Abstract
Clavibacter michiganensis subsp. michiganensis (Cmm) is a gram-positive actinomycete, causing bacterial wilt and canker disease in tomato (Solanum lycopersicum). Host responses to gram-positive bacteria and molecular mechanisms associated with the development of disease symptoms caused by Cmm in tomato are largely unexplored. To investigate plant responses activated during this compatible interaction, we used microarray analysis to monitor changes in host gene expression during disease development. This analysis was performed at 4 d postinoculation, when bacteria were actively multiplying and no wilt symptoms were yet visible; and at 8 d postinoculation, when bacterial growth approached saturation and typical wilt symptoms were observed. Of the 9,254 tomato genes represented on the array, 122 were differentially expressed in Cmm-infected plants, compared with mock-inoculated plants. Functional classification of Cmm-responsive genes revealed that Cmm activated typical basal defense responses in the host, including induction of defense-related genes, production and scavenging of free oxygen radicals, enhanced protein turnover, and hormone synthesis. Cmm infection also induced a subset of host genes involved in ethylene biosynthesis and response. After inoculation with Cmm, Never ripe (Nr) mutant plants, impaired in ethylene perception, and transgenic plants with reduced ethylene synthesis showed significant delay in the appearance of wilt symptoms, compared with wild-type plants. The retarded wilting in Nr plants was a specific effect of ethylene insensitivity, and was not due to altered expression of defense-related genes, reduced bacterial populations, or decreased ethylene synthesis. Taken together, our results indicate that host-derived ethylene plays an important role in regulation of the tomato susceptible response to Cmm.
The gram-positive bacterium Clavibacter michiganensis subsp. michiganensis (Cmm) is the causal agent of canker and wilt disease in tomato (Solanum lycopersicum; Davis et al., 1984). This disease causes severe economic losses that may become devastating to tomato production worldwide. Cmm bacteria penetrate the plant through wounds or natural openings, find their way into the xylem, and develop a massive systemic infection (Jahr et al., 1999). Cmm colonization of xylem vessels leads to the appearance of typical disease symptoms that include unilateral wilting of leaves, leaflet necrosis, and development of canker lesions on the stem.
Although little is known about the virulence strategies utilized by Cmm bacteria during infection, several hypotheses have been proposed to explain the mechanism by which Cmm induces bacterial wilt (Gartemann et al., 2003). Early claims that exopolysaccharides (EPS) are responsible for the wilt symptoms caused by Cmm, either by plugging xylem vessels or through a phytotoxic effect (Van Alfen et al., 1987; Van den Bulk et al., 1989), were ruled out (Jahr et al., 1999). Endophytic growth of certain Cmm strains, without the appearance of disease symptoms, indicated that there is an active enzymatic or other activity that leads to the formation of symptoms (Bermpohl et al., 1996). This was also confirmed by the finding that genes encoded by the endogenous plasmids pCM1 and pCM2 are important virulence determinants (Meletzus et al., 1993). One of these genes, celA, is located on the pCM1 plasmid and encodes a secreted cellulase with endo-β-1,4 glucanase activity (Jahr et al., 2000). CelA and other extracellular enzymes produced by Cmm were hypothesized to cause degradation of cell walls in xylem vessels, resulting in impairment of water transport and consequent wilting (Jahr et al., 2000; Gartemann et al., 2003). An additional Cmm pathogenicity determinant is the pat-1 gene, which is located on the pCM2 plasmid and encodes a secreted, cell-wall-bound protease (Dreier et al., 1997). However, the exact molecular function of Pat-1, and its possible plant protein targets remain to be determined.
Host responses to Cmm infection, and the molecular mechanisms associated with the development of disease symptoms caused by this pathogen are largely unexplored. The attempt of the plant to counteract pathogen invasion is opposed by the virulence mechanisms of the pathogen that contribute to disease development. Upon pathogen infection, recognition of extracellular pathogen-associated molecular patterns (PAMPs) by transmembrane pattern recognition receptors (PRRs) activates basal defense responses in the host plant (Nurnberger et al., 2004; Zipfel and Felix, 2005). These responses have a profound effect on plant cellular activities and they include production of reactive oxygen species, alkalinization of the medium, activation of mitogen-activated protein kinase cascades, and induction of pathogenesis-related genes (Asai et al., 2002; Nurnberger et al., 2004; Zipfel et al., 2004, 2006). For their part, bacterial pathogens have evolved a variety of virulence determinants that subvert host defenses and act to derive nutrients from the plant (Abramovitch et al., 2006). For example, bacterial phytotoxins, plant hormones, EPS, and, for most gram-negative bacteria, effectors of the type III secretion system have been shown to manipulate host physiological processes and contribute to the development of disease (Abramovitch et al., 2006).
Several lines of evidence indicate that during compatible plant-pathogen interactions the host, too, plays an important role in the development of disease symptoms. For example, various Arabidopsis (Arabidopsis thaliana) mutants were reported to form disease-like lesions spontaneously (Pilloff et al., 2002; Yao and Greenberg, 2006). In addition, inhibition of cell death in tomato plants infected with bacterial pathogens limits the appearance of disease symptoms and reduces pathogen growth (Lincoln et al., 2002; del Pozo et al., 2004). Interestingly, the plant hormones ethylene, jasmonic acid (JA), and salicylic acid (SA), which are known to be involved in plant defense responses (Feys and Parker, 2000; Kunkel and Brooks, 2002), have also been reported to play a role in disease development (O'Donnell et al., 2003). Arabidopsis and tomato mutants impaired in ethylene perception develop reduced disease symptoms in response to Pseudomonas and Xanthomonas pathogens (Bent et al., 1992; Lund et al., 1998; Cohn and Martin, 2005). There is also evidence that when tomatoes become infected by Xanthomonas bacteria ethylene promotes SA production, which is critical for the necrosis caused by this pathogen (O'Donnell et al., 2001, 2003). In addition to ethylene and SA, host-derived JA actively contributes to the virulence of phytopathogens. In fact, signaling pathways activated in response to JA are critical to the ability of Pseudomonas bacteria to cause disease on Arabidopsis and tomato (Feys et al., 1994; Zhao et al., 2003).
