Abstract
In traditional mutant screening approaches, genetic variants are tested for one or a small number of phenotypes. Once bona fide variants are identified, they are typically subjected to a limited number of secondary phenotypic screens. Although this approach is excellent at finding genes involved in specific biological processes, the lack of wide and systematic interrogation of phenotype limits the ability to detect broader syndromes and connections between genes and phenotypes. It could also prevent detection of the primary phenotype of a mutant. As part of a systems biology approach to understand plastid function, large numbers of Arabidopsis thaliana homozygous T-DNA lines are being screened with parallel morphological, physiological, and chemical phenotypic assays (www.plastid.msu.edu). To refine our approaches and validate the use of this high-throughput screening approach for understanding gene function and functional networks, approximately 100 wild-type plants and 13 known mutants representing a variety of phenotypes were analyzed by a broad range of assays including metabolite profiling, morphological analysis, and chlorophyll fluorescence kinetics. Data analysis using a variety of statistical approaches showed that such industrial approaches can reliably identify plant mutant phenotypes. More significantly, the study uncovered previously unreported phenotypes for these well-characterized mutants and unexpected associations between different physiological processes, demonstrating that this approach has strong advantages over traditional mutant screening approaches. Analysis of wild-type plants revealed hundreds of statistically robust phenotypic correlations, including metabolites that are not known to share direct biosynthetic origins, raising the possibility that these metabolic pathways have closer relationships than is commonly suspected.
Identification and analysis of mutants has played an important role in understanding biological processes of all types and in a wide variety of organisms. Traditionally this approach involves screening through large numbers of individuals for the small subset that have a change in a specific class of phenotype. A common approach is to use visual identification of variants with altered morphology under standard conditions (Bowman et al., 1989; Pyke and Leech, 1991), or following growth under altered environment (Glazebrook et al., 1996; Landry et al., 1997). Mutant screens can also be conducted using more specific molecular phenotypic outputs, ranging from changes in expression of specific genes (Susek et al., 1993) to direct analysis of metabolites (Benning, 2004; Jander et al., 2004; Valentin et al., 2006).
Once mutants are identified from a narrow screen detailed studies typically are performed to reveal secondary phenotypes. This deeper analysis is useful for several reasons. First, it can separate mutants into different classes and suggest novel relationships between the genes responsible for the phenotypic traits. Second, these studies can lead to a deeper understanding of the gene(s) responsible for the first phenotype discovered, and can reveal the underlying mechanism for the original phenotype (Conklin et al., 1996). Third, knowledge of secondary phenotypes can be useful in more rapidly identifying additional related mutants and genes and help to generate a complete understanding of a complex physiological trait or pathway (Conklin et al., 1999, 2000, 2006; Laing et al., 2007; Linster et al., 2007).
Until recently, mutant identification was performed either by ‘forward’ or ‘reverse’ genetic analysis (Alonso and Ecker, 2006). Forward genetics is the traditional approach where groups of randomly generated mutants (often at saturating mutational density; Jander et al., 2003) are screened based on their phenotype, and the gene responsible for the phenotype is then identified from the mutant (Jander et al., 2002). A strong advantage of forward genetics is that no prior assumptions need be made about the types of mutant genes that would generate the phenotype, making this unbiased approach very useful in identifying roles for genes of previously unknown function. In reverse genetics, mutants in specific genes (McCallum et al., 2000; Alonso et al., 2003) are analyzed, typically with a limited number of phenotypic assays. This approach allows more facile association of mutant phenotype with the affected gene and offers the possibility that a broader array of phenotypes can be run against the mutants than in a forward genetics screen (Lahner et al., 2003; Messerli et al., 2007).
As biology moves increasingly away from reductionism to systems thinking, there are several reasons why one phenotype or one gene/gene family at a time reverse genetic approaches hamper creation of large and durable genetic data sets. First, a limited number of genes are tested and phenotypes assayed in any given study, and protocols for screens are rarely consistent within or across laboratory groups. Second, the lack of common germplasm across different studies hampers comparisons. Finally, the tried and true approaches to data analysis and presentation in published articles, on laboratory Web sites, and community databases, with inconsistent descriptions of experiments and other metadata, make it difficult to discover all relevant data sets and to mine the data once discovered.
With the sequencing of an increasing number of plant genomes, accurately and efficiently assessing the function of the tens of thousands of genes that are annotated of unknown function or whose annotation is based upon similarity to genes from other organisms becomes an increasingly high priority. Tools for genome-wide analysis of mRNA and proteins have advanced very rapidly in recent years, enabling facile placement of genes into regulatory networks (Li et al., 2004; Schmid et al., 2005). However, changes in mRNA expression often do not accurately predict regulation of protein activity (Gibon et al., 2004; Wakao and Benning, 2005), metabolites (Kaplan et al., 2007), or the functional importance of those genes (Giaever et al., 2002). As a result, achieving high-confidence predictions of complex biological networks necessary for a systems understanding (Sweetlove et al., 2003) will require large-scale analysis of gene function through high-throughput mutant analysis.
Changes in technology are creating new opportunities to perform systematic phenotypic studies. Eukaryotic model organisms offer an increasing number of mutants defective in known genes identified through classical genetic screening and collections of sequenced insertion mutants (Winzeler et al., 1999; Alonso et al., 2003) or high-throughput gene-silencing approaches (Sönnichsen et al., 2005; Schwab et al., 2006). Software improvements permit rapid creation of laboratory information management systems, allowing large numbers of samples to be processed with minimal tracking error. Screening a large and enduring collection of mutant germplasm with many phenotypic assays would also permit the detection of syndromes of mutant phenotypes and allow the detection of genetic networks (Roessner et al., 2001; Schauer et al., 2006; Messerli et al., 2007).
We describe a pilot study performed to create a high-throughput and parallel-mutant screening and analysis pipeline (www.plastid.msu.edu). This study employed approximately 100 wild-type Arabidopsis (Arabidopsis thaliana) plants and three to six replicates each of 13 previously characterized mutants (Table I). These plants were analyzed using 10 phenotypic screens, many of which provided multiple phenotypic outputs (for example, a liquid chromatography-tandem mass spectrometry [LC-MS/MS] assay that captured data for 25 protein amino acids and related compounds), for a total of 85 data points per plant line. Analysis of the data permitted assessment of phenotypic variability within a genotype and evaluation of statistical and data display methods. It also revealed unexpected phenotypic signatures and relationships for the characterized mutants, which would not have been detected if fewer mutants and phenotypic characteristics were assessed.
Table I.
Arabidopsis mutants used in this study
| Genotype | Gene Locus | Ecotype | Mutagen | Annotation | Published Phenotype | References |
|---|---|---|---|---|---|---|
| 5-fcl | At5g13050 | Col | T-DNA | 5-Formyltetrahydrofolate cycloligase (EC 6.3.3.2) | High leaf Gly, Ser | Goyer et al., 2005 |
| arc10 | At5g55280 | Ws | T-DNA | FtsZ1 allele, tubulin-like protein | Reduced number of larger chloroplasts in mature leaf cells | Yoder et al., 2007 |
| arc12 | At1g69390 | Col | X-ray | Arabidopsis homolog of bacterial MinE1 | Reduced number of larger chloroplasts in mature leaf mesophyll cells | Glynn et al., 2007 |
| ats1-1a, act1-1 | At1g32200 | Col | EMS | Glycerol-3-P acyltransferase (EC 2.3.1.15) | Low 16:3 and low overall ratio of C16-C18 chains | Kunst et al., 1988; Xu et al., 2006 |
| dpe2-1 | At2g40840 | Ws | T-DNA | Disproportionating enzyme 2 (EC 2.4.1.25) | Excess leaf starch | Lu et al., 2004 |
| fatb-ko | At1g08510 | Ws | T-DNA | FATB acyl-carrier protein thioesterase (EC 3.1.2.14) | Low saturated fatty acids in leaves | Bonaventure et al., 2003 |
| lkr-sdh | At4g33150 | Ws | T-DNA | Lys ketoglutarate reductase (EC 1.5.1.8)/saccharopine dehydrogenase (EC 1.5.1.9) | High seed Lys | Zhu et al., 2001 |
| npq1-2 | At1g08550 | Col | EMS | Violaxanthin deepoxidase (EC 1.10.99.3) | Low NPQ | Niyogi et al., 1998 |
| pig1-1 | Phe insensitive growth | Ws | EMS | High total plantlet amino acid content on agar plates | Voll et al., 2004 | |
| sex1-1 | At1g10760 | Col | EMS | Glucan water dikinase 1(EC 2.7.9.4) | Excess leaf starch | Yu et al., 2001 |
| sex4-5 | At3g52180 | Col | T-DNA | Laforin-like carbohydrate phosphatase | Excess leaf starch | Niittylä et al., 2006; Sokolov 2006; Gentry et al., 2007 |
| tha1-1 | At1g08630 | Col | EMS | Thr aldolase 1 (EC 4.1.2.5) | High seed Thr | Jander et al., 2004 |
| tt7-1b | At5g07990 | Ler | EMS | Flavonoid 3′-hydroxylase (EC 1.14.13.21) | Pale brown seeds | Schoenbohm et al., 2000 |
| tt7-3 | At5g07990 | Col | T-DNA | Flavonoid 3′-hydroxylase (EC 1.14.13.21) | Pale brown seeds | Abrahams et al., 2002; Salaita et al., 2005 |
The act1-1 mutant was biochemically and physiologically characterized by Kunst et al. (1988) and was later renamed ats1-1 (Xu et al., 2006).
The tt7-1 mutant was not included in the pilot study; it was only used to investigate the association between the lack of tannins in the seed coat and excess seed coat starch.
RESULTS
Analysis of Mutants and Wild Type with High-Throughput Screens
Because the long-term goal of the project is to identify functions for genes involved in chloroplast physiology, the project incorporated a variety of efficient phenotypic assays that interrogate chloroplast function as well as the general growth and development of the plant from our laboratories or the literature. Chloroplast morphology and chlorophyll fluorescence screens were included as direct measures of the development and function of the chloroplast. Three classes of metabolites were assayed because they include pathways operating entirely or partly within the plastid: qualitative assays were performed for leaf and seed starch whereas quantitative assays were done for leaf and seed amino acids and leaf fatty acids. Finally, vegetative-stage plant morphology, seed morphology, and a quantitative assay for seed total carbon (C) and nitrogen (N) composition were chosen to assess the overall health of the plants and to look for correlations between leaf and seed physiology.
The phenotypic assays were adapted from established methods to a pipeline process, with the goal of minimizing variability in growth conditions and assays, and discovering a wide variety of relevant morphological and physiological traits. Leaf tissues were harvested in a set process, with each assay (morning starch, amino acids, fatty acids, etc.) sampled in the identical order, on the equivalent leaf (judged by order of leaf emergence) starting at the same time of day after the same number of days of growth. Biological replicates of mutants were grown in separate flats along with large numbers of each wild-type ecotype. A laboratory information management system was designed to increase the speed and accuracy of each planting and harvesting step. Whenever possible, phenotypic data were captured directly to the database. All sample collection and processing was performed with anonymous bar code identifiers, and the technicians who recorded the data did not know the genotype of the plants.
One goal of the pilot study was to assess how well the phenotypic assays were working in the relatively high-throughput environment of the project. Three related issues were addressed: the ability of the assays to detect phenotypic changes, the variability of the assays, and the accuracy of plant and sample tracking. To this end, eight known mutants of ecotype Columbia of Arabidopsis (Col) and five known mutants of ecotype Wassilewskija of Arabidopsis (Ws; Table I) were planted in 6-fold replication along with 114 wild-type plants (72 Col and 42 Ws ecotypes). Seeds were harvested from the plants that survived to maturity and these were assayed for seed phenotypes and plants were grown to assay vegetative traits. The majority of the quantitative data from Col and Ws wild-type samples were found to be normally distributed (Shapiro-Wilk test, p > 0.01; Shapiro and Wilk, 1965). Amino acids of low concentration (for example, Cys) and amino acids with poor ionization during HPLC-MS/MS (for example, Gly; Gu et al., 2007) tended not to be normally distributed. The effect of detection limit on the distribution of metabolite concentration also applies to the fatty acid assay. Fatty acids 14:0 and 18:1d11 are not abundant and their concentrations in Col wild-type plants are not normally distributed.
As detailed below, in every case relevant phenotypes described in the literature were identified in this blind study (Table I), validating that the mutants were correct and that our assays can accurately track large numbers of samples and discover a wide variety of targeted phenotypes. Dunnett's test, a method developed for multiple comparisons involving a control (Dunnett, 1955), was used to compare means of the mutants and their corresponding wild type (Bucciarelli et al., 2006). Differences between a mutant and the wild type were considered statistically significant when the p value was <0.05 in Dunnett's test, unless otherwise indicated.
The data on amino acids in leaves and seeds of the previously described mutants confirmed that the LC-MS/MS assay accurately reported levels of these metabolites (Tables II–V; Supplemental Tables S1–S4). The 5-fcl mutant, defective in folate metabolism, had substantially higher Gly content (6- to 10-fold increase) with an approximately 2-fold increase of Ser content in leaves (Table II; Supplemental Table S1), as previously reported (Goyer et al., 2005). The Lys ketoglutarate reductase/saccharopine dehydrogenase knock-out mutant (lkr-sdh), defective in seed Lys catabolism, had significantly higher seed Lys (Table V; Supplemental Table S4), as described (Zhu et al., 2001). The leaf total free amino acid content (nmol/g fresh weight [FW]) was somewhat higher in the pig1-1 mutant (Student's t test, p < 0.05; Supplemental Table S2), as reported by Voll et al. (2004). Finally, Thr aldolase-deficient tha1-1 mutant seeds had >12-fold higher mol % Thr content (Table IV), as described in Jander et al. (2004).
Table II.
