Abstract
Superoxide dismutases (SODs) are ubiquitous metalloenzymes that catalyze the dismutation of superoxide radicals. Chloroplasts have two isozymes, copper/zinc SOD (Cu/ZnSOD) and iron SOD (FeSOD), encoded by nuclear genes. Because bryophytes are considered as the earliest land plants, they are one of the most interesting plant models for adaptation against oxidative stress. In a previous study, we found that the FeSOD gene was expressed under Cu-deficient conditions and repressed under high-Cu-supply conditions; on the other hand, the Cu/ZnSOD gene was induced by Cu in a moss, Barbula unguiculata. The expression of Cu/ZnSOD and FeSOD is coordinately regulated at the transcriptional level depending on metal bioavailability. Here, using transgenic moss plants, we determined that the GTACT motif is a negative cis-acting element of the moss FeSOD gene in response to Cu. Furthermore, we found that a plant-specific transcription factor, PpSBP2 (for SQUAMOSA promoter-binding protein), and its related proteins bound to the GTACT motif repressed the expression of the FeSOD gene. The moss FeSOD gene was negatively regulated by Cu in transgenic Nicotiana tabacum plants, and the Arabidopsis thaliana FeSOD gene promoter containing the GTACT motif was repressed by Cu. Our results suggested that molecular mechanisms of GTACT motif-dependent transcriptional suppression by Cu are conserved in land plants.
Reactive oxygen species (ROS), such as superoxide and hydrogen peroxide (H2O2), are constantly produced during metabolic processes in all living species. Excessive ROS accumulation leads to cellular injury, such as damage to DNA, protein, and the lipid membrane (Mittler, 2002). Because of their potential harmful effects, excessive ROS must be promptly eliminated from the cells by a variety of antioxidant defense mechanisms, including superoxide dismutase (SOD), catalase, and various peroxidases.
SODs are ubiquitous metalloenzymes that catalyze the dismutation of superoxide radicals to H2O2 and molecular oxygen (2O2− + 2H+ → H2O2 + O2; Mittler et al., 2004). Plants possess three types of SODs, classified according to their prosthetic metal: iron SOD (FeSOD), manganese SOD (MnSOD), and copper/zinc SOD (Cu/ZnSOD). FeSOD and MnSOD are likely to have arisen from a common ancestor, whereas Cu/ZnSOD appears to have evolved separately, because it exhibits only limited sequence similarity to FeSOD and MnSOD (Steinman and Hill, 1973; Stallings et al., 1983; Alscher et al., 2002; Fink and Scandalios, 2002). FeSOD is considered to be the most primitive enzyme, because it occurs in anaerobic bacteria. Many algae contain FeSOD but lack Cu/ZnSOD; however, evolved green algae, Charales and Conjugales, do have Cu/ZnSOD (Asada et al., 1977; Kanematsu and Asada, 1989). The SOD isoforms differ in their subcellular location; FeSODs are generally located in chloroplasts, MnSODs are located in mitochondria and peroxisomes, and Cu/ZnSODs are found in the cytosol, chloroplasts, peroxisomes, and apoplasts (Alscher et al., 2002).
When life was first forming on the earth, oxygen in the atmosphere was almost nonexistent. Fe was probably the first metal used as a metal cofactor at the active site of the ancient SOD because of an abundance of Fe in a soluble Fe (II) form at that time. As the levels of O2 in the environments increased by oxygenic photosynthesis, the mineral components in the biosphere were oxidized. The decrease in soluble Fe (II) in the biosphere caused a shift to the use of a more available metal, Mn (III), in the evolution of detoxification. When the atmosphere was completely replenished with oxygen, Fe (II) was almost completely unavailable, and insoluble Cu (I) in the ancient Earth was converted into soluble Cu (II). At this stage, Cu (II) began to be used as the metal cofactor at the active sites of SODs (Egami, 1975; Lumsden and Hall, 1975; Asada et al., 1977; Alscher et al., 2002).
Because bryophytes are considered as the earliest land plants (Waters, 2003), they are one of the most interesting plant models for adaptation against oxidative stress. Bryophytes contain three classes: liverworts, hornworts, and mosses. The liverwort Marchantia paleacea var diptera has only FeSOD in chloroplasts (Tanaka et al., 1995), but the moss Barbula unguiculata, which has been placed closer to vascular plants than to liverworts in the phylogenetic tree (Kenrick and Crane, 1997; Wellman et al., 2003), has both FeSOD and Cu/ZnSOD in chloroplasts (Yamahara et al., 1999), as do most vascular plants. Depending on metal bioavailability, various organisms coordinately regulate the alternative use of Fe- versus Cu-containing SODs with completely different apoproteins.
We previously found that chloroplastic FeSOD was replaced with Cu/ZnSOD when Cu was available in the moss B. unguiculata (Shiono et al., 2003). The expression of Cu/ZnSOD was repressed in a Cu-deficient medium and induced by the addition of Cu. On the other hand, FeSOD activity was increased under Cu-deficient conditions and decreased in a Cu-containing medium. This coordinated regulation of SODs by Cu has also been observed in the fern Matteuccia struthiopteris (Murao et al., 2004) and higher plants such as tobacco (Nicotiana tabacum; Kurepa et al., 1997) and Arabidopsis (Arabidopsis thaliana; Abdel-Ghany et al., 2005b). However, the molecular mechanisms of transcriptional regulation by Cu are still largely unknown.
In this study, we identified the negative cis-acting element, a GTACT motif, responsible for Cu-responsive transcriptional repression of the moss FeSOD gene. The DNA fragment containing GTACTs conferred the actin promoter with a remarkable property of Cu responsiveness. The cis-acting element of moss is also functional in higher plants. Furthermore, overexpression of a transcription factor that binds to the GTACT motif repressed the expression of the FeSOD gene in transgenic moss plants. These results suggested that the molecular mechanisms for transcriptional regulation by Cu are ancient and evolutionally conserved.