In recent years, gene expression studies have facilitated significant progress in the characterization of plant responses to virulent and avirulent gram-negative bacteria and to their PAMPs (e.g. Mysore et al., 2002; Tao et al., 2003; Gibly et al., 2004; Navarro et al., 2004; Thilmony et al., 2006; Zipfel et al., 2006). However, host responses to infection by gram-positive bacteria still await elucidation. In this study, we hypothesized that Cmm infection activates tomato basal defense responses and that defense-related hormones are involved in the regulation of such responses and in the development of disease symptoms. To test these hypotheses, we used microarray analysis to identify transcriptional changes in the stems of a susceptible tomato host, in response to Cmm. In addition, we used tomato lines defective in ethylene synthesis or perception to test whether ethylene plays a role in regulation of wilt disease caused by Cmm. Our study provided a detailed description of the main cellular responses triggered by this gram-positive bacterium in tomato stems. Moreover, our findings demonstrate that both synthesis and perception of ethylene play important roles in the development of wilt symptoms caused by Cmm in tomato plants.
RESULTS
Disease Progress and Bacterial Growth in Tomato Plants Infected with Cmm
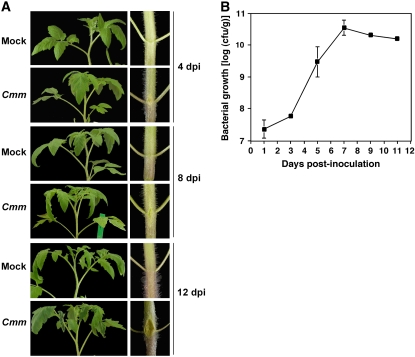
An experimental system was developed to achieve coordinated and reproducible infection of tomato plants by Cmm. Plants of the tomato line Rio Grande were inoculated by injecting a suspension of Cmm bacteria at a titer of 108 colony forming units (cfu)/mL into the stem region between the cotyledons. A mock-inoculation treatment was also included as control, and the plants were monitored for development of canker and wilt disease symptoms. As shown in Figure 1A, at 4 d postinoculation (dpi), canker lesions were visible at the inoculation site in the stem of about 50% of the Cmm-infected plants, whereas wilt symptoms were not yet apparent. Wilting of leaves started to appear at 6 dpi. At 8 dpi about 80% of the Cmm-inoculated plants showed initial wilt symptoms, and the canker lesions had enlarged by spreading in both directions from the inoculation site, and appeared on the stems of all the Cmm-inoculated plants. At 12 dpi, advanced wilt symptoms and large canker lesions were observed in the infected plants. Mock-inoculated plants remained symptom-free throughout the course of the experiment. To establish a correlation between the appearance of disease symptoms in infected plants and the size of bacterial populations, Cmm growth in stem tissues above the inoculation site was monitored for 11 d. As shown in Figure 1B, bacterial multiplication was linear during the first days after infection and approached saturation at 7 dpi. In typical experiments Cmm populations colonized stem tissues with a titer of up to 1010 cfu/g. Taken together, these results show that in our experimental system initial canker lesions appear when Cmm is still growing in infected stems, whereas leaf wilting and formation of enlarged cankers is observed when the bacterial growth has already reached saturation.
Figure 1.
Disease symptoms and bacterial growth in tomato plants infected with Cmm. Four-week-old plants of the tomato line Rio Grande were inoculated with a suspension of Cmm bacteria at a titer of 108 cfu/mL in the stem region between the cotyledons or mock inoculated. A, Leaves and stems photographed at 4, 8, and 12 dpi. B, Bacterial populations estimated from stem samples harvested at different time points during an 11-d period after inoculation. Data represent the mean ± se (n = 3).
Tomato Transcriptional Changes in Response to Cmm Infection
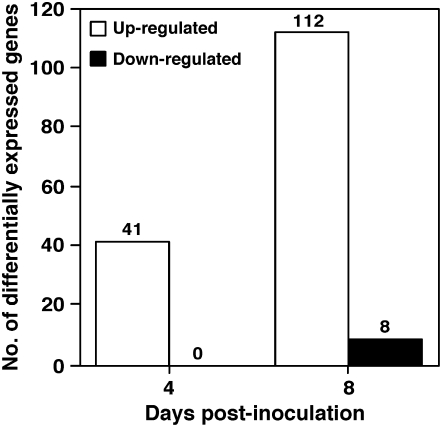
To identify host responses caused by Cmm infection, we analyzed the expression profiles of 9,254 genes represented on the Affymetrix Tomato Genome Array GeneChip. Plants were inoculated with Cmm or mock inoculated, and total RNA was extracted from stem samples taken at two time points: at 4 dpi, when bacterial growth in infected stems was linear and the only disease symptom detected was the initial appearance of canker lesions; and at 8 dpi, when bacterial growth had already reached saturation and distinctive symptoms were visible in stems and leaves (Fig. 1). Gene expression levels were analyzed at the two time points in stems of tomato plants inoculated with Cmm, and were compared with those in mock-inoculated plants by using two independent biological replicates. Genes were considered as differentially expressed if they showed at least a 2-fold change that was significant at P < 0.05 in both replicates. According to these criteria, a total of 122 genes exhibited differential expression in at least one time point (Supplemental Table S1). At 4 dpi, 41 transcripts were up-regulated, whereas none of the genes assayed was down-regulated (Fig. 2). At 8 dpi, 112 genes were up-regulated and eight were down-regulated. Remarkably, 39 out of the 41 genes that were up-regulated at 4 dpi were also up-regulated at 8 dpi, and their changes in expression level were higher at the late time point than at the early one.
Figure 2.
Differential gene expression in tomato stems infected with Cmm. Numbers of genes differentially expressed in tomato stems at 4 and 8 d after inoculation with Cmm bacteria. Genes were considered as differentially expressed if they showed a change by a factor of at least 2, and withstood a statistical Student's t test at P < 0.05, in both biological repeats. White and black bars represent up- and down-regulated genes, respectively.
To validate the microarray results, transcript levels of randomly selected genes that were differentially expressed at 8 dpi were determined by quantitative reverse transcription (RT)-PCR (qRT-PCR) analysis in stems of Cmm- and mock-inoculated plants. These experiments were performed with specific primers for 11 up-regulated and four down-regulated genes (Supplemental Table S2), and in three biological replicates that were independent of those used for microarray analysis. As summarized in Table I, the qRT-PCR data correlated well with the microarray results, confirming the trends of up- or down-regulation of all the analyzed genes. Although differences in the magnitudes of changes were observed between the qRT-PCR and microarray results, 13 out of 15 genes tested by qRT-PCR showed at least a 2-fold change, which was used as the threshold value for differential expression in the microarray analysis.
Table I.
