Abstract
The MADS-domain transcriptional regulator AGAMOUS-LIKE15 (AGL15) has been reported to enhance somatic embryo development when constitutively expressed. Here we report that loss-of-function mutants of AGL15, alone or when combined with a loss-of-function mutant of a closely related family member, AGL18, show decreased ability to produce somatic embryos. If constitutive expression of orthologs of AGL15 is able to enhance somatic embryo development in other species, thereby facilitating recovery of transgenic plants, then AGL15 may provide a valuable tool for crop improvement. To test this idea in soybean (Glycine max), a full-length cDNA encoding a putative ortholog of AGL15 was isolated from soybean somatic embryos. Subsequently, the corresponding genomic region of the gene was obtained. This gene, designated GmAGL15, encodes a protein with highest similarity to AGL15 from Arabidopsis (Arabidopsis thaliana) and Brassica napus that accumulates to its highest amount in embryos in these species. Like Arabidopsis and Brassica AGL15, GmAGL15 was preferentially expressed in developing embryos. When ectopically overexpressed the soybean protein was able to enhance somatic embryo development in soybean.
Somatic embryo systems provide a more easily accessible tissue for studies on embryogenesis than does zygotic embryo development that occurs embedded within maternal layers and involves small numbers of cells at the early stages. Somatic embryogenesis (SE) is also important as a means to regenerate plants after transformation or to propagate commercially valuable genotypes. However, early events in somatic embryo development remain a mystery and the ability of an explant to form somatic embryos either directly or indirectly depends on many factors including in some cases particular genotype (Vogel, 2005; Rose and Nolan, 2006; Namasivayam, 2007). In most systems, treatment of the explant with auxin, usually the synthetic auxin 2,4-dichlorophenoxyacetic acid (2,4-D), is necessary for induction of somatic embryo development (Jiménez, 2005; Vogel, 2005). Whether 2,4-D acts as a hormone or a stressor or both remains open to investigation. Often, but not always, the auxin level must be decreased to allow continued embryo development.
A number of genes have been identified in Arabidopsis (Arabidopsis thaliana) that when ectopically expressed promote somatic embryo development. These include the key embryo transcriptional regulators LEAFY COTYLEDON1 (LEC1) and LEC2 that when ectopically expressed will result in a fraction of seedlings that will then produce somatic embryo tissue without need for any hormone treatment. Likewise, ectopic expression of the B3 domain transcription factor FUS3 in the L1 layer via the ML1 promoter causes lateral organs to develop with embryonic features rather than as postgerminative vegetative tissue (Gazzarrini et al., 2004). LEC2 and FUS3 act at least in part by control of GA biosynthesis through regulation of AtGA3ox2, the product of which converts inactive GA to biologically active forms. Because LEC2 and FUS3 repress AtGA3ox2, less active GA is present, impacting on the GA-to-abscisic acid ratio that in turn determines whether an embryonic or adult leaf develops (Curaba et al., 2004; Gazzarrini et al., 2004; Lumba and McCourt, 2005). LEC1, LEC2, and FUS3 are essential for induction of somatic embryo development (Gaj et al., 2005).
The APETALA2 domain transcription factor BABYBOOM can also promote SE when ectopically expressed (Boutilier et al., 2002) as can the homeodomain transcription factor WUSCHEL (WUS; Zuo et al., 2002). In some situations the effect of ectopic expression of WUS depends on other proteins or hormones: With LEAFY, floral development was induced whereas embryogenesis required increased auxin (Gallois et al., 2004). SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 (SERK1) enhances somatic embryo development from the shoot apical region of seedlings that are allowed to complete germination in liquid culture media (Hecht et al., 2001). Likewise, the MADS-domain protein AGAMOUS-LIKE15 (AGL15) promotes somatic embryo development in this system and enhances production of secondary embryonic tissue from cultured zygotic embryos (Harding et al., 2003). AGL15 was initially identified using differential display of mRNA as an embryo expressed gene as well as during characterization of MADS-box genes in Arabidopsis (Heck et al., 1995; Rounsley et al., 1995). Although the gene is expressed and the protein accumulates to its highest level in developing embryos, AGL15 is expressed in subsets of cells, generally at lower levels after the completion of germination (Heck et al., 1995; Rounsley et al., 1995; Perry et al., 1996; Fernandez et al., 2000). MADS-domain proteins are a family of transcriptional regulatory factors found in eukaryotic organisms. In plants, MADS-domain proteins are central players in many developmental processes, including control of flowering time, homeotic regulation of floral organogenesis, fruit development, and seed pigmentation (Parenicová et al., 2003, and refs. therein). Interestingly and perhaps relevant for SE, AGL15 has been identified as a component of a SERK1 protein complex (Karlova et al., 2006), and both SERK1 and AGL15, as well as FUS3, are expressed in response to auxin treatment (Nolan et al., 2003; Gazzarrini et al., 2004; Zhu and Perry, 2005). Also intriguing are recent results that indicate that LEC2 may directly induce expression of AGL15 (Braybrook et al., 2006). Like LEC2 and FUS3, AGL15 impacts upon bioactive GA accumulation, but AGL15 mediates its effect at least in part by directly inducing expression of AtGA2ox6 (At1g02400) that encodes a GA 2-oxidase that catabolizes biologically active GA (Wang et al., 2004). Expression of this GA 2-oxidase affects somatic embryo development from the shoot apical meristem (SAM) of liquid culture grown seedlings in the presence of 2,4-D (Wang et al., 2004). We report here that loss-of-function alleles of AGL15, in some cases when combined with loss of function of AGL18, a closely related family member that is redundant to AGL15 in control of flowering time (Adamczyk et al., 2007), significantly impairs the ability to form somatic embryos in two systems in Arabidopsis.
Although immunoreactive proteins could be detected using AGL15-specific antibodies in a variety of embryos or embryonic tissues from angiosperms (Heck et al., 1995; Perry et al., 1996, 1999; S.E. Perry, unpublished data), little is known about the molecular nature of AGL15 orthologs in higher plants other than Arabidopsis and Brassica napus. We report on a putative ortholog of AGL15 from soybean (Glycine max) and document that, as found in Arabidopsis, ectopic expression of GmAGL15 can enhance SE.
RESULTS
Loss of Function of AGL15 Leads to Decreased Frequency of Somatic Embryo Development
We previously reported that when developing zygotic embryos are removed from the seed at the green bent cotyledon stage and placed into culture on media lacking any exogenous hormones, embryonic foci will appear on the embryos within 2 to 3 weeks (Harding et al., 2003). Embryos from plants carrying a 35S:AGL15 transgene produce secondary embryo tissue at higher frequency than wild type (approximately 40% of the cultured 35S:AGL15 embryos compared to approximately 18% for Wassilewskija [Ws] wild type), and when embryonic foci are subcultured, 35S:AGL15 tissue continues development in embryonic mode for extended periods of time (over 11 years to date), whereas wild-type tissue does not maintain embryonic development. Furthermore, a 35S:MIK transgene that is predicted to function as a dominant negative shows reduced ability to produce secondary embryonic tissue compared to wild type (Harding et al., 2003).
