JA, LIGHT, AND AUXIN INTERACTIONS
As discussed above, protein degradation pathways play essential roles not only in JA but also in light and auxin signaling. Not surprisingly, therefore, most cross talk among these pathways revolves around the SCF E3 ubiquitin ligase and the COP9 signalosome (CSN) complexes. For instance, mutations in CULLIN1/AUXIN RESISTANT6 (AXR6) component of the SCF ubiquitin ligase and CSN complexes compromise auxin, JA, and light responses. The axr6 mutant shows reduced sensitivity to JA and auxin (Feng et al., 2003; Ren et al., 2005) and hypersensitivity to far-red light (Quint et al., 2005). A direct interaction between CSN and SCFCOI1 has been shown. CSN reduction-of-function plants show a JA-insensitive root elongation phenotype and reduced expression from JA-responsive genes (Feng et al., 2003). AXR1, which encodes a subunit of the RUB1-activating enzyme that regulates the protein degradation activity of SCF complexes, regulates both SCFTIR1 and SCFCOI1 involved in auxin and JA signaling, respectively (Schwechheimer et al., 2002). The axr1 mutant shows reduced sensitivity to both JA and auxin (Tiryaki and Staswick, 2002). SUPPRESSOR OF THE G2 ALLELE OF skp1-4b is required for both SCFTIR-mediated auxin and SCFCOI1-mediated JA responses (Gray et al., 2003). Auxin and JA pathways are also interlinked at the level of ARFs. At least two ARFs, ARF6 and ARF8, are required for JA biosynthesis and flower fertility (Fig. 1). In addition to defective auxin responses, the arf6/arf8 double mutant shows JA deficiency, aberrant flower development, and reduced expression of several JA biosynthesis genes in flowers (Nagpal et al., 2005). JA activates expression from auxin biosynthesis genes (Dombrecht et al., 2007), while auxin activates expression of JA biosynthesis genes (Tiryaki and Staswick, 2002). These examples clearly illustrate that auxin and JA signaling are intimately interlinked. The Arabidopsis phytochrome mutants hy1 and hy2 show a JA overproduction phenotype and constitutive activation of the JA-inducible and SCFCOI1-dependent defense genes (Zhai et al., 2007). Importantly, JIN1/MYC2 acts as a negative regulator of blue light-mediated photomorphogenic growth and blue and far-red light-regulated gene expression in Arabidopsis (Yadav et al., 2005).
JA-ABSCISIC ACID INTERACTIONS
Both antagonistic and synergistic interactions occur between abscisic acid (ABA) and JA signaling in Arabidopsis. Both ABA and MeJA induce stomatal closure, most likely by triggering the production of reactive oxygen species (ROS) in stomatal guard cells (Munemasa et al., 2007). The coi1 mutation disrupts only MeJA-mediated ROS production without influencing ABA-mediated ROS production, suggesting that COI1 acts upstream from the convergence of ABA and MeJA signaling pathways.
JIN1/MYC2, a negative regulator of JA-dependent pathogen defense gene expression, positively regulates ABA-dependent drought responses (Anderson et al., 2004). A recent study has shown that endogenous ABA had positive and negative regulatory effects on JA-responsive insect and pathogen defense genes, respectively. In the ABA-deficient mutant aba2-1, the insect-responsive expression of VSP2 was reduced while that of PDF1.2 was increased. Consistent with this, the Spodeptera littoralis larvae had a higher weight gain on the aba2-1 mutant than on wild-type plants (Bodenhausen and Reymond, 2007).
Nevertheless, JA and ABA activate a large subset of genes also activated by the pathogenic oomycete Pythium irregulare. This effect of ABA is proposed to be due to the effect of this root-infecting pathogen to impose water stress in plants by clogging the vasculature (Adie et al., 2007). ABA also appears to induce JA biosynthesis in Arabidopsis, and increased JA levels found in ABA-treated plants were proposed to be a reason behind the reduced SA defense gene expression by ABA (Adie et al., 2007).
CONCLUSIONS AND FUTURE PROSPECTS
As exemplified throughout this article, the genetic and genomic resources available in Arabidopsis have been a driving force behind the recent discoveries made regarding how JA signals are transmitted. Some of the JA signaling components that have been identified in Arabidopsis have also been functionally analyzed in a few other dicot species, such as tobacco (Paschold et al., 2007; Rayapuram and Baldwin, 2007; Wang et al., 2008) and tomato (Boter et al., 2004; Li et al., 2004b; Thines et al., 2007), where they have been shown to be largely conserved. However, currently there is very little evidence regarding the actual roles of the Arabidopsis JA signaling genes in monocots. In crop plants that are not particularly amenable to functional studies (e.g. due to genetic redundancy and/or technical difficulties associated with genetic transformation), functional homology to Arabidopsis JA signaling genes was established based on the successful restoration of the wild-type phenotype when the cloned crop gene of interest was introduced into the well-characterized Arabidopsis mutant (e.g. coi1) defective for the same JA signaling component (Wang et al., 2005).
