Abstract
Pterin-4a-carbinolamine dehydratases (PCDs) recycle oxidized pterin cofactors generated by aromatic amino acid hydroxylases (AAHs). PCDs are known biochemically only from animals and one bacterium, but PCD-like proteins (COG2154 in the Clusters of Orthologous Groups [COGs] database) are encoded by many plant and microbial genomes. Because these genomes often encode no AAH homologs, the annotation of their COG2154 proteins as PCDs is questionable. Moreover, some COG2154 proteins lack canonical residues that are catalytically important in mammalian PCDs. Diverse COG2154 proteins of plant, fungal, protistan, and prokaryotic origin were therefore tested for PCD activity by functional complementation in Escherichia coli, and the plant proteins were localized using green fluorescent protein fusions. Higher and lower plants proved to have two COG2154 proteins, a mitochondrial one with PCD activity and a noncanonical, plastidial one without. Phylogenetic analysis indicated that the latter is unique to plants and arose from the former early in the plant lineage. All 10 microbial COG2154 proteins tested had PCD activity; six of these came from genomes with no AAH, and six were noncanonical. The results suggested the motif [EDKH]-x(3)-H-[HN]-[PCS]-x(5,6)-[YWF]-x(9)-[HW]-x(8,15)-D as a signature for PCD activity. Organisms having a functional PCD but no AAH partner include angiosperms, yeast, and various prokaryotes. In these cases, PCD presumably has another function. An ancillary role in molybdopterin cofactor metabolism, hypothesized from phylogenomic evidence, was supported by demonstrating significantly lowered activities of two molybdoenzymes in Arabidopsis thaliana PCD knockout mutants. Besides this role, we propose that partnerless PCDs support the function of as yet unrecognized pterin-dependent enzymes.
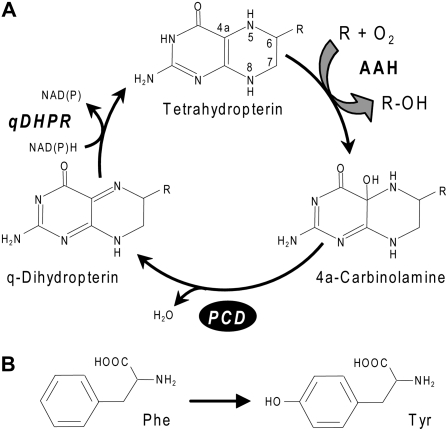
Pterin-4a-carbinolamine dehydratase (PCD; EC 4.2.1.96) is a small protein that mediates the first of two reactions in the recycling of tetrahydropterins, the cofactors of aromatic amino hydroxylases (AAHs; Thöny et al., 2000). PCD catalyzes the dehydration of the pterin-4a-carbinolamine formed in the hydroxylase reaction, giving a quinonoid (q)-dihydropterin that is then reduced to the tetrahydro level by q-dihydropterin reductase (Fig. 1A). The dehydration reaction can also occur spontaneously at a low rate (Thöny et al., 2000). PCDs are best known from mammals but have also been cloned and functionally verified from fruit flies (Seong et al., 1998), apicomplexan protists (Wang et al., 2006), and the bacterium Pseudomonas aeruginosa (Zhao et al., 1994).
Figure 1.
The role of PCD in regenerating the tetrahydropterin cofactor of AAHs. A, The cofactor regeneration cycle. R = Phe, Tyr, or Trp. B, The Phe → Tyr hydroxylation reaction mediated by Phe hydroxylase, a representative tetrahydropterin-dependent AAH. In AAH reactions, the tetrahydropterin cofactor is oxidized to a 4a-carbinolamine. The carbinolamine is dehydrated via the action of PCD to give a q-dihydropterin, which is then reduced to the tetrahydro level by an NAD(P)H-linked q-dihydropterin reductase (qDHPR). The natural cofactor for AAHs is tetrahydrobiopterin in mammals (Thöny et al., 2000) and may be tetrahydromonapterin in bacteria (Guroff and Rhoads, 1969).
PCDs have no established metabolic role beyond supporting the function of AAHs by regenerating their cofactors, and this role is substantiated by genetic evidence (Thöny et al., 1998; Song et al., 1999). In animals, PCD supports three different AAHs that act on Phe, Tyr, or Trp (Hufton et al., 1995), while in P. aeruginosa and related bacteria, PCD supports a single Phe hydroxylase that is induced by Phe (Nakata et al., 1979; Song et al., 1999). Phe hydroxylases in animals and P. aeruginosa serve a catabolic function, converting Phe to Tyr (Fig. 1B) that can then be degraded via the homogentisate pathway (Gu et al., 1998; Arias-Barrau et al., 2004; Moran, 2005).
Crystal structures, sequences, and site-directed mutagenesis of animal and P. aeruginosa PCDs point to a canonical catalytic motif, [DE]-x(3)-H-H-P-x(5)-[YW]-x(9)-H-x(8)-D (PROSITE syntax; Hulo et al., 2006), in which the three His residues are particularly critical (Cronk et al., 1996; Köster et al., 1996, 1998; Suck and Ficner, 1996; Rebrin et al., 1998). Crystal structures also establish that mammalian PCD is a tetramer with a “dimers of dimers” configuration, whereas bacterial PCDs are dimers (Suck and Ficner, 1996; Rose et al., 2004). The tetramerization relates to a second function of mammalian PCD as a transcriptional coactivator that binds to, and enhances activity of, HNF1 transcription factors; this nonmetabolic function is termed Dimerization Cofactor of HNF1 (DCoH; Suck and Ficner, 1996). DCoH activity is unrelated to PCD activity; the HNF1 and pterin binding sites are separate, and complex formation with HNF1 does not affect PCD activity (Rhee et al., 1997). The DCoH function, like HNF1 proteins, is confined to mammals and other vertebrates and is thought to have coevolved with tetramerization in the vertebrate lineage (Rose et al., 2004). Bacterial PCD has no role in transcription (Song et al., 1999).
This classical picture of PCD distribution and function, based on very few organisms, has been challenged by the advent of hundreds of sequenced genomes. Inspection of these genomes indicates that PCD-like sequences (COG2154 in the Clusters of Orthologous Groups [COGs] database; Tatusov et al., 2003) occur widely in plants, fungi, protists, archaea, and bacteria. However, such inspection also shows that COG2154 sequences in many plants and other organisms occur without an AAH (Wang et al., 2006), and that in some of them the inferred catalytic motif is not conserved. These observations cast doubt on whether all COG2154 proteins have PCD activity, making their annotation as PCDs (as is common in current databases) unwarranted without further evidence. Another reason for caution in annotating COG2154 proteins is that known PCDs share as little as 20% to 30% amino acid identity (Suck and Ficner, 1996; Seong et al., 1998).
This situation led us to conduct a comprehensive comparative genomic (phylogenomic) analysis of the COG2154 proteins of plants and microorganisms to assess their diversity and possible function. This analysis guided experiments to test the PCD activity of representative COG2154 proteins and to localize them in plants. The results established that plants have functional PCDs and strongly implied that PCDs have other metabolic roles besides supporting the function of AAHs.
RESULTS
Phylogenomic Analysis of COG2154 in Plants and Microbes
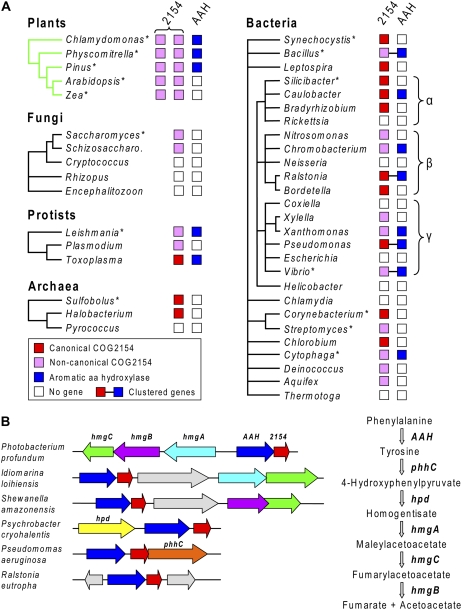
We first systematically analyzed the distribution of COG2154 sequences in relation to that of AAH sequences in plants, fungi, protists, and prokaryotes. COG2154 sequences were identified using the National Center for Biotechnology Information (NCBI) Conserved Domain (CD) search tool (Marchler-Bauer et al., 2005). For prokaryotes, the enzymes of the Tyr-catabolizing homogentisate pathway were included in the analysis because of the known catabolic role of P. aeruginosa Phe hydroxylase and PCD (Gu et al., 1998). The results are summarized in Figure 2 and given in detail in Supplemental Table S1. Five points emerged from the data.
Figure 2.