RESULTS
FeSOD Gene Is Transcriptionally Regulated by Cu
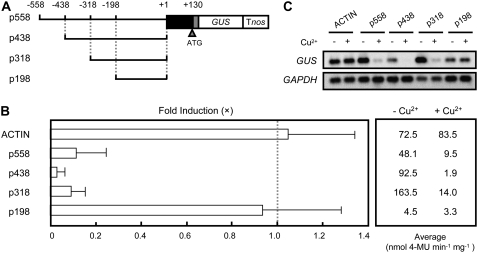
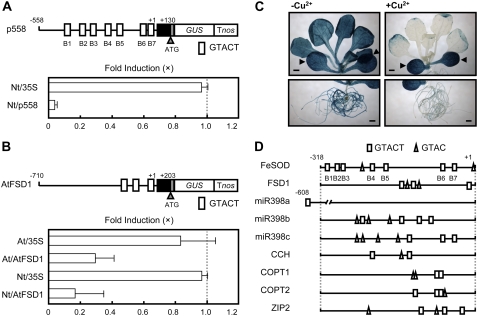
To investigate the molecular mechanisms of transcriptional control of the FeSOD gene of B. unguiculata by Cu, we cloned the promoter region of the FeSOD gene (GenBank accession no. AB370198) by thermal asymmetric interlaced (TAIL)-PCR. The 5′ RACE analysis showed that the transcription start site of FeSOD was 122 bp upstream of the translation initiation site (data not shown). To define the transcriptional regulatory region of the FeSOD promoter by Cu, we generated transgenic moss plants (Physcomitrella patens subsp. patens) carrying a series of 5′ deletions of the FeSOD promoter fused to a GUS reporter gene (Fig. 1A). We examined the effect of Cu on the expression of the FeSOD promoter-GUS fusion genes in transgenic moss plants. The application of Cu decreased the GUS activities in the p558 construct that contains from −558 to +130 nucleotides, showing that the fusion gene was under the control of Cu (Fig. 1B). Other 2.0 μm metals, including Zn2+, Co2+, Mn2+, Ni2+, and Fe2+, had only a limited effect on the expression of p558 (data not shown), suggesting that down-regulation of FeSOD is specific to Cu. The control rice (Oryza sativa) actin promoter was not regulated by Cu treatment. Deletion of 240 bp from −558 to −319 (p318) of the FeSOD promoter did not affect the responsiveness to Cu. Further deletion of 120 bp from −318 to −199 (p198) resulted in a loss of repression by Cu. We confirmed these results at the mRNA levels by reverse transcription (RT)-PCR. Consistent with the GUS activities, the GUS mRNAs were repressed by Cu in p558, p438, and p318 but not in p198 (Fig. 1C). Therefore, there should be one or more cis-elements responsible for the repression by Cu between −318 and −199 in the FeSOD promoter.
Figure 1.
Identification of cis-regions for Cu responsiveness. A, Schematic diagram of constructs. A plasmid construct containing the 5′ flanking region (solid line), 5′ untranslated region (black box), and the coding region (gray box; three amino acids) of FeSOD connected to the GUS gene was introduced into P. patens subsp. patens by polyethylene glycol-mediated transformation. Nucleotide positions are indicated in base pairs from the transcription initiation site. Tnos, Nopaline synthase terminator. B, GUS activities of the transgenic moss plants treated with or without CuSO4. Left, The repression of the GUS activities by Cu is indicated as the relative value. The bars represent the GUS activities of transgenic moss plants cultured in the presence of 2.0 μm CuSO4. The GUS activities of the transgenic plants cultured in the absence of CuSO4 were arbitrarily set to 1.0 (dotted line). For each construct, at least three independent transgenic plants were examined. The bars indicate ± sd. Right, The average GUS activities are expressed in nanomoles of 4-methylumbelliferone (4-MU) produced per milligram of extract protein per minute. The GUS activities of transgenic moss plants containing the promoterless-GUS construct were below 2 × 10−2 nmol 4-MU min−1 mg−1 and were not affected by Cu (data not shown). C, RT-PCR analysis of GUS gene expression in transgenic moss plants cultured with (+) or without (−) CuSO4. After amplification with specific primers, the products were detected by DNA gel-blot hybridization. GAPDH was used as an internal control of RT-PCR.
Multiple cis-Elements Are Located between −318 and −199
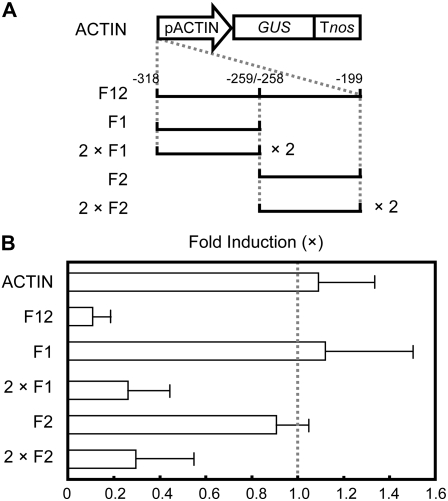
To examine the function of the 120-bp sequence between −318 and −199 in Cu responsiveness, we carried out a gain-of-function experiment. The 120-bp DNA fragment of the FeSOD promoter was cloned upstream of the constitutive rice actin promoter (F12; Fig. 2A). The 120-bp DNA fragment endowed the reporter with a remarkable property of Cu responsiveness (Fig. 2B), suggesting that the sequences are sufficient for transcriptional repression by Cu. To further define the cis-elements for the Cu responsiveness of the FeSOD promoter, the function of two dissected DNA fragments of the 120-bp sequence was examined using an actin promoter. The Cu-dependent repression of GUS activities was not observed with the 60-bp DNA fragment between −318 and −259 (F1) or between −258 and −199 (F2), whereas Cu responsiveness was detected with the tandem repeats of the 60-bp DNA fragment between −318 and −259 (2 × F1) and that between −258 and −199 (2 × F2), respectively (Fig. 2, A and B). These results suggested that both F1 and F2 contain the Cu-responsive element and that multiple copies of the cis-element are necessary for Cu-responsive transcriptional repression.
Figure 2.
Gain-of-function analysis for Cu responsiveness in transgenic moss plants. A, Schematic diagram of constructs. The fragments of the FeSOD promoter were inserted into upstream of rice actin promoter (pACTIN). F12 contains 120-bp DNA (−318 to −199). F1 and F2 contain 60-bp DNAs from −318 to −259 and from −258 to −199, respectively. 2 × F1 and 2 × F2 contain two tandem copies of 60-bp fragments (−318 to −259 or −258 to −199), respectively. B, GUS activities of the transgenic moss plants treated with or without CuSO4. The bars represent the GUS activities of transgenic moss plants cultured in the presence of 2.0 μm CuSO4. The GUS activities of the transgenic plants cultured in the absence of CuSO4 were arbitrarily set to 1.0 (dotted line). The actin promoter-GUS (ACTIN) was used as a control. For each construct, at least three independent transgenic plants were examined. The bars indicate ± sd.
GTACT Is Necessary for Cu-Responsive Repression
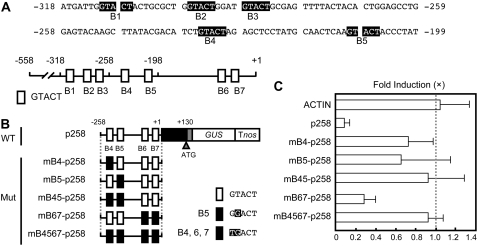
When we analyzed the DNA sequences of the FeSOD promoter, five copies of GTACT sequences were found between −318 and −199 (Fig. 3A). Two additional GTACT sequences are found between −198 and +1. Because the p198 construct that contains two copies of GTACT sequences did not respond to Cu (Fig. 1), we hypothesized that multiple copies of GTACT sequences are necessary for the transcriptional repression of the FeSOD promoter by Cu. If this is correct, the expression of a p258 construct that contains four copies of GTACT (Fig. 3B) might be repressed by Cu. As we expected, the GUS activities decreased with an application of Cu in p258 transgenic moss plants (Fig. 3C).
Figure 3.
The GTACT motif is required for Cu responsiveness. A, Sequences of the region (−318 to −199) that are involved in Cu responsiveness. The GTACT sequences are printed in white on black. The bottom diagram represents the location of GTACT sequences. GTACTs are shown as boxes (B1–B7) and numbered from the 5′ end of the FeSOD promoter. B, Mutagenesis of GTACT motifs. The mutated GTACT sequences are indicated by black boxes in the schematic diagram of constructs. The mutated bases in B4 to B7 are highlighted. Wild type (WT) containing −258 to +130 was used as a positive control. C, Effect of mutations in GTACT motifs on Cu responsiveness. The bars represent the GUS activities of transgenic moss plants cultured in the presence of 2.0 μm CuSO4. The GUS activities of the transgenic plants cultured in the absence of CuSO4 were arbitrarily set to 1.0 (dotted line). The actin promoter-GUS (ACTIN) was used as a control. For each construct, at least three independent transgenic plants were examined. The bars indicate ± sd.