The recent discovery of JAZ repressors in Arabidopsis and tomato has not only revealed new mechanical insights into JA signaling but also reinforced the notion that signal-mediated degradation of repressors is a common theme used in plant hormone signaling. The JAZ family contains at least 12 members in Arabidopsis (Vanholme et al., 2007). So far, the involvement of at least three (JAZ1, JAI3/JAZ3, and JAS1.3/JAZ10.3) with JA signaling has been demonstrated. Many members of this gene family also show JA, wound, and herbivore inducibility (Vanholme et al., 2007; Chung et al., 2008), although currently their roles in JA signaling are not clear. It is also possible that JAZ repressors may have additional roles in JA signaling. Publicly available expression data show that a number of JAZ genes, including JAI3/JAZ3, are strongly repressed by SA, indicating their possible involvement in SA-JA cross talk.
The discovery of JAZ repressors has also led to the proposal that the complexes between SCFCOI1 and different JAZ proteins might be the sites of reception of different JA signals (Parry and Estelle, 2006; Chini et al., 2007; Farmer, 2007). Although this seems to be a plausible proposal, particularly in light of SCFTIR being an auxin receptor, the biochemical evidence for SCFCOI1 or SCFCOI-JAZ complexes being JA receptors is still missing. One of the criteria for a hormone receptor is reversible and high affinity binding to hormone or its derivatives. At the time of writing this article, no such ligand-receptor relationships between SCFCOI1-JAZ complexes and any JA signal have been demonstrated.
Given the redundancy of receptors for other plant hormones, it would not be surprising that multiple JA receptors exist in plants. Indeed, not all JA responses are SCFCOI1 dependent (Devoto et al., 2005), suggesting that SCFCOI1 either is not a JA receptor or is functionally redundant. Although at least six SCFCOI1 homologs are found in the Arabidopsis genome, their possible function(s) in JA or other physiological processes has not yet been characterized. An intriguing question is whether the commonalities identified between JAs and prostaglandins can be extended into their perceptions. After synthesis, prostaglandins are transported out of cells and bind to plasma membrane-located, G-protein-coupled receptors (Hata and Breyer, 2004). In Arabidopsis, about 25 putative membrane-located, G-protein-coupled receptors have so far been identified (Grill and Christmann, 2007). The ligands for most, if not all, of these putative receptors are unknown.
It is becoming evident that signaling cascades regulated by protein phosphorylation/dephosphorylation have roles in regulating JA signaling, although JA signaling components phosphorylated/dephosphorylated by these pathways are mostly unknown. Despite observations of extensive interactions between JA and other hormonal signaling pathways, our knowledge on the molecular mechanisms involved in these interactions is also still rudimentary. Nevertheless, this complex interaction among signaling networks is a testament to the plant's ability to integrate diverse signals from multiple sources so expediently that a finely tuned output can be produced and thereby provide adaptation to its environment. We expect that the elucidation of the intricate interactions between JA and other signaling pathways will continue to be a fertile area for future research.
Acknowledgments
Owing to space limitations, not all relevant work on this topic could be cited. We thank Bruno Dombrecht, Louise Thatcher, and Brendan Kidd for useful discussions, Louise Thatcher and Brendan Kidd for critical manuscript reading, and two anonymous reviewers for useful comments.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Kemal Kazan (kemal.kazan@csiro.au).