Validation of microarray data by quantitative RT-PCR for 11 up-regulated and four down-regulated genes
| GenBank Accession | Gene Product | Array Ratioa (Cmm/Mock) | qRT-PCR Ratiob (Cmm/Mock) |
|---|---|---|---|
| U89256 | Pti5, ERF/AP2 transcription factor | 163 | 193 |
| AF272366 | Verticillium wilt disease R protein | 88 | 3 |
| AI776170 | Cys protease | 81 | 116 |
| AJ133600 | Extensin | 20 | 105 |
| BG629612 | Chitinase | 55 | 317 |
| AY359965 | EIX receptor 1 | 10 | 9 |
| AW219676 | Wound-induced protein Sn-1 | 8 | 21 |
| AY093595 | Osmotin-like protein | 10 | 2 |
| AW032581 | KNOTTED1 transcription factor | 9 | 24 |
| BG629373 | Zeatin glucosyl transferase | 6 | 6 |
| CN385925 | Patatin-like protein 3 | 5 | 4 |
| AW622607 | Nitrate reductase | 0.5 | 0.6 |
| AW647641 | Peroxidase | 0.5 | 0.9 |
| BG630282 | Probable protein kinase | 0.4 | 0.4 |
| BF176599 | Pro-rich protein | 0.2 | 0.2 |
Ratio between expression in Cmm- and mock-inoculated stems as estimated by microarray analysis at 8 dpi. Data represent the mean of two biological replicates
Ratio between expression in Cmm- and mock-inoculated stems as estimated by qRT-PCR at 8 dpi. Data represent the mean of three biological replicates.
Host Cellular Processes Affected by Cmm Infection
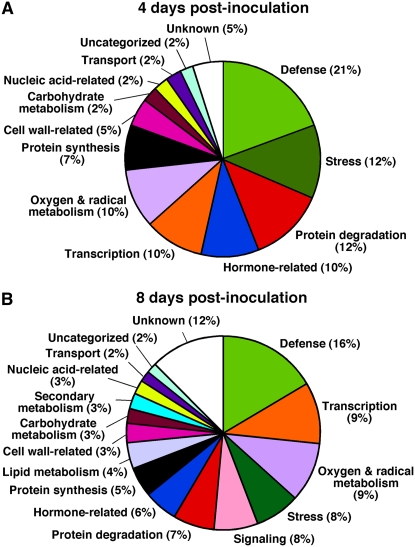
To explore host cellular processes affected by Cmm infection, differentially expressed genes were classified into 16 functional categories, based on annotations of the tomato databases of the Solanaceae Genomics Network and the Institute for Genomic Research (Fig. 3). At the early stage of infection (4 dpi) the most represented category was defense (21%), followed by stress (12%), protein degradation (12%), hormone-related (10%), transcription (10%), and oxygen and radical metabolism (10%; Fig. 3A). Intermediate categories (5%–7%) included genes related to protein synthesis (7%) and cell wall (5%). At the later stage of infection (8 dpi), major categories were defense (16%), transcription (9%), oxygen and radical metabolism (9%), stress (8%), and signaling (8%; Fig. 3B). Intermediate functional groups were: protein degradation (7%), hormone-related (6%), and protein synthesis (5%). It is interesting to note that both at 4 and 8 dpi the functional groups of defense, oxygen and radical metabolism, stress, and hormone-related were significantly enriched (P < 0.05) as compared to their estimated frequency in the tomato genome (Supplemental Table S3). Differentially expressed genes that did not exhibit significant similarity to known genes in the database were classified under the category of unknown function. This category represented 5% and 12% of the Cmm-modulated genes at 4 and 8 dpi, respectively.
Figure 3.
Classification into functional categories of genes differentially expressed between stem tissues of tomato plants infected with Cmm and those of mock-inoculated ones. A and B, Pie charts show classification and percentage of functional categories for 41 and 120 genes differentially expressed at 4 and 8 dpi, respectively.
Prominent in the category of Cmm-responsive genes classified as related to defense were pathogenesis-related genes of various classes, and other genes induced during R gene-mediated resistance, such as a Pto-responsive gene and an Avr9/Cf9 rapidly elicited gene (Supplemental Table S1). This group also contained the EDS1 gene, which is required for resistance mediated by certain classes of disease resistance (R) proteins, and is involved in basal defense against virulent biotrophic fungi and bacterial strains (Wiermer et al., 2005). The up-regulation of two extensin genes, which were categorized in the cell-wall-related category, is also consistent with a defense function. Extensins are, in fact, Hyp-rich glycoproteins that are hypothesized to form cross-linked networks that strengthen the plant cell wall, and so create a barrier to pathogen attack (Wei and Shirsat, 2006). A significant number of genes related to the production and neutralization of reactive oxygen species were found to be up-regulated in this study. The genes of this group showing the highest differential expression were oxidoreductase, disulfide isomerases, and peroxidase.
Because basal defense responses are triggered by the plant surveillance system upon recognition of bacterial PAMPs (Jones and Dangl, 2006), it is interesting that a group of genes encoding cell-surface receptors were differentially expressed during Cmm infection. These proteins include several receptor-like kinases and the putative extracellular glycoproteins EIX1 (Ron and Avni, 2004) and Ve1 (Kawchuk et al., 2001). The gene expression levels of transcription factors of several classes were altered by Cmm infection. There was remarkably high induction of the Pti5 transcription factor, which belongs to the ERF/AP2 class and is involved in transcriptional activation of pathogenesis-related genes (Gu et al., 2002), and there was also high up-regulation of the expression of three genes encoding members of the WRKY family of transcription factors that are well known for their involvement in plant basal defense and disease resistance (Asai et al., 2002; Eulgem, 2005). Genes encoding three zinc finger transcription factors of the C2H2 type, the homeobox KNOTTED1, and a protein with an NAC domain were also differentially expressed. A large proportion of the Cmm-modulated genes were related to protein degradation at both of the analyzed time points. For example, an AAA-type ATPase, which is a component of the 26S proteasome complex, a Cys protease, an ubiquitin-conjugating enzyme, and subtilin-like proteases were found to be highly up-regulated.