We have now examined effects of insertional mutations into AGL15 on production of embryonic tissue in this system. Insertional alleles agl15-3 and agl15-4 are in the Columbia (Col) ecotype and were compared to wild-type Col and 35S:AGL15 introduced into Col for production of secondary embryonic tissue from cultured zygotic embryos. We found that Col wild type was much more efficient at production of secondary embryo tissue than was Ws wild type. For wild-type Col, 47% of cultured embryos had secondary embryonic foci present at 3 weeks of culture (Fig. 1A) compared to 18.2% of Ws as reported by Harding et al. (2003). These numbers represent averages from experiments performed over a number of years and not at the same time (before 2003 for Ws and 2005–2006 for Col). To examine ecotype effect directly, one experiment compared the two ecotypes at the same time and we found that in this particular experiment 50% of wild-type Col produced secondary embryonic tissue compared to only 22% for Ws (data not shown). The fraction of Ws embryos developing secondary foci in this experiment agrees well with that reported in 2003.
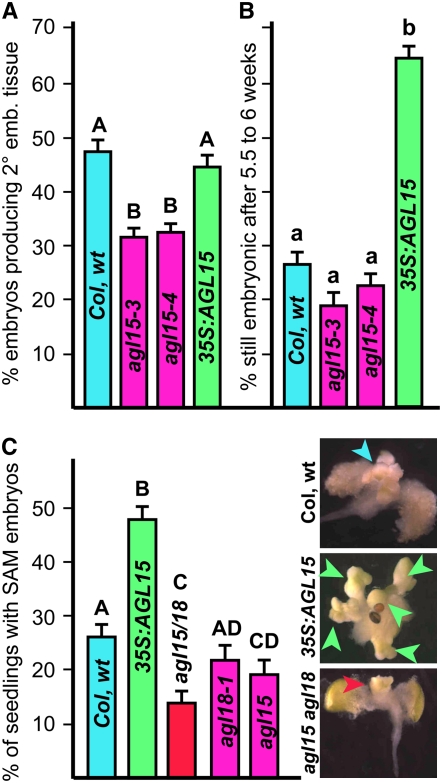
Figure 1.
Effect of AGL15/AGL18 accumulation on somatic embryo tissue production. A, Percentage of cultured zygotic embryos of the indicated genotypes that produce secondary embryonic tissue. Results shown are the means and se of the mean derived from 15 to 16 plates with 81 to 140 embryos per plate (with the exception of agl15-4 for which there were only six plates). Different letters indicate significance of P < 0.01. wt, Wild type. B, Percentage of embryonic foci of the indicated genotypes that were subcultured at approximately 3 weeks and continued to produce embryonic tissue approximately 2.5 to 3 weeks after subculture. Mean and se of the mean are shown. Different letters indicate significance at P < 0.01. C, Percentage of seedlings that showed somatic embryo development from the SAM when seeds of the indicated genotypes were allowed to compete germination in liquid media containing 2,4-D. Mean and se of the mean are shown. Different letters indicate significance at P < 0.05. Images are representative seedlings of the indicated genotypes that have SAM somatic embryo development.
Both insertional alleles into AGL15, agl15-3 and agl15-4, showed a decreased ability to produce secondary embryo tissue compared to Col wild type that was significant at P < 0.01 (Fig. 1A). Production of secondary embryos by the two insertional alleles was not different from one another. An insertional allele in the Ws ecotype, agl15-2, was also significantly impaired in production of secondary embryonic tissue (12% of cultured embryos produced secondary embryonic tissue compared to 22% for Ws wild type; this difference is significant at P < 0.05, data not shown). Unlike in the Ws ecotype where introduction of a 35S:AGL15 transgene significantly enhanced production of embryonic tissue (Harding et al., 2003), the 35S:AGL15 transgene in Col did not increase the percentage of cultured embryos with secondary embryonic foci (Fig. 1A). However, as previously reported in Ws, when embryonic foci are subcultured, the 35S:AGL15 transgene significantly enhances maintenance of embryonic development with 26% of wild type developing embryonic foci continuing development in embryonic mode at 3 weeks after initial subculture (5–6 weeks total in culture) compared to 64% of 35S:AGL15 in the Col background (Fig. 1B). The agl15 mutant tissue was not significantly different at P < 0.01 than wild type in this regard with 22% of the foci that initiated this development continuing production of embryonic tissue at 3 weeks after the first subculture. 35S:AGL15 in Col has now continuously produced embryonic tissue for over 1 year to date whereas the 35S:AGL15 in Ws that we reported on in 2003 has continued development in embryonic mode for over 11 years.
We also previously reported that a 35S:AGL15 transgene enhanced production of somatic embryo tissue from SAMs of seedlings in liquid culture (Harding et al., 2003). When seeds are allowed to complete germination in liquid media containing the synthetic auxin 2,4-D, the shoot apical region can produce somatic embryo tissue and mutants in which the SAM is enlarged are more efficient at producing this tissue (Mordhorst et al., 1998). Because a very young SAM is one place where nuclear accumulation of AGL15 can be detected, but this detectable accumulation is transient (Fernandez et al., 2000; Harding et al., 2003), we tested whether accumulation of AGL15, in terms of level or persistence, would impact on the ability to form somatic embryos. Previous work demonstrated that a 35S:AGL15 transgene in the Ws ecotype of Arabidopsis or in the amp1 mutant that has an enlarged meristem increased the fraction of seedlings with somatic embryo tissue compared to the wild-type/amp1 background alone (Harding et al., 2003). However, because neither Ws nor amp1 efficiently produced somatic embryos in our hands, we were unable to test loss of function.
Because many of the insertional mutants are in the Col ecotype, including two of the agl15 alleles, we tested the effect of accumulation of AGL15 on SAM SE in the Col ecotype. As shown in Figure 1C, for experiments performed with several seed lots and over a 2-year period, an average of 26% of Col wild-type seedlings had somatic embryo tissue at the apex in this system (7,427 seedlings total, n = 29). While over all experiments, agl15-3 and agl15-4 (5,114 seedlings total scored, n = 19) did show a significant reduction in production of SAM somatic embryos at P < 0.05 with 19% of seedlings showing this development, whether the difference was significant or not in individual experiments varied and the difference was not significant at P < 0.01. Because AGL18 is the closest family member to AGL15 and is expressed in overlapping developmental context (Lehti-Shiu et al., 2005), we tested an agl15 agl18 double mutant in this system. While agl18-1 did not significantly decrease production of SAM somatic embryos (2,162 seedlings scored, n = 13), the double mutant showed significant reduction compared to wild type (14% of agl15-3 agl18-1 and agl15-4 agl18-1 seedlings had SAM somatic embryo tissue, 4,916 seedlings, n = 27; this is significant at P < 0.01; Fig. 1C). Conversely, prolonged or increased accumulation of AGL15 as provided by the 35S:AGL15 transgene increased the fraction of seedlings with SAM somatic embryo development (48% of 2,666 seedlings scored, n = 19; Fig. 1C). Not only did an increased fraction of seedlings have somatic embryo tissue, but the extent of somatic embryo tissue development was also on average greater than found for Col wild type (inset images in Fig. 1C).
