Abstract
Molecular genetic studies suggest that FLORICAULA (FLO)/LEAFY (LFY) orthologs function to control compound leaf development in some legume species. However, loss-of-function mutations in the FLO/LFY orthologs result in reduction of leaf complexity to different degrees in Pisum sativum and Lotus japonicus. To further understand the role of FLO/LFY orthologs in compound leaf development in legumes, we studied compound leaf developmental processes and characterized a leaf development mutant, single leaflet1 (sgl1), from the model legume Medicago truncatula. The sgl1 mutants exhibited strong defects in compound leaf development; all adult leaves in sgl1 mutants are simple due to failure in initiating lateral leaflet primordia. In addition, the sgl1 mutants are also defective in floral development, producing inflorescence-like structures. Molecular cloning of SGL1 revealed that it encodes the M. truncatula FLO/LFY ortholog. When properly expressed, LFY rescued both floral and compound leaf defects of sgl1 mutants, indicating that LFY can functionally substitute SGL1 in compound leaf and floral organ development in M. truncatula. We show that SGL1 and LFY differed in their promoter activities. Although the SGL1 genomic sequence completely rescued floral defects of lfy mutants, it failed to alter the simple leaf structure of the Arabidopsis thaliana plants. Collectively, our data strongly suggest that initiation of lateral leaflet primordia required for compound leaf development involves regulatory processes mediated by the SGL1 function in M. truncatula.
Leaves are determinate organs initiated from the periphery of the pluripotent shoot apical meristem (SAM). The class I KNOTTED1-like (KNOX1) homeobox genes are involved in acquisition and maintenance of the meristem activity of SAM (Long et al., 1996). Initiation of lateral organs such as leaves requires down-regulation of KNOX1 genes at the incipient leaf primordia, and subsequent acquisition of determinacy in developing organs further requires continued suppression of KNOX1 gene expression (Smith et al., 1992; Sinha et al., 1993; Jackson et al., 1994). Thus, development of lateral organs from the periphery of the SAM requires fine-tuned interactions of multiple regulators in SAM, some maintaining the meristem fate, such as the KNOX1 genes, and others promoting determinacy by suppressing expression of meristem genes, such as ROUGH SHEATH2 (RS2) and ASYMMETRIC LEAVES1 (AS1) in maize (Zea mays) and Arabidopsis (Arabidopsis thaliana), respectively (Timmermans et al., 1999; Tsiantis et al., 1999; Byrne et al., 2000; Ori et al., 2000; Hay et al., 2006).
In compound-leafed species such as tomato (Solanum lycopersicum), the expression of KNOX1 genes is initially excluded from the incipient leaf primordia but subsequently reactivated in developing leaf primordia (Kim et al., 2003). Ectopic expression of KNOX1 in tomato results in ramification of compound leaves, suggesting that KNOX1 genes may play a role in compound leaf development (Hareven et al., 1996). Direct evidence that supports the role for KNOX1 genes in compound leaf development comes from studies of Cardamine hirsuta, a small crucifer related to the simple-leafed model plant Arabidopsis. In transgenic C. hirsuta lines, in which expression of an endogenous KNOX1 gene SHOOTMERISTEMLESS is down-regulated, leaflet initiation is reduced (Hay and Tsiantis, 2006). Thus, development of compound leaves involves reactivation of genes that promote indeterminacy in leaf primordia in compound-leafed species (Bharathan et al., 2002; Hay and Tsiantis, 2006; Jasinski et al., 2007; Ori et al., 2007).
The involvement of KNOX1 genes in compound leaf development is evidenced in a number of compound-leafed species including legumes (Bharathan et al., 2002; Hay and Tsiantis, 2006; Champagne et al., 2007). However, in some cases, the final leaf forms are not entirely correlated with the expression of KNOX1 genes in leaf primordia, because some complex leaf primordia in which KNOX1 genes are expressed may mature into simple leaves (Bharathan et al., 2002). In a large subclade of legumes, the inverted repeat-lacking clade (IRLC), including pea (Pisum sativum) and alfalfa (Medicago sativa), expression of KNOX1 genes is excluded from the initiating leaf primordia (Gourlay et al., 2000; Hofer et al., 2001; Champagne et al., 2007). Thus, KNOX1 genes are not likely correlated with compound leaf development in this group of legume plants (Hofer et al., 2001; Champagne et al., 2007).
Molecular genetic studies indicate that the legume orthologs of the floral meristem (FM) identity gene FLORICAULA (FLO) from snapdragon (Antirrhinum majus) and LEAFY (LFY) from Arabidopsis, and their coregulators FIMBRIATA (FIM) and UNUSUAL FLORAL ORGAN (UFO), respectively, play a role in compound leaf development in compound-leafed legumes (Hofer et al., 1997; Hofer and Ellis, 1998; DeMason and Schmidt, 2001; Taylor et al., 2001; Dong et al., 2005). The pea unifoliata (uni) and stamina pistilloida (stp) mutants exhibit inflorescence and floral defects that are similar to that of the snapdragon flo and fim mutants, and Arabidopsis lfy and ufo mutants, respectively, and in addition reduced compound leaf phenotypes (Hofer et al., 1997; Taylor et al., 2001). In Lotus japonicus, a legume species that is outside of the IRLC, proliferating floral meristem (pfm) mutants of the FLO/LFY ortholog also exhibit reduced compound leaf phenotypes (Dong et al., 2005). In transgenic soybean (Glycine max; outside of the IRLC) lines in which the endogenous LFY genes are down-regulated, the leaflet number is moderately reduced (Champagne et al., 2007). Taken together, these data support a significant role for the FLO/LFY and STP/UFO orthologs in compound leaf development in some legumes and a minor role in others.
Here, we describe compound leaf developmental processes in the model legume species Medicago truncatula and report characterization of a leaf development mutant, single leaflet1 (sgl1) isolated from a tobacco (Nicotiana tabacum) Tnt1 retrotransposon-tagged mutant population of M. truncatula. Our genetic analyses of four loss-of-function alleles of the sgl1 mutant indicate that SGL1 plays a key role in initiation of lateral leaflet primordia during an early stage of leaf development resulting in a complete conversion of compound leaves into simple leaves. We also show that SGL1 controls the development of petioles along the proximodistal axis of compound leaves at a late developmental stage and FM identity during the reproductive phase of growth. Sequence analyses indicate that SGL1 encodes a plant-specific protein related to the snapdragon FLO, Arabidopsis LFY, and pea UNI transcription factors. We discuss possible mechanisms by which FLO/LFY orthologs regulate compound leaf development in legumes.
RESULTS
Compound Leaf Development in M. truncatula
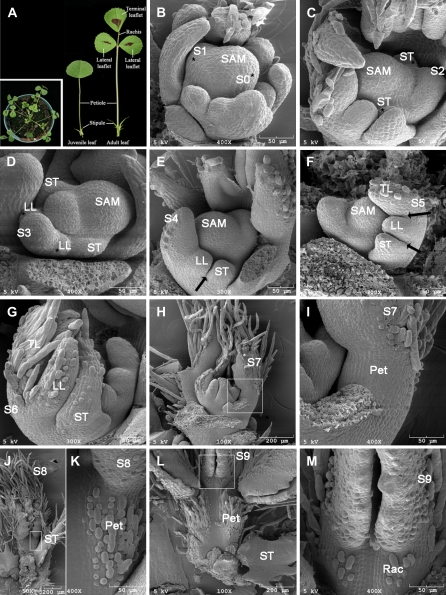
In pea, a legume species forming odd-pinnate compound leaves with distal tendrils, development of leaflet primordia is acropetal during early stages of leaf development. However, development of distal tendrils follows a basipetal pattern, giving rise to the unique compound leaf structure of pea plants (Hofer and Ellis, 1998). By contrast, the order of leaflet primordial development is basipetal in L. japonicus and other compound-leafed species (Hofer and Ellis, 1998; Gourlay et al., 2000; Luo et al., 2005). To facilitate characterization of leaf mutants in M. truncatula, we investigated leaf developmental processes with both visual and scanning electron microscopic (SEM) analyses. Similar to pea, L. japonicus, and other eudicot species, leaf development in M. truncatula is heteroblastic. However, the degree of heteroblasty is much simpler in M. truncatula than in other species, i.e. a single juvenile leaf with simple leaf morphology develops on the first node of a developing M. truncatula plant, and all other leaves that develop subsequently are in trifoliolate adult form, consisting of a pair of lateral (or proximal) leaflets and a terminal leaflet at the distal end of a petiole subtended by a pair of stipules (Fig. 1A). In M. truncatula, development of petioles was also heteroblastic, i.e. in 5-week-old plants, adult leaves developed on the third and fourth nodes had the longest petioles (Fig. 2H).
Figure 1.