Our microarray analysis also revealed differential expression of genes related to plant hormones, examples of which include genes involved in the biosynthesis of JA (allene oxide synthase and lipoxygenase), and polyamines (arginase 2). JA is involved in wound responses and resistance to necrotrophic pathogens (Browse, 2005), whereas polyamines are involved in responses to abiotic and biotic stresses (Walters, 2003; Groppa and Benavides, 2008). Also intriguing was the up-regulation of the SlACO1 gene, which encodes 1-aminocyclopropane-1-carboxylic acid (ACC) oxidase (ACO), a key enzyme of ethylene biosynthesis (Broekaert et al., 2006). Additional genes associated with ethylene signaling and response were identified in other functional categories (Table II). These included: genes encoding transcription factors of the ERF/AP2 family, which bind to a cis-acting element commonly found in the promoter of ethylene-inducible genes (Gutterson and Reuber, 2004); pathogenesis-related genes, e.g. chitinases of various classes, PR1a, and osmotin; and other defense-related genes (Schenk et al., 2000; Zhong and Burns, 2003). Because ethylene is a principal modulator of host-pathogen interactions and was shown to regulate the susceptible response to pathogen infection in tomato plants (Lund et al., 1998; Broekaert et al., 2006), the role of this phytohormone in bacterial canker and wilt disease of tomato was further investigated.
Table II.
Cmm-modulated tomato genes related to ethylene synthesis and response
| GenBank Accession | Gene Product | Expression Ratioa
|
|
|---|---|---|---|
| 4 dpi | 8 dpi | ||
| U89256 | Pti5, ERF/AP2 transcription factor | 21.6 | 163.2 |
| K03291 | Wound-induced protease inhibitor II | 18.1 | 24 |
| BT012691 | 2-Oxoglutarate-dependent dioxygenase E8 | 3.9 | 22.5 |
| CN384809 | ACO1 | 4.9 | 17.6 |
| M69247 | Pathogenesis-related protein PR1a (P4) | 1.8 | 16.2 |
| M69248 | Pathogenesis-related protein 6 (P6) | 3.6 | 13.5 |
| U30465 | Chitinase class 2 | 2.2 | 11.4 |
| AY093595 | Osmotin-like protein | 3.1 | 10.1 |
| Z15141 | Acidic 26-kD endochitinase precursor | 4.5 | 10 |
| BM956714 | Protease inhibitor/lipid transfer protein | 2.6 | 5.8 |
| CN385772 | ERF/AP2 transcription factor | 1.6 | 4.3 |
| BT013355 | Pathogenesis-related protein 2 (P2) | 1.9 | 3.4 |
Ratio between expression in Cmm- and mock-inoculated stems as estimated by microarray analysis at 4 and 8 dpi.
Cmm Infection Promotes Ethylene Production in Tomato Stems and Specific SlACO1 Up-Regulation
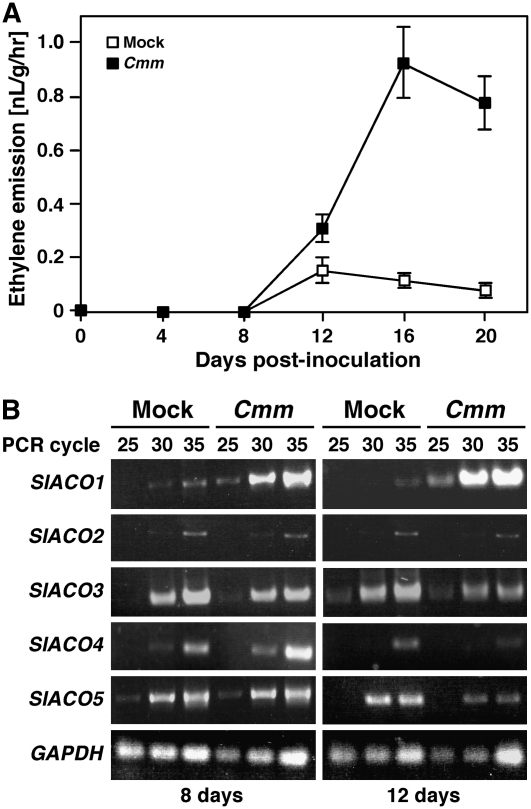
The up-regulation of ethylene-related genes by Cmm prompted us to monitor ethylene production in tomato stem tissues after Cmm infection. As shown in Figure 4A, small, yet significant levels of ethylene were released by Cmm-infected stems, starting at 12 dpi and peaking at 16 dpi when leaf wilting and canker were clearly visible in the plants (Figs. 1A and 4A). Although at 4 and 8 dpi differential expression of ethylene-related genes was already evident, at these time points ethylene accumulation was not detected in Cmm-infected stems (Fig. 4A). Despite the failure to detect ethylene release from stem tissues before the appearance of disease symptoms, we cannot exclude the possibility that at these time points ethylene was produced at levels that are biologically significant but below the detection limit of our experimental system.
Figure 4.
Ethylene emission and expression analysis of the ACO gene family in stems infected with Cmm. Four-week-old Rio Grande tomato plants were inoculated with Cmm or mock inoculated. A, Ethylene emission was measured in inoculated plants at the indicated time points for a period of 20 d. Gas chromatography was performed as described in “Materials and Methods”. Data represent the mean ± se (n = 3) and are representative of two independent experiments. B, Total RNA was extracted from samples of inoculated stems harvested at 8 and 12 dpi, and used for semiquantitative RT-PCR analysis with gene-specific primers designed to amplify individual ACO gene family members or GAPDH, as control. PCR products were sampled at the indicated cycles, separated on a 1% agarose gel, and visualized by ethidium bromide staining.
Ethylene production in response to Cmm infection was suggested by up-regulation of the ACO-encoding gene SlACO1. ACO is encoded in tomato by at least five family members that show different expression patterns during plant development and in response to biotic and abiotic challenges (Barry et al., 1996; Tuomainen et al., 1997; Moeder et al., 2002; Cohn and Martin, 2005). Interestingly, SlACO1 and SlACO2 were recently shown to be up-regulated during a compatible interaction of tomato plants and the bacterial pathogen Pseudomonas syringae pv tomato, whereas other SlACO genes remained unaffected (Cohn and Martin, 2005). In our microarray analysis, although all five SlACO family members were represented on the slides, only the SlACO1 gene was differentially expressed in response to Cmm infection. To confirm that SlACO1 was specifically up-regulated and that transcriptional control of other family members did not contribute to the regulation of ethylene production during the tomato-Cmm interaction, we used semiquantitative RT-PCR to determine the transcript levels of SlACO1, SlACO2, SlACO3, SlACO4, and SlACO5 in stems of Cmm- and mock-inoculated plants. For this analysis we used stem samples harvested either at 8 dpi, before ethylene emission was detected, or at 12 dpi, when ethylene accumulation was already at measurable levels. As shown in Figure 4B, SlACO1 was up-regulated at both time points, whereas transcript abundance of the other family members did not increase significantly upon Cmm infection. Taken together with the SlACO1 differential expression observed in microarray analysis at 4 and 8 dpi, these results indicate that SlACO1 is specifically up-regulated in tomato stems throughout the entire course of infection, whereas ethylene emission from stem tissues reaches measurable levels only concomitantly with the appearance of well-developed disease symptoms.