Isolation of an Ortholog of AGL15 from Soybean
Would ectopic expression of AGL15 enhance SE in other species, perhaps providing a tool to facilitate recovery of transgenic plants via SE? To address this question, we cloned a soybean MADS-box gene from somatic embryo tissue. Isolation of sequences encoding GmAGL15 was initiated by searching the GenBank EST database for possible candidates. One entry (accession no. AW756465) was found annotated as “similar to…AGL15.” BLASTX program (http://www.ncbi.nlm.nih.gov/blast) confirmed this EST represented the N-terminal 37 amino acid residues of the conserved MADS domain. To obtain the full-length cDNA, RNA was isolated from a soybean somatic embryo culture (Reddy et al., 2001). Oligonucleotide primers were designed based on the EST sequence, and 3′-RACE PCR was performed as described by Ausubel et al. (1998). After sequencing this partial cDNA, additional primers were designed for 5′-RACE PCR to recover the full-length cDNA. This gene, designated GmAGL15, had highest sequence similarity to the previously published AGL15s.
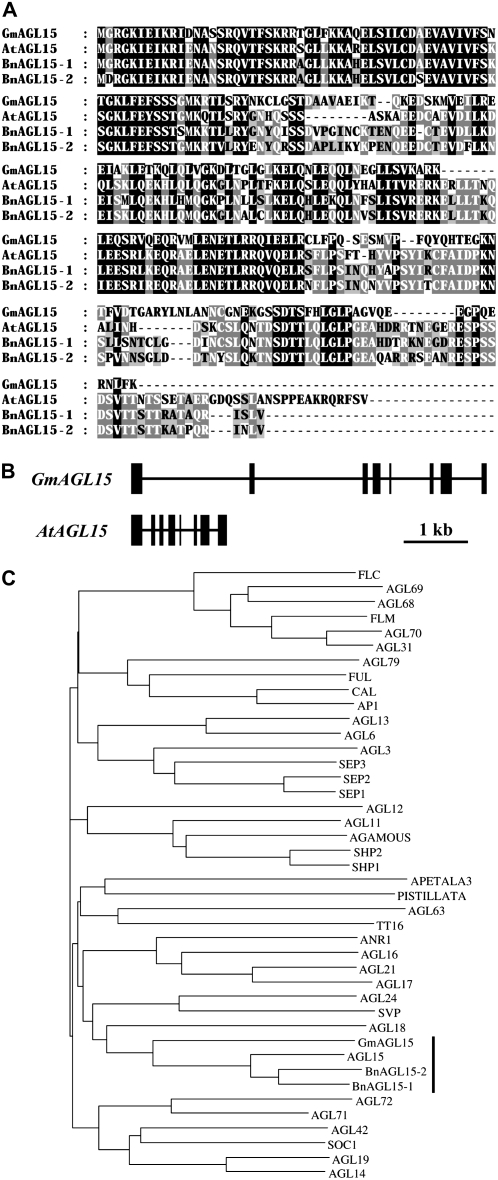
The longest cDNA clone (GenBank accession no. AY370659) consisted of an approximately 270-bp 5′-untranslated region (UTR), an approximately 260-bp 3′-UTR plus polyA tail, and an open reading frame of 708 bp, which encodes a protein of 235 amino acid residues. A BLASTP search was performed using the protein sequence. The highest scoring matches were to Arabidopsis AGL15 (AtAGL15), Brassica type I and type II AGL15 (BnAGL15-1 and -2; accession nos. NP_196883, Q39295, and AAB03807, respectively), and to proteins from cotton (Gossypium hirsutum), petunia (Petunia hybrida), and grape (Vitis vinifera) that when used to search the Arabidopsis AGI protein database had highest similarity to AGL15. Pairwise comparison of GmAGL15 protein with the other three AGL15s revealed an identity of approximately 50%. Multiple sequence alignment showed a moderate conservation among these proteins (Fig. 2A). This was not unexpected, because even between Arabidopsis and B. napus, two closely related species, considerable divergence in AGL15 exists. Nevertheless, the soybean sequence displayed overall homology to the AGL15 proteins, including divergent domains outside the conserved MADS domain. In addition, the soybean protein contained several signature sequences that are rarely found in MADS-domain proteins other than AGL15, such as the C-terminal LENETLRRQI/VE/QELR and LGLP motifs. Therefore, it is likely that GmAGL15 is the soybean ortholog of AGL15.
Figure 2.
A, GeneDoc (www.pac.edu/biomed/genedoc) sequence alignment between GmAGL15, AtAGL15, BnAGL15-1, and BnAGL15-2. The shade levels represent conservation degrees. B, Schematic representation of the gene structures of GmAGL15 and AtAGL15. The numbers and positions of the introns/exons were conserved between these two species. Boxes, exons; lines, introns. C, A phylogenetic tree generated from GmAGL15, BnAGL15s, and all 39 MIKC-type Arabidopsis MADS-domain proteins. Neighbor-joining method with a bootstrap number of 1,000 was used (ClustalX 1.81, http://www-igbmc.u-strasbg.fr/BioInfo/ClustalX). AGL15 group is indicated.
The 5.8-kb genomic region of GmAGL15 (accession no. AY370660) was amplified from ‘Jack’ soybean genomic DNA, using primers corresponding to the UTRs of the cDNA. An alignment of the genomic and cDNA sequences revealed that GmAGL15 contained eight exons and seven introns (Fig. 2B). The introns were longer than found in Arabidopsis. Nevertheless, the exon-intron boundary locations appeared to be identical between the two species, as often observed among evolutionarily conserved orthologs. This further suggested GmAGL15 was the soybean counterpart of AGL15.
A phylogenetic tree was constructed using protein sequences of GmAGL15, BnAGL15s, and all 39 Arabidopsis MIKC-type MADS-domain proteins (Parenicová et al., 2003). GmAGL15 was grouped more closely to the AGL15s (Fig. 2C).
Expression of GmAGL15 in Soybean
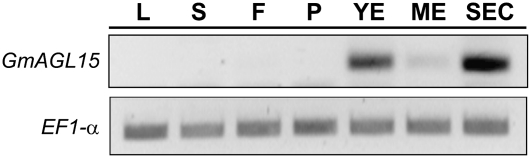
To investigate the expression pattern of GmAGL15, semiquantitative reverse transcription (RT)-PCR was performed on RNAs isolated from various tissues. As shown in Figure 3, GmAGL15 transcript was not detected in the vegetative tissues (leaves, stems), or in the open flowers. GmAGL15 mRNA was not detectable at very early stages of seed pod development, but was present in young developing embryos, and the level declined after maturation. This pattern was consistent with that previously reported for AGL15 from Arabidopsis and Brassica (Heck et al., 1995; Rounsley et al., 1995). Notably, the highest level of GmAGL15 mRNA accumulation was detected in the somatic embryo culture. AGL15-specific antiserum (Perry et al., 1999) detected immunoreactive protein in nuclear extracts prepared from soybean somatic embryos (data not shown). Additionally, reaction of the antiserum against the product of the cloned GmAGL15 was tested by expressing the soybean gene in Escherichia coli. The AGL15-specific antiserum recognized the E. coli produced protein (Fig. 5B).
Figure 3.
Expression pattern of GmAGL15. Semiquantitative RT-PCR was performed on RNA derived from various tissues of ‘Jack’ soybean. The coding region of EF1-α was amplified for normalization. L, Young leaves; S, stems; F, open flowers; P, seed pods containing very young embryos; YE, young embryos (average length 2 mm); ME, mature green embryos; SEC, somatic embryo culture. The color of the image is inverted for clarity.