The ontogeny of compound leaf development in wild-type M. truncatula. A, Morphology of M. truncatula ‘Jemalong’ A17. Shown in inset were juvenile (left) and adult (right) leaves. B to M, SEM analysis of compound leaf development. B, Sites of incipient leaf primordia were specified at the periphery of the SAM at S0, albeit no morphological changes were visible at this stage. At S1, a common leaf primordium was initiated as a strip of cells outgrown along the periphery of SAM (asterisk). C, A pair of stipule primordia (ST) was initiated from the proximal end of the common leaf primordium at S2. D, At S3, a pair of lateral leaflet primordia (LL) emerged between the stipule and common leaf primordia. E, Boundaries (arrow) between the stipule and lateral leaflet primordia formed, and the common leaf primordium differentiated into a terminal leaflet primordium (TL) as indicated by development of trichomes from the abaxial surface at S4. F, At S5, boundaries (arrows) formed between the lateral and terminal leaflet primordia. G, While trichomes developed from the abaxial surface of both stipule and lateral leaflet primordia, the terminal leaflet primordium folded as a result of outgrowth of the abaxial surface at S6. H, At S7, a petiole primordium (Pet) formed between the stipule and lateral leaflet primordia. I, A close-up view of H. J, The lateral and terminal leaflet primordia folded due to outgrowth of the abaxial surface at S8. Trichomes developed from the adaxial surface of the petiole primordium at this stage. K, A close-up view of J. L, A rachis primordium (Rac) formed between the lateral and terminal leaflet primordia and trichomes developed from its adaxial surface at S9. M, A close-up view of L. Scale bars, 50 or 200 μm as indicated.
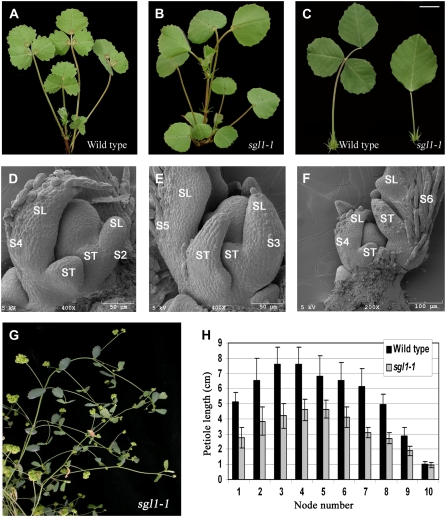
Figure 2.
Phenotypes of M. truncatula sgl1 mutants. Three-week-old wild-type M. truncatula (ecotype R108; A) and sgl1-1 mutant (B) exhibited compound and simple leaf forms, respectively. C, Close-up views of adult leaves of wild type R108 (left) and sgl1-1 mutant (right). In S2 and S4 (D), S3 and S5 (E), and S4 and S6 (F) leaf primordia of sgl1-1 mutant, lateral leaflet primordia did not form at the proximal end of common leaf primordia. G, Morphology of a mature sgl1-1 mutant plant, exhibiting simple leaf and floral homeotic phenotypes. H, Petiole length was significantly reduced in sgl1-1 mutant plants compared to the wild-type R108 plants, but no significant differences in petiole length were found in newly developed leaves of wild-type and sgl1-1 mutant (node 10). SL, Single leaflet; ST, stipule. Scale bars, 10 mm (A–C); 50 μm; or 100 μm as indicated (D–F). [See online article for color version of this figure.]
Morphological changes during compound leaf primordial development can be divided into 10 distinct stages in M. truncatula. At Stage 0 (S0), cells along the periphery of SAM were recruited and became an incipient leaf primordium, albeit no signs of outgrowth were visible at this stage (Fig. 1B). At S1, a common leaf primordium formed as a strip of cells outgrew along the periphery of SAM (Fig. 1B). At the subsequent S2, a pair of stipule primordia emerged as small bumps of cells that grew out of the proximal end of the common leaf primordium (Fig. 1C). At S3, a pair of lateral leaflet primordia emerged between the stipule and common leaf primordia (Fig. 1D). Because no additional primordia developed after this stage, the common leaf primordium differentiated into a terminal leaflet primordium. Subsequently, at S4, the stipule and lateral leaflet primordia were separated away from each other so that boundaries were established between the stipule and lateral leaflet primordia (Fig. 1E). Adaxial-abaxial specification of the terminal leaflet primordium was apparent at this stage, because trichomes initiated as spherical outgrowth from the abaxial surface of the terminal leaflet primordium, indicating that the adaxial-abaxial polarity first established in the terminal leaflet primordium. Following S4, the lateral and terminal leaflet primordia grew away from each other, and boundaries formed between them at S5 (Fig. 1F). At this stage, it was apparent that trichomes differentiated further as tubular trichomes elongated from the abaxial surface of the terminal leaflet primordium (Fig. 1F). At S6, trichomes developed from the abaxial surface of the stipule and lateral leaflet primordia. Furthermore, at this stage, the abaxial surface of the terminal leaflet primordium outgrew the adaxial surface such that the terminal leaflet primordium became folded (Fig. 1G). At the subsequent stage (S7), the region between stipule and lateral leaflet primordia expanded to become a petiole as a result of cell division and cell expansion (Fig.1, H and I). At S8, differentiation of petioles was apparent as trichomes initiated from the adaxial surface of petioles. Furthermore, at this stage, the region between lateral and terminal leaflet primordia expanded to form a rachis (Fig. 1, J–M). And, at the last stage (S9), the proximal regions of lateral and terminal leaflet primordia expanded to form petiolules (Fig. 1, L and M; data not shown).
Isolation and Characterization of M. truncatula sgl1 Mutants
We screened and isolated a leaf development mutant with four alleles from a M. truncatula mutant collection generated by tobacco Tnt1 retrotransposon insertion mutagenesis (d'Erfurth et al., 2003; Tadege et al., 2005; Tadege et al., 2008). These mutants were named sgl1-1 to sgl1-4, because all adult leaves are simple in these mutants, resembling the first leaf (juvenile leaf) developed in the wild-type plants (Figs. 1A and 2, A–C). Flowers developed in sgl1 mutants were abnormal and infertile, lacking petals and stamens and producing many more flowers within flowers with cauliflower-like morphology (Figs. 2G and 3, A–C). Because of their infertility, the sgl1 mutants were maintained as heterozygotes. Progenies from self-pollination of heterozygous lines segregated wild-type-like and mutant plants in a 3:1 ratio, suggesting that the mutant phenotype was linked to a single recessive locus (Supplemental Table S1).
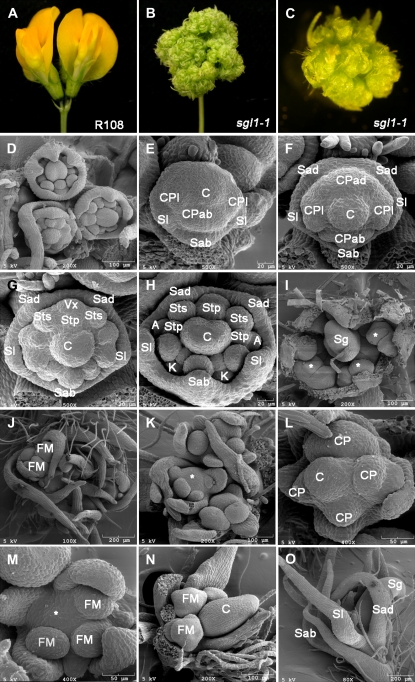
Figure 3.
Flower phenotypes of M. truncatula sgl1 mutants. Two mature flowers developed on a single spike in wild type (R108). The bilateral symmetry along the dorsal-ventral axis was shown. B, Defective flowers developed on a single spike in sgl1-1 mutant. C, A close-up view of a single flower of the sgl1-1 mutant. D to I, Flower development in wild type (R108). D, Three FMs were initiated from I2. E, In S3, the abaxial (Sab) and two lateral (Sl) sepal primordia were initiated. F, In S4, two adaxial sepal primordia (Sad) were initiated. The carpel primordium (C) and the abaxial (CPab) and two lateral (CPl) common primordia formed. G, In S5, four common primordia differentiated into petal and stamen primordia along the abaxial to adaxial axis. Subsequently, the adaxial common primordia produced one inner antepetal (Stp) and two outer antesepal (Sts) stamens and the standard primordium (Vexillum or Vx). H, At subsequent stages, all floral organ primordia formed. A, Alae petals; K, keel petals. I, Stamen primordia differentiated anthers at S8 (asterisk). Sg, Stigma. J to O, Flower development in sgl1 mutants. J, Multiple FM developed from a single I2 inflorescence meristem in sgl1-1 mutant. K, A close-up view of an I2 with multiple FM. L and M, An S5 FM in sgl1-1 mutant initiated three to six common primordia (CP) between sepal (S) and carpel (C) primordia. The carpel primordium was occasionally missing from the center of the FMs (asterisks in K and M). M, Secondary FM were initiated from the second whorl of FMs in sgl1-1 mutant. N, An S7 FM in sgl1-1 mutant with its carpel primordium (C) started to fold. Secondary FM started to differentiate. O, Secondary floral-like meristems developed between the carpel and sepal primordia and gave rise to proliferating structures with elongated sepals (S). Scale bars as indicated. [See online article for color version of this figure.]