References
- Adie BA, Perez-Perez J, Perez-Perez MM, Godoy M, Sanchez-Serrano JJ, Schmelz EA, Solano R (2007) ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant Cell 19 1665–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JP, Badruzsaufari E, Schenk PM, Manners JM, Desmond OJ, Ehlert C, Maclean DJ, Ebert PR, Kazan K (2004) Antagonistic interaction between abscisic acid and JA-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 16 3460–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasson E, Jenkins T, Brodersen P, Thorgrimsen S, Petersen NH, Zhu S, Qiu JL, Micheelsen P, Rocher A, Petersen M, et al (2005) The MAP kinase substrate MKS1 is a regulator of plant defense responses. EMBO J 24 2579–2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenhausen N, Reymond P (2007) Signaling pathways controlling induced resistance to insect herbivores in Arabidopsis. Mol Plant Microbe Interact 20 1406–1420 [DOI] [PubMed] [Google Scholar]
- Bonaventure G, Gfeller A, Proebsting WM, Hortensteiner S, Chetelat A, Martinoia E, Farmer EE (2007) A gain-of-function allele of TPC1 activates oxylipin biogenesis after leaf wounding in Arabidopsis. Plant J 49 889–898 [DOI] [PubMed] [Google Scholar]
- Boter M, Ruiz-Rivero O, Abdeen A, Prat S (2004) Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev 18 1577–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broekaert WF, Delauré SL, De Bolle MF, Cammue BP (2006) The role of ethylene in host-pathogen interactions. Annu Rev Phytopathol 44 393–416 [DOI] [PubMed] [Google Scholar]
- Browse J (2005) Jasmonate: an oxylipin signal with many roles in plants. Vitam Horm 72 431–456 [DOI] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernandez G, Adie B, Chico JM, Lorenzo O, Garcia-Casado G, Lopez-Vidriero I, Lozano FM, Ponce MR, et al (2007) The JAZ family of repressors is the missing link in jasmonate signaling. Nature 448 666–671 [DOI] [PubMed] [Google Scholar]
- Chung HS, Koo AJK, Gao X, Jayanty S, Thines B, Jones AD, Howe GA (2008) Regulation and function of Arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiol 146 952–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos M, Van Oosten VR, Van Poecke RM, Van Pelt JA, Pozo MJ, Mueller MJ, Buchala AJ, Metraux JP, Van Loon LC, Dicke M, et al (2005) Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol Plant Microbe Interact 18 923–937 [DOI] [PubMed] [Google Scholar]
- Devoto A, Ellis C, Magusin A, Chang HS, Chilcott C, Zhu T, Turner JG (2005) Expression profiling reveals COI1 to be a key regulator of genes involved in wound- and methyl JA-induced secondary metabolism, defence, and hormone interactions. Plant Mol Biol 58 497–513 [DOI] [PubMed] [Google Scholar]
- Dombrecht B, Xue GP, Sprague SJ, Kirkegaard JA, Ross JJ, Reid JB, Fitt GP, Sewelam N, Schenk PM, Manners JM, et al (2007) MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19 2225–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis C, Karafyllidis I, Wasternack C, Turner JG (2002) The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell 14 1557–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer EE (2007) Jasmonate perception machines. Nature 448 659–660 [DOI] [PubMed] [Google Scholar]
- Feng S, Ma L, Wang X, Xie D, Dinesh-Kumar SP, Wei N, Deng XW (2003) The COP9 signalosome interacts physically with SCF COI1 and modulates jasmonate responses. Plant Cell 15 1083–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys B, Benedetti CE, Penfold CN, Turner JG (1994) Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl JA, and resistant to a bacterial pathogen. Plant Cell 6 751–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Muskett PR, Chuang HW, Parker JE (2003) Arabidopsis SGT1b is required for SCF(TIR1)-mediated auxin response. Plant Cell 15 1310–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill E, Christmann A (2007) Botany. A plant receptor with a big family. Science 315 1676–1677 [DOI] [PubMed] [Google Scholar]
- Hata AN, Breyer RM (2004) Pharmacology and signaling of prostaglandin receptors: multiple roles in inflammation and immune modulation. Pharmacol Ther 103 147–166 [DOI] [PubMed] [Google Scholar]
- Katou S, Yoshioka H, Kawakita K, Rowland O, Jones JD, Mori H, Doke N (2005) Involvement of PPS3 phosphorylated by elicitor-responsive mitogen-activated protein kinases in the regulation of plant cell death. Plant Physiol 139 1914–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie-Berry N, Joardar V, Street IH, Kunkel BN (2006) The Arabidopsis thaliana JASMONATE INSENSITIVE 1 gene is required for suppression of salicylic acid-dependent defenses during infection by Pseudomonas syringae. Mol Plant Microbe Interact 19 789–800 [DOI] [PubMed] [Google Scholar]