Impaired Perception and Synthesis of Ethylene Delay the Development of Wilt Symptoms in Tomato
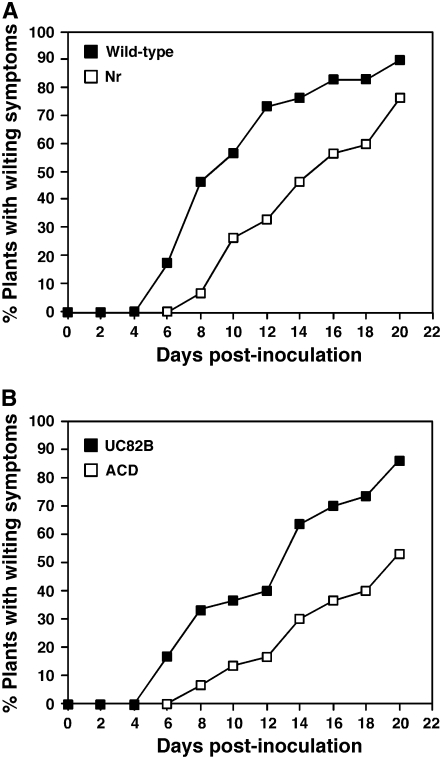
Ethylene previously has been shown to play an important role in symptom development during compatible interactions of tomato plants with phytopathogenic gram-negative bacteria and fungi (Lund et al., 1998; Cohn and Martin, 2005). We therefore tested whether ethylene perception is involved in the regulation of Cmm-induced symptoms. For these experiments we used the ethylene insensitive tomato mutant Never ripe (Nr), which carries a mutation in a member of the ethylene receptor gene family, which renders plants ethylene insensitive in all tissues examined (Lanahan et al., 1994). Nr mutants in the Ailsa Craig background and wild-type plants were inoculated with a Cmm suspension and scored for the development of wilt symptoms during a period of 20 d. In these experiments, to highlight differences between wild-type and mutant plants, we used 6-week-old plants that develop disease symptoms more slowly than the 4-week-old plants used for the microarray analysis. As shown in Figure 5A, in Nr plants the development of wilt symptoms was significantly delayed as compared with wild-type plants. The wilting index, which was defined elsewhere as the number of days after infection at which 50% of the plants displayed the first wilt symptoms (Meletzus et al., 1993), was 9 d for wild-type plants and 15 d for the Nr mutant. Similar results were obtained by comparing the development of disease symptoms in the Nr mutant in the Pearson background with that in wild-type plants (data not shown).
Figure 5.
Effect of impaired ethylene sensitivity or production on development of wilt symptoms in tomato plants infected with Cmm. Six-week-old plants were infected with a Cmm suspension (108 cfu/mL) and examined for development of wilt symptoms during a 20-d period. The percentage of plants showing wilt symptoms was calculated in a group of at least 30 plants for each line. A, The ethylene-insensitive mutant Nr and wild-type Ailsa Craig plants. B, The ethylene-deficient transgenic tomato line ACD and progenitor line UC82B. Data shown are representative of at least two independent experiments for each genetic background.
To substantiate further the involvement of ethylene in the plant host response to Cmm, we tested whether the impairment of ethylene production in the host affected the appearance of wilt symptoms. In these experiments we used a transgenic tomato line expressing the ACD (bacterial ACC deaminase) gene under the control of a constitutive promoter (Klee et al., 1991). ACD converts ACC, the substrate of ACO, into α-ketobutyric acid, and its overexpression in plant tissues inhibits ethylene biosynthesis. As previously observed in the Nr mutant, the appearance of wilt symptoms after inoculation with Cmm was slower in the ACD line than in the UC82B progenitor line (Fig. 5B). The wilting index was determined to be 13 and 20 d for the UC82B and ACD lines, respectively. Ethylene emission was undetectable in stems of Cmm-infected ACD plants (data not shown), in agreement with the reported 95% reduction in ethylene production in leaves and fruits of the ACD line as compared with that in UC82B (Klee et al., 1991). Taken together, these data indicate that both perception and production of ethylene play important roles in the onset of Cmm-induced disease development.
Delayed Appearance of Wilt Symptoms in the Nr Mutant Does Not Correlate with Altered Expression of Defense-Related Genes, Reduced Bacterial Growth, or Decreased Ethylene Emission
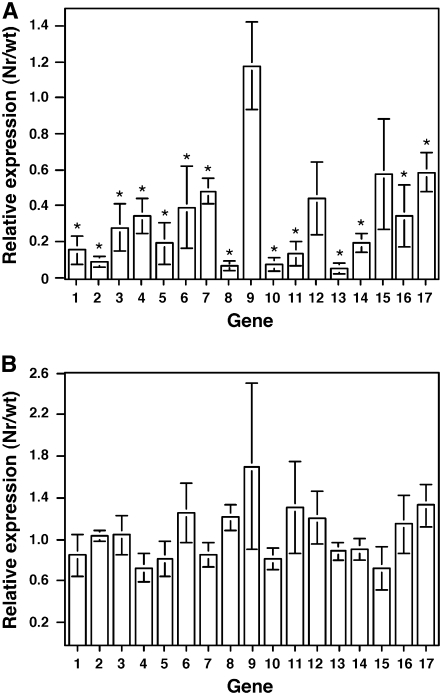
We next tested whether the retarded-wilting phenotype observed in Cmm-inoculated Nr plants resulted specifically from ethylene insensitivity, or was a result of one of the following factors: altered expression of defense-related genes, reduction in the bacterial populations, or decrease of ethylene emission in the stem. To estimate the extent of basal defense responses activated by Cmm infection in the Nr mutant, qRT-PCR analysis was used to compare expression levels of 17 Cmm-modulated defense genes in Nr mutant and wild-type Ailsa Craig plants. Plants were inoculated with Cmm or a mock solution, as previously performed with Rio Grande plants for the microarray experiments, and expression levels of the selected genes were monitored in stems of treated plants at 8 dpi. Induction levels of the analyzed defense-related genes in the Ailsa Craig line correlated well with those previously observed in Rio Grande plants (Supplemental Table S4). The expression of most of these genes (14 of 17) was significantly lower (P < 0.05) in the mock-inoculated Nr mutant than in wild-type plants (Fig. 6A). However, upon Cmm infection expression levels of all monitored genes did not significantly differ between mutant and wild-type plants (Fig. 6B). These results suggest that the Nr mutation affects expression levels of defense-related genes in unchallenged but not in Cmm-infected plants.
