Abstract
Strawberry (Fragaria × ananassa) fruit contains several anthocyanins that give the ripe fruits their attractive red color. The enzyme that catalyzes the formation of the first stable intermediate in the anthocyanin pathway is anthocyanidin-3-O-glucosyltransferase. A putative glycosyltransferase sequence (FaGT1) was cloned from a strawberry fruit cDNA library and the recombinant FaGT1 transferred UDP-glucose to anthocyanidins and, to a lesser extent, flavonols, generating the respective 3-O-glucosides. Quantitative polymerase chain reaction revealed that transcripts of FaGT1 were almost undetectable in green fruits, but gene expression increased dramatically in both turning and ripe red fruit, corresponding closely to the accumulation of anthocyanins during fruit ripening. The expression of FaGT1 is fruit associated and negatively regulated by auxin. To elucidate the in planta function of FaGT1, Agrobacterium tumefaciens cells harboring an intron-hairpin construct of a partial FaGT1 sequence were injected into midsized ripening fruits. In about one-third of the injected fruits, this led to significant down-regulation of FaGT1 transcript levels that corresponded to reduced concentrations of anthocyanin pigments in ripe strawberry fruits. In contrast, significant levels of epiafzelechin—formed by anthocyanidin reductase (ANR) from pelargonidin—were identified in FaGT1-silenced fruits, indicating competition of FaGT1 and FaANR for the common anthocyanidin substrate. Thus, FaGT1 represents an important branching-point enzyme because it is channeling the flavonoid pathway to anthocyanins. These results demonstrate a method to redirect the anthocyanin biosynthesis into flavan-3-ol production to increase the levels of bioactive natural products or modify pigments in plant tissues.
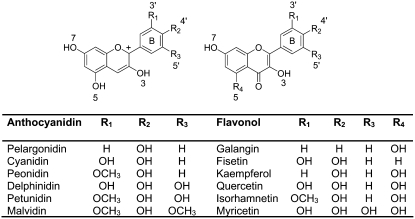
Strawberry (Fragaria × ananassa) is one of the most popular fruit crops worldwide and has emerged as a model for nonclimacteric fruit ripening (Giovannoni, 2001). The strawberry fruit is an aggregate composed of a swollen receptacle and the numerous achenes that are the true fruit (Hancock, 1999). In addition to its flavor, much of the popularity of strawberry fruit is due to the attractive, deep red color caused by anthocyanin pigments. Fruit ripening is triggered by a decline in the levels of auxins released by the achenes in early developmental stages (Perkins-Veazie, 1995). The major pigments of the strawberry cultivar ‘Elsanta’ are pelargonidin 3-O-glucoside (92%), followed by cyanidin 3-O-glucoside (4%) and minor amounts of other pelargonidin derivatives (Fig. 1; Bakker et al., 1994). More recently, metabolite analyses revealed that pelargonidin- and cyanidin-3-O-glucoside-malonate as well as anthocyanin-flavanol conjugates such as (epi)afzelechin-pelargonidin 3-O-glucoside also occur in strawberries (Fossen et al., 2004; Yoshida and Tamura, 2005). Interestingly, the achenes and the receptacle differ significantly in their anthocyanin composition (Aaby et al., 2005; Yoshida and Tamura, 2005). Achenes have higher concentrations of malonylated pigments and contain almost equal amounts of cyanidin- and pelargonidin-derived anthocyanins. Quercetin and kaempferol are the major flavonols in strawberry fruits and occur as 3-O-glucosides and 3-O-glucuronides (Ryan, 1971; Häkkinen et al., 1999). It has been reported that anthocyanins are undetectable in green and white fruits and that their levels increase rapidly in turning and red stages (Given et al., 1988; Cheng and Breen, 1991). In contrast, the concentrations of polyphenols and non-tannin flavonoids reach their maximum in green fruits and decrease afterward (Cheng and Breen, 1991; Wang and Lin, 2000).
Figure 1.
Anthocyanidins and flavonols are substrates of FaGT1 in vitro. Strawberries contain only pelargonidin- and cyanidin-derived pigments, mostly 3-O-glucosides (Bakker et al., 1994). Glycosides of quercetin and kaempferol are the main flavonols in strawberry fruit (Ryan, 1971; Häkkinen et al., 1999).
Whereas the chemical composition of the anthocyanins has been studied in detail, genetic and biochemical information about the last steps in anthocyanin biosynthesis in strawberry fruit is still limited (Almeida et al., 2007). The first stable product of the anthocyanin pathway is formed when a glycosyltransferase attaches a sugar molecule to the hydroxyl group at position 3 on the anthocyanidin aglycone. It has been reported that anthocyanin concentration and flavonoid-3-O-glucosyltransferase activity increase in parallel during fruit ripening (Given et al., 1988). Cheng et al. (1994) isolated a glucosyltransferase from strawberry fruit that preferentially glucosylates biochanin A and several flavonols, but does not accept any anthocyanidins. Recently, an anthocyanidin glucosyltransferase gene has been isolated from strawberry fruits and the recombinant protein preliminarily characterized (Almeida et al., 2007).
From grape (Vitis vinifera), another commercially important fruit crop, a UDP-Glc:flavonoid 3-O-glycosyltransferase (VvGT1) has been cloned that preferentially accepts anthocyanidins in vitro, whereas flavonols are less favored (Ford et al., 1998). Comparing glycosylation activities of recombinant VvGT1 and total enzyme extracts from different tissues, the authors concluded that VvGT1 is mainly responsible for the glucosylation of anthocyanidins, whereas additional glycosyltransferases would account for the formation of flavonol glycosides. This is in agreement with earlier findings that VvGT1 mRNA, unlike transcripts of other flavonoid pathway genes, can only be detected in the skin of red grape varieties and that gene expression of VvGT1 is induced after véraison (Boss et al., 1996a, 1996b). Recently, the three-dimensional structure of VvGT1 has been reported, which enabled the identification of key amino acids involved in substrate binding and catalytic activity (Offen et al., 2006).
Anthocyanidin 3-O-glucosyltransferases have been isolated from flowers of many ornamental plants in which anthocyanins are the major determinants of flower color, including Gentiana triflora, Petunia hybrida, and Iris hollandica (Tanaka et al., 1996; Yamazaki et al., 2002; Yoshihara et al., 2005). The genome of the model plant Arabidopsis (Arabidopsis thaliana) contains over 100 putative glycosyltransferase sequences, and most of the respective proteins have been expressed heterologously (Li et al., 2001; Ross et al., 2001). Of 91 glycosyltransferases tested, 29 have been reported to accept the flavonol quercetin (Lim et al., 2004). However, there is only one report on an enzyme from Arabidopsis that glycosylates anthocyanidins (Tohge et al., 2005).
Here, we report the cloning and biochemical characterization of a glucosyltransferase involved in anthocyanin biosynthesis in strawberry fruit. We provide data on the ripening-related and auxin-controlled expression of this gene. Whereas other techniques have been used to verify the function of glycosyltransferases in planta, this article reports on the RNA interference (RNAi)-mediated down-regulation of an anthocyanidin-3-O-glycosyltransferase gene in a commercially important fruit crop, thus confirming its function in planta.
RESULTS
Phylogenetic Analysis
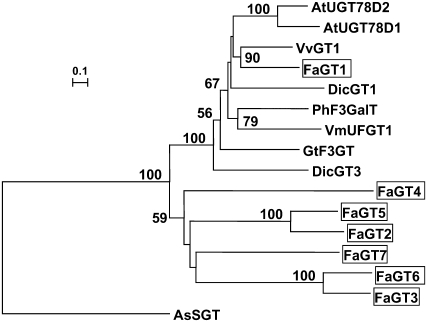
FaGT1 and three other putative glycosyltransferase genes (FaGT2–FaGT4) were identified among a set of 1,100 strawberry ESTs (Aharoni and O'Connell, 2002) and the full-length sequences were successfully cloned (Lunkenbein et al., 2006a). Further screening of other strawberry cDNA libraries and subsequent application of RACE led to the identification of three additional strawberry glycosyltransferase genes (FaGT5–FaGT7). Phylogenetic analysis (Fig. 2) revealed the relationship between FaGT1 and other strawberry as well as biochemically characterized plant glycosyltransferases. FaGT1 was most similar to the UDP-Glc:flavonoid 3-O-glycosyltransferase (VvGT1) from grape, which preferentially catalyzes the transfer of UDP-Glc to anthocyanidins (Ford et al., 1998). Similarities to glycosyltransferases from Dianthus caryophyllus (DicGT1, DicGT3; Ogata et al., 2004), P. hybrida (PhF3GalT; Miller et al., 1999), Vigna mungo (VmUFGT1; Mato et al., 1998), and G. triflora (GtF3GT; Tanaka et al., 1996) were also found, all of which transfer either UDP-Glc or UDP-Gal to anthocyanidins or flavonols. FaGT1 also shows close homology to UGT78D2 and UGT78D1 from Arabidopsis and can be allocated to group F of Arabidopsis glycosyltransferases. All genes from this group contain intron 2, which is the most widespread intron among the Arabidopsis glycosyltransferases (Li et al., 2001). A 166-bp intron inserted after position 493 of the FaGT1 open reading frame is consistent with the Arabidopsis intron. UGT78D2 has been expressed heterologously and has shown activity with anthocyanidins and flavonols using UDP-Glc as the sugar donor (Tohge et al., 2005). Whereas UGT78D1 was first reported to catalyze the transfer of UDP-Rha to position 3 of kaempferol and quercetin (Jones et al., 2003), it was later shown that UGT78D1 also accepts UDP-Glc (Lim et al., 2004). UGT78D2 has been identified as UDP-Glc:flavonoid 3-O-glucosyltransferase (Lee et al., 2005).