Phylogenomic analysis of COG2154 proteins and AAH homologs in plants and microorganisms. A, Distribution of canonical and noncanonical COG2154 proteins and AAH homologs. COG2154 sequences taken as canonical contain the motif [DE]-x(3)-H-H-P-x(5)-[YW]-x(9)-H-x(8)-D. Major groups of plants and microorganisms are represented by single genera. For plant groups, the genus is typical of all members of the group. For microorganism groups, the genus has a distribution pattern that is common in the group but not necessarily universal. Gene clustering in prokaryotes is symbolized by connecting lines. The major groups of proteobacteria (α, β, γ) are indicated. Asterisks denote organisms whose COG2154 proteins were tested for PCD activity in this study. Schizosaccharo., Schizosaccharomyces. B, Clustering of bacterial genes encoding COG2154 and AAH with each other and with genes of the homogentisate pathway of Tyr degradation. Matching colors correspond to orthologous genes; gray arrows are unrelated genes. The steps in the homogentisate pathway and the corresponding genes are shown on the right.
First, COG2154 sequences of two types were found in all plant taxa examined (chlorophyte algae, mosses, gymnosperms, and angiosperms). Second, a single COG2154 sequence was found in all protists surveyed and in many but not all fungi, bacteria, and archaea. Third, COG2154 sequences are not accompanied by an AAH sequence in many organisms, including angiosperms, fungi, and a diverse array of prokaryotes. Fourth, in bacteria with both COG2154 and AAH, these genes are often adjacent and sometimes clustered with homogentisate pathway genes. Fifth, certain COG2154 sequences lack one or more of the canonical His residues or other putatively crucial residues or show atypical spacing of the latter. These points are explored below in more detail. For simplicity and clarity, hereafter in the text we designate test organisms by their genus name only.
Plant COG2154 Sequences
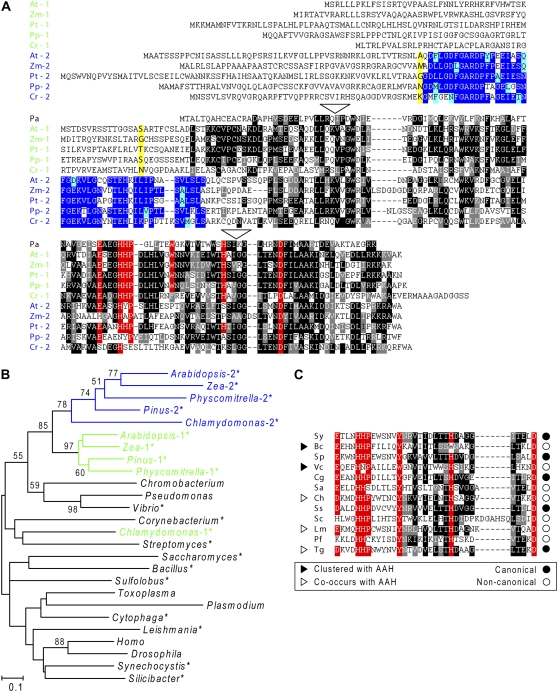
Both types of COG2154 sequences in plants have N-terminal extensions of approximately 60 to 100 amino acids compared to Pseudomonas PCD but are otherwise clearly distinct from each other (Fig. 3A). The first type (type 1) has an almost canonical catalytic motif, departing only in having an extra residue between the H-H-P and [YW] (i.e. Y or W) elements. This type is typically predicted by three algorithms (TargetP, Predotar, and PSORT) to be targeted to mitochondria. The second type of sequence (type 2) departs radically from the canon; most notably, the [YW] element is invariably absent, and, in all cases except Pinus, one or more of the His residues are missing. Moreover, a key feature of type 2 sequences not shared with type 1 or with any other COG2154 sequences is a conserved, approximately 45-residue domain just upstream of the core COG2154 domain (Fig. 3A). Type 2 sequences are most often predicted to be plastidial.
Figure 3.
Primary structures and phylogeny of COG2154 proteins. A, Multiple sequence alignment of representative plant type 1 and type 2 COG2154 proteins. At, Arabidopsis thaliana; Zm, Zea mays; Pt, Pinus taeda; Pp, Physcomitrella patens; Cr, Chlamydomonas reinhardtii. P. aeruginosa (Pa) PCD is included for comparison and defines the approximate extent of the core COG2154 domain common to all sequences in the alignment. The domain unique to type 2 proteins is in blue. Identical residues are shaded in black or dark blue, similar residues in gray or light blue. Residues of the catalytic motif [DE]-x(3)-H-H-P-x(5)-[YW]-x(9)-H-x(8)-D are shaded in red. Residues bolded and shaded in yellow were changed to an initiation codon in the constructs used in complementation assays. Dashes are gaps introduced to maximize alignment. White triangles show positions of two introns present in type 1 and type 2 proteins from Arabidopsis and Physcomitrella and the type 2 protein from Chlamydomonas, but not in other eukaryotic proteins. B, Unrooted neighbor-joining tree for COG2154 proteins from plants and representative species of animals, protists, fungi, and prokaryotes. The sequences analyzed were those tested in this study (asterisked) and those known or inferred from the literature to have PCD activity. Bootstrap values are indicated only for nodes with >50% support. Evolutionary distances are in units of the number of amino acid substitutions per site. C, Sequence alignment of the catalytic motif region of selected non-plant COG2154 proteins from the phylogenetic analysis. Shading is as in part A. Sy, Synechocystis sp.; Bc, Bacillus cereus; Sp, Silicibacter pomeroyi; Vc, Vibrio cholerae; Cg, Corynebacterium glutamicum; Sa, Streptomyces avermitilis; Ch, Cytophaga hutchinsonii; Ss, Sulfolobus solfataricus; Sc, Saccharomyces cerevisiae; Lm, Leishmania major; Pf, Plasmodium falciparum; Tg, Toxoplasma gondii.
Phylogenetic analysis of representative COG2154 sequences from plants and other organisms placed plant type 1 and type 2 proteins in two clades that branched together, with high bootstrap values, implying a shared evolutionary history (Fig. 3B). An exception was the Chlamydomonas type 1 sequence, which did not cluster with the rest. Due to the diverged nature of COG2154 sequences, other branching patterns were poorly resolved, reflected by low bootstrap values. The close relationship between plant type 1 and 2 sequences was corroborated by a comparison of intron positions: type 1 and 2 genes were found to share two introns not found in other eukaryotes (Fig. 3A). Chlamydomonas type 1 COG2154 was again unusual because it lacked both of these introns. Taken together, the above data basically suggest that an archetypal type 2 sequence arose early within the plant lineage from a type 1 sequence and acquired a novel domain.
Although all plant groups have COG2154 sequences, not all have AAH sequences. As indicated in Figure 2A, Chlamydomonas, Physcomitrella, and Pinus have proteins very like animal and bacterial AAHs (35%–45% amino acid identity), but Arabidopsis and Zea lack proteins with any detectable similarity to AAHs. Data from other plant genomes and ESTs (not shown) confirmed the generality of this pattern: nonflowering plants have AAH sequences, angiosperms do not.
Microorganism COG2154 Sequences
COG2154 sequences were found in all protists surveyed, in ascomycete fungi, and in diverse groups of bacteria and archaea (Fig. 2A), generally as a single copy. Of 467 complete bacterial and archaeal genomes analyzed, 211 genomes (45%) encoded COG2154 proteins and represented almost all the major taxa. Many of the bacteria lacking COG2154 proteins are obligate intracellular organisms, such as Rickettsia, Coxiella, and Chlamydia (Fig. 2A) that have ceded various metabolic functions to the host. Of the 211 genomes with COG2154, fewer than one-half (90) have an AAH sequence, so that the “partnerless COG2154” situation noted above for angiosperms is very common among prokaryotes.
The 90 COG2154 genes that co-occur with AAH can reasonably be presumed a priori to encode active PCDs. Moreover, as in Pseudomonas (Zhao et al., 1994), these COG2154 genes are often adjacent to an AAH gene in an operonic structure, providing additional circumstantial evidence for their functionality (Fig. 2B). Yet further support for this inference is the fairly frequent clustering of COG2154 and AAH genes with one or more genes of the homogentisate pathway (Fig. 2B; Supplemental Table S1).
Given the strong probability of PCD activity that is implied by association evidence (i.e. co-occurrence and clustering), it is noteworthy that microbial COG2154 proteins that are associated in these ways with AAH quite often lack a canonical catalytic motif (Fig. 3C). This implies that the current motif (based only on PCDs from animals and one bacterium) is too narrowly defined. If so, it would follow that the various noncanonical microbial and plant COG2154 proteins that have no AAH partner could also be active PCD enzymes.
Tests of PCD Activity
The above issues led us to test a range of plant and prokaryote COG2154 proteins for PCD activity and, in the process, to reexamine the catalytic motif. We used a functional complementation assay in Escherichia coli (Song et al., 1999; Wang et al., 2006) rather than in vitro assays because the latter involve artificial pterin-4a-carbinolamine substrates (which may not necessarily be attacked by all PCDs) or lack sensitivity due to a background rate of chemical dehydration (Citron et al., 1992; Rebrin et al., 1995).