Figure 6.
Expression of defense-related genes in the ethylene-insensitive Nr mutant relative to that in wild-type Ailsa Craig plants. Four-week-old Nr and wild-type plants were inoculated with a suspension of Cmm bacteria (108 cfu/mL) or mock inoculated. Total RNA was extracted from leaf samples collected at 8 dpi and used to perform qRT-PCR analysis using primers specific for the following 17 Cmm-modulated defense genes (Supplemental Table S2): 1, PR P6; 2, chitinase class 2; 3, acidic 26-kD endochitinase; 4, protease inhibitor; 5, Pti5; 6, wound-induced protease inhibitor II; 7, Verticillium wilt R protein; 8, cysteine protease; 9, extensin; 10, dioxygenase E8; 11, PR P4; 12, PR P2; 13, chitinase; 14, EIX receptor 1; 15, wound-induced protein Sn-1; 16, osmotin-like protein; 17, patatin-like protein 3. Gene expression levels in Nr plants were calculated relative to those in wild-type plants for mock-inoculated (A) and Cmm-infected plants (B). Values are average ± se (n = 4). Genes, whose expression was significantly different (P < 0.05) in Nr than in wild-type plants according to a two-sided one sample t test, are marked with an asterisk.
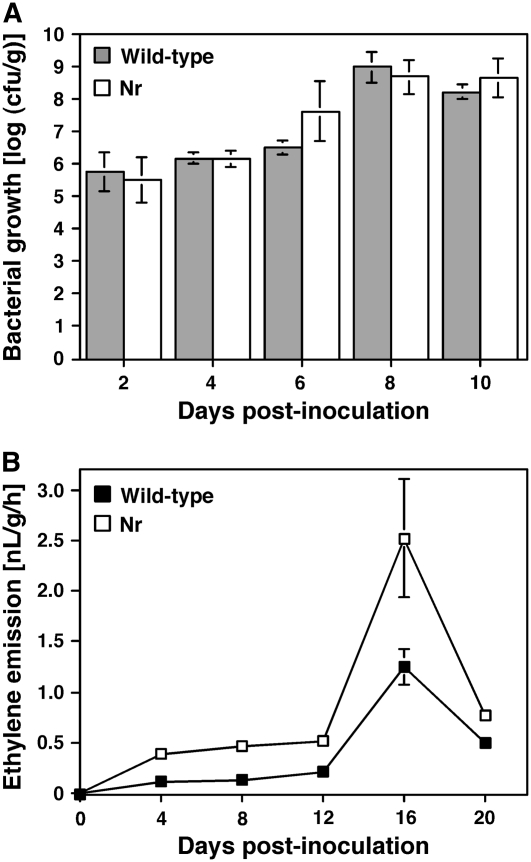
Bacterial population sizes were monitored in stem tissues for 10 d after inoculation of 6-week-old plants of the wild-type Ailsa Craig line, and Nr mutants with a Cmm suspension (106 cfu/mL). As shown in Figure 7A, for the entire course of the experiment the bacterial populations were nearly equivalent in stem extracts derived from wild-type and from Nr plants. In these experiments, which were carried out in the Ailsa Craig background, the bacterial populations at saturation (109 cfu/g) were lower that those observed when Rio Grande plants were infected (Figs. 1B and 7A). This difference can be attributed to differing susceptibility of the two lines and/or to the fact that the Rio Grande plants were used at earlier developmental stages, so that there was a higher ratio between bacterial count in the xylem vessels and the amount of stem tissue utilized for preparation of the extracts. Bacterial populations were also compared between wild-type and Nr mutant plants of the Pearson background, and no significant difference was detected (data not shown), which provides further support for the observations related to the Ailsa Craig background.
Figure 7.
Effect of ethylene insensitivity on bacterial growth and ethylene emission in tomato plants infected with Cmm. Six-week-old plants of the ethylene-insensitive mutant Nr and wild-type Ailsa Craig plants were infected with a Cmm suspension (106 cfu/mL). A, Bacterial populations were estimated in stems of the infected plants from samples harvested during a 10-d period. B, Ethylene emission from stems of infected plants measured at the indicated numbers of days after infection with Cmm. Gas chromatography was performed as described in “Materials and Methods”. A and B, Data represent the mean ± se (n = 3) and are representative of two independent experiments.
As shown in Figure 7B, ethylene emission from stem tissues was detected in both Ailsa Craig wild-type and Nr plants, starting 4 d after Cmm infection, before wilt symptoms were visible, and reached their highest levels at 16 dpi, concomitantly with the appearance of well-developed disease symptoms. Emission of greater amounts of ethylene was observed from the stems of the Nr mutant than from those of the wild-type plants (Fig. 7B). This is consistent with enhanced ethylene emission previously observed in leaves of Nr plants and ascribed to an autoinhibition mechanism that regulates ethylene synthesis and is impaired by ethylene insensitivity (Lund et al., 1998). Taken together, these results indicate that the delayed appearance of wilt symptoms in Nr plants is a specific effect of ethylene insensitivity rather than increased resistance to Cmm.
DISCUSSION
In this study, to get molecular insights into the response of susceptible tomato plants to the gram-positive bacterium Cmm, we analyzed gene expression profiles of 9,254 tomato genes in Cmm-infected stem tissues. This analysis was performed during the early endophytic stage of infection (4 dpi) and again at a later stage (8 dpi) when wilt symptoms in the plant were already visible. We identified a total of 122 genes that were differentially expressed in at least one time point and represent about 1.3% of the genes analyzed. Alterations of gene expression in various compatible plant-pathogen interactions have been reported to typically involve a larger number of genes representing up to 13% of the plant genome (Wise et al., 2007). For example, 643 of 7,680 analyzed genes (13%) were found to be differentially expressed in potato (Solanum tuberosum) plants infected by the fungal pathogen Phytophtora infestans (Restrepo et al., 2005), whereas 1,396 of 24,000 analyzed genes (5.8%) were induced or repressed in susceptible Arabidopsis plants challenged with P. syringae (Thilmony et al., 2006). It should be noted that the number of tomato genes differentially expressed in response to Cmm infection is probably larger than that identified in our study due to the fact that the array we used includes approximately one-fourth (9,254 genes) of the total genes (35,000) that are estimated to be present in the tomato genome (Van der Hoeven et al., 2002). Moreover, the impact of Cmm infection on the tomato transcriptome might have been underestimated, because of the low proportion of stem tissues that directly sense and respond to this bacterium, which specifically colonizes xylem vessels.