To examine the functional importance of GTACT sequences on the Cu-responsive transcriptional repression of the FeSOD promoter, we constructed mutant versions of p258 in which mutations were introduced to four GTACT sequences (Fig. 3B). Although the mutations in B6 and B7 (mB67-p258) did not abolish the Cu responsiveness, those in B4 (mB4-p258) or B5 (mB5-p258) apparently reduced Cu-responsive transcriptional repression in the transgenic moss plants (Fig. 3C). The Cu responsiveness was completely eliminated by the mutations in B4 and B5 (mB45-p258) and by those in B4, B5, B6, and B7 (mB4567-p258). These results indicated that multiple copies of GTACT sequences are necessary for the maximum transcriptional repression of the FeSOD promoter in response to Cu and that B4 and B5 play important roles in the regulation.
Expression Pattern of FeSOD
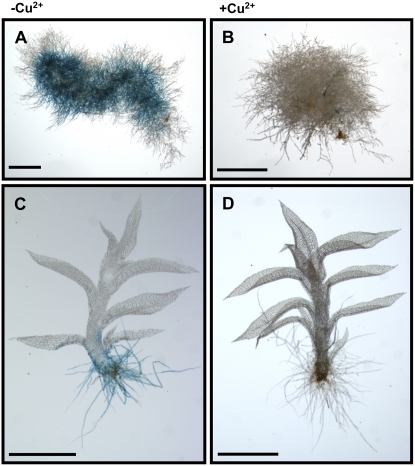
To examine the spatial and temporal pattern of FeSOD expression, p558 transgenic plants were analyzed by histochemical staining of GUS activities. GUS activities were detected in protonema before the development of the gametophore (Fig. 4A) and in the rhizoids except for the gametophore (Fig. 4C) under Cu-deficient conditions. The application of Cu resulted in the loss of GUS activities in whole plants (Fig. 4, B and D), indicating that the molecular mechanisms of Cu-responsive transcriptional repression of FeSOD gene were operative in all tissues in which the FeSOD gene was expressed in P. patens.
Figure 4.
Histochemical analysis of p558 transgenic moss plants cultured without or with CuSO4. Transgenic moss plants precultured with 0.2 μm CuSO4 for 7 d were treated without (A and C) or with (B and D) 2.0 μm CuSO4 for 3 d. More than six independent transgenic moss plant lines were investigated. A and B, Protonema cells; C and D, gametophore. Scale bars, 1 mm.
Transcription Factor PpSBP2 Regulates the Expression of FeSOD
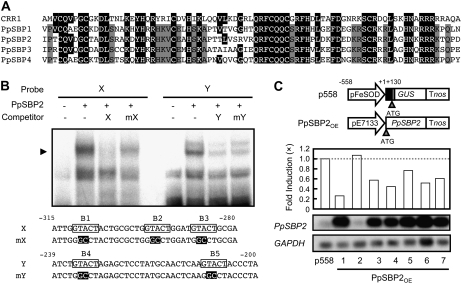
The unicellular green alga Chlamydomonas reinhardtii activates the transcription of the Cyc6 and the Cpx1 genes (encoding cytochrome c6 and coprogen oxidase) in response to Cu deficiency to replace the Cu-dependent function of the plastocyanin function by a heme-containing cytochrome (Merchant and Bogorad, 1986; Hill et al., 1991; Hill and Merchant, 1995; Quinn et al., 1999). The GTAC sequences of the promoters are critical for Cu responsiveness in C. reinhardtii (Quinn et al., 2000). The COPPER RESPONSE REGULATOR1 (CRR1) is a putative transcription factor that is required for the activation of the Cyc6 and Cpx1 genes under Cu-deficient conditions of C. reinhardtii, with a SQUAMOSA promoter-binding protein (SBP) domain, ankyrin repeats, and a C-terminal Cys-rich region (Kropat et al., 2005). Furthermore, the consensus DNA sequence bound by the SBP domain contains a GTAC core (Birkenbihl et al., 2005). These observations prompted us to examine whether a transcription factor with an SBP domain of moss is involved in the Cu-responsive transcriptional repression of the FeSOD gene.
When we started our work, the registered P. patens proteins with an SBP domain were PpSBP1 to 4. The amino acid sequence of these proteins exhibited high similarities in the SBP domain of C. reinhardtii CRR1 (Fig. 5A). In contrast, the similarities of the amino acid sequences were limited in regions outside the SBP domain for these sequences. We examined the expression of PpSBP1 to 4 by RT-PCR. Although these genes were expressed in moss plants, their expression was not affected by the application of Cu (data not shown). Because neither the PpSBP2 gene nor the Crr1 gene has an intron within the DNA encoding the SBP domain, while other PpSBP genes carry an intron at the conserved position within the SBP domain, PpSBP2 was chosen for further analysis. We examined whether PpSBP2 binds to the cis-regulatory sequences for the Cu-responsive transcriptional repression of the FeSOD gene. A gel retardation assay showed that the PpSBP2 protein prepared by in vitro translation formed a specific complex with both the 32P-labeled fragment X (−315 to −280) and Y (−239 to −200) containing GTACT sequences (Fig. 5B). To determine whether the GTACT sequences are important for binding to PpSBP2, we used mutant variants of fragments X and Y as competitors for PpSBP2 binding in gel shift assays. The formation of a complex of the probes with PpSBP2 was not efficiently inhibited in the presence of an excess amount of the mutated sequences. These results indicated that GTACT sequences are important for PpSBP2 binding.
Figure 5.
Binding of transcription factor PpSBP2 to the GTACT motif. A, Alignment of amino acid sequences of the SBP domain of CRR1 and PpSBPs. The highlighted residues indicate amino acids that are identical to those of CRR1. The gray residues indicate amino acids that are identical among PpSBPs. B, Gel retardation assay with in vitro-translated PpSBP2. The specific PpSBP2-DNA complexes are indicated by an arrowhead. Oligonucleotides containing the GTACT motif were used as the probes. The GTACT motif is boxed, and the mutated bases are highlighted. +, Present; −, absent. C, The overexpression of PpSBP2 repressed the expression of the FeSOD gene. Top, Schematic diagram of constructs. The p558 is the reporter construct containing −558 to +130 of the FeSOD gene. The effector construct PpSBP2OE expresses PpSBP2 cDNA under the control of the constitutive pE7133 promoter. Middle, Effect of overexpression PpSBP2 on GUS activities of p558. The bars represent the GUS activities of transgenic moss plants cultured without CuSO4. The GUS activities of the mother transgenic lines containing only the reporter construct were arbitrarily set to 1.0 (dotted line). Bottom, RT-PCR analysis of PpSBP2 expression in transgenic moss plants. After amplification with specific primers, the products were detected by DNA gel-blot hybridization. GAPDH was used as an internal control of RT-PCR.