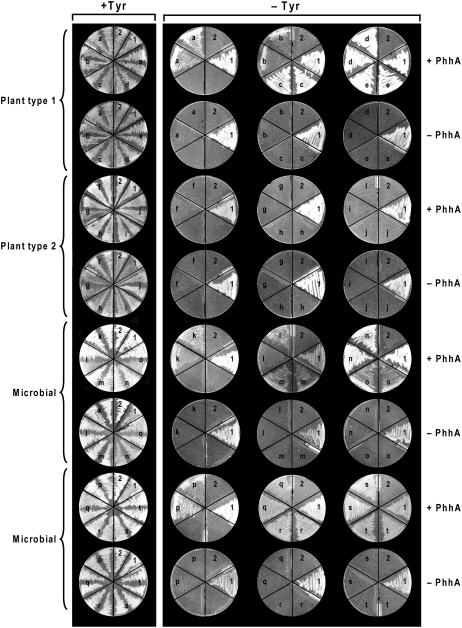
The complementation assay uses an E. coli Tyr auxotroph. Because E. coli lacks both Phe hydroxylase and PCD but has q-dihydropterin reductase activity (Fig. 1A), Tyr prototrophy can be restored by coexpression of foreign Phe hydroxylase and PCD genes; neither gene alone suffices (Zhao et al., 1994; Song et al., 1999). The Pseudomonas Phe hydroxylase gene (phhA) and selected COG2154 sequences were accordingly cloned into compatible plasmids and introduced into the mutant. Transformants were then tested for growth in the presence or absence of Tyr (Fig. 4). The COG2154 proteins chosen included plant types 1 and 2 from five major taxa (green algae, mosses, gymnosperms, monocots, and eudicots; Fig. 3A) and 10 microbial proteins that were either noncanonical or from genomes lacking AAH or both (Fig. 3C), these being potentially the most informative. The microbial proteins were also selected for taxonomic diversity; they represented fungi, protists, archaea, and five phyla of bacteria. The plant proteins were truncated at the positions shown in Figure 3A to remove their N-terminal extensions.
Figure 4.
Assay of PCD activity by functional complementation in E. coli. A Tyr auxotroph (strain JP2255) was transformed with pACYC177 containing P. aeruginosa phhA or with pACYC177 (with the Ap gene inactivated) alone, plus pBluescript SK− alone or containing P. aeruginosa PCD (phhB) or COG2154 sequences from plants (a–j) or microbes (k–t). The doubly transformed cells were plated on medium plus or minus Tyr. Each plate included a positive control (sector 1, phhA + P. aeruginosa PCD) and a negative control (sector 2, phhA + pBluescript). Key to COG2154 genes: a, Arabidopsis thaliana 1 (At1g29810); b, Chlamydomonas reinhardtii 1; c, Physcomitrella patens 1; d, Pinus taeda 1; e, Zea mays 1; f, A. thaliana 2 (At5g51110); g, C. reinhardtii 2; h, P. patens 2; i, P. taeda 2; j, Z. mays 2; k, Corynebacterium glutamicum; l, Streptomyces avermitilis; m, Cytophaga hutchinsonii; n, Vibrio cholerae; o, Bacillus cereus; p, Sulfolobus solfataricus; q, Silicibacter pomeroyi; r, Synechocystis; s, Saccharomyces cerevisiae; t, Leishmania major.
Among the plant proteins, all type 1 sequences (whose catalytic motif is close to canonical) had PCD activity, whereas all the type 2 sequences lacked activity (Fig. 4, top four rows). All the microbial proteins proved to be active (Fig. 4, bottom four rows). While we cannot exclude the possibility that the truncation point used for the plant type 2 proteins caused loss of activity, we consider it unlikely because this point lay well upstream of the core COG2154 domain (Fig. 3A).
Subcellular Localization of Plant Proteins
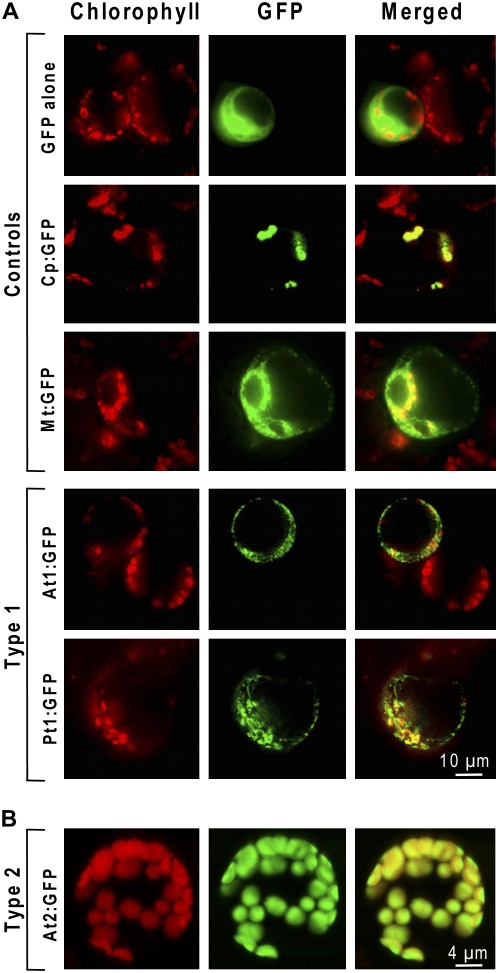
To test whether the predicted organellar targeting sequences of type 1 and 2 proteins are functional in planta, selected COG2154 coding sequences were fused to GFP and expressed in Arabidopsis. Subcellular localization in protoplasts was visualized by epifluorescence or confocal laser scanning microscopy. The Arabidopsis and Pinus type 1 proteins gave many small, punctate GFP signals that did not coincide with chloroplasts, indicating a mitochondrial location (Fig. 5A). In contrast, the Arabidopsis type 2 protein typically gave a strong GFP signal that precisely overlapped with chlorophyll fluorescence (Fig. 5B). Our results for the Arabidopsis type 2 protein agree with a brief report, based on a yellow fluorescent protein fusion, that this protein is chloroplastic (Valkai, 2004).
Figure 5.
Organellar targeting of plant types 1 and 2 COG2154 proteins fused to GFP. Chlorophyll autofluorescence (left) and GFP fluorescence (middle) are merged in the right-hand segments. A, Transient expression in Arabidopsis protoplasts of GFP fused to the type 1 protein of Arabidopsis (At1g29810, At1, fourth row) or Pinus (Pt1, fifth row). The top three rows show controls: cytosolic expression of GFP alone (top row), chloroplastic expression of GFP fused to the transit peptide of the Rubisco small subunit (second row), and mitochondrial expression of GFP fused to hydroxymethyldihydropterin pyrophosphokinase-dihydropteroate synthase (third row). Fluorescence was observed with an epifluorescence microscope. B, Confocal laser scanning micrographs of mesophyll protoplasts isolated from leaves of transgenic Arabidopsis plants expressing GFP fused to the type 2 protein of Arabidopsis (At5g51110, At2).
Essentiality Data and Functional Predictions
The puzzling occurrence of COG2154 proteins with PCD activity in genomes with no AAH led us first to mine the microbial literature for data on COG2154 knockout mutants. Five reports were found, involving four organisms, three of which lack an AAH (Table I). In no case was the COG2154 gene classified as essential, although in the fission yeast Schizosaccharomyces, which has no AAH, deletants showed defective spore wall formation (Kakihara et al., 2003). In Pseudomonas, which has AAH, inactivating PCD abolished the ability to grow on Phe, as would be expected (Song et al., 1999). Relative to these findings of nonessentiality, it should be noted that PCD mutations in mammals give mild phenotypes. This may be partly due to nonenzymatic dehydration of the pterin-4a-carbinolamine (Thöny et al., 2000). The nonessentiality of PCD clearly constrains our ability to hypothesize or test its functions in organisms that lack AAH.
Table I.
Literature reports of COG2154 knockouts in microorganisms
| Organism | AAH | Essential | Phenotype | References |
|---|---|---|---|---|
| Schizosaccharomyces pombe | No | No | Defective spore walls | Kakihara et al. (2003) |
| Mycobacterium tuberculosis | No | No | Sassetti et al. (2003) | |
| Methylobacterium extorquens | No | No | Kalyuzhnaya et al. (2005) | |
| P. aeruginosa | Yes | No | Unable to grow on Phe | Song et al. (1999); Liberati et al. (2006) |
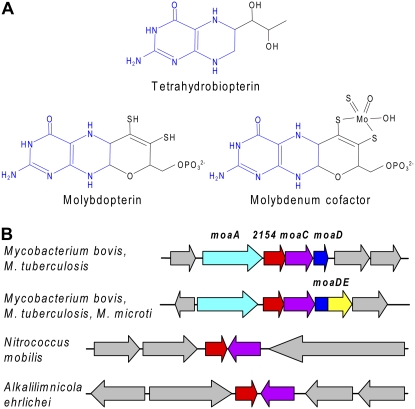
However, given that a pterin-4a-carbinolamine can be formed by chemical oxidation of a tetrahydropterin (Moore et al., 2002), one hypothesis is that PCD facilitates, but is not essential for, regeneration of chemically oxidized pterins. Besides AAH cofactors, another common type of pterin is the molybdenum cofactor (Moco) and its precursors (e.g. molybdopterin). Moco occurs in all life's kingdoms and is essential for about 40 enzymes, four of which occur in plants (Schwarz and Mendel, 2006). It is very sensitive to oxidation (Rajagopalan and Johnson, 1992). Moco has a ring structure that is electronically equivalent to a tetrahydropterin (Fig. 6A) and has some chemical characteristics of tetrahydropterins as well as others due to the third ring (Enemark and Garner, 1997; Nieter Burgmayer et al., 2004). Both substrate specificity studies (Rebrin et al., 1995) and the crystal structure of PCD complexed with a substrate analog (Cronk et al., 1996) indicate that PCD recognizes the pterin ring rather than its side chains. Thus, if Moco or its precursors form 4a-carbinolamines, they could be PCD substrates. Furthermore, some genomic evidence suggests a role for COG2154 proteins in Moco metabolism. Analysis of the genomic context of COG2154 genes in bacteria with no AAH reveals one embedded in molybdopterin synthesis gene clusters in Mycobacterium species and translationally coupled to moaC (Fig. 6B). A COG2154 gene is also next to moaC in some γ-proteobacteria (Fig. 6B). Indeed, COG2154 sequences in GenBank are occasionally annotated as “putative molybdopterin biosynthesis protein.”