To characterize leaf development defects in sgl1 mutants, SEM analyses of leaf development were carried out. These analyses indicated that in sgl1 mutants, leaf development was initially normal until it reached S3, in which the pair of lateral leaflet primordia failed to initiate between the stipule and common leaf primordia (Fig. 2, D–F). All four alleles of the sgl1 mutant exhibited identical defects. The defect in the initiation of lateral leaflet primordia was persistent throughout subsequent developmental stages, resulting in the formation of simple adult leaves in sgl1 mutants (Fig. 2, A–G).
We also examined development of petioles in sgl1 mutants and found that the length of petioles on newly developed leaves at node 10 was not significantly different between sgl1 mutants and the wild-type plants (Fig. 2H). However, petioles on older leaves were significantly shorter in the sgl1 mutants than in the wild-type plants (Fig. 2H), indicating that SGL1 also plays a role in petiole development.
M. truncatula flowers have pentamerous organs in the outermost four whorls (sepals, petals, and outer and inner stamens) and a single carpel in the center. The development of M. truncatula flowers has been previously described (Benlloch et al., 2003). To characterize floral defects of sgl1 mutants, we compared floral development of sgl1 mutants with that of wild-type plants. Consistent with previous studies, we observed that during the reproductive phase of growth, one to three FMs were produced from a secondary inflorescence meristem (I2) in wild-type plants (Fig. 3, A and D). From FMs, floral organ primordia were unidirectionally initiated along the abaxial-adaxial axis in a bilateral symmetric pattern (Fig. 3, E–G). At late S3 development, the abaxial and two lateral sepal primordia (Sl) were initiated from the outermost layer (Fig. 3E). At the subsequent stage, the abaxial common primordium and the central carpel primordium were initiated, followed by the initiation of two lateral common primordia and the adaxial common primordium (Fig. 3, E and F). Between S4 and S5, the four common primordia in the second whorl differentiated to give rise to petal and stamen primordia along the abaxial-adaxial axis (Fig. 3G; alae petal, keel petal, inner antepetal stamen, and outer antesepal stamen). At late S5, all floral primordia were developed (Fig. 3H). Later, at S7, the stigma of the central carpel was folded and the stamen primordia differentiated filaments and anther locules (Fig. 3I; asterisk). By contrast, in sgl1 mutants, three to five incomplete FMs were initiated from I2 (Fig. 3, B, C, J, and K). Furthermore, even though sepals and the central carpel occasionally formed (Fig. 3, K, L, and O), petals and stamens were missing in sgl1 mutants (Figs. 3, N and O). Instead, the common primordia developed from the second whorl formed three to five incomplete FMs, which gave rise to defective flowers of similar inflorescence-like morphology (Fig. 3, J–N).
Molecular Cloning of the SGL1 Gene
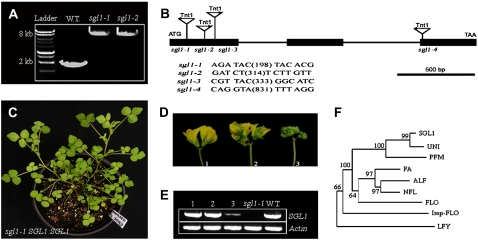
The floral homeotic defects of the sgl1 mutants resemble that of uni and stp mutants. PCR amplification of M. truncatula STP genomic sequence from sgl1 mutants and wild-type plants yielded identical products, indicating that the M. truncatula STP gene is not interrupted in the sgl1 mutants (data not shown). On the contrary, PCR amplification of M. truncatula FLO/LFY/UNI genomic sequence from the sgl1 mutants and the wild-type plants exhibited a difference of 5.3 kb, suggesting that each of the four sgl1 alleles carried a single Tnt1 insert (Fig. 4A; data not shown).
Figure 4.
Molecular cloning of SGL1 gene and functional complementation of sgl1-1 mutant. A, PCR amplification of SGL1gene from wild-type M. truncatula (ecotype R108), sgl1-1, and sgl1-2 mutants. A single insertion of tobacco Tnt1 retrotransposon was detected as a shift in molecular weights of the amplified SGL1gene from each sgl1 mutant allele. B, The intron and exon structure of SGL1 and positions and orientation of Tnt1 insertions in sgl1 mutants. Tnt1 was inserted in the first exon at positions 198 bp, 314 bp, and 333 bp, and in the third exon at the position 831 bp downstream from the translation initiation codon of SGL1 in sgl1-1 to sgl1-4 mutants, respectively. C, A representative sgl1-1 transgenic line transformed with the SGL1 genomic sequence (SGL1:SGL1) exhibited completely rescued wild type-like leaves. D, Representative flowers of three independent sgl1 SGL1:SGL1 transgenic lines, showing completely rescued (line 1), partially rescued (line 2), and nonrescued (line 3) flowers, albeit leaves from all three lines were rescued. E, RT-PCR analysis of SGL1 expression level in developing flowers of three independent sgl1 SGL1:SGL1 transgenic lines (lanes 1–3) compared with that of the sgl1-1 mutant (lane 4) and wild type (lane 5). An Actin gene was used as an internal control (bottom). F, Phylogenetic analysis of SGL1 and its putative orthologs: FLO of snapdragon, LFY of Arabidopsis, NFL of tobacco, UNI of pea, ALF of petunia, Imp-FLO of impatiens, FA of tomato, and PFM of L. japonicus. Bootstrap supports above 50% from 1,000 replicates were shown. [See online article for color version of this figure.]
Flanking sequence analyses indicated that Tnt1 was inserted in the corresponding SGL1 gene in the first exon at positions 198, 314, and 333 bp, and in the third exon at the position 831 bp downstream from the translation initiation codon ATG in sgl1-1 to sgl1-4 alleles, respectively (Fig. 4B). Furthermore, the orientation of Tnt1 in sgl1-1 and sgl1-4 was opposite from that in sgl1-2 and sgl1-3 alleles (Fig. 4B).
Cosegregation and Genetic Complementation Analysis
All four sgl1 alleles were infertile. Segregation analysis of an F2 population of the sgl1-1 allele indicated that 34 out of a total of 139 individuals were homozygous for Tnt1 insertion and exhibited both simple leaf and floral homeotic defects, suggesting that Tnt1 insertion in the corresponding SGL1 gene cosegregated with the mutant phenotype (Supplemental Table S1).
To confirm that we have cloned the SGL1 gene, we carried out a genetic complementation test in sgl1-1 mutant using Agrobacterium tumefaciens-mediated stable transformation with a wild-type SGL1 genomic sequence. Phenotypic analysis of transgenic plants indicated that three lines were completely rescued, exhibiting both wild-type-like compound leaves and flowers (Figs. 4C; data not shown). The other two lines were only rescued for the compound leaf phenotype (data not shown), the floral phenotype of which was only partially rescued in one line but not at all in the other line (Fig. 4D).
We examined the expression level of the introduced SGL1 gene in these stable transgenic lines using reverse transcription (RT)-PCR. The results indicated that the introduced SGL1 gene was expressed in all five transgenic lines (Fig. 4E). However, the expression level was much higher in lines that exhibited fully and partially rescued flowers than in the line with nonrescued flowers (Fig. 4E).
Thus, the interruption of the same gene in four independent lines together with the genetic complementation data unambiguously confirmed that the SGL1 gene is the M. truncatula FLO/LFY/UNI ortholog.
Comparison of the genomic and full-length complementary DNA (cDNA) sequences of SGL1 indicate that SGL1 consists of three exons and two introns (Fig. 4B). Furthermore, the intron and exon structure of SGL1 was very similar to that of FLO, LFY, and UNI. Phylogenetic analysis of selected FLO/LFY orthologs from a diverse group of species placed SGL1 in close proximity to UNI from pea and PFM from L. japonicus (Fig. 4F). Together, they formed a cluster that is distantly related to FLO from snapdragon (Coen et al., 1990), LFY from Arabidopsis (Weigel et al., 1992), and FLO/LFY orthologs from other species: Imp-FLO from impatiens (Impatiens balsamina; Pouteau et al., 1998), NFL1 and NFL2 from tobacco (Kelly et al., 1995), ABBERANT LEAF AND FLOWER (ALF) from petunia (Petunia hybrida; Souer et al., 1998), and FALSIFLORA (FA) from tomato (Molinero-Rosales et al., 1999).
Tissue-Specific Expression of SGL1
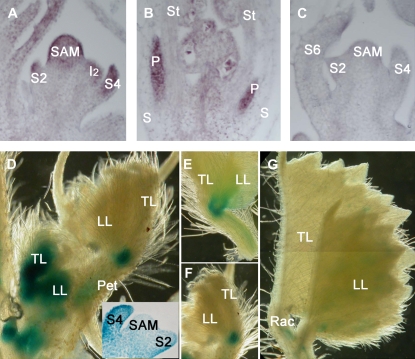
RNA in situ hybridization data revealed that SGL1 was expressed in the SAM and emerging leaf primordia (Fig. 5A). The highest expression was detected in the distal region of leaf primordia (Fig. 5A). During the reproductive phase of growth, transcripts of SGL1 were detected in the I2 (Fig. 5A). SGL1 transcripts were also detected in the peripheral region of FMs (data not shown). During S7 of floral development (Benlloch et al., 2003), SGL1 transcripts were detected in petal primordia (Fig. 5B).