Mol % of amino acids in leaves of Col wild type and mutants
The asterisk indicates a significant difference of mol % of amino acid between the mutant and Col wild type (Dunnett's test, *, p < 0.05; **, p < 0.01; ***, p < 0.001).
| Amino Acida | Col Wild Type | 5-fcl | arc12 | ats1-1 | npq1-2 | sex1-1 | sex4-5 | tha1-1 | tt7-3 |
|---|---|---|---|---|---|---|---|---|---|
| Ala | 12 ± 0.4 | 10 ± 1 | 10 ± 0.2 | 9.4 ± 1.1 | 12 ± 1 | 9.6 ± 1.0 | 9.3 ± 1.1 | 11 ± 2 | 17 ± 1** |
| Arg | 0.27 ± 0.03 | 0.15 ± 0.02 | 0.15 ± 0.01 | 0.15 ± 0.02 | 0.26 ± 0.05 | 0.31 ± 0.04 | 0.09 ± 0.01 | 0.14 ± 0.01 | 0.39 ± 0.08 |
| Asn | 0.70 ± 0.01 | 0.62 ± 0.06 | 0.78 ± 0.04 | 0.60 ± 0.03 | 0.68 ± 0.02 | 2.2 ± 0.4*** | 0.72 ± 0.06 | 0.72 ± 0.05 | 0.66 ± 0.03 |
| Asp | 13 ± 0.2 | 14 ± 1 | 14 ± 0.5 | 16 ± 1** | 13 ± 1 | 17 ± 1*** | 14 ± 1 | 16 ± 1.0* | 11 ± 1 |
| Cys | 0.041 ± 0.001 | 0.039 ± 0.003 | 0.040 ± 0.005 | 0.037 ± 0.004 | 0.039 ± 0.003 | 0.054 ± 0.004 | 0.056 ± 0.012 | 0.044 ± 0.007 | 0.042 ± 0.005 |
| Gln | 8.0 ± 0.2 | 7.4 ± 0.4 | 7.5 ± 0.3 | 6.2 ± 0.3** | 8.5 ± 0.3 | 7.8 ± 0.7 | 6.0 ± 0.4** | 6.8 ± 0.4 | 8.5 ± 0.2 |
| Glu | 41 ± 1 | 33 ± 2** | 42 ± 1 | 45 ± 1 | 40 ± 2 | 36 ± 2 | 41 ± 2 | 41 ± 1 | 37 ± 1 |
| Gly | 1.8 ± 0.03 | 11 ± 3*** | 1.9 ± 0.1 | 1.9 ± 0.1 | 1.9 ± 0.1 | 2.8 ± 0.2 | 2.1 ± 0.1 | 1.9 ± 0.1 | 2.0 ± 0.1 |
| His | 0.54 ± 0.02 | 0.32 ± 0.05*** | 0.51 ± 0.02 | 0.44 ± 0.03 | 0.54 ± 0.03 | 0.49 ± 0.08 | 0.28 ± 0.02*** | 0.45 ± 0.02 | 0.46 ± 0.03 |
| Ile | 0.22 ± 0.01 | 0.21 ± 0.02 | 0.22 ± 0.01 | 0.26 ± 0.02 | 0.21 ± 0.05 | 0.33 ± 0.12 | 0.22 ± 0.01 | 0.20 ± 0.01 | 0.16 ± 0.02 |
| Leu | 0.46 ± 0.03 | 0.34 ± 0.07 | 0.33 ± 0.03 | 0.45 ± 0.08 | 0.46 ± 0.09 | 0.35 ± 0.12 | 0.25 ± 0.02 | 0.32 ± 0.03 | 0.44 ± 0.04 |
| Lys | 0.61 ± 0.01 | 0.47 ± 0.05 | 0.51 ± 0.04 | 0.62 ± 0.09 | 0.63 ± 0.05 | 0.52 ± 0.11 | 0.40 ± 0.02** | 0.52 ± 0.05 | 0.61 ± 0.03 |
| Met | 0.31 ± 0.01 | 0.19 ± 0.03*** | 0.28 ± 0.03 | 0.24 ± 0.02 | 0.34 ± 0.01 | 0.13 ± 0.01*** | 0.17 ± 0.01*** | 0.26 ± 0.03 | 0.33 ± 0.02 |
| Phe | 0.91 ± 0.03 | 0.78 ± 0.05 | 0.75 ± 0.05 | 0.83 ± 0.03 | 0.99 ± 0.06 | 0.86 ± 0.12 | 0.78 ± 0.03 | 0.55 ± 0.03** | 1.2 ± 0.1* |
| Pro | 2.4 ± 0.2 | 2.0 ± 0.1 | 2.1 ± 0.1 | 2.1 ± 0.2 | 2.3 ± 0.1 | 1.9 ± 0.2 | 2.2 ± 0.6 | 1.9 ± 0.1 | 2.6 ± 0.1 |
| Ser | 5.0 ± 0.2 | 8.7 ± 0.5*** | 5.3 ± 0.3 | 4.8 ± 0.5 | 5.2 ± 0.8 | 7.8 ± 1.2** | 7.6 ± 2.1* | 6.0 ± 0.5 | 4.7 ± 0.1 |
| Thr | 8.7 ± 0.2 | 8.2 ± 0.4 | 9.3 ± 0.5 | 7.9 ± 0.5 | 7.9 ± 0.5 | 9.4 ± 1.1 | 12 ± 2** | 9.2 ± 1.3 | 8.5 ± 0.6 |
| Trp | 0.10 ± 0.01 | 0.097 ± 0.018 | 0.086 ± 0.006 | 0.13 ± 0.01 | 0.11 ± 0.03 | 0.15 ± 0.05 | 0.11 ± 0.01 | 0.097 ± 0.012 | 0.078 ± 0.011 |
| Tyr | 0.17 ± 0.01 | 0.16 ± 0.02 | 0.15 ± 0.01 | 0.19 ± 0.02 | 0.19 ± 0.02 | 0.15 ± 0.03 | 0.12 ± 0.01 | 0.16 ± 0.02 | 0.18 ± 0.01 |
| Val | 1.3 ± 0.05 | 1.1 ± 0.1 | 1.2 ± 0.1 | 1.1 ± 0.1 | 1.3 ± 0.1 | 1.2 ± 0.3 | 1.0 ± 0.01 | 1.1 ± 0.1 | 1.6 ± 0.1 |
Data are presented as mean ± se (n = 3–6 for mutants, n = 71 for Col wild type). GABA, Ho-Ser, and Hyp were included in calculating the mol % of above amino acids.
Table III.
Mol % of amino acids in leaves of Ws wild type and mutants
The asterisk indicates a significant difference of mol % of amino acid between the mutant and Ws wild type (Dunnett's test, *, p < 0.05; **, p < 0.01; ***, p < 0.001).
| Amino Acida | Ws Wild Type | arc10 | dpe2-1 | fatb-ko | lkr-sdh | pig1-1 |
|---|---|---|---|---|---|---|
| Ala | 16 ± 0.5 | 16 ± 1 | 16 ± 1 | 8.2 ± 0.6*** | 16 ± 3 | 13 ± 1* |
| Arg | 0.85 ± 0.07 | 0.47 ± 0.12 | 0.38 ± 0.08 | 0.15 ± 0.03*** | 0.68 ± 0.35 | 0.49 ± 0.10 |
| Asn | 0.96 ± 0.02 | 0.87 ± 0.03 | 1.3 ± 0.1*** | 0.78 ± 0.07* | 1.1 ± 0.04 | 0.83 ± 0.04 |
| Asp | 12 ± 0.3 | 12 ± 0.4 | 8.9 ± 0.7* | 17 ± 1*** | 11 ± 1 | 13 ± 1 |
| Cys | 0.032 ± 0.002 | 0.036 ± 0.004 | 0.042 ± 0.007 | 0.045 ± 0.004* | 0.037 ± 0.009 | 0.040 ± 0.005 |
| Gln | 8.7 ± 0.3 | 8.0 ± 0.5 | 12 ± 1*** | 9.7 ± 0.7 | 8.8 ± 1.1 | 8.1 ± 0.5 |
| Glu | 35 ± 1 | 38 ± 2 | 31 ± 0.4 | 40 ± 1* | 34 ± 1 | 39 ± 2 |
| Gly | 2.1 ± 0.1 | 1.9 ± 0.2 | 6.5 ± 0.8*** | 2.0 ± 0.1 | 2.3 ± 0.4 | 2.0 ± 0.2 |
| His | 0.53 ± 0.03 | 0.41 ± 0.05 | 0.58 ± 0.05 | 0.44 ± 0.04 | 0.51 ± 0.10 | 0.71 ± 0.12 |
| Ile | 0.15 ± 0.02 | 0.14 ± 0.03 | 0.14 ± 0.03 | 0.20 ± 0.01 | 0.15 ± 0.05 | 0.20 ± 0.06 |
| Leu | 0.52 ± 0.04 | 0.43 ± 0.06 | 0.39 ± 0.04 | 0.31 ± 0.05 | 0.34 ± 0.05 | 0.77 ± 0.37 |
| Lys | 0.81 ± 0.04 | 0.68 ± 0.08 | 0.55 ± 0.05 | 0.51 ± 0.07* | 0.65 ± 0.10 | 0.83 ± 0.12 |
| Met | 0.43 ± 0.02 | 0.38 ± 0.05 | 0.33 ± 0.02 | 0.26 ± 0.02** | 0.35 ± 0.04 | 0.35 ± 0.02 |
| Phe | 1.2 ± 0.1 | 1.1 ± 0.1 | 2.0 ± 0.2*** | 0.73 ± 0.03* | 1.0 ± 0.1 | 1.3 ± 0.2 |
| Pro | 2.9 ± 0.2 | 2.4 ± 0.2 | 2.4 ± 0.1 | 2.1 ± 0.1 | 3.4 ± 0.8 | 3.1 ± 0.2 |
| Ser | 6.2 ± 0.2 | 6.2 ± 0.5 | 5.3 ± 0.3 | 7.0 ± 0.4 | 6.8 ± 1.2 | 4.2 ± 0.4** |
| Thr | 6.3 ± 0.2 | 7.5 ± 0.8 | 7.9 ± 0.6 | 7.7 ± 1.2 | 7.8 ± 1.7 | 8.2 ± 0.8 |
| Trp | 0.073 ± 0.005 | 0.083 ± 0.008 | 0.17 ± 0.02** | 0.084 ± 0.005 | 0.079 ± 0.016 | 0.14 ± 0.06* |
| Tyr | 0.24 ± 0.01 | 0.20 ± 0.02 | 0.25 ± 0.02 | 0.14 ± 0.02* | 0.19 ± 0.03 | 0.26 ± 0.07 |
| Val | 2.2 ± 0.1 | 1.8 ± 0.2 | 2.0 ± 0.1 | 0.96 ± 0.06*** | 2.0 ± 0.4 | 1.9 ± 0.4 |
Data are presented as mean ± se (n = 3–6 for mutants, n = 42 for Ws wild type). GABA, Ho-Ser, and Hyp were included in calculating the mol % of above amino acids.
Table IV.
Mol % of amino acids in seeds of Col wild type and mutants
The asterisk indicates a significant difference of amino acid content between the mutant and Col wild type (Dunnett's test, *, p < 0.05; **, p < 0.01; ***, p < 0.001).
| Amino Acida | Col Wild Type | 5-fcl | arc12 | ats1-1 | npq1-2 | sex1-1 | sex4-5 | tha1-1 | tt7-3 |
|---|---|---|---|---|---|---|---|---|---|
| Ala | 4.7 ± 0.1 | 1.6 ± 0.3*** | 5.1 ± 0.2 | 4.8 ± 0.3 | 4.8 ± 0.3 | 5.3 ± 0.2 | 5.4 ± 0.1 | 3.2 ± 0.2*** | 8.4 ± 0.8*** |
| Arg | 1.5 ± 0.1 | 2.7 ± 0.2** | 1.5 ± 0.1 | 1.6 ± 0.1 | 1.9 ± 0.2 | 1.7 ± 0.1 | 1.4 ± 0.1 | 1.5 ± 0.2 | 2.6 ± 0.3*** |
| Asn | 5.7 ± 0.1 | 1.3 ± 0.2*** | 6.3 ± 0.5 | 6.3 ± 0.7 | 5.5 ± 0.5 | 4.9 ± 0.2 | 4.7 ± 0.4 | 3.5 ± 0.3*** | 6.7 ± 1.0 |
| Asp | 9.0 ± 0.2 | 0.52 ± 0.03*** | 8.3 ± 0.3 | 8.0 ± 0.4 | 8.0 ± 0.9 | 7.9 ± 0.7 | 7.7 ± 0.8 | 3.4 ± 0.3*** | 6.3 ± 0.6*** |
| Cys | 0.043 ± 0.001 | 0.030 ± 0.004 | 0.040 ± 0.004 | 0.046 ± 0.005 | 0.041 ± 0.001 | 0.044 ± 0.004 | 0.048 ± 0.004 | 0.22 ± 0.01*** | 0.032 ± 0.002* |
| Gln | 2.9 ± 0.1 | 2.1 ± 0.3 | 2.7 ± 0.3 | 3.6 ± 0.8 | 3.4 ± 0.5 | 2.7 ± 0.2 | 2.9 ± 0.2 | 2.9 ± 0.5 | 3.0 ± 0.3 |
| Glu | 33 ± 1 | 4.5 ± 0.9*** | 34 ± 3 | 34 ± 4 | 29 ± 4 | 28 ± 2 | 32 ± 3 | 14 ± 1*** | 23 ± 2*** |
| Gly | 4.4 ± 0.1 | 41 ± 2*** | 4.3 ± 0.2 | 3.9 ± 0.1 | 4.2 ± 0.2 | 4.3 ± 0.1 | 3.9 ± 0.2 | 2.5 ± 0.2*** | 4.9 ± 0.4 |
| His | 1.6 ± 0.04 | 0.75 ± 0.07*** | 1.8 ± 0.2 | 1.8 ± 0.4 | 1.6 ± 0.04 | 2.0 ± 0.2 | 1.5 ± 0.1 | 1.5 ± 0.1 | 1.6 ± 0.1 |
| Ile | 2.0 ± 0.03 | 0.57 ± 0.05*** | 2.0 ± 0.1 | 2.2 ± 0.2 | 2.2 ± 0.3 | 2.1 ± 0.1 | 2.1 ± 0.1 | 1.3 ± 0.2*** | 3.0 ± 0.1*** |
| Leu | 2.6 ± 0.1 | 0.65 ± 0.05*** | 2.7 ± 0.2 | 2.7 ± 0.3 | 3.0 ± 0.5 | 2.9 ± 0.2 | 2.8 ± 0.1 | 1.7 ± 0.3* | 3.4 ± 0.1** |
| Lys | 1.4 ± 0.03 | 1.1 ± 0.2 | 1.5 ± 0.1 | 1.4 ± 0.2 | 1.7 ± 0.2* | 1.7 ± 0.1* | 1.5 ± 0.1 | 1.0 ± 0.05 | 1.6 ± 0.04 |
| Met | 0.52 ± 0.01 | 0.14 ± 0.01*** | 0.64 ± 0.03* | 0.58 ± 0.07 | 0.51 ± 0.06 | 0.52 ± 0.03 | 0.60 ± 0.05 | 0.28 ± 0.03*** | 0.57 ± 0.05 |
| Phe | 2.6 ± 0.03 | 0.74 ± 0.03*** | 2.7 ± 0.1 | 2.7 ± 0.1 | 2.9 ± 0.2 | 2.7 ± 0.1 | 2.7 ± 0.1 | 1.6 ± 0.1*** | 2.5 ± 0.1 |
| Pro | 12 ± 1 | 1.9 ± 0.1* | 9.8 ± 3.1 | 10 ± 4 | 14 ± 3 | 16 ± 3 | 13 ± 4 | 7.4 ± 2.6 | 13 ± 2 |
| Ser | 4.4 ± 0.1 | 36 ± 2*** | 5.0 ± 0.2 | 4.3 ± 0.1 | 4.5 ± 0.3 | 5.0 ± 0.4 | 5.1 ± 0.2 | 2.5 ± 0.1*** | 5.4 ± 0.2** |
| Thr | 3.8 ± 0.1 | 2.5 ± 0.1 | 4.2 ± 0.3 | 3.7 ± 0.4 | 4.0 ± 0.2 | 4.1 ± 0.2 | 3.9 ± 0.2 | 48 ± 3*** | 4.0 ± 0.3 |
| Trp | 2.6 ± 0.1 | 0.34 ± 0.01*** | 2.8 ± 0.1 | 2.4 ± 0.1 | 2.6 ± 0.2 | 2.8 ± 0.2 | 3.6 ± 0.3*** | 1.2 ± 0.1*** | 3.1 ± 0.3 |
| Tyr | 0.73 ± 0.01 | 0.20 ± 0.01*** | 0.82 ± 0.04 | 0.79 ± 0.02 | 0.81 ± 0.03 | 0.78 ± 0.01 | 0.77 ± 0.05 | 0.45 ± 0.03*** | 0.80 ± 0.04 |
| Val | 3.3 ± 0.1 | 1.1 ± 0.1*** | 3.1 ± 0.1 | 3.5 ± 0.1 | 3.8 ± 0.5 | 4.0 ± 0.2* | 3.2 ± 0.2 | 1.9 ± 0.4*** | 5.1 ± 0.2*** |
Data are presented as mean ± se (n = 3–6 for mutants, n = 67 for Col wild type). GABA, Ho-Ser, and Hyp were included in calculating the mol % of above amino acids.