Figure 6.
Structure of Moco and clustering of COG2154 genes with Moco biosynthesis genes in bacterial genomes. A, The structures of Moco and its precursor molybdopterin compared to that of a typical tetrahydropterin. The tetrahydropterin ring structure is colored blue. The form of Moco shown occurs in xanthine dehydrogenase and aldehyde oxidase. B, Clustering of COG2154 genes with up to four genes of Moco biosynthesis (moaA, moaC, moaD, and a moaD-moaE fusion). Matching colors correspond to orthologous genes; gray arrows are unrelated genes. Note that the genomes of Mycobacterium bovis and Mycobacterium tuberculosis each have two clusters and that the genes flanking the COG2154-moaC pair in Nitrococcus mobilis and Alkalilimnicola ehrlichei are different, so that only the pair is conserved.
A Link between Arabidopsis PCD and Moco Metabolism
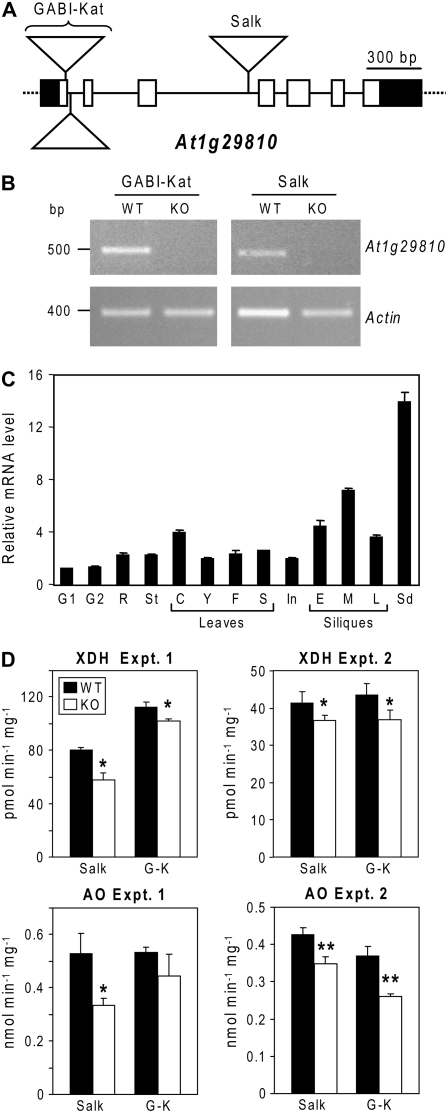
To explore a connection between PCD and Moco, two independent insertional mutants in the Arabidopsis gene (At1g29810) encoding the type 1 protein were isolated and authenticated; the mutants came from the Salk and GABI-Kat collections (Fig. 7, A and B). Both mutations were knockouts, as judged by absence of detectable At1g29810 mRNA (Fig. 7B). The expression of the At1g29810 gene in wild-type plants was examined and found to be constitutive, with levels (relative to actin) highest in seeds (Fig. 7C). Mutant homozygotes grew normally compared to wild type in soil or when cultured in vitro (not shown).
Figure 7.
Insertional knockout and expression of the At1g29810 gene and the effect of knockouts on activities of Moco-dependent enzymes. A, The At1g29810 gene showing the positions of the double T-DNA insert in the GABI-Kat and the single insert in the Salk line. Introns are represented as solid lines and exons by boxes; 5′ and 3′ untranslated regions are in black. B, RT-PCR validation that the insertions in both lines eliminate a functional At1g29810 message. Leaf RNA was used as RT-PCR template. Note the absence of an At1g29810 amplicon in both mutant lines and the presence of the control (actin) amplicon. C, Expression of At1g29810 in organs of Arabidopsis ecotype Columbia: G1 and G2, seedlings at 3 and 7 d of germination, respectively; R, root; St, stem; C, cauline leaves; Y, F, S, young, fully expanded, and senescent rosette leaves; In, inflorescence; E, M, L, siliques at early, mid, and late (yellowing) stages of development; Sd, dry seeds. D, Activities of xanthine dehydrogenase (XDH) and aldehyde oxidase (AO) in in vitro cultured plantlets of homozygous Salk and GABI-Kat (G-K) knockout lines (KO) and the corresponding wild-type segregants (WT). The data are from two separate experiments, with three (experiment 1) or four to five (experiment 2) independent replicates. Error bars are se. Asterisks indicate activities in the knockout that are significantly lower (*, P < 0.05; **, P < 0.01) than in the corresponding wild type.
Plantlets grown in vitro were tested for the activities of the four molybdoenzymes, nitrate reductase, sulfite oxidase, xanthine dehydrogenase, and aldehyde oxidase. As these enzymes lose activity in plants with Moco synthesis defects (Schwarz and Mendel, 2006), we reasoned that there should also be some activity loss if PCD facilitates salvage of 4a-carbinolamines formed by oxidation of Moco or its precursors. Given the normal phenotype of the mutant plants, the activity loss was predicted to be modest, mandating high precision (low variability) in measuring enzyme activities. Pilot experiments showed that nitrate reductase and sulfite oxidase activities were low and too variable (coefficient of variation among replicates up to approximately 40%) to allow detection of subtle differences. We accordingly focused on xanthine dehydrogenase and aldehyde oxidase and developed sensitive fluorometric HPLC assays for both enzymes (see “Materials and Methods” and Supplemental Figure S1). In two separate experiments, xanthine dehydrogenase activity was significantly (P < 0.05) lower in both knockout lines than in the corresponding wild-type controls, the activity loss being between 10% and 28% (Fig. 7D). In the same experiments, aldehyde oxidase activity was also consistently lower in the knockout lines (by 17%–36%), the differences being statistically significant (P < 0.05 or <0.01) in all but one case (Fig. 7D).
DISCUSSION
The results of in vivo tests of COG2154 sequences from plants and diverse microorganisms (Fig. 4; Wang et al., 2006) allow definition of a new catalytic motif that can be used as a signature for PCD activity in any organism: [EDKH]-x(3)-H-[HN]-[PCS]-x(5,6)-[YWF]-x(9)-[HW]-x(8,15)-D. This motif, which is far broader than the one based solely on mutational and structural studies of mammalian and Pseudomonas PCDs, can be used to more reliably annotate COG2154 genes from plants and other organisms. By assuming that all microbial COG2154 genes co-occurring with AAH genes (Supplemental Table S1) encode active PCDs, the motif can be broadened slightly further to [EDKHQN]-x(3)-H-[HN]-[PCS]-x(5,6)-[YWFH]-x(9)-[HW]-x(8,15)-D.
Our study demonstrates that COG2154 genes specifying active PCDs occur in the absence of AAH genes in many microorganisms and in angiosperms. While this surprising situation in angiosperms could be due to an evolutionarily recent loss of AAH that has not yet been followed by loss of the redundant PCD, this explanation seems improbable in light of the prevalence of PCDs in microorganisms with no AAH. It is more plausible to suppose that PCD has at least one other function besides recycling the pterin cofactors of AAHs and that this function is common to plants, fungi, bacteria, and archaea. What could this function be?