- Li J, Brader G, Palva ET (2004. a) The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16 319–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Zhao Y, McCaig BC, Wingerd BA, Wang J, Whalon ME, Pichersky E, Howe GA (2004. b) The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell 16 126–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O, Chico JM, Sánchez-Serrano JJ, Solano R (2004) JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16 1938–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O, Piqueras R, Sanchez-Serrano JJ, Solano R (2003) ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15 165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandaokar A, Thines B, Shin B, Lange BM, Choi G, Koo YJ, Yoo YJ, Choi YD, Choi G, Browse J (2006) Transcriptional regulators of stamen development in Arabidopsis identified by transcriptional profiling. Plant J 46 984–1008 [DOI] [PubMed] [Google Scholar]
- Mao P, Duan M, Wei C, Li Y (2007) WRKY62 transcription factor acts downstream of cytosolic NPR1 and negatively regulates jasmonate-responsive gene expression. Plant Cell Physiol 48 833–842 [DOI] [PubMed] [Google Scholar]
- Miao Y, Zentgraf U (2007) The antagonist function of Arabidopsis WRKY53 and ESR/ESP in leaf senescence is modulated by the jasmonic and salicylic acid equilibrium. Plant Cell 19 819–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munemasa S, Oda K, Watanabe-Sugimoto M, Nakamura Y, Shimoishi Y, Murata Y (2007) The coronatine-insensitive 1 mutation reveals the hormonal signaling interaction between abscisic acid and methyl jasmonate in Arabidopsis guard cells. Specific impairment of ion channel activation and second messenger production. Plant Physiol 143 1398–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mur LA, Kenton P, Atzorn R, Miersch O, Wasternack C (2006) The outcomes of concentration-specific interactions between salicylate and jasmonate signaling include synergy, antagonism, and oxidative stress leading to cell death. Plant Physiol 140 249–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal P, Ellis CM, Weber H, Ploense SE, Barkawi LS, Guilfoyle TJ, Hagen G, Alonso JM, Cohen JD, Farmer EE, et al (2005) Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 132 4107–4118 [DOI] [PubMed] [Google Scholar]
- Ndamukong I, Abdallat AA, Thurow C, Fode B, Zander M, Weigel R, Gatz C (2007) SA-inducible Arabidopsis glutaredoxin interacts with TGA factors and suppresses jasmonate-responsive PDF1.2 transcription. Plant J 50 128–139 [DOI] [PubMed] [Google Scholar]
- Overmyer K, Brosché M, Kangasjärvi J (2003) Reactive oxygen species and hormonal control of cell death. Trends Plant Sci 8 335–342 [DOI] [PubMed] [Google Scholar]
- Quint M, Ito H, Zhang W, Gray WM (2005) Characterization of a novel temperature-sensitive allele of the CUL1/AXR6 subunit of SCF ubiquitin-ligases. Plant J 43 371–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry G, Estelle M (2006) Auxin receptors: a new role for F-box proteins. Curr Opin Cell Biol 18 152–156 [DOI] [PubMed] [Google Scholar]
- Paschold A, Halitschke R, Baldwin IT (2007) Co(i)-ordinating defenses: NaCOI1 mediates herbivore- induced resistance in Nicotiana attenuata and reveals the role of herbivore movement in avoiding defenses. Plant J 51 79–91 [DOI] [PubMed] [Google Scholar]
- Penninckx IA, Thomma BP, Buchala A, Métraux JP, Broekaert WF (1998) Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10 2103–2113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen M, Brodersen P, Naested H, Andreasson E, Lindhart U, Johansen B, Nielsen HB, Lacy M, Austin MJ, Parker JE, et al (2000) Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell 103 1111–1120 [DOI] [PubMed] [Google Scholar]
- Rayapuram C, Baldwin IT (2007) Increased SA in NPR1-silenced plants antagonizes jasmonate and jasmonate-dependent direct and indirect defenses in herbivore-attacked Nicotiana attenuata in nature. Plant J 52 700–715 [DOI] [PubMed] [Google Scholar]
- Ren C, Pan J, Peng W, Genschik P, Hobbie L, Hellmann H, Estelle M, Gao B, Peng J, Sun C, et al (2005) Point mutations in Arabidopsis Cullin1 reveal its essential role in jasmonate response. Plant J 42 514–524 [DOI] [PubMed] [Google Scholar]
- Schwechheimer C, Serino G, Deng XW (2002) Multiple ubiquitin ligase-mediated processes require COP9 signalosome and AXR1 function. Plant Cell 14 2553–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweighofer A, Kazanaviciute V, Scheikl E, Teige M, Doczi R, Hirt H, Schwanninger M, Kant M, Schuurink R, Mauch F, et al (2007) The PP2C-type phosphatase AP2C1, which negatively regulates MPK4 and MPK6, modulates innate immunity, jasmonic acid, and ethylene levels in Arabidopsis. Plant Cell 19 2213–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikata M, Takemura M, Yokota A, Kohchi T (2003) Arabidopsis ZIM, a plant-specific GATA factor, can function as a transcriptional activator. Biosci Biotechnol Biochem 67 2495–2497 [DOI] [PubMed] [Google Scholar]