Table V.
Mol % of amino acids in seeds of Ws wild type and mutants
The asterisk indicates a significant difference of amino acid content between the mutant and Ws wild type (Dunnett's test, *, p < 0.05; **, p < 0.01; ***, p < 0.001).
| Amino Acida | Ws Wild Type | arc10 | dpe2-1 | fatb-ko | lkr-sdh | pig1-1 |
|---|---|---|---|---|---|---|
| Ala | 5.7 ± 0.1 | 5.5 ± 0.2 | 7.0 ± 0.5* | 5.3 ± 0.6 | 6.9 ± 0.5 | 3.7 ± 0.5*** |
| Arg | 1.6 ± 0.1 | 1.7 ± 0.1 | 1.7 ± 0.2 | 2.2 ± 0.1** | 1.8 ± 0.2 | 1.2 ± 0.1 |
| Asn | 4.4 ± 0.1 | 4.3 ± 0.2 | 3.6 ± 0.3 | 5.9 ± 0.4*** | 5.0 ± 0.3 | 5.4 ± 0.3** |
| Asp | 9.1 ± 0.2 | 8.2 ± 0.4 | 8.0 ± 0.7 | 6.5 ± 0.3*** | 7.5 ± 0.4 | 6.8 ± 0.6** |
| Cys | 0.052 ± 0.003 | 0.058 ± 0.005 | 0.045 ± 0.004 | 0.050 ± 0.003 | 0.049 ± 0.003 | 0.046 ± 0.004 |
| Gln | 2.9 ± 0.2 | 2.9 ± 0.5 | 5.0 ± 1.3 | 2.5 ± 0.1 | 2.7 ± 0.3 | 10 ± 2*** |
| Glu | 36 ± 1 | 35 ± 2 | 32 ± 1 | 31 ± 2 | 28 ± 1* | 23 ± 1*** |
| Gly | 4.1 ± 0.1 | 4.0 ± 0.3 | 3.8 ± 0.5 | 4.2 ± 0.2 | 4.7 ± 0.4 | 4.4 ± 0.4 |
| His | 1.2 ± 0.02 | 1.2 ± 0.1 | 1.3 ± 0.1 | 1.8 ± 0.1*** | 1.4 ± 0.03 | 3.0 ± 0.3*** |
| Ile | 2.4 ± 0.04 | 2.6 ± 0.1 | 2.3 ± 0.1 | 2.3 ± 0.1 | 2.7 ± 0.1 | 6.0 ± 0.4*** |
| Leu | 2.8 ± 0.1 | 3.0 ± 0.2 | 3.4 ± 0.2 | 2.9 ± 0.2 | 3.5 ± 0.2* | 4.8 ± 0.2*** |
| Lys | 1.4 ± 0.04 | 1.4 ± 0.1 | 1.7 ± 0.1 | 1.7 ± 0.1 | 3.5 ± 0.04*** | 1.4 ± 0.1 |
| Met | 0.60 ± 0.02 | 0.65 ± 0.03 | 0.58 ± 0.05 | 0.68 ± 0.04 | 0.59 ± 0.07 | 0.39 ± 0.06*** |
| Phe | 2.8 ± 0.03 | 2.9 ± 0.1 | 3.1 ± 0.2 | 3.2 ± 0.1** | 3.0 ± 0.1 | 2.4 ± 0.2** |
| Pro | 5.7 ± 0.9 | 4.8 ± 0.9 | 4.1 ± 0.3 | 11 ± 3 | 7.4 ± 1.4 | 8.0 ± 0.8 |
| Ser | 5.0 ± 0.1 | 4.9 ± 0.2 | 4.8 ± 0.5 | 5.7 ± 0.4* | 5.6 ± 0.1 | 3.6 ± 0.2*** |
| Thr | 3.3 ± 0.1 | 3.1 ± 0.1 | 3.8 ± 0.3 | 3.4 ± 0.2 | 3.8 ± 0.2 | 5.3 ± 0.4*** |
| Trp | 6.2 ± 0.2 | 8.8 ± 0.3*** | 7.9 ± 0.7 | 5.4 ± 0.4 | 6.4 ± 0.7 | 1.4 ± 0.1*** |
| Tyr | 0.95 ± 0.01 | 0.99 ± 0.03 | 1.1 ± 0.05 | 0.79 ± 0.03*** | 0.89 ± 0.01 | 1.0 ± 0.1 |
| Val | 3.5 ± 0.05 | 3.4 ± 0.1 | 3.6 ± 0.2 | 3.1 ± 0.03 | 3.5 ± 0.2 | 6.7 ± 0.8*** |
Data are presented as mean ± se (n = 3–6 for mutants, n = 33 for Ws wild type). GABA, Ho-Ser, and Hyp were included in calculating the mol % of above amino acids.
Leaf samples from the ats1-1 and fatb-ko mutants (deficient in glycerol-3-P acyltransferase and acyl-acyl carrier protein thioesterase, respectively) were used to validate the fatty acid screening method. In ats1-1 mutants both the mol % of 16:3 (carbons in chain:number of double bonds) and overall proportion of C16 (Cnumber of carbons) relative to C18 chains were significantly reduced (Tables VI and VII), as described previously (Kunst et al., 1988; Xu et al., 2006). The fatb-ko leaves had significantly higher mol % of the unsaturated fatty acids cis-16:1, 16:2, 18:1d9, 18:1d11, and 18:2, and significantly lower mol % of saturated fatty acids, 16:0 and 18:0 (all p < 0.001; Table VI), as reported by Bonaventure et al. (2003). The fatb-ko mutant also showed a strongly significant (p < 0.001) reduction in seed C/N ratio, consistent with the fatty acid biosynthetic defect in seeds (Bonaventure et al., 2003).
Table VI.
Mol % of fatty acids in wild-type and mutant leaves
The asterisk indicates a significant difference of mol % of fatty acid between the mutant and corresponding wild type (Col or Ws; Dunnett's test, *, p < 0.05; **, p < 0.01; ***, p < 0.001). Myristic acid (14:0) was included in calculating the mol % of fatty acids.
| Genotypea | 16:0 | cis-16:1 | trans- 16:1d3 | 16:2 | 16:3 | 18:0 | 18:1d9 | 18:1d11 | 18:2 | 18:2 Dicarboxylic Acid | 18:3 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Col wild type | 12.8 ± 0.1 | 0.55 ± 0.01 | 3.6 ± 0.05 | 0.88 ± 0.01 | 13.2 ± 0.1 | 0.96 ± 0.01 | 3.6 ± 0.03 | 0.26 ± 0.01 | 13.6 ± 0.1 | 1.05 ± 0.02 | 48.8 ± 0.1 |
| 5-fcl | 12.7 ± 0.2 | 0.71 ± 0.21* | 3.7 ± 0.2 | 0.82 ± 0.02 | 13.2 ± 0.2 | 0.94 ± 0.04 | 3.3 ± 0.1 | 0.35 ± 0.05** | 13.3 ± 0.2 | 1.15 ± 0.03 | 49.1 ± 0.3 |
| arc12 | 12.4 ± 0.1 | 0.49 ± 0.02 | 3.8 ± 0.1 | 0.79 ± 0.04 | 13.1 ± 0.2 | 0.92 ± 0.05 | 3.6 ± 0.2 | 0.23 ± 0.01 | 14.0 ± 0.3 | 1.02 ± 0.04 | 48.9 ± 0.4 |
| ats1-1 | 11.1 ± 0.1*** | 0.16 ± 0.01*** | 3.3 ± 0.1 | 0.18 ± 0.01*** | 0.63 ± 0.05*** | 1.14 ± 0.04** | 6.7 ± 0.3*** | 0.40 ± 0.05*** | 18.3 ± 0.2*** | 1.30 ± 0.04*** | 55.5 ± 0.6*** |
| npq1-2 | 12.4 ± 0.2 | 0.51 ± 0.03 | 3.7 ± 0.1 | 0.90 ± 0.03 | 13.5 ± 0.2 | 0.93 ± 0.01 | 3.5 ± 0.1 | 0.24 ± 0.02 | 13.5 ± 0.2 | 1.03 ± 0.04 | 49.0 ± 0.3 |
| sex1-1 | 12.7 ± 0.3 | 0.55 ± 0.02 | 3.7 ± 0.2 | 0.84 ± 0.05 | 13.6 ± 0.4 | 1.11 ± 0.11* | 3.9 ± 0.2 | 0.33 ± 0.04 | 16.1 ± 0.2*** | 0.98 ± 0.01 | 45.0 ± 0.8*** |
| sex4-5 | 13.0 ± 0.3 | 0.50 ± 0.02 | 3.8 ± 0.2 | 0.81 ± 0.03 | 13.2 ± 0.3 | 0.96 ± 0.03 | 3.6 ± 0.1 | 0.25 ± 0.02 | 14.3 ± 0.5* | 1.13 ± 0.05 | 47.3 ± 0.3** |
| tha1-1 | 13.4 ± 0.2 | 0.51 ± 0.02 | 3.0 ± 0.1* | 0.79 ± 0.03 | 11.8 ± 0.1*** | 1.21 ± 0.06*** | 3.7 ± 0.1 | 0.19 ± 0.005 | 14.6 ± 0.2*** | 1.16 ± 0.05 | 49.0 ± 0.4 |
| tt7-3 | 12.8 ± 0.2 | 0.67 ± 0.08 | 4.0 ± 0.2 | 0.96 ± 0.03 | 12.9 ± 0.2 | 0.84 ± 0.03 | 3.8 ± 0.1 | 0.24 ± 0.01 | 14.3 ± 0.2** | 1.10 ± 0.06 | 47.8 ± 0.3* |
| Ws wild type | 12.3 ± 0.1 | 0.50 ± 0.01 | 3.6 ± 0.1 | 0.64 ± 0.01 | 14.6 ± 0.1 | 0.95 ± 0.02 | 2.9 ± 0.1 | 0.30 ± 0.005 | 13.7 ± 0.1 | 0.76 ± 0.04 | 48.9 ± 0.1 |
| arc10 | 12.5 ± 0.2 | 0.48 ± 0.02 | 3.5 ± 0.2 | 0.57 ± 0.03 | 14.5 ± 0.3 | 0.93 ± 0.04 | 2.8 ± 0.2 | 0.31 ± 0.02 | 13.4 ± 0.2 | 0.74 ± 0.10 | 49.4 ± 0.4 |
| dpe2-1 | 12.4 ± 0.2 | 0.55 ± 0.02 | 4.4 ± 0.2** | 0.77 ± 0.03* | 13.0 ± 0.1*** | 1.07 ± 0.03* | 3.8 ± 0.1*** | 0.26 ± 0.01 | 14.7 ± 0.1*** | 1.01 ± 0.06 | 47.2 ± 0.3*** |
| fatb-ko | 8.4 ± 0.2*** | 1.05 ± 0.06*** | 3.3 ± 0.3 | 0.90 ± 0.04*** | 13.8 ± 0.2* | 0.46 ± 0.02*** | 7.4 ± 0.2*** | 0.63 ± 0.08*** | 19.7 ± 0.3*** | 0.97 ± 0.03 | 42.7 ± 0.3*** |
| lkr-sdh | 12.3 ± 0.1 | 0.47 ± 0.04 | 4.2 ± 0.4 | 0.59 ± 0.05 | 15.2 ± 0.3 | 0.98 ± 0.04 | 2.9 ± 0.1 | 0.30 ± 0.02 | 13.2 ± 0.3 | 0.70 ± 0.09 | 48.2 ± 0.7 |
| pig1-1 | 12.9 ± 0.1** | 0.54 ± 0.02 | 3.5 ± 0.1 | 0.67 ± 0.03 | 14.0 ± 0.2 | 0.89 ± 0.03 | 3.6 ± 0.1** | 0.33 ± 0.02 | 13.4 ± 0.1 | 1.07 ± 0.03** | 48.3 ± 0.2 |
Data are presented as mean ± se (n = 3–6 for mutants, n = 71 for Col wild type, n = 42 for Ws wild type).
Table VII.