Figure 2.
Phylogenetic analysis of selected plant secondary product glycosyltransferases and putative strawberry glycosyltransferases (boxed). The neighbor-joining tree was calculated with the Treecon software package (van de Peer and de Wachter, 1994). Distance calculation was performed with Poisson correction and insertions/deletions were not taken into account. The tree was rooted using a sterol glycosyltransferase from Avena sativa (AsSGT) as an outgroup. Branch lengths indicate the number of substitutions per site. Bootstrap analysis was performed with 100 replicates and only values above 50% are shown. GenBank accession numbers and sources for the respective protein sequences are AtUGT78D2 (NP_197207; Arabidopsis), AtUGT78D1 (NP_564357; Arabidopsis), VvGT1 (AAB81682; grape), FaGT1 (AAU09442; strawberry), DicGT1 (BAD52003; D. caryophyllus), PhF3GalT (AAD55985; P. hybrida), VmUFGT1 (BAA36972; V. mungo), GtF3GT (BAA12737; G. triflora), DicGT3 (BAD52005; D. caryophyllus), FaGT4 (AAU09445; strawberry), FaGT5 (ABB92747; strawberry), FaGT2 (AAU09443; strawberry), FaGT7 (ABB92749; strawberry), FaGT6 (ABB92748; strawberry), FaGT3 (AAU09444; strawberry), and AsSGT (CAB06081; A. sativa).
Biochemical Characterization
The full-length open reading frame of FaGT1 was cloned into the expression vector pET-29a(+) for heterologous protein expression in Escherichia coli. FaGT1 encodes a protein of 50.5 kD consisting of 466 amino acids. The recombinant protein was partially affinity purified on a Ni2+-charged resin that binds the protein's C-terminal His-tag. The presence of target protein was confirmed by SDS-PAGE and western blots using anti-His antibodies (data not shown). Initial activity tests were performed with UDP-Glc and various anthocyanidins and flavonols. Assays were analyzed by liquid chromatography (LC)-electrospray ionization (ESI)-mass spectrometry (MSn) for the formation of glycosylated products. FaGT1 activity could be readily detected with all tested anthocyanidins and flavonols, except for 3-hydroxyflavone and morin (3,5,7,2′,4′-pentahydroxyflavone; Fig. 3). Subsequently, the substrate screening was extended to other flavonoid subgroups, such as flavan-3-ols (catechin, epicatechin), flavanones (naringenin), dihydroflavonols (taxifolin), flavones (5-hydroxyflavone, 7-hydroxyflavone, chrysin, apigenin), flavonol glucosides (quercetin 3-O-glucoside, kaempferol 3-O-glucoside), and anthocyanins (pelargonidin 3-O-glucoside). However, no activity was detected with any of these substrates. The same applied for betanidin, 4-hydroxy-2,5-dimethyl-3(2H)-furanone, benzoic acid, cinnamic acid, and several hydroxycoumarins. These results strongly suggest that FaGT1 acts exclusively on anthocyanidins and flavonols in vitro. No product was formed when UDP-Gal and UDP-GlcUA were used as sugar donors with pelargonidin or quercetin as acceptor molecules.
Figure 3.
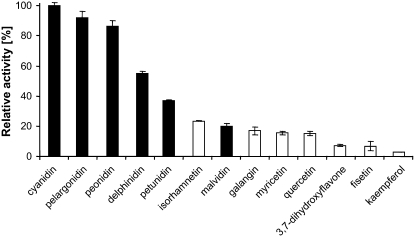
Flavonoid substrate preference of recombinant FaGT1. Enzymatic activity was measured in a liquid scintillation counter with a range of anthocyanidins (black bars) and flavonols (white bars) along with the radioactive donor substrate [6-3H]UDP-Glc.
FaGT1 displayed a broad pH optimum in Tris-HCl buffer with maximal activity occurring between pH 7.0 and pH 8.0. After storage at 4°C for 1 week, the enzyme still showed 70% of its initial activity. The temperature optimum was located at 30°C and product formation was linear for 60 min under these conditions. Under optimal conditions, the maximal enzyme activity was with the substrates pelargonidin and cyanidin, both of which are endogenous strawberry anthocyanidins (Fig. 3; Bakker et al., 1994). Peonidin was also among the preferred substrates, whereas flavonols were poor substrates. It is noteworthy that anthocyanidins lacking substitution at position 5′ (pelargonidin, cyanidin, and peonidin) were better substrates than those with a hydroxyl or methoxy group at this position. The substitution of a hydroxyl group by a methoxy group seems to further reduce product formation (Fig. 3). Kinetic constants were determined for UDP-Glc and pelargonidin because it is the most common strawberry anthocyanidin. The Km for pelargonidin was 30 μm and Vmax was 21 nkat/mg, whereas UDP-Glc had a Km of 1.1 mm and a Vmax of 73 nkat/mg.
Identification of Reaction Products
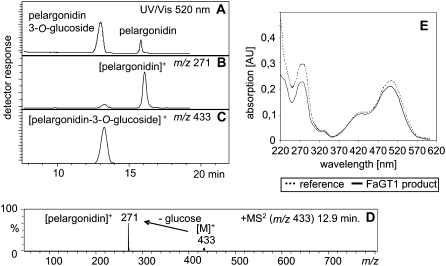
FaGT1 formed only a single monoglucoside from each substrate (Fig. 4, A–C). Authentic reference compounds were used to identify the product derived from pelargonidin and the flavonols quercetin and kaempferol. The products and the reference compounds were analyzed by HPLC-diode array detector (DAD) and LC-ESI-MSn. Anthocyanins exhibit characteristic UV/Vis spectra and the recorded spectrum of the FaGT1 reaction product was identical to that of authentic pelargonidin 3-O-glucoside (Fig. 4E). In LC-ESI-MSn analyses, both the retention times and mass spectra of pelargonidin 3-O-glucoside, quercetin 3-O-glucoside, and kaempferol 3-O-glucoside corresponded closely to those of the respective assay products. Furthermore, the formation of glycosylated products could be confirmed in product ion experiments because the mass spectra of all FaGT1 reaction products were characterized by the loss of one Glc moiety to yield the respective aglycons (Fig. 4D). Together, the findings confirm that FaGT1 forms anthocyanidin and flavonol 3-O-glucosides in vitro.
Figure 4.
Identification of reaction products. Pelargonidin was incubated with recombinant FaGT1 and UDP-Glc followed by LC-ESI-MSn (A–D) and HPLC-DAD (E) analyses. Both the chromatogram at 520 nm (A) and the ion traces show that pelargonidin (m/z 271; B) is converted into a single glucosylated product (m/z 433; C). The MS2 spectrum (D) shows that this product readily loses a Glc moiety upon fragmentation. HPLC-DAD (E) revealed identical UV/Vis spectra for both reaction product and authentic pelargonidin 3-O-glucoside (reference).