The extent of differential gene expression differed between the two analyzed time points: it affected 41 and 120 genes at 4 and 8 dpi, respectively. The vast majority of the genes modulated by Cmm infection at the early time point were also differentially expressed at the later infection stage, which hampered the identification of genes specific to the very first stages of infection. Conversely, a group of 81 genes was specific for the later time point and these genes are possibly associated with physiological processes related to the development of disease symptoms. An unexpected result of our analysis was the very small number of down-regulated genes at each time point. A typical response to various pathogens, which is qualitatively similar in compatible and incompatible interactions, is a shift from housekeeping to defense metabolism, which is associated with down-regulation of genes related to cellular processes such as chloroplast organogenesis, chlorophyll biosynthesis, and carbohydrate metabolism (Mysore et al., 2002; Gibly et al., 2004; Restrepo et al., 2005; Zou et al., 2005). The almost complete absence of gene down-regulation observed during the tomato-Cmm interaction in this study may be related to derepression by Cmm of general metabolic processes that limit the allocation of cellular resources to defense responses.
We anticipated that changes in gene expression that occur during the compatible interaction between tomato and Cmm would reflect the interplay between, on the one hand, the attempt of the plant to counteract pathogen invasion and, on the other hand, the activity of bacterial virulence determinants that facilitate the establishment of disease. Our finding that a large number of the Cmm-modulated genes were related to defense, stress, oxygen metabolism, and protein degradation strongly supports the notion that basal defense responses are activated in tomato plants during their interaction with Cmm. In fact, activation of pathogenesis-related genes, an oxidative burst, and derepression of defense responses by enhanced protein turnover are typical cellular responses activated in plants by recognition of PAMPs (Nurnberger et al., 2004). In addition, classes of genes similar to those differentially expressed during Cmm infection were shown to be modulated in Arabidopsis by perception of the bacterial PAMPs flagellin and EF-Tu (Navarro et al., 2004; Zipfel et al., 2004, 2006).
An important question to be addressed is: what are the Cmm PAMPs perceived by tomato plants and their corresponding PRRs? Cold-shock protein from gram-positive bacteria and various microbial patterns of gram-negative bacteria have been shown to act as PAMPs in plants (Nurnberger et al., 2004). Similarly, Cmm cold shock protein or several cell wall components, which are known to be gram-positive-derived PAMPs in animal systems (Aderem and Ulevitch, 2000), may act as PAMPs during the tomato-Cmm interaction. Additional possible Cmm PAMPs are EPS, which are produced in large amounts by the bacterium (Van den Bulk et al., 1989), or the numerous extracellular cell-wall-degrading enzymes secreted by Cmm, and products of their hydrolytic activity. Only a few of the PRRs responsible for PAMP recognition have been identified to date (Ingle et al., 2006). Interestingly, we found several tomato genes encoding proteins with characteristics of cell-surface receptors that were differentially expressed in response to Cmm infection. These proteins, which can be considered candidate PRRs, include two receptor-like kinases, the Ve1 R protein, which confers resistance in tomato to the vascular disease Verticillium wilt (Kawchuk et al., 2001), and a homolog of the EIX receptor (Ron and Avni, 2004).
The identity of plant targets of Cmm virulence is still elusive, and it is still not clear whether some of the Cmm-modulated genes were differentially expressed as a result of the activity of Cmm pathogenicity determinants. Whereas type III effectors of gram-negative bacteria have been shown to have a large impact on the transcriptome of the host plant (Cohn and Martin, 2005; Thilmony et al., 2006; Truman et al., 2006), the effect of virulence determinants of gram-positive bacteria on plant gene expression is largely unknown. An important observation, derived from our microarray analysis and that may be related to the virulence activity of the pathogen, is the up-regulation of a significant number of genes related to ethylene response, as well as differential expression of the ACO1 gene encoding the ethylene biosynthetic enzyme ACO (Table II). Measurements of ethylene emission confirmed that ethylene production in tomato stem tissues was stimulated by Cmm infection. However, although ethylene-related genes were already up-regulated at 4 and 8 dpi, ethylene release reached detectable levels only at 12 dpi. In addition, it should be noted that ethylene levels were significantly lower than those previously reported to be released from leaves inoculated with foliar bacterial pathogens (Lund et al., 1998; Cohn and Martin, 2005). The late detection and the relatively low levels of ethylene emitted may be ascribed to the lifestyle of Cmm: it is possible that during the early stages of infection ethylene synthesis was localized and restricted to the cells immediately surrounding the xylem vessels where Cmm bacteria proliferate, whereas later in the infection development, maceration of stem parenchymatic cells further enhanced ethylene production.
Our microarray and semiquantitative RT-PCR analyses revealed that transcriptional control of SlACO1, but not of other SlACO family members, may contribute to the regulation of ethylene production during the tomato-Cmm interaction. Interestingly, the SlACO1 and SlACO2 genes were recently shown to be up-regulated by the type III effectors AvrPto and AvrPtoB from Pst (Cohn and Martin, 2005), prompting us to hypothesize that expression of a specific subset of ACO genes may be modulated by virulence determinants of different phytopathogenic bacteria. ACC synthase (ACS) is an additional key enzyme in ethylene biosynthesis, that could be subject to transcriptional regulation (Broekaert et al., 2006). However, none of the six genes encoding ACS present in our microarray were differentially expressed during the tomato-Cmm interaction. Our results correlate with previous reports that ACS gene family members were not up-regulated in tomato challenged with Pst (Cohn and Martin, 2005). These findings together with the evidence that certain members of the ACS family can be regulated at the posttranslational level (Liu and Zhang, 2004) suggest that transcriptional regulation of ACS does not play a critical role in ethylene production during plant-pathogen compatible interactions.