Ratios of mol % fatty acids in leaves and ratio of C to N in seeds
The asterisk indicates a significant difference of ratio between the mutant and corresponding wild type (Col or Ws; Dunnett's test, *, p < 0.05; **, p < 0.01; ***, p < 0.001).
| Genotypea | (16:3+trans-16:1d3)/(18:0+18:2) in Leaves | 16:3/18:2 in Leaves | 16:0/(18:1d9+18:1d11) in Leaves | C/N in Seeds |
|---|---|---|---|---|
| Col wild type | 1.16 ± 0.01 | 0.98 ± 0.01 | 3.29 ± 0.02 | 14.0 ± 0.1 |
| 5-fcl | 1.19 ± 0.03 | 0.99 ± 0.02 | 3.49 ± 0.08 | 12.6 ± 0.4*** |
| arc12 | 1.14 ± 0.04 | 0.94 ± 0.03 | 3.26 ± 0.14 | 13.2 ± 0.2* |
| ats1-1 | 0.20 ± 0.005*** | 0.034 ± 0.003*** | 1.58 ± 0.04*** | 13.5 ± 0.2 |
| npq1-2 | 1.20 ± 0.03 | 1.00 ± 0.02 | 3.31 ± 0.05 | 13.4 ± 0.1 |
| sex1-1 | 1.01 ± 0.05** | 0.85 ± 0.03** | 3.04 ± 0.16* | 12.4 ± 0.3*** |
| sex4-5 | 1.13 ± 0.06 | 0.93 ± 0.05 | 3.38 ± 0.15 | 13.5 ± 0.1 |
| tha1-1 | 0.94 ± 0.02*** | 0.81 ± 0.02*** | 3.49 ± 0.06 | 12.4 ± 0.2*** |
| tt7-3 | 1.12 ± 0.03 | 0.91 ± 0.02 | 3.16 ± 0.05 | 12.3 ± 0.1*** |
| Ws wild type | 1.24 ± 0.01 | 1.07 ± 0.01 | 3.90 ± 0.07 | 14.5 ± 0.1 |
| arc10 | 1.26 ± 0.03 | 1.09 ± 0.04 | 4.01 ± 0.19 | 15.2 ± 0.1** |
| dpe2-1 | 1.11 ± 0.02* | 0.89 ± 0.01*** | 3.08 ± 0.06*** | 13.7 ± 0.4** |
| fatb-ko | 0.85 ± 0.04*** | 0.70 ± 0.02*** | 1.04 ± 0.03*** | 12.6 ± 0.2*** |
| lkr-sdh | 1.37 ± 0.04 | 1.15 ± 0.05 | 3.89 ± 0.14 | 13.8 ± 0.1* |
| pig1-1 | 1.23 ± 0.03 | 1.05 ± 0.03 | 3.32 ± 0.09* | 14.8 ± 0.3 |
Data are presented as mean ± se (n = 3–6 for mutants, n = 71 for Col wild-type leaves, n = 67 for Col wild-type seeds, n = 42 for Ws wild-type leaves, n = 33 for Ws wild-type seeds).
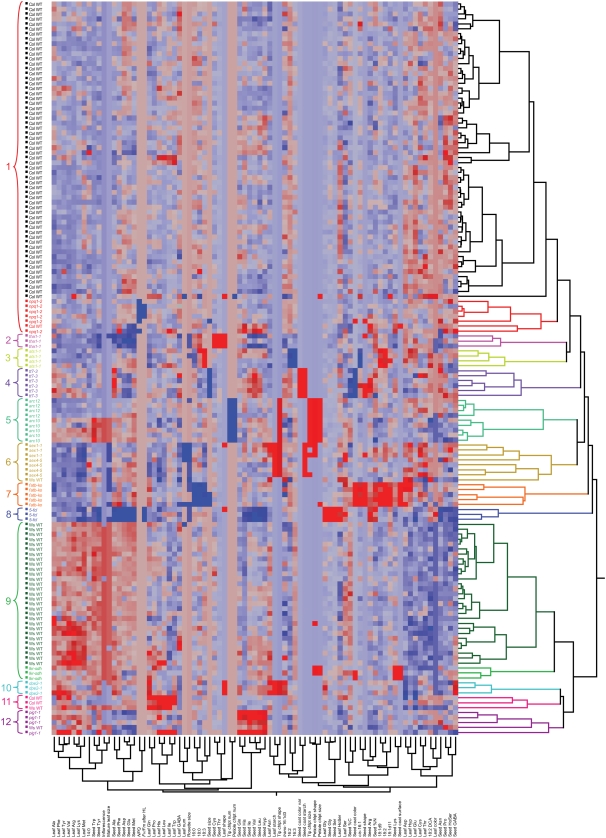
We also confirmed the phenotypes of mutants included to validate the qualitative assays. The arc10 and arc12 mutants (deficient in chloroplast division proteins AtFtsZ1 and AtMinE1, respectively) had fewer chloroplasts in the mesophyll cells from expanded leaf tips: the arc10 mutant often contained one greatly enlarged chloroplast and some smaller chloroplasts (Fig. 1, F and N) and the arc12 mutant had a single giant chloroplast (Fig. 1, B and J), as reported (Glynn et al., 2007; Yoder et al., 2007). The glucan-water dikinase-deficient sex1-1 mutant, laforin-like carbohydrate phosphatase-deficient sex4-5 mutant, and disproportionating enzyme-deficient dpe2-1 mutant had higher leaf starch (Fig. 2, clusters 6 and 10), as previously published (Yu et al., 2001; Lu and Sharkey, 2004; Niittylä et al., 2006; Sokolov et al., 2006; Gentry et al., 2007). The violaxanthin deepoxidase mutant npq1-2 had lower nonphotochemical quenching (NPQ; Fig. 2, cluster 1), as described (Niyogi et al., 1998). Finally, the tt7-3 and tt7-1 mutants, carrying mutations in the gene encoding flavonoid 3′-hydroxylase, had pale brown seed coats as expected (Schoenbohm et al., 2000; Abrahams et al., 2002; Salaita et al., 2005).
Figure 1.
Morphological and physiological phenotypes of the mutants. A to H, Light micrographs representing chloroplast morphology in expanded leaf tips from Col wild type (A), arc12 (B), sex1-1 (C), sex4-5 (D), Ws wild type (E), arc10 (F), dpe2-1 (G), and lkr-sdh (H). I to P, Light micrographs representing chloroplast morphology in expanded leaf petioles from Col wild type (I), arc12 (J), sex1-1 (K), sex4-5 (L), Ws wild type (M), arc10 (N), dpe2-1 (O), and lkr-sdh (P). A to P, Bars are 20 μm. Q to U, Light micrographs representing iodine-stained dry seeds from Col wild type (Q), sex1-1 (R), sex4-5 (S), tt7-3 (T), and tt7-1 (U). Q and U, Bars are 500 μm. DNA sequence analysis confirmed that both tt7-3 and tt7-1 mutants had the expected lesions in the TT7 locus. V to X, False-color images representing Fv/Fm after high light in Col wild type (V), 5-fcl (W), and npq1-2 (X). A red image indicates Fv/Fm after high light for the plant is below the cutoff value. For the 5-fcl mutant, all six plants had a mutant phenotype; for npq1-2, three out of six images were of mutant phenotype.
Figure 2.
Hierarchical clustering of 148 samples by 81 variables using Ward's minimum variance methods. Data from different variables were standardized so that all variables have equal impact on the computation of distance. Traits with a positive z-score or numeric code are shown in red squares; traits with a negative z-score or numeric code are shown in blue squares. The 12 clusters are color coded by JMP 6.0, and shown in similar text color. Sixty of the 63 Col wild-type plants and the npq1-2 plants form one cluster, which is made of two subclusters: Col wild type and npq1-2. The Col wild-type subcluster is in black text. Twenty-nine of the 32 Ws wild-type plants and the lkr-sdh plants form one cluster, which is made of two subclusters: Ws wild type and lkr-sdh. The subcluster of Ws wild-type plants is in dark green text. Chlpt, Chloroplast; HL, high light; num, number; var, variation.
Parallel Assays Reveal Phenotypic Networks
Typical forward genetics and reverse genetics strategies suffer from the interrogation of each mutant with a limited number of phenotypic assays. This has two related consequences: it limits the likelihood that the full effects of a mutation will be discovered, and blinds us from discovering unexpected relationships between genes. The mutants included in this study were previously characterized (and except for pig1-1, the affected gene published), and have diverse primary physiological defects. This allowed us to look for unexpected secondary phenotypes and syndromes of effects.
The 5-fcl mutant is defective in an enzyme that recycles 5-formyltetrahydrofolate, which is implicated as an inhibitor of mitochondrial Ser hydroxymethyltransferase, a key enzyme in photorespiration (Goyer et al., 2005). This mutant showed an especially large number of previously unreported phenotypes. The large number of alterations in free amino acids in seeds is especially striking for this mutant, with 11 of 20 protein amino acids showing statistically significant changes based on nmol/g FW (Supplemental Table S3) and 16 of 20 based on mol % (Table IV). The theme of changes in seed composition is also seen for total C and N in seed. The C/N ratio of 5-fcl was significantly lower than that in Col wild-type seeds (p < 0.001; Table VII). This is in contrast to leaves, where Gly and Ser are the only amino acids showing >2-fold differences in total content (Supplemental Table S1).
In addition to these striking seed phenotypes, the 5-fcl mutant has previously unreported changes in leaf biochemistry and physiology. First, there are modest, but statistically significantly higher contents (in both mol % and nmol/g FW) of the unsaturated fatty acids cis-16:1 and 18:1d11 in total leaf lipids (Table VI; Supplemental Table S5). After high light treatment, all six 5-fcl mutant plants tested had lower maximum photochemical efficiency of PSII (Fv/Fm) than Col wild type (displayed in red in false-color image in Fig. 1W). The only other mutant to show this chlorophyll fluorescence phenotype is npq1-2 (Fig. 1X). This mutant was previously shown to be defective in NPQ due to an inability to convert violaxanthin to zeaxanthin under conditions of excessive light (Niyogi et al., 1998).
Because of the central role of starch in chloroplast biochemistry, three previously characterized excess leaf starch mutants, sex1-1, sex4-5, and dpe2-1, were phenotypically analyzed, and each was found to have pleiotropic phenotypes. The accumulation of starch resulted in wrinkled chloroplasts in the leaf tips of each mutant (Fig. 1, C, D, and G), presumably due to excess amounts of starch stored in the chloroplast. In contrast, wrinkled petiole cell chloroplasts were only seen in sex1-1 mutant (Fig. 1, compare K to L and O). This is unlike the arc10 and arc12 mutants, which have dramatically altered leaf tip and petiole cell chloroplast morphology (Fig. 1, J and N).
An interesting example of phenotypic diversity was seen for leaf starch-excess mutants. Our iodine-staining assay indicates that mature and dried sex1-1 and sex4-5 seeds have excess starch in their seed coats (Fig. 1, R and S). In contrast, dried seeds of the leaf starch-excess mutants dpe2-1 and dpe2-2 did not stain positive with iodine solution (Fig. 2, cluster 10). In Ws wild-type Arabidopsis seeds, starch accumulates in the outer integument during the early stage of development, and is degraded later in development (Baud et al., 2002). We hypothesize that sex1-1 and sex4-5 mutants do not fully degrade the starch transiently accumulated in the seed coat early in development. The lack of excess starch in dpe2 mutant seeds is consistent with the hypothesis that transitory starch degradation in leaves and seed coats may share some enzymes at the earlier steps and differ at later steps.
A variety of other metabolic differences were seen in the three starch mutants, although the changes from wild type and from one another were small compared with the dramatic changes in C metabolism and chloroplast morphology. Although sex1-1 had altered seed C/N ratio (p < 0.001), the other two high-starch mutants were unaffected for seed C/N ratio. There were statistically significant differences in mol % levels of leaf and seed free amino acids in each of the three mutants compared with wild type, though in only two cases was the change 3-fold or more (Tables II–V). Similarly, statistically significant changes in mol % and absolute quantities of leaf fatty acids were observed in the three mutants, though the magnitude of the changes was quite low compared with the biosynthetic mutants fatb-ko and ats1-1 (Table VI; Supplemental Table S5).
Two classes of mutants altered in amino acid homeostasis were chosen for this study. The first, originally found to have changes in metabolism of specific amino acids, is represented by the lkr-sdh and tha1-1 mutants, which are deficient in seed Lys catabolism (Zhu et al., 2001) and seed Thr catabolism (Jander et al., 2004; Joshi et al., 2006), respectively. Our results indicate that these pathway-specific changes in seed amino acid metabolism have a limited effect on the range of phenotypes analyzed. The lkr-sdh mutant had no other substantial changes in leaf or seed metabolites, and the only phenotypic change noted was the occurrence of larger dumbbell-shaped chloroplasts in all three plants tested (Fig. 1P). Further work would be required to test whether this phenotype is caused by the lkr-sdh insertion allele. The tha1-1 mutant also had fairly minor pleiotropic effects. In addition to the >25-fold increase in nmol/g FW seed free Thr, a previously unreported reproducible >10-fold increase in nmol/g FW seed Cys was also found (Supplemental Table S3; note that for a mutant with such a dramatic change in one or more metabolites, mol % is a less useful metric for analysis than concentration, as seen in Table IV). Subtle changes were observed for several other amino acids and 18:0 fatty and 18:2 dicarboxylic acid (Supplemental Table S5) in the tha1-1 mutant, as was a significant decrease in seed C/N ratio (p < 0.001; Table VII).
The pig1-1 mutant was chosen for this study because it was found to have more global changes in amino acid homeostasis; it was reported to have abnormal levels of multiple free amino acids and an approximately 2-fold increase in total soluble amino acids in 2-week-old plate-grown seedlings (Voll et al., 2004). Although the published phenotypic analysis focused on seedlings, the most striking pig1-1 phenotypic changes observed were for free amino acids in seeds (Supplemental Table S4): 12 amino acids had statistically significant differences compared with wild-type Ws, with six of these compounds showing 3- to 6-fold increases, and a >70% increase in total seed free amino acids (significant at the p < 0.001 level). The situation was notably different for leaf samples, where only five amino acids showed statistically significant increases (at the p < 0.05 or p < 0.01 significance level) and all but one was <2-fold increased (Supplemental Table S2). These data highlight an inherent strength of measuring multiple phenotypes in parallel because the pronounced difference in seed compared with 5-week-old plant leaf amino acids was missed when a single developmental stage was assayed.
The tt7-3 mutant, deficient in flavonoid 3′-hydroxylase, represents another example of strong pleiotropy in seed phenotypes without dramatic effects in the leaf. It was originally included in the study because it has a subtle pale brown seed coat and smaller seeds than Col wild type. Surprisingly the seeds stain very dark purple-black with iodine solution, suggesting that the line may have excess seed coat starch (Fig. 1T). Consistent with their pleiotropic seed morphology and iodine staining, tt7-3 had statistically significant increases in nine amino acids (p < 0.001 for eight; Supplemental Table S3) and seed C/N ratio (p < 0.001; Table VII). These abnormalities are confined to the seed because tt7-3 has relatively normal leaf amino acid (Supplemental Table S1) and fatty acid (Supplemental Table S5) content. It is unclear whether the tt7-3 lesion is responsible for the pleiotropic phenotypes in this mutant because tt7-1 ecotype Landsberg erecta of Arabidopsis (Ler) seeds did not stain dark with iodine solution (Fig. 1U), and unstained seeds were rounder, lighter, and more evenly colored than tt7-3. Both lines have the expected mutations, and each should produce a protein truncated within the first half of the coding sequence, as previously published (Schoenbohm et al., 2000; Salaita et al., 2005). Whether or not the flavonoid pathway lesion causes the battery of secondary phenotypes, this result is an example of a broad syndrome of effects on seed morphology and biochemistry.
Systematic Data Analysis
Although examination of differences between individual mutants and the progenitor wild-type ecotype was useful in looking for specific phenotypes or syndromes of changes in the mutant, other approaches are necessary to reveal more complex relationships between genotype and phenotype inherent in the data set. Two general approaches were followed: clustering and principal component analysis (PCA; Quackenbush, 2001; Schauer et al., 2006) to visualize phenotypic patterns correlated with genotypes, and correlation analysis to discover relationships among the phenotypic traits in the wild-type ecotypes.