Figure 5.
Expression pattern of SGL1 gene. A to C, RNA in situ hybridization analysis of SGL1 gene expression. A, SGL1 gene expression was detected in SAM, developing leaf primordia (S2 and S4), and I2. B, SGL1 gene expression was detected in developing floral organs in S7 flowers of wild-type plants. P, Petal; S, sepal; St; stamen. C, SGL1 sense probes were used as a negative control, no hybridization signal was detected in SAM and leaf primordia (S2, S4, and S6). D to G, SGL1:GUS histochemical staining pattern. D, GUS staining was restricted to the SAM (inset), developing leaf primordia at early stages (inset), vascular tissues of petioles, and the basal regions of leaflets at late stages. E and F, Close-up views of GUS staining patterns at the basal region of leaflets. G, GUS staining in mature leaves.
To gain a better spatial and temporal resolution, we fused the SGL1 promoter to the Escherichia coli uidA gene encoding GUS and introduced the resulting SGL1:GUS construct into M. truncatula wild-type plants (R108 ecotype). Out of four independent transgenic lines obtained, three showed strong and consistent GUS staining patterns, and the remaining one did not show any detectable GUS activity. We found that GUS staining from the GUS positive lines was restricted to the SAM and emerging leaf anlagen during early stages of leaf development (Fig. 5D, inset), consistent with our RNA in situ hybridization data. Interestingly, we observed that strong GUS staining was in the entire young leaflets (Fig. 5D, inset). In older leaves, GUS staining was detected at the proximal region of leaflets and regions in rachis where leaflets were attached (Figs. 5, D–F). GUS staining in leaves was gradually reduced and eventually disappeared when leaves aged (Fig. 5G). In petioles and rachis, GUS staining was mainly detected in the vascular tissues (Figs. 5, D–F).
Functional Conservation between LFY and SGL1
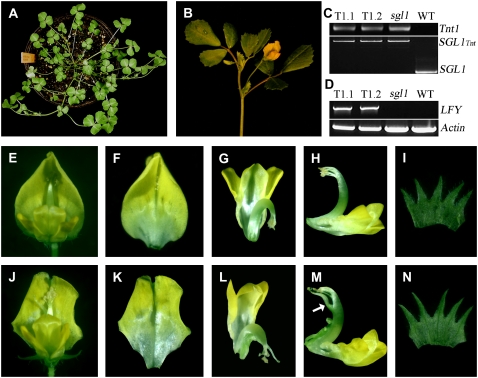
Previous studies have shown that FLO or LFY encodes a plant-specific transcription factor that controls FM identity and plant architecture (Coen et al., 1990; Weigel et al., 1992; Yoon and Baum, 2004). The role of FLO/LFY and their orthologs from angiosperm species in FM identity control is highly conserved (Maizel et al., 2005). Other studies suggest that mutations in LFY coding and/or 5′ upstream sequences are involved in driving morphological variations in closely related species during evolution (Yoon and Baum, 2004). Thus, it is not clear whether development of different leaf forms between Arabidopsis and M. truncatula is attributed to differences in sequences between LFY and SGL1. To test this, we carried out reciprocal genetic complementation experiments in Arabidopsis lfy and M. truncatula sgl1 mutants. First, we transformed homozygous sgl1 mutants with a full-length LFY cDNA driven by the native SGL1 promoter (SGL1:LFY). Out of six stable transgenic lines obtained, all showed rescued phenotypes: wild-type-like compound leaves and flowers (Fig. 6, A and B, E–N). RT-PCR analyses indicated that the introduced LFY gene was expressed in these transgenic lines (Figs. 6, C and D). Furthermore, development of floral organs was also rescued in the transgenic lines (Fig. 6, E–N). Although LFY driven by the native SGL1 promoter rescued both leaf and floral defects of sgl1-1 mutant, we observed some minor developmental defects in the transgenic lines, i.e. the central carpel appeared to elongate slightly faster than the staminal tube such that it often curled and protruded out of the staminal tube in the transgenic lines (Fig. 6, E–N, arrow).
Figure 6.
Genetic complementation of sgl1 mutants by LFY. A, Homozygous sgl1-1 mutant plants transformed with SGL1:LFY, all adult leaves exhibiting wild-type compound leaf morphology. B, Wild-type-like flowers were developed from secondary inflorescence from axils of compound leaves of sgl1 SGL1:LFY transgenic lines. C, PCR-based genotyping of sgl1 SGL1:LFY transgenic lines indicated that three independent transgenic lines were homozygous for Tnt1 insertion in SGL1 gene (lanes 1–3) in contrast to the wild-type M. truncatula (ecotype R108), where no Tnt1 insertion was detected in SGL1 gene (lane 4). Top segment for detecting Tnt1 inserts; bottom for detecting Tnt1 insertion in SGL1. D, RT-PCR analysis of expression of LFY gene in sgl1 SGL1:LFY transgenic lines. Shown were LFY expression in two independent sgl1 SGL1:LFY transgenic lines (lanes 1 and 2), sgl1-1 mutant (lane 3), and wild-type M. truncatula (lane 4). Expression of an Actin gene was used as an internal loading control (bottom). E, A typical mature flower of wild-type M. truncatula (ecotype R108), exhibiting bilateral symmetric zygomorphic morphology. F to I, Floral organs of wild-type M. truncatula were dissected. Shown were a top view of vexillum (F), top (G), and side (H) views of keel and alae, the central carpel enclosed by the staminal tube, and a top view of dissected sepals (I). J, A representative mature flower of sgl1 SGL1:LFY transgenic lines, exhibiting wild-type-like morphology. K to N, Floral organs of sgl1 SGL1:LFY transgenic lines were dissected. Shown were a top view of vexillum (K), top (L) and side (M) views of keel and alae, the central carpel, and a top view of dissected sepals (N).
Second, homozygous lfy mutant plants were transformed with the full-length SGL1 cDNA driven by the LFY promoter. Sixteen out of 17 transgenic lines expressed the SGL1 gene and rescued the lfy floral defects (Figs. 7, A–C, E and F; Supplemental Table S2). The remaining line did not express the SGL1 gene to a detectable level and did not rescue the lfy phenotype (data not shown). However, both rosette and cauline leaves of lfy LFY:SGL1 transgenic lines remained to be simple (Fig. 7C; data not shown). These data indicate that SGL1 and LFY are also functionally conserved in control of FM identity and that expression of SGL1 driven by the LFY promoter was not sufficient to alter the simple leaf structure of the Arabidopsis plants.
Figure 7.
Genetic complementation of lfy mutants by SGL1 and comparison of expression patterns of LFY:GUS and SGL1:GUS in Arabidopsis. A, Inflorescence of homozygous lfy (left) and lfy transformed with LFY:SGL1 (right). B to D, Two-week-old wild-type (B), lfy LFY:SGL1 (C), and lfy SGL1:SGL1 (D) seedlings; insets, mature flowers. E, Morphology of lfy mutant flowers. F, RT-PCR analysis of SGL1 expression in wild-type M. truncatula (lane 1), lfy (lane 2), and three independent transgenic lfy lines transformed with LFY:SGL1 (lanes 3–5). G, RT-PCR analysis of SGL1 expression in M. truncatula (lane 1), Arabidopsis (lane 2), and two independent transgenic lfy lines transformed with the SGL1 genomic sequence (SGL1:SGL1; lanes 3 and 4). An Actin gene was used as an internal loading control (F and G; bottom). H and L, Histochemical staining of LFY:GUS in mature and young seedlings. Strong GUS staining was restricted to the base of flowers, vascular tissues of inflorescence stems, and the shoot apex but not of pedicels and leaves. M to Q, Histochemical staining of SGL1:GUS in mature and young seedlings. Strong GUS staining was detected in style, but not at the base of flowers, and in vascular tissues of inflorescence stems, pedicels, sepals, carpel, cauline, and rosette leaves, and the shoot apex. In cauline and rosette leaves, a high level of GUS staining was localized to the distal margins (arrows).
M. truncatula SGL1 Genomic Sequence Functionally Rescued lfy Floral Defects But Failed to Alter the Simple Leaf Structure of the Arabidopsis Plants
Even though LFY was expressed at the periphery of vegetative SAM, its expression was largely excluded from developing leaves and pedicels (Weigel et al., 1992; Yoon and Baum, 2004; Fig. 7, H–L). By contrast, the M. truncatula SGL1 gene was expressed in the entire SAM, including the central rib region, the initiating leaflet primordia, and developing leaflets (Fig. 5, A, D–G). In transgenic Arabidopsis lines, the introduced SGL1:GUS reporter gene was also expressed in vascular bundles and at margins of developing leaves, and in sepals, pedicels, inflorescence stems, and styles of flowers (Fig. 7, M–Q).