We obtained evidence to support the intriguing scenario, suggested by phylogenomics, that PCD has a role in the metabolism of Moco or its precursors, at least in Arabidopsis. Such a role would necessarily be ancillary, not central, to Moco formation because: (1) no genes are missing from the Moco biosynthesis pathway (Schwarz and Mendel, 2006); (2) various Moco-producing bacteria (e.g. E. coli) lack PCD; (3) PCD mutants are viable in Arabidopsis and microorganisms; and (4) ablating PCD activity reduced molybdoenzyme activity only modestly. One possibility is that PCD helps salvage 4a-carbinolamines formed by oxidation of Moco biosynthesis intermediates or Moco itself. As the first steps in Moco synthesis in plants may be mitochondrial (Schwarz and Mendel, 2006), PCD, a mitochondrial enzyme, would have ready access to a 4a-carbinolamine formed in the early part of the synthesis pathway. However, later biosynthesis steps, and all four Moco-containing enzymes, are extramitochondrial in plants, so that a role for PCD at these levels would require trafficking of 4a-carbinolamines and pterins in and out of mitochondria. Although carbinolamine oxidation products of Moco precursors have not so far been detected (Santamaria-Araujo et al., 2004), their formation is not chemically unreasonable. In this connection, it is worth noting that even for the best known pterin, tetrahydrobiopterin, whose chemical oxidation has received considerable attention for 30 years, definitive evidence for carbinolamine formation during hydrogen peroxide-mediated oxidation emerged only quite recently (Moore et al., 2002). Lastly, it should be noted that, while reduced cofactor activity is the simplest explanation for the parallel activity reductions in two Moco enzymes in Arabidopsis PCD knockouts, our data do not exclude the possibility that this is due to correlated reductions in apoprotein levels.
There are several other possible functions for PCDs. First, they might facilitate recycling of chemically oxidized tetrahydropterins. However, this still leaves the question of why such hypothetical tetrahydropterins would need to be recycled if there is no AAH. A second possibility, that PCDs recycle chemically oxidized tetrahydrofolates (which have a tetrahydropterin ring), might seem attractive because one common folate, 5-methyltetrahydrofolate, is known to form a 4a-hydroxy derivative upon oxidation (Gregory, 2007). However, this compound is not a likely substrate for PCD because this enzyme's action is considered to require a proton at the N5 position (Fig. 1A; Rebrin et al., 1998) and a methyl group replaces this proton in 5-methyltetrahydrofolate. A closely related possibility, albeit only in some prokaryotes, is that PCD recycles oxidized tetrahydromethanopterin (a tetrahydrofolate analog confined to methanogens and methylotrophs). Favoring this, PCD genes in the genomes of aerobic methylotrophic bacteria cluster with genes of tetrahydromethanopterin synthesis and metabolism (Kalyuzhnaya et al., 2005).
A third possibility is that genomes with a PCD but no AAH have other pterin-dependent enzymes that generate 4a-carbinolamines. We view this as probable because mammals are known to have a pterin-dependent glyceryl ether monooxygenase (Taguchi and Armarego, 1998) that has not been cloned and therefore cannot be recognized in genomes. Moreover, other pterin-dependent monooxygenases have been reported from microorganisms, but again not cloned (Reddy and Vaidyanathan, 1975; Bhat and Vaidyanathan, 1976). Another formal possibility is that PCD proteins participate in transcriptional regulation, as in mammals. However, there is experimental evidence against this in Pseudomonas (Song et al., 1999), and such a role is hard to reconcile with the distribution pattern of COG2154 genes in bacteria, where some species of a genus may have COG2154 and others not (e.g. Bacillus and Corynebacterium; Supplemental Table S1). Furthermore, a transcriptional role like that in mammals would not involve the PCD catalytic site (Rhee et al., 1997), and so is inconsistent with the conservation of catalytic residues and of enzyme activity in COG2154 proteins that have no AAH partner. Lastly, neither plant type 1 nor type 2 proteins were apparently targeted to the nucleus, which a transcriptional role would require.
Our results show that plants, unlike other organisms surveyed, have two distinct types of COG2154 protein. Type 1 is canonical (according to the catalytic motif just defined), has PCD activity, and is located in mitochondria. Type 2 is unique to plants, lacks PCD activity, is localized in plastids, and apparently arose from type 1 early in plant evolution. Apart from lacking the catalytic motif, type 2 proteins have a characteristic subterminal domain that includes the motif [GE]-[DN]-[FL]-G-A-R-D-P-x(3)-E-x(4)-F-G-[DE]K (Fig. 3A), which can be used for positive identification. That the type 2 protein is not a functional PCD, yet is apparently evolved from the type 1 enzyme, implies that it has taken on a different role. The presence in type 2 proteins of the subterminal domain fits with this idea. In an exploratory Arabidopsis study, overexpressing the type 2 gene (At5g51110) had no apparent phenotypic effect, and its suppression by RNA interference resulted in a small (approximately 10%) but significant reduction in leaf pigment content and a larger reduction (approximately 30%) in chloroplast number per mesophyll cell (Plume, 2002). The function of type 2 proteins is thus unclear. We propose that they be annotated as “PCD-like protein” until more is known.
For the plant mitochondrial PCD to participate in pterin recycling would require the existence of a q-dihydropterin reductase (Fig. 1A), which, given the lability of dihydropterins, seems likely also to be mitochondrial. q-Dihydropterin reductases in mammals, protists, and certain bacteria belong to the large and diverse short chain dehydrogenase-reductase family (Lye et al., 2002; Wilquet et al., 2004). Although Arabidopsis has some 86 short chain dehydrogenase-reductase proteins, only eight have predicted mitochondrial targeting peptides, and none of the eight yet has a known function (Noiriel et al., 2007). One or more of these could thus well have q-dihydropterin reductase activity.
MATERIALS AND METHODS
Bioinformatics
Microbial genomes were analyzed using the SEED database and its tools (Overbeek et al., 2005) at http://anno-3.nmpdr.org/anno/FIG/subsys.cgi. Results are available in full in the “Pterin carbinolamine dehydratase subsystem” on the public SEED server at http://theseed.uchicago.edu/FIG/ and in summary form in Supplemental Table S1. Plant genomes and/or ESTs were sought using BLAST algorithms and the NCBI (http://www.ncbi.nlm.nih.gov/), Department of Energy Joint Genome Institute (http://www.jgi.doe.gov/), and Chlamydomonas (http://www.chlamy.org/chlamydb.html) databases. PCD-like sequences were identified using the CD database and search tool (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml), which includes COG and pfam classifications as well as CD classifications. As all three classifications are equivalent in the case of PCD-like proteins, we used the COG identifier (COG2154) in preference to the CD (cd00488) or pfam (pfam01329) identifier because the COG system is widely used in genome annotation. Other resources were the integrated microbial genomes system (http://img.jgi.doe.gov/cgi-bin/pub/main.cgi) and the COG database (http://www.ncbi.nlm.nih.gov/COG/). Sequence alignments (Fig. 3, A and C) were made using MultAlin (http://bioinfo.genopole-toulouse.prd.fr/multalin/multalin.html). Phylogenetic analyses were made with MEGA 4 (Tamura et al., 2007).
Microbial COG2154 Genes
Genomic DNA of Bacillus cereus strain NRS 248, Corynebacterium glutamicum strain 534, Silicibacter pomeroyi strain DSS-3, Streptomyces avermitilis strain MA-4680, Synechocystis sp. strain PCC 6803, and Cytophaga hutchinsonii strain NCIB 9469 was purchased from the American Type Culture Collection. For Sulfolobus solfataricus strain DSM 1617 and Saccharomyces cerevisiae strain 971/6c (a gift from M.L. Agostini Carbone, Università di Milano), DNA was extracted from cells; a S. cerevisiae colony was suspended in 0.5 mL of water and microwaved for 3 min, and lyophilized S. solfataricus cells were suspended in 0.5 mL of water and boiled for 10 min. Extracts then were centrifuged for 10 min at 10,000g. The COG2154 gene of Vibrio cholerae (GenBank accession no. NC_002506) was synthesized by GenScript, adding 5μ NdeI and 3μ KpnI sites. The COG2154 coding sequences of Synechocystis and S. cerevisiae were PCR-amplified using KOD HiFi polymerase (Novagen) and primers harboring 5μ BamHI and 3μ KpnI sites. Primer sequences for COG2154 constructs are given in Supplemental Table S2. The forward primers contained a BamHI site, a stop codon in frame with LacZ, a Shine-Dalgarno sequence, and an NdeI site that included the start codon. The amplicons were cloned between the BamHI and KpnI sites of pBluescript SK− and introduced into Escherichia coli strain DH5α. The sequence-verified construct containing the Synechocystis gene was the parent of all other COG2154 constructs except that from Leishmania major. COG2154 coding sequences were PCR-amplified from genomic DNA using Taq (Invitrogen) or KOD HiFi polymerase; dimethyl sulfoxide 6% (v/v) was added to the reaction mixture in the case of C. glutamicum. The control PCD gene phhB from Pseudomonas aeruginosa was amplified from plasmid pJZ9-4 (Song et al., 1999). Amplicons were digested with NdeI and KpnI and then ligated into the Synechocystis construct from which the COG2154 sequence had been excised with NdeI and KpnI. The L. major strain FV1 COG2154 gene was amplified from genomic DNA using Phusion DNA polymerase (New England BioLabs), a forward primer containing an XbaI site, a stop codon in frame with LacZ, a Shine-Dalgarno sequence, and a reverse primer containing an EcoRI site. The amplicon was cloned into pGEM-T Easy (Promega). The insert was then excised with XbaI and EcoRI and cloned into the matching sites in pBluescript SK−. pBluescript SK− constructs were introduced into E. coli strain DH5α or DH10B and sequenced.