Ethylene plays distinct roles in different plant-pathogen interactions: it contributes to resistance in some interactions but is involved in disease development in others (Kunkel and Brooks, 2002). For example, the ethylene-insensitive Arabidopsis mutant ein2 exhibited increased susceptibility to Botrytis cinerea and Erwinia carotovora (Thomma et al., 1999; Norman-Setterblad et al., 2000). Conversely, Arabidopsis and tomato mutants of ethylene synthesis or perception showed decreased symptoms when infected with virulent strains of Pst and Xcv (Bent et al., 1992; Lund et al., 1998; O'Donnell et al., 2003; Cohn and Martin, 2005). The significant delay in the appearance of wilt symptoms that was observed following Cmm infection of tomato mutants impaired in ethylene synthesis or perception is important evidence that during the Cmm-tomato compatible interaction host-derived ethylene is a major signal that regulates disease progression. In addition, although defense-related genes were expressed at lower levels in mock-inoculated Nr mutant plants than in the wild type, their expression upon Cmm infection was similar in the two genetic backgrounds. This evidence, together with the similar bacterial growth observed in Nr and wild-type plants, suggests that ethylene perception is not important for the regulation of basal defenses against Cmm infection. The delayed appearance of wilt symptoms in the mutant plants started to be evident at 6 dpi, when ethylene emission was barely detectable in wild-type and mutant plants. This observation, along with the differential expression of ethylene signaling and biosynthetic genes as early as 4 dpi, suggest that ethylene, albeit at levels not measurable in our experimental system, plays a physiological function during the initial stages of Cmm infection, before the development of visible disease symptoms.
How ethylene contributes to the development of wilt symptoms during Cmm infection of tomato plants remains to be determined. Host ethylene perception and synthesis have been previously shown to be an important factor in development of wilt symptoms elicited by fungal vascular pathogens (Van der Molen et al., 1983; Lund et al., 1998; Robison et al., 2001). Interestingly, ethylene treatment of various plant species causes xylem occlusion, which leads to the wilt phenotype of vascular diseases (Van der Molen et al., 1983; Perez-Donoso et al., 2007). In addition, it was shown that ethylene produced by galls of Agrobacterium-infected tomato plants decreases the diameter of xylem vessels in the stem adjacent to the tumor (Aloni et al., 1998). In light of these observations, it is tempting to speculate that ethylene emission by Cmm-infected tissues may contribute to the development of wilt symptoms by affecting physical properties of xylem vessels in the host plant.
In conclusion, our study provides a comprehensive view of host responses associated with the infection of tomato plants by a gram-positive bacterium, and reveals that typical basal defense responses and hormone biosynthesis are activated during tomato infection by Cmm. In addition, our findings demonstrate that ethylene plays an important role in the development of Cmm-induced wilt symptoms, and represent a first step in understanding the molecular mechanisms involved in the progression of bacterial wilt disease. This and future investigations will improve our ability to mitigate the effects of this highly detrimental disease.
MATERIALS AND METHODS
Plant Materials and Bacterial Strains
The tomato (Solanum lycopersicum) lines used were: Rio Grande (Pedley and Martin, 2003); Nr mutant and its background lines Pearson (Lanahan et al., 1994) and Ailsa Craig (Alba et al., 2005); and a transgenic line expressing ACD and its background line UC82B (Klee et al., 1991). Plants were grown using standard greenhouse practices (25°C ± 2°C and 16-/8-h light/dark regime). The bacterial strain 42 of Clavibacter michiganensis subsp. michiganensis (Cmm), isolated in Israel, was used throughout this study.
Inoculation Procedures and in Planta Bacterial Growth
Cmm bacteria were grown overnight at 28°C with shaking, in Luria-Bertani broth medium supplemented with rifampicin at 100 mg/L. Bacteria were pelleted by centrifugation at 5,400g for 20 min, washed twice, and diluted to the desired titer in 10 mm MgCl2, as indicated for each experiment. Bacterial suspensions or a 10 mm MgCl2 mock solution (25–50 μL) were injected into the stem region between the cotyledons of 4- to 6-week-old plants with a syringe fitted with a 30-gauge needle. In planta bacterial growth was measured for 10 d at 2-d intervals by grinding three stem pieces (1 cm in length) in 10 mm MgCl2 and plating serially diluted samples on Luria-Bertani broth medium plates supplemented with rifampicin at 100 mg/L. Each stem piece was derived from an independent plant and was cut at 1 cm above the inoculation site. After incubation of the plates at 28°C for 5 d, the number of colony-forming units per gram of tissue (cfu/g) was determined for each sample.
DNA Microarray and Data Analysis
Microarray experiments were performed with the GeneChip Tomato Genome Array (Affymetrix), which contains 10,038 probe sets representing 9,254 transcripts. The procedure followed the Minimum Information about a Microarray Experiment guidelines for international standardization and quality control of microarray experiments (Brazma et al., 2001). Two independent biological repeats were performed for each time point analyzed. For each treatment, a pool of 16 1-cm-long stem pieces, cut from independent plants at 1 cm above the inoculation site, was used as a source for extraction of total RNA with the RNeasy Plant Mini Kit (QIAGEN) according to the manufacturer's instructions. Preparation of biotinylated target complementary RNA (cRNA), hybridization, and imaging procedures were performed according to Affymetrix protocols (http://www.affymetrix.com/support/technical/manuals.affx). Briefly, double-stranded cDNA was synthesized from 5 μg of total RNA and in vitro transcription was performed with biotinylated UTP and CTP, to generate biotinylated target cRNA. The labeled target cRNA was purified, fragmented, and hybridized to the GeneChip Tomato Genome Array for 16 h in a Hybridization Oven 640(Affymetrix). The GeneChips were washed and stained with streptavidin-phycoerythrin using a GeneChip Fluidics Station 400 (Affymetrix) and were then scanned with a GeneArray scanner (Hewlett-Packard). Gene expression data were analyzed with the Affymetrix Microarray Suite (MAS 5.0) software. Output from all GeneChip hybridizations was globally normalized such that its average intensity was equal to an arbitrary target intensity of 800, to enable comparisons between GeneChips. The expression levels of the genes were measured according to the average difference in intensity between the perfect-match probes and mismatch probes. According to their expression level and statistical analysis results, a “present”, “marginal”, or “absent” call was assigned to each probe set. A list of 8,322 “valid” probe sets (out of 10,038 present on the GeneChip), which were probe sets with signals higher than 20 that were detected as present in at least one sample, was generated. Gene expression levels were then compared between sample and control chips from the same biological replicate, to produce a signal log2 ratio. The software was then used to determine whether the signal log2 ratios represented a genuine change in mRNA level (change call I for increase, D for decrease), and to determine the change call significance levels (P values). Variability of the biological repeats was estimated by applying the linear regression method and by calculating correlation coefficient (R2) