Abstract
Previous research has demonstrated that AtPHR1 plays a central role in phosphate (Pi)-starvation signaling in Arabidopsis thaliana. In this work, two OsPHR genes from rice (Oryza sativa) were isolated and designated as OsPHR1 and OsPHR2 based on amino acid sequence homology to AtPHR1. Their functions in Pi signaling in rice were investigated using transgenic plants. Our results showed that both OsPHR1 and OsPHR2 are involved in Pi-starvation signaling pathway by regulation of the expression of Pi-starvation-induced genes, whereas only OsPHR2 overexpression results in the excessive accumulation of Pi in shoots under Pi-sufficient conditions. Under Pi-sufficient conditions, overexpression of OsPHR2 mimics Pi-starvation stress in rice with enhanced root elongation and proliferated root hair growth, suggesting the involvement of OsPHR2 in Pi-dependent root architecture alteration by both systematic and local pathways. In OsPHR2-overexpression plants, some Pi transporters were up-regulated under Pi-sufficient conditions, which correlates with the strongly increased content of Pi. The mechanism behind the OsPHR2 regulated Pi accumulation will provide useful approaches to develop smart plants with high Pi efficiency.
AtPHR1 plays a key role in the phosphorus (P) signaling system in Arabidopsis (Arabidopsis thaliana). AtPHR1 is a transcription factor with an MYB domain and a predicted coiled-coil (CC) domain defined as a member of the MYB-CC family. AtPHR1 as a dimer binds a cis-element with an imperfect palindromic sequence (GNATATNC; Rubio et al., 2001). The function loss of AtPHR1 reduces expression of several phosphate (Pi)-starvation-inducible genes, such as AtIPS1, AtRNS1, and At4, and accumulation of anthocyanins in leaves, which is one of the most conspicuous symptoms of low Pi (LP) stress in Arabidopsis (Raghothama, 1999). It has been found that many Pi-starvation-inducible genes contain the cis-element (Franco-Zorrilla et al., 2004); therefore, AtPHR1 has been considered a key regulator in the Pi-starvation signaling pathway.
A regulation system for the Pi-starvation signaling pathway in Arabidopsis has been proposed (Schachtman and Shin, 2007). In this regulation system, PHR1 is sumoylated by an AtSIZ1-dependent process (Miura et al., 2005). AtSIZ1 is a plant small ubiquitin-like modifier (SUMO) E3 ligase that is a focal controller of Pi-starvation-dependent responses. Downstream of AtPHR1, miRNA399 as a PHR1 target is specifically induced by Pi starvation. miRNA399 reciprocally regulates the gene PHO2 at the transcriptional level (Miura et al., 2005). PHO2 functions as a ubiquitin-conjugating E2 enzyme (UBC24; Aung et al., 2006; Bari et al., 2006), and loss of function of PHO2/UBC24 will lead to excessive accumulation of Pi in the shoot tissue (Fujii et al., 2005; Chiou et al., 2006).
Plants have evolved a wide range of adaptive strategies to adapt to P deficiency and improve P mobilization and uptake from the soil (Raghothama, 1999). Alterations in root architecture are important to enable the plant to more efficiently explore and exploit the insoluble P in soils. In Arabidopsis, the shortened primary root, proliferated lateral roots and root hairs toward the apical root, and accumulation of anthocyanins are the typical traits of a response to Pi starvation (Williamson et al., 2001; Linkohr et al., 2002). Function loss of AtSIZ1 causes Arabidopsis to exhibit exaggerated prototypical Pi-starvation responses, including cessation of primary root growth, extensive lateral root and root hair development, increased root-shoot mass ratio, and greater anthocyanin accumulation. AtSIZ1 targets PHR1, whereas the phr1 mutants do not exhibit these root architectural changes. Therefore, the function of PHR1 as a key regulator in the Pi-signaling system in the developmental and physiological adaptation in plants to Pi-starvation stress remains to be elucidated. In addition, AtPHR1 is a member of a large gene family consisting of 11 PHR-like genes in Arabidopsis (Todd et al., 2004). Whether different PHR1 homologous genes have different functions in the regulation of Pi signaling is also unknown.
Compared with Arabidopsis, knowledge about the functions of PHR-like genes in monocot crops is limited. Based on the protein sequence similarity of AtPHR1, we isolated two homologous genes in rice (Oryza sativa) as single copies, respectively, designated as OsPHR1 and OsPHR2. To investigate the function of OsPHR1/OsPHR2 in the Pi-signaling regulation system, we developed transgenic lines with reduction and overexpression of OsPHR1/OsPHR2 in rice for Pi-signaling and Pi-uptake analysis. Our results indicate that both OsPHR1 and OsPHR2 are involved in the Pi-signaling pathway, whereas only the overexpression of OsPHR2 results in Pi accumulation in shoots of rice.
RESULTS
Cloning and Characterization of OsPHR1 and OsPHR2 Genes
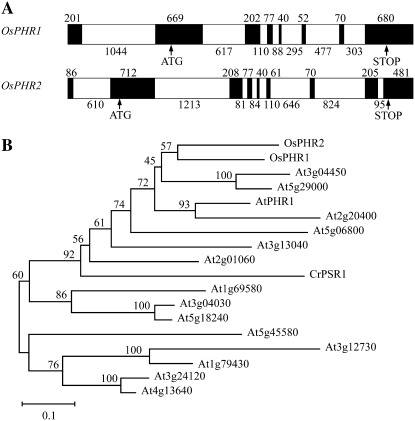
By using the protein sequence of AtPHR1 (NP_194590) as a query, we identified two homologous genes in rice through a TBLASTN search in the National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov/BLAST) located on chromosome 3 (120,024–124,948) and chromosome 7 (24,959–30,561) with a single copy, respectively. The identified genes were designated as OsPHR1 (AK063486) and OsPHR2 (AK100065) according to their amino acid sequence identity (SI) to AtPHR1 with 51.7% and 45.5%, respectively. OsPHR1 and OsPHR2 share 43.6% SI with each other. The full-length complementary DNAs (cDNAs) of the two genes were obtained from rice (‘Nipponbare’), followed by PCR amplification using primers predicted from the cDNA sequences. Based on sequencing verification, the obtained cDNAs were identical to the sequence data released from NCBI. OsPHR1 and OsPHR2 are 1,991- and 1,941-bp long, respectively, and contain an open reading frame (ORF) encoding a predicted protein of 428 and 426 amino acids, respectively. The gene structure analysis showed that both genes have a similar splicing pattern, with the exception of one more exon and intron present in OsPHR2 (Fig. 1A).
Figure 1.
Structures of the OsPHR1 and OsPHR2 genes and phylogenetic analysis with other related MYB-CC proteins in Arabidopsis and Chlamydomonas reinhardtii. A, Structures of the OsPHR1 and OsPHR2 genes. Exons are indicated as black boxes and introns as white boxes. Numbers indicate the length of each exon and intron. ATG and stop codon are shown with arrowheads. B, Phylogram of proteins sharing the MYB and predicted CC domains constructed using the Clustal (Thompson et al., 1997) program and the neighbor-joining method. The numbers above the lines refer to bootstrap values (of 100 samples). Scale bar, 0.1 substitutions per site.
AtPHR1 and CrPSR1 belong to a large gene family (including another 14 proteins in Arabidopsis) with a conserved MYB DNA-binding domain (BD) and a predicted CC domain in each family member (Rubio et al., 2001). Multiple sequence alignment of OsPHR1 and OsPHR2 with the MYB-CC family members using the ClustalW program shows that OsPHR1 and OsPHR2 are two novel members of the MYB-CC gene family with high conservation in both of the domains (Supplemental Fig. S1). Phylogenetic analysis based on the neighbor-joining method and bootstrap analysis (n = 100) within this family reveals that two main groups can be distinguished; OsPHR1 and OsPHR2 belong to the same subgroup with AtPHR1 and CrPSR1 and are more closely related to AtPHR1 (Fig. 1B).
Expression Patterns of OsPHR1 and OsPHR2
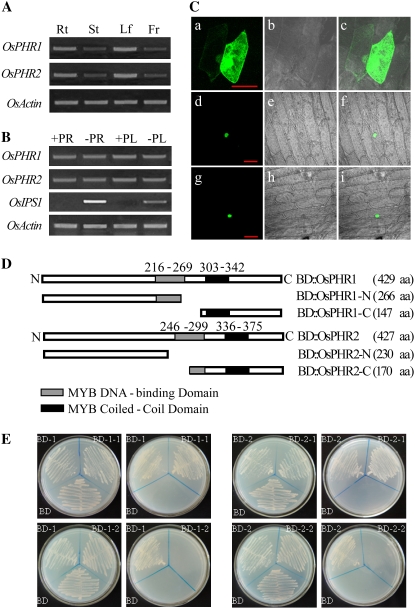
To investigate the spatial expression patterns of OsPHR1 and OsPHR2 in rice, semiquantitative reverse transcription (RT)-PCR analysis was performed in roots, leaves, stems, and panicles of 90-d-old seedlings. Both OsPHR1 and OsPHR2 showed a similar constitutive expression pattern in all tissues with higher expression level in roots and leaves (Fig. 2A). Like AtPHR1, the steady expression of OsPHR1 and OsPHR2 in both roots and shoots was not very responsive to Pi deprivation, but the expression of the PSI (Pi-starvation induced) gene OsIPS1 was dramatically induced under Pi-starvation conditions (Fig. 2B).
Figure 2.
Expression patterns and transcriptional activities of the OsPHR1 and OsPHR2 genes. A, Tissue-specific expression of OsPHR1 and OsPHR2 by RT-PCR analysis. B, The expression of OsPHR1 and OsPHR2 responsive to P starvation. RT-PCR was performed on total RNAs from leaf (L) and root (R) of 21-d-old seedlings after transfer to grow under Pi-sufficient (+P) and -deficient (−P) conditions for 7 d. The expression of OsIPS1 was tested as a systematic control for the Pi-starvation induction. C, The subcellular localization of OsPHR1 and OsPHR2 proteins. Confocal images of onion epidermis cells under the GFP channel showing the constitutive localization of Ubi∷GFP (a), nuclear localization of Ubi∷OsPHR1-GFP (d), and Ubi∷OsPHR2-GFP (g). The confocal images (b, e, and h) are of the same cells in a, d, and g with transmitted light. The merged images (c, f, and i) are of a and b; d and e; and g and h, respectively. Bar = 100 μm. D, The diagram of protein fusion constructs used for transcriptional activity assay. The black and gray boxes indicate the MYB DNA-BD and the CC domain, respectively. The numbers indicate their positions in OsPHR1 and OsPHR2 proteins. N and C refer to the N terminus and C terminus of the protein. E and F, The growth of transformed yeast strain AH109 with constructs of OsPHR1 (E) and OsPHR2 (F) under SD/-Trp and SD/-Trp-His-A nutrition-deficient medium. For abbreviations, BD-1, BD∷OsPHR1; BD-1-1, BD∷OsPHR1-1; BD1-2, BD∷OsPHR1-2 (constructs indicated in D). BD-2, BD∷OsPHR2; BD-2-1, BD∷OsPHR2-1; and BD-2-2, BD∷OsPHR2-2 (constructs indicated in D). BD refers to the pGBKT7 vector. which serves as the negative control.
OsPHR1 and OsPHR2 Are Transcription Activators
To test whether OsPHR1 and OsPHR2 are transcription factors like AtPHR1, the full-size ORFs of the two genes were in-frame fused to the N terminus of the coding region of the GFP5 (green fluorescence protein 5). The fusion proteins and the GFP protein alone were transiently expressed in the onion (Allium spp.) epidermal cells under the control of the ubiquitin promoter. The fluorescence of the OsPHR1- and OsPHR2-fused GFP protein was only found in the nucleus, whereas the fluorescence of the GFP alone as a control was distributed throughout the cell (Fig. 2C), suggesting that both OsPHR1 and OsPHR2 are nuclear proteins.
To analyze the transcription activation ability of OsPHR1 and OsPHR2, an autonomous gene activation test was performed in the yeast system. The full-length fragments of the two proteins and their separated N-terminal and C-terminal fragments were fused to the DNA-BD of the yeast transcription factor GAL4. These six clones were designated as BD∷OsPHR1, BD∷OsPHR1-N (1–266 amino acids in the N terminus), BD∷OsPHR1-C (last 147 amino acids in the C terminus), BD∷OsPHR2, BD∷OsPHR2-N (1–230 amino acids in the N terminus), and BD∷OsPHR2-C (last 170 amino acids in the C terminus), respectively, and transformed to yeast strain AH109 (Fig. 2D). On the minimal synthetic dextrose (SD) medium lacking Trp, all of the yeast strains with each of the six vectors could grow well, indicating a successful transformation of these vectors (Fig. 2, E and F, left). However, on the triple nutrition-deficient SD medium (SD/-Trp-His-A), only the strains with the full-length form or the N terminus of both of the proteins could grow well (Fig. 2, E and F, right). The transcription activities of these strains were also measured by quantifying MEL1 gene expression via the α-galactosidase assay. The N-terminal peptides of both proteins had almost the same transcription activities as the full-length proteins, whereas the C-terminal peptides showed activities similar to those of the negative controls (data not shown). These results indicate that the full-length OsPHR1 and OsPHR2 proteins have transcription-activation abilities, and the activation domain of both proteins is located in the N-terminal peptide.
Overexpression of OsPHR2 Caused Excessive Pi Accumulation in Shoots of Transgenic Plants
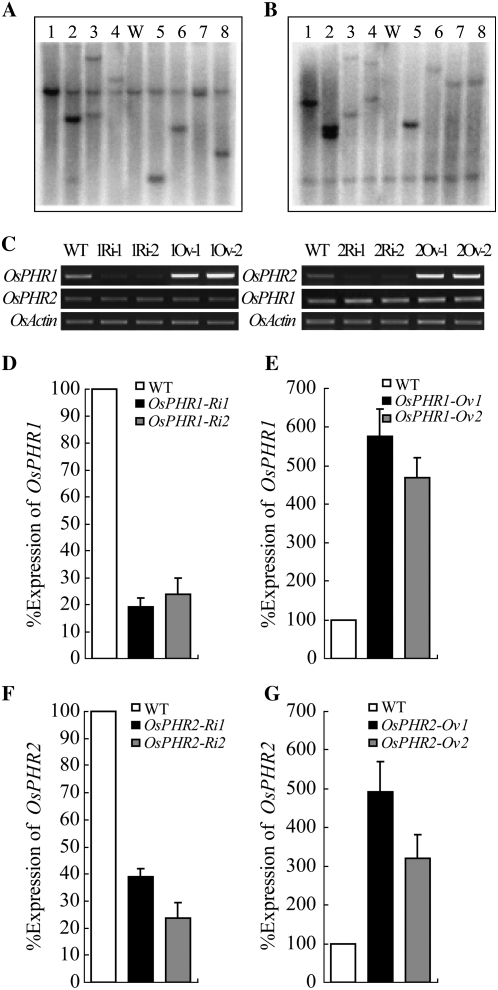
To investigate the functions of OsPHR1 and OsPHR2, we developed transgenic plants with overexpressed or depressed expression of the two genes, using cauliflower mosaic virus 35S promoter-controlled overexpression and RNA interference (RNAi) constructs. Two transgenic lines of each construct with strongest up- or down-regulation of OsPHR1 or OsPHR2 were selected for Southern-blot analysis, indicating that the transgenic lines obtained from the same construct are independent from each other (Fig. 3, A and B). The levels of the OsPHR1 and OsPHR2 transcripts in these transgenic lines were detected by RT-PCR (Fig. 3C) and qRT-PCR (Fig. 3, D–G). The OsPHR1 and OsPHR2 transcripts were elevated in the OsPHR1 and OsPHR2 overexpressing lines (designated as OsPHR1-Ov1, OsPHR1-Ov2 and OsPHR2-Ov1, OsPHR2-Ov2), and repressed in the OsPHR1 and OsPHR2 RNAi lines (designated as OsPHR1-Ri1/2 and OsPHR2-Ri1/2). And there are no cross-effects of overexpression or RNAi of one OsPHR gene on the proper expression of the other one (Fig. 3C). The transgenic lines were subjected to hydroponic culture with 0.5 L of solution for each plant at a high Pi (HP) level (10 mg Pi/L) and an LP level (1 mg Pi/L) for phenotype observation. After 30 d of growth at the HP level, both of the two overexpressing lines of OsPHR2 (OsPHR2-Ov1 and OsPHR2-Ov2) displayed Pi toxicity, as indicated by smaller plants with chlorosis or necrosis on the leaf margins, predominantly in mature leaves (Fig. 4, A and B). Significant differences in tiller number and shoot and root biomass were also observed between wild-type and transgenic plants. No obvious phenotypic variations were observed in RNAi lines of OsPHR2 (Supplemental Fig. S4D) and other types of transgenic lines (data not shown).
Figure 3.
The expression levels of OsPHR1 and OsPHR2 in RNAi and overexpression transgenic lines. A, Southern-blot analysis for the wild type (WT; ‘Nipponbare’), two lines of OsPHR1-Ri (1 and 2), two lines of 35S-OsPHR1 (3 and 4), two lines of OsPHR2-Ri (6 and 7), and two lines of 35S-OsPHR2 (8 and 9) using the hygromycin gene as probe. Five micrograms of genomic DNA of each sample were digested by EcoRI (A) and HindIII (B), and separated by agarose gel. C, Semiquantitative RT-PCR analysis of the expression levels of OsPHR1 (left) and OsPHR2 (right) in the leaf of RNAi and overexpression transgenic lines by using the specific primers for OsPHR1 and OsPHR2. OsActin was used as the loading control. D to G, Real-time qRT-PCR analysis of the RNAi and overexpression transgenic lines of OsPHR1 (D and E) and OsPHR2 (F and G). The relative expression levels were shown in percentages as compared with wild type as 100% expression.
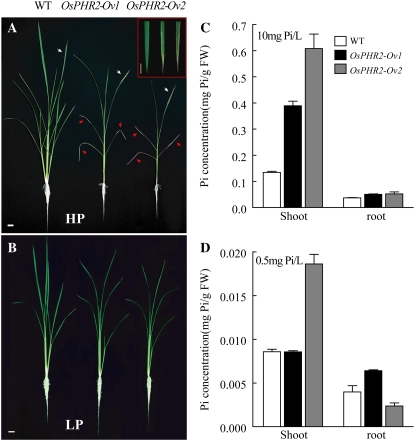
Figure 4.
Phenotype of OsPHR2-overexpressing transgenic lines under HP and LP conditions. A, Pi toxic phenotype shown as smaller plant, reduced tiller number, and decreased shoot and root biomass of two independent OsPHR2-overexpressing transgenic lines designated as OsPHR2-Ov1 and OsPHR2-Ov2 after being grown under HP conditions (10 mg/L Pi, 0.5 L/plant) for 30 d. The red arrowheads indicate the severe chlorosis and necrosis that appeared in the old leaves of the two OsPHR2-overexpressing lines. The inset in A shows a close-up of young leaves of wild type (left), OsPHR2-Ov1 (middle), and OsPHR2-Ov2 (right). Bar = 1 cm. B, Phenotype of transgenic plants after being grown under LP conditions (1 mg/L Pi, 0.5 L/plant) for 30 d. Bar = 1 cm. C, Pi concentration in the shoots and roots of wild type, OsPHR2-Ov1, and OsPHR2-Ov2 from A. Error bars indicate the sd (n = 5). D, Pi concentration in the shoots and roots of wild type, OsPHR2-Ov1, and OsPHR2-Ov2 from B. Error bars indicate the sd (n = 5). [See online article for color version of this figure.]
Pi concentrations in shoots of the OsPHR2-overexpressing plants grown at a HP level were 2.0- to 2.5-fold higher than those in wild-type plants (Fig. 4C), whereas in roots, the Pi concentration was similar to the wild type (Fig. 4C). At the LP level, the significant difference in plant growth between wild-type and transgenic plants was effaced. Higher Pi concentrations of shoots of the transgenic plants were also observed, although the absolute concentration was much lower than that at the HP level (Fig. 4D). But no significant changes were found of Pi concentration between wild-type and RNAi lines of OsPHR1 and OsPHR2 (Supplemental Fig. S4, B and C).
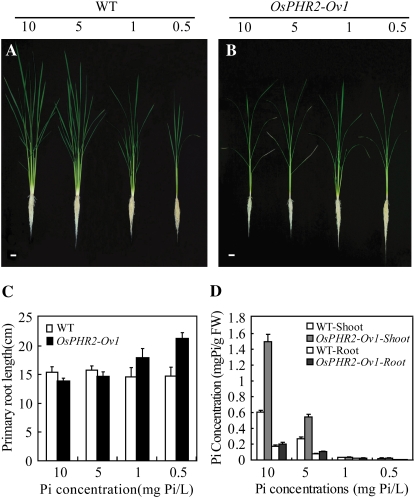
We further checked the phenotype of the transgenic line OsPHR2-Ov2 under different Pi levels. After 30 d of growth in hydroponic solution, transgenic plants displayed severe toxicity phenotypes at the high (10 mg Pi/L) and moderately high (5 mg Pi/L) Pi levels. The toxicity phenotype could be alleviated gradually along with a decrease in Pi levels (Fig. 5, A and B). At LP levels (1 and 0.5 mg Pi/L), the root length of the transgenic plants was longer than that of wild-type plants, indicating that the transgenic plants were more sensitive to LP induction compared to the wild-type plants (Fig. 5C). The shoot Pi concentrations of the transgenic plants grown at different Pi levels were correlated with their phenotypes (Fig. 5D).
Figure 5.
Growth performances of wild type (WT) and the OsPHR2-Ov1 line at different Pi levels in a solution culture. A and B, Growth of 30-d-old seedlings of wild type and OsPHR2-Ov1 in hydroponic solutions with 10, 5, 1, and 0.5 μg mL−1 Pi. Scale bars in A and B represent 2.5 cm. C, Primary root length of wild type and OsPHR2-Ov1 at different Pi levels. Error bars indicate se (n = 8). D, Pi concentrations in shoots and roots of wild-type and OsPHR2-Ov1 plants. Error bars indicate se (n = 5). [See online article for color version of this figure.]
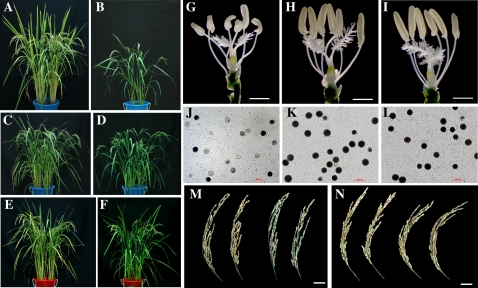
Soil pot experiments were carried out in a greenhouse for phenotype observation during the whole-plant growth period. LP acid red soil (pH 4.3, soil:water = 1:1) was used with four Pi levels designated as high (200 mg Pi/kg soil, Bray-II method, Pi = 38.2 mg/kg soil), moderately high (120 mg Pi/kg soil, Bray-II method, Pi = 19.89 mg/kg soil), low (60 mg Pi/kg soil, Bray-II method, Pi = 8.15 mg/kg soil), and extremely low (30 mg Pi/kg soil, Bray-II method, Pi = 4.50 mg/kg soil). Forty days after planting, transgenic plants at the HP level showed Pi toxicity phenotypes like that in hydroponic culture (Supplemental Fig. S2). From moderate- to LP levels, the toxicity phenotypes were gradually recovered. After 110 d at harvest stage, growth parameters, including plant height, panicle length, maximum tiller number, seed-setting tiller number, seed-setting rate, grain number/panicle, and chlorophyll content were measured (Fig. 6, A–F; Supplemental Fig. S3). At the HP level, most parameters of transgenic plants were suppressed in wild-type plants, especially maximum tiller number (1.9-fold lower), seed-setting tiller number (8-fold lower), seed-setting rate (4-fold lower), and grain number (4.6-fold lower). Along with the decreased Pi level in the soil, these parameters in transgenic plants were recovered to almost the same level as wild-type plants. The male reproductive organs of transgenic plants were disordered with twisted anther structures, few pollen grains, and low pollen viability when grown in HP soil. In the extremely LP soil, the development of the male reproductive organs was normal, like that of wild-type plants (Fig. 6, H–K).
Figure 6.
Growth performances of wild type and the OsPHR2-Ov1 line at different Pi levels in a pot experiment. A and B, Growth performances of wild type (A) and OsPHR2-Ov1 (B) at HP level (200 mg Pi/kg soil). C and D, Growth performances of wild type (C) and OsPHR2-Ov1 (D) at moderate Pi level (120 mg Pi/kg soil). E and F, Growth performances of wild type (E) and OsPHR2-Ov1 (F) at LP level (60 mg Pi/kg soil). G to I, Anther structure of OsPHR2-Ov1 (G) and wild type (H) at HP level (200 mg Pi/kg soil), and OsPHR2-Ov1 under LP level (I; 60 mg Pi/kg soil). J to L, Pollen viability of OsPHR2-Ov1 (J) and wild type (K) at HP level (200 mg Pi/kg soil), and OsPHR2-Ov1 under LP level (L; 60 mg Pi/kg soil). The scale bars represent 1 mm for anthers, 50 μm for pollens. M, Panicle filling rate of wild type (left) and OsPHR2-Ov1 (right) at HP level (200 mg Pi/kg soil). N, Panicle filling rate of wild type (left) and OsPHR2-Ov1 (right) at LP level (60 mg Pi/kg soil).
OsPHR2 Is Involved in the Root Architectural Alteration in Response to Pi Starvation
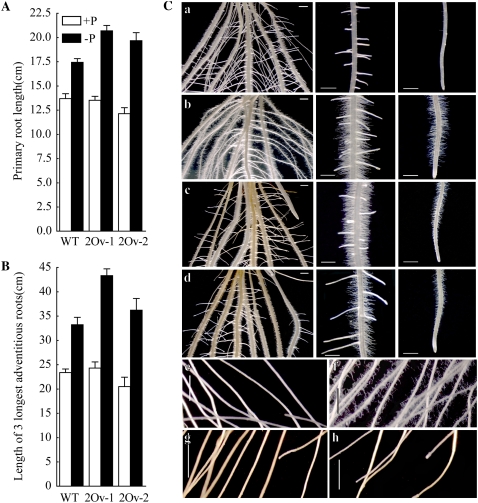
Different with Arabidopsis, under flooding conditions the elongation of rice primary and adventitious roots are the typical traits stimulated by Pi starvation, but proliferated root hairs toward the apical root is the same Pi-starvation response with Arabidopsis (Wissuwa, 2003; Yi et al., 2005). Two transgenic lines with overexpressing OsPHR2 were subjected to hydroponic culture with or without Pi supply (10 or 0 mg Pi/L) for 10 d. The primary root length and total length of the three longest adventitious roots of wild-type, OsPHR2-Ov1, and OsPHR2-Ov2 plants were measured. Under Pi-sufficient conditions, no significant differences were observed in the primary and adventitious root lengths between wild-type plants and the transgenic lines (Fig. 7, A and B). The proliferated root hairs toward the apical root were observed in the OsPHR2-overexpressing lines under Pi-sufficient conditions compared to the wild type (Fig. 7C, a–d). The abundant, long root hair growth was also observed in lateral roots of the OsPHR2-overexpressing lines (Fig. 7C, e–h). These results suggest that the OsPHR2-overexpressing lines were hypersensitive to Pi-starvation stress compared to wild-type plants and that OsPHR2 may be involved in a P-mediated regulatory pathway in root hair growth. But no significant changes were found of primary root length (data not shown) and root hairs between wild-type and RNAi lines of OsPHR1 and OsPHR2 (Supplemental Fig. S4A).
Figure 7.
Root performances of wild-type (WT) and OsPHR2-overexpressing seedlings under Pi-sufficient (+P) and Pi-deficient (−P) conditions. A and B, Primary root length and adventitious root length (total length of three longest adventitious roots) of 10-d-old seedlings of wild-type and OsPHR2-overexpressing lines (2Ov-1 and 2Ov-2). Values are mean ± sd (n = 10). C, Root hair proliferation of OsPHR2-overexpressing line in the regions of root base (left), elongation zoon (middle), and primary root tip (right) grown under Pi-supplied (b) and Pi-deficient (d) conditions compared with wild type (a and c). The hair proliferation on the lateral root of the OsPHR2-overexpressing line, under Pi-supplied (f) and Pi-deficient (h) conditions compared with wild type (e and g). The scale bars in the panels represent 1 mm.
OsPHR1 and OsPHR2 Are Involved in the Pi-Starvation Signaling Pathway with Different Functions
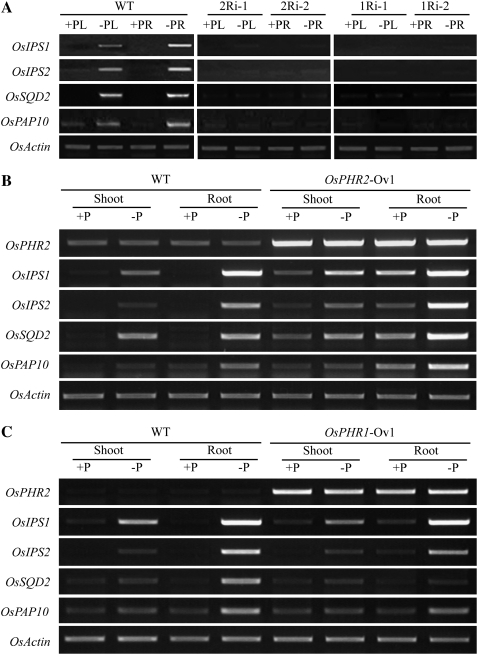
The expression patterns of Pi-starvation-inducible genes in rice were examined by RT-PCR analysis in the OsPHR1/2-overexpressing and RNAi transgenic lines under both Pi-sufficient (+P) and -deficient (−P) conditions. RT-PCR analysis showed that suppression of OsPHR1 and OsPHR2 reduced the Pi-starvation-inducible expression of two members of the Mt4/TPSI1 family in rice, OsIPS1 and OsIPS2 (Wasaki et al., 2003; Hou et al., 2005), and OsSQD2 and OsPAP10, rice homologs for Arabidopsis genes encoding a sulfolipid synthase and a purple acid phosphatase, respectively (Essigmann et al., 1998; Yu et al., 2002; Wang et al., 2006; Fig. 8A), indicating that both genes are involved in Pi-starvation signaling. The expression of these four genes was up-regulated in the lines with overexpressing OsPHR2 (Fig. 8B). The Pi-starvation induction of OsIPS1, OsIPS2, and OsPAP10 was increased in both shoots and roots compared with wild-type plants. For OsSQD2, the Pi-starvation induction was profoundly increased in roots but not in shoots. Moreover, the expression of all of these genes was induced in both shoots and roots of the OsPHR2-overexpressing line even under Pi-sufficient conditions. However, in the OsPHR1-overexpressing line, these Pi-starvation-induced genes were not greatly induced and even repressed in OsSQD2 (Fig. 8C). These results indicate that both OsPHR1 and OsPHR2 are involved in the P-starvation signaling pathway in rice but have different transcriptional activities in downstream gene expression, which may account for different consequences between overexpression of OsPHR1 and OsPHR2.
Figure 8.
Effect of OsPHR1 and OsPHR2 overexpression and reduction on the expression of PSI genes. A, Expression of OsIPS1, OsIPS2, OsSQD2, and OsPAP10 in wild-type (WT), OsPHR1 RNAi, and OsPHR2 RNAi plants (+PL, HP shoot; −PL, Non-Pi shoot; +PR, HP root; −PL, Non-Pi root). B, Expression of OsIPS1, OsIPS2, OsSQD2, and OsPAP10 in wild type and the OsPHR2-overexpressing (OsPHR2-Ov1) line. C, Expression of OsIPS1, OsIPS2, OsSQD2, and OsPAP10 in wild type and the OsPHR1-overexpressing (OsPHR1-Ov1) line. For all the RT-PCR analysis, shoots and roots were sampled separately from 10-d-old seedlings grown in Pi-sufficient (+P, 10 mg/L Pi) and Pi-deficient (−P, 0 mg/L Pi) conditions. OsActin was used as the loading control. The primers of the PSI genes used in this assay are shown in Supplemental Table S1.
Overexpression of OsPHR2 Up-Regulates Several Pi Transporters
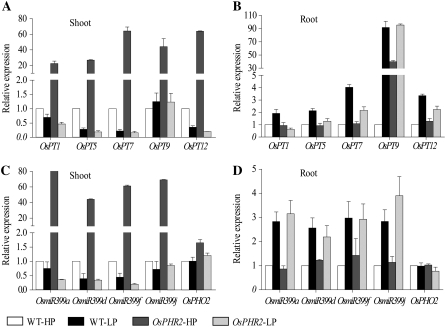
To investigate whether the Pi transport (PT) system is regulated by OsPHR2 for the excessive Pi accumulation in shoots of OsPHR2-overexpressing lines, we examined the expression of five members of rice high-affinity Pi transporters (PHT; Paszkowski et al., 2002). Shoot and root samples isolated at HP (10 mg Pi/L, 20 seeds/3 L) and LP (1 mg Pi/L, 20 seeds/3 L) levels were subjected to quantitative real-time PCR analysis (Fig. 9, A and B). In both shoots and roots of the wild type, all the examined members of the OsPHT family had detectable expression at HP levels. Under LP levels, induced expression of these genes was observed in roots of wild-type plants but not in the shoots. In OsPHR2-overexpressing plants, a remarkable up-regulation of these genes was observed at HP levels in shoots compared with wild-type plants, whereas in roots, only OsPT9 was up-regulated to about 40-fold higher than wild-type plants. Expression of these genes at HP levels was reduced in the roots of the OsPHR2-overexpressing plant except OsPT9. Thus, the enhanced expression of these OsPHT genes, especially OsPT9, in shoots and roots of an OsPHR2-overexpressing plant at HP levels may be associated with increased Pi translocation into shoots.
Figure 9.
Expression of five members of the rice PHT family, OsmiR399 family, and OsPHO2 in wild-type (WT) and OsPHR2-overexpressing plants under high and LP conditions. A and B, Effect of overexpression of OsPHR2 on transcript levels of PHT family members in shoots (A) and roots (B) when grown at HP (10 mg/L Pi) and LP (1 mg/L Pi) levels compared with wild type. Results are shown for OsPT1, OsPT5, OsPT7, OsPT9, and OsPT12. Gene accession numbers, specific primers, and UPL probes are listed in Supplemental Table S2. C and D, Levels of miR399 PTs and OsPHO2 transcript in shoots (C) and roots (D) of OsPHR2-overexpressing plants grown at HP (10 mg/L Pi) and LP (1 mg/L Pi) levels compared with wild type. Results are shown for miR399a, miR399d, miR399f, miR399j, and OsPHO2. miR399 amplification are listed in Supplemental Table S3 and primers and UPL probes for OsPHO2 are listed in Supplemental Table S2. For all qRT-PCR analysis, relative expression levels are given after being normalized to the expression level of OsActin and compared with the expression level of wild type under HP conditions (represent as 1-fold). The results are shown as mean ± sd (n = 3).
OsPHR2 Positively Controls Expression of OsmiR399
In Arabidopsis, function loss of PHO2/UBC24 leads to a Pi toxicity phenotype similar to that which results from overexpression of microRNA399 (Fujii et al., 2005; Chiou et al., 2006). Based on the Pi-starvation-inducible nature of miR399 and its strong repression in a phr1 mutant, it is plausible to place miR399 and PHO2 in a branch of the Pi-signaling network downstream of PHR1 (Bari et al., 2006). To determine if this is the same case in rice, we checked the expression patterns of four OsmiR399 genes in wild-type and OsPHR2-overexpressing plants, which are also responsive to Pi starvation in rice. OsPHO2, the potential ortholog of AtPHO2 in rice (Bari et al., 2006), was also analyzed. We found that the expression of the four examined OsmiR399 (OsmiR399a, OsmiR399d, OsmiR399f, and OsmiR399j) primary transcripts in shoots of OsPHR2-overexpressing plants was dramatically increased at HP levels compared with wild-type plants (Fig. 9C). No significant differences were found in roots between wild-type and OsPHR2-overexpressing plants at either high or LP levels (Fig. 9D). Moreover, the OsPHO2 transcript abundance at the HP level was not changed between the OsPHR2-overexpressing plants and wild-type plants; i.e. no reciprocal expression of OsmiR399 and OsPHO2 was observed. These results suggest that between rice and Arabidopsis, there may be a different regulatory mechanism downstream of microRNA399 in controlling Pi homeostasis.
DISCUSSION
Specific Function of OsPHR2 in P Homeostasis in Rice
In rice, two PHR1-like genes were found based on the rice genome database search. They are quite similar in gene structure except that the last exon of OsPHR1 is divided into two exons in OsPHR2. This feature implies that OsPHR2 may be derived from OsPHR1 via an evolutionary event. Although both OsPHR1 and OsPHR2 are localized to the nucleus and have transcription activities in yeast, they differ in the regulation of some downstream genes in rice, and only OsPHR2-overexpressing plants showed Pi excessive accumulation in shoots, resulting in Pi toxicity. Different functions of AtPHR1 homologs suggest that the MYB-CC gene family may not be redundant among its members (Todd et al., 2004). High expression of OsPHR2 leads to increased Pi accumulation in shoot under Pi-sufficient conditions and is in accordance with recent studies in Arabidopsis by overexpression of AtPHR1 in wild type and in the phr1 mutant. This strongly suggests that OsPHR2 is a functional homology of AtPHR1, and the key component of P homeostasis regulatory network may conserve in both monocot and dicot species (Nilsson et al., 2007).
Altered Expression of PSI Genes and PHT Genes in OsPHR2-Overexpressing Plants
Based on studies of the transcriptional responses to P deprivation, an increasing number of genes have been identified in plant adaptation to Pi deficiency (Hammond et al., 2003; Wu et al., 2003; Misson et al., 2005). These PSI genes are involved in various processes such as transcriptional regulation, metabolic pathways, and ion transport, and many of them are activated by AtPHR1 through the conserved P1BS element presented in their promoters (Rubio et al., 2001). All the PSI genes selected in this study belong to the PHR1 regulon, which contains one or more P1BS elements in its 2,000-bp upstream region. The non-protein-coding gene AtIPS1, which belongs to the Mt4/TPSI1 family, is the target gene of AtPHR1 (Rubio et al., 2001). Its function in control of P homeostasis by inhibiting the cleavage of PHO2 by miR399 was recently identified (Franco-Zorrilla et al., 2007). OsIPS1 and OsIPS2 are two AtIPS1 homologs in rice and also have a rapid and specific response to P starvation (Wasaki et al., 2003; Hou et al., 2005), but their functions in rice remain unclear. In the OsPHR2-overexpressing plant, the Pi-starvation induction of both OsIPS1 and OsIPS2 increased in shoots and roots, and their expression was induced even under Pi-sufficient conditions. Although the functions of OsIPS1 and OsIPS2 were not identified, it could be suggested that they may have the same function as AtIPS1 because they also share the 23-nt nucleotide motif that was functional in AtIPS1 due to its complementarity with miR399, which is interrupted by a mismatched loop at the predicted miR399 cleavage site (Hou et al., 2005; Franco-Zorrilla et al., 2007). However, their inhibition in Pi accumulation in shoots through sequestering miR399 seems to be counter to this idea because of the substantial up-regulation of miR399 in shoots by overexpression of OsPHR2 under Pi-supplied conditions.
When plants encounter Pi limitation, scavenging systems can be activated to recover Pi from lipids. As a result, phospholipids are substituted to galacto- and sulfolipids for saving Pi reserves. In Arabidopsis, the expression of SQD2, a gene involved in the second step of sulfolipid biosynthesis, responded specifically to Pi starvation (Yu et al., 2002). The correspondence homolog of SQD2 in rice is also strongly induced under Pi starvation (Wang et al., 2006). In this work, this gene was highly expressed in both shoots and roots of OsPHR2-overexpressing plants even when Pi was sufficient. This result is an example of the bypassing of the Pi metabolism pathway activated in OsPHR2-overexpressing plants under Pi-sufficient conditions. Thus, the reduced use of Pi in cells may possibly lead to the accumulation of excess Pi in cytoplasts. Although in the RNAi lines of both OsPHR1 and OsPHR2, the induction of all the four PSI genes were repressed compared to wild type when Pi was deficient, no significant changes in Pi homeostasis and root system architecture were observed. These results may be due to the incomplete interference of the mRNA level of OsPHR1 and OsPHR2. And their remaining expression can still slightly induce the induction of OsIPS1 and OsIPS2 after a long period of Pi starvation (data not shown).