A variety of data transformations and tests were performed to make meaningful comparisons between qualitative and quantitative phenotypes, as detailed in “Materials and Methods”. For example, the controlled vocabulary text descriptions associated with individual morphological or qualitative traits were systematically coded into numerical form as summarized in Table VIII. Before raw quantitative data from different flats of plants and assay plates were merged, O'Brien's test was conducted to confirm the homogeneity of variance across flats and plates (O'Brien, 1979). The normality of the quantitative data from Col and Ws wild-type data was tested using the Shapiro-Wilk test (Shapiro and Wilk, 1965). To allow comparisons of different types of quantitative data derived from plants grown in different microenvironments, data for mol % of fatty acids, mol % of free amino acids, seed %C, seed %N, seed C/N ratio, and fatty acid ratios were converted to z-scores. The merged data set contains 148 samples, 85 variables, and three types of data—continuous (data that can fall into an infinite number of values such as concentration of a metabolite), ordinal (ordered categorical data such as smaller, normal, and larger), and dichotomous (data divided into two categories such as inflorescence present or absent).
Table VIII.
Numeric codes used for morphological traits and qualitative traits
| Trait | Numeric Code
|
||||
|---|---|---|---|---|---|
| −3 | −2 | −1 | 0 | 1 | |
| Whole plant morphology | |||||
| Rosette size (cm in diameter) | <1.3 | 1.3–2.4 | 2.5–3.9 | ≥4.0 | |
| Inflorescence | Not visible | Visible | |||
| Leaf color | Lighter | Normal | Darker | ||
| Leaf color variation | Evenly colored | Color variation | |||
| Leaf number | Less | Normal | More | ||
| Leaf shape | Normal | Abnormala | |||
| Mature leaf size | Smaller | Normal | Larger | ||
| Trichomes | Absent | Present | |||
| Chloroplast morphology in expanded leaf petiole | |||||
| Chloroplast number | Less | Normal | More | ||
| Chloroplast shape | Normal | Abnormalb | |||
| Chloroplast size | Smaller | Normal | Largerc | ||
| Chloroplast morphology in expanded leaf tip | |||||
| Chloroplast number | Less | Normal | More | ||
| Chloroplast shape | Normal | Abnormalb | |||
| Chloroplast size | Smaller | Normal | Largerc | ||
| Seed morphology | |||||
| Seed coat color | Lighter | Normal | Darker | ||
| Seed coat color variation | Evenly colored | Color variation | |||
| Seed coat surface | Normal | Abnormald | |||
| Seed shape | Normal | Abnormale | |||
| Seed size | Smaller | Normal | Larger | ||
| Leaf starchf | Less | Normal | Excess | ||
| Seed coat starch | Normal | Excess | |||
| Chlorophyll fluorescence | |||||
| Fv/Fm before high light | Lower | Normal | |||
| Fv/Fm after high light | Lower | Normal | |||
| Fv/Fm after recovery | Lower | Normal | |||
| NPQ | Lower | Normal | |||
Examples for leaves of abnormal shape include curled, flat, narrow, rolled, round, serrated, succulent, wilted, or wrinkled leaves, or leaves with a pointed apex or a short petiole.
Examples for chloroplasts of abnormal shape include amorphous, dumbbell-shaped, elongated, heterogeneously shaped, or wrinkled chloroplasts.
Examples for larger chloroplasts include larger or heterogeneously larger chloroplasts.
Examples for seed coat of abnormal surface include dull or shiny seed coat.
Examples for seeds of abnormal shape include aborted, elongated, round, or wrinkled seeds.
Results from leaf discs harvested at the beginning of the light period and 8 h after light period begins were combined.
Classification of Mutants via Clustering Analysis and PCA
Hierarchical clustering analysis (HCA) was performed using Ward's minimum variance method to systematically analyze and visualize the full set of qualitative data and z-scores from the quantitative assays. As shown in Figure 2, this method resulted in 12 clusters and, in the vast majority of cases, biological replicates of each genotype clustered together. Notably, 29/32 Ws and 60/63 Col plants were in the same clusters, showing that the biological and process variations were substantially lower than the phenotypic differences between genotypes. The mutants clustered with or near the wild-type lines from which they were derived, indicating that the general clustering pattern was influenced by a suite of phenotypic traits, and was not simply caused by the strong outlier phenotypes associated with the mutations. For example, npq1-2, tha1-1, ats1-1, and tt7-3 clustered near Col wild-type lines whereas lkr-sdh, dpe2-1, and pig1-1 clustered near Ws wild-type lines (Fig. 2). The clustering of these mutants with their wild-type ecotypes extends the results described by Fiehn et al. (2000) for the mutants dgd1 and sdd1. When z-scores of amino acids and fatty acids were calculated from corresponding nmol/g FW data, similar groupings were obtained (Y. Lu, unpublished data). Clustering patterns resulted from other HCA approaches, including average linkage, centroid method, single linkage, and complete linkage, were not as discrete as that from Ward's method (Fig. 2), although biological replicates of some genotypes tended to cluster together.
The robustness of these clusters was tested in several ways. To study the impact of individual variables (i.e. phenotypes) on clustering, individual phenotypic variables were removed one by one, and the remaining data reclustered using HCA. Removal of most variables individually and reclustering with HCA did not dramatically alter the groupings (Y. Lu, unpublished data). The npq1-2 mutant was an exception, consistent with the hypothesis that decreased Fv/Fm after high light and NPQ are the only traits distinguishing it from Col. The three Col and three Ws wild-type plants that initially did not cluster with the majorities were sometimes relocated to a different cluster when one variable was removed (Y. Lu, unpublished data). This indicates that these unusually behaving wild-type samples (in clusters 1, 6, 11, and 12 of Fig. 2) were at cluster boundaries.
To test the contribution of qualitative versus quantitative data to the discrimination of genotypes, HCA was performed after removing each full set of phenotypes individually. Removal of all the qualitative variables changed the groupings for half of the clusters, in ways not seen when individual traits were removed. The two subclusters containing large numbers of wild-type samples became less well differentiated from the arc and lkr-sdh knockout individuals. This emphasizes the importance of chloroplast morphology in creating the clusters containing these mutants. The npq1-2 subcluster also became unresolved from the Col cluster because of removal of the chlorophyll fluorescence phenotypes. Reclustering without the quantitative z-score data also changed the groupings for about half of the clusters, whereas six clusters did not change: tt7-3, arc10 and arc12, sex1-1 and sex4-5, fatb-ko, 5-fcl, and dpe2-1. Three clusters in Figure 2 had some substantial changes: Col wild type and npq1-2 (cluster 1), ats1-1 (cluster 3), and pig1-1 (cluster 12). Four ats1-1 plants, four pig1-1 plants, and one Ws wild-type plant became mixed with Col wild-type plants. Taken together, these results strongly reinforce the value of using a combination of qualitative and quantitative traits to detect phenotypic relationships and differences.
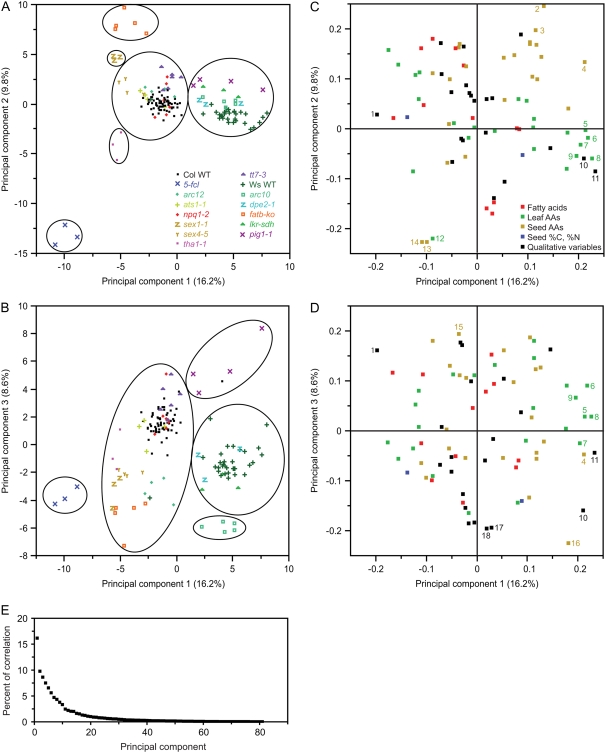
To facilitate graphical interpretation of the differences and the similarities among the mutants and wild-type plants and to look for variables with significant impacts on clustering results, the same data set was analyzed by PCA. Eighty-one principal components were extracted and, as expected, clustering with the entire set of 81 principal components resulted in clusters identical to that shown in Figure 2. Although the first, second, and third principal components together explained only 35% of the variation within the entire data set (Fig. 3E), the overall similarity of mutants in the same background to each other and to their isogenic wild type was well reflected in these dimensions (Fig. 3, A and B), consistent with the clustering results of HCA (Fig. 2). When plotting the dimensions of the first and second principal components or the first and third principal components, ats1-1, npq1-2, and tt7-3 clustered around Col wild-type plants whereas dpe2-1 and lkr-sdh mutants clustered around Ws wild-type plants (Fig. 3, A and B). Six of the 12 mutants formed distinct clusters in one or both of the graphs. Many variables have significant weightings in PCA (Fig. 3, C and D), indicating that the clustering of biological replicates of the same genotype is due to changes in many phenotypic traits, consistent with the results from HCA. The top 18 variables with significant weightings (>0.19 or <−0.19) include six leaf amino acids (Arg, Gly, Lys, Met, Tyr, and Val), seven seed amino acids (Gly, His, Leu, Phe, Ser, Trp, and Tyr), and five qualitative traits.
Figure 3.
PCA of 148 samples by 81 variables. A and B, Each point represents one biological sample, which is color- and symbol-coded by genotype. A, Scores plot of genotypes visualized in the dimensions of the first and second principal components. B, Scores plot of genotypes visualized in the dimensions of the first and third principal components. C, Loading plot for the first and second principal components. The distance from the origin indicates the relative importance of each phenotypic character in determining the separation in A. D, Loading plot for the first and third principal components. The distance from the origin indicates the relative importance of each phenotypic character in determining the separation in B. C and D, Different types of data are color-coded. Examples of variables with absolute value of weighting larger than 0.19 for the first, second, and third components are numbered: 1, leaf color; 2, seed Phe; 3, seed Leu; 4, seed Tyr; 5, leaf Met; 6, leaf Lys; 6, leaf Lys; 7, leaf Arg; 8, leaf Val; 9, leaf Tyr; 10, inflorescence; 11, mature leaf size; 12, leaf Gly; 13, seed Ser; 14, seed Gly; 15, seed His; 16, seed Trp; 17, petiole chloroplast size; 18, petiole chloroplast shape. E, Scree plot of all principal components and the percent of correlation they explain within the entire data set.
Correlations among Traits in Wild-Type Plants
Having a large set of phenotypic observations on multiple wild-type plant and seed samples permits the detection of minor phenotypic changes that are due to small differences in the physiological state of each plant. We took advantage of this biological variability to look for associations between the various phenotypic and morphological traits. The data set of 63 Col and 32 Ws samples assayed for the full set of 85 variables was analyzed by nonparametric Spearman's ρ correlation. A total of 1,327 significant Spearman's correlations (p < 0.05) were identified, nearly equally divided between negative and positive correlations (Supplemental Fig. S1). Data from mutants were not included to avoid correlations influenced by phenotypes of outlier individuals. The 364 pairs of variables with correlation coefficient | ρ | > 0.50 are listed in Supplemental Table S6.
To ask whether any of the identified correlations were due to the large differences in phenotypic patterns observed between the Col and Ws ecotypes (Figs. 2 and 3), the data set of 63 Col wild-type samples was analyzed separately by Spearman's correlation. Only 429 significant correlations (p < 0.05) were identified: approximately one-third as many as those identified in the dataset with both Col and Ws wild-type samples. Among the 364 correlations listed in Supplemental Table S6, 161 were still significant with the Col-only data set (| ρ | > 0.50; Supplemental Table S7). Presumably, many of the correlations that disappeared when Ws data were excluded (indicated by superscript b in Supplemental Table S6) either reflected phenotypic differences between the two ecotypes or reduction in sample size (63 Col samples versus 95 total Col + Ws samples).
The impact of using mol % on correlation analysis was investigated by merging z-scores calculated from nmol/g FW of amino acids and fatty acids with numeric codes from qualitative assays. Spearman's ρ correlation analysis was performed on the new data set of Col and Ws wild-type samples. A total number of 1,468 significant correlations were identified: about 26% of them are negative correlations and 74% are positive correlations. Overall, fewer positive correlations were identified when mol % of amino acids and fatty acids were employed, consistent with the fact that mol % of individual amino acids or fatty acids are reciprocally dependent upon each other.
To identify correlations reflecting intrinsic mechanisms of metabolic pathways, we sought strong and significant correlations (| ρ | > 0.5, p < 0.0001) identified from Col wild-type samples (Supplemental Table S7). Those correlations seen both with z-scores calculated from mol % and from nmol/g FW were of special interest because they might represent particularly robust examples (Table IX). Correlations that are not caused by mathematical reasons (for example between metabolites and ratios that include those metabolites) are shown in Table IX and reported below.
Table IX.
Spearman's correlation of quantitative traits in Col wild-type plants with | ρ | > 0.5 in both mol % and nmol/g FW
| Variablea | By Variable | ρ | ρ |
|---|---|---|---|
| mol %b | nmol/g (FW)c | ||
| 18:0 | 16:0 | 0.5351 | 0.7031 |
| 18:2 | 18:0 | 0.5766 | 0.6768 |
| 18:2 | 18:1d9 | 0.5995 | 0.7750 |
| (16:3+trans-16:1d3)/(18:0+18:2) | 16:0 | −0.6186 | −0.6448 |
| (16:3+trans-16:1d3)/(18:0+18:2) | 18:1d9 | −0.6878 | −0.6795 |
| 16:3/18:2 | 16:0 | −0.5967 | −0.5840 |
| 16:3/18:2 | 18:0 | −0.5921 | −0.6402 |
| 16:3/18:2 | 18:1d9 | −0.6558 | −0.6146 |
| 18:1d9 | 16:0 | 0.6114 | 0.8005 |
| 18:1d9 | 18:0 | 0.6465 | 0.7252 |
| Leaf Gln | Leaf Asn | 0.5260 | 0.7975 |
| Leaf Glu | Leaf Arg | −0.5698 | −0.7091 |
| Leaf Glu | Leaf Asp | 0.6154 | 0.8835 |
| Leaf Ho-Ser | Leaf Glu | 0.5138 | 0.6884 |
| Leaf Ho-Ser | Leaf Pro | 0.5180 | 0.6545 |
| Leaf Hyp | Leaf Arg | −0.5439 | −0.5183 |
| Leaf Leu | Leaf His | 0.5398 | 0.6526 |
| Leaf Lys | Leaf His | 0.6326 | 0.6074 |
| Leaf Phe | Leaf Leu | 0.6762 | 0.7545 |
| Leaf Pro | Leaf Asn | 0.5201 | 0.6643 |
| Leaf Pro | Leaf Gln | 0.5508 | 0.6741 |
| Leaf Pro | Leaf Met | 0.7480 | 0.7686 |
| Leaf Ser | Leaf Leu | 0.5431 | 0.6718 |
| Leaf Tyr | Leaf His | 0.5107 | 0.5637 |
| Leaf Tyr | Leaf Leu | 0.5735 | 0.6829 |
| Leaf Val | Leaf Ala | 0.6425 | 0.5860 |
| Leaf Val | Leaf His | 0.5226 | 0.5930 |
| Leaf Val | Leaf Leu | 0.6979 | 0.7558 |
| Leaf Val | Leaf Phe | 0.7422 | 0.8111 |
| Leaf Val | Leaf Ser | 0.5365 | 0.6230 |
| Seed Asn | Seed Arg | 0.5771 | 0.7939 |
| Seed Asp | Seed Asn | 0.5442 | 0.6198 |
| Seed GABA | Seed Gln | 0.5175 | 0.6348 |
| Seed GABA | Seed Leu | 0.5793 | 0.7808 |
| Seed GABA | Seed Pro | 0.5419 | 0.7193 |
| Seed GABA | Seed Thr | 0.5632 | 0.7485 |
| Seed GABA | Seed Val | 0.5402 | 0.7751 |
| Seed Glu | Seed Asn | 0.5632 | 0.6248 |
| Seed Glu | Seed Asp | 0.8114 | 0.7402 |
| Seed His | Seed Gln | 0.5398 | 0.7005 |
| Seed Ho-Ser | Seed Pro | 0.5031 | 0.7932 |
| Seed Hyp | Seed His | 0.5602 | 0.7730 |
| Seed Hyp | Seed Ile | 0.5173 | 0.7832 |
| Seed Ile | Seed His | 0.5180 | 0.8048 |
| Seed Leu | Seed Ile | 0.7248 | 0.8344 |
| Seed Lys | Seed Leu | 0.5237 | 0.8278 |
| Seed Met | Seed Glu | 0.6861 | 0.5942 |
| Seed Phe | Seed Gln | 0.5239 | 0.7362 |
| Seed Phe | Seed Leu | 0.7467 | 0.8647 |
| Seed Pro | Seed Ile | 0.6247 | 0.8214 |
| Seed Thr | Seed Leu | 0.5172 | 0.7976 |
| Seed Tyr | Seed Lys | 0.5177 | 0.8406 |
| Seed Tyr | Seed Met | 0.5043 | 0.6753 |
| Seed Val | Seed His | 0.5760 | 0.8229 |
| Seed Val | Seed Ile | 0.8065 | 0.9291 |
| Seed Val | Seed Leu | 0.6910 | 0.8129 |
| Seed Val | Seed Phe | 0.5358 | 0.9002 |
| Seed Val | Seed Pro | 0.6617 | 0.8292 |
Correlations due to mathematical reasons were not listed.
Spearman's ρ correlations were calculated from the table containing z-scores of mol % of amino acids and fatty acids, z-scores of %C and %N, z-scores of fatty acid, and C/N ratios. Only data from Col wild-type plants were used. All the correlations are significant (p < 0.0001).
Spearman's ρ correlations were calculated from the table containing z-scores of nmol/g FW of amino acids and fatty acids, z-scores of %C and %N, z-scores of fatty acid, and C/N ratios. Only data from Col wild-type plants were used. All the correlations are significant (p < 0.0001).
Fatty acids 16:0, 18:0, 18:1d9, and 18:2 showed strong positive correlation with each other (Table IX). This is consistent with our understanding that 16:0, 18:0, and 18:1d9 are consecutive intermediates in fatty acid biosynthesis and precursors to the most abundant fatty acid, 18:3 (Somerville et al., 2000). Specific lipid classes in leaf subcellular organelles have distinct fatty acid compositions. For example, 16:3 and trans-16:1d3 are almost exclusively found in plastidial galactolipids and phosphatidylglycerol whereas 18:0 and 18:2 are enriched in extraplastidial membrane lipids. Therefore, the two ratios (16:3+trans-16:1d3)/(18:0+18:2) and 16:3/18:2 provide a representation of the abundance of thylakoid and extraplastidial membrane fatty acids. Both ratios are negatively correlated with 16:0 and 18:1 and these correlations could be indicative of altered ratios of thylakoid to extraplastidial membranes across the sample set. Further work would be required to study the significance of these correlations.
The data in Table IX contain examples of metabolically related amino acids that show positive correlations using both nmol/g FW and mol % data (Coruzzi and Last, 2000). For example, Glu, Gln, Asp, and Asn play a variety of important roles in plants in N transport and metabolism and these amino acids showed robust patterns of coaccumulation consistent with their metabolic relationships (Table IX). In leaves the amide compounds Asn and Gln were positively correlated as were the amino donors Asp and Glu. Even in dry seeds, Asn, Asp, and Glu were positively correlated with each other. Accumulation of all pairs of the branched-chain amino acids Ile, Leu, and Val was correlated in seeds, presumably reflecting their shared biosynthetic pathways (with four enzymatic steps in common). Correlations between biosynthetically related amino acids were also found in the fruit of tomato chromosomal substitution lines (Schauer et al., 2006).
Of greater interest is the number of strongly correlated metabolites that are not known to share a direct biosynthetic origin. For example, the branched chain amino acid Leu is correlated with the aromatic amino acids Phe and Tyr in leaf, whereas seed Phe is correlated with Leu and Val. His, which is derived from the relatively unusual precursor 5-phosphoribosyl-1-pyrophosphate, shows correlation to a variety of biosynthetically unrelated amino acids in leaf (Leu, Lys, Tyr, and Val) and seed (Ile and Val). γ-Aminobutyric acid (GABA), which is synthesized from Glu and thought to be involved in N-homeostasis, N-transport, and stress responses (Bouché and Fromm, 2004), is correlated with five biosynthetically diverse amino acids in the seed (Gln, Leu, Pro, Thr, and Val). These varied examples of correlated metabolites are consistent with the hypothesis that expression of the amino acid biosynthetic enzymes might be coregulated, or that these pathways have closer relationships than is apparent from their two-dimensional renderings in textbooks (Sweetlove et al., 2003).
DISCUSSION
To go beyond one-mutant-at-a-time analysis of complex biological processes requires systematic analysis of genomes and the networks that operate within complex organisms. This project had multiple goals aimed at enabling systematic analysis of Arabidopsis mutants. The first was to set up a relatively high-throughput plant growth and phenotypic assay process facilitated by a laboratory information management system. Second was evaluation of how well this pipeline could be used to identify mutants altered in a variety of phenotypes. A third goal was to explore the extent to which unknown mutant phenotypes could be discovered by parallel phenotypic analysis and to assess the level of pleiotropy in previously characterized mutants. Finally, we analyzed the large data set to look for correlations between phenotypes, both in mutant and wild-type plants.
Previously unknown phenotypes were detected by subjecting the mutants to a large number of phenotypic assays. The 5-fcl mutant is an example of a mutant with a far more complex phenotype than previously reported (Goyer et al., 2005). In addition to the documented increase in leaf Gly and Ser under normal growth conditions, we discovered statistically significant changes in concentration of more than half of the seed free amino acids (Supplemental Table S3) as well as a decrease in the maximum photochemical efficiency of PSII parameters following exposure to high light conditions for 3 h (Fv/Fm; Fig. 1W). The high Gly and Ser contents are indicative of a defect in the photorespiratory pathway (Bräutigam et al., 2007). The reduction in Fv/Fm after high light treatment indicates an increase in photoinhibition of PSII (Takahashi et al., 2007). The cooccurrence of high Gly and Ser contents and low Fv/Fm after high light in the 5-fcl mutant is consistent with the hypothesis that impairment of the photorespiratory pathway accelerates photoinhibition of PSII by suppressing the repair of photodamaged PSII (Takahashi et al., 2007).
The theme of differences in leaf and seed phenotypes was seen in other mutants. The pig1-1 mutant was altered in 12 seed amino acids (six with very large changes) and had a >70% increase in total free seed amino acids (Supplemental Table S4), whereas leaf amino acid changes were fewer and smaller in magnitude (Supplemental Table S2). Although the tha1-1 mutant was found to have an increase in seed Cys levels not previously reported (due to use of an improved analytical assay; Gu et al., 2007), tha1-1 and lkr-sdh plants did not show dramatic differences in leaf amino acids.
As this and other parallel multiphenotype data are accumulated for a larger set of mutants, it should be possible to discover emergent patterns associated with different classes of mutants. For example, our results show that all three starch-excess mutants tested have similar chloroplast abnormalities. Now that this is known, high-starch mutants could not only be found by screening directly for leaf or seed starch, but could also be identified by analysis of data from screens for changes in chloroplast morphology or leaf free amino acids. The fact that the detailed phenotypic patterns vary across mutants (in this case sex1, sex4, and dpe2) will also be very useful in detailed studies of gene function. For instance, assembly of such a data set for all high-starch mutants (or any other set of mutants of interest that have multiple phenotypes) would help place the gene products into pathways of action and may allow the deduction of functions for unknown genes (Messerli et al., 2007).
Although strong pleiotropy was observed for some mutants, others showed remarkably restricted phenotypic changes. Despite impressive changes in chloroplast number and morphology (Fig. 1, B, F, J, and N), arc10 and arc12 mutants were wild type for all other phenotypes measured, including chlorophyll fluorescence and metabolite accumulation (Fig. 2, compare to Col and Ws, respectively). This indicates that Arabidopsis has a remarkable resilience to large changes in chloroplast morphology, and that the pleiotropy observed for starch-excess mutants is not the default condition when chloroplast function is impaired. Because such a large number of phenotypic traits were measured, we regard the small number of defined phenotypes for mutants such as arc10, arc12, lkr-sdh, npq1-2, and tha1-1 as noteworthy.
Inclusion of a large number of wild-type lines allowed evaluation of the variability of each assay and discovery of traits that covaried; 126 strong correlations were identified when Spearman's ρ correlation analysis was used to analyze the Col-only data (| ρ | > 0.5; p < 0.0001; Supplemental Table S7). We asked whether these correlations would persist in a larger data set derived from screening >600 homozygous Col background T-DNA insertion lines from our mutant analysis pipeline (www.plastid.msu.edu). A total of 843 significant Spearman's ρ correlations were identified from the T-DNA mutant data and compared with those from Col wild-type samples in the pilot study. Among the 126 strong correlations (| ρ | > 0.5) identified in the pilot study, 90% were identified as significant (p < 0.05) and in the same direction in the pipeline data (Supplemental Table S7; all those not marked with superscript d), demonstrating the reproducibility of the correlation results.
The identified correlations allow the creation of hypotheses about regulatory and biosynthetic relationships that might exist between seemingly disparate metabolic pathways. One set of examples is the positive correlations between branched chain amino acids Leu and Val and aromatic amino acids Phe and Tyr. A plausible explanation is that the branched-chain amino acids are derived from pyruvate, whereas aromatic amino acid synthesis requires phosphoenolpyruvate. Recently published work indicates that phosphoenolpyruvate conversion to pyruvate by plastidial pyruvate kinase disrupts seed oil accumulation (Andre et al., 2007), suggesting the hypothesis that the plastidial phosphoenolpyruvate pool might be limiting for both branched-chain and aromatic amino acids. Mining of the data for other correlations should yield other testable hypotheses and yield insights into a variety of physiological processes.
CONCLUSION
This study demonstrates the strong utility of parallel phenotypic measurements on mutant and wild-type plants, and argues that this mode of mutant analysis has strong advantages over the traditional one-phenotype-at-a-time approach. The study benefited from participation of a large group of collaborators with complementary technical expertise in biology, chemistry, informatics, and statistics. This diverse know-how allowed us to create a robust experimental pipeline and to interpret the complex phenotypic results. Similar industrial scale mutant analysis approaches have been proposed and performed for gene discovery in industry and academia, reinforcing the general utility of this approach (Boyes et al., 2001; Fernie et al., 2004; Schauer et al., 2006).
For functional genomics to maximally impact systems biology will require extension of this idea to a larger germplasm (for instance, a broader set of sequence indexed insertion mutants or ethylmethanesulfonate (EMS) mutants, ecotypes, and recombinant inbred or introgression lines) and more diverse sets of phenotypic assays under a broader set of environmental conditions. Because of the clear value of creation of a vast phenotypic data set that would be of long-term utility (similar to GenBank for DNA sequence and AtGenExpress for gene expression; Schmid et al., 2005), we propose a community-wide project that would collaboratively expand the range of germplasm and phenotypic assays employed. Success of such a project would require careful germplasm selection, close collaboration of laboratories with expertise in the different areas of biology and technology, adherence to well-defined methods for growing plants and assaying phenotypes, and direct deposit of the data into a common relational database. Combining these results with other functional genomics data such as protein interaction (Geisler-Lee et al., 2007; Cui et al., 2008), mRNA and protein expression would create a powerful data set for plant systems biology. Although it is arguable that such a mega-genetics project would be as challenging from a sociological viewpoint as it would be scientifically, the payoff would greatly justify the effort.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) mutants used in the study are summarized in Table I. Seeds were sown in 3.5-inch deep 2.5- × 2.5-inch pots in 1- × 2-foot flats (32 pots per flat) using Redi-earth plug and seedling mix (Hummert International) topped with a thin layer of vermiculite. One pot of each mutant, 12 pots of wild-type Col, and seven pots of wild-type Ws were randomly placed in each flat. Sown seeds were stratified at 4°C in the dark for 3 to 4 d before they were moved to the same controlled environment chamber at a 16-h light/8-h dark photoperiod. The first set of 96 pots was moved to the growth chamber on the third day and the last set on the fourth day to facilitate rapid harvesting of tissue. The irradiance was 100 μmol m−2 s−1 photosynthetic photon flux density (PPFD) using a mix of cool-white fluorescent and incandescent bulbs, the temperature was 21°C, and the relative humidity was set to 50%. After 7 d in the growth chamber, seedlings were thinned to one plant per pot. Seeds harvested from plants under the 16-/8-h photoperiod were used for seed assays and were sown for growth in a 12-/12-h photoperiod, under the same light conditions as for seed bulk-up. These plants were used for leaf assays when they were 4 to 5 weeks old. Full sets of assays were obtained for leaf and seed from 148 lines; these constitute samples in our analyses as described in “Results” and in “Materials and Methods” below. Plants for chlorophyll fluorescence analysis were grown separately, as described below. To maximize accuracy in data tracking, every seed stock, flat, pot, and sample container was bar-coded and the associations among them and the phenotypic data tracked in a relational database. Leaf samples for different assays were harvested in the following order: morning starch assay (for high-starch mutants), amino acid assay, fatty acid assay, afternoon starch assay (for low-starch phenotype), and chloroplast morphology.