- Spoel SH, Johnson JS, Dong X (2007) Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proc Natl Acad Sci USA 104 18842–18847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel SH, Koornneef A, Claessens SM, Korzelius JP, Van Pelt JA, Mueller MJ, Buchala AJ, Metraux JP, Brown R, Kazan K, et al (2003) NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 15 760–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley D (2006) Prostaglandins and other eicosanoids in insects: biological significance. Annu Rev Entomol 51 25–44 [DOI] [PubMed] [Google Scholar]
- Staswick PE, Tiryaki I (2004) The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell 16 2117–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Callis J (2007) Ubiquitin ligases mediate growth and development by promoting protein death. Curr Opin Plant Biol 10 624–632 [DOI] [PubMed] [Google Scholar]
- Takahashi F, Yoshida R, Ichimura K, Mizoguchi T, Seo S, Yonezawa M, Maruyama K, Yamaguchi-Shinozaki K, Shinozaki K (2007) The mitogen-activated protein kinase cascade MKK3-MPK6 is an important part of the jasmonate signal transduction pathway in Arabidopsis. Plant Cell 19 805–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taki N, Sasaki-Sekimoto Y, Obayashi T, Kikuta A, Kobayashi K, Ainai T, Yagi K, Sakurai N, Suzuki H, Masuda T, et al (2005) 12-Oxo-phytodienoic acid triggers expression of a distinct set of genes and plays a role in wound-induced gene expression in Arabidopsis. Plant Physiol 139 1268–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J (2007) JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signaling. Nature 448 661–665 [DOI] [PubMed] [Google Scholar]
- Tiryaki I, Staswick PE (2002) An Arabidopsis mutant defective in jasmonate response is allelic to the auxin-signaling mutant axr1. Plant Physiol 130 887–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme B, Grunewald W, Bateman A, Kohchi T, Gheysen G (2007) The tify family previously known as ZIM. Trends Plant Sci 12 239–244 [DOI] [PubMed] [Google Scholar]
- Vick BA, Zimmerman DC (1984) Biosynthesis of jasmonic acid by several plant species. Plant Physiol 75 458–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Amornsiripanitch N, Dong X (2006) A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog 2 e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Allmann S, Wu J, Baldwin IT (2008) Comparisons of LIPOXYGENASE3- and JASMONATE-RESISTANT4/6-silenced plants reveal that jasmonic acid and jasmonic acid-amino acid conjugates play different roles in herbivore resistance of Nicotiana attenuata. Plant Physiol 146 904–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Dai L, Jiang Z, Peng W, Zhang L, Wang G, Xie D (2005) GmCOI1, a soybean F-box protein gene, shows ability to mediate jasmonate-regulated plant defense and fertility in Arabidopsis. Mol Plant Microbe Interact 18 1285–1295 [DOI] [PubMed] [Google Scholar]
- Wasternack C (2007) Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann Bot (Lond) 100 681–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward AW, Bartel B (2005) Auxin: regulation, action, and interaction. Ann Bot (Lond) 95 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Wang L, Baldwin IT (2008) Methyl jasmonate-elicited herbivore resistance: Does MeJA function as a signal without being hydrolyzed to JA? Planta (in press) [DOI] [PMC free article] [PubMed]
- Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG (1998) COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280 1091–1094 [DOI] [PubMed] [Google Scholar]
- Yadav V, Mallappa C, Gangappa SN, Bhatia S, Chattopadhyay S (2005) A basic helix-loop-helix transcription factor in Arabidopsis, MYC2, acts as a repressor of blue light-mediated photomorphogenic growth. Plant Cell 17 1953–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Stolz S, Chételat A, Reymond P, Pagni M, Dubugnon L, Farmer EE (2007) A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 19 2470–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Q, Li CB, Zheng W, Wu X, Zhao J, Zhou G, Jiang H, Sun J, Lou Y, Li C (2007) Phytochrome chromophore deficiency leads to overproduction of jasmonic acid and elevated expression of jasmonate-responsive genes in Arabidopsis. Plant Cell Physiol 48 1061–1071 [DOI] [PubMed] [Google Scholar]