Plant COG2154 cDNAs
For complementation tests, plant COG2154 type 1 and 2 cDNAs were truncated, using PCR to replace their N-terminal putative targeting sequences by a start codon. Primers (Supplemental Table S2) harbored NdeI and KpnI sites as above. Type 1 and 2 cDNAs were amplified from the following ESTs: GenBank numbers BAC41856 and U13619 for Arabidopsis (Arabidopsis thaliana); BU647613 and BI999118 for Chlamydomonas reinhardtii; BJ203205 and BQ039368 for Physcomitrella patens; DT635299 and CO168958 for Pinus taeda; and DN218430 and EC883040 for Zea mays.
Functional Complementation Tests
Plasmid pJS11, consisting of the P. aeruginosa phhA cloned into pACYC177 (Song et al., 1999), provided the phhA gene for complementation tests. The matching vector-only control plasmid (pACYCΔAp) was constructed by digesting pACYC177 with ScaI and FspI to delete an internal fragment of the ampicillin-resistance gene (Ap) and religating. Electroporation was used to simultaneously transform E. coli strain JP2255 (aroF363 pheA361 pheO352 tyrA382 thi-1 strR712 lacY1 xyl-15; Zhao et al., 1994) with each COG2154 construct or pBluescript SK− and pJS11 or pACYCΔAp. Complementation tests were made by streaking transformed cells on plates of minimal medium containing M9 salts (Sambrook et al., 1989), 0.1 mm CaCl2, 0.5 mm MgSO4, trace elements (Sherman, 1991), 0.4% (w/v) Glc, thiamine (17 μg mL−1), isopropylthio-β-galactoside (0.5 or 1 mm), ampicillin (100 μg mL−1), streptomycin (100 μg mL−1), kanamycin (100 μg mL−1), Phe (50 μg mL−1), plus or minus Tyr (54 μg mL−1). The agar concentration in plates was 15 g L−1. Plates were incubated at room temperature (22°C) for 2 to 7 d.
Construction and Expression in Arabidopsis of GFP Fusion Proteins
Full-length cDNAs of two plant type 1 COG2154 sequences (At1g29810 and its P. taeda ortholog) were amplified using primers At1g29810-GFP-Fwd and At1g29810-GFP-Rev and PtPCD-GFP-Fwd and PtPCD-GFP-Rev, respectively (Supplemental Table S3). The PCR products were digested with SalI and NcoI and cloned in-frame upstream of the GFP sequence in the pTH2 plasmid (Niwa, 2003). Transient expression in protoplasts, controls for GFP targeting, and epifluorescence microscopy were as previously described (Pinon et al., 2005).
The coding region of type 2 COG2154 sequence At5g51110 was cloned into pAVA393 in frame with the GFP-coding region under the control of the cauliflower mosaic virus 35S promoter. The pAVA393 plasmid is based on pAVA319 (von Arnim et al., 1998) but contains the GFP5 variant (Siemering et al., 1996). The cauliflower mosaic virus 35S promoter-At5g51110-gfp5-35S terminator cassette from this vector was subcloned into the binary vector pSLJ755I5, a derivative of pRK290 (Jones et al., 1992). Arabidopsis (ecotype Columbia) plants were transformed by the floral dip method (Clough and Bent, 1998). Herbicide selection was performed by spraying seedlings growing in soil with 0.4% glufosinate (Basta). Protoplasts for confocal laser scanning microscopy were isolated from leaves of single-copy, homozygous T3 plants by incubation at 30°C for 1 h in a solution containing 400 mm d-mannitol, 8 mm CaCl2·2H2O, 5 mm MES-KOH, pH 5.6, 2% (w/v) SERVA Onozuka cellulase R-10, and 0.5% (w/v) SERVA macerozyme R-10. Protoplasts were analyzed using a Bio-Rad Radiance 2000 microscope with blue excitation (DAPI filter set) and green (fluorescein isothiocyanate filter set; GFP) or red (tetramethylrhodamine isothiocyanate filter set; chlorophyll autofluorescence) emission.
Isolation of Arabidopsis At1g29810 Mutants
Two T-DNA insertional mutant lines (ecotype Columbia) were identified: line 648F10 was from the GABI-Kat collection and line SALK_121474 from the Salk collection. Wild-type or homozygote mutant segregants from each line were identified by PCR screening using At1g29810 gene-specific primers located 5′ and 3′ of the insertion site and primers located on the T-DNA (Supplemental Table S3). The insertion sites were confirmed by sequencing the amplicons obtained from mutant homozygotes. The kanamycin resistance gene was shown by PCR with primers nptB and nptD (Supplemental Table S3) to cosegregate with the insertion, indicating the absence of insertions at other loci. Line 648F10 was shown to have duplicate T-DNA insertions at the same locus in opposite orientation (Fig. 7A). Homozygous mutants and wild-type segregants were selfed and the progeny used for experiments. For reverse transcription (RT)-PCR experiments to demonstrate absence of a functional At1g29810 mRNA in the mutant lines, RNA was prepared from plantlets 4 weeks old and treated as described below. Primers used to analyze At1g29810 expression were located in the first and last exons. These primers and those for the Actin7 gene (At5g09810) are given in Supplemental Table S3. For PCR, 30 ng of cDNA was used per reaction. After 5 min at 95°C, the reaction was carried out with 35 cycles of 95°C for 30 s, 45°C for 30 s and 72°C for 50 s, and a final step at 72°C for 10 min.
RNA Isolation and Analysis
Tissues were ground in liquid N2. Total RNA from the following tissues was isolated using RNeasy plant mini kits (Qiagen): seedlings 3 or 7 d old, roots from hydroponic culture, rosette leaves from plants at 16, 30, or 42 d of age, stems and cauline leaves from plants at 42 d of age, inflorescences, and siliques in early development. Total RNA from siliques at mid or late (yellowing) stages of development and from dry seeds was extracted using a LiCl precipitation method (Vicient and Delseny, 1999). The samples were dissolved in ribonuclease-free water and DNase treated. For real-time PCR, 1 μg of RNA was reverse transcribed using a RevertAid first-strand cDNA synthesis kit (Fermentas) with random hexamers. The mixture was then diluted 5-fold with water. PCR primers for At1g29810 and for the reference gene Actin7 (At5g09810) are given in Supplemental Table S3; amplicon lengths were 69 and 71 bp, respectively. The At1g29810 amplicon spanned the third and fourth introns. PCR was performed using the MyiQ version 2.0 Real-Time PCR Detection system (Bio-Rad) using the SYBR Green PCR master mix (Applied Biosystems) in 25-μL mixtures containing 5 μL of the diluted cDNA solution and 300 nm of each primer. After 10 min at 95°C, the PCR reaction was carried out with 40 cycles of 95°C for 15 s and 60°C for 60 s. Data were analyzed using MyiQ software. Expression values were normalized to that of Actin7, calculating the difference between the crossing threshold of At1g29810 and that of Actin7 for each sample (Livak and Schmittgen, 2001). The results presented are for cDNA pools from three independent RNA extractions, each analyzed in triplicate. The identity of the PCR products was confirmed by sequencing.
Culture of Arabidopsis Plantlets for Molybdoenzyme Assays
Arabidopsis seeds were surface sterilized and germinated on plates containing agar and 0.33× Murashige and Skoog medium. At 12 d, seedlings were transferred aseptically to 250-mL flasks (seven per flask) containing 100 mL of 0.33× Murashige and Skoog medium containing 10 g L−1 Suc and cultured for 17 d. Flasks were shaken at 80 rpm. Temperature was 23°C to 28°C. Daylength was 12 h; photosynthetic photon flux density was 80 μE m−2 s−1. Harvested plantlets from each flask (constituting one replicate sample) were frozen in liquid N2 and stored at −80°C.
Extraction and Assay of Molybdoenzymes
Protein extracts for assay of xanthine dehydrogenase, aldehyde oxidase, and sulfite oxidase were prepared as follows. Samples were ground in liquid N2 and the resulting powder was extracted in ice-cold buffer (1:3, w/v) containing 100 mm potassium phosphate, pH 7.5, 0.1 mm Na2MoO4, 1 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, 1 mm dithiothreitol, and 10% (w/v) polyvinylpolypyrrolidone. Extracts were centrifuged (30,000g, 20 min, 4°C), and supernatants were fractionated with ammonium sulfate (0%–60% saturation), stirring for 30 min at 4°C. After centrifuging (40,000g, 25 min, 4°C), pellets were resuspended in 1 to 2 mL of 100 mm potassium phosphate, pH 7.5, and 0.2-mL aliquots were desalted on 1-mL Sephadex G25 spin columns equilibrated with 100 mm potassium phosphate, pH 7.5, or, for sulfite oxidase assays, 20 mm Tris-acetate, pH 8.0, 0.1 mm EDTA.