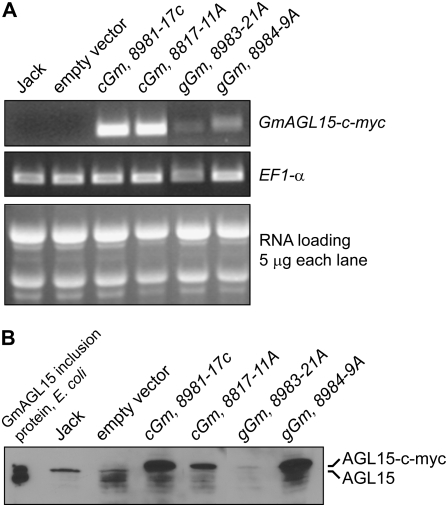
Figure 5.
Evaluation of expression of 35S:AGL15 in soybean. A, RT-PCR was performed using a GmAGL15 forward primer and a c-myc reverse primer on RNA extracted from young leaves. RT-PCR with EF1-α was performed as a normalization control. Total RNA was also analyzed on an agarose gel to evaluate integrity. B, Protein gel-blot analysis to evaluate accumulation of AGL15 in transgenic soybean. Protein extracts were prepared from flowers of the indicated genotypes and proteins were separated on a polyacrylamide gel, blotted to membrane, and probed using anti-AGL15 serum. Inclusion protein containing GmAGL15 (without a c-myc tag) was obtained by expression in E. coli and used as a positive control. All lanes within a section are from the same gel or blot.
Ectopic Expression of GmAGL15 Enhances SE in Soybean
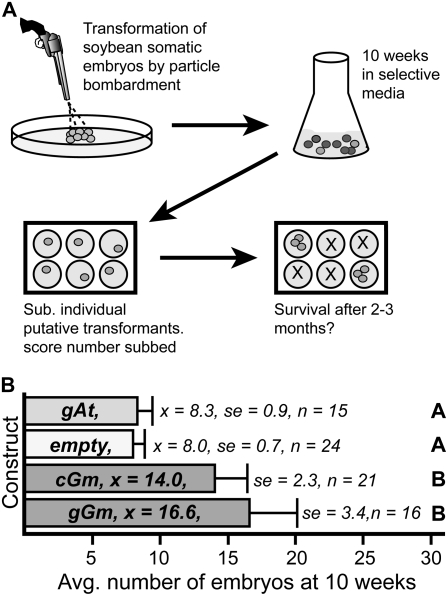
To test whether ectopic expression of AGL15 via the 35S promoter would enhance SE in soybean, biolistic transformation of ‘Jack’ soybean somatic embryo tissue was performed with 35S:gGmAGL15 (includes introns), 35S:cGmAGL15 (lacks introns), 35S:AtAGL15 (Arabidopsis AGL15 containing the first three introns), or empty vector control. After recovery for 7 d, the bombarded tissue was cultured for 10 weeks in selective media that contained hygromycin, changing media at 7- to 8-d intervals. After 10 weeks, putative transformants were identified by their green color and characteristic shape and subcultured individually in six-well plates (schematic showing steps involved is shown in Fig. 4A). The number of putative transformants per bombardment was scored and is shown in Figure 4B. Transformation with either the 35S:gGmAGL15 and 35S:cGmAGL15 transgenes showed enhanced recovery of putative transformants compared to the empty vector control (16.6 and 14.0 putative transformants moved to individual subculture for the genomic and for the cDNA transformations, respectively, compared to 8.0 putative transformants for the empty vector). Transformation with a 35S:AtAGL15 transgene did not significantly increase the number of potential transformants recovered at 10 weeks compared to the empty vector control (Fig. 4B).
Figure 4.
Effects of overexpression of AGL15 on SE in soybean. A, Schematic of the transformation and scoring scheme for soybean. Please see “Materials and Methods” for further details. B, Transformation of soybean with soybean genomic (gGm) or cDNA (cGm) versions of GmAGL15 driven by the 35S cauliflower mosaic virus promoter produced more potential transformants that were individually subcultured than did the empty vector control. However, expression of Arabidopsis AGL15 did not result in an increase in potential transformants. Different letters indicate significant difference at P < 0.05.
Constitutive expression of GmAGL15 also impacted on survival in culture. After individual transformants were subcultured in six-well plates, they were allowed to continue development for an additional 2.5 months under selection. Some putative transformants will proliferate, whereas others die during this period. Individual putative transformants were scored for survival after approximately 2.5 months. For the empty vector control, 21% survived, compared to 33% for 35S:cGmAGL15, 26% for 35S:gGmAGL15, and 25% for 35S:AtAGL15. Promotion of SE and survival in culture thus resulted in a net of 1.7 transformants per bombardment progressing to maturation media for the empty vector control compared to 4.6 for 35S:cGmAGL15, 4.3 for 35S:gGmAGL15, and 2.1 for 35S:AtAGL15.
Regeneration of Transgenic Soybean Plants
35S:AGL15 in Arabidopsis cultured embryos leads to long-term (over 11 years) maintenance of development as embryo tissue (Harding et al., 2003; data not shown). Although no attempt was made in Arabidopsis to switch the tissue from embryonic to postgerminative development, a concern for use of orthologs of AGL15 in other species such as soybean may be that overexpression of AGL15 via a constitutive promoter would not allow for maturation and plantlet development of the transgenic somatic embryos. However, Arabidopsis seed containing 35S:AGL15 undergoes maturation, desiccation, and can complete germination to give a seedling.
Although 35S:GmAGL15 transgenes enhanced recovery of putative transformants, perhaps by promoting somatic embryo production, the somatic embryos obtained in soybean underwent maturation and differentiation, and produced plantlets that were transplanted to soil. The plants continued development and appeared relatively normal considering the culture process. Of 17 separate events that were recovered as plantlets and subsequently tested, 88% were verified as containing the transgene by PCR. We tested a number of plants for expression and verified transcript accumulation from the transgenes by RT-PCR (using a GmAGL15 forward primer and an oligonucleotide primer to the C-terminal c-myc tag as a reverse primer). As shown in Figure 5A, transcript from the transgene was detected in young leaf tissue of potential transgenics but not in untransformed ‘Jack’ soybean or in the empty vector transgenics.
To determine if transgenic plants accumulated AGL15, nuclei were isolated from open flowers, extract prepared and analyzed by SDS-PAGE, transfer to membrane and probing with anti-AGL15 serum. As shown in Figure 5B, the different transgenics exhibited a range of AGL15 accumulation from no detectable accumulation from the transgene (data not shown), low levels of accumulation (gGm, 8983-21A), to high levels of accumulation (cGm, 8981-17C; and gGm, 8984-9A). The slightly lower band in ‘Jack’ and in the empty vector control likely represents endogenous AGL15 that is expressed, at least in Arabidopsis, at low levels in flowers (Fernandez et al., 2000).
