Abstract
Although the nonphotosynthetic NAD-malic enzyme (NAD-ME) was assumed to play a central role in the metabolite flux through the tricarboxylic acid cycle, the knowledge on this enzyme is still limited. Here, we report on the identification and characterization of two genes encoding mitochondrial NAD-MEs from Arabidopsis (Arabidopsis thaliana), AtNAD-ME1 and AtNAD-ME2. The encoded proteins can be grouped into the two clades found in the plant NAD-ME phylogenetic tree. AtNAD-ME1 belongs to the clade that includes known α-subunits with molecular masses of approximately 65 kD, while AtNAD-ME2 clusters with the known β-subunits with molecular masses of approximately 58 kD. The separated recombinant proteins showed NAD-ME activity, presented comparable kinetic properties, and are dimers in their active conformation. Native electrophoresis coupled to denaturing electrophoresis revealed that in vivo AtNAD-ME forms a dimer of nonidentical subunits in Arabidopsis. Further support for this conclusion was obtained by reconstitution of the active heterodimer in vitro. The characterization of loss-of-function mutants for both AtNAD-MEs indicated that both proteins also exhibit enzymatic activity in vivo. Neither the single nor the double mutants showed a growth or developmental phenotype, suggesting that NAD-ME activity is not essential for normal autotrophic development. Nevertheless, metabolic profiling of plants completely lacking NAD-ME activity revealed differential patterns of modifications in light and dark periods and indicates a major role for NAD-MEs during nocturnal metabolism.
The enzymatic oxidation of l-malate to pyruvate and CO2 is catalyzed by two classes of malic enzymes (MEs) with the general requirement for divalent cations: NADP-dependent ME (NADP-ME; EC 1.1.1.40) and NAD-dependent ME (NAD-ME; EC 1.1.1.39 or 1.1.1.38, depending on the ability to decarboxylate oxaloacetate [OAA]). In plants, NADP-ME isoforms function in chloroplasts and the cytosol (Drincovich et al., 2001; Gerrard Wheeler et al., 2005), while NAD-ME isoforms are found in mitochondria (Winning et al., 1994). Photosynthetic NADP-ME isoforms are involved in metabolism in chloroplasts (C4 plants) or the cytosol (Crassulacean acid metabolism [CAM] plants; Drincovich et al., 2001), while plastidic nonphotosynthetic isoforms have been suggested to play a role in plant defense responses and lipid biosynthesis (Smith et al., 1992; Eastmond et al., 1997; Casati et al., 1999; Maurino et al., 2001). Several biological roles were proposed for the cytosolic NADP-ME isoforms, including participation in plant defense responses, lignin biosynthesis, and control of cytosolic pH (Martinoia and Rentsch, 1994).
In some C4 and CAM plants, NAD-MEs provide CO2 for the Calvin cycle during photosynthetic metabolism (Hatch and Kagawa, 1974; Artus and Edwards, 1985). In some C4 plant species, NAD-ME is present at an activity of around 50 times that found in C3 plants and functions in bundle sheath mitochondria (Hatch and Carnal, 1992). Apart from the photosynthetic role, NAD-ME is also involved in malate respiration, a universal role that takes place in the mitochondria (Grover et al., 1981). Two sources of respiratory substrates are furnished to plant mitochondria: pyruvate and malate. Depending on the metabolic demands, it is possible that dark CO2 fixation diverts much phosphoenolpyruvate from pyruvate to malate formation, with malate entering the mitochondria and fulfilling two roles, replenishment of the tricarboxylic acid (TCA) cycle pool and furnishing part of the carbon used for mitochondrial respiration (Day and Hanson, 1977). In mitochondria, malate can be metabolized by malate dehydrogenase (MDH) or NAD-ME. In the latter case, malate is decarboxylated to pyruvate and this, in turn, is converted to acetyl-CoA, which by condensation with oxalacetate forms citrate, allowing repeated cycling of carbon skeletons through the TCA cycle. By providing a means of generating acetyl-CoA and thus ATP and carbon skeletons, NAD-ME was assumed to play a central role in the management of flux through the TCA cycle (Grover et al., 1981). It was postulated that the pH is an important modulator of this branch point: at pH 6.5, malate would be mainly oxidized by the NAD-ME, while at pH 7.5, where NAD-ME is inactive, malate is oxidized by the mitochondrial MDH (Palmer et al., 1982; Agius et al., 2001).
Nonplant NAD-MEs are typically homotetrameric proteins (Caldes et al., 1978; Allen and Harris, 1981; Nagel and Sauer, 1982; Moreadith and Lehninger, 1984; Davisson and Schulz, 1985) and belong to the EC 1.1.1.38 subtype. On the other hand, plant NAD-ME, although very few have been characterized, belong to the EC 1.1.1.39 subtype, as they are not able to decarboxylate oxalacetate (Grover et al., 1981; Wedding and Black, 1983). One intriguing property of plant NAD-ME is that they are composed of two dissimilar subunits (α and β) at a 1:1 molar radio, at least in some species, e.g. Crassula argentea (CAM plant; Willeford and Wedding, 1987), potato (Solanum tuberosum; C3 plant; Grover and Wedding, 1982; Willeford and Wedding, 1987), Urochloa panicoides (C4-PEPCK plant; Burnell, 1987), and Amaranthus hypochondriacus (C4-NAD-ME plant; Long et al., 1994). Although the α- and β-subunits contain all the motifs required for a functional NAD-ME (Winning et al., 1994; Long et al., 1994), no activity was associated with the separated subunits, but activity could be found in a reconstituted system in potato and C. argentea (Willeford and Wedding, 1987). The β-subunit was suggested to play a regulatory role (Long et al., 1994). The cDNAs encoding for both subunits were isolated from potato and amaranth (Long et al., 1994; Winning et al., 1994). These subunits share 65% sequence identity at the amino acid level and are immunologicaly different, as antisera raised against the α-subunit did not cross react with the β-subunit (Long et al., 1994). The enzyme is active as a dimer, tetramer, and octamer, with the tetramer being the most active form (Grover and Wedding, 1982). On the other hand, the enzyme isolated from Eleusine coracana, Panicum dichotomiflorum, and Amaranthus tricolor (all C4-NAD-ME plants) was postulated to form an octamer of identical subunits (Oshugi and Murata, 1980; Murata et al., 1989).
The genome of Arabidopsis (Arabidopsis thaliana) possesses two genes encoding putative NAD-MEs. Due to the assumed central role of NAD-ME in plant mitochondria, the aim of this work was to characterize the AtNAD-ME gene family at the biochemical and functional levels. Based on the biochemical characterization of recombinant proteins and metabolite profiles of loss-of-function mutants, novel properties for plant NAD-MEs are described.
RESULTS
Cloning and Sequence Analysis of AtNAD-ME1 and AtNAD-ME2
Two putative AtNAD-ME genes are present in the Arabidopsis genome, AtNAD-ME1 (At2g13560) and AtNAD-ME2 (At4g00570). The full-length cDNAs encoding AtNAD-ME1 and AtNAD-ME2 were cloned by reverse transcription (RT)-PCR and sequenced. The deduced proteins have molecular masses of 69.6 and 66.6 kD, respectively, share 63% identity at the amino acid level and are both predicted to contain a mitochondrial targeting peptide by four different prediction programs (ARAMEMNON, http://aramemnon.botanik.uni-koeln.de/).
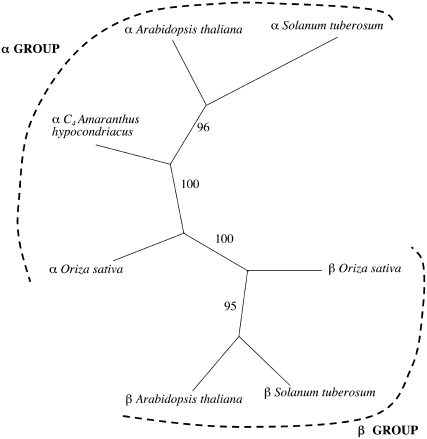
A phylogenetic analysis based on an alignment of the available plant NAD-ME full-length protein sequences indicated that NAD-MEs are divided into two clades: (1) the α-NAD-MEs, which include proteins of approximately 62 kD; and (2) the β-NAD-MEs, which include proteins of approximately 58 kD. AtNAD-ME1 and AtNAD-ME2 belong to the α- and β-NAD-ME clades, respectively (Fig. 1).
Figure 1.
Phylogenetic tree of plant NAD-MEs. Mature proteins were aligned using the ClustalW (1.81) multiple alignment program (Thompson et al., 1994), and the alignment obtained was modified by visual inspection to exclude the sites containing gaps. The phylogenetic tree was constructed by the neighbor-joining method using the Phylip software package (Felsenstein, 1989). Statistical significance of each branch of the tree was evaluated by bootstrap analysis by 100 iterations of bootstrap samplings and reconstruction of trees by the neighbor-joining method. The topology obtained by this method is shown along with statistical significance of each branch. The following sequences are included: α-subunits from A. hypochondriacus (U01162; photosynthetic NAD-ME), Arabidopsis (At2g13560), potato (Z23023), and Oryza sativa (NM_001066235), and β-subunits from Arabidopsis (At4g00570), O. sativa (NM_001071533), and potato (Z23002).