Xanthine dehydrogenase was assayed with pterin as substrate (Mest et al., 1992; Montalbini, 1998). Pterin (Sigma) and isoxanthopterin (Schircks) were dissolved in 0.1 m NaOH and the solutions were titered spectrophotometrically using extinction coefficients of 21,380 m−1 cm−1 at 251 nm and 13,800 m−1 cm−1 at 339 nm, respectively (Pfleiderer, 1985). Assays (50 μL) contained 20 μL of protein extract, 0.1 mm pterin, and 1 mm 2,6-dichlorophenolindophenol in 100 mm potassium phosphate, pH 7.5. Triplicate assays were run at 30°C for 0, 30, and 60 min and stopped by adding 5 μL of 1 n HCl. After centrifuging, a portion of the supernatant was injected onto a 4-μm, 250- × 4.6-mm Synergi Fusion RP 80 column (Phenomenex), which was eluted isocratically (1.5 mL min−1) for 20 min using 20 mm Gly, pH 2.5, or 10 mm sodium phosphate, pH 6.0. The column was washed for 5 min with a 0% to 50% acetonitrile gradient then re-equilibrated in separation buffer for 5 min. Peaks were quantified by fluorescence (350 nm excitation, 450 nm emission). Aldehyde oxidase was assayed with indole-3-carboxaldehyde as substrate (Seo et al., 1998). Indole-3-carboxaldehyde and indole-3-carboxylic acid (Aldrich) were dissolved in water. Assays (50 μL) contained 20 μL of protein extract and 0.17 mm indole-3-carboxaldehyde in 100 mm potassium phosphate, pH 7.5. Triplicate assays were run at 30°C for 0, 30, and 60 min and stopped, processed, and analyzed by HPLC as above, using 10 mm sodium phosphate, pH 6.0, as eluant. The indole-3-carboxylic acid product was quantified by fluorescence (280 nm excitation, 360 nm emission). Xanthine dehydrogenase and aldehyde oxidase activities were measured in two separate experiments. Experiment 1 had three independent knockout and wild-type replicates. Experiment 2 had four to five such replicates, and knockout and wild-type samples were extracted and analyzed in pairs. Statistically significant differences were evaluated using one-tailed Student's t tests for independent (experiment 1) or paired (experiment 2) samples.
Sulfite oxidase assays (100 μL) contained 20 to 50 μL of desalted extract and 0.4 mm K3Fe(CN)6, in 20 mm Tris-acetate, pH 8.0, containing 0.1 mm EDTA. Sodium sulfite (2 μL, 20 mm) was added and the reaction was monitored spectrophotometrically by following reduction of ferricyanide (extinction coefficient 1,020 m−1 cm−1) at 420 nm, correcting for the base-line rate in absence of sulfite (Garrett and Rajagopalan, 1994; Eilers et al., 2001).
Nitrate reductase activity was extracted by grinding tissue (0.1–0.5 g) in a mortar with ice-cold 100 mm HEPES-KOH, pH 7.5, 2 mm EDTA, 7 mm Cys (1:5, w/v). After centrifugation at 14,000g for 10 min at 4°C, 60-μL samples of the supernatant were immediately added to assay mixtures (700 μL final volume) containing 100 mm HEPES-KOH, pH 7.5, 2 mm EDTA, 0.2 mm NADH, and 5 mm KNO3 (Jonassen et al., 2008). Assays were run in triplicate at 22°C for up to 40 min and stopped by adding 700 μL of 1% sulfanilamide and 0.02% N-(naphthyl)-ethylene-diamine dihydrochloride in 1.5 m HCl. The color was allowed to develop for 30 min before absorbance was measured at 540 nm.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Xanthine dehydrogenase and aldehyde oxidase assays by HPLC.
Supplemental Table S1. Distribution and clustering of microbial genes encoding COG2154.
Supplemental Table S2. Synthetic oligonucleotides used to amplify COG2154 sequences for complementation tests.
Supplemental Table S3. Synthetic oligonucleotides used to construct GFP fusion constructs, to screen segregating populations, and to study At1g29810 gene expression.
Supplementary Material
Acknowledgments
We thank the following organizations for providing ESTs: the University of Leeds (UK), the RIKEN Bio Resource Center (Tsukuba-shi, Japan), the Carnegie Institute (Stanford, CA), the Schnable laboratory, Iowa State University (Ames, IA), the University of Georgia (Athens, GA), the J. Craig Venter Institute (Rockville, MD), and the Arizona Genomics Institute (Tucson, AZ).
This work was supported by the U.S. Department of Energy (grant no. DE–FG02–07ER64498 to V.C. and A.D.H.), by the National Institutes of Health (grant no. AI 21903 to S.M.B.), and by an endowment from the C.V. Griffin Sr. Foundation.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Andrew D. Hanson (adha@ufl.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Arias-Barrau E, Olivera ER, Luengo JM, Fernandez C, Galan B, Garcia JL, Diaz E, Minambres B (2004) The homogentisate pathway: a central catabolic pathway involved in the degradation of L-phenylalanine, L-tyrosine, and 3-hydroxyphenylacetate in Pseudomonas putida. J Bacteriol 186 5062–5077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat SG, Vaidyanathan CS (1976) Involvement of 4-hydroxymandelic acid in the degradation of mandelic acid by Pseudomonas convexa. J Bacteriol 127 1108–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citron BA, Davis MD, Milstien S, Gutierrez J, Mendel DB, Crabtree GR, Kaufman S (1992) Identity of 4a-carbinolamine dehydratase, a component of the phenylalanine hydroxylation system, and DCoH, a transregulator of homeodomain proteins. Proc Natl Acad Sci USA 89 11891–11894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Cronk JD, Endrizzi JA, Alber T (1996) High-resolution structures of the bifunctional enzyme and transcriptional coactivator DCoH and its complex with a product analogue. Protein Sci 5 1963–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers T, Schwarz G, Brinkmann H, Witt C, Richter T, Nieder J, Koch B, Hille R, Hänsch R, Mendel RR (2001) Identification and biochemical characterization of Arabidopsis thaliana sulfite oxidase. J Biol Chem 276 46989–46994 [DOI] [PubMed] [Google Scholar]
- Enemark JH, Garner CD (1997) The coordination chemistry and function of the molybdenum centres of the oxomolybdoenzymes. J Biol Inorg Chem 2 817–822 [Google Scholar]
- Garrett RM, Rajagopalan KV (1994) Molecular cloning of rat liver sulfite oxidase. J Biol Chem 269 272–276 [PubMed] [Google Scholar]
- Gregory JF III (2007) Vitamins. In S Damodaran, K Parkin, OR Fennema, eds, Fennema's Food Chemistry, Ed 4. CRC Press, Boca Raton, FL, pp 439–521
- Gu W, Song J, Bonner CA, Xie G, Jensen RA (1998) PhhC is an essential aminotransferase for aromatic amino acid catabolism in Pseudomonas aeruginosa. Microbiology 144 3127–3134 [DOI] [PubMed] [Google Scholar]
- Guroff G, Rhoads CA (1969) Phenylalanine hydroxylation by Pseudomonas species (ATCC 11299a). Nature of the cofactor. J Biol Chem 244 142–146 [PubMed] [Google Scholar]
- Hufton SE, Jennings IG, Cotton RGH (1995) Structure and function of the aromatic amino acid hydroxylases. Biochem J 311 353–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulo N, Bairoch A, Bulliard V, Cerutti L, De Castro E, Langendijk-Genevaux PS, Pagni M, Sigrist CJ (2006) The PROSITE database. Nucleic Acids Res 34 D227–D230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonassen EM, Lea US, Lillo C (2008) HY5 and HYH are positive regulators of nitrate reductase in seedlings and rosette stage plants. Planta 227 559–564 [DOI] [PubMed] [Google Scholar]
- Jones JD, Shlumukov L, Carland F, English J, Scofield SR, Bishop GJ, Harrison K (1992) Effective vectors for transformation, expression of heterologous genes, and assaying transposon excision in transgenic plants. Transgenic Res 1 285–297 [DOI] [PubMed] [Google Scholar]
- Kakihara Y, Nabeshima K, Hirata A, Nojima H (2003) Overlapping omt1+ and omt2+ genes are required for spore wall maturation in Schizosaccharomyces pombe. Genes Cells 8 547–558 [DOI] [PubMed] [Google Scholar]
- Kalyuzhnaya MG, Korotkova N, Crowther G, Marx CJ, Lidstrom ME, Chistoserdova L (2005) Analysis of gene islands involved in methanopterin-linked C1 transfer reactions reveals new functions and provides evolutionary insights. J Bacteriol 187 4607–4614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köster S, Stier G, Ficner R, Hölzer M, Curtius HC, Suck D, Ghisla S (1996) Location of the active site and proposed catalytic mechanism of pterin-4a-carbinolamine dehydratase. Eur J Biochem 241 858–864 [DOI] [PubMed] [Google Scholar]
- Köster S, Stier G, Kubasch N, Curtius HC, Ghisla S (1998) Pterin-4a-carbinolamine dehydratase from Pseudomonas aeruginosa: characterization, catalytic mechanism and comparison to the human enzyme. Biol Chem 379 1427–1432 [DOI] [PubMed] [Google Scholar]