To investigate the function of PpSBP2 in the Cu-responsive transcriptional repression of the FeSOD gene, PpSBP2 was overexpressed under the control of the constitutive pE7133 promoter derived from the cauliflower mosaic virus (CaMV) promoter (Mitsuhara et al., 1996) in transgenic P. patens carrying the p558 construct (Fig. 5C, top). Because the mutations in the GTACT sequences eliminated both the Cu-responsive repression of the FeSOD promoter (Fig. 3) and PpSBP2 binding (Fig. 5), we anticipated that the overexpression of PpSBP2 would decrease the expression of the p558 construct in the moss plants. As expected, GUS activities were reduced by the overexpression of PpSBP2 in the absence of Cu (Fig. 5C, middle), suggesting that PpSBP2 and PpSBP2-related proteins function as transcriptional repressors of the FeSOD promoter.
The Mechanisms of Cu-Responsive Repression Are Conserved in Higher Plants
To investigate whether the mechanisms of Cu-responsive transcriptional repression are conserved in higher plants, we generated transgenic tobacco plants carrying the moss p558 construct (Nt/p558; Fig. 6A). The application of Cu reduced the GUS activities of transgenic tobacco plants, suggesting that GTACT sequences are also functional in the transcriptional repression by Cu in higher plants. It has been reported that the expression of the chloroplastic FeSOD genes of Arabidopsis (Abdel-Ghany et al., 2005b) and tobacco (Kurepa et al., 1997) is repressed by Cu at the transcriptional level. Three copies of GTACT sequences were found within −200 bp of the promoter of the Arabidopsis chloroplastic FeSOD gene FSD1, whereas the chloroplastic Cu/ZnSOD gene CSD2, which is not regulated by Cu, has no GTACT sequences within −1,000 bp of the promoter region. To examine whether the Arabidopsis FSD1 promoter is regulated by Cu, we generated transgenic Arabidopsis and tobacco plants carrying the fusion gene of the FSD1 promoter-GUS (Fig. 6B). Cu decreased GUS activities in the transgenic Arabidopsis and tobacco plants, showing that Arabidopsis FSD1 is regulated by Cu in the same way as the moss FeSOD gene. These results suggested that the molecular mechanisms of Cu-responsive transcriptional suppression are conserved between bryophytes and higher plants.
Figure 6.
The mechanisms for Cu responsiveness are conserved in higher plants. A, The moss FeSOD promoter was repressed by Cu in the transgenic tobacco plants. Top, Schematic diagram of the p558 construct. The white boxes represent GTACT sequences. Bottom, GUS activities of the transgenic tobacco plants. The bars represent the GUS activities of transgenic plants cultured in the presence of 1.0 μm CuSO4. The GUS activities of the transgenic plants cultured in the absence of CuSO4 were arbitrarily set to 1.0 (dotted line). The transgenic tobacco plants containing the CaMV 35S promoter fused to the GUS gene were used as a negative control (Nt/35S). The bars indicate ± sd. B, The Arabidopsis FeSOD gene FSD1 is regulated by Cu. Top, Schematic diagram of the construct. The site of transcription initiation is indicated (+1). Black box, 5′ Untranslated region; gray box, coding region (three amino acids). Bottom, GUS activities of transgenic Arabidopsis (At/35S and At/AtFSD1) and tobacco plants (Nt/35S and Nt/AtFSD1). The 35S-GUS construct was used as a negative control. The bars represent the GUS activities of transgenic plants cultured in the presence of 1.0 μm CuSO4. The GUS activities of the transgenic plants cultured in the absence of CuSO4 were arbitrarily set to 1.0 (dotted line). The bars indicate ± sd. C, Histochemical analysis of transgenic Arabidopsis plants carrying the AtFSD1-GUS construct cultured without or with CuSO4. Transgenic Arabidopsis seedlings precultured with 0.1 μm CuSO4 for 7 d were treated without (left; −Cu2+) or with 1.0 μm CuSO4 (right; +Cu2+) for 7 d. The arrowheads indicate the cotyledons. More than three independent transgenic lines were investigated. Scale bars, 1 mm. D, Schematic diagrams of the promoter region of Arabidopsis genes regulated by Cu. FeSOD, moss FeSOD (AB370198); FSD1, FeSOD (At4g25100); miR398a, microRNA 398a (At2g03445); miR398b, microRNA 398b (At5g14545); miR398c, microRNA 398c (At5g14565); CCH, Cu chaperone (At3g56240); COPT1, Cu transporter (At5g59030); COPT2, Cu transporter (At3g46900); ZIP2, ZRT/IRT-like protein (At5g59520). The white boxes and triangles represent GTACT sequences and GTAC sequences, respectively.
We examined whether Cu affects the pattern of expression of the FSD1 gene or simply decreases the levels of expression in those same organs using transgenic Arabidopsis carrying the FSD1 promoter-GUS. Although no expression of FeSOD was observed in aerial parts in bryophyte plants (Fig. 4), the expression of Arabidopsis FSD1 was detected in all organs under Cu-deficient conditions (Fig. 6C). Cu decreased GUS activities in the roots and rosette leaves but not in the cotyledons of the transgenic Arabidopsis, suggesting that the Cu responsiveness was lost in cotyledons. Alternatively, the accumulation of Cu in cotyledons could be of a low level to repress the FSD1 gene, because the absorbed nutrients from roots are preferentially transported to young leaves through the vascular system.
DISCUSSION
As sessile organisms, plants have acquired plastic developmental programs to adapt to fluctuating environments throughout their life cycles. Transition metals, including Cu, Mn, Fe, and Zn, are essential for life, because they act as effective electron acceptors and donors in the active sites of many proteins involved in oxidation and reduction reactions (Shcolnick and Keren, 2006). The acquisition of these metals is a nutritional problem for plants, because bioavailability has changed in the course of evolution owing to the increase of oxygen in the atmosphere and the movement of life from aquatic to terrestrial habitats. Plants change the expression patterns of three types of SODs (i.e. FeSOD, MnSOD, and Cu/ZnSOD) depending on the internal and external availability of the transition metals. Therefore, elucidating the transcriptional regulation of SOD is crucial to identify the molecular mechanisms involved in plant adaptation to changes in their mineral environments.
In this article, we have identified the GTACT motif as a cis-acting element that is involved in the Cu-responsive transcriptional repression of the moss FeSOD gene. Seven copies of the GTACT motif (B1 to B7) were located within 400 nucleotides upstream of the transcription start site of the FeSOD promoter. We found that both B4 and B5, but not B6 and B7, are necessary for repression of the FeSOD gene (Fig. 3), suggesting that two copies of the GTACT sequences could repress the transcription in a Cu-dependent manner and that the GTACT sequences of B4 to B7 are not functionally equivalent. The latter could be due to the effect of surrounding sequences of each GTACT motif and/or the position of the GTACT motif on the promoter DNA sequences. Furthermore, a tandem repeat of the 60-bp DNA fragment between −258 and −199 of FeSOD that contains B4 and B5 conferred Cu responsiveness to the actin promoter, whereas the solo 60-bp DNA fragment did not (Fig. 2). This implied that the copy number of the GTACT motif that is required for Cu-responsive repression varied with the primary structure of the promoter. Taken together, our results suggested that the GTACT motif plays a central role in the Cu-responsive transcriptional repression of the FeSOD gene and that its function is affected by the surrounding DNA context.