Specific Expression of AtNAD-ME Genes
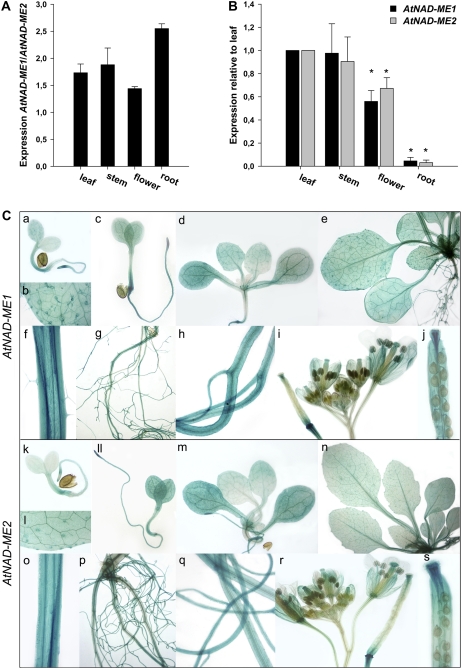
To study the tissue-specific expression of AtNAD-ME genes, quantitative real-time (qRT)-PCR experiments were performed. Transcripts for both AtNAD-ME1 and AtNAD-ME2 were detected in all the organs tested. In all cases, the transcript levels of AtNAD-ME1 were higher than that of AtNAD-ME2 (Fig. 2A). The comparison of the abundance of each transcript relative to the expression in leaves indicated that both genes have the same relative level of expression in all mature organs (Fig. 2B). The expression was similar in leaves and stems (100%), while the expression in flowers and roots accounted for 60% and 4% of the level in leaves, respectively (Fig. 2B).
Figure 2.
Expression analysis of AtNAD-ME in different tissues. A, Expression of AtNAD-ME1 transcript relative to AtNAD-ME2 mRNA levels for each organ analyzed by qRT-PCR. B, Relative expression of AtNAD-ME1 and AtNAD-ME2 in different organs with respect to leaf analyzed by qRT-PCR. The y axis refers to the fold-difference of a particular AtNAD-ME transcript level relative to its amount found in leaf. The asterisks indicate that the expression values obtained were statistically significantly different from the ones obtained for leaf as determined by the Student's t test (P < 0.05). C, Analysis of AtNAD-ME∷GUS expression. a to j, AtNAD-ME1∷GUS; k to s, AtNAD-ME2∷GUS. a to d and k to m, Seedlings at 3, 5, and 12 DAI. e and n, Three-week-old rosette. f and o, Longitudinal section through the stem. g and p, Root system of a 3-week-old plant. h and q, Close-ups of the roots. i and r, Inflorescence with flowers at different stages. j and s, Siliques.
To investigate more precisely the organ- and tissue-specific expression of both AtNAD-ME genes, transgenic Arabidopsis plants expressing the GUS reporter gene driven by the AtNAD-ME promoters were generated and analyzed throughout development. About eight transgenic AtNAD-ME∷GUS lines were analyzed in detail, most of them showing a similar tissue-specific pattern of AtNAD-ME1 and AtNAD-ME2 expression. The pattern of GUS activity was very similar for the AtNAD-ME1∷GUS and AtNAD-ME2∷GUS plants (Fig. 2C). In both cases, GUS expression could be observed 2 d after imbibition (DAI) in the cotyledons, hypocotyls, and root tip (data not shown). At 3 DAI, the roots were completely stained (Fig. 2C, a and k) and in the case of AtNAD-ME1-promoter∷GUS plants, the root tip was highly stained (Fig. 2C, a). A high expression was observed in trichomes and trichome basal cells, especially in AtNAD-ME1∷GUS plants (Fig. 2C, b and l). At 5 DAI, both root tips were highly stained, and expression in all seedling tissues was maintained (Fig. 2C, c and ll). At 12 DAI, GUS expression was very low in the new leaves but became higher with maturation (Fig. 2C, d, e, m, and n). At all developmental stages, the expression in leaves was observed in the mesophyll and the cells that surround the vascular bundles (bundle sheet cells; Supplemental Fig. S1). Stems (Fig. 2C, f and o) and roots (Fig. 2C, g, h, p, and q) presented high expression in all tissues in both AtNAD-ME∷GUS plants. It is interesting to note that longitudinal sections of stems revealed a high level of activity of both promoters around the vascular system (Fig. 2C, f and o). In the reproductive organs of both lines, GUS expression was detected in the apical part of the gynoecium, stigmatic papillae, the filaments, and sepals (Fig. 2C, i and r). In developing siliques, expression was high in the apical part and the abscission zone (Fig. 2C, i, j, r, and s). It is worth mentioning that the GUS expression driven by the AtNAD-ME1-promoter was always stronger than by AtNAD-ME2-promoter in all lines tested. It should also be noted that the observations described above are consistent with AtGenExpress data from the Genevestigator microarray database (Zimmermann et al., 2004; http://www.genevestigator.ethz.ch/).
Biochemical and Structural Properties of Recombinant AtNAD-MEs
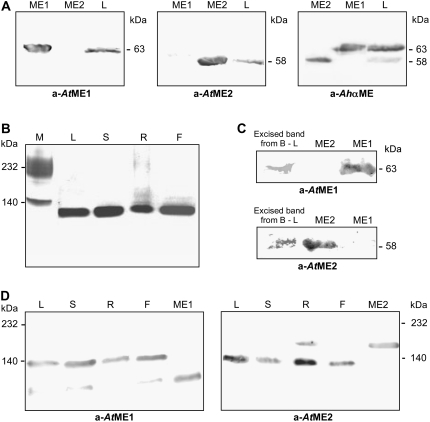
To assess whether the two predicted AtNAD-MEs are enzymatically active proteins, AtNAD-ME1 and AtNAD-ME2 cDNAs were cloned and expressed as recombinant proteins. The prediction of the length of the mitochondrial targeting sequences was performed by sequence comparison with potato NAD-ME and the assistance of prediction programs (ARAMEMNON, http://aramemnon.botanik.uni-koeln.de/). After eliminating mitochondrial targeting sequences of 38 and 31 amino acid residues for AtNAD-ME1 and AtNAD-ME2, respectively, the mature proteins were expressed in Escherichia coli. Following induction of the expression by isopropyl-β-thiogalactopyranoside or lactose, proteins with the expected molecular masses of 80 and 76 kD (AtNAD-ME1 and AtNAD-ME2, respectively) were purified by affinity chromatography (Fig. 3A). After enterokinase digestion to remove the His tag used for purification, products of 63 and 58 kD were obtained for AtNAD-ME1 and AtNAD-ME2, respectively (Fig. 3A). Both recombinant proteins were recognized by the anti-A. hypochondriacus α-NAD-ME antibody, although AtNAD-ME1 displayed a higher reactivity (Figs. 3C and 4A). Antibodies raised against AtNAD-ME1 and AtNAD-ME2 reacted with the respective proteins but did not show cross reactivities (Fig. 4A). These results indicate antigenic differences between both AtNAD-ME proteins.
Figure 3.
Recombinant Arabidopsis NAD-ME isoforms analyzed by gel electrophoresis. A, Coomassie-stained SDS-PAGE of recombinant NAD-ME isoforms. Five micrograms of purified recombinant AtNAD-ME1 and AtNAD-ME2 before (1) and after (2) enterokinase digestion was loaded in each case. The estimated molecular mass of the purified proteins is indicated on the right. B, Native-PAGE stained for NAD-ME activity. Approximately 20 milliunits of AtNAD-ME1 and AtNAD-ME2 were loaded, as well as a mixture of equal amounts of both proteins. A mitochondrial leaf crude extract (L, 20 milliunits) was also loaded in the gel. C, Western blot of native-PAGE using antibodies against A. hypochondriacus α-NAD-ME. Approximately 5 μg of NAD-ME1 and AtNAD-ME2 were loaded, as well as a mixture of equal amounts of both proteins. A mitochondrial leaf crude extract (L, 30 μg) was also loaded in the gel. Molecular mass markers (M) were run in parallel and stained with Coomassie Blue.
Figure 4.
SDS- and native-PAGE of mitochondrial extracts analyzed for activity or by western blot. A, SDS-PAGE of leaf mitochondrial extracts (L, 50 μg of total protein) analyzed by western blot using antibodies against AtNAD-ME1 (a-AtME1) or AtNAD-ME2 (a-AtME2), or, alternatively, against A. hypochondriacus α-NAD-ME (a-AhαME). As control, recombinant AtNAD-ME1 (3 μg, ME1) and AtNAD-ME2 (3 μg, ME2) were loaded. B, Native-PAGE of Arabidopsis mitochondrial extracts stained for NAD-ME activity. Approximately 20 milliunits of NAD-ME from mitochondrial crude extracts from leaf (L), stem (S), root (R), and flower (F) was loaded in each lane. Molecular mass markers were run in parallel and stained with Coomassie Blue. C, The active band from leaf mitochondrial crude extracts (excised band from B–L) was excised and analyzed by SDS-PAGE followed by western-blot analysis using antibodies against AtNAD-ME1 (a-AtME1) or AtNAD-ME2 (a-AtME2). As controls, recombinant purified AtNAD-ME1 and AtNAD-ME2 (3 μg, ME1 and ME2) were loaded. D, Western-blot analysis of native-PAGE of mitochondrial crude extracts (30 μg) from leaf (L), stem (S), root (R), and flower (F) using antibodies against AtNAD-ME1 (a-AtME1) or AtNAD-ME2 (a-AtME2). As control, recombinant purified AtNAD-ME1 and AtNAD-ME2 (3 μg, ME1 and ME2) were loaded. The molecular masses of the marker proteins run in parallel are indicated.