Vegetative and Seed Morphology Assays
Plant, chloroplast, and seed morphology were assessed using controlled vocabulary descriptions (detailed in Table VIII), and captured by photography (see Fig. 1 for examples), with both types of data stored in the database. Plants under the 16-/8-h photoperiod and the 12-/12-h photoperiod were photographed after 23 and 30 d in the growth chamber, respectively. Morphology data from plants grown under the 12-/12-h photoperiod were used in this study.
Chloroplast morphology was assessed by harvesting petioles and tips from mature expanded leaves at the beginning of the light period. Leaf tissues were fixed and macerated as previous described (Osteryoung et al., 1998). Samples were photographed with a DMI3000B inverted microscope (Leica Microsystems), using polarization contrast optics and a 40× HCX PL FLUOTAR objective (Leica Microsystems).
Seeds were visually inspected with a MZ12.5 high-performance stereomicroscope (Leica Microsystems), using a polarizing lens. Images were captured by computer using a SPOT Insight Color 3.2.0 digital camera and SPOT advanced imaging software (Diagnostic Instruments).
Leaf and Seed Coat Starch Assay
Leaf discs (5.5-mm diameter) were harvested from leaf numbers 8 and 9 (counting from the newest visible leaf) at the beginning of the light period and 8 h after the light period began, respectively. Leaf discs obtained with a number 2 cork borer were harvested into a chilled microtiter plate and stained with iodine solution as previously described (Yu et al., 2001). Leaf discs harvested at the beginning of the light period were scored by eye as starch normal or starch excess. Leaf discs harvested 8 h after the light period begins were scored as starch normal or starchless.
Aliquots of seeds were placed into a 96-well microtiter plate and stained with iodine solution with 0.67% (w/v) iodine and 3.33% (w/v) of potassium iodide using the same protocol as with leaf discs.
Leaf and Seed Free Amino Acid Analysis by HPLC-MS/MS
To prepare leaf samples for amino acid analysis, leaf number 7 (counting from the newest visible leaf) was harvested beginning 1 h after the light period started, weighed, placed into 2-mL microfuge tubes containing a single 3-mm stainless steel ball. Leaf samples were immediately frozen with dry ice and then ground frozen to a fine powder for 1 min on a S2200 paint shaker (Hero Products Group). Samples were suspended in 0.4 mL of extraction solution containing 1 μm of L-Phe-α,β,β,2,3,4,5,6-d8 (Phe-d8; Cambridge Isotope Laboratories) and 10 μm of 1,4-dithiothreitol (Roche Applied Science) in dH2O. The extracts were incubated at 85°C for 5 min and were centrifuged at 3,220g at 4°C for 5 min. The supernatant was transferred to a prewetted MultiScreen Solvinert filter plate (Millipore) and centrifuged at 2,000g at 4°C for 5 min to remove insoluble materials. Ninety microliters of filtrate were transferred to a 96-well plate and mixed with 10 μL of 9 μm of L-Val-2,3,4,4,4,5,5,5-d8 (Val-d8; Cambridge Isotope Laboratories). Phe-d8 and Val-d8 were used to normalize extraction and loading accuracy, respectively (Jander et al., 2004; Gu et al., 2007).
To prepare seed samples for the amino acid assay, approximately 7 mg aliquots of seeds were placed into a deep-well microplate (VWR International) containing a single 3-mm stainless steel ball in each well. The same extraction solution as used for leaf was added to the seeds and the samples were ground for 5 min on the paint shaker. Further processing of the seed samples was the same as described above for leaf samples except that the seed samples were centrifuged at 2,000g at 4°C for 50 min to remove insoluble materials prior to filtration.
Leaf and seed extracts were analyzed with HPLC coupled with tandem mass spectrometry as described (Gu et al., 2007). For quantification, mixtures of L-isomers of 20 protein amino acids, GABA, anthranilate, homo-Ser (Ho-Ser), Hyp, and S-methyl Met of varying concentrations plus Phe-d8 and Val-d8 of 0.9 μm were run along with the plant samples.
Leaf Fatty Acid Assay by Gas Chromatography-Flame Ionization Detector
For measurement of leaf fatty acid contents, two leaves (numbers 5 and 6 counting from the newest visible leaf) were harvested from each plant beginning 5 h after the light period started. Fatty acids were transmethylated in 1 mL of 1 n methanolic HCl containing 5 μg/mL pentadecanoic acid (15:0) standard and 10 μg/mL butylated hydroxytoluene at 80°C for 30 min. One milliliter of 0.9% NaCl and 0.15 mL of heptane were added to the methylated samples. One microliter of the heptane phase were separated using a J & W DB-23 capillary column on an Agilent 6890 series gas chromatography system with a flame ionization detector (Agilent; Bonaventure et al., 2003).
In Arabidopsis leaves, 16:3 mainly exists in plastidial lipids, such as monogalactosyldiacylglycerol, and trans-16:1d3 is exclusively present in plastidial phosphatidylglycerol, whereas 18:0 and 18:2 are more abundant in extraplastidial lipids, such as phosphatidylethanolamine (Bonaventure et al., 2003). Thus, (16:3+trans-16:1d3)/(18:0+18:2) and 16:3/18:2 were calculated to estimate the ratio between plastidial and extraplastidial membrane lipids. Also, in Arabidopsis leaves, 16:0 and 18:1 are the major products of plastidial fatty acid synthesis exported to the cytosol (Bonaventure et al., 2003). Ratio 16:0/(18:1d9+18:1d11) was calculated to assess and to identify mutants with altered ratio between saturated fatty acids and unsaturated fatty acids among fatty acids that were exported from chloroplasts to the cytosol.
Seed C and N Assay
Seeds harvested from plants grown under the 16-/8-h photoperiod were desiccated under vacuum for 48 h, weighed with an AP110 Analytical Plus balance (Ohaus), and packed into tin capsules (CE Elantech). Approximately 10 to 12 mg of desiccated seeds was analyzed by the Duke Environmental Stable Isotope Laboratory (http://www.biology.duke.edu/jackson/devil/). The C and N contents in the seeds were quantified by combusting the seeds at 1,200°C in an elemental analyzer in the presence of chemical catalysts. Seed C/N ratio was calculated to estimate the relative abundance of storage oil and storage protein in seeds because, in Arabidopsis seeds, approximately 90% of N is in protein and more than 50% of C exists in oil (Baud et al., 2002).
Chlorophyll Fluorescence Assay
Plants used for chlorophyll fluorescence assay were grown for 3 weeks in one flat of eight 13- × 13-cm subflats (12 pots per subflat, 1 plant per pot) so that each subflat could be analyzed with the MAXI version of the IMAGING-PAM M-Series chlorophyll fluorescence system (Heinz-Walz Instruments). The system was equipped with an AVT Dolphin camera (Allied Vision Technologies). The growth conditions were the same as those for leaf assays (see above). The plants were dark-adapted for 20 min before measurement. Maximum photochemical efficiency of PSII (Fv/Fm) before high light, and NPQ, i.e. (F°m − F′m)/F′m, were determined at the beginning of the light period according to Maxwell and Johnson (2000). The plants were then treated with high light (1,600 μmol m−2 s−1 PPFD) for 3 h and Fv/Fm after high light was determined. The plants were returned to the growth chamber under standard growth conditions (100 μmol m−2 s−1 PPFD) for 2 d and Fv/Fm after recovery was then determined. False-color images were recorded, stored, and compared with the ImagingWin software provided with the instrument. Cutoff values were determined for each flat empirically: Fv/Fm before high light, 0.765; Fv/Fm after high light, 0.482; Fv/Fm after recovery, 0.745; NPQ, 0.302 to 0.349. Any plants with one or more values below the corresponding parameter cutoff value were considered a putative hit.
Data Analysis
All statistical analyses were performed with JMP 6.0 statistical software (SAS Institute).
Before data from different assays were combined for analysis, morphological traits and qualitative traits were systematically coded into numeric form (Bucciarelli et al., 2006), with the details summarized in Table VIII. Results from morphological and other qualitative assays contain two types of data: dichotomous data and ordinal data. Traits with dichotomous data include inflorescence, leaf color variation, leaf shape, trichomes, chloroplast shape, seed coat color variation, seed coat surface, seed shape, seed coat starch, Fv/Fm before high light, Fv/Fm after high light, Fv/Fm after recovery, and NPQ. Traits with ordinal data include rosette size, leaf color, leaf number, mature leaf size, chloroplast number, chloroplast size, seed coat color, seed size, and leaf starch.
Before raw quantitative data from different flats or plates were merged, O'Brien's test was conducted to test the homogeneity of variance across flats and plates (O'Brien, 1979). This analysis showed that variance across flats or plates was not significantly different (p > 0.01) for 19 protein amino acids in leaves and seeds, leaf fatty acids, and seed C and N contents. The only exception was Met, which did not show uniform variance across the two plates. Based upon this analysis, data from different flats and plates were combined.
The Shapiro-Wilk test (Shapiro and Wilk, 1965) was used to test the normality of quantitative data from Col and Ws wild-type samples, i.e. leaf amino acid composition, leaf fatty acid composition and ratios, seed amino acid composition, and seed C and N composition and ratio. The data from each ecotype were analyzed separately. The α value was set to 0.01.
To minimize the problem of multiple comparisons involving a control, Dunnett's test, instead of the more commonly employed Student's t test, was used to compare means between the mutants and their corresponding Col or Ws wild type (Dunnett, 1955; Bucciarelli et al., 2006). Unless otherwise addressed, differences were stated as significant when the p value was <0.05.
To allow comparisons of results between plants grown in the microenvironments of different flats, quantitative data (mol % of fatty acids, mol % of amino acids, and C/N ratio) were converted to z-scores (Schmid et al., 2005). Because mean and sd are sensitive to extreme phenotypes found in some of the mutants analyzed, median and median absolute deviation (MAD) of each flat were used in calculating z-scores (Rousseeuw and Croux, 1993). MAD is given by the equation MAD = 1.482 × medi (|xi − median|), where xi is the value of each individual measurement and medi is the median of the n absolute values of the deviations about the median. The z-score is calculated by the equation zi = (xi − median)/MAD. After conversion to z-scores, the homogeneity of variance was retested with O'Brien's method. This analysis showed that the variance was not significantly different (p > 0.05) across different flats or plates for each metabolite. Numeric codes and z-scores from different assays were merged with JMP 6.0 statistical software (SAS Institute). Data for plants with one or more missing values were not used in the analyses. Three seed stocks for the lkr-sdh mutant appeared to be wild-type contaminants and data from these three lines were deleted.
Methods for Visualizing Phenotypic Patterns
Clustering analyses were used to identify complex phenotypic relationships in the data. Heterogeneous data, including dichotomous and ordinal data from morphological and qualitative assays and continuous data from z-scores of amino acids, fatty acids, and C/N ratios, were merged by the unique barcode identifiers for the plant pots and seed stocks with JMP 6.0 statistical software (SAS Institute). The fatty acid ratios (16:3+trans-16:1d3)/(18:0+18:2), 16:3/18:2, and 16:0/(18:1d9+18:1d11) were excluded because individual fatty acids were included in the analysis. Seed C/N ratio was excluded because seed %C and seed %N were included. Data from different variables were standardized so that all variables have equal impact on the computation of distance. The final data table was analyzed by hierarchical clustering using Ward's minimum variance method and by k-means clustering (Quackenbush, 2001). Ward's method tends to join clusters with a small number of observations (Milligan, 1980), which is appropriate to identify mutants with a small number of replicates. This is in contrast to other hierarchical clustering methods, such as average linkage method, also known as the unweighted pair group method with arithmetic mean.
To identify mutants that are distinctly different from wild-type plants in multiple assays, the final data table was analyzed by principal components using the correlation matrix (Quackenbush, 2001).
Parametric Pearson product-moment correlation and nonparametric Spearman's ρ correlation were performed to determine the degree of correlation between pairs of traits among the complete set of data (148 samples and 85 phenotypic variables; Schmid et al., 2005). These methods were also used to test the contribution of individual mutants to the observed correlations. Multivariate outliers were detected using the Jackknife technique.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Distribution plot of significant Spearman's correlation coefficients.
Supplemental Figure S2. Impact of individual mutants on Pearson product-moment correlation between metabolic phenotypes.
Supplemental Figure S3. Hierarchical clustering of 148 samples by 81 variables without standardization.
Supplemental Table S1. Amino acid content (nmol/g FW) in leaves of Col wild type and mutants.
Supplemental Table S2. Amino acid content (nmol/g FW) in leaves of Ws wild type and mutants.
Supplemental Table S3. Amino acid content (nmol/g FW) in seeds of Col wild type and mutants.
Supplemental Table S4. Amino acid contents (nmol/g FW) in seeds of Ws wild type and mutants.
Supplemental Table S5. Fatty acids (nmol/g FW) in wild-type and mutant leaves.
Supplemental Table S6. Spearman's correlation of traits in wild-type plants with | ρ | > 0.5.
Supplemental Table S7. Spearman's correlation of traits in Col wild-type plants with | ρ | > 0.5.
Supplemental Table S8. k-means clustering analysis of mutants and wild-type plants in this study.
Supplementary Material
Acknowledgments
The authors thank Matt Larson and Drew Bomhof of the Michigan State University Research Technology Support Facility Bioinformatics Core for creation of the laboratory information management system. We are grateful to A. Daniel Jones of the Michigan State University Mass Spectrometry Facility for his advice on the amino acid assay, Mike Pollard for consulting on the fatty acid methyl ester analysis, and Joonyul Kim and Betsy Ampofo for help in configuring the chlorophyll fluorescence assay. We thank Jessica Reif, Amanda Charbonneau, Ardian Coku, David Hall, and Kayla Kerr for technical assistance. We thank Cathy Laurie for comments on the manuscript and Sandra E. Herman of Michigan State University, Center for Statistical Training and Consulting, for her statistical advice.