A total of 13 putative, high-affinity Pi transporters in rice have been identified, and they share 38.1% to 87.4% SI (Goff et al., 2002; Paszkowski et al., 2002). In this study, we selected five of the 13 members, OsPT1, OsPT5, OsPT7, OsPT9, and OsPT12 from each phylogenetic clade of these genes (Paszkowski et al., 2002). The expression of these genes was greatly up-regulated in shoots of OsPHR2-overexpressing plants under Pi sufficiency, but in the roots, they showed similar levels to wild-type plants, except OsPT9. Accumulation of OsPT9 was hardly detected in Pi-sufficient shoots and roots of wild-type plants, but a substantial increase was detected in Pi-sufficient shoots and roots of OsPHR2-overexpressing plants. Phylogenetic analysis showed that OsPT9 is most closely related to the Arabidopsis Pi transporters Pht1;8 and Pht1;9, which can be clearly phylogenetically separated from the other seven family members (Poirier and Bucher, 2002). In the pho2 mutant, the high expression of Pht1;8 and Pht1;9 under Pi-sufficient conditions contributed to the establishment of the pho2 leaf Pi accumulation phenotype; however, out of the 11 Arabidopsis PHT Pi transporters, only Pht1;8 and Pht1;9 were greatly changed in the pho2 mutant (Aung et al., 2006; Bari et al., 2006). In our case, all of the tested PHT transporter genes were up-regulated in Pi-sufficient shoots or both shoots and roots. They are possibly under the direct control of OsPHR2 in addition to regulation by OsPHO2, if this is also the case in rice. The transcript level of PHT transporters did not change markedly under Pi-deficient conditions.
The altered expression of these PSI genes and Pi transporters indicates that OsPHR2 plays an important role in the primary P-starvation signaling pathway in rice. It seems that the overexpression of OsPHR2 stimulated the Pi-starvation signaling and triggered the Pi-starvation responses even though plants were not deprived of Pi.
OsPHR2 Is Involved in Regulating Root Growth under Pi Deprivation
The phr1 mutants showed an impaired root-shoot growth ratio and a decrease in plant growth under Pi starvation conditions, which were specific for systematic Pi-starvation stress. But no effect of the mutations of PHR1 was observed for the Pi-starvation-induced proliferation of root hair, which is governed by local Pi status (Bates and Lynch, 1996; Rubio et al., 2001). In our observations, the OsPHR2-overexpressing plants showed higher sensitivity to Pi starvation with an enhanced induction rate of primary root and adventitious roots than wild-type plants, which strongly suggests that OsPHR2 is involved in root architectural alteration in response to Pi starvation. Moreover, OsPHR2 also affected root hair patterning, which is controlled by the local Pi-signaling pathway. Studies of mutants in Arabidopsis revealed that phospholipase D (PLD) is involved in root elongation during Pi limitation (Li et al., 2006). The possible genetic link between PHR1 and PLDs needs to be elucidated in further studies because the expression of PLDs increases in response to Pi limitation. It would be interesting to find OsPHR2 and its downstream genes integrated into the local Pi signal for root hair formation.
Though the ectopic expression of OsPHR2 caused excessive Pi accumulation in shoots, which retarded whole-plant growth when Pi was sufficient, the increased root growth response to Pi starvation could provide a potentially useful tool for engineering plants that require less Pi fertilizer. Our data indicated that the excessive Pi accumulation may result from enhanced expression of PTs in shoots driven by overexpression of OsPHR2 (Fig. 9). Another possible reason for excessive Pi accumulation may be due to the activation of metabolic bypasses favoring Pi-releasing reactions and impeding Pi-consuming reactions in the primary metabolism, which may be under the regulation of OsPHR2. In that case the increased Pi cannot be used efficiently and causes Pi toxicity inversely. Therefore, if OsPHR2 can be overexpressed only in roots and the Pi metabolism systems still remain normal in shoots, the enhanced Pi uptake by root may lead to better growth. In that case, a root-specific promoter would be implied.
Shoot Pi Transfer in Rice May Be Controlled by a PHO2-Independent Pathway
Recent research work with Arabidopsis found that the mutant pho2 has a similar Pi toxicity phenotype to that resulting from overexpression of microRNA399, which can recognize and cleave the 5′-untranslated region of the ubiquitin-conjugating E2 enzyme UBC24 mRNA (Fujii et al., 2005; Bari et al., 2006; Chiou et al., 2006). Based on the Pi-deprivation-inducible nature of miR399 and its strongly repressed expression in the Pi-deprived phr1 mutant, it is reasonable to place miR399 and PHO2 in a branch of the Pi-signaling network downstream of PHR1. In rice, Pi-regulated miR399 expression and PHO2 gene structure are also conserved, suggesting that the same signaling pathway may function in control of rice Pi homeostasis (Bari et al., 2006). Thus, we tested the primary transcripts of four members of OsmiR399 (OsmiR399a, OsmiR399d, OsmiR399f, and OsmiR399j) and the putative PHO2 homolog OsPHO2 (Os5g48390). The expression levels of all of the tested OsmiR399 members were up-regulated in Pi-sufficient shoots of OsPHR2-overexpressing plants compared with wild-type plants, but no major differences were found in Pi-deficient shoots or roots at either HP or LP levels between wild-type and OsPHR2-overexpressing plants. The Pi-inducible expression of OsmiR399 could be observed in roots of both wild-type and OsPHR2-overexpressing plants; in shoots, however, OsmiR399 expression could not be induced when Pi was low. This result is consistent with previously reported findings (Bari et al., 2006) when wild type was treated under LP conditions; the expression of miR399 in shoot was quite similar or even a little bit repressed compared to the expression under full Pi conditions, whereas in root the expression of miR399 remained at relative high induction.
Under LP levels, the expression of miR399 in OsPHR2-overexpressing plants could not be induced like it was under HP levels. There was an interesting correlation between the internal Pi status change and the expression level of miR399. In other words, the internal Pi content of the OsPHR2-overexpressing plant is similar to wild type, and a similar expression pattern for miR399 was found. This implies miR399 may interplay with internal Pi levels and OsPHR2 may not be the only regulator of miR399.
As for OsPHO2, the expected OsmiR399-mediated transcript degradation was not found; transcription abundance of OsPHO2 was stable despite the change in OsmiR399 levels. Our data suggest that OsPHR2 may positively regulate OsmiR399, but OsPHO2 may not be the target of OsmiR399 in controlling Pi homeostasis in rice; it is also possible that its activity may be controlled by another level of posttranscriptional regulation that needs to be further studied.
MATERIALS AND METHODS
Gene Cloning
Using the AtPHR1 protein sequence (NP_194590) as the query, a TBALSTN search was performed on the web pages of the NCBI to identify sequences containing AtPHR1 orthologs in the rice genomic database. This resulted in the identification of two cDNA clones AK063486 (designated as OsPHR1) and AK100065 (designated as OsPHR2). Full-length cDNAs were amplified from the leaf cDNA template of a Japonica variety of rice (Oryza sativa ‘Nipponbare’), using the primers at the end of the cDNA sequence, and then cloned into pUCm-T vector (Takara) for sequencing verification.
Sequence Extraction and Alignments
Gene structures for OsPHR1 and OsPHR2 were assembled by alignment of cDNAs with genomic sequences (chromosome 3 clone OSJNBa0052J20 and chromosome 7 PAC clone P0443H10) via Megalign program. Multiple sequence alignment of the MYB and the predicted CC conserved domains were conducted using the ClustalX 1.81 program (Thompson et al., 1997) with default multiple alignment parameters and viewed by GeneDoc 3.2 with default BLOSUM score. The phylogenetic analysis was carried out by the neighbor-joining method. The phylogenetic tree was constructed using PHYLIP version 3.5c (http://bioweb.pasteur.fr) with a bootstrap analysis of 100 resampling replications. The protein sequences for alignment were extracted from NCBI.
Construction of Overexpression and RNAi Vectors and Plant Transformation
The overexpression vector was constructed as follows. First, the cauliflower mosaic virus 35S promoter was subcloned between the EcoRI and SacI sites of pCAMBIA1301. Second, the poly(A) addition sequence of pea (Pisum sativum) small subunit of Rubisco E9 (Coruzzi et al., 1984) was inserted into the sites between HindIII and PstI. The resulting vector was named as 35S-pCAMBIA1301. And then the ORF of OsPHR1 and OsPHR2 were inserted into 35S-pCAMBIA1301 using XbaI and SmaI, KpnI and SalI, respectively.
For the RNAi construct, a 283-bp fragment of OsPHR1 and a 207-bp fragment of OsPHR2 were cloned in both orientations in 35S-pCAMBIA1301, separated by the second intron of NIR1 of maize (Zea mays) to form a hairpin structure. All the constructs were transformed into mature embryos developed from seeds of wild type (‘Nipponbare’) via Agrobacterium tumefaciens EHA105 as in the previously described method (Chen et al., 2003).
Subcellular Localization of OsPHR1 and OsPHR2
The mGFP5 fragment was obtained from the pCAMBIA1302 vector with SpeI and BstEII digestion, and then cloned into the pCAMBIA1390 vector, which has the ubiquitin promoter inserted into the PstI site. The coding regions of OsPHR1 and OsPHR2 without stop codon were cloned into the SpeI site with in-frame fused to mGFP5. The fusion sites were verified by sequencing, and the resulting constructs were purified and used for transient expression in onion (Allium spp.) epidermal cells via shotgun bombardment (PDS–1000/He; Bio-Rad). The GFP signal was visualized by an LSM 510 laser-scanning microscope (Zeiss).
Activation Domain Analysis
The Matchmaker yeast two-hybrid system (Clontech) was used to detect the activation domain in OsPHR1 and OsPHR2. The deduced amino acid sequences of these two genes and their divided fragments (OsPHR1-N [1–266 amino acids in N terminus], OsPHR1-C [last 147 amino acids in C terminus], OsPHR2-N [1–230 amino acids in N terminus], OsPHR2-C [last 170 amino acids in C terminus]) were inserted into pGBKT7 in-frame fused with the GAL4 DNA-BD. These fusion constructs were transformed into yeast strain AH109 and selected on the minimal medium SD/-Trp and SD/-Trp-His-A to examine the reporter gene expression. α-Galactosidase quantitative assay was performed by using the p-nitrophenyl-α-galactosidase method described in the Clontech Yeast Protocols Handbook. The interaction between the pGBKT7 (BD)-p53 and pGADT7 (AD)-SV40 large T-antigen (Matchmaker) served as a positive control, whereas the empty pGBKT7 (BD) vector was used as a negative control.
Hydroponic and Soil Pot Experiments
Hydroponic experiments were conducted using normal rice culture solution with 10 mg Pi L−1 (Yoshida et al., 1976) and Pi-deficient solution (0.5 mg Pi L−1). For Pi gradient experiments another two solutions with 5 mg Pi L−1 and 1 mg Pi L−1 were added. The pH of the culture solution was adjusted to 5.0 using 1 m NaOH and checked every week. In all the hydroponic experiments, seeds were directly grown in each kind of solution culture (3 L) after being germinated in water under a photosynthetic photon flux density of approximately 200 μmol photons m−2 s−1 with a 12-h light (30°C)/12-h dark (28°C) photoperiod. Humidity was controlled at approximately 70%; 10-d-old seedlings were observed for phenotype or transferred to a corresponding solution culture (0.5 L plant−1) for further growth; 30-d-old seedlings were observed for phenotype or sampled for Pi and total P concentration measurement. Soil pot experiments were conducted using fine-clay acid red soil (pH 4.3; water:soil, 1:1, fine-clay kaolinitic thermic typic plinthudults), with three Pi-supplied levels (use KH2PO4 as Pi source): 200 mg Pi/kg soil (Bray-II method, Pi = 38.2 mg/kg soil), 120 mg Pi/kg soil (Bray-II method, Pi = 19.89 mg/kg soil), and 60 mg Pi/kg soil (Bray-II method, Pi = 8.15 mg/kg soil). Each pot contained 8 kg of air-dried soil with two plants; 4-d-old seedlings of wild-type and OsPHR2 overexpressing plants were transplanted in pots after being germinated in water. The pots were randomly arranged with three replications in each treatment (six plants per treatment) and grown in a greenhouse for the whole growth period.
Measurements of Total P and Pi Content in Plants and Soils
Samples were frozen after fresh weight measurement or dried at 80°C for 3 d to determine the dry weight. Inorganic Pi measurement followed the previously described method (Nanamori et al., 2004). A frozen sample was homogenized in 1 mL 10% (w/v) of perchloric acid, using an ice-cold mortar and pestle. The homogenate was then diluted 10 times with 5% (w/v) perchloric acid and placed on ice for 30 min. After centrifugation at 10,000g for 10 min at 4°C, the supernatant was used for Pi measurement via the molybdenum blue method: 0.4% (w/v) ammonium molybdate melted in 0.5 m H2SO4 (solution A) was mixed with 10% ascorbic acid (solution B; A:B = 6:1). Two milliliters of this work solution was added to 1 mL of the sample solution, and incubated in a water bath at 42°C for 20 min. After being cooled on ice, the absorbance was measured at 820-nm wavelength. Pi concentration was calculated by normalization of fresh weight. Plant total P content was analyzed by the molybdenum blue method after digested with H2SO4-H2O2 at 300°C, and normalized by dry weight. Soil Pi concentration was determined by the Bray-II method according to the previously described method (Bray and Kurtz, 1945) and the data was listed in Supplemental Table S1.
Southern-Blot Analysis of Transgenic Plants
Genomic DNA was isolated using the SDS method (Murray and Thompson, 1980) and was digested with two restriction enzymes EcoRI and HindIII. Five micrograms of digested DNA were separated on 0.8% agarose gel. After electrophoresis, the digested DNA was transferred to Hybond-N+ nylon membrane (Amersham Pharmacia) and hybridized with a 32P-dCTP-labeled hygromycin-resistant gene probe. The blots were washed at 65°C under stringent conditions and analyzed using Typhoon-8600.
Semiquantitative RT-PCR
Total RNA was extracted using the Trizol D0410 reagent, according to the manufacturer's instructions (Invitrogen). The first-strand cDNA was synthesized from 5 μg of DNaseI-treated total RNA using SuperScript II reverse transcriptase (Invitrogen). Semiquantitative RT-PCR was performed using the gene-specific primers designed by PRIMEREXPRESS software (Applied Biosystems) and listed in Supplemental Table S2. The PCR products were loaded on 1% agarose gels and imaged by a CCD camera. The gene expression levels were compared by the band density after normalization of initial variation in sample concentration by the density of the housekeeping gene Actin.
Real-Time qRT-PCR Analysis
Real-time qRT-PCR was performed by using the FastStart DNA Master SYBR Green I Kit or the Universal Probe Library (UPL) and LightCycler 480 Probes Master Kit on the LightCycler480 machine (Roche), following the manufacturer's instructions. The amplification program for SYBR Green I was performed at 95°C for 10 s, 58°C for 10 s, and 72°C for 20 s. The program for UPL was performed at 95°C for 10 s, 60°C for 25 s, and 72°C for 1 s. Triplicate quantitative assays were performed on each cDNA sample. The relative quantification of each sample was determined by normalization to the amount of actin cDNA detected in the same sample. Relative expression level was calculated by the formula 2−Δ(ΔCp). The primers for OsmiR399 were the same as Bari et al. (2006) described and listed in Supplemental Table S3. The gene-specific primers for OsPTs and the corresponding UPL probe were designed using the online analysis center of the Roche Web site (http://www.universalprobelibrary.com) and given in Supplemental Table S4.
Statistical Analysis of Data
A Student's t test was performed for mean comparisons of plant growth parameters and Pi content between wild-type and transgenic plants using the algorithm incorporated into the Excel software program (Microsoft).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alignment of the MYB (A) and predicted CC (B) conserved domains constructed by use of the ClustalX 1.81 program (Thompson et al., 1997) and colored by use of the GeneDoc 3.2 program with default BLOSUM score.
Supplemental Figure S2. Growth performance of 45-d-old wild-type (WT) and one line of OsPHR2-overexpressing plants (OsPHR2-Ov-1) in a pot experiment using acidic red soil supplied with three Pi levels.
Supplemental Figure S3. Growth parameters measured from wild-type (white column) and OsPHR2-Ov-1 (black column) plants in the soil pot experiment with four Pi levels.
Supplemental Figure S4. Root hair proliferation of OsPHR1-RNAi and OsPHR2-RNAi lines.
Supplemental Table S1. Primers of PSI genes used for RT-PCR analysis.
Supplemental Table S2. Primers of Pi transporter genes used for qRT-PCR analysis.
Supplemental Table S3. Primers of OsmiRNA399 and OsPHO2 used for qRT-PCR analysis.
Supplementary Material
This work was supported by the National Basic Research and Development Program of China (2005CB120900) and the China Rice Functional Genomics Program.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: P. Wu (clspwu@zju.edu.cn).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Aung K, Lin SI, Wu CC, Huang YT, Su CL, Chiou TJ (2006) pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiol 141 1000–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R, Datt Pant B, Stitt M, Scheible WR (2006) PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol 141 988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates TR, Lynch JP (1996) Stimulation of root hair elongation in Arabidopsis thaliana by low phosphorus availability. Plant Cell Environ 19 529–538 [Google Scholar]
- Bray RH, Kurtz LT (1945) Determination of total, organic, and available forms of phosphorus in soils. Soil Sci 59 39–45 [Google Scholar]
- Chen SY, Jin WZ, Wang MY, Zhang F, Zhou J, Jia QJ, Wu YR, Liu FY, Wu P (2003) Distribution and characterization of over 1000 T-DNA tags in rice genome. Plant J 36 105–113 [DOI] [PubMed] [Google Scholar]
- Chiou TJ, Aung K, Lin SI, Wu CC, Chiang SF, Su CL (2006) Regulation of phosphate homeostasis by microRNA in Arabidopsis. Plant Cell 18 412–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coruzzi G, Broglie R, Edwards C, Chua NH (1984) Tissue-specific and light-regulated expression of a pea nuclear gene encoding the small subunit of ribulose-1,5-bisphosphate carboxylase. EMBO J 3 1671–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essigmann B, Guler S, Narang RA, Linke D, Benning C (1998) Phosphate availability affects the thylakoid lipid composition and the expression of SQD1, a gene required for sulfolipid biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 95 1950–1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Gonzalez E, Bustos R, Linhares F, Leyva A, Paz-Ares J (2004) The transcriptional control of plant responses to phosphate limitation. J Exp Bot 55 285–293 [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, Garcia JA, Paz-Ares J (2007) Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet 39 1033–1037 [DOI] [PubMed] [Google Scholar]
- Fujii H, Chiou TJ, Lin SI, Aung K, Zhu JK (2005) A miRNA involved in phosphate-starvation response in Arabidopsis. Curr Biol 15 2038–2043 [DOI] [PubMed] [Google Scholar]
- Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296 92–100 [DOI] [PubMed] [Google Scholar]
- Hammond JP, Bennett MJ, Bowen HC, Broadley MR, Eastwood DC, May ST, Rahn C, Swarup R, Woolaway KE, White PJ (2003) Changes in gene expression in Arabidopsis shoots during phosphate starvation and the potential for developing smart plants. Plant Physiol 132 578–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou XL, Wu P, Jiao FC, Jia QJ, Chen HM, Yu J, Song XW, Yi KK (2005) Regulation of the expression of OsIPS1 and OsIPS2 in rice via systemic and local Pi signalling and hormones. Plant Cell Environ 28 353–364 [Google Scholar]
- Li M, Qin C, Welti R, Wang X (2006) Double knockouts of phospholipases Dzeta1 and Dzeta2 in Arabidopsis affect root elongation during phosphate-limited growth but do not affect root hair patterning. Plant Physiol 140 761–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkohr BI, Williamson LC, Fitter AH, Leyser HM (2002) Nitrate and phosphate availability and distribution have different effects on root system architecture of Arabidopsis. Plant J 29 751–760 [DOI] [PubMed] [Google Scholar]
- Misson J, Raghothama KG, Jain A, Jouhet J, Block MA, Bligny R, Ortet P, Creff A, Somerville S, Rolland N, et al (2005) A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc Natl Acad Sci USA 102 11934–11939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Rus A, Sharkhuu A, Yokoi S, Karthikeyan AS, Raghothama KG, Baek D, Koo YD, Jin JB, Bressan RA, et al (2005) The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc Natl Acad Sci USA 102 7760–7765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8 4321–4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanamori M, Shinano T, Wasaki J, Yamamura T, Rao IM, Osaki M (2004) Low phosphorus tolerance mechanisms: phosphorus recycling and photosynthate partitioning in the tropical forage grass, Brachiaria hybrid cultivar Mulato compared with rice. Plant Cell Physiol 45 460–469 [DOI] [PubMed] [Google Scholar]
- Nilsson L, Muller R, Nielsen TH (2007) Increased expression of the MYB-related transcription factor, PHR1, leads to enhanced phosphate uptake in Arabidopsis thaliana. Plant Cell Environ 30 1499–1512 [DOI] [PubMed] [Google Scholar]
- Paszkowski U, Kroken S, Roux C, Briggs SP (2002) Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci USA 99 13324–13329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier Y, Bucher M (2002) Phosphate transport and homeostasis in Arabidopsis. In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, doi: 10.1199/tab.0009, www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed]
- Raghothama KG (1999) Phosphate acquisition. Annu Rev Plant Physiol Plant Mol Biol 50 665–693 [DOI] [PubMed] [Google Scholar]
- Rubio V, Linhares F, Solano R, Martin AC, Iglesias J, Leyva A, Paz-Ares J (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15 2122–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachtman DP, Shin R (2007) Nutrient sensing and signaling: NPKS. Annu Rev Plant Biol 58 47–69 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd CD, Zeng P, Huete AM, Hoyos ME, Polacco JC (2004) Transcripts of MYB-like genes respond to phosphorous and nitrogen deprivation in Arabidopsis. Planta 219 1003–1009 [DOI] [PubMed] [Google Scholar]
- Wang XM, Yi KK, Tao Y, Wang F, Wu ZC, Jiang DA, Chen X, Zhu LH, Wu P (2006) Cytokinin represses phosphate-starvation response through increasing of intracellular phosphate level. Plant Cell Environ 29 1924–1935 [DOI] [PubMed] [Google Scholar]
- Wasaki J, Yonetani R, Shinano T, Kai M, Osaki M (2003) Expression of the OsPI1 gene, cloned from rice roots using cDNA microarray, rapidly responds to phosphorus status. New Phytol 158 239–248 [Google Scholar]
- Williamson LC, Ribrioux SP, Fitter AH, Leyser HM (2001) Phosphate availability regulates root system architecture in Arabidopsis. Plant Physiol 126 875–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissuwa M (2003) How do plants achieve tolerance to phosphorus deficiency? Small causes with big effects. Plant Physiol 133 1947–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Ma LG, Hou XL, Wang MY, Wu YR, Liu FY, Deng XW (2003) Phosphate starvation triggers distinct alterations of genome expression in Arabidopsis roots and leaves. Plant Physiol 132 1260–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi K, Wu Z, Zhou J, Du L, Guo L, Wu Y, Wu P (2005) OsPTF1, a novel transcription factor involved in tolerance to phosphate starvation in rice. Plant Physiol 138 2087–2096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Forno DA, Cock JH, Gomez KA (1976) Laboratory Manual for Physiological Studies of Rice, Ed 3. The International Rice Research Institute, Manila, The Philippines
- Yu B, Xu C, Benning C (2002) Arabidopsis disrupted in SQD2 encoding sulfolipid synthase is impaired in phosphate-limited growth. Proc Natl Acad Sci USA 99 5732–5737 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.