We showed that the overexpression of the transcription factor PpSBP2 resulted in the repression of the FeSOD gene in transgenic moss plants (Fig. 5); however, the effect was moderate, i.e. overexpression of PpSBP2 is not sufficient for full repression of the FeSOD gene. This suggests the involvement of the posttranslational modification of the transcription factor. One possible mechanism for the functional regulation of PpSBPs is the interaction with accessory proteins, including other transcriptional repressors, activators, general transcription factors, coactivators, chaperones, or chromatin-remodeling factors that mobilize nucleosomes. In this context, we observed that PpSBP1 bound to GTACT sequences and repressed the expression of the FeSOD gene as well as PpSBP2 (our unpublished data). Heterodimer formation of PpSBP2 with PpSBP1 or with other PpSBPs could enhance the repression activity of PpSBP2. Alternatively, covalent modification of PpSBPs, including acetylation, methylation, and phosphorylation, could be involved in the transcriptional regulation of the FeSOD gene in response to Cu.
The molecular mechanisms for Cu-responsive transcriptional repression seemed to be conserved in higher plants. The Arabidopsis chloroplastic FeSOD gene FSD1 containing three copies of GTACT sequences was regulated by Cu at the transcriptional level (Fig. 6). On the other hand, the mRNA levels of the chloroplastic Cu/ZnSOD gene CSD2 of Arabidopsis have been shown to be up-regulated by Cu (Abdel-Ghany et al., 2005b). Although Cu did not activate the transcription of the CSD2 gene, it promoted the expression of the CSD2 gene at the posttranscriptional level (Sunkar et al., 2006; Yamasaki et al., 2007). MicroRNAs are a class of regulatory small RNAs that posttranscriptionally regulate gene expression by directing mRNA cleavage or translational inhibition (Bonnet et al., 2004; Jones-Rhoades and Bartel, 2004). MicroRNA miR398 targeted CSD2 mRNA and negatively regulated the expression of the CSD2 gene (Sunkar and Zhu, 2004; Sunkar et al., 2006; Yamasaki et al., 2007). The transcription of miR398 was repressed by Cu (Sunkar et al., 2006), and multiple copies of GTACT sequences were found in MIR398 promoter sequences (Fig. 6D). This suggests that Cu promotes the expression of the Cu/ZnSOD gene CSD2 posttranscriptionally by repressing the transcription of miR398 through GTACT sequence motifs. Thus, GTACT sequences play key roles in the replacement of FeSOD by Cu/ZnSOD in response to Cu by directly repressing the transcription of the FeSOD gene and indirectly inducing the transcription of the Cu/ZnSOD gene in Arabidopsis. Moreover, it has been reported that high Cu availability reduced the expression of several Cu-related genes of Arabidopsis, including COPT1, COPT2 encoding Cu transporters, ZIP2 encoding divalent metal transporters, and CCH encoding Cu chaperones (Himelblau et al., 1998; Sancenón et al., 2003; Wintz et al., 2003). It is worth noting that the GTACT motifs are commonly found in the promoter of these genes (Fig. 6D). The expression of the Arabidopsis gene, CCS, encoding the Cu chaperone for SOD, is up-regulated by Cu (Abdel-Ghany et al., 2005a); however, its molecular mechanisms are unknown at present.
Cu is one of the toxic, but essential, metals for all aerobic organisms, and its metabolism is tightly controlled (Balamurugan and Schaffner, 2006; Puig et al., 2007). For example, in Saccharomyces cerevisiae, a transcription factor Ace1p regulates Cu homeostasis by activating Cu-sequestering proteins under Cu-excess conditions (Thiele, 1988; Gralla et al., 1991). Cu directly binds to Ace1p and promotes Ace1 binding to DNA and transcriptional activation (Fürst et al., 1988; Dameron et al., 1991). In addition, a transcription factor Mac1p is required for the up-regulation of the expression of Cu transporters under Cu-deficient conditions (Labbé et al., 1997; Keller et al., 2005). The target sequences of Ace1p and Mac1p are TNNNGCTG and TTTGCTC (Huibregtse et al., 1989; Evans et al., 1990; Gralla et al., 1991; Labbé et al., 1997), respectively, clearly different from that of the Cu-responsive motif containing the GTAC core sequence of C. reinhardtii, moss, and higher plants. Furthermore, occurrence of SBP-related transcription factors that bind to the GTAC motif is limited to the plant kingdom. These results suggest the existence of plant-specific mechanisms of Cu-responsive signaling and transcriptional regulation.
In C. reinhardtii, the Cu-responsive element, together with its transcription factor, CRR1, positively regulates the transcription of the Cyc6 and Cpx1 genes (Kropat et al., 2005). However, our results suggested that the Cu-responsive element gene and the transcription factor PpSBPs negatively regulate the transcription of the FeSOD gene in response to Cu in moss. This discrepancy may be attributable to the functional difference of transcription factors, namely, a transcriptional activator for CRR1 and a repressor for PpSBP2. In this context, the similarities in the amino acid sequence of C. reinhardtii CRR1 and moss PpSBPs were very limited in regions outside the DNA-binding SBP domain (Fig. 5). Because the expression of both the moss FeSOD promoter and the Arabidopsis FSD1 promoter is repressed by Cu in transgenic Arabidopsis (Fig. 6), the mechanisms for Cu-dependent negative transcriptional regulation may be conserved between moss and higher plants. Collectively, the molecular mechanisms of Cu responsiveness seem to be more sophisticated during the evolution of plants from green algae to vascular plants, keeping the GTAC core cis-regulatory sequence for Cu responsiveness through adaptation to terrestrial habitats and differentiation from higher plants. Further investigation regarding how the function of PpSBPs is controlled by Cu will help to reveal the molecular mechanisms behind the fine transcriptional regulation by Cu and provide insights into the evolution of SODs, which constitute the first line of defense against oxidative stress and signal the emergence of land plants.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Physcomitrella patens Bruch & Schimp subsp. patens Tan (Ashton and Cove, 1977) was cultured at 25°C under continuous light in a BCDATG agar medium (Nishiyama et al., 2000). For vegetative propagation, the plants were ground with a pestle and mortar in sterile water and soaked in the BCDATG agar medium overlaid with a layer of cellophane.
For the GUS assay and histochemical analysis, the transgenic plants were precultured in a BCDATG agar medium containing 0.2 μm CuSO4 for 7 d at 25°C and then transferred to a BCDATG liquid medium with (2.0 μm CuSO4) or without CuSO4 for 3 d at 25°C under continuous light (<50 μmol m−2 s−1).
The Arabidopsis (Arabidopsis thaliana) lines used were in the Columbia-0 background. Seeds were sterilized and sown in a Murashige and Skoog medium (Murashige and Skoog, 1962) containing 0.8% agar (Wako) and 3% Suc. The plates were incubated in the dark for 3 d at 4°C and subsequently transferred to a long-day condition (18 h of light/6 h of dark) at 22°C (<100 μmol m−2 s−1). For the Cu treatment, 7-d-old seedlings were transferred to plates with (1.0 μm CuSO4) or without CuSO4 for 7 d at 22°C. The tobacco (Nicotiana tabacum) lines used were in the ‘Petit Havana SR1’ background. Seeds were sterilized and sown in the same medium composition as Arabidopsis. The plates were cultured at 28°C under continuous light (<100 μmol m−2 s−1). For the Cu treatment, 7-d-old seedlings were transferred to the plates with (1.0 μm CuSO4) or without CuSO4 for 7 d at 28°C.