To address the question whether differences in the expression patterns between LFY and SGL1 underlie development of different forms of leaves in Arabidopsis and M. truncatula, we generated homozygous lfy transgenic lines transformed with the SGL1 genomic sequence. Out of 17 transgenic lines obtained, all properly expressed the SGL1 gene and rescued the lfy floral defects (Fig. 7, D and G; Supplemental Table S2), indicating that the SGL1 genomic sequence is sufficient to rescue the lfy floral defects, despite differences in the promoter activities between SGL1 and LFY. However, both rosette and cauline leaves in transgenic lfy SGL1:SGL1 lines remained to be simple (Fig. 7D; data not shown). These data strongly suggest that expression of the M. truncatula SGL1 gene driven by the SGL1 promoter, which is active in the initiating leaf primordia and developing leaves, was not sufficient to alter the simple leaf structure in Arabidopsis.
DISCUSSION
The Role of FLO/LFY Orthologs in Compound Leaf Development
In most compound-leafed species, activation of KNOX1 gene expression in initiating leaf primordia is correlated with development of compound leaves. In simple-leafed species, however, expression of the KNOX1 genes is permanently down-regulated in the initiating leaf primordia in a process that requires MYB domain transcription repressors RS2 and AS1 in maize and Arabidopsis, respectively, and involves hormonal signaling (Smith et al., 1992; Lincoln et al., 1994; Nishimura et al., 1999; Timmermans et al., 1999; Tsiantis et al., 1999; Byrne et al., 2000; Ori et al., 2000; Sakamoto et al., 2001; Hay et al., 2002; Hay et al., 2006; Uchida et al., 2007; Guo et al., 2008). Molecular genetic studies suggest that regulatory processes leading to compound leaf development are more complex than that required for simple leaf development and involve multiple regulators such as the KNOX1 homeobox genes, the TCP class transcription factor Lanceolate, whose activity is in turn regulated by miR319 and perhaps other yet-to-be-identified regulators (Bharathan and Sinha, 2001; Bharathan et al., 2002; Brand et al., 2007; Jasinski et al., 2007; Ori et al., 2007).
In compound-leafed legume species such as pea and alfalfa, in which the KNOX1 genes are excluded from the leaf primordia and thus are not likely correlated with compound leaf development in these plants, the FLO/LFY orthologs appear to function in place of the KNOX1 genes in control of compound leaf development (Champagne et al., 2007). Additional support for the role of FLO/LFY in compound leaf development comes from studies of tomato fa mutants and L. japonicus pfm mutants, in which the corresponding FLO/LFY orthologs are mutated (Molinero-Rosales et al., 1999; Dong et al., 2005), and from studies of down-regulation of two LFY genes in soybean (Champagne et al., 2007). In tomato fa mutants, the number of small intercalary leaflets is reduced, although the number of primary leaflets is not affected (Molinero-Rosales et al., 1999). In L. japonicus pfm mutants, 99% of compound leaves are lacking one or two basal leaflets (Dong et al., 2005). In transgenic soybean lines in which the endogenous LFY genes are down-regulated, the leaflet number is slightly reduced (Champagne et al., 2007). Although these results collectively support a role for FLO/LFY in compound leaf development in tomato (Solanaceae species) and in L. japonicus and soybean (two legume species), the weaker leaf phenotypes of the fa and pfm mutants suggest that FA and PFM may play a minor role in compound leaf development. Because the weak leaf phenotype in LFY RNA interference transgenic soybean lines may be attributed to partial down-regulation of the endogenous genes, whether the two soybean FLO/LFY orthologs play a significant role in compound leaf development in soybean is not clear so far (Champagne et al., 2007).
By contrast, M. truncatula sgl1 mutants exhibit strong compound leaf defects with all adult leaves changed to simple ones (Fig. 2), indicating that the M. truncatula FLO/LFY ortholog SGL1 plays a significant role in the initiation of lateral leaflet primordia required for compound leaf development (Figs. 1 and 2). Similar to sgl1 mutants, pea uni mutants also exhibit strong compound leaf defects, i.e. the number of leaflets is greatly reduced to one to three, and the terminal tendrils are completely missing. Genetic analyses of uni, afila (af), and tendril-less (tl) pea mutants lead to a hypothesis that UNI functions as a determinant of a central-fated shoot-like structure (Hofer and Ellis, 1998).
It is well established that FLO/LFY plays a key role in vegetative to reproductive transition, leading to flower development (Blazquez et al., 1997). This function is quite conserved across angiosperm species. Mutants of FLO/LFY orthologous genes are unable to complete the developmental transition, thus forming inflorescence-like structures in place of flowers (Coen et al., 1990; Weigel et al., 1992; Weigel and Nilsson, 1995; Hofer et al., 1997; Souer et al., 1998; Molinero-Rosales et al., 1999; Dong et al., 2005). The involvement of FLO/LFY in transition from development of indeterminate to determinate structures during the reproductive phase of growth suggests that FLO/LFY plays a role in promoting determinacy in inflorescence. The role of SGL1 in the reproductive phase of growth appears to be contradictory with its role in compound leaf development in M. truncatula, because our data suggest that it promotes a transient phase of indeterminate growth (initiation of leaflet primordia). Future experiments are required to reconcile the discrepancy in the function of SGL1 in floral and leaf development in M. truncatula. Interestingly, our hypothesis seems to be consistent with an earlier model, which postulates that UNI functions as a determinant for an indeterminate structure (central-fated shoot-like structure) during compound leaf development in garden pea (Hofer and Ellis, 1998). Furthermore, our hypothesis is consistent with the expression of FLO/LFY orthologs in leaf marginal blastozones (marginal areas with organogenetic activities) in a number of compound-leafed species, from which the leaflet primordia initiate (Hofer et al., 1997; Molinero-Rosales et al., 1999; Busch and Gleissberg, 2003). The transient nature of SGL1 expression in developing leaves and the shortened petiole phenotype observed in sgl1, uni, and pfm mutants, as well as in transgenic soybean lines in which expression of two endogenous LFY genes are compromised, further support the role for FLO/LFY in promoting a transient phase of indeterminate growth, which is required for development of determinate structures, i.e. leaflets (Hofer et al., 1997; Dong et al., 2005; Champagne et al., 2007).
Regulatory Aspects of Compound Leaf and Floral Development
The fact that proper expression of the Arabidopsis LFY rescued compound leaf and floral defects of the sgl1 mutants and proper expression of the M. truncatula SGL1 rescued the floral defects but did not alter the simple leaf structure of the lfy mutants unambiguously demonstrates that LFY and SGL1 are functional orthologs and that differences in leaf forms between the simple-leafed Arabidopsis and compound-leafed M. truncatula are not due to functional differences between LFY and SGL1. Rather, our results support a hypothesis that regulatory processes mediated by the SGL1 function are involved in initiation of leaflet primordia and development of compound leaves in M. truncatula.
Alterations in cis-regulatory sequences, e.g. 5′-upstream sequences, are also implicated in morphological variations during plant evolution (Busch and Gleissberg, 2003; Yoon and Baum, 2004; Baum et al., 2005; Maizel et al., 2005). Our data indicate that SGL1 differs in its promoter activity from LFY, because only SGL1 is highly expressed in developing leaves (Fig. 7). Additionally, SGL1 is expressed in the entire SAM, including the central region, in contrast to LFY, whose expression is restricted to the periphery of SAM and excluded from the center of SAM (Weigel et al., 1992). A large variation in LFY expression pattern in the vegetative SAM has been reported across vascular plants (Busch and Gleissberg, 2003; Yoon and Baum, 2004; Baum et al., 2005). Currently, it is not clear whether variations in the expression of LFY orthologs in the vegetative SAM contribute to morphological variations during plant evolution. However, it was reported that prolonged expression of LFY in developing leaves appears to be associated with species with compound leaves (Busch and Gleissberg, 2003), consistent with the expression pattern of SGL1 during compound leaf development in M. truncatula (Fig. 5).
In contrast to its role in compound leaf development being prominent only in legume species, particularly in the IRLC studied to date, the role of FLO/LFY in inflorescence and FM identity control has been shown to be highly conserved in flowering plants. In loss-of-function mutants of FLO/LFY orthologs, flowers exhibit homeotic defects, including lack of petals and stamens, and FMs being converted into inflorescence-like structures (Coen et al., 1990; Weigel et al., 1992; Hofer et al., 1997; Molinero-Rosales et al., 1999). The development of bilateral symmetric (zygomorphic) flowers in legume species, including M. truncatula, is quite different from development of Arabidopsis flowers with ancestral radial symmetry. In M. truncatula and many other species in the Papilionoideae subfamily of legumes, floral organs are initiated in the following order: sepals, carpel, petals, outer stamens, and inner stamens. Within each whorl, organs are initiated unidirectionally from the abaxial to adaxial direction. Furthermore, heterogeneous organs are initiated in the same whorl, and homogeneous organs are initiated in different whorls (Benlloch et al., 2003; Dong et al., 2005; Feng et al., 2006). By contrast, Arabidopsis and many other eudicot species initiate floral organs in a centripetal pattern in the following direction: sepals, petals, stamens, and carpel (Coen et al., 1990; Smyth et al., 1990; Bowman and Meyerowitz, 1991; Krizek and Fletcher, 2005). Despite dramatic differences in floral organ development among these diverse species, loss-of-function mutants of FLO/LFY orthologs exhibit similar homeotic defects. The observations that proper expression of LFY orthologous genes from flowering plants driven by the native LFY promoter functionally rescued lfy flower defects and expression of LFY rescued sgl1 floral defects strongly support that FLO/LFY orthologs function in FM identity control during reproductive phase of growth (Coen et al., 1990; Weigel et al., 1992; Molinero-Rosales et al., 1999; Dong et al., 2005). Future work intended to dissect regulatory processes mediated by the function of FLO/LFY orthologs promise to provide new insights on compound leaf development in legumes and molecular mechanisms that underlie morphological variations of vascular plants in general.