DISCUSSION
Previous work demonstrated that ectopic expression of AGL15 could enhance production of somatic embryos from cultured zygotic embryos and from the SAMs of seeds that complete germination in liquid media that contains 2,4-D (Harding et al., 2003). However, at the time, no loss of function of AGL15 was available. In lieu of a knockout allele of AGL15, a form of AGL15 missing the C-terminal domain (35S:MIK) and predicted to function as a dominant negative was tested. Although a decrease in production of secondary embryos from cultured zygotic embryos was observed, it was not possible to judge the effect on the SAM because wild-type seedlings of the Arabidopsis ecotype Ws did not produce somatic embryos efficiently in this system (Harding et al., 2003).
The Arabidopsis ecotype Col was more efficient at somatic embryo production than was Ws in both the cultured embryo and the SAM somatic embryo systems. Other work has demonstrated differences between ecotypes in the ability to produce somatic embryos. Specifically, Luo and Koop (1997) found that while Col was moderately efficient at producing somatic embryos from zygotic embryos cultured in liquid media with 2,4-D, Ws was the one ecotype tested that did not form somatic embryos. Ecotype effects on stress-induced SE have also been reported (Ikeda-Iwai et al., 2003). The increased competence for SE in wild-type Col, combined with the fact that agl15 and agl18 insertional alleles in Col were available (Alonso et al., 2003), allowed us to test the loss of function of these proteins on somatic embryo production.
Two different loss-of-function alleles of AGL15 in the Col ecotype significantly decreased secondary embryo production when zygotic embryos were cultured on media lacking exogenous hormones (Fig. 1A). A loss-of-function allele of AGL15 (agl15-2) in the Ws ecotype also significantly decreased the fraction of cultured zygotic embryos producing secondary embryo tissue in this system (data not shown). Although the double mutant of agl15 with the closest related family member agl18 (Col) was not tested in this system, the agl18 single mutant alone did not show a decrease in fraction of cultured zygotic embryos that produce secondary embryo tissue compared to wild type (data not shown). However, expression of a transgene that produces a form of AGL15 that is predicted to function as a dominant negative (35S:MIK), and could interfere with not only AGL15 but other MADS or proteins that interact with MADS factors, had a more severe effect on production of secondary embryos than did the agl15-2 mutant in the Ws background (2.4% compared to 12%; Harding et al., 2003; data not shown). This may indicate redundancy of AGL15 with other factors, perhaps including AGL18. Recent work has indicated that AGL15 can bind not only to its own regulatory regions but also to regulatory regions of AGL18, and can repress AGL18 expression (Hill et al., 2008). Therefore, although the single agl18 mutant did not decrease somatic embryo formation, it may still have an effect when tested as a higher order mutant with agl15. The 35S:AGL15 transgene did not cause an increase in the fraction of cultured embryos that initially produce secondary embryo tissue in the Col ecotype compared to wild-type Col, as previously reported for Ws (Fig. 1A; Harding et al., 2003). However, the 35S:AGL15 transgene still promoted continued development as embryo tissue for extended time periods in Col as previously found for Ws (Fig. 1B; Harding et al., 2003). It would be interesting to determine whether there are differences in accumulation of AGL15 in later embryo development in these two ecotypes to perhaps explain the differences observed during the early culture period.
There exist systems by which Arabidopsis cultured embryos are used to generate somatic embryo tissue in the presence of 2,4-D and sometimes other hormones (Wu et al., 1992; Pillon et al., 1996; Luo and Koop, 1997; Mordhorst et al., 1998; Ikeda-Iwai et al., 2002). However, 35S:AGL15 embryos can produce secondary embryo tissue in the absence of any exogenous hormone and then maintain development as embryo tissue for very extended time periods (Harding et al., 2003; this study). Interestingly, expression of AGL15 is induced by auxin, both 2,4-D and indole-3-acetic acid (Zhu and Perry, 2005). AGL18 is also induced somewhat by auxin treatment (S.E. Perry, unpublished data).
In the SAM SE system, where seeds are allowed to complete germination in liquid media with 2,4-D, the single agl15 mutant alleles did not show a completely consistent significant reduction in production of SAM somatic embryos for every experiment. However, in some experiments there was such a reduction and when the data collected over more than a 2-year period is considered as a whole, the single agl15 mutants are significantly different from wild type in production of SAM somatic embryos. The single agl18 mutant showed some reduction in frequency of SAM SE compared to wild type but this was not significant. Data as a whole shows significant reduction of SAM SE for the agl15 agl18 double mutant compared to wild type at P < 0.01, and in almost all cases individual experiments showed a significant reduction. It is interesting to note that often the tips of the cotyledons for the double mutant remained green and uncallused (Fig. 1C). This was also observed in the Ws ecotype containing the dominant negative construct 35S:MIK (Harding et al., 2003). This could reflect an altered response to auxin. Because the SAM SE system, unlike the cultured embryo system, requires auxin treatment for embryo formation, other factors responsive to auxin must be involved.
To expand our work to plants that are agriculturally relevant, we cloned a gene and genomic region encoding a potential AGL15 ortholog from soybean. Sequence similarity, gene structure, and expression pattern support GmAGL15 as the likely soybean ortholog to previously reported AGL15s from Arabidopsis and B. napus. In addition, ectopic expression of either the cDNA or the intron-containing version of GmAGL15 via the 35S promoter enhanced recovery of transformants via SE. In the Arabidopsis Ws ecotype, ectopic expression of AGL15 promoted the initial production of secondary foci and then enhanced continued development in this mode. Similarly, in soybean, more potential transformants were recovered at 10 weeks, and then, at least for the cDNA version of the transgene, there was a significant enhancement in survival after subculturing the individual putative transformants. However, it was possible to switch the tissue out of embryogenic mode and recover plantlets. Although we have not attempted purposefully to switch the Arabidopsis embryogenic culture tissue that has been developing in embryonic mode for over 11 years, into a postembryonic state, it is interesting that we have observed increased leaf-like development when plates are not subcultured for lengthy times and begin to dry (S.E. Perry, unpublished data). The recovered transgenic soybean plants constitutively expressing AGL15 appear relatively normal, with some plants showing perhaps increased branching or darker green color combined with reduced seed set and some meristem abnormalities. All of these potential phenotypes would be consistent with phenotypes observed with ectopic expression of AGL15 in Arabidopsis (Fernandez et al., 2000; Wang et al., 2004). However, because the plants thus far are from tissue culture, it is premature to make any conclusions about effects of the transgene on plant morphology or development at this time. Transgenic plants were found to accumulate AGL15 and the slightly higher mass compared to endogenous GmAGL15 is due to a c-myc tag on the carboxyl-terminal end of the transgenic form (Fig. 5B). It is interesting to note that when AGL15-c-myc accumulates, there may be a reduction in the endogenous AGL15. This could indicate that AGL15 may negatively autoregulate its own expression in soybean as has been reported in Arabidopsis (Zhu and Perry, 2005).
GmAGL15, like Arabidopsis AGL15, appears to promote development as embryonic tissue and may be a valuable tool for recovery of transgenics in this and other species recalcitrant to regeneration by SE. However, ectopic expression of AtAGL15 did not enhance embryogenesis in soybean. The 35S:GmAGL15 constructs were also stably transformed into Arabidopsis and the transgenic plants recovered did not show phenotypes typical of overexpression of AtAGL15 in this plant (i.e. lack of abscission and senescence of floral organs after fertilization; Fernandez et al., 2000). Although it is possible that there may be differences in protein accumulation in the heterologous system, codon usage in Arabidopsis and soybean is similar (Wang and Roossinck, 2006). Recently we found that a LxLxL motif that is conserved between Arabidopsis and B. napus AGL15s and that is also found in a number of potential orthologs of AGL15 identified in other species, acts to repress gene expression and the mechanism by which this occurs may be via recruitment of SAP18 (for SIN3-associated polypeptide of 18 kD) and subsequently histone deacetylase components (Hill et al., 2008). Intriguingly, the first Leu in this motif is replaced by a Phe in GmAGL15. Perhaps differences in protein-protein interactions may be responsible for the lack of effect of expression of AGL15 in a heterologous system. An EST encoding a probable SAP18 ortholog is in the EST database (EV274062) that is 76% identical and 87% similar to AtSAP18 (At2g45640). It will be intriguing to test interactions between AGL15 and SAP18 between and within species.
The experiments performed in this report utilized the cultivar ‘Jack’ that has been reported as highly embryogenic as well as uniformly responsive to induction of SE across diverse locations (Meurer et al., 2001). It would be interesting to determine if transformation of a less consistently embryogenic cultivar such as ‘Stonewall’ or ‘Defiance’ would enhance subsequent SE in these cultivars, thereby facilitating transformation with other constructs. If so, this could address a priority outlined in the Soybean Genomic Research Strategic Plan for 2008 to 2012 to expand the number of genotypes that can undergo SE (Specht et al., 2007).
MATERIALS AND METHODS
Plant Material
Arabidopsis (Arabidopsis thaliana) ecotypes Ws and Col wild type, loss-of-function mutants with insertional alleles (agl15-2 in Ws; agl15-3, agl15-4, and agl18-1 in Col), and transgenic plants were sown on germination medium (GM; Murashige and Skoog, 1962) with 50 μg mL−1 kanamycin for transgenic seed, chilled for 2 to 3 d at 4°C, and transferred to a growth room with a 23°C to 24°C, 16-h-light/8-h-dark regime. The loss-of-function insertional mutants are described by Lehti-Shiu et al. (2005). After 7 to 10 d, seedlings were transplanted to potting mix (ProMix BX; Premier Brands, Inc.) and grown under a 16-h-light (20°C)/8-h-dark (18°C) cycle in a growth chamber. To stage siliques, flowers were tagged on the day that they opened.
Soybean (Glycine max Merr. ‘Jack’) plants were grown in greenhouse at a temperature of approximately 27°C and 15-h-light regime as described by Parrott et al. (1989) in a steam-sterilized mixture of 2:2:1 of soil:Promix:sand and fed weekly with Peters 20:20:20 (The Scotts Company).
Arabidopsis Somatic Embryo Systems
The embryo culture system has been described previously (Harding et al., 2003). Briefly, developing embryos were removed at 10 to 12 d after flowering (green bent cotyledon stage) and placed into culture on GM. After about 3 weeks, embryos were scored for presence of secondary embryo tissue that was then subcultured on GM and scored again after an additional 2.5 to 3 weeks for continued development in embryo mode.
The SAM somatic embryo system is as described by Mordhorst et al. (1998). Briefly, seed was surface sterilized as done by Harding et al. (2003), chilled for 2 d at 4°C, and put into liquid culture media containing 2,4-D (Mordhorst et al., 1998). Cultures were incubated at 23°C to 24°C on a rotary shaker under a 23-h-light/1-h-dark regime. Cultures were scored at 21 d.
Plasmid Constructs
Two different versions of the cDNA and of the genomic GmAGL15 constructs were generated. One version was untagged, while the other contained a c-myc tag at the carboxyl-terminal end. For the untagged versions, the coding region lacking (cDNA) or containing introns (gDNA) were inserted downstream of the cauliflower mosaic virus 35S promoter in the pBIMC vector (gift from Dr. D. Falcone, University of Massachusetts Lowell). A nopaline synthase (Nos) terminator was present after the stop codon. The 35S-AGL15-Nos expression cassettes were subsequently cloned into pCambia 1301 that also constitutively expresses a GUS gene and confers hygromycin B resistance.
To add a c-myc tag onto the C-terminal end of cGm and gGmAGL15, the respective forms of AGL15 were PCR amplified from the original pCambia 1301-based plasmid constructs, using oligonucleotides that introduced restriction sites SpeI at the 5′ end and SacI at the 3′ end. The reverse 3′-oligonucleotide corresponded to the sequence before the stop codon and engineered in sequences encoding a c-myc epitope. The sequence of the forward and reverse oligonucleotides was 5′-GACTAGTCCATGGGTCGAGGGAAAATCGAG-3′ and 5′-GAGCTCTCACAGGTCCTCCTCTGAGATCAGCTTCTGCTCCTCTTTGAAAAGGTTTCTTTCTTGGGGGCC-3′, respectively. The SpeI and NcoI (forward) and SacI (reverse) sites are underlined, the sequence encoding the c-myc epitope is in italics, and the engineered stop codon is in bold. For amplification, high-fidelity Taq polymerase (Ex Taq; Takara Bio Inc.) was used following the manufacturer's instructions. The PCR-amplified fragments were gel purified and cloned into the pGEM-T Easy vector system (Promega Corporation) for sequence verification. For cloning into a binary vector system, the SpeI/SacI fragment was recovered from pGEM-T and cloned into pBIMC such that the AGL15-c-myc sequences were flanked by the 35S promoter and Nos terminator. The 35S:cGmAGL15-cmyc:NOS cassette was then restricted with EcoRI/HindIII and the purified fragment was ligated into the pC1301 vector resulting in vector pcGmAGL15-cmyc. To transfer gGmAGL15-cmyc from pBIMC, the plasmid was restricted with SalI and SacI and the purified insert was ligated with vector obtained by digestion of pcGmAGL15-cmyc with the same restriction enzymes. The finished vector 35S:gGmAGL15-cmyc:NOS cassette, i.e. pgGmAGL15-cmyc, was confirmed by sequencing and used for biolistic transformation of soybean.
Transformation of Soybean
Somatic embryos were generated from the cultivar ‘Jack’ (soybean Merr.). Immature pods (approximately 15–20 d after anthesis) were harvested, rinsed with distilled water containing a few drops of liquinox, and surface sterilized by immersing for 30 s in 70% isopropanol, 12 min in 25% Clorox solution with a few drops of liquinox, and washing twice for 5 min with sterile distilled water. Immature zygotic embryos of 4.0 ± 1 mm were aseptically excised and the embryonic axis removed by cutting through both cotyledons. About 25 individual cotyledons were cultured abaxial side down toward the media, on D40 induction media plates under diffused light (Murashige and Skoog salt containing B5 vitamins, 3% Suc, 40 mg L−1 2,4-D, 0.2% phytagel [Sigma], pH 7.0). After 6 to 8 weeks of incubation, the embryos induced on the cotyledons from D40 medium were transferred to the D20 media (Murashige and Skoog salt containing B5 vitamins, 3% Suc, 20 mg L−1 2,4-D, 0.2% phytagel, pH 5.8). Good quality proliferating embryos clusters were subcultured after every 4 weeks on fresh D20 media plates for no more than four cycles.
Four to five days before transformation, equal amounts of somatic embryo tissue was placed at the center of a plate containing D20 media. Gold microcarriers (5 mg of 0.6 μm; Bio-Rad) were washed with ethanol (100%, 10 s), incubated on ice for 30 s, centrifuged at approximately 2,300g, and the liquid removed. Ethanol (35 μL of 100%) was added, microcarriers gently vortexed for 1 min, and pelleted (approximately 2,000g, 10 s). One milliliter of water was added and carriers pelleted at 400g, 5 min. The supernatant was removed, and DNA in the order as follows (3 μg at 0.5 μg μL−1, 220 μL sterile water, 250 μL of 2.5 m CaCl2, and 100 μL of 0.1 m spermidine) were added while mixing gently (vortex speed 3), incubated on ice for 2 min, and then mixed gently by vortex, 10 min at 0°C to 4°C. Carriers were pelleted by centrifugation 5 min, approximately 100g, liquid removed, and carriers washed by adding ethanol (600 μL, 100%) and gentle vortexing, 1 min. Wash was removed by pelleting carriers as before, followed by resuspension in 36 μL ethanol and incubation on ice for 1 h. Microcarriers were resuspended by pipetting and 10 μL used per macrocarrier (Bio-Rad, catalog no. 1652335), for bombardment using a DuPont BioListic particle delivery system (model PSD-1000; Bio-Rad), and 1,100-psi rupture discs (Inbio Gold). Bombardment of embryos with each construct was performed in triplicate for each experiment.
After bombardment, plates were sealed with parafilm and tissue let recover by inverting plates and incubating at room temperature under diffused light for 7 d. Tissue was then transferred to 25 mL of FN-Lite (Finner and Nagasawa, 1988) media containing 20 mg L−1 hygromycin B (Invitrogen) in a 125-mL flask and incubated at room temperature, 200 rpm, and under diffused light. The media was changed periodically after every week for 8 to 10 weeks. After 10 weeks of culturing the potential transformants per bombardment were scored by green color and morphology and were transferred to individual wells of six-well tissue culture plates (MidSci) containing FN-Lite media supplemented with hygromycin for an additional month. Surviving cultures were transferred to 125-mL flasks containing 25 mL of FN-Lite media without hygromycin and allowed to continue development for additional 2 months, replacing the media at 1 month and separating the larger embryonic masses into several pieces. The numbers of surviving green calli for each construct were also counted after 2 months.
Transgenic plants were recovered by disruption of tissue and transfer to soybean histodifferentiation and maturation media as described by Schmidt et al. (1995), for 4 to 5 weeks with constant agitation (200 rpm), followed by desiccation in petri plates sealed with parafilm for 7 to10 d and then transfer to MSO lite media (Murashige and Skoog salts, B5 vitamins, 1.5% Suc, 0.2% phytagel, pH 5.8) in 100- × 20-mm petri dishes at room temperature to allow shoot and root development. Healthy plants (branched roots and elongating shoots) were transferred to 0.5× B5 media in phyta-trays (Sigma). These plantlets were then allowed to grow for 2 to 3 weeks and subsequently transplanted to soil.
RT-PCR
For detection of accumulation of c-myc-tagged AGL15 transcript in soybean transgenic plants, RNA was extracted from leaf tissue using Trizol (Invitrogen Life Technologies). The AMV RT system (Promega) was used for cDNA synthesis following the manufacturer's protocol. The primers for PCR were 5′-GGTCCCATTCCAATACCAAC-3′ corresponding to GmAGL15 and 5′-CAGGTCCTCCTCGTAGATCAGCTT-3′ corresponding to c-myc epitope tag. Control primers amplified EF1-α (5′-ACGCTCTACTTGCTTTCACC-3′ and 5′-GCACCGTTCCAATACCACC-3′).
Protein Gel Blot
Crude nuclear extracts were prepared from approximately 75 mg of open flowers following the protocol described by Busk and Pagès (1997), except 50 μL of low salt buffer and 25 μL of high salt buffer were used per sample and the proteinase inhibitors aprotinin, leupeptin, and pepstatin (1 mm each) were added to buffers. After extraction by vortexing, the proteins (60 μg per lane) were separated on a 12% polyacrylamide gel, blotted to Immobilon polyvinylidene difluoride (Millipore), and probed with anti-AGL15 antiserum as described by Heck et al. (1995).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AtAGL15, At5g13790; GmAGL15, AY370659 (cDNA); and AY370660 (genomic).
Acknowledgments
We thank Kay McAllister, Jeanne Prather, and Carl Redmond for providing the soybean somatic embryo culture, and Dr. Randy Dinkins and Dr. Glenn Collins and his lab for training and assistance with soybean transformation. We thank Dr. Donna Fernandez for the loss-of-function mutants. This article (no. 07–06–139) is published with the approval of the Director of the Kentucky Agricultural Experiment Station.
This work was supported by a National Research Initiative Competitive Grant (grant no. 2005–35301–15699) from the U.S. Department of Agriculture Cooperative State Research, Education, and Extension Service, by the National Science Foundation (grant no. IBN–9984274), and by the University of Kentucky.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Sharyn E. Perry (sperr2@uky.edu).
Open Access articles can be viewed online without a subscription.
References
- Adamczyk BJ, Lehti-Shiu MD, Fernandez DE (2007) The MADS-domain factors AGL15 and AGL18 act redundantly as repressors of the floral transition in Arabidopsis. Plant J 50 1007–1019 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen HM, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors (1998) Current Protocols in Molecular Biology. John Wiley & Sons, Cambridge, MA
- Boutilier K, Offringa R, Sharma VK, Kieft H, Ouellet T, Zhang L, Hattori J, Liu CM, van-Lammeren AAM, Miki BLA, et al (2002) Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 14 1737–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braybrook SA, Stone SL, Park S, Bui AQ, Lee BH, Fischer RL, Goldberg RB, Harada JJ (2006) Genes directly regulated by LEAFY COTYLEDON2 provide insight into the control of embryo maturation and somatic embryogenesis. Proc Natl Acad Sci USA 103 3468–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busk PK, Pagès M (1997) Microextraction of nuclear proteins from single maize embryos. Plant Mol Biol Rep 15 371–376 [Google Scholar]
- Curaba J, Moritz T, Blervaque R, Parcy F, Raz V, Herzog M, Vachon G (2004) AtGA3ox2, a key gene responsible for bioactive gibberellin biosynthesis, is regulated during embryogenesis by LEAFY COTYLEDON2 and FUSCA3 in Arabidopsis. Plant Physiol 136 3660–3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez DE, Heck GR, Perry SE, Patterson SE, Bleecker AB, Fang SC (2000) The embryo MADS domain factor AGL15 acts postembryonically: inhibition of perianth senescence and abscission via constitutive expression. Plant Cell 12 183–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finner JJ, Nagasawa A (1988) Development of an embryogenic suspension culture of soybean (Glycine max Merrill.). Plant Cell Tissue Organ Cult 15 125–136 [Google Scholar]
- Gaj MD, Zhang SB, Harada JJ, Lemaux PG (2005) LEAFY COTYLEDON genes are essential for induction of somatic embryogenesis of Arabidopsis. Planta 222 977–988 [DOI] [PubMed] [Google Scholar]
- Gallois JL, Nora FR, Mizukami Y, Sablowski R (2004) WUSCHEL induces shoot stem cell activity and developmental plasticity in the root meristem. Genes Dev 18 375–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzarrini S, Tsuchiya Y, Lumba S, Okamoto M, McCourt P (2004) The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the hormones gibberellin and abscisic acid. Dev Cell 7 373–385 [DOI] [PubMed] [Google Scholar]
- Harding EW, Tang W, Nichols KW, Fernandez DE, Perry SE (2003) Expression and maintenance of embryogenic potential is enhanced through constitutive expression of AGAMOUS-Like 15. Plant Physiol 133 653–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht V, Vielle-Calzada JP, Hartog MV, Schmidt ED, Boutilier K, Grossniklaus U, de Vries SC (2001) The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE 1 gene is expressed in developing ovules and embryos and enhances embryogenic competence in culture. Plant Physiol 127 803–816 [PMC free article] [PubMed] [Google Scholar]
- Heck GR, Perry SE, Nichols KW, Fernandez DE (1995) AGL15, a MADS domain protein expressed in developing embryos. Plant Cell 7 1271–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K, Wang H, Perry SE (2008) A transcriptional repression motif in the MADS factor AGL15 is involved in recruitment of histone deacetylase complex components. Plant J 53 172–185 [DOI] [PubMed] [Google Scholar]
- Ikeda-Iwai M, Satoh S, Kamada H (2002) Establishment of a reproducible tissue culture system for the induction of Arabidopsis somatic embryos. J Exp Bot 53 1575–1580 [DOI] [PubMed] [Google Scholar]
- Ikeda-Iwai M, Umehara M, Satoh S, Kamada H (2003) Stress-induced somatic embryogenesis in vegetative tissues of Arabidopsis thaliana. Plant J 34 107–114 [DOI] [PubMed] [Google Scholar]
- Jiménez VM (2005) Involvement of plant hormones and plant growth regulators on in vitro somatic embryogenesis. Plant Growth Regul 47 91–110 [Google Scholar]
- Karlova R, Boeren S, Russinova E, Aker J, Vervoort J, de Vries S (2006) The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 protein complex includes BRASSINOSTEROID-INSENSITIVE1. Plant Cell 18 626–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehti-Shiu MD, Adamczyk BJ, Fernandez DE (2005) Expression of MADS-box genes during the embryonic phase in Arabidopsis. Plant Mol Biol 58 89–107 [DOI] [PubMed] [Google Scholar]
- Lumba S, McCourt P (2005) Preventing leaf identify theft with hormones. Curr Opin Plant Biol 8 501–505 [DOI] [PubMed] [Google Scholar]
- Luo YK, Koop HU (1997) Somatic embryogenesis in cultured immature zygotic embryos and leaf protoplasts of Arabidopsis thaliana ecotypes. Planta 202 387–396 [DOI] [PubMed] [Google Scholar]
- Meurer CA, Dinkins RD, Redmond CT, McAllister KP, Tucker DT, Walker DR, Parrott WA, Trick HN, Essig JS, Frantz HM, et al (2001) Embryogenic response of multiple soybean [Glycine max (L.) Merr.] cultivars across three locations. In Vitro Cell Dev Biol Plant 37 62–67 [Google Scholar]
- Mordhorst AP, Voerman KJ, Hartog MV, Meijer EA, van Went J, Koornneef M, de Vries SC (1998) Somatic embryogenesis in Arabidopsis thaliana is facilitated by mutations in genes repressing meristematic cell divisions. Genetics 149 549–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15 473–497 [Google Scholar]
- Namasivayam P (2007) Acquisition of embryogenic competence during somatic embryogenesis. Plant Cell Tissue Organ Cult 90 1–8 [Google Scholar]
- Nolan KE, Irwanto RR, Rose RJ (2003) Auxin up-regulates MtSERK1 expression in both Medicago truncatula root-forming and embryogenic cultures. Plant Physiol 133 218–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parenicová L, de Folter S, Kieffer M, Horner DS, Favalli C, Busscher J, Cook HE, Ingram RM, Kater MM, Davies B, et al (2003) Molecular and phylogenetic analysis of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. Plant Cell 15 1538–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrott WA, Williams EG, Hildebrand DF, Collins GB (1989) Effect of genotype on somatic embryogenesis from immature cotyledons of soybean. Plant Cell Tissue Organ Cult 16 15–21 [Google Scholar]
- Perry SE, Lehti MD, Fernandez DE (1999) The MADS-domain protein AGAMOUS-like 15 accumulates in embryonic tissues with diverse origins. Plant Physiol 120 121–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry SE, Nichols KW, Fernandez DE (1996) The MADS domain protein AGL15 localizes to the nucleus during early stages of seed development. Plant Cell 8 1977–1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillon E, Terzi M, Baldan B, Mariani P, Schiavo FL (1996) A protocol for obtaining embryonic cell lines from Arabidopsis. Plant J 9 573–577 [DOI] [PubMed] [Google Scholar]
- Reddy MSS, Ghabrial SA, Redmond CT, Dinkins RD, Collins GB (2001) Resistance to Bean pod mottle virus in transgenic soybean lines expressing the capsid polyprotein. Phytopathology 91 831–838 [DOI] [PubMed] [Google Scholar]
- Rose RJ, Nolan KE (2006) Invited review: genetic regulation of somatic embryogenesis with particular reference to Arabidopsis thaliana and Medicago truncatula. In Vitro Cell Dev Biol Plant 42 473–481 [Google Scholar]
- Rounsley SD, Ditta GS, Yanofsky MF (1995) Diverse roles for MADS box genes in Arabidopsis development. Plant Cell 7 1259–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt MA, Tucke DM, Cahoon EB, Parrott WA (1995) Towards normalization of soybean somatic embryo maturation. Plant Cell Rep 24 383–391 [DOI] [PubMed] [Google Scholar]
- Specht J, Shoemaker R, Jackson S, Stacey G, Cregan P (2007) Soybean genomics research: a strategic plan for 2008-2012. USDA CSREES. www.csrees.usda.gov/nea/plants/pdfs/soybean_genomics.pdf (January 3, 2008)
- Vogel G (2005) What Don't We Know? How does a single somatic cell become a whole plant. Science 309 86. [DOI] [PubMed] [Google Scholar]
- Wang H, Caruso LV, Downie AB, Perry SE (2004) The embryo MADS-domain protein AGAMOUS-Like 15 directly regulates expression of a gene encoding an enzyme involved in gibberellin metabolism. Plant Cell 16 1206–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Roossinck MJ (2006) Comparative analysis of expressed sequences reveals a conserved pattern of optimal codon usage in plants. Plant Mol Biol 61 699–710 [DOI] [PubMed] [Google Scholar]
- Wu Y, Haberland G, Zhou C, Koop HU (1992) Somatic embryogenesis, formation of morphogenetic callus and normal development in zygotic embryos of Arabidopsis thaliana in vitro. Protoplasma 169 89–96 [Google Scholar]
- Zhu C, Perry SE (2005) Control of expression and autoregulation of AGL15, a member of the MADS-box family. Plant J 41 583–594 [DOI] [PubMed] [Google Scholar]
- Zuo JR, Niu QW, Frugis G, Chua NH (2002) The WUSCHEL gene promotes vegetative-to-embryonic transition in Arabidopsis. Plant J 30 349–359 [DOI] [PubMed] [Google Scholar]