Spatial and Developmental Expression
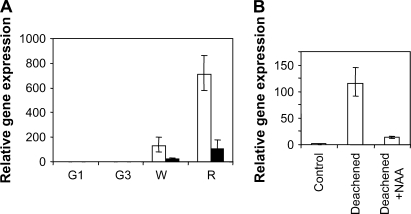
The spatial and temporal expression pattern of FaGT1 was studied with quantitative PCR (qPCR). RNA was extracted from vegetative tissue (flowers, roots, runners, leaves, and crowns) as well as from receptacles and achenes of small-sized green (G1), full-sized green (G3), white (W), and red (R) fruits. Following reverse transcription (RT), the respective mRNA levels were quantified using sequence-specific primers. The generated amplicon was sequenced and melting point determination, as well as gel electrophoresis, was applied to every sample to ensure the specificity of the qPCR analyses. FaGT1 clearly showed ripening-related expression in both achenes and receptacles, with the highest transcript levels being detected in fully ripe red receptacles (Fig. 5A). The expression in receptacles was more than an order of magnitude higher than in achenes. FaGT1 transcripts could also be detected in runners, leaves, flowers, and crowns, but only at very low levels similar to the level in green receptacles (data not shown). These results clearly indicate that the expression of FaGT1 is fruit associated and highly ripening related.
Figure 5.
FaGT1 expression is ripening induced and auxin repressed. A, Ripening-related gene expression of FaGT1 in receptacle tissue (white bars) and achenes (black bars). RNA was extracted from pooled fruits at small-size green (G1), full-size green (G3), white (W), and red (R) stages. Transcript levels were analyzed by qRT-PCR as described in “Materials and Methods.” Gene expression is calibrated against expression in receptacle tissue from full-size green fruit (G3). B, Hormonal control of FaGT1 expression. Achenes were carefully removed at the midsize green stage (G2) and the fruits were harvested after 5 d. One set of de-achened green fruits was then treated with a lanolin paste containing the synthetic auxin NAA. FaGT1 expression was analyzed by qRT-PCR as described in “Materials and Methods” and normalized against the expression in untreated strawberries with attached achenes.
Hormonal Control of FaGT1 and Developmental Expression
To investigate whether FaGT1 is under the control of auxin, midsized green fruits were carefully de-achened and gene expression was studied after 5 d. Additionally, de-achened fruits were treated with a lanolin paste containing naphthalene acetic acid (NAA), a synthetic auxin. The expression of FaGT1 increased substantially after 5 d in de-achened fruits and could be largely reversed by the auxin treatment (Fig. 5B). These results show that the expression of FaGT1 is ripening related and negatively regulated by auxin.
Metabolite Studies in Ripening Strawberry Fruit
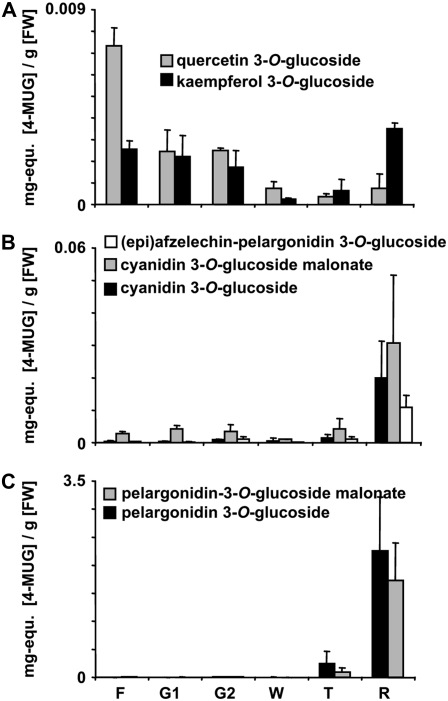
To study the content of individual flavonoids during strawberry fruit ripening, small-sized green (G1), midsized green (G2), white (W), turning (T), and ripe red (R) fruits were extracted with methanol and analyzed by LC-ESI-MSn (Fig. 6). Only small amounts of pelargonidin and cyanidin 3-O-glucoside and pelargonidin 3-O-glucoside-malonate were detected in fruits of the early developmental stages. These compounds showed the highest concentration in ripe red fruits. Unlike its potential precursors, cyanidin 3-O-glucoside-malonate displays two concentration maxima during strawberry fruit ripening. High levels of both kaempferol 3-O-glucoside and quercetin 3-O-glucoside were found in small green fruits and their levels decreased in further developmental stages. However, both flavonol glucosides exhibit a second maximum at late-ripening stages.
Figure 6.
Relative metabolite levels at different ripening stages. The levels of flavonol glucosides (kaempferol 3-O-glucoside, quercetin 3-O-glucoside; A), minor anthocyanins [cyanidin 3-O-glucoside, cyanidin 3-O-glucoside-malonate, (epi)afzelechin-pelargonidin 3-O-glucoside; B], and major anthocyanins (pelargonidin 3-O-glucoside, pelargonidin 3-O-glucoside-malonate; C) were determined in flowers (F), small-size green (G1), midsize green (G2), white (W), turning (T), and ripe red (R) fruits. Samples were extracted with methanol and quantified as mg-equ 4-methylumbelliferyl β-d-glucuronide (MUG) per g fresh weight by LC-ESI-MSn.
Transient Gene Silencing in Ripening Strawberry Fruit
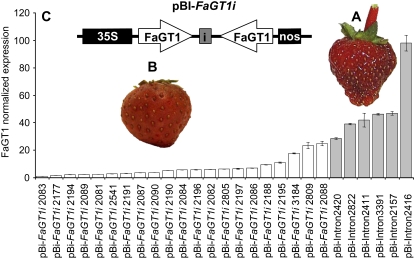
To elucidate the function of FaGT1 in planta, the expression of FaGT1 was down-regulated in strawberry fruit by RNAi using Agrobacterium tumefaciens cells harboring an intron-hairpin construct consisting of two inverted repeats of the FaGT1 sequence (Hoffmann et al., 2006). About one-third (21 of 65) of the fruits injected with pBI-FaGT1i were phenotypically different from fruits injected with the control construct pBI-Intron. Their color was generally less intense and of a different hue compared to the bright red fruits of the controls (Fig. 7, A and B). In all fruits that showed a phenotypic change, a significant down-regulation of FaGT1 expression was determined by quantitative PCR in comparison to the transcript levels in control fruits (Fig. 7C).
Figure 7.
Down-regulation of FaGT1 in fruits injected with Agrobacterium cells harboring a FaGT1 intron-hairpin construct (pBI-FaGT1i). A and B, Phenotype of control fruits and fruits infiltrated with pBI-FaGT1i. Control fruits (A) infiltrated with Agrobacterium containing the control construct (pBI-Intron) are deeply red colored, whereas FaGT1-silenced fruits (B) show a less intense red pigmentation with a different hue. Photographs were taken 14 d after injection. C, qRT-PCR analysis of FaGT1 expression in strawberry fruits. cDNA was generated from total RNA isolated from single fruits infiltrated with Agrobacterium containing either the silencing pBI-FaGT1i (white bars) or the control pBI-Intron vector (gray bars). qRT-PCR was performed using FaGT1 and FaActin gene-specific primers, the latter used as internal control for normalization.
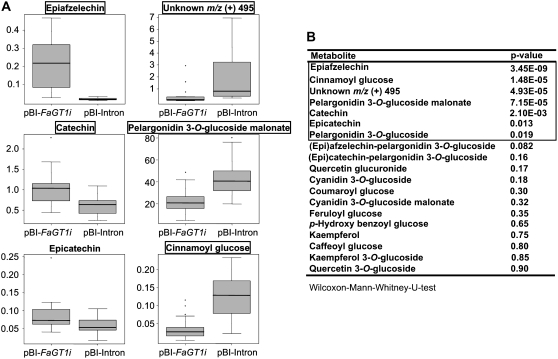
The effect of FaGT1 silencing on the metabolite concentrations was analyzed by LC-ESI-MSn. At first, levels of potential metabolites downstream of anthocyanidins and flavonols and the concentrations of phenylpropanoid Glc esters were determined. Due to biological variation and heterogeneous silencing effects, the variance of each compound within a sample group was relatively high. Therefore, statistical methods were applied to identify metabolites with significantly altered concentrations. Because the levels of the individual compounds were not normally distributed, we used the Wilcoxon-Mann-Whitney U test (Hart, 2001; Hoffmann et al., 2006). Only the levels of pelargonidin 3-O-glucoside malonate and pelargonidin 3-O-glucoside decreased significantly (P < 0.05) in fruits injected with pBI-FaGT1i, whereas changes in the levels of other anthocyanin derivatives, flavonol glucosides, and phenylpropanoid Glc esters were not significant, except for cinnamoyl Glc, whose level was higher in the controls (Fig. 8, A and B). To explore also unexpected effects, we applied the software packages MZmine and XCMS for differential analyses of the LC-ESI-MSn data (Katajamaa et al., 2006; Smith et al., 2006). Both computer programs performed untargeted metabolite profiling and uncovered that the levels of epiafzelechin, catechin, and an unknown compound with a pseudomolecular ion m/z 495 were also significantly different in fruits injected with pBI-FaGT1i when compared with control fruits (Supplemental Figs. S1 and S2). The Wilcoxon-Mann-Whitney U test confirmed the findings (Fig. 8, A and B). These results show that pelargonidin is a substrate of FaGT1 in planta, whereas quercetin and kaempferol are probably glucosylated by different enzymes in strawberry fruit. The effect of FaGT1 down-regulation on the levels of upstream metabolites is much stronger than that on downstream natural products.
Figure 8.
Effect of FaGT1 gene silencing on the metabolite level. Fourteen days after pollination, strawberry fruits were infiltrated with Agrobacterium containing pBI-FaGT1i or control pBI-Intron vector. Levels of metabolites were determined by LC-ESI-MSn 14 d after infiltration in FaGT1-silenced (pBI-FaGT1i; n = 21) and control (pBI-Intron; n = 14) fruits. Identities of the compounds were confirmed with references and literature data. Box-whisker plots (A) show the median (horizontal line) and the first and third quartiles, whereas the whiskers extend to the data extremes. The Wilcoxon-Mann-Whitney U test (B) was used for the nonparametric analysis of between-group comparisons of data from FaGT1-silenced and control fruits. Metabolites showing statistically significantly reduced levels (P < 0.05) in the pBI-FaGT1i-infiltrated fruits are boxed.
DISCUSSION
Biochemical Characterization
FaGT1 exhibited a rather broad substrate preference, accepting all tested anthocyanidins and flavonols in vitro (Fig. 3). However, aglycones, belonging to chemically similar subgroups, such as flavones, flavanones, and dihydroflavonols, were not converted. VvGT1, the enzyme from grape that shows greatest similarity to FaGT1, exhibited a comparable substrate spectrum and was also unable to convert morin at a reasonable rate (Ford et al., 1998). This might be due to morin's unusual hydroxyl group at position 2′, which may cause a sterical hindrance for the deprotonation of hydroxyl group 3 by His-20 of VvGT1 (Offen et al., 2006). In accordance with published results, FaGT1 showed highest activity with pelargonidin and cyanidin, the endogenous substrates in strawberries (Almeida et al., 2007). FaGT1 activity decreased along with the substitution pattern (H > OH > OCH3) at the 5′ position of the anthocyanidin molecule. Replacement of a B-ring hydroxyl group with one methoxy group (e.g. cyanidin to peonidin) decreased the reactivity slightly, but dimethoxylated malvidin was the poorest substrate. These substitutions seem to hinder the efficient binding of hydroxyl group 3 near the His-22 residue of FaGT1. It is worth noting that there are no anthocyanins with a 5′ substitution in strawberries (Bakker et al., 1994). Contrary to its broad substrate tolerance for the flavonoid aglycones, FaGT1 only accepted UDP-Glc as a sugar donor.
In contrast to FaGT1, the VvGT1 from grape showed weak activity with pelargonidin (Ford et al., 1998), which is not an endogenous substrate in grape (Mazza and Miniati, 1993). However, the obtained Km values for anthocyanidins and UDP-Glc are within the same order of magnitude for both enzymes.
The attachment of the sugar was highly regiospecific because FaGT1 formed only anthocyanidin and flavonol 3-O-glucosides. This is consistent with the fact that 3,7-dihydroxyflavone, but not chrysin (5,7-hydroxyflavone), could serve as substrate for FaGT1. In accordance, plant secondary product glycosyltransferases have been reported to exhibit strict regioselectivity toward the position of the attached sugar (Vogt and Jones, 2000).
Gene Expression Studies
Key enzymes of the flavonoid pathway exhibit two maxima in enzymatic activity during strawberry fruit development (Halbwirth et al., 2006). This includes flavonoid 3-O-glycosyltransferase activity when measured with quercetin as the substrate, and Phe ammonia lyase (PAL) activity (Cheng and Breen, 1991). At the transcript level, it has been reported that the dihydroflavonol 4-reductase (DFR), putative chalcone synthase (CHS), and flavonoid 3-hydoxylase (F3H) genes show two peaks in gene expression at early and late developmental stages (Manning, 1998; Moyano et al., 1998), whereas glycosyltransferase genes highly homologous to FaGT1 were only expressed at late developmental stages (Manning, 1998; Almeida et al., 2007).
In this article, we have confirmed the fruit-associated and ripening-related expression of FaGT1. The substantial increases in FaGT1 transcript levels in turning and ripe red fruit agree with our findings (Fig. 6, B and C) and earlier reports that both anthocyanin concentration and flavonoid 3-O-glycosyltransferase activity with malvidin increase during fruit development (Given et al., 1988). In contrast, the levels of polyphenols, non-tannin flavonoids, and cyanidin 3-O-glucoside-malonate are at a maximum shortly after flower opening, peaking again in the late ripening stages (Fig. 6, A and B; Cheng and Breen, 1991; Wang and Lin, 2000). Thus, it can be concluded that at least two different enzymes are involved in the glucosylation of anthocyanidins and flavonols in strawberry: a transferase, which forms the glucosides found at the early ripening stages, and FaGT1, which catalyzes the Glc transfer in ripe strawberry fruits.
In grape berries, most anthocyanin pathway genes are expressed up to 4 weeks after anthesis and after véraison mainly in the skin, whereas VvGT1 transcripts are only detected in the skin of red grapes after véraison (Boss et al., 1996a, 1996b). Thus, VvGT1 seems to be regulated differently than other anthocyanin pathway genes and appears to be the major control point in anthocyanin biosynthesis. Interestingly, the expression of VvGT1 can be delayed by application of a synthetic auxin (Davies et al., 1997). Therefore, FaGT1 and VvGT1 share negative regulation by auxin, which plays an important role in nonclimacteric fruit ripening.
Function of FaGT1 in Planta
To demonstrate the role of FaGT1 in planta, we used a recently developed transient gene-silencing approach based on RNAi to down-regulate FaGT1 expression in strawberry fruit (Hoffmann et al., 2006). A large number of glycosyltransferases involved in plant secondary metabolism have already been cloned and heterologously expressed (Bowles et al., 2005). However, only in a few cases have their biological products been analyzed in planta (Chong et al., 2002; Jones et al., 2003; Morita et al., 2005; Sepúlveda-Jiménez et al., 2005).
In about one-third of the fruits, the injection of pBI-FaGT1i caused a different phenotype with less intense color (Fig. 7A). In contrast, all fruits injected with A. tumefaciens carrying the pBI-FaCHSi construct showed reduced levels of anthocyanins and FaCHS transcripts (Hoffmann et al., 2006). The reason for the lower efficiency of FaGT1 down-regulation remains unknown, but we suspect that it depends on the part of the gene-specific sequence chosen for the cloning of the construct. The average FaGT1 transcript levels in pBI-FaGT1i fruits were about 15% of the levels in fruits injected with a control construct (Fig. 7C). When stable strawberry antisense lines of CHS were analyzed, only fruits with <25% of the original transcript level showed a detectable change in pigmentation (Lunkenbein et al., 2006b). Although the content of pelargonidin 3-O-glucoside in these fruits was reduced to 7.5% of control levels, the fruits were still colored orange and pink.
A principal problem in silencing glycosyltransferases in plants is the occurrence of enzymes with redundant functions. In our study, the presence of other glycosyltransferases active on anthocyanidins and not silenced by pBI-FaGT1i can account for the partial phenotype observed. The large number of glycosyltransferase sequences in the genomes of Arabidopsis (Li et al., 2001), Oryza sativa (Ko et al., 2006), and Medicago truncatula (Achnine et al., 2005), and the presence of at least seven glycosyltransferase genes (Fig. 2) in strawberry, suggest that strawberries should also contain a large multigene family of glycosyltransferases. This consideration is further supported by the fact that the octoploid background of the cultivated strawberry genotype used in this study leads to a theoretical 4-fold increase of genes. One of these enzymes could be compensating for down-regulated FaGT1. In a similar case in Arabidopsis, the ugt78d2 mutant, lacking UGT78D2 function closely related to FaGT1 activity, still accumulated small amounts of anthocyanins (21% of the wild type), leading the authors to suggest the activity of other 3-O-glucosyltransferases (Tohge et al., 2005). Thus, the simultaneous silencing of several glycosyltransferase genes is likely to be necessary to obtain a clear white phenotype.
Down-regulation of FaGT1 is accompanied by significantly (P < 0.05) reduced levels of the strawberry pigments pelargonidin 3-O-glucoside malonate and pelargonidin 3-O-glucoside, but the concentrations of other anthocyanins did not change significantly (Fig. 8, A and B). A second glucosyltransferase (e.g. the one that glucosylates cyanidin at early developmental stages and provides the precursor for the malonated derivative) could compensate for FaGT1 silencing in the case of cyanidin 3-O-glucoside (Fig. 6B). The strongly reduced concentrations of pelargonidin 3-O-glucoside-malonate in pBI-FaGT1i fruits are consistent with the idea that the first modification of the anthocyanidin structure is the attachment of a Glc molecule followed by esterification with malonic acid, and shows that pelargonidin is a substrate of FaGT1 in the late ripening stages in vivo. Weak enzymatic activity of the malonyl transferase at low pelargonidin 3-O-glucoside levels would lead to strongly reduced amounts of the malonylated derivative, whereas the level of pelargonidin 3-O-glucoside remains almost constant. This scenario would explain—at least for the pelargonidin derivatives—why the level of the 3-O-glucoside-malonate is stronger affected than the level of the glucoside.
Surprisingly, the level of cinnamoyl Glc was also significantly reduced in fruits injected with pBI-FaGT1i, even though FaGT1 showed no activity toward cinnamic acid in vitro. Because we observed a similar effect when FaCHS was transiently silenced, we reason that manipulation of the flavonoid pathway modulates PAL activity through transcriptional and posttranscriptional mechanisms as has been shown by antisense expression of an alfalfa (Medicago sativa) cinnamic acid 4-hydroxylase (Supplemental Fig. S3; Blount et al., 2000; Hoffmann et al., 2006). The modulation of PAL activity by the metabolites of the phenylpropanoid pathway has already been reported and could cause the reduced level of the cinnamic acid derivative (Shirsat and Nair, 1986).
Based on in vitro substrate preference and additional biochemical data, it has been proposed that VvGT1 is primarily responsible for the glucosylation of anthocyanidins in vivo (Ford et al., 1998). In contrast, in the homozygous Arabidopsis ugt78d2 mutant, the total level of anthocyanins (consisting exclusively of cyanidin glucoside derivatives) and the levels of four flavonol 3-O-glycosides were reduced (Tohge et al., 2005). Thus, because recombinant UGT78D2 glucosylates anthocyanidins (cyanidin, pelargonidin, and delphindin) and flavonols (kaempferol, quercetin, and myricetin) in vitro, it was concluded that the native enzyme catalyzes the glucosylation of both cyanidin and flavonols at the 3 position as UDP-Glc:flavonoid 3-O-glucosyltransferase in planta. Our data provide experimental evidence that FaGT1 is involved only in the formation of anthocyanins in planta, whereas flavonols can be excluded as in vivo substrates. This conclusion is based on the significantly reduced levels of pelargonidin 3-O-glucoside malonate and pelargonidin 3-O-glucoside and the strongly increased concentrations of flavan-3-ols (epiafzelechin, catechin, and epicatechin) in fruits in which FaGT1 transcript level had been genetically down-regulated. Because epiafzelechin is produced from pelargonidin by the action of anthocyanidin reductase (ANR), increased amounts of the ANR product can only be explained by enhanced availability of the pelargonidin substrate because average FaANR transcript levels were unaffected in pBI-FaGT1i fruits (Fig. 9; Supplemental Fig. S4). Thus, the increased level of epiafzelechin confirms the efficient and specific down-regulation of FaGT1 activity and provides further evidence for pelargonidin as in planta substrate of FaGT1. The successful redirection of the flavonoid biosynthesis also suggests that the reciprocal regulation of ANR and FaGT1 is important in directing the metabolic flux to either anthocyanins or flavan-3-ols (Lee et al., 2005). But FaGT1 silencing also provides enhanced levels of cyanidin and its precursor leucocyanidin, which are converted by FaANR and leucoanthocyanidin 4-reductase (LAR) to epicatechin and catechin, respectively (Fig. 9). The amounts of both flavan-3-ols increased as a consequence of FaGT1 down-regulation (Fig. 8). Because LAR from Fragaria shows strict substrate specificity toward leucocyanidin, the lack of afzelechin in strawberry is conceivable (Almeida et al., 2007). The unknown metabolite with a pseudomolecular ion of m/z 495 might be an anthocyanin derivative (downstream metabolite) according to its mass spectral data (data not shown) and reduced level in pBI-FaGT1i fruits.
Figure 9.
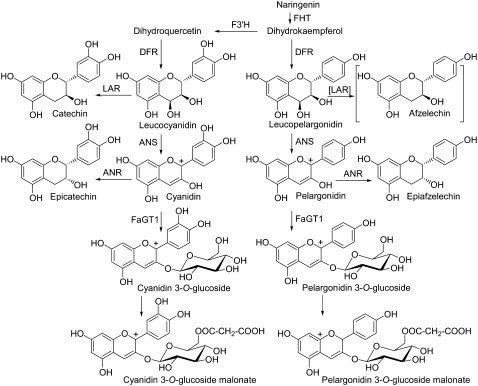
Biosynthetic pathway leading to anthocyanins and flavan-3-ols. FHT, Flavanone 3-hydroxylase; ANS, anthocyanidin synthase. Because of the strict specificity of Fragaria LAR for leucocyanidin, afzelechin is not found in strawberry fruit (Almeida et al., 2007).
In addition to the findings in Arabidopsis, this article reports on gene silencing of an anthocyanidin 3-O-glucosyltransferase from a commercial fruit crop. We could clearly demonstrate that FaGT1 acts on anthocyanidins (pelargonidin and cyanidin), whereas other glycosyltransferases should be responsible for the glucosylation of flavonols (kaempferol and quercetin) in the receptacle. The redirection of the metabolic flux toward flavan-3-ols through down-regulation of FaGT1 offers a new method to increase the levels of these bioactive metabolites in fruit crops. The findings emphasize the necessity of in planta experiments for the elucidation of biological functions and enable novel insights into the flux control of the flavonoid pathway.
MATERIALS AND METHODS
Chemicals
Except when otherwise stated, all chemicals, solvents, and reference compounds were obtained from Sigma, Aldrich, Fluka, Riedel de Haën, Merck, or Roth. Anthocyanidins were purchased from Polyphenols Laboratories. Radiolabeled [6-3H]UDP-Glc (1 mCi/mL, 60 Ci/mmol) was obtained from American Radiolabeled Compounds.
Cloning of FaGT1 Genomic Sequence
DNA used as a template for PCR amplification was isolated from strawberry (Fragaria × ananassa ‘Elsanta’) leaves using a commercial kit (Qiagen). Products obtained with primers located at the 5′- and the 3′-end of the full-length sequences were cloned into the pGEM-T vector (Promega) and sequenced by a commercial sequencing service (MWG Biotech).
Construction of FaGT1 Expression Plasmid
The cDNA library construction and sequence analysis of the ESTs have been described (Aharoni and O'Connell, 2002). The FaGT1 full-length open reading frame was subcloned in the pGEM-T vector (Promega). For the FaGT1 construct, 5′-TGAATTCATGGCACCAGTATCAAACC-3′ was used as the forward primer and 5′-TCTCGAGATTGGTTGTAGTCATTTCC-3′ as the reverse primer, introducing EcoRI and XhoI restriction sites that were used for subsequent cloning into the pET-29a(+) expression vector (Novagen). The insert was cloned in frame with a coding region for a C-terminal His-tag. The identity of the cloned gene was confirmed by sequencing the complete insert (MWG Biotech).
Heterologous Protein Expression
Escherichia coli BL21 (DE3) pLysS cells (Novagen) were transformed with the pET-29a(+) expression vector containing the FaGT1 open reading frame. Cultures were grown overnight at 37°C in Luria-Bertani medium containing 25 μg/mL kanamycin and 34 μg/mL chloramphenicol. The next day, the cultures were diluted to an OD600 of 0.06 with Luria-Bertani medium containing the appropriate antibiotics in a final volume of 800 mL. This culture was grown at 37°C to an OD600 of 0.4 to 0.6, cooled to 16°C, and 1 mm isopropylthio-β-galactoside was added to induce protein expression. After overnight incubation at 16°C to 18°C, cells were harvested by centrifugation and stored at −20°C.
Cell Lysis and Protein Purification
His-tagged protein was affinity purified using the His Bind Quick 900 cartridges (Novagen) as recommended by the manufacturer. Briefly, cells were resuspended in binding buffer and sonicated three times on ice. Viscosity was reduced through incubation with benzonase nuclease (Novagen) and the protein raw extract was applied to the cartridges. Recombinant protein was eluted with a buffer containing 1 m imidazol and kept on ice. Protein concentration was determined (Bradford, 1976) and the presence of recombinant protein was confirmed by SDS-PAGE and western blot using anti-His antibodies (Novagen). The amount of FaGT1 present in the partially purified eluate was quantified using the Alpha Imager 2200 documentation and analysis system and Alpha Ease software (Alpha Innotech). Aliquots of purified protein were stored at −20°C.
Enzyme Assays
Enzyme activity was assayed in a buffer containing 100 mm Tris-HCl (pH 7.0), 10% glycerol, and 10 mm β-mercaptoethanol. Standard assays contained 5 mm UDP-Glc, 200 μm substrate, and protein raw extract with approximately 0.25 μg of recombinant protein in a total volume of 250 μL. Enzyme assays were incubated for 30 min at 30°C and stopped by the addition of 200 μL of 5% HCl (anthocyanidin substrates) or 50 μL of acetic acid (all other substrates). As a control, BL21 (DE3) pLysS cells were transformed with an empty pET29-a(+) vector and the resulting protein extract was assayed under the same conditions. As an additional control, assays were conducted with heat-inactivated enzyme solution (5 min at 95°C). Initial enzyme activity was monitored by detecting the reaction products with LC-ESI-MSn. The identity of the FaGT1 glycosylation product was also confirmed by HPLC-DAD.
Enzyme Kinetics
The biochemical characterization was carried out with radioactively labeled UDP-Glc allowing for detection of the reaction product by liquid scintillation counting. Assay conditions were essentially the same, except only 0.1 μg of affinity-purified recombinant protein was used in a total volume of 200 μL. The UDP-Glc was a mixture of 1 μL of 0.016 mm [6-3H]UDP-Glc (1 mCi/mL) and 99 μL of unlabeled 101 mm UDP-Glc. Assays were incubated at 30°C for 30 min and extracted with 1 mL of water-saturated n-butanol. The radioactivity of the products was determined by liquid scintillation counting (LKB Rackbeta 1219) after the addition of 4 mL of Ultima Gold XR LSC cocktail (Perkin-Elmer). The kinetic constants were calculated with SigmaPlot 8.0 software (Systat Software) assuming single-site saturation binding.
HPLC-DAD
The instrument used was a LaChrom HPLC (Merck-Hitachi) equipped with a DAD. HPLC separation was performed with a Phenomenex Luna C-8 column (150 mm long × 4.6 mm i.d., particle size 3 μm) applying a gradient that went from 100% A (0.05% formic acid in water) to 100% B (acetonitrile) in 30 min at a flow rate of 1 mL/min. Spectra and chromatograms were acquired with the Chromatography Data Station software (Merck-Hitachi).
LC-ESI-MSn
A Bruker Daltonics esquire 3000plus ion trap mass spectrometer (Bruker Daltonics) connected to an Agilent 1100 HPLC system (Agilent Technologies) equipped with a quaternary pump and a variable wavelength detector was utilized for all experiments. Components were separated with a Phenomenex Luna C-18 column (150 mm long × 2.0 mm i.d., particle size 5 μm) that was held at 25°C. Enzyme assays were analyzed using a linear gradient that went from 100% A (0.1% formic acid in water) to 100% B (acetonitrile) in 30 min with a flow rate of 0.2 mL/min. For metabolite analyses in strawberry fruit extracts, the gradient went from 100% A to 40% B in 40 min, then to 100% B in 5 min. The detection wavelength was either 520 (anthocyanidins) or 280 nm (other substrates and metabolite analyses). The ESI voltage of the capillary was set to −4,000 V and the end plate to −500 V. Nitrogen was used as dry gas at a temperature of 300°C and a flow rate of 10 L/min. The full-scan mass spectra were measured in a scan range from 50 to 800 m/z with a scan resolution of 13,000 m/z/s until the ICC target reached 20,000 or 200 ms, whichever was achieved first. Tandem MS was carried out using helium as the collision gas (3.56 × 10−6 mbar) with 1-V collision voltage. Spectra were acquired in the positive and negative ionization mode. Data analysis was performed using the DataAnalysis 3.1 software (Bruker Daltonics).
Auxin Treatment
The hormone treatment was performed as described previously (Medina-Escobar et al., 1997). Briefly, achenes from midsized green fruits were removed carefully with a scalpel blade. One set of fruits was treated with a lanolin paste containing 1 mm NAA in 1% (v/v) dimethyl sulfoxide. The other set of de-achened fruits was treated with the same paste without the synthetic auxin NAA. Fruits were harvested after 5 d and immediately frozen in liquid nitrogen.
Developmental Analyses of FaGT1 Expression
Total RNA was isolated from pools of six to seven strawberry fruits from each ripening stage as described (Asif et al., 2000). RNA was treated with DNase I (Amersham Bioscience) to remove any DNA contamination prior to cDNA synthesis. RT was carried out using 1 μg of total RNA and the iScript cDNA synthesis kit (Bio-Rad) as recommended by the manufacturer. qRT-PCR analysis was performed using the iCycler system (Bio-Rad) as described previously (Benítez-Burraco et al., 2003; Raab et al., 2006). FaGT1-specific primers were designed (5′-TGCGGTTGGAACTCGTGCGGTTGGAACTCGGTGCTTGCCTGTTGTGCGAGTTGTTTTGTGCTT-3′ and 5′-GCCTGTTGTGCGAGTTGTTTT-3′) and tested with cDNA from different ripening stages. The obtained amplification products were cloned and subsequently sequenced. The qRT-PCR data for FaGT1 were normalized against the expression levels of an interspacer 26S-18S RNA housekeeping gene (5′-ACCGTTGATTCGCACAATTGGTCATACTGCGGGTCGGCAATCGGACGTCG-3′ and 5′-TACTGCGGGTCGGCAATCGGACG-3′). PCR reactions contained 2 mm MgCl2, 0.2 mm dNTPs, 0.2 μm of the respective primers, 2.5 μL SYBR-Green I (diluted 1:15,000), 2 μL cDNA, and 0.5 units Taq polymerase (Biotools) in a total volume of 25 μL. The thermal cycling conditions were as follows: 2 min at 94°C, followed by 40 cycles of 95°C for 15 s, 55°C for 30 s, and 72°C for 30 s. Melting-point analysis and agarose gel electrophoresis were performed on every reaction to ensure that only the desired amplicon had been generated. Each reaction was performed at least in triplicate and relative expression levels were calculated by calibrating the results with receptacle expression levels from full-sized green fruits (G3) or untreated controls with attached achenes (Livak and Schmittgen, 2001).
Construction of pBI-FaGT1i
A blunt-end PCR product containing the full-length sequence of FaGT1 was generated using a high-fidelity polymerase (Finnzymes) and the same primers used to subclone the expression vector. This amplicon was then digested with SpeI yielding a fragment of approximately 500 bp, which was used for ligation into the binary vector pBI121 that contained XbaI/NheI and SpeI/SacI (Ecl136II) restriction sites separated by an intron from strawberry (Hoffmann et al., 2006). To do this, the vector was cut with XbaI and a high-fidelity polymerase was used to create a blunt end. After cutting the vector with NheI, the 500-bp fragment was ligated in the sense direction. Second, the vector was digested with SpeI and Ecl136II and the 500-bp fragment was inserted in the antisense direction, yielding the intron-hairpin construct pBI-FaGT1i.
Transfection of Strawberries by Agroinfiltration
Strawberry cultivar ‘Elsanta’ plants were grown under standard conditions at 25°C and a 16-h photoperiod. Transfection was carried out as described previously (Hoffmann et al., 2006). Briefly, Agrobacterium tumefaciens strain AGL0 cells containing the pBI-FaGT1i construct were grown at 28°C until the OD600 reached approximately 0.8. Cells were harvested and resuspended in modified MacConkey agar medium and injected evenly in fruits still attached to the plant about 14 d after pollination. Ripe fruits with dark achenes were harvested approximately 14 d after injection and stored at −80°C. Control experiments were carried out by injecting strawberry fruits with A. tumefaciens AGL0 cells harboring a pBI-Intron control construct (Hoffmann et al., 2006).
RNA Extraction and Expression Analyses Used for Transfected Fruits
Agroinfiltrated fruits were individually freeze dried in a lyophilizer (Christ ALPHA 1–4) and ground to a fine powder. Fifty milligrams of the powder from each fruit was used for RNA extraction, followed by DNase I treatment and RT, as described before. Five nanograms of cDNA was used for qRT-PCR experiments, carried out with FaGT1, FaANR, and FaActin gene-specific primers (Almeida et al., 2007) using an ABI 7900 thermocycler (Applied Biosystems) and Platinum SYBR-Green kit (Invitrogen) according to the manufacturer's instructions. The FaActin gene, having constant expression levels, was used to normalize raw data and calculate relative FaGT1 and FaANR transcript levels.
Metabolite Analysis
Fifty milligrams of freeze-dried strawberry powder was extracted with 250 μL of methanol containing 0.2 mg/mL 4-methylumbelliferyl β-d-glucuronide as an internal standard. Methanol was removed in a rotary vacuum concentrator (Christ RVC 2–18) and the extract was redissolved in 35 μL of water for analysis by LC-ESI-MSn. For metabolite analyses during fruit development, 500-mg samples were extracted with 500 μL of methanol, 0.05 mg of internal standard was added, centrifuged (5,000g, 10 min), and analyzed by LC-ESI-MSn. Metabolite quantification was performed using QuantAnalysis 1.5 software (Bruker Daltonics) normalizing all results against the internal standard. Each analysis was performed in triplicate. Levels of metabolites determined in the RNAi experiment were displayed as box and whisker plots using the software SigmaPlot 8.0 (Systat Software). Statistical significance levels were calculated with the Wilcoxon-Mann-Whitney U Test (Hart, 2001) using the software package R (www.r-project.org).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AAU09442 (FaGT1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Comparative HPLC-ESI-MS analysis of the metabolites visualized by MZmine.
Supplemental Figure S2. Extracted ion trace m/z 273 visualized by XCMS.
Supplemental Figure S3. Effect on the cinnamoyl glucose level by transient silencing of FaCHS.
Supplemental Figure S4. Quantitative PCR analysis of FaANR expression in strawberry fruits.
Supplementary Material
Acknowledgments
We thank Christian Landmann for helpful discussions and Heather Coiner for correcting the manuscript.
This work was supported by Degussa AG.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Wilfried Schwab (schwab@wzw.tum.de).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Aaby K, Skrede G, Wrolstad RE (2005) Phenolic composition and antioxidant activities in flesh and achenes of strawberries (Fragaria ananassa). J Agric Food Chem 53 4032–4040 [DOI] [PubMed] [Google Scholar]
- Achnine L, Huhman DV, Farag MA, Sumner LW, Blount JW, Dixon RA (2005) Genomics-based selection and functional characterization of triterpene glycosyltransferases from the model legume Medicago truncatula. Plant J 41 875–887 [DOI] [PubMed] [Google Scholar]
- Aharoni A, O'Connell AP (2002) Gene expression analysis of strawberry achene and receptacle maturation using DNA microarrays. J Exp Bot 53 2073–2087 [DOI] [PubMed] [Google Scholar]
- Almeida JRM, D'Amico E, Preuss A, Carbone F, de Vos RCH, Deiml B, Mourgues F, Perrotta G, Fischer TC, Bovy AG, et al (2007) Characterization of major enzymes and genes involved in flavonoid and proanthocyanidin biosynthesis during fruit development in strawberry (Fragaria × ananassa). Arch Biochem Biophys 465 61–71 [DOI] [PubMed] [Google Scholar]
- Asif MH, Dhawan P, Nath P (2000) A simple procedure for the isolation of high quality RNA from ripening banana fruit. Plant Mol Biol Rep 18 109–115 [Google Scholar]
- Bakker J, Bridle P, Bellworthy SJ (1994) Strawberry juice colour: a study of the quantitative and qualitative pigment composition of juices from 39 genotypes. J Sci Food Agric 64 31–37 [Google Scholar]
- Benítez-Burraco A, Blanco-Portales R, Redondo-Nevado J, Bellido ML, Moyano E, Caballero JL, Munoz-Blanco J (2003) Cloning and characterization of two ripening related strawberry (Fragaria x ananassa cv. Chandler) pectate lyase genes. J Exp Bot 54 633–645 [DOI] [PubMed] [Google Scholar]
- Blount JW, Korth KL, Masoud SA, Rasmussen S, Lamb C, Dixon RA (2000) Altering expression of cinnamic acid 4-hydroxylase in transgenic plants provides evidence for a feedback loop at the entry point into the phenylpropanoid pathway. Plant Physiol 122 107–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boss PK, Davies C, Robinson SP (1996. a) Analysis of the expression of anthocyanin pathway genes in developing Vitis vinifera L. cv Shiraz grape berries and the implications for pathway regulation. Plant Physiol 111 1059–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boss PK, Davies C, Robinson SP (1996. b) Expression of anthocyanin biosynthesis pathway genes in red and white grapes. Plant Mol Biol 32 565–569 [DOI] [PubMed] [Google Scholar]
- Bowles D, Isayenkova J, Lim EK, Poppenberger B (2005) Glycosyltransferases: managers of small molecules. Curr Opin Plant Biol 8 254–263 [DOI] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72 248–254 [DOI] [PubMed] [Google Scholar]
- Cheng GW, Breen PJ (1991) Activity of phenylalanine ammonia-lyase (PAL) and concentration of anthocyanins and phenolics in developing strawberry fruit. J Am Soc Hortic Sci 116 865–869 [Google Scholar]
- Cheng GW, Malencik DA, Breen PJ (1994) UDP-glucose:flavonoid O-glucosyltransferase from strawberry fruit. Phytochemistry 35 1435–1439 [Google Scholar]
- Chong J, Baltz R, Schmitt C, Beffa R, Fritig B, Saindrenan P (2002) Down-regulation of a pathogen-responsive tobacco UDP-Glc:phenylpropanoid glucosyltransferase reduces scopoletin glucoside accumulation, enhances oxidative stress, and weakens virus resistance. Plant Cell 14 1093–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies C, Boss PK, Robinson SP (1997) Treatment of grape berries, a nonclimacteric fruit with a synthetic auxin, retards ripening and alters the expression of developmentally regulated genes. Plant Physiol 115 1155–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford CM, Boss PK, Høj PB (1998) Cloning and characterization of Vitis vinifera UDP-glucose:flavonoid 3-O-glucosyltransferase, a homologue of the enzyme encoded by the maize Bronze-1 locus that may primarily serve to glucosylate anthocyanidins in vivo. J Biol Chem 273 9224–9233 [DOI] [PubMed] [Google Scholar]
- Fossen T, Rayyan S, Andersen ØM (2004) Dimeric anthocyanins from strawberry (Fragaria ananassa) consisting of pelargonidin 3-glucoside covalently linked to four flavan-3-ols. Phytochemistry 65 1421–1428 [DOI] [PubMed] [Google Scholar]
- Giovannoni J (2001) Molecular biology of fruit maturation and ripening. Annu Rev Plant Physiol Plant Mol Biol 52 725–749 [DOI] [PubMed] [Google Scholar]
- Given NK, Venis MA, Grierson D (1988) Phenylalanine ammonia-lyase activity and anthocyanin synthesis in ripening strawberry fruit. J Plant Physiol 133 25–30 [Google Scholar]
- Häkkinen SH, Kärenlampi SO, Heinonen M, Mykkänen HM, Törrönen AR (1999) Content of the flavonols quercetin, myricetin, and kaempferol in 25 edible berries. J Agric Food Chem 47 2274–2279 [DOI] [PubMed] [Google Scholar]
- Halbwirth H, Puhl I, Haas U, Jezik K, Treutter D, Stich K (2006) Two-phase flavonoid formation in developing strawberry (Fragaria x ananassa) fruit. J Agric Food Chem 54 1479–1485 [DOI] [PubMed] [Google Scholar]
- Hancock JF (1999) Strawberries. CABI Publishing, New York
- Hart A (2001) Mann-Whitney test is not just a test of medians: differences in spread can be important. BMJ 323 391–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann T, Kalinowski G, Schwab W (2006) RNAi-induced silencing of gene expression in strawberry fruit (Fragaria x ananassa) by agroinfiltration: a rapid assay for gene function analysis. Plant J 48 818–826 [DOI] [PubMed] [Google Scholar]
- Jones P, Messner B, Nakajima J-I, Schäffner AR, Saito K (2003) UGT73C6 and UGT78D1, glycosyltransferases involved in flavonol glycoside biosynthesis in Arabidopsis thaliana. J Biol Chem 278 43910–43918 [DOI] [PubMed] [Google Scholar]
- Katajamaa M, Miettinen J, Orešič M (2006) MZmine: toolbox for processing and visualization of mass spectrometry based molecular profile data. Bioinformatics 22 634–636 [DOI] [PubMed] [Google Scholar]
- Ko JH, Kim BG, Hur HG, Lim Y, Ahn JH (2006) Molecular cloning, expression and characterization of a glycosyltransferase from rice. Plant Cell Rep 25 741–746 [DOI] [PubMed] [Google Scholar]
- Lee Y, Yoon HR, Paik YS, Liu JR, Chung WI, Choi G (2005) Reciprocal regulation of Arabidopsis UGT78D2 and BANYULS is critical for regulation of the metabolic flux of anthocyanidins to condensed tannins in developing seed coats. J Plant Biol 48 356–370 [Google Scholar]
- Li Y, Baldauf S, Lim EK, Bowles DJ (2001) Phylogenetic analysis of the UDP-glucosyltransferase multigene family of Arabidopsis thaliana. J Biol Chem 276 4338–4343 [DOI] [PubMed] [Google Scholar]
- Lim EK, Ashford DA, Hou B, Jackson RG, Bowles DJ (2004) Arabidopsis glycosyltransferases as biocatalysts in fermentation for regioselective synthesis of diverse quercetin glucosides. Biotechnol Bioeng 87 623–631 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25 402–408 [DOI] [PubMed] [Google Scholar]
- Lunkenbein S, Bellido ML, Aharoni A, Salentijn EMJ, Kaldenhoff R, Coiner HA, Munoz-Blanco J, Schwab W (2006. a) Cinnamate metabolism in ripening fruit. Characterization of a UDP-glucose:cinnamate glucosyltransferase from strawberry. Plant Physiol 140 1047–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunkenbein S, Coiner H, de Vos RCH, Schaart JG, Boone MJ, Krens FA, Schwab W, Salentijn EMJ (2006. b) Molecular characterization of a stable antisense chalcone synthase phenotype in strawberry (Fragaria x ananassa). J Agric Food Chem 54 2145–2153 [DOI] [PubMed] [Google Scholar]
- Manning K (1998) Isolation of a set of ripening-related genes from strawberry: their identification and possible relationship to fruit quality traits. Planta 205 622–631 [DOI] [PubMed] [Google Scholar]
- Mato M, Ozeki Y, Itoh Y, Higeta D, Yoshitama K, Teramoto S, Aida R, Ishikura N, Shibata M (1998) Isolation and characterization of a cDNA clone of UDP-galactose:flavonoid 3-O-galactosyltransferase (UF3GaT) expressed in Vigna mungo seedlings. Plant Cell Physiol 39 1145–1155 [DOI] [PubMed] [Google Scholar]
- Mazza G, Miniati E (1993) Anthocyanins in Fruits, Vegetables, and Grains. CRC Press, Boca Raton, FL
- Medina-Escobar N, Cárdenas J, Moyano E, Caballero JL, Munoz-Blanco J (1997) Cloning, molecular characterization and expression pattern of a strawberry ripening-specific cDNA with sequence homology to pectate lyase from higher plants. Plant Mol Biol 34 867–877 [DOI] [PubMed] [Google Scholar]
- Miller KD, Guyon V, Evans JNS, Shuttleworth WA, Taylor LP (1999) Purification, cloning, and heterologous expression of a catalytically efficient flavonol 3-O-galactosyltransferase expressed in the male gametophyte of Petunia hybrida. J Biol Chem 274 34011–34019 [DOI] [PubMed] [Google Scholar]
- Morita Y, Hoshino A, Kikuchi Y, Okuhara H, Ono E, Tanaka Y, Fukui Y, Saito N, Nitasaka E, Noguchi H, et al (2005) Japanese morning glory dusky mutants displaying reddish-brown or purplish-gray flowers are deficient in a novel glycosylation enzyme for anthocyanin biosynthesis, UDP-glucose:anthocyanidin 3-O-glucoside-2″-O-glucosyltransferase, due to 4-bp insertions in the gene. Plant J 42 353–363 [DOI] [PubMed] [Google Scholar]
- Moyano E, Portero-Robles I, Medina-Escobar N, Valpuesta V, Munoz-Blanco J, Caballero JL (1998) A fruit specific putative dihydroflavonol 4-reductase gene is differentially expressed in strawberry during the ripening process. Plant Physiol 117 711–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offen W, Martinez-Fleites C, Yang M, Kiat-Lim E, Davis BG, Tarling CA, Ford CM, Bowles DJ, Davies GJ (2006) Structure of a flavonoid glucosyltransferase reveals the basis for plant natural product modification. EMBO J 25 1396–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata J, Itoh Y, Ispida M, Yoshida H, Ozeki Y (2004) Cloning and heterologous expression cDNAs encoding flavonoid glucosyltransferases from Dianthus caryophyllus. Plant Biotechnol J 21 367–375 [Google Scholar]
- Perkins-Veazie P (1995) Growth and ripening of strawberry fruit. Hortic Rev (Am Soc Hortic Sci) 17 267–297 [Google Scholar]
- Raab T, López-Ráez JA, Klein D, Caballero JL, Moyano E, Schwab W, Munoz-Blanco J (2006) FaQR, required for the biosynthesis of the strawberry flavor compound 4-hydroxy-2,5-dimethyl-3(2H)-furanone, encodes an enone oxidoreductase. Plant Cell 18 1023–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J, Li Y, Lim EK, Bowles DJ (2001) Higher plant glycosyltransferases. Genome Biol 2 3004.1–3004.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan JJ (1971) Flavonol glycosides of the cultivated strawberry. J Food Sci 36 867–870 [Google Scholar]
- Sepúlveda-Jiménez G, Rueda-Benítez P, Porta H, Rocha-Sosa M (2005) A red beet (Beta vulgaris) UDP-glucosyltransferase gene induced by wounding, bacterial infiltration and oxidative stress. J Exp Bot 56 605–611 [DOI] [PubMed] [Google Scholar]
- Shirsat SG, Nair PM (1986) The mode of inhibition of the biosynthesis of phenylalanine ammonia lyase by its product cinnamic acid in aging potato parenchyma tissue. J Biosci 10 393–402 [Google Scholar]
- Smith CA, Want EJ, Tong GC, Abagyan R, Siuzdak G (2006) XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem 78 779–787 [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Yonekura K, Fukuchi-Mizutani M, Fukui Y, Fujiwara H, Ashikari T, Kusumi T (1996) Molecular and biochemical characterization of three anthocyanin synthetic enzymes from Gentiana triflora. Plant Cell Physiol 37 711–716 [DOI] [PubMed] [Google Scholar]
- Tohge T, Nishiyama Y, Hirai MY, Yano M, Nakajima JI, Awazuhara M, Inoue E, Takahashi H, Goodenowe DB, Kitayama M, et al (2005) Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing a MYB transcription factor. Plant J 42 218–235 [DOI] [PubMed] [Google Scholar]
- van de Peer Y, de Wachter Y (1994) TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Appl Biosci 10 569–570 [DOI] [PubMed] [Google Scholar]
- Vogt T, Jones P (2000) Glycosyltransferases in plant natural product synthesis: characterization of a supergene family. Trends Plant Sci 5 380–386 [DOI] [PubMed] [Google Scholar]
- Wang SY, Lin HS (2000) Antioxidant activity in fruits and leaves of blackberry, raspberry, and strawberry varies with cultivar and developmental stage. J Agric Food Chem 48 140–146 [DOI] [PubMed] [Google Scholar]
- Yamazaki M, Yamagishi E, Gong Z, Fukuchi-Mizutani M, Fukui Y, Tanaka Y, Kusumi T, Yamaguchi M, Saito K (2002) Two flavonoid glucosyltransferases from Petunia hybrida: molecular cloning, biochemical properties and developmentally regulated expression. Plant Mol Biol 48 401–411 [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Tamura H (2005) Variation in concentration and composition of anthocyanins among strawberry cultivars. J Jpn Soc Hort Sci 74 36–41 [Google Scholar]
- Yoshihara N, Imayama T, Fukuchi-Mizutani M, Okuhara H, Tanaka Y, Ino I, Yabuya T (2005) cDNA cloning and characterization of UDP-glucose:anthocyanidin 3-O-glucosyltransferase in Iris hollandica. Plant Sci 169 496–501 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.