MATERIALS AND METHODS
Plant Materials and Growth Condition
Medicago truncatula ecotypes R108 and ‘Jemalong’ A17 were used in this study. NF740 (sgl1-1), NF1240 (sgl1-2), NF2138 (sgl1-3), and NF2703 (sgl1-4) alleles were isolated from a tobacco (Nicotiana tabacum) Tnt1 retrotransposon tagged mutant collection of M. truncatula (d'Erfurth et al., 2003; Tadege et al., 2005, 2008). Initially, M. truncatula genotype R108 was transformed with a construct containing the complete Tnt1 retroelement of tobacco as described (d'Erfurth et al., 2003). Tnt1 tagging was carried out using one of these transgenic lines as starting material and tissue culture to activate Tnt1 transposition. Mutagenized plants that contain multiple independent Tnt1 inserts were regenerated from leaf explants of the starter line via somatic embryogenesis as described (Tadege et al., 2005, 2008). The four sgl1 alleles were identified from screening of a segregating population of approximately 3,000 independent R1 lines. Arabidopsis (Arabidopsis thaliana) lfy heterozygous lines were isolated from a bulked population of SALK_057202 T-DNA insertion line. Plants were grown in MetroMix 350 soil mix in a greenhouse or a growth chamber with the following conditions: 24°C day/20°C night temperature; 16-h day/8-h night photoperiod; 70% to 80% relative humidity, and 150 μmol/m2/s and 80 μmol/m2/s light intensity for M. truncatula and Arabidopsis, respectively.
SEM
Shoot apical and FMs were collected from shoot apices of wild-type and mutant plants 2 to 4 weeks and 2 to 3 months postgermination, respectively. Plant tissues were fixed by vacuum infiltration with 3.0% glutaraldehyde in 25 mm phosphate buffer, pH 7.0, for 1 h and then incubated in 4°C overnight. Plant tissues were further fixed with 1.0% osmium tetroxide in the same phosphate buffer overnight and then dehydrated in a graded ethanol series. Before electron scanning microscopic observations, tissues were critical-point dried in liquid CO2, mounted on aluminum stubs, dissected, and sputter-coated with gold. Specimens were then examined under a Zeiss DSM-960A SEM (Carl Zeiss) at an accelerating voltage of 5 kV. Digital photographs were collected and assembled using Photoshop.
Molecular Cloning of Full-Length SGL1 and MtSTP Genes
To clone full-length M. truncatula SGL1 (MtUNI) and MtSTP genes, we first searched all available M. truncatula sequences in GenBank against UNI and STP. The genomic sequence was available for SGL1 in the database. Therefore, sequence alignments of the SGL1 genomic sequence with UNI/LFY/FLO coding sequences allowed us to define the open reading frame of SGL1. In the case of MtSTP, no sequences were available in the database. Therefore, oligonucleotide primers were designed based on consensus sequences derived from UFO, FIM, and STP sequences. Total RNA was prepared from shoot apices of 2-week-old wild-type plants (‘Jemalong’ A17). RT-PCR was carried out by following the manufacturer's instructions (Invitrogen) using the following oligonucleotide primers: SGL1-forward: 5′-GCTTACCATGGATCCCGACGCATT-3′; SGL1-reverse: 5′-TAACTTAAAAAGGAAGGTGAGCAGTTC-3′; MtSTP-forward: 5′-ATGGAAGGTTTTCACCCATCTATGA-3′; and MtSTP-reverse: 5′-CATAAGCAAAACCATGCAACTCAAAC-3′. PCR products were cloned into pGEM-T Easy vector (Promega) and sequences were confirmed by sequencing and deposited into the GenBank (AY928184 and AY932821).
We also PCR amplified and sequenced the SGL1 genomic sequence from M. truncatula R108 ecotype. The corresponding sequence was also deposited into the GenBank (DQ672589).
Identification of Tnt1 Insertion Sites in SGL1
Genomic DNA samples were prepared from wild-type M. truncatula (R108), and sgl1-1, sgl1-2, sgl1-3, and sgl1-4 alleles using a standard protocol. PCR amplification was carried out using either SGL1 or MtSTP primers listed above. PCR amplification gave rise to a product 5.3 kb larger from sgl1-1 to -4 alleles than from the wild type when SGL1 primers were used. We PCR amplified the Tnt1 flanking sequences from sgl1-1 to sgl1-4 alleles using a combination of SGL1 primers (see above) and Tnt1 primers (Tnt1-upstream, 5′-CTCCAGACATTTTTATTTTTCACCAAG-3′; Tnt1-downstream, 5′-GCATTCAAACTAGAAGACAGTGCTACC-3′). These PCR products were fully sequenced.
Generation of Plant Transformation Constructs
To generate the SGL1 genomic clone for functional complementation of M. truncatula sgl1 mutants, we first PCR amplified SGL1 promoter sequence from wild-type M. truncatula (‘Jemalong’ A17) with the following primers: (SGL1 promoter-forward) 5′-AATTGAATTCAAAAATGGTGTACCAAACATGAGGTAGAA-3′ (an EcoRI site was introduced as underlined) and (SGL1 promoter-reverse) 5′-CGTCGGGATCCATGGTAAGCAATG- 3′ (a unique BamHI site was highlighted). The PCR product was digested by EcoRI and BamHI and cloned into the EcoRI-BamHI sites of pCAMBIA3300 binary vector (the resulting construct was labeled as pCAMBIA3300-pSGL1). Similarly, we PCR amplified the SGL1 coding sequence and 3′ untranslated region from the wild type with the following primers: (SGL1-forward) 5′-AGTTTCATTGCTTACCATGGATCC-3′ (a unique BamHI site was highlighted) and (SGL1-reverse) 5′-AATTCTGCAGAAAAATTAGCCTTTCCCATGCTAAACTTC-3′ (a PstI site was introduced as underlined). The PCR product was digested by BamHI and PstI and cloned into the BamHI-PstI sites of the pCAMBIA3300-pSGL1 construct. The resulting construct pCAMBIA3300-SGL1 was sequenced and confirmed to contain no errors and was then introduced into disarmed Agrobacterium tumefaciens EHA105 and AGL1 strains via electroporation.
To generate the Arabidopsis LFY clone for complementation of M. truncatula sgl1 mutants, we first recloned the SGL1 promoter into the EcoRI-BamHI sites of pCAMBIA3301. We then PCR amplified the full-length LFY cDNA from pDW123 (Blazquez et al., 1997) using the following primers: (LFY-forward) 5′-TTATGGATCCTGAAGGTTTCACGAG-3′ (a unique BamHI site was highlighted) and (LFY-reverse) 5′-AATTGGTCACCCTAGAAACGCAAGTCGTCGC-3′ (the underlined sequence was an introduced BstEII site). The PCR product was digested by BamHI and BstEII and subcloned into the BamHI-BstEII sites of the pCAMBIA3301-pSGL1 construct. The resulting construct pCAMBIA3301-pSGL1∷LFY was introduced into A. tumefaciens EHA105 and AGL1 strains via electroporation.
We generated pCAMBIA3301-pSGL1∷GUS construct by restriction enzyme digestion of the pCAMBIA3301-pSGL1 with NcoI and followed by religation, which removed cauliflower mosaic virus 35S promoter between SGL1 promoter and GUS gene from the vector. The resulting construct CAMBIA3301-pSGL1∷GUS was introduced into A. tumefaciens EHA105 and AGL1 strains.
To clone the full-length SGL1 cDNA for genetic complementation of Arabidopsis lfy mutants, we amplified the Arabidopsis LFY promoter from Columbia-0 wild-type plants by PCR using the following primers: (LFY promoter-forward) 5′-TATAGAATTCGTAATGGGCTGACCGAGAAGATATAAA-3′ (an EcoRI site was introduced as underlined) and (LFY promoter-reverse) 5′-CGTGAAACCTTCAGGATCCATAATCTA-3′(a unique BamHI site was highlighted). The PCR product was digested with EcoRI and BamHI and ligated into the EcoRI-BamHI sites of pCAMBIA3301, resulting in pCAMBIA3301-pLFY. The full-length SGL1 cDNA was reamplified using the following primers: (SGL1-forward) 5′-AGTTTCATTGCTTACCATGGATCC-3′ (a unique BamHI site was highlighted) and (SGL1-reverse2) 5′-AATTGGTCACCTAACTTAAAAAGGAAGGTGAGCAGTTC-3′ (a BstEII site was introduced as underlined). The PCR product was digested by BamHI and BstEII and ligated into the BamHI-BstEII sites of pCAMBIA3301-pLFY. The resulting construct, pCAMBIA3301-pLFY:SGL1, was confirmed by sequencing and introduced into A. tumefaciens GV3101 strain.
Stable Plant Transformation
M. truncatula sgl1-1 (R108 ecotype) and R108 wild type were transformed with A. tumefaciens EHA105 or AGL1 strain harboring the various binary constructs described, using a transformation-regeneration protocol previously described (Chabaud et al., 1996; Trinh et al., 1998; Cosson et al., 2006). Transgenic T1 plants were genotyped using PCR with the following primers for amplification of the selection marker gene: BAR-forward, 5′-CCGTACCGAGCCGCAGGAAC-3′ and BAR-reverse, 5′-CAGATCTCGGTGACGGGCAGGAC-3′.
For Arabidopsis transformation, heterozygous (lfy +/−) plants (SALK_057202) were transformed by A. tumefaciens GV3101 strain harboring various binary constructs described, using the floral dipping method as previously described (Clough and Bent, 1998; Bent, 2006). T1 plants were selected on one-half strength Murashige and Skoog minimal organic medium supplemented with 1% agar and 5 μg/mL glufosinolate (PPT).
RT-PCR
Total RNA was prepared using Tri Reagent (Molecular Research Center). Genomic DNA was removed using a DNA-free kit (Ambion). cDNA synthesis was performed using SuperScript II reverse transcriptase (Invitrogen) starting with 2 μg of total RNA in a 20-μL reaction with oligo(dT)15 primers (Promega) at 42°C for 1 h. Primers 5′-AGACGCCTTGATGAAGAGGAAATTAA-3′ and 5′-TAGCAATTGCTTGAACCTGAATCAAG-3′ were designed to amplify 424 bp of the internal coding region of SGL1. Primers 5′-TCTTACTCTCAAGTACCCCATTGAGC-3′ and 5′-GTGGGAGTGCATAACCTTCATAGATT-3′ were designed to amplify 329 bp M. truncatula Actin as an internal loading control. Primers 5′-CAGTGTCTGGATCGGAGGAT-3′ and 5′-TGAACAATCGATGGACCTGA-3′ were designed to amplify the Arabidopsis Actin gene (At5G09810) as an internal loading control. The PCR was performed as follows: 2 min at 95°C, 25 cycles of 30 s at 94°C, 30 s at 60°C and 1 min at 72°C, and 5 min at 72°C. Amplified products were separated by electrophoresis according to their molecular weights.
Phylogenetic Analysis
Phylogenetic analysis was performed using PAUP version 4 software (paup.csit.fsu.edu/). A maximum parsimony criterion was used to generate the single most parsimonious tree, with bootstrap values from 1,000 replicates and greater than 50% shown above the relevant branches.
RNA in Situ Hybridization
RNA in situ hybridization was essentially carried out as previously described (Coen et al., 1990) with minor modifications. Briefly, SGL1 transcripts were generated from a cDNA clone corresponding to the nucleotide 179 to 1,189 of the coding sequence and labeled with digoxigenin. Then 10-μm sections from shoot apices of 2-week-old R108 and sgl1 mutants were processed and hybridized with digoxigenin-labeled sense and antisense probes followed by detection under the same condition.
Histochemical GUS Staining
GUS staining was carried out as previously described (Shin et al., 2005) with modifications. Briefly, tissue samples were incubated in a GUS staining buffer (100 mm sodium phosphate, pH 7.0, 1 mm EDTA, 0.05% Triton X-100, 1 mm potassium ferricyanide and potassium ferrocyanide, and 1 mg/mL X-glucuronide) at 37°C overnight. Samples were then cleared in 50%, 70%, and 100% ethanol, rehydrated with 70% and 50% ethanol, and transferred to glycerol:ethanol solution (1:1, v/v) before microscopic examination. Images of GUS staining patterns were collected from a digital camera mounted on a dissection microscope (SMZ1500, Nikon) and assembled using Adobe Photoshop Elements 4.0 package.
Sequence data from this article can be found in the EMBL/GenBank data libraries under accession numbers AY928184, AY932821, and DQ672589.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Amino acid sequence alignments of SGL1 and its orthologs.
Supplemental Table S1. Genetic segregation analysis of the sgl1-1 mutant suggests that sgl1-1 segregates as a single recessive locus.
Supplemental Table S2. Distribution of the floral organ number in homozygous lfy transgenic plants compared with wild-type Columbia-0 plants.
Supplementary Material
Acknowledgments
We thank members of the Chen laboratory for helpful discussions, Kelly Craven and Neelima Sinha for comments on a previous version of the manuscript, Detlef Weigel for providing plasmids, Zeng-yu Wang and Elaine Wright for advice on M. truncatula transformation, Bill Chisso and Preston Larson for assistance with SEM, Kelly Craven and Nikki Charlton for phylogenetic analysis, the Arabidopsis Stock Center at The Ohio State University for providing Arabidopsis lines, and Kuihua Zhang, Xirong Xiao, and Shuirong Zhang for technical assistance.
This work was supported by The Samuel Roberts Noble Foundation and by the European Union (EU FP6–GLIP project).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Rujin Chen (rchen@noble.org).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Baum DA, Yoon HS, Oldham RL (2005) Molecular evolution of the transcription factor LEAFY in Brassicaceae. Mol Phylogenet Evol 37 1–14 [DOI] [PubMed] [Google Scholar]
- Benlloch R, Navarro C, Beltran JP, Canas LA (2003) Floral development of the model legume Mediago truncatula: ontogeny studies as a tool to better characterize homeotic mutations. Sex Plant Reprod 15 231–241 [Google Scholar]
- Bent A (2006) Arabidopsis thaliana floral dip transformation method. Methods Mol Biol 343 87–103 [DOI] [PubMed] [Google Scholar]
- Bharathan G, Goliber TE, Moore C, Kessler S, Pham T, Sinha NR (2002) Homologies in leaf form inferred from KNOXI gene expression during development. Science 296 1858–1860 [DOI] [PubMed] [Google Scholar]
- Bharathan G, Sinha NR (2001) The regulation of compound leaf development. Plant Physiol 127 1533–1538 [PMC free article] [PubMed] [Google Scholar]
- Blazquez MA, Soowal LN, Lee I, Weigel D (1997) LEAFY expression and flower initiation in Arabidopsis. Development 124 3835–3844 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Meyerowitz EM (1991) Genetic control of pattern formation during flower development in Arabidopsis. Symp Soc Exp Biol 45 89–115 [PubMed] [Google Scholar]
- Brand A, Shirding N, Shleizer S, Ori N (2007) Meristem maintenance and compound-leaf patterning utilize common genetic mechanisms in tomato. Planta 226 941–951 [DOI] [PubMed] [Google Scholar]
- Busch A, Gleissberg S (2003) EcFLO, a FLORICAULA-like gene from Eschscholzia californica is expressed during organogenesis at the vegetative shoot apex. Planta 217 841–848 [DOI] [PubMed] [Google Scholar]
- Byrne ME, Barley R, Curtis M, Arroyo JM, Dunham M, Hudson A, Martienssen RA (2000) Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408 967–971 [DOI] [PubMed] [Google Scholar]
- Chabaud M, Larsonneau C, Marmouget C, Huguet T (1996) Transformation of barrel medic (Medicago truncatula Gaertn) by Agrobacterium tumefaciens and regeneration via somatic embryogenesis of transgenic plants with the MtENOD12 nodulin promoter fused to the gus reporter gene. Plant Cell Rep 15 305–310 [DOI] [PubMed] [Google Scholar]
- Champagne CE, Goliber TE, Wojciechowski MF, Mei RW, Townsley BT, Wang K, Paz MM, Geeta R, Sinha NR (2007) Compound leaf development and evolution in the legumes. Plant Cell 19 3369–3378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Coen ES, Romero JM, Doyle S, Elliott R, Murphy G, Carpenter R (1990) Floricaula: a homeotic gene required for flower development in Antirrhinum majus. Cell 63 1311–1322 [DOI] [PubMed] [Google Scholar]
- Cosson V, Durand P, d'Erfurth I, Kondorosi A, Ratet P (2006) Medicago truncatula transformation using leaf explants. Methods Mol Biol 343 115–127 [DOI] [PubMed] [Google Scholar]
- DeMason DA, Schmidt RJ (2001) Roles of the Uni gene in shoot and leaf development of pea (Pisum sativum); phenotypic characterization and leaf development in the uni and uni-tac mutants. Int J Plant Sci 162 1033–1051 [Google Scholar]
- d'Erfurth I, Cosson V, Eschstruth A, Lucas H, Kondorosi A, Ratet P (2003) Efficient transposition of the Tnt1 tobacco retrotransposon in the model legume Medicago truncatula. Plant J 34 95–106 [DOI] [PubMed] [Google Scholar]
- Dong ZC, Zhao Z, Liu CW, Luo JH, Yang J, Huang WH, Hu XH, Wang TL, Luo D (2005) Floral patterning in Lotus japonicus. Plant Physiol 137 1272–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X, Zhao Z, Tian Z, Xu S, Luo Y, Cai Z, Wang Y, Yang J, Wang Z, Weng L, et al (2006) Control of petal shape and floral zygomorphy in Lotus japonicus. Proc Natl Acad Sci USA 103 4970–4975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourlay CW, Hofer JM, Ellis TH (2000) Pea compound leaf architecture is regulated by interactions among the genes UNIFOLIATA, cochleata, afila, and tendril-lessn. Plant Cell 12 1279–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Thomas J, Collins G, Timmermans MC (January 18, 2008) Direct repression of KNOX loci by the ASYMMETRIC LEAVES1 complex of Arabidopsis. Plant Cell http://www.plantcell.org/cgi/doi/10.1105/tpc.1107.056127 [DOI] [PMC free article] [PubMed]
- Hareven D, Gutfinger T, Parnis A, Eshed Y, Lifschitz E (1996) The making of a compound leaf: genetic manipulation of leaf architecture in tomato. Cell 84 735–744 [DOI] [PubMed] [Google Scholar]
- Hay A, Barkoulas M, Tsiantis M (2006) ASYMMETRIC LEAVES1 and auxin activities converge to repress BREVIPEDICELLUS expression and promote leaf development in Arabidopsis. Development 133 3955–3961 [DOI] [PubMed] [Google Scholar]
- Hay A, Kaur H, Phillips A, Hedden P, Hake S, Tsiantis M (2002) The gibberellin pathway mediates KNOTTED1-type homeobox function in plants with different body plans. Curr Biol 12 1557–1565 [DOI] [PubMed] [Google Scholar]
- Hay A, Tsiantis M (2006) The genetic basis for differences in leaf form between Arabidopsis thaliana and its wild relative Cardamine hirsuta. Nat Genet 38 942–947 [DOI] [PubMed] [Google Scholar]
- Hofer J, Ellis TH (1998) The genetic control of patterning in pea leaves. Trends Plant Sci 3 439–444 [Google Scholar]
- Hofer J, Gourlay C, Michael A, Ellis TH (2001) Expression of a class 1 knotted1-like homeobox gene is down-regulated in pea compound leaf primordia. Plant Mol Biol 45 387–398 [DOI] [PubMed] [Google Scholar]
- Hofer J, Turner L, Hellens R, Ambrose M, Matthews P, Michael A, Ellis N (1997) UNIFOLIATA regulates leaf and flower morphogenesis in pea. Curr Biol 7 581–587 [DOI] [PubMed] [Google Scholar]
- Jackson D, Veit B, Hake S (1994) Expression of maize KNOTTED 1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development 120 405–413 [Google Scholar]
- Jasinski S, Kaur H, Tattersall A, Tsiantis M (2007) Negative regulation of KNOX expression in tomato leaves. Planta 226 1255–1263 [DOI] [PubMed] [Google Scholar]
- Kelly AJ, Bonnlander MB, Meeks-Wagner DR (1995) NFL, the tobacco homolog of FLORICAULA and LEAFY, is transcriptionally expressed in both vegetative and floral meristems. Plant Cell 7 225–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Pham T, Hamidi A, McCormick S, Kuzoff RK, Sinha N (2003) Reduced leaf complexity in tomato wiry mutants suggests a role for PHAN and KNOX genes in generating compound leaves. Development 130 4405–4415 [DOI] [PubMed] [Google Scholar]
- Krizek BA, Fletcher JC (2005) Molecular mechanisms of flower development: an armchair guide. Nat Rev Genet 6 688–698 [DOI] [PubMed] [Google Scholar]
- Lincoln C, Long J, Yamaguchi J, Serikawa K, Hake S (1994) A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell 6 1859–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JA, Moan EI, Medford JI, Barton MK (1996) A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379 66–69 [DOI] [PubMed] [Google Scholar]
- Luo JH, Yan J, Weng L, Yang J, Zhao Z, Chen JH, Hu XH, Luo D (2005) Different expression patterns of duplicated PHANTASTICA-like genes in Lotus japonicus suggest their divergent functions during compound leaf development. Cell Res 15 665–677 [DOI] [PubMed] [Google Scholar]
- Maizel A, Busch MA, Tanahashi T, Perkovic J, Kato M, Hasebe M, Weigel D (2005) The floral regulator LEAFY evolves by substitutions in the DNA binding domain. Science 308 260–263 [DOI] [PubMed] [Google Scholar]
- Molinero-Rosales N, Jamilena M, Zurita S, Gomez P, Capel J, Lozano R (1999) FALSIFLORA, the tomato orthologue of FLORICAULA and LEAFY, controls flowering time and floral meristem identity. Plant J 20 685–693 [DOI] [PubMed] [Google Scholar]
- Nishimura A, Tamaoki M, Sato Y, Matsuoka M (1999) The expression of tobacco knotted1-type class 1 homeobox genes correspond to regions predicted by the cytohistological zonation model. Plant J 18 337–347 [DOI] [PubMed] [Google Scholar]
- Ori N, Cohen AR, Etzioni A, Brand A, Yanai O, Shleizer S, Menda N, Amsellem Z, Efroni I, Pekker I, et al (2007) Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat Genet 39 787–791 [DOI] [PubMed] [Google Scholar]
- Ori N, Eshed Y, Chuck G, Bowman JL, Hake S (2000) Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 127 5523–5532 [DOI] [PubMed] [Google Scholar]
- Pouteau S, Tooke F, Battey N (1998) Quantitative control of inflorescence formation in Impatiens balsamina. Plant Physiol 118 1191–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, Kamiya N, Ueguchi-Tanaka M, Iwahori S, Matsuoka M (2001) KNOX homeodomain protein directly suppresses the expression of a gibberellin biosynthetic gene in the tobacco shoot apical meristem. Genes Dev 15 581–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H, Shin HS, Guo Z, Blancaflor EB, Masson PH, Chen R (2005) Complex regulation of Arabidopsis AGR1/PIN2-mediated root gravitropic response and basipetal auxin transport by cantharidin-sensitive protein phosphatases. Plant J 42 188–200 [DOI] [PubMed] [Google Scholar]
- Sinha NR, Williams RE, Hake S (1993) Overexpression of the maize homeo box gene, KNOTTED-1, causes a switch from determinate to indeterminate cell fates. Genes Dev 7 787–795 [DOI] [PubMed] [Google Scholar]
- Smith LG, Greene B, Veit B, Hake S (1992) A dominant mutation in the maize homeobox gene, Knotted-1, causes its ectopic expression in leaf cells with altered fates. Development 116 21–30 [DOI] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM (1990) Early flower development in Arabidopsis. Plant Cell 2 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souer E, van der Krol A, Kloos D, Spelt C, Bliek M, Mol J, Koes R (1998) Genetic control of branching pattern and floral identity during Petunia inflorescence development. Development 125 733–742 [DOI] [PubMed] [Google Scholar]
- Tadege M, Ratet P, Mysore KS (2005) Insertional mutagenesis: a Swiss Army knife for functional genomics of Medicago truncatula. Trends Plant Sci 10 229–235 [DOI] [PubMed] [Google Scholar]
- Tadege M, Wen J, He J, Tu H, Kwak Y, Eschstruth A, Cayrel A, Endre G, Zhao P, Chabaud M, et al (2008) Large scale insertional mutagenesis using Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J (in press) [DOI] [PubMed]
- Taylor S, Hofer J, Murfet I (2001) Stamina pistilloida, the Pea ortholog of Fim and UFO, is required for normal development of flowers, inflorescences, and leaves. Plant Cell 13 31–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmermans MC, Hudson A, Becraft PW, Nelson T (1999) ROUGH SHEATH2: a Myb protein that represses knox homeobox genes in maize lateral organ primordia. Science 284 151–153 [DOI] [PubMed] [Google Scholar]
- Trinh TH, Ratet P, Kondorosi E, Durand P, Kamate K, Bauer P, Kondorosi A (1998) Rapid and efficient transformation of diploid Medicago truncatula and Medicago sativa ssp falcata lines improved in somatic embryogenesis. Plant Cell Rep 17 345–355 [DOI] [PubMed] [Google Scholar]
- Tsiantis M, Brown MI, Skibinski G, Langdale JA (1999) Disruption of auxin transport is associated with aberrant leaf development in maize. Plant Physiol 121 1163–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsiantis M, Schneeberger R, Golz JF, Freeling M, Langdale JA (1999) The maize rough sheath2 gene and leaf development programs in monocot and dicot plants. Science 284 154–156 [DOI] [PubMed] [Google Scholar]
- Uchida N, Townsley B, Chung KH, Sinha N (2007) Regulation of SHOOT MERISTEMLESS genes via an upstream-conserved noncoding sequence coordinates leaf development. Proc Natl Acad Sci USA 104 15953–15958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM (1992) LEAFY controls floral meristem identity in Arabidopsis. Cell 69 843–859 [DOI] [PubMed] [Google Scholar]
- Weigel D, Nilsson O (1995) A developmental switch sufficient for flower initiation in diverse plants. Nature 377 495–500 [DOI] [PubMed] [Google Scholar]
- Yoon HS, Baum DA (2004) Transgenic study of parallelism in plant morphological evolution. Proc Natl Acad Sci USA 101 6524–6529 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.