Plasmid Construction
The FeSOD gene promoter was isolated by TAIL-PCR using a random primer, 5′-NGTCGA(G/C)(A/T) GANA(A/T) GAA-3′, and gene-specific primers, 5′-GAGACTTCTCCACCATCAACTTG-3′, 5′-CGAGCTCCGTCCCCTCGATCTGC-3′, and 5′-TCGTCAAAGGCAACGGAACGGCAGCC-3′. Genomic DNA was used as the template for TAIL-PCR. The amplified product was cloned into a pGEM-T Easy vector (Promega). A series of 5′ deletion constructs was generated by PCR using this plasmid as a template with primers, generating a SalI site at the 5′ and the 3′ ends. The primer pairs were as follows: for p558, 5′-GGGGGTCGACAGAAGAAGTGAAGAC-3′ and 5′-GGGGGTCGACGGCAGCCATCCTGGTG-3′ (primer-R); for p438, 5′-GGGGGTCGACACGGAGAAACTGATATCAG-3′ and primer-R; for p318, 5′-GGGGGTCGACATGATTGGTACTAC-3′ and primer-R; for p258, 5′-GGGGGTCGACGAGTACAAGCTTATACGAC-3′ and primer-R; and for p198, 5′-GGGGGTCGACCATCCAAAAACTCAC-3′ and primer-R. The amplified DNA fragment was digested with SalI and cloned into SalI of pGUSmutNPTII, which contained the coding sequence of GUS, the nopaline synthase polyadenylation signal, and the NPTII cassette (NPTII; Nishiyama et al., 2000). To make the gain-of-function experimental constructs, the PCR-amplified fragment was cloned into KpnI of pCRII-ACTIN, which contained the rice (Oryza sativa) actin promoter derived from the pTN90 plasmid, the coding sequence of GUS, the nopaline synthase polyadenylation signal, and the NPTII cassette. The primer pairs were as follows: for F12, 5′-GGGGGGTACCATGATTGGTACTACTGCGC-3′ (primer-F12-F) and 5′-GGGGGGTACCATAGGGTAGTACTTGAGTTG-3′ (primer-F12-R); for F1, primer-F12-F and 5′-GGGGGGTACCCAGGCTCCAGTGTAGTAAAAC-3′; and for F2, 5′-GGGGGGTACCGAGTACAAGCTTATACGAC-3′ and primer-F12-R. The mutant version constructs were generated by PCR using primers including specific mutations. The PpSBP2 was amplified by a PCR reaction from P. patens cDNA library with primers 5′-GGGGGCCCATGTCTGCCGTTGACC-3′ and 5′-GGGGGCCCCTACAGATGCAGGGACAC-3′. To make the PpSBP2OE construct, the PCR-amplified fragment was cloned into ApaI of the pCMAK1 plasmid, which contained pE7133 elements derived from the CaMV 35S promoter and some enhancers (Mitsuhara et al., 1996). The pGUSmutNPTII-derived constructs (p558, p438, p318, and p198), pCRII-ACTIN-derived constructs (F12, F1, and F2), and PpSBP2OE construct were linearized with SpeI-XhoI, ApaI, and NotI, respectively; the subsequent polyethylene glycol-mediated transformation followed the method of Hiwatashi et al. (2001). To make the p558 construct for transgenic Arabidopsis and tobacco plants, the SalI-digested promoter DNA fragment from the p558 construct was cloned into the pBI101 vector to generate GUS fusion genes. The promoter of FSD1 was amplified by a PCR reaction from Arabidopsis genomic DNA with primers 5′-GGAAGCTTACTGTATAAGATACAAGGTG-3′ and 5′-GGGGATCCAGCAGCCATTCTTTGTAATTG-3′. The amplified DNA fragment was digested with HindIII-BamHI and cloned into the pBI101 vector. The integrity of all constructs was confirmed by DNA sequencing. These constructs were introduced into Arabidopsis and tobacco by way of Agrobacterium tumefaciens-mediated transformation (Fukazawa et al., 2000; Matsushita et al., 2007).
RT-PCR
Total RNA was isolated from plants using the RNeasy Plant Mini kit (QIAGEN). For the RT-PCR studies, total RNA was treated with DNase using a TURBO DNA-free kit (Ambion), and then 1 μg of RNA was converted into cDNAs with a mixture of the dT20 primer using SuperScript III reverse transcriptase (Invitrogen). PCR was performed using TaKaRa Ex Taq DNA polymerase (TaKaRa). The primer pairs were as follows: for GUS, 5′-CTGTGGGCATTCAGTCT-3′ and 5′-CGGATTCACCACTTGCA-3′; for glyceraldehyde 3-phosphate dehydrogenase (GAPDH), 5′-CGACAGCAGGTCAAGCATCT-3′ and 5′-AACATGAACGCTGGCGATGG-3′; and for PpSBP2, 5′-GACTCATCTAGAGATGTCACG-3′ and 5′-TGGCCCTACTAAGATCAGC-3′. The products by RT-PCR were electrophoresed on 1% (w/v) agarose gels, transferred onto Hybond-N+ membranes (GE Healthcare), and hybridized with DNA probes. The chemiluminescence signal was detected with an imaging system (LAS1000 plus; Fujifilm).
Fluorometric GUS Enzyme Analysis
The transgenic plants were homogenized in the extraction buffer described by Jefferson (1987). After the cell debris was removed by centrifugation, GUS activities were determined with 1 mm 4-methylumbelliferyl glucuronide as a substrate at 37°C.
Histochemical Staining
The histochemical GUS activities were assayed according to Nishiyama et al. (2000) with slight modifications. The transgenic plants were stained with 50 mm sodium phosphate, pH 7.0, 0.5 mm 5-bromo-4-chloro-3-indolyl-β-d-GlcA, 0.5 mm K3Fe(CN)6, 0.5 mm K4Fe(CN)6, and 0.05% Triton X-100 and then incubated for 0.5 to 2 h at 37°C for staining. After the incubation, the transgenic plants were fixed in 5% (v/v) formalin for 10 min and soaked in 5% (v/v) acetic acid for 10 min. To facilitate observation, the specimens were treated successively with 30% ethanol, 50% ethanol, 70% ethanol, and 100% ethanol to remove chlorophyll in the tissues. Photomicrographs were taken with a stereoscopic microscope (Leica MZ16F; Leica Microsystems) under light field illumination.
Gel Mobility Shift Assay
The PpSBP2 protein was prepared by in vitro transcription (Epicentre Technologies) and a rabbit reticulocyte lysate translation system (GE Healthcare; Matsushita et al., 2007). The nucleotide sequences of the double-stranded oligonucleotides used for the gel mobility shift assay were X (5′-GAGATTGGTACTACTGCGCTGGTACTGGATGTACTGCGA-3′ and 5′-GTGTCGCAGTACATCCAGTACCAGCGCAGTAGTACCAAT-3′), mX (5′-GAGATTGGGCCTACTGCGCTGGGCCTGGATGGCCTGCGA-3′ and 5′-GTGTCGCAGGCCATCCAGGCCCAGCGCAGTAGGCCCAAT-3′), Y (5′-GAGATCTGTACTAGAGCTCCTATGCAACTCAAGTACTACCCTA-3′ and 5′-GTGTAGGGTAGTACTTGAGTTGCATAGGAGCTCTAGTACAGAT-3′), and mY (5′-GAGATCTGGCCTAGAGCTCCTATGCAACTCAAGGCCTACCCTA-3′ and 5′-GTGTAGGGTAGGCCTTGAGTTGCATAGGAGCTCTAGGCCAGAT-3′). The oligonucleotides were annealed and then labeled with [α-32P]dCTP and the Klenow fragment of DNA polymerase I. The binding mixtures contained 1 pmol of a labeled probe, in vitro-translated proteins, and 2 μg of poly(dI-dC). DNA competitors were used at 10-fold molar excess. The binding buffer consisted of 15 mm Tris-HCl, pH 7.5, 75 mm NaCl, 1.5 mm EDTA, 7.5% glycerol, 1.5 mm dithiothreitol, 0.3% nonidet P-40, and 1 mg/mL bovine serum albumin. Reactions were incubated for 30 min at room temperature and loaded onto 4% polyacrylamide gels containing 44.5 mm Tris, 44.5 mm borate, 1 mm EDTA, and 2.5% glycerol.
The DDBJ/EMBL/GenBank accession numbers of the sequences used in this study are: B. unguiculata FeSOD, AB370198; PpSBP2, AJ968403.
Acknowledgments
We are grateful to Dr. Jutarou Fukazawa, Dr. Tsuyoshi Furumoto, Dr. Hironori Deguchi, and Dr. Toshio Satoh for technical assistance and their participation in helpful discussions during the course of this work. We also thank Dr. Tomoaki Nishiyama, Dr. Kaoru Hashimoto, Dr. Rumiko Kofuji, and Dr. Mitsuyasu Hasebe, all of the National Institute of Basic Biology, Okazaki, Japan, for plasmids pTN90, pCMAK1, and pGUSmutNPTII.
This work was supported by the Japan Society for the Promotion of Science (grant no. 18657017 to Y.T.) and by the Ministry of Education, Culture, Sports, Science and Technology (grant no. 17054029 to Y.T.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instruction for Authors is: Yohsuke Takahashi (ytakahas@hiroshima-u.ac.jp).
References
- Abdel-Ghany SE, Burkhead JL, Gogolin KA, Andrés-Colás N, Bodecker JR, Puig S, Peñarrubia L, Pilon M (2005. a) AtCCS is a functional homolog of the yeast copper chaperone Ccs1/Lys7. FEBS Lett 579 2307–2312 [DOI] [PubMed] [Google Scholar]
- Abdel-Ghany SE, Muller-Moulé P, Niyogi KK, Pilon M, Shikanai T (2005. b) Two P-type ATPases are required for copper delivery in Arabidopsis thaliana chloroplasts. Plant Cell 17 1233–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alscher RG, Erturk N, Heath LS (2002) Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. J Exp Bot 53 1331–1341 [PubMed] [Google Scholar]
- Asada K, Kanematsu S, Uchida K (1977) Superoxide dismutase in photosynthetic organisms: absence of the cuprozinc enzyme in eukaryotic algae. Arch Biochem Biophys 179 243–256 [DOI] [PubMed] [Google Scholar]
- Ashton NW, Cove DJ (1977) The isolation and preliminary characterization of auxotrophic and analogue resistant mutants in the moss Physcomitrella patens. Mol Gen Genet 154 87–95 [Google Scholar]
- Balamurugan K, Schaffner W (2006) Copper homeostasis in eukaryotes: teetering on a tightrope. Biochim Biophys Acta 1763 737–746 [DOI] [PubMed] [Google Scholar]
- Birkenbihl RP, Jach G, Saedler H, Huijser P (2005) Functional dissection of the plant-specific SBP-domain: overlap of the DNA-binding and nuclear localization domains. J Mol Biol 352 585–596 [DOI] [PubMed] [Google Scholar]
- Bonnet E, Wuyts J, Rouzé P, Van de Peer Y (2004) Evidence that micro RNA precursors, unlike other non-coding RNAs, have lower folding free energies than random sequences. Bioinformatics 20 2911–2917 [DOI] [PubMed] [Google Scholar]
- Dameron CT, Winge DR, George GN, Sansone M, Hu S, Hamer D (1991) A copper-thiolate polynuclear cluster in the ACE1 transcription factor. Proc Natl Acad Sci USA 88 6127–6131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egami F (1975) Origin and early evolution of transition element enzymes. J Biochem 77 1165–1169 [PubMed] [Google Scholar]
- Evans CF, Engelke DR, Thiele DJ (1990) ACE1 transcription factor produced in Escherichia coli binds multiple regions within yeast metallothionein upstream activation sequences. Mol Cell Biol 10 426–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink RC, Scandalios JG (2002) Molecular evolution and structure-function relationship of the superoxide dismutase gene families in angiosperms and their relationship to other eukaryotic and prokaryotic superoxide dismutase. Arch Biochem Biophys 399 19–36 [DOI] [PubMed] [Google Scholar]
- Fukazawa J, Sakai T, Ishida S, Yamaguchi I, Kamiya Y, Takahashi Y (2000) REPRESSION OF SHOOT GROWTH, a bZIP transcriptional activator, regulates cell elongation by controlling the level of gibberellins. Plant Cell 12 901–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fürst P, Hu S, Hackett R, Hamer D (1988) Copper activates metallothionein gene transcription by altering the conformation of a specific DNA binding protein. Cell 55 705–717 [DOI] [PubMed] [Google Scholar]
- Gralla EB, Thiele DJ, Silar P, Valentine JS (1991) ACE1, a copper-dependent transcription factor, activates expression of the yeast copper, zinc superoxide dismutase gene. Proc Natl Acad Sci USA 88 8558–8562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill KL, Li HH, Singer J, Merchant S (1991) Isolation and structural characterization of the Chlamydomonas reinhardtii gene for cytochrome c6. J Biol Chem 266 15060–15067 [PubMed] [Google Scholar]
- Hill KL, Merchant S (1995) Coordinate expression of coproporphyrinogen oxidase and cytochrome c6 in the green alga Chlamydomonas reinhardtii in response to changes in copper availability. EMBO J 14 857–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himelblau E, Mira H, Lin S-J, Culotta VC, Peñarrubia L, Amasino RM (1998) Identification of a functional homolog of the yeast copper homeostasis gene ATX1 from Arabidopsis. Plant Physiol 117 1227–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiwatashi Y, Nishiyama T, Fujita T, Hasebe M (2001) Establishment of gene-trap and enhancer-trap systems in the moss Physcomitrella patens. Plant J 28 105–116 [DOI] [PubMed] [Google Scholar]
- Huibregtse JM, Engelke DR, Thiele DJ (1989) Copper-induced binding of cellular factors to yeast metallothionein upstream activation sequences. Proc Natl Acad Sci USA 86 65–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep 5 387–405 [Google Scholar]
- Jones-Rhoades MW, Bartel DP (2004) Computational identification of plant micro RNAs and their targets, including a stress-induced miRNA. Mol Cell 14 787–799 [DOI] [PubMed] [Google Scholar]
- Kanematsu S, Asada K (1989) CuZn-superoxide dismutases from the fern Equisetum arvense and the green alga Spirogyra sp.: occurrence of chloroplast and cytosol types of enzyme. Plant Cell Physiol 30 717–727 [Google Scholar]
- Keller G, Bird A, Winge DR (2005) Independent metalloregulation of Ace1 and Mac1 in Saccharomyces cerevisiae. Eukaryot Cell 4 1863–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenrick P, Crane PR (1997) The origin and early evolution of plants on land. Nature 389 33–39 [Google Scholar]
- Kropat J, Tottey S, Birkenbihl RP, Depége N, Huijser P, Merchant S (2005) A regulator of nutritional copper signaling in Chlamydomonas is an SBP domain protein that recognizes the GTAC core of copper response element. Proc Natl Acad Sci USA 102 18730–18735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurepa J, Van Montagu M, Inzé D (1997) Expression of sodCp and sodB genes in Nicotiana tabacum: effects of light and copper excess. J Exp Bot 48 2007–2014 [Google Scholar]
- Labbé S, Zhu Z, Thiele DJ (1997) Copper-specific transcriptional repression of yeast genes encoding critical components in the copper transport pathway. J Biol Chem 272 15951–15958 [DOI] [PubMed] [Google Scholar]
- Lumsden J, Hall DO (1975) Superoxide dismutase in photosynthetic organisms provides an evolutionary hypothesis. Nature 257 670–672 [DOI] [PubMed] [Google Scholar]
- Matsushita A, Furumoto T, Ishida S, Takahashi Y (2007) AGF1, an AT-hook protein, is necessary for the negative feedback of AtGA3ox1 encoding GA 3-oxidase. Plant Physiol 143 1152–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant S, Bogorad L (1986) Regulation by copper of the expression of plastocyanin and cytochrome c552 in Chlamydomonas reinhardtii. Mol Cell Biol 6 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuhara I, Ugaki M, Hirochika H, Ohshima M, Murakami T, Gotoh Y, Katayose Y, Nakamura S, Honkura R, Nishimiya S, et al (1996) Efficient promoter cassettes for enhanced expression of foreign genes in dicotyledonous and monocotyledonous plants. Plant Cell Physiol 37 49–59 [DOI] [PubMed] [Google Scholar]
- Mittler R (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 7 405–410 [DOI] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Gollery M, Van Breusegem F (2004) Reactive oxygen gene network of plants. Trends Plant Sci 9 490–498 [DOI] [PubMed] [Google Scholar]
- Murao K, Takamiya M, Ono K, Takano H, Takio S (2004) Copper deficiency induced expression of Fe-superoxide dismutase gene in Matteuccia struthiopteris. Plant Physiol Biochem 42 143–148 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15 437–497 [Google Scholar]
- Nishiyama T, Hiwatashi Y, Sakakibara K, Kato M, Hasebe M (2000) Tagged mutagenesis and gene-trap in the moss, Physcomitrella patens by shuttle mutagenesis. DNA Res 7 9–17 [DOI] [PubMed] [Google Scholar]
- Puig S, Andrés-Colás N, García-Molina A, Peñarrubia L (2007) Copper and iron homeostasis in Arabidopsis: responses to metal deficiencies, interactions and biotechnological applications. Plant Cell Environ 30 271–290 [DOI] [PubMed] [Google Scholar]
- Quinn JM, Barraco P, Eriksson M, Merchant S (2000) Coordinate copper- and oxygen-responsive Cyc6 and Cpx1 expression in Chlamydomonas is mediated by the same element. J Biol Chem 275 6080–6089 [DOI] [PubMed] [Google Scholar]
- Quinn JM, Nakamoto SS, Merchant S (1999) Induction of coproporphyrinogen oxidase in Chlamydomonas chloroplasts occurs via transcriptional regulation of Cpx1 mediated by copper response elements and increased translation from a copper deficiency-specific form of the transcript. J Biol Chem 274 14444–14454 [DOI] [PubMed] [Google Scholar]
- Sancenón V, Puig S, Mira H, Thiele DJ, Peñarrubia L (2003) Identification of a copper transporter family in Arabidopsis thaliana. Plant Mol Biol 51 577–587 [DOI] [PubMed] [Google Scholar]
- Shcolnick S, Keren N (2006) Metal homeostasis in cyanobacteria and chloroplasts. Balancing benefits and risks to the photosynthetic apparatus. Plant Physiol 141 805–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiono T, Nakata M, Yamahara T, Matuzaki M, Deguchi H, Satoh T (2003) Repression by Cu of the expression of Fe-superoxide dismutase of the chloroplasts in the moss Barbula unguiculata but not in the liverwort Marchantia paleacea var. diptera. J Hattori Bot Lab 93 141–153 [Google Scholar]
-
Stallings WC, Powers TB, Pattridge KA, Fee JA, Ludwig ML (1983) Iron superoxide dismutase from Escherichia coli at 3.1-
 resolution: a structure unlike that of copper/zinc protein at both monomer and dimmer levels. Proc Natl Acad Sci USA 80 3884–3888 [DOI] [PMC free article] [PubMed] [Google Scholar]
resolution: a structure unlike that of copper/zinc protein at both monomer and dimmer levels. Proc Natl Acad Sci USA 80 3884–3888 [DOI] [PMC free article] [PubMed] [Google Scholar] - Steinman HM, Hill RL (1973) Sequence homologies among bacterial and mitochondrial superoxide dismutases. Proc Natl Acad Sci USA 70 3725–3729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Kapoor A, Zhu JK (2006) Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 18 2066–2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Zhu JK (2004) Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16 2001–2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Takio S, Satoh T (1995) Inactivation of the cytosolic Cu/Zn-superoxide dismutase induced by copper deficiency in suspension-cultured cells of Marchantia paleacea var. diprera. J Plant Physiol 146 361–365 [Google Scholar]
- Thiele DJ (1988) ACE1 regulates expression of the Saccharomyces cerevisiae metallothionein gene. Mol Cell Biol 8 2745–2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters ER (2003) Molecular adaptation and the origin of land plants. Mol Phylogenet Evol 29 456–463 [DOI] [PubMed] [Google Scholar]
- Wellman CH, Osterloff PL, Mohiuddin U (2003) Fragments of the earliest land plants. Nature 425 282–285 [DOI] [PubMed] [Google Scholar]
- Wintz H, Fox T, Wu YY, Feng V, Chen W, Chang HS, Zhu T, Vulpe C (2003) Expression profiles of Arabidopsis thaliana in mineral deficiencies reveal novel transporters involved in metal homeostasis. J Biol Chem 278 47644–47653 [DOI] [PubMed] [Google Scholar]
- Yamahara T, Haraguchi T, Amakawa K, Ono K, Takio S, Tanaka K, Deguchi H, Satoh T (1999) Superoxide dismutase suggests phylogenetic relationships among bryophytes. J Hattori Bot Lab 87 309–313 [Google Scholar]
- Yamasaki H, Abdel-Ghany SE, Cohu CM, Kobayashi Y, Shikanai T, Pilon M (2007) Regulation of copper homeostasis by micro-RNA in Arabidopsis. J Biol Chem 282 16369–16378 [DOI] [PubMed] [Google Scholar]