This work was supported by the National Science Foundation 2010 Project (grant no. MCB–0519740).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Robert L. Last (lastr@msu.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Abrahams S, Tanner GJ, Larkin PJ, Ashton AR (2002) Identification and biochemical characterization of mutants in the proanthocyanidin pathway in Arabidopsis. Plant Physiol 130 561–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen HM, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Ecker JR (2006) Moving forward in reverse: genetic technologies to enable genome-wide phenomic screens in Arabidopsis. Nat Rev Genet 7 524–536 [DOI] [PubMed] [Google Scholar]
- Andre C, Froehlich JE, Moll MR, Benning C (2007) A heteromeric plastidic pyruvate kinase complex involved in seed oil biosynthesis in Arabidopsis. Plant Cell 19 2006–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S, Boutin JP, Miquel M, Lepiniec L, Rochat C (2002) An integrated overview of seed development in Arabidopsis thaliana ecotype WS. Plant Physiol Biochem 40 151–160 [Google Scholar]
- Benning C (2004) Genetic mutant screening by direct metabolite analysis. Anal Biochem 332 1–9 [DOI] [PubMed] [Google Scholar]
- Bonaventure G, Salas JJ, Pollard MR, Ohlrogge JB (2003) Disruption of the FATB gene in Arabidopsis demonstrates an essential role of saturated fatty acids in plant growth. Plant Cell 15 1020–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouché N, Fromm H (2004) GABA in plants: just a metabolite? Trends Plant Sci 9 110–115 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM (1989) Genes directing flower development in Arabidopsis. Plant Cell 1 37–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Görlach J (2001) Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräutigam A, Gagneul D, Weber APM (2007) High-throughput colorimetric method for the parallel assay of glyoxylic acid and ammonium in a single extract. Anal Biochem 362 151–153 [DOI] [PubMed] [Google Scholar]
- Bucciarelli B, Hanan J, Palmquist D, Vance CP (2006) A standardized method for analysis of Medicago truncatula phenotypic development. Plant Physiol 142 207–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin PL, Williams EH, Last RL (1996) Environmental stress sensitivity of an ascorbic acid-deficient Arabidopsis mutant. Proc Natl Acad Sci USA 93 9970–9974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin PL, Norris SR, Wheeler GL, Williams EH, Smirnoff N, Last RL (1999) Genetic evidence for the role of GDP-mannose in plant ascorbic acid (vitamin C) biosynthesis. Proc Natl Acad Sci USA 96 4198–4203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin PL, Saracco SA, Norris SR, Last RL (2000) Identification of ascorbic acid-deficient Arabidopsis thaliana mutants. Genetics 154 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin PL, Gatzek S, Wheeler GL, Dowdle J, Raymond MJ, Rolinski S, Isupov M, Littlechild JA, Smirnoff N (2006) Arabidopsis thaliana VTC4 encodes L-galactose-1-P phosphatase, a plant ascorbic acid biosynthetic enzyme. J Biol Chem 281 15662–15670 [DOI] [PubMed] [Google Scholar]
- Coruzzi GM, Last RL (2000) Amino acids. In RB Buchanan, W Gruissem, R Jones, eds, Biochemistry and Molecular Biology of Plants. American Society of Plant Physiology Press, Rockville, MD, pp 358–410
- Cui J, Li P, Li G, Xu F, Zhao C, Li Y, Yang Z, Wang G, Yu Q, Li Y, et al (2008) AtPID: Arabidopsis thaliana protein interactome database an integrative platform for plant systems biology. Nucleic Acids Res 36 D999–D1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunnett CW (1955) A multiple comparison procedure for comparing several treatments with a control. J Am Stat Assoc 50 1096–1121 [Google Scholar]
- Fernie AR, Trethewey RN, Krotzky AJ, Willmitzer L (2004) Metabolite profiling: from diagnostics to systems biology. Nat Rev Mol Cell Biol 5 763–769 [DOI] [PubMed] [Google Scholar]
- Fiehn O, Kopka J, Dörmann P, Altmann T, Trethewey RN, Willmitzer L (2000) Metabolite profiling for plant functional genomics. Nat Biotechnol 18 1157–1161 [DOI] [PubMed] [Google Scholar]
- Geisler-Lee J, O'Toole N, Ammar R, Provart NJ, Millar AH, Geisler M (2007) A predicted interactome for Arabidopsis. Plant Physiol 145 317–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentry MS, Dowen RH, Worby CA, Mattoo S, Ecker JR, Dixon JE (2007) The phosphatase laforin crosses evolutionary boundaries and links carbohydrate metabolism to neuronal disease. J Cell Biol 178 477–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Véronneau S, Dow S, Lucau-Danila A, Anderson K, André B, et al (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature 418 387–391 [DOI] [PubMed] [Google Scholar]
- Gibon Y, Blaesing OE, Hannemann J, Carillo P, Höhne M, Hendriks JHM, Palacios N, Cross J, Selbig J, Stitt M (2004) A robot-based platform to measure multiple enzyme activities in Arabidopsis using a set of cycling assays: comparison of changes of enzyme activities and transcript levels during diurnal cycles and in prolonged darkness. Plant Cell 16 3304–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J, Rogers EE, Ausubel FM (1996) Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics 143 973–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn JM, Miyagishima S, Yoder DW, Osteryoung KW, Vitha S (2007) Chloroplast division. Traffic 8 451–461 [DOI] [PubMed] [Google Scholar]
- Goyer A, Collakova E, de la Garza RD, Quinlivan EP, Williamson J, Gregory JF, Shachar-Hill Y, Hanson AD (2005) 5-Formyltetrahydrofolate is an inhibitory but well tolerated metabolite in Arabidopsis leaves. J Biol Chem 280 26137–26142 [DOI] [PubMed] [Google Scholar]
- Gu L, Jones AD, Last RL (2007) LC-MS/MS assay for protein amino acids and metabolically related compounds. Anal Chem 79 8067–8075 [DOI] [PubMed] [Google Scholar]
- Jander G, Norris SR, Rounsley SD, Bush DF, Levin IM, Last RL (2002) Arabidopsis map-based cloning in the post-genome era. Plant Physiol 129 440–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jander G, Baerson SR, Hudak JA, Gonzalez KA, Gruys KJ, Last RL (2003) Ethylmethanesulfonate saturation mutagenesis in Arabidopsis to determine frequency of herbicide resistance. Plant Physiol 131 139–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jander G, Norris SR, Joshi V, Fraga M, Rugg A, Yu SX, Li LL, Last RL (2004) Application of a high-throughput HPLC-MS/MS assay to Arabidopsis mutant screening; evidence that threonine aldolase plays a role in seed nutritional quality. Plant J 39 465–475 [DOI] [PubMed] [Google Scholar]
- Joshi V, Laubengayer KM, Schauer N, Fernie AR, Jander G (2006) Two Arabidopsis threonine aldolases are nonredundant and compete with threonine deaminase for a common substrate pool. Plant Cell 18 3564–3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan F, Kopka J, Sung DY, Zhao W, Popp M, Porat R, Guy CL (2007) Transcript and metabolite profiling during cold acclimation of Arabidopsis reveals an intricate relationship of cold-regulated gene expression with modifications in metabolite content. Plant J 50 967–981 [DOI] [PubMed] [Google Scholar]
- Kunst L, Browse J, Somerville C (1988) Altered regulation of lipid biosynthesis in a mutant of Arabidopsis deficient in chloroplast glycerol-3-phosphate acyltransferase activity. Proc Natl Acad Sci USA 85 4143–4147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahner B, Gong JM, Mahmoudian M, Smith EL, Abid KB, Rogers EE, Guerinot ML, Harper JF, Ward JM, McIntyre L, et al (2003) Genomic scale profiling of nutrient and trace elements in Arabidopsis thaliana. Nat Biotechnol 21 1215–1221 [DOI] [PubMed] [Google Scholar]
- Laing WA, Wright MA, Cooney J, Bulley SM (2007) The missing step of the L-galactose pathway of ascorbate biosynthesis in plants, an L-galactose guanyltransferase, increases leaf ascorbate content. Proc Natl Acad Sci USA 104 9534–9539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry LG, Stapleton AE, Lim J, Hoffman P, Hays JB, Walbot V, Last RL (1997) An Arabidopsis photolyase mutant is hypersensitive to ultraviolet-B radiation. Proc Natl Acad Sci USA 94 328–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Armstrong CM, Bertin N, Ge H, Milstein S, Boxem M, Vidalain P-O, Han J-DJ, Chesneau A, Hao T, et al (2004) A map of the interactome network of the metazoan C. elegans. Science 303 540–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linster CL, Gomez TA, Christensen KC, Adler LN, Young BD, Brenner C, Clarke SG (2007) Arabidopsis VTC2 encodes a GDP-L-galactose phosphorylase, the last unknown enzyme in the Smirnoff-Wheeler pathway to ascorbic acid in plants. J Biol Chem 282 18879–18885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Sharkey TD (2004) The role of amylomaltase in maltose metabolism in the cytosol of photosynthetic cells. Planta 218 466–473 [DOI] [PubMed] [Google Scholar]
- Maxwell K, Johnson GN (2000) Chlorophyll fluorescence—a practical guide. J Exp Bot 51 659–668 [DOI] [PubMed] [Google Scholar]
- McCallum CM, Comai L, Greene EA, Henikoff S (2000) Targeted screening for induced mutations. Nat Biotechnol 18 455–457 [DOI] [PubMed] [Google Scholar]
- Messerli G, Nia VP, Trevisan M, Kolbe A, Schauer N, Geigenberger P, Chen JC, Davison AC, Fernie AR, Zeeman SC (2007) Rapid classification of phenotypic mutants of Arabidopsis via metabolite fingerprinting. Plant Physiol 143 1484–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan GW (1980) An examination of the effect of 6 types of error perturbation on 15 clustering algorithms. Psychometrika 45 325–342 [Google Scholar]
- Niittylä T, Comparot-Moss S, Lue WL, Messerli G, Trevisan M, Seymour MDJ, Gatehouse JA, Villadsen D, Smith SM, Chen JC, et al (2006) Similar protein phosphatases control starch metabolism in plants and glycogen metabolism in mammals. J Biol Chem 281 11815–11818 [DOI] [PubMed] [Google Scholar]
- Niyogi KK, Grossman AR, Björkman O (1998) Arabidopsis mutants define a central role for the xanthophyll cycle in the regulation of photosynthetic energy conversion. Plant Cell 10 1121–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien RG (1979) A general ANOVA method for robust tests of additive models for variances. J Am Stat Assoc 74 877–880 [Google Scholar]
- Osteryoung KW, Stokes KD, Rutherford SM, Percival AL, Lee WY (1998) Chloroplast division in higher plants requires members of two functionally divergent gene families with homology to bacterial ftsZ. Plant Cell 10 1991–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke KA, Leech RM (1991) Rapid image analysis screening procedure for identifying chloroplast number mutants in mesophyll cells of Arabidopsis thaliana (L.) Heynh. Plant Physiol 96 1193–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quackenbush J (2001) Computational analysis of microarray data. Nat Rev Genet 2 418–427 [DOI] [PubMed] [Google Scholar]
- Roessner U, Luedemann A, Brust D, Fiehn O, Linke T, Willmitzer L, Fernie AR (2001) Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell 13 11–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseeuw PJ, Croux C (1993) Alternatives to the median absolute deviation. J Am Stat Assoc 88 1273–1283 [Google Scholar]
- Salaita L, Kar RK, Majee M, Downie AB (2005) Identification and characterization of mutants capable of rapid seed germination at 10°C from activation-tagged lines of Arabidopsis thaliana. J Exp Bot 56 2059–2069 [DOI] [PubMed] [Google Scholar]
- Schauer N, Semel Y, Roessner U, Gur A, Balbo I, Carrari F, Pleban T, Perez-Melis A, Bruedigam C, Kopka J, et al (2006) Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nat Biotechnol 24 447–454 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37 501–506 [DOI] [PubMed] [Google Scholar]
- Schoenbohm C, Martens S, Eder C, Forkmann G, Weisshaar B (2000) Identification of the Arabidopsis thaliana flavonoid 3′-hydroxylase gene and functional expression of the encoded P450 enzyme. Biol Chem 381 749–753 [DOI] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro SS, Wilk MB (1965) An analysis of variance test for normality (complete samples). Biometrika 52 591–611 [Google Scholar]
- Sokolov LN, Dominguez-Solis JR, Allary AL, Buchanan BB, Luan S (2006) A redox-regulated chloroplast protein phosphatase binds to starch diurnally and functions in its accumulation. Proc Natl Acad Sci USA 103 9732–9737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville C, Browse J, Jaworski JG, Ohlrogge J (2000) Lipids. In RB Buchanan, W Gruissem, R Jones, eds, Biochemistry and Molecular Biology of Plants. American Society of Plant Physiology Press, Rockville, MD, pp 456–527
- Sönnichsen B, Koski LB, Walsh A, Marschall P, Neumann B, Brehm M, Alleaume AM, Artelt J, Bettencourt P, Cassin E, et al (2005) Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature 434 462–469 [DOI] [PubMed] [Google Scholar]
- Susek RE, Ausubel FM, Chory J (1993) Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74 787–799 [DOI] [PubMed] [Google Scholar]
- Sweetlove LJ, Last RL, Fernie AR (2003) Predictive metabolic engineering: a goal for systems biology. Plant Physiol 132 420–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Bauwe H, Badger M (2007) Impairment of the photorespiratory pathway accelerates photoinhibition of photosystem II by suppression of repair but not acceleration of damage processes in Arabidopsis. Plant Physiol 144 487–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin HE, Lincoln K, Moshiri F, Jensen PK, Qi QG, Venkatesh TV, Karunanandaa B, Baszis SR, Norris SR, Savidge B, et al (2006) The Arabidopsis vitamin E pathway gene5-1 mutant reveals a critical role for phytol kinase in seed tocopherol biosynthesis. Plant Cell 18 212–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voll LM, Allaire EE, Fiene G, Weber APM (2004) The Arabidopsis phenylalanine insensitive growth mutant exhibits a deregulated amino acid metabolism. Plant Physiol 136 3058–3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakao S, Benning C (2005) Genome-wide analysis of glucose-6-phosphate dehydrogenases in Arabidopsis. Plant J 41 243–256 [DOI] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, et al (1999) Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285 901–906 [DOI] [PubMed] [Google Scholar]
- Xu CC, Yu B, Cornish AJ, Froehlich JE, Benning C (2006) Phosphatidylglycerol biosynthesis in chloroplasts of Arabidopsis mutants deficient in acyl-ACP glycerol-3-phosphate acyltransferase. Plant J 47 296–309 [DOI] [PubMed] [Google Scholar]
- Yoder DW, Kadirjan-Kalbach D, Olson BJ, Miyagishima SY, DeBlasio SL, Hangarter RP, Osteryoung KW (2007) Effects of mutations in Arabidopsis FtsZ1 on plastid division, FtsZ ring formation and positioning, and FtsZ filament morphology in vivo. Plant Cell Physiol 48 775–791 [DOI] [PubMed] [Google Scholar]
- Yu TS, Kofler H, Hausler RE, Hille D, Flügge UI, Zeeman SC, Smith AM, Kossmann J, Lloyd J, Ritte G, et al (2001) The Arabidopsis sex1 mutant is defective in the R1 protein, a general regulator of starch degradation in plants, and not in the chloroplast hexose transporter. Plant Cell 13 1907–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XH, Tang GL, Granier F, Bouchez D, Galili G (2001) A T-DNA insertion knockout of the bifunctional lysine-ketoglutarate reductase/saccharopine dehydrogenase gene elevates lysine levels in Arabidopsis seeds. Plant Physiol 126 1539–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.