Interestingly, recombinant purified AtNAD-ME1 and AtNAD-ME2 showed both enzymatic activities and could thus be characterized with respect to their kinetic properties (Table I). Both isoforms displayed similar kcat values, with AtNAD-ME2 having a 1.3-fold higher kcat value than AtNAD-ME1 (Table I). Comparing the apparent Km values for NAD and malate, AtNAD-ME1 and AtNAD-ME2 exhibited very similar affinities toward both compounds, while AtNAD-ME2 presented the highest catalytic efficiency (kcat/Km; Table I). It is worth mentioning that the kinetic behavior obtained for AtNAD-ME1 with respect to both NAD and malate was nonhyperbolic, presenting some kind of sigmoidicity and thus probable cooperative binding of the ligand in both cases (Table I). The pH optimum for both isoforms was very similar, at about pH 6.5 (Table I). Neither AtNAD-ME1 nor AtNAD-ME2 was able to decarboxylate OAA, even when using high protein concentrations for the assay (data not shown). Thus, both AtNAD-MEs clearly belong to the EC 1.1.1.39 subtype.
Table I.
Kinetic properties of recombinant Arabidopsis NAD-ME isoforms
The indicated values, obtained by nonlinear regression, are the average of at least three different measurements ± se.
| Parameter | NAD-ME1 | NAD-ME2 |
|---|---|---|
| pH optimum | 6.4 | 6.6 |
| kcat (s−1) | 31.1 ± 1.7 | 44.1 ± 1.2 |
| Km NAD (mm) | 0.50 ± 0.2a | 0.50 ± 0.1 |
| kcat/Km NAD | 60.2 | 88.2 |
| Km malate (mm) | 3.0 ± 0.7b | 3.0 ± 0.2 |
| kcat/Km malate | 10.3 | 14.7 |
S0.5 (Hill coefficient = nH = 1.3).
S0.5 (nH = 1.9).
Native electrophoresis of the purified recombinant AtNAD-MEs was analyzed by activity staining and western blot (Fig. 3, B and C). An active band compatible with a dimeric oligomeric state for the recombinant AtNAD-ME2 was detected by activity staining assays (Fig. 3B). This active band also reacted with the anti-A. hypochondriacus α-NAD-ME-antibody (Fig. 3C). Interestingly, the recombinant AtNAD-ME1 could not be detected by activity staining (even loading up to 40 milliunits of the purified enzyme), although a band with a higher mobility than that of AtNAD-ME2 was obtained by western blot using the anti-A. hypochondriacus α-NAD-ME- antibody (Fig. 3C). As the purified AtNAD-ME1 exhibited enzymatic activity in solution, it is possible that the protein lost its activity during electrophoresis.
Size-exclusion chromatography was used to estimate the native molecular masses for both recombinant AtNAD-MEs. The calculated molecular masses for the purified AtNAD-ME1 and AtNAD-ME2 proteins were 120.0 ± 6 and 117.5 ± 6.5 kD, respectively. Thus, although AtNAD-ME1 and AtNAD-ME presented different mobilities by native electrophoresis, probably due to differences in the charge, both proteins obviously assembled as dimers in solution.
AtNAD-ME1 and AtNAD-ME2 Form Both Homo- and Heterooligomers in Vitro and in Vivo
NAD-ME activity was measured in different organs of mature Arabidopsis plants. Crude extracts from roots showed the highest activity expressed on the basis of total protein concentration (0.038 ± 0.03 units/mg), while leaves, stems, and flowers displayed between 58% and 65% of the activity measured in roots (Fig. 5C). When extracts of isolated mitochondria from each organ were analyzed by SDS-PAGE followed by western-blot analysis, two well-separated bands could be detected using the anti-A. hypochondriacus α-NAD-ME-antibody (Fig. 4A). On the other hand, while the antibodies raised against AtNAD-ME1 recognize the 63-kD band, the ones raised against AtNAD-ME2 recognized the 58-kD band (Fig. 4A). The molecular masses of the immunoreactive bands in crude extracts correlated well with those of the recombinant purified AtNAD-ME1 and AtNAD-ME2 proteins (Fig. 4A). Interestingly, native PAGE of mitochondrial extracts from different Arabidopsis organs stained for NAD-ME activity showed a unique band of approximately 120 kD (Fig. 4B). To analyze the composition of this activity-containing band, gel slices including the band with activity from the leaf mitochondrial extract were excised from the native PAGE and subjected to SDS-PAGE coupled to western blot using specific antibodies against AtNAD-ME1 or AtNAD-ME2. As shown in Figure 4C, reactive bands corresponding to AtNAD-ME1 (63 kD) and AtNAD-ME2 (58 kD) could be detected. Considering the molecular masses of the separated AtNAD-MEs, these results clearly indicate that the active native band of approximately 120 kD is composed of AtNAD-ME1 and AtNAD-ME2, most probably in a 1:1 ratio.
Figure 5.
Identification of nad-me insertion lines. A, AtNAD-ME gene structure showing the locations of the T-DNA insertions in the knockout mutants. The orientation of the T-DNA insertion is indicated as left border (LB). B, Semiquantitative RT-PCR showing the absence of the corresponding AtNAD-ME transcript in the single (nad-me1 and nad-me2) and double (1 × 2) T-DNA knockout lines. PCR products of 750 (NAD-ME1) and 928 (NAD-ME2) bp were amplified using 35 cycles. As loading control, a 521-bp Actin2 cDNA fragment was amplified by 29 cycles. C, NAD-ME activity (units/mg) in different organs of Arabidopsis wild type and the T-DNA insertion lines. The bars indicate the sd of the measurements from three different crude extract preparations. The activity measurement was performed three independent times with each crude extract preparation, with <5% sd within each preparation.
On the other hand, native PAGE of mitochondrial extracts from different Arabidopsis organs analyzed by western blot showed a band of approximately 120 kD using both specific antibodies directed against AtNAD-ME1 or AtNAD-ME2, while the recombinant proteins differ in electrophoretic mobilities and show specific reactions against their own antibodies (Fig. 4D). Moreover, a second immunoreactive band with the same mobility as the recombinant protein was detected in the mitochondrial extracts (Fig. 4D), indicating the possible existence of homodimers in vivo.
The interaction of AtNAD-ME1 and AtNAD-ME2 observed in mitochondrial extracts was further tested in vitro using recombinant AtNAD-ME. Purified recombinant AtNAD-ME1 and AtNAD-ME2 were mixed in an equimolar ratio and subsequently analyzed by native PAGE. As shown in Figure 3, B and C, the mixture has active and immunoreactive bands as the isolated recombinant proteins, and, additionally, it has a band with similar mobility to that observed in the mitochondrial extracts. Taken together, these results revealed that the separated AtNAD-ME1 and AtNAD-ME2 assemble as active dimers and associate to form a heterodimer in vitro and in vivo.
Isolation and Characterization of T-DNA Insertion Mutants of AtNAD-ME1 and AtNAD-ME2
Arabidopsis insertion mutants that contained T-DNA elements inserted in the AtNAD-ME1 and AtNAD-ME2 genes were isolated from the Sail-lines (Fig. 5A). Homozygous lines for each mutant were confirmed by PCR and designated nad-me1.1 (Sail-374-A02) and nad-me2.1 (Sail-291-C05), respectively. The sites of the insertions were analyzed by sequencing the PCR products obtained after amplifying both ends of the T-DNA insertion and the flanking genomic DNA. The knockout line nad-me1.1 had an insertion in exon 4 (at position +833 bp), and in line nad-me2.1 the insertion was localized in exon 5 (at position +1,030 bp). The mutant lines showed no expression of the corresponding genes as analyzed by RT-PCR (Fig. 5B), and the absence of the corresponding protein was confirmed by western blot using specific antibodies (data not shown). A second allelic mutant for each gene (Sail-374-A02, nad-me1.2 and Salk-131720, nad-me2.2) was also isolated and characterized in parallel, but the lines were not included in this work. As growth and development of all the single mutants analyzed did not show visual differences with respect to the wild type, homozygous double mutants were generated between nad-me.1.1 and nad-me2.1 by crossing. The absence of transcripts and proteins for both AtNAD-MEs in the double mutants was confirmed by RT-PCR and western blot (Fig. 5B; data not shown).
Figure 5C shows residual NAD-ME activities measured in different organs from nad-me1.1 and nad-me2.1. The results indicated that in vivo, both AtNAD-ME1 and AtNAD-ME2 exhibit enzymatic activity and that AtNAD-ME2 possesses the highest specific activity, results that correlate well with those obtained for the recombinant proteins. The double mutant did not show any NAD-ME activity, indicating that AtNAD-ME1 and AtNAD-ME2 are solely responsible for the NAD-ME activities measured in crude extracts. Native PAGE conducted with mitochondria extracted from both single mutants showed an immunoreactive band corresponding to the remaining functional AtNAD-ME protein, while in the double knockout extracts, no immunoreactive band was detected (data not shown). It is worth mentioning that transcript levels of the remaining intact AtNAD-ME gene in the organs of each single mutant did not show significant difference relative to the expression observed in the wild-type organs when analyzed by qRT-PCR (data not shown).
The germination, development, vegetative growth, and flowering time of the double knockouts were very similar to that of the wild type and the single knockout mutants when grown either at moderate (100 μmol m−2 s−1) or high (500 μmol m−2 s−1) light intensities. The data obtained indicated that there are no statistical differences in rosette diameter or dry weight between the wild type and homozygous mutants grown in both conditions (Table II; data not shown). Moreover, the photosynthetic parameters maximum quantum efficiency of PSII and electron transport rate (ETR; Table II) and the qP and qN values (data not shown) indicated no differences between the knockout mutants and the wild type.
Table II.
Growth and photosynthetic parameters of the T-DNA insertional lines and the wild type grown for 5 weeks in long days at a photon flux density of 100 μmol m−2 s−1
DW, Dry weight; RD, rosette diameter; ETR measured at 800 μmol m−2 s−1. Values are means ± sd of determinations made on eight plants.
| Wild Type | nad1.1 | nad2.1 | nad1.1 × 2.1 | |
|---|---|---|---|---|
| DW (mg) | 6.75 ± 0.08 | 6.76 ± 0.05 | 6.76 ± 0.07 | 6.78 ± 0.03 |
| RD (cm) | 6.86 ± 0.64 | 7.09 ± 0.73 | 7.09 ± 0.76 | 6.97 ± 0.46 |
| ETR (μmol m−2 s−1) | 80.9 ± 3.3 | 76.1 ± 8.8 | 83.4 ± 1.9 | 81.2 ± 3.0 |
| Maximum quantum efficiency of PSII | 0.713 ± 0.008 | 0.713 ± 0.003 | 0.726 ± 0.005 | 0.720 ± 0.002 |
AtNAD-ME Is More Active during the Night Period, and Mutant Lines Completely Lacking NAD-ME Activity Display Altered Steady-State Levels of Sugars and Amino Acids
To gain further information about the extent of physiological disturbances generated by the lack of total NAD-ME activity, metabolite profiling analyses using gas chromatography-mass spectrometry (GC-MS; Fahnenstich et al., 2007) were performed using whole rosettes of mature plants harvested at the end of the light and the dark period. This study revealed distinct alterations in the metabolic status of the double loss-of-function mutant depending on the light/dark period. While both the double knockout mutant and the wild type showed the same accumulation of starch by the end of both periods (data not shown), the contents of a range of other metabolites were altered in the mutant compared to the wild type (Table III; the complete data set of metabolites is available in Supplemental Table S1). At the end of the light period, the double knockout mutant showed an accumulation of mono- and disaccharides, such as Fru, Gal, Glc, and Suc (Table III). The levels of intermediates of the TCA cycle and of amino acids were comparable to those of the wild type, except citrate was decreased (Table III). In contrast, at the end of the night period, the carbohydrate levels were invariable with respect to the wild type, while many amino acids, principally those derived from 2-oxoglutarate and OAA, accumulated in the double knockout mutant (Table III). Moreover, the contents of the TCA metabolites were also altered. In the double knockout mutant, the levels of 2-oxoglutarate and succinate were higher, while that of citrate and fumarate were lower, and those of malate, pyruvate, and OAA were invariable compared to the wild type.
Table III.
Metabolite content determined by GC-MS of plants lacking NAD-ME activity
Values are mean ± se of four different biological replicas, each consisting of four determinations made on eight plants. The values are relative to the respective wild type (each metabolite = 1; the corresponding complete data set of metabolites is available in Supplemental Table S1). Those values that are significantly different from the respective wild type as determined by the Student's t test (P < 0.05) are in bold type.
| nad1 × 2
|
||
|---|---|---|
| End of the Day | End of the Night | |
| Ala | 0.98 ± 0.21 | 1.34 ± 0.13 |
| Asn | 0.88 ± 0.16 | 2.52 ± 0.35 |
| Asp | 0.87 ± 0.09 | 1.29 ± 0.10 |
| γ-Aminobutyrate | 1.00 ± 0.09 | 1.91 ± 0.22 |
| Glu | 0.92 ± 0.07 | 1.40 ± 0.12 |
| Gln | 0.89 ± 0.15 | 3.51 ± 0.40 |
| Gly | 0.87 ± 0.11 | 1.78 ± 0.23 |
| His | 1.13 ± 0.26 | 1.63 ± 0.18 |
| Ile | 0.87 ± 0.11 | 1.15 ± 0.15 |
| Leu | 0.81 ± 0.11 | 1.15 ± 0.16 |
| Lys | 1.26 ± 0.18 | 1.89 ± 0.29 |
| Met | 1.26 ± 0.08 | 1.32 ± 0.10 |
| Phe | 0.86 ± 0.08 | 1.25 ± 0.11 |
| Pro | 0.81 ± 0.12 | 1.63 ± 0.40 |
| Ser | 0.92 ± 0.09 | 1.96 ± 0.20 |
| Thr | 1.13 ± 0.07 | 1.47 ± 0.14 |
| Val | 1.07 ± 0.09 | 1.23 ± 0.15 |
| Citrate | 0.60 ± 0.13 | 0.72 ± 0.07 |
| 2-Oxoglutarate | 0.75 ± 0.06 | 1.30 ± 0.05 |
| Succinate | 0.82 ± 0.06 | 1.43 ± 0.15 |
| Fumarate | 0.84 ± 0.11 | 0.57 ± 0.08 |
| Malate | 0.84 ± 0.12 | 1.13 ± 0.07 |
| OAA | 1.18 ± 0.13 | 0.87 ± 0.14 |
| Pyruvate | 0.76 ± 0.11 | 0.91 ± 0.13 |
| Ara | 1.08 ± 0.07 | 0.92 ± 0.08 |
| Ascorbate | 1.43 ± 0.25 | 1.03 ± 0.30 |
| Fru | 1.43 ± 0.18 | 1.06 ± 0.22 |
| Gal | 1.77 ± 0.19 | 0.71 ± 0.10 |
| Glc | 1.57 ± 0.12 | 1.12 ± 0.17 |
| Man | 1.33 ± 0.17 | 0.73 ± 0.11 |
| Rib | 0.83 ± 0.16 | 1.64 ± 0.24 |
| Suc | 1.42 ± 0.13 | 0.98 ± 0.06 |
| 3-PGA | 1.14 ± 0.20 | 1.11 ± 0.10 |
| DHAP | 0.76 ± 0.12 | 1.41 ± 0.20 |
| Lactate | 1.01 ± 0.17 | 1.47 ± 0.27 |
| Glycerate | 0.98 ± 0.09 | 1.14 ± 0.15 |
| Glycerol | 1.26 ± 0.08 | 1.39 ± 0.22 |
| Glycolate | 1.56 ± 0.39 | 1.09 ± 0.43 |
| Glyoxylate | 1.22 ± 0.14 | 1.07 ± 0.11 |
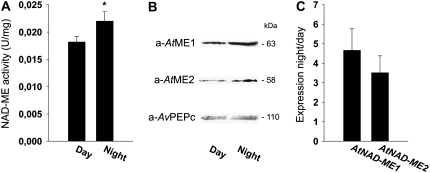
Due to the differences found in the metabolic profile in the double loss-of-function mutant, the total NAD-ME activity, NAD-ME protein amount and the expression levels of both genes were analyzed at the end of the light and night periods in the wild type. Leaf crude extracts contained about 20% higher NAD-ME specific activities at the end of the night period than at the end of the day period (Fig. 6A). In line with this result, western-blot analysis of these extracts showed that both AtNAD-ME1 and AtNAD-ME2 were more abundant during the night period (Fig. 6B). Quantification of the amount of immunoreactive protein of three biological replicates indicated that AtNAD-ME1 and AtNAD-ME2 were enhanced approximately 3- and 2.5-fold in the night period with respect to the day period, respectively. Moreover, the expression of both genes, evaluated by qRT-PCR, was also enhanced by the end of the night (Fig. 6C). Taken together, Arabidopsis leaf extracts presented higher NAD-ME activity during the night period as a result of protein accumulation due to enhanced gene expression.
Figure 6.
Diurnal changes in NAD-ME activity and expression level in wild-type leaves. A, Total NAD-ME activity at the end of the day and night periods. The bars indicate the sd of the measurements from three different crude extracts preparations. The activity measurement was performed three independent times with each crude extract preparation. The asterisk indicates that the NAD-ME activity determined at the end of the night period was statistically significantly higher than the one at the end of the light period as determined by the Student's t test (P < 0.05). B, SDS-PAGE of leaf crude extracts (50 μg of total protein) prepared at the end of the light and night periods and analyzed by western blot using antibodies against AtNAD-ME1 (a-AtME1) or AtNAD-ME2 (a-AtME2). As control of the amount of protein loaded, antibodies against A. viridis PEPc (a-AvPEPc) were also used for detection. The estimated molecular mass of the immunoreactive bands is indicated on the left. C, Expression of AtNAD-ME1 and AtNAD-ME2 transcripts by the end of the night period relative to the expression by the end of the day period analyzed by qRT-PCR. The bars indicate the sd of measurements from three different biological replicates. For both genes, the relative expression values obtained at the end of the night period were statistically significantly higher than the ones at the end of the light period as determined by the Student's t test (P < 0.05).
DISCUSSION
Arabidopsis Possesses a Heterodimeric NAD-ME, and Both Subunits Exhibit Enzymatic Activities in Vitro and in Vivo
AtNAD-ME1 and AtNAD-ME2 show homologies to the α- and β-subunits of other plant NAD-MEs and are localized to mitochondria, as previously revealed by a mitochondrial proteomic study (Haezlewood et al., 2004). A multiple alignment showed that these two proteins possess the conserved motifs present in all members of the NAD(P)-ME family (Winning et al., 1994; Drincovich et al., 2001). In line with this, recombinant individual AtNAD-ME1 and AtNAD-ME2 subunits displayed NAD-ME activities. This is in contrast to a previous study of potato and C. argentea NAD-MEs that showed that the individual subunits do not possess NAD-ME activities (Willeford and Wedding, 1987). This discrepancy can be reconciled when considering that a urea-based procedure was used to separate these subunits, most probably causing denaturation of the proteins.
A heteromeric composition of plant NAD-MEs has been reported for some species (Grover and Wedding, 1982; Burnell, 1987; Willeford and Wedding, 1987; Long et al., 1994). In Arabidopsis, several lines of evidence indicate that AtNAD-ME1 and AtNAD-ME2 are subunits of a heteromeric enzyme. A recombinant protein mixture of both AtNAD-MEs revealed an additional band with NAD-ME activity and similar mobility to that exhibited by the active and inmunoreactive bands observed in mitochondrial extracts (Fig. 3, B and C). Moreover, the active band found in the extracts contained two dissimilar and immunoreactive proteins that corresponded to the recombinant monomers (Fig. 4C). In addition, our results clearly show that AtNAD-ME1 and AtNAD-ME2 are functional enzymes in vivo, as the single insertion mutants exhibited residual NAD-ME activities, whereas NAD-ME activities were completely absent in the double knockout. In the active conformational state, the separated AtNAD-ME1 and AtNAD-ME2 assemble as dimers in vitro and in vivo. These observations lead to the suggestion that in vivo, AtNAD-ME1 and AtNAD-ME2 can form not only a heterodimer but also a homodimer.
The kinetic analyses performed showed that AtNAD-ME2 displayed a slightly higher specific activity in comparison to AtNAD-ME1 (Table I). Nevertheless, the pH optimum and the affinity for NAD and malate were almost identical for both proteins, and this data is in agreement with previous reports on plant NAD-ME (Artus and Edwards, 1985). Although AtNAD-ME2 displays hyperbolic kinetics with respect to both substrates, AtNAD-ME1 presents sigmoidicity, which indicates differences in the substrate binding and/or cooperation between the subunits in AtNAD-ME1 (Table I). AtNAD-ME isoforms exhibit low affinity to the cofactor and higher specific activity at low pH values (below pH 7.0). Moreover, plant NAD-MEs display the weakest affinity for NAD and the lowest sensitivity to NADH inhibition of all NAD-linked dehydrogenases described in plant mitochondria (Pascal et al., 1990).
Finally, in accordance to the universal role in plant mitochondria, the AtNAD-ME1 and AtNAD-ME2 genes have very similar levels of expression in all mature organs. It is worth mentioning that the level of expression of the AtNAD-ME1 transcript was higher than that of AtNAD-ME2 in all the organs analyzed; nevertheless, this does not necessarily imply a higher level of AtNAD-ME1 protein.
Enhanced NADP-ME Activity during the Night Period and Distinctly Modified Leaf Metabolic Profiles during the Day and Night Periods of Mutants Lacking NAD-ME Activity Indicate a Major Participation of NAD-ME in Nocturnal Metabolism
As NAD-ME was assumed to play a central role in the management of flux through the TCA cycle by providing a means of generating acetyl-CoA, and thus, ATP and carbon skeletons (Grover et al., 1981), we aimed to evaluate the relevance of NAD-ME in plant metabolism. For this purpose, T-DNA insertion mutants for each AtNAD-ME gene were isolated and double mutants were produced and characterized. Neither the single nor double loss-of function mutants showed differences in growth or lack of fitness when grown at moderate or high light intensities. Also, leaf photosynthesis was invariant compared to the wild-type, indicating that NAD-ME activity is not essential for normal autotrophic growth in a C3 plant. Jenner et al. (2001) showed that a reduction in NAD-ME activity in potato by antisense repression had no significant effects on plant morphology, tuber fresh weight, or number of tubers. Moreover, no effects on mitochondrial function associated with any changes in the levels of malate or citrate were found. The authors attributed this negligible impact of reduced NAD-ME activities on metabolism to compensation by activation of the residual NAD-ME activity or compensation via other metabolic processes. The Arabidopsis double knockout mutants showed no residual NAD-ME and thus, the small changes in metabolism found at the end of the light period can only be attributed to compensation through other metabolic processes that can supply pyruvate to the mitochondria. Neither malate nor pyruvate, the substrate and product of the NAD-ME reaction, was altered in the plants completely lacking the NAD-ME activity. As NAD-ME is not the sole source of pyruvate in mitochondria, it is not unexpected that the lack of NAD-ME activity resulted in no considerable effects on diurnal plant metabolism. In accordance with this, two genes encoding mMDH in Arabidopsis (At1g53240 and At3g47520) are induced by light (Price et al., 2004; Thum et al., 2004; Rasmusson and Escobar, 2007). Moreover, our results showed that NAD-ME expression is lower during the day than during the night period. In this way, it is possible that during the light period, the activity of NAD-ME is less important to fuel respiration.
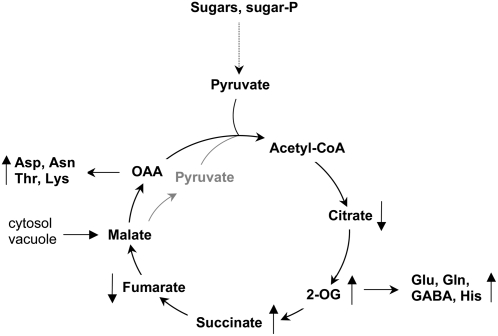
On the contrary, at the end of night, the levels of amino acids derived from OAA and 2-oxoglutarate were increased in the mutants completely lacking NAD-ME activity, particularly Asn and Gln. At the same time, these plants showed enhanced levels of the TCA cycle intermediates 2-oxoglutarate and succinate compared to the wild type. Interestingly, the levels of amino acids derived from pyruvate, e.g. Ala, Val, and Ile, were invariable in the mutant. In Arabidopsis, the malate level increases during the day, and this organic acid is accumulated in the vacuole until metabolic demands are sensed (Gout et al., 1993; Fahnenstich et al., 2007). During night, vacuolar malate is utilized in the mitochondria to furnish part of the TCA cycle and to replenish the TCA cycle pool. This function can be fulfilled through the concerted action of MDH and NAD-ME. During the night period, the activity of MDH is lower than during the light period (Price et al., 2004; Thum et al., 2004; Rasmusson and Escobar, 2007) but on the contrary, the activity of the NAD-ME was found to be higher during the night period (Fig. 6). Moreover, the nad-me loss-of-function mutant showed specifically accumulation of amino acids derived from intermediates of the TCA cycle. Taken together, it could be concluded that in plants lacking NAD-ME activity, the excess of mitochondrial malate occurring in the night period is diverted to the synthesis of amino acids from intermediates of the TCA cycle. Thus, the described differential patterns of modifications of the metabolic profile reveal a major participation of NAD-ME during the night period. In this way, we propose a role for the NAD-ME in the coordination of the carbon and nitrogen metabolisms in Arabidopsis. When carbon in the form of malate cannot be converted into pyruvate through the activity of NAD-ME and thus cannot completely flow through the TCA cycle, the flux from TCA intermediates toward the synthesis of amino acids is increased (Fig. 7).
Figure 7.
Scheme representing the flux of metabolites in Arabidopsis lacking NAD-ME activities. The direction of the arrows indicates accumulation or depletion of the respective metabolite. During the night period, mitochondrial pyruvate derives from glycolysis and from vacuolar malate reserves through the action of NAD-ME in leaves.
It was a matter of debate whether or not mitochondrial NAD-ME could compensate for limited capacity for pyruvate transport across the mitochondrial membrane by providing the TCA cycle with pyruvate under conditions of high-energy demands (Day and Hanson, 1977; Brailsford et al., 1986; Hill et al., 1994). It would thus be interesting to challenge the nad-me double knockout mutant to different conditions in order to prove this hypothesis.
On the other hand, high activities of NAD-ME and NADP-ME were reported in cells around the vascular bundles in tobacco (Hibberd and Quick, 2002), where they were proposed to participate in the decarboxylation of malate derived from the respiratory activity of heterotrophic tissues. In C4 plants, MEs in vascular bundles decarboxylate malate derived from the C4 pathway. We show here that both AtNAD-ME genes are strongly expressed in bundle sheath cells of stems and petioles. Similarly, high GUS activity driven by the promoters of AtNADP-ME1, AtNADP-ME2, and AtNADP-ME4 was described for cells surrounding the vasculature of stems in Arabidopsis (Gerrard Wheeler et al., 2005). These results suggest a possible universal and specific function for these decarboxylases in the bundle sheath cells of C3 plants that still awaits elucidation.
MATERIALS AND METHODS
Isolation of T-DNA Insertion Lines and Plant Growth Conditions
The T-DNA insertion lines Sail-374-A02 (nad-me1.1) and Sail-291-C05 (nad-me2.1) were obtained from the Nottingham Arabidopsis Stock Center (http://www.arabidopsis.info/). The genotype of the lines was determined using genomic DNA of individual plants as template for PCR amplifications of the wild-type and nad-me alleles. The primers used to amplify the wild-type alleles were as follows: NAD-ME1wtF (5′-ACGATGACGGAGAGAATCGT-3′) and NAD-ME1wtR (5′-ATGTTCAATGATGATGTCCAG-3′), and NAD-ME2wtF (5′-GACCTGTGTACAGCAATGTGATCG-3′) and NAD-ME2wtR (5′-GGTCTTGTCACCACGGAGAGGACA-3′). To amplify the nad-me alleles, the primers NAD-ME1wtF and NAD-ME2wtF were combined separately in a PCR with the primer SailLB (5′-TAGCATCTGAATTTCATAACCAATCTCGATACAC-3′). The location of the inserts was verified by amplifying and sequencing the T-DNA flanking genomic DNA. Seeds of Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 and the transformant lines were placed on soil and kept in darkness for 4 d at 4°C to synchronize germination. After 2 weeks, the seedlings were transferred to pots (one per pot) containing three parts of soil (Gebr. Patzer KG) and one part of vermiculite (Basalt Feuerfest). Plants were grown under a 16-/8-h photoperiod at 100 or, alternatively, at 500 μmol m−2 s−1 and 21°C/18°C (day/night) temperatures and 65% relative humidity in a controlled growth cabinet. Alternatively, Arabidopsis seeds were sterilized and sown in Murashige and Skoog agar plates containing 1% Suc.
Cloning of Full-Length AtNAD-ME1 and AtNAD-ME2
Arabidopsis full-length cDNAs encoding NAD-ME1 and NAD-ME2 were amplified by RT-PCR using RNA extracted from leaves and the TRizol reagent (Gibco-BRL). Amplification was conducted using SuperScript II reverse transcriptase (Invitrogen) and specific primers. In the case of AtNAD-ME1, the oligonucleotide pair NAD-ME1GWF (5′-CACCATGGGAATAGCCAATAAGCTCCGGCT-3) and NAD-ME1GWF (5′-GAGTACCCGACTTTGGTCTACAAGGATGAC-3′) was used. AtNAD-ME2 was amplified with the primer pair NAD-ME2GWF (5′-CACCATGTGGAAGAACATTGCTGGGTTGTC-3′) and NAD-ME2GWR (5′-CCTGTTTACAGCCCTCTCGTTCACGAGAAA-3′). The PCR products were cloned into pENTR/D-TOPO (Invitrogen) and completely sequenced.
Heterologous Expression of the Mature AtNAD-MEs and Purification of the Recombinant Proteins
To amplify the cDNA fragments corresponding to the mature AtNAD-MEs, a PCR reaction was conducted using as template the full-length cDNAs cloned as described above and the following primer pairs: NAD-ME1F (5′-GGATCCCCCACCATCGTTCATAAA-3′) and NAD-ME1R (5′-GTCTACAAGGATGACTAAGTCGAC-3′), and NAD-ME2F (5′-GGATCCTGCATCGTCCACAAGCGT-3′) and NAD-ME2R (5′-ACGCTTGTGGACGATGCAGGATCC-3′). The primers were designed to introduce unique BamHI and SalI sites at the 5′ and 3′ ends, respectively, to facilitate the subcloning into the pET32 expression vector. In each pET32 vector containing the inserts of AtNAD-ME1 and AtNAD-ME2 (pET-NAD-ME1 and pET-NAD-ME2), the NAD-MEs are fused in-frame to a His tag to facilitate purification of the expressed fusion protein by a nickel-containing His-Bind column (Novagen). The induction and purification of the fusion proteins were performed as previously described for the Arabidopsis NADP-ME isoforms (Gerrard Wheeler et al., 2005). The fusion proteins were then concentrated on Centricon YM-30 (Amicon) using buffer MMG (50 mm MES-NaOH, pH 6.5, 5 mm MnCl2, and 10% [v/v] glycerol). Purified fusion NAD-ME proteins were then incubated with 0.05 to 0.075 units of enterokinase (EK-Max; Invitrogen) per milligram of protein at 16°C for 2 h to remove the N terminus coded for by the expression vector. The proteins were further purified using a Sephadex G-50 column equilibrated with buffer A (50 mm MES-NaOH, pH 6,5, 5 mm MnCl2, 5 mm dithiothreitol, and 20% [v/v] glycerol). Purified AtNAD-ME1 and AtNAD-ME2 were stored at −80°C in buffer A (with 50% glycerol) for further studies.
Gel Filtration Chromatography
Molecular masses of recombinant native AtNAD-ME1 and AtNAD-ME2 were evaluated by gel filtration chromatography on a FPLC system using a Superdex 200 10/300 GL column (Amersham Biosciences). The column was equilibrated with 25 mm Tris-HCl, pH 7.5, or with 50 mm MES-NaOH, pH 6.5, and calibrated using molecular mass standards. The sample and the standards were applied separately in a final volume of 50 μL at a constant flow rate of 0.5 mL/min.
Preparation of Antibodies against AtNAD-ME1 and AtNAD-ME2
Polyclonal antibodies against recombinant AtNAD-ME1 and AtNAD-ME2 were obtained by immunization of rabbits with 200 μg of the purified proteins in four subcutaneous injections of 50 μg at 15-d intervals. The antibodies against the recombinant AtNAD-MEs were further purified from the crude antiserum (Plaxton, 1989), concentrated, and used (1:200 dilution) for western-blot analysis.
Protein Crude Extract Preparations
Different Arabidopsis organs (leaf, stem, flowers, and roots) of 6-week-old wild-type and T-DNA insertion lines were ground in N2, and the resulting powder was suspended in buffer B (50 mm MES-NaOH pH 6.5, 5 mm MnCl2, 1 mm EDTA, 10 mm 2-mercaptoethanol, 0.05% Triton X-100, 20% glycerol, and 1 mm phenylmethylsulfonyl fluoride). The homogenates were clarified by centrifugation. The supernatants were desalted using a Sephadex G-50 column equilibrated with buffer A and separated for activity measurements or subjected to electrophoresis.
Isolation of Mitochondria from Different Tissues
Mitochondria from leaves, stems, roots, and flowers were prepared by a modification of the method previously described by Keech et al. (2005). The tissue (approximately 2–5 g) was homogenized in a mortar with grinding buffer and the homogenate was filtered through two layers of muslin and centrifuged at 2,500g for 5 min. The supernatant was subjected to a second round of centrifugation at 12,000g for 15 min. The pellet containing the mitochondria was washed and resuspended with buffer C (50 mm HEPES, pH 6.5, 5 mm MnCl2, 1 mm EDTA, 10 mm 2-mercaptoethanol, 0.05% Triton X-100, 20% glycerol, and 1 mm phenylmethylsulfonyl fluoride). After three freeze cycles, the sample was centrifuged at 12,000g for 10 min and used for further analysis.
Enzyme Activity Measurements
NAD-ME activity in crude extracts (whole plant tissues or isolated mitochondria) was measured spectrophotometrically using a standard reaction mixture containing 50 mm MES-NaOH, pH 6.5, 4 mm NAD, 10 mm malate, 5 mm dithiothreitol; 10 mm MnCl2, and 10 units of MDH. There was a rapid but small increase of the A340 as the reaction catalyzed by the MDH reached the equilibrium. With the assay system specified above, the subsequent steady increase of A340 was attributable to the decarboxylation of l-malate by the NAD-ME (Chapman and Hatch, 1977).
In the case of purified recombinant AtNAD-ME, enzymatic activity was determined spectrophotometrically using a standard reaction mixture containing 50 mm HEPES, pH 6.4 or 6.6 (for AtNAD-ME1 and AtNAD-ME2, respectively), 10 mm MnCl2, 4 mm NAD, and 10 mm l-malate in a final volume of 0.5 mL. The reaction was started by the addition of l-malate. Initial velocity studies were performed by varying the concentration of one of the substrates around its Km value while keeping the other substrates concentrations at saturating levels. All kinetic parameters were calculated at least by triplicate determinations and adjusted to nonlineal regression using free concentrations of all substrates. OAA decarboxylation was monitored by measuring the OAA disappearance at 260 nm (ɛ260 nm = 850 m−1 cm−1) in an assay medium containing 50 mm MES-NaOH, pH 5.5, 1 mm OAA, and 10 mm MnCl2.
One unit is defined as the amount of enzyme that catalyzes the formation of 1 μmol of NADH min−1 under the specified conditions. Protein concentration was determined by the method of Sedmak and Grossberg (1977) using bovine serum albumin as standard.
PAGE and Western-Blot Analysis
Denaturing PAGE (SDS-PAGE) was performed in 10% (w/v) or 7.5–15% (w/v) linear gradient polyacrylamide gels according to Laemmli (1970). Proteins were visualized with Coomassie Blue or electroblotted onto a nitrocellulose membrane for immunoblotting. Antibodies against Amaranthus hypochondriacus α-NAD-ME (provided by Dr. J.O. Berry, SUNY Buffalo, Buffalo, NY), against phosphoenolcarboxylase (PEPc) from Amaranthus viridis (Colombo et al., 1998), or against AtNAD-ME1 or AtNAD-ME2 were used for detection. Bound antibodies were visualized by linking to alkaline phosphatase-conjugated goat anti-rabbit IgG according to the manufacturer's instructions (Sigma). Alkaline phosphatase activity was detected colorimetrically or by using a chemiluminescent kit (Immun-Star; Bio-Rad).
Native PAGE was performed using a 6% (w/v) acrylamide separating gel. Electrophoresis was run at 150 V at 10°C. Gels were assayed for NAD-ME activity by incubating the gel in a solution containing 50 mm HEPES, pH 6.5, 60 mm malate, 4 mm NAD, 10 mm MnCl2, 35 μg/mL nitroblue tetrazolium, and 10 μg/mL phenazine methosulfate at 30°C. Alternatively, native gels were electroblotted onto a nitrocellulose membrane and subjected to western-blot analysis.
Semiquantitative RT-PCR
To evaluate the expression of the AtNAD-ME genes in the T-DNA insertional mutants, total RNA from leaves were isolated from 100 mg tissue using the TRIzol reagent (Gibco-BRL). RNA was converted into first-strand cDNA using the SuperScriptII Reverse Transcriptase (Invitrogen). PCR reactions were conducted in a final volume of 10 μL using 1 μL of the transcribed product and Taq DNA polymerase (Qiagen). The pairs of primers used were NAD-ME1wtF and NAD-ME1wtR and NAD-ME2wtF and NAD-ME2wtR. Amplification conditions were as follows: 3 min denaturation at 94°C; 35 cycles at 94°C for 30 s, 53 to 55°C for 40 s and 72°C for 30 s, followed by 5 min at 72°C. As control, the actin2 gene was amplified by 28 cycles and the following primers were used: actin2-F (5′-TAACTCTCCCGCTATGTATGTCGC-3′) and actin2-R (5′-GTACGGTAACATTGTGCTCAGTGG-3′).
qRT-PCR
Relative gene expression was determined by performing qRT-PCR in an iCycler iQ detection system and the Optical System Software version 3.0a (Bio-Rad), using the intercalation dye SYBRGreen I (Invitrogen) as a fluorescent reporter, with 2.5 mm MgCl2, 0.5 μm of each primer, and 0.04 units/μL GoTaq (Promega). A 2-fold dilution of cDNA was used as template. PCR controls were performed in the absence of added reverse transcriptase to ensure that RNA samples were free of DNA contamination. Cycling parameters were as follows: initial denaturation at 94°C for 2 min; 40 cycles of 96°C for 10 s, and 56°C for 15 s; 72°C for 1 min; and 72°C for 10 min. Melting curves for each PCR reaction were determined by measuring the decrease of fluorescence with increasing temperature (from 65°C to 98°C). The specificity of the PCR reactions was confirmed by melting curve analysis using the software as well as by agarose gel electrophoresis of the products. Relative gene expression was calculated using the comparative 2−ΔΔCT method (Livak and Schmittgen, 2001) and polyubiquitin 10 (At4g05320) as reference gene. Each RNA sample was run in triplicate and repeated in at least two independent biological samples. The oligonucleotide primers pairs used for AtNAD-ME1 and AtNAD-ME2 were NAD1left (5′-GCACGAATGTTGGGAAATAC-3′) and NAD1right (5′-AAACCAAGAAGCACATCAGG-3′), and NAD2left (5′-GGCATCCTTTACCCTTCAAT-3′) and NAD2right (5′-ACCACATGTTGCGTGTAATG-3′). In the case of ubiquitin, the primers used were UBQleft (5′-AAGCAGCTTGAGGATGGAC-3′) and UBQright (5′-AGATAACAGGAACGGAAACATAGT-3′).
Construction of NAD-ME∷GUS Gene Fusions, Plant Transformation, and Histochemical Analysis of GUS Activity
For the generation of promoter-GUS constructs, fragments containing a 1.7-kb promoter region upstream the ATG start codon, the first exon and intron and part of the second exon of both AtNAD-ME genes were amplified by PCR from genomic DNA. The following primer pairs were used: pNAD-ME1GWF (5′-CACCTCGAGAGTTCTTAGCTAAACAATCT-3′) and pNAD-ME1GWR (5′-GGGACTGCGTTTACGATGACGGAGAGA-3′), and pNAD-ME2GWF (5′-CACCATGGGTTGGAGCAGATGGATT-3′) and pNAD-ME2GWR (5′-ATACGAGGCTTGCTTCCTCCTCGT-3′). The amplified products were sequenced and cloned into pGWB3, a gateway-compatible binary vector that carries the GUS gene and the kanamycin resistance gene (provided by T. Nakagawa, Shimane University, Izumo, Japan). The resulting constructs were introduced into Arabidopsis by A. tumefaciens transformation using the vacuum infiltration method. Transgenic lines were selected on Murashige and Skoog plates containing kanamycin (50 μg/mL). The histochemical localization of GUS activity was conducted as described by Maurino et al. (2006). Rosette leaves to be sectioned were stained as described above and fixed in 4% glutaraldehyde overnight at 4°C. Fixed tissues were dehydrated through an ethanol series and embedded in Paraplast (Roth). Embedded material was cut into 10-μm sections using a microtome, dewaxed with Rotihistol (Merk), mounted on glass slides with Entellan New (Merk), and observed under a microscope (Nikon Eclipse E800) equipped with a digital camera (ky-F1030; JVC).
Chlorophyll Fluorescence Parameters
Chlorophyll fluorescence measurements were performed with a PAM-2000 pulse amplitude modulated chlorophyll fluorometer (Walz). At the start of each measurement, a plant was dark adapted for 10 min. Basal fluorescence was measured with modulated weak red light and maximal fluorescence was induced with a saturating white light pulse (5,000 μmol m−2 s−1; duration 0.8 s).
Metabolic Analysis by GC-MS
Whole rosettes were harvested at the end of the light and dark periods and transferred into liquid nitrogen in less than 10 s. At least 8 rosettes were combined per sample. The leaves were homogenized using liquid nitrogen and stored at −80°C until use. The extraction and GC-MS analysis was conducted as described by Fahnenstich et al. (2007).
Statistical Analysis
Significance was determined according to the Student's t test using Excel software (Microsoft).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Cross section of rosette leaves of AtNAD-ME∷GUS plants. Scale bar = 50 μm.
Supplemental Table S1. Complete data set of metabolites of rosettes at the end of the light and dark period.
Supplementary Material
Acknowledgments
We thank Andreas Weber for helpful discussions and Ulrike Hebbeker and Claudia Nothelle for technical assistance.
This work was supported by the Deutsche Forschungsgemeinschaft (to V.G.M.), and by CONICET and Agencia Nacional de Promoción de Actividades Científicas y Tecnológicas (to M.F.D. and C.S.A.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Verónica G. Maurino (v.maurino@uni-koeln.de).
The online version of this article contains Web-only data.
References
- Agius SC, Rasmusson AG, Moller IM (2001) NAD(P) turnover in plant mitochondria. Aust J Plant Physiol 28 461–470 [Google Scholar]
- Allen BL, Harris BG (1981) Purification of malic enzyme from Ascaris suum using NAD+-agarose. Mol Biochem Parasitol 2 367–372 [DOI] [PubMed] [Google Scholar]
- Artus NN, Edwards GE (1985) Properties of leaf NAD-malic enzyme from the inducible crassulacean acid metabolism species Mesembryanthemum crystallinum. Plant Cell Physiol 26 341–350 [Google Scholar]
- Brailsford MA, Thompson AG, Kaderbhai N, Beechey RB (1986) Pyruvate metabolism in castor-bean mitochondria. Biochem J 239 355–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnell JN (1987) Photosynthesis in phosphoenolpyruvate carboxykinase-type C4 species: properties of NAD-malic enzyme from Urochloa panicoides. Aust J Plant Physiol 14 517–525 [Google Scholar]
- Casati P, Andreo CS, Edwards GE (1999) Characterization of NADP-malic enzyme from two species of Chenopodiaceae: Haloxylon persicum (C4) and Chenopodium album (C3). Phytochemistry 52 985–992 [Google Scholar]
- Chapman KSR, Hatch MD (1977) Regulation of mitochondrial NAD-malic enzyme involved in C4 pathways photosynthesis. Arch Biochem Biophys 184 298–306 [DOI] [PubMed] [Google Scholar]
- Caldes T, Fatania HR, Dalziel K (1978) Purification of malic enzyme from bovine heart mitochondria by affinity chromatography. Anal Biochem 100 299–303 [DOI] [PubMed] [Google Scholar]
- Colombo SL, Andreo CS, Chollet R (1998) The interaction of shikimic acid and protein phosphorylation with PEP carboxylase from the C4 dicot Amaranthus viridis. Phytochemistry 48 55–59 [DOI] [PubMed] [Google Scholar]
- Davisson VJ, Schulz AR (1985) The purification and steady-state kinetic behaviour of rabbit heart mitochondrial NAD(P)+ malic enzyme. Biochem J 225 335–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day AD, Hanson JB (1977) Pyruvate and malate transport and oxidation in corn mitochondrial. Plant Physiol 59 630–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drincovich M, Casati P, Andreo CS (2001) NADP-malic enzyme from plants: a ubiquitous enzyme involved in different metabolic pathways. FEBS Lett 490 1–6 [DOI] [PubMed] [Google Scholar]
- Eastmond PJ, Dennis DT, Rawsthorne S (1997) Evidence that a malate/inorganic phosphate exchange translocators imports carbon across the leucoplast envelope for fatty acid synthesis in developing castor seed endosperm. Plant Physiol 114 851–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahnenstich H, Saigo M, Niessen M, Zanor MI, Andreo CS, Fernie A, Drincovich MF, Flügge UI, Maurino VG (2007) Alteration of organic acid metabolism in Arabidopsis overexpressing the maize C4-NADP-malic enzyme causes accelerated senescence during extended darkness. Plant Physiol 145 640–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J (1989) PHYLIP: phylogeny inference package (version 3.2). Cladistics 5 164–166 [Google Scholar]
- Gerrard Wheeler MC, Tronconi MA, Drincovich MF, Andreo CS, Flügge UI, Maurino VG (2005) A comprehensive analysis of the NADP-malic enzyme gene family of Arabidopsis. Plant Physiol 139 39–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gout E, Bligny R, Pascal N, Douce R (1993) 13C Nuclear magnetic resonance studies of malate and citrate synthesis and compartmentation in higher plant cells. J Biol Chem 268 3986–3992 [PubMed] [Google Scholar]
- Grover SD, Canellas PF, Wedding RT (1981) Purification of NAD malic enzyme from potato and investigation of some physiological and kinetic properties. Arch Biochem Biophys 209 396–407 [DOI] [PubMed] [Google Scholar]
- Grover SD, Wedding RT (1982) Kinetic ramifications of the association-dissociation behaviour of NAD malic enzyme. Plant Physiol 70 1169–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haezlewood JL, Tonti-Filippini JS, Gout AM, Day DA, Whelan J, Millar AH (2004) Experimental analysis of the Arabidopsis mitochondrial proteome highlights signalling and regulatory components, provides assessment of targeting prediction programs, and indicates plant-specific mitochondrial proteins. Plant Cell 16 241–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch MD, Carnal NW (1992) The role of mitochondria in C4 photosynthesis. In H Lambers, LHW van der Plas, eds, Molecular, Biochemical and Physiological Aspects of Plant Respiration. Academic Publishing, The Hague, The Netherlands, pp 135–148
- Hatch MD, Kagawa T (1974) Activity, location and role of NAD malic enzyme in the leaves of C4 photosynthesis. Aust J Plant Physiol 1 357–369 [Google Scholar]
- Hibberd JM, Quick WP (2002) Characteristics of C4 photosynthesis in ítems and petiols of C3 flowering plants. Nature 415 451–454 [DOI] [PubMed] [Google Scholar]
- Hill SA, Bryce JH, Leaver CJ (1994) Pyruvate metabolism in mitochondria from cucumber cotyledons during early seedling development. J Exp Bot 45 1489–1491 [Google Scholar]
- Jenner HL, Winning BM, Millar H, Tomlinson KL, Leaver CJ, Hill SA (2001) NAD malic enzyme and the control of carbohydrate metabolism in potato tubers. Plant Physiol 126 1139–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keech O, Dizengremel P, Gardeström P (2005) Preparation of leaf mitochondria from Arabidopsis. Physiol Plant 124 403–409 [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using realtime quantitative PCR and the 2ΔΔCt method. Methods 25 402–408 [DOI] [PubMed] [Google Scholar]
- Long LL, Wang JL, Berry JO (1994) Cloning and analysis of the C4 NAD-dependent malic enzyme of amaranth mitochondria. J Biol Chem 269 2827–2833 [PubMed] [Google Scholar]
- Martinoia E, Rentsch D (1994) Malate compartmentation: responses to a complex metabolism. Annu Rev Plant Physiol Plant Mol Biol 45 447–467 [Google Scholar]
- Maurino VG, Grube E, Zielinski J, Schild A, Fischer K, Flügge UI (2006) Identification and expression analysis of twelve members of the Nucleobase-Ascorbate Transporter (NAT) gene family in Arabidopsis. Plant Cell Physiol 48 1381–1393 [DOI] [PubMed] [Google Scholar]
- Maurino VG, Saigo M, Andreo CS, Drincovich MF (2001) Non-photosynthetic malic enzyme from maize: a constitutively expressed enzyme that responds to plant defense inducers. Plant Mol Biol 45 409–420 [DOI] [PubMed] [Google Scholar]
- Moreadith RW, Lehninger AL (1984) Purification, kinetic behavior, and regulation of NAD(P)+ malic enzyme of tumor mitochondria. J Biol Chem 259 6222–6227 [PubMed] [Google Scholar]
- Murata T, Oshugi R, Matsuoka M, Nakamoto M (1989) Purification and characterization of NAD ME from leaves of Eleusine coracana and Panicum dichotomiflorum. Plant Physiol 89 316–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel WO, Sauer LA (1982) Mitochondrial malic enzymes. Purification and properties of the NAD(P)- dependent malic enzyme from canine small intestinal mucosa. J Biol Chem 257 12405–12411 [PubMed] [Google Scholar]
- Oshugi R, Murata T (1980) Leaf anatomy, post-illumination CO2 burst and NAD-malic enzyme activity in Panicum dichotomiflorum. Plant Cell Physiol 21 1329–1333 [Google Scholar]
- Palmer JM, Schwitzguel JP, Moller IM (1982) Regulation of malate oxidation in plant mitochondria. Responses to rotenone and exogenous NAD+. Biochem J 208 703–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascal N, Dumas R, Douce R (1990) Comparison of the kinetic behavior toward pyridine nucleotides of NAD+-linked dehydrogenases from plant mitochondria. Plant Physiol 94 189–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaxton WC (1989) Molecular and immunological characterization of plastid and cytosolic pyruvate kinase isozymes from castor-oil-plant endosperm and leaf. Eur J Biochem 181 443–451 [DOI] [PubMed] [Google Scholar]
- Price J, Laxmi A, St Martin SK, Jang JC (2004) Global transcription profiling reveals multiple sugar signal transduction mechanisms in Arabidopsis. Plant Cell 16 2128–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmusson AG, Escobar MA (2007) Light and diurnal regulation of plant respiratory gene expression. Physiol Plant 129 57–67 [Google Scholar]
- Sedmak J, Grossberg S (1977) A rapid, sensitive, and versatile assay for protein using Coomassie Brilliant Blue G-250. Anal Biochem 79 544–552 [DOI] [PubMed] [Google Scholar]
- Smith RG, Gauthier DA, Dennis DT, Turpin DH (1992) Malate- and pyruvate-dependent fatty acid synthesis in leucoplasts from developing castor endosperm. Plant Physiol 89 1233–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thum KE, Shin MJ, Palenchar PM, Kouranov A, Coruzzi GM (2004) Genome-wide investigation of light and carbon signalling interations in Arabidopsis. Genome Biol 5 R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedding RT, Black MK (1983) Physical and kinetic properties and regulation of the NAD malic enzyme purified from leaves of Crasula argentea. Plant Physiol 72 1021–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willeford KO, Wedding RT (1987) Evidence for a multiple subunit composition of plant NAD malic enzyme. J Biol Chem 262 8423–8429 [PubMed] [Google Scholar]
- Winning BM, Bourguignon J, Leaver CJ (1994) Plant mitochondrial NAD+-dependent malic enzyme. cDNA cloning, deduced primary structure of the 59- and 62-kDa subunit, import, gene complexity and expression analysis. J Biol Chem 269 4780–4786 [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol 136 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.