- Liberati NT, Urbach JM, Miyata S, Lee DG, Drenkard E, Wu G, Villanueva J, Wei T, Ausubel FM (2006) An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci USA 103 2833–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25 402–408 [DOI] [PubMed] [Google Scholar]
- Lye LF, Cunningham ML, Beverley SM (2002) Characterization of quinonoid-dihydropteridine reductase (QDPR) from the lower eukaryote Leishmania major. J Biol Chem 277 38245–38253 [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Cherukuri PF, DeWeese-Scott C, Geer LY, Gwadz M, He S, Hurwitz DI, Jackson JD, Ke Z, et al (2005) CDD: a Conserved Domain Database for protein classification. Nucleic Acids Res 33 D192–D196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mest SJ, Kosted PJ, van Kuijk FJ (1992) 2,6-Dichlorophenolindophenol is a competitive inhibitor for xanthine oxidase and is therefore not usable as an electron acceptor in the fluorometric assay. Free Radic Biol Med 12 189–192 [DOI] [PubMed] [Google Scholar]
- Montalbini P (1998) Purification and some properties of xanthine dehydrogenase from wheat leaves. Plant Sci 134 89–102 [Google Scholar]
- Moore J, Wood JM, Schallreuter KU (2002) H2O2-mediated oxidation of tetrahydrobiopterin: Fourier transform Raman investigations provide mechanistic implications for the enzymatic utilization and recycling of this essential cofactor. J Raman Spectrosc 33 610–617 [Google Scholar]
- Moran GR (2005) 4-Hydroxyphenylpyruvate dioxygenase. Arch Biochem Biophys 433 117–128 [DOI] [PubMed] [Google Scholar]
- Nakata H, Yamauchi T, Fujisawa H (1979) Phenylalanine hydroxylase from Chromobacterium violaceum. Purification and characterization. J Biol Chem 254 1829–1833 [PubMed] [Google Scholar]
- Nieter Burgmayer SJ, Pearsall DL, Blaney SM, Moore EM, Sauk-Schubert C (2004) Redox reactions of the pyranopterin system of the molybdenum cofactor. J Biol Inorg Chem 9 59–66 [DOI] [PubMed] [Google Scholar]
- Niwa Y (2003) A synthetic green fluorescent protein gene for plant biotechnology. Plant Biotechnol 20 1–11 [Google Scholar]
- Noiriel A, Naponelli V, Bozzo GG, Gregory JF III, Hanson AD (2007) Folate salvage in plants: pterin aldehyde reduction is mediated by multiple non-specific aldehyde reductases. Plant J 51 378–389 [DOI] [PubMed] [Google Scholar]
- Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, de Crécy-Lagard V, Diaz N, Disz T, Edwards R, et al (2005) The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res 33 5691–5702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfleiderer W (1985) Chemistry of naturally occurring pterins. In RL Blakley, SJ Benkovic, eds, Folates and Pterins, Vol 2. Wiley, New York, pp 43–114
- Pinon V, Ravanel S, Douce R, Alban C (2005) Biotin synthesis in plants. The first committed step of the pathway is catalyzed by a cytosolic 7-keto-8-aminopelargonic acid synthase. Plant Physiol 139 1666–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plume AM (2002) Functional characterisation of Arabidopsis DRGs: clues from the DRG2 interactor PDL1. PhD Thesis, University of Queensland, Brisbane, Australia
- Rajagopalan KV, Johnson JL (1992) The pterin molybdenum cofactors. J Biol Chem 267 10199–10202 [PubMed] [Google Scholar]
- Rebrin I, Bailey SW, Boerth SR, Ardell MD, Ayling JE (1995) Catalytic characterization of 4a-hydroxytetrahydropterin dehydratase. Biochemistry 34 5801–5810 [DOI] [PubMed] [Google Scholar]
- Rebrin I, Thony B, Bailey SW, Ayling JE (1998) Stereospecificity and catalytic function of histidine residues in 4a-hydroxy-tetrahydropterin dehydratase/DCoH. Biochemistry 37 11246–11254 [DOI] [PubMed] [Google Scholar]
- Reddy CC, Vaidyanathan CS (1975) Purification, properties and induction of a specific benzoate-4-hydroxylase from Aspergillus niger (UBC 814). Biochim Biophys Acta 384 46–57 [DOI] [PubMed] [Google Scholar]
- Rhee KH, Stier G, Becker PB, Suck D, Sandaltzopoulos R (1997) The bifunctional protein DCoH modulates interactions of the homeodomain transcription factor HNF1 with nucleic acids. J Mol Biol 265 20–29 [DOI] [PubMed] [Google Scholar]
- Rose RB, Pullen KE, Bayle JH, Crabtree GR, Alber T (2004) Biochemical and structural basis for partially redundant enzymatic and transcriptional functions of DCoH and DCoH2. Biochemistry 43 7345–7355 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Santamaria-Araujo JA, Fischer B, Otte T, Nimtz M, Mendel RR, Wray V, Schwarz G (2004) The tetrahydropyranopterin structure of the sulfur-free and metal-free molybdenum cofactor precursor. J Biol Chem 279 15994–15999 [DOI] [PubMed] [Google Scholar]
- Sassetti CM, Boyd DH, Rubin EJ (2003) Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol 48 77–84 [DOI] [PubMed] [Google Scholar]
- Schwarz G, Mendel RR (2006) Molybdenum cofactor biosynthesis and molybdenum enzymes. Annu Rev Plant Biol 57 623–647 [DOI] [PubMed] [Google Scholar]
- Seo M, Akaba S, Oritani T, Delarue M, Bellini C, Caboche M, Koshiba T (1998) Higher activity of an aldehyde oxidase in the auxin-overproducing superroot1 mutant of Arabidopsis thaliana. Plant Physiol 116 687–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong C, Jeong S, Park D, Yoon J, Oh Y, Yim J, Han K, Baek K (1998) Molecular characterization of the Drosophila melanogaster gene encoding the pterin 4alpha-carbinolamine dehydratase. Biochim Biophys Acta 1388 273–278 [DOI] [PubMed] [Google Scholar]
- Sherman F (1991) Getting started with yeast. Methods Enzymol 194 3–21 [DOI] [PubMed] [Google Scholar]
- Siemering KR, Golbik R, Sever R, Haseloff J (1996) Mutations that suppress the thermosensitivity of green fluorescent protein. Curr Biol 6 1653–1663 [DOI] [PubMed] [Google Scholar]
- Song J, Xia T, Jensen RA (1999) PhhB, a Pseudomonas aeruginosa homolog of mammalian pterin 4a-carbinolamine dehydratase/DCoH, does not regulate expression of phenylalanine hydroxylase at the transcriptional level. J Bacteriol 181 2789–2796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suck D, Ficner R (1996) Structure and function of PCD/DCoH, an enzyme with regulatory properties. FEBS Lett 389 35–39 [DOI] [PubMed] [Google Scholar]
- Taguchi H, Armarego WL (1998) Glyceryl-ether monooxygenase [EC 1.14.16.5]. A microsomal enzyme of ether lipid metabolism. Med Res Rev 18 43–89 [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24 1596–1599 [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al (2003) The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thöny B, Auerbach G, Blau N (2000) Tetrahydrobiopterin biosynthesis, regeneration and functions. Biochem J 347 1–16 [PMC free article] [PubMed] [Google Scholar]
- Thöny B, Neuheiser F, Kierat L, Rolland MO, Guibaud P, Schluter T, Germann R, Heidenreich RA, Duran M, de Klerk JB, et al (1998) Mutations in the pterin-4alpha-carbinolamine dehydratase (PCBD) gene cause a benign form of hyperphenylalaninemia. Hum Genet 103 162–167 [DOI] [PubMed] [Google Scholar]
- Valkai I (2004) 28D, a new component of the phytochrome B signal transduction, in Arabidopsis thaliana. Acta Biol Szeged 48 87 [Google Scholar]
- Vicient CM, Delseny M (1999) Isolation of total RNA from Arabidopsis thaliana seeds. Anal Biochem 268 412–413 [DOI] [PubMed] [Google Scholar]
- von Arnim AG, Deng XW, Stacey MG (1998) Cloning vectors for the expression of green fluorescent protein fusion proteins in transgenic plants. Gene 221 35–43 [DOI] [PubMed] [Google Scholar]
- Wang Q, Hauser V, Read M, Wang P, Hanson AD, Sims PF, Hyde JE (2006) Functional identification of orthologous genes encoding pterin recycling activity in Plasmodium falciparum and Toxoplasma gondii. Mol Biochem Parasitol 146 109–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilquet V, Van de Casteele M, Gigot D, Legrain C, Glansdorff N (2004) Dihydropteridine reductase as an alternative to dihydrofolate reductase for synthesis of tetrahydrofolate in Thermus thermophilus. J Bacteriol 186 351–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G, Xia T, Song J, Jensen RA (1994) Pseudomonas aeruginosa possesses homologues of mammalian phenylalanine hydroxylase and 4 alpha-carbinolamine dehydratase/DCoH as part of a three-component gene cluster. Proc Natl Acad Sci USA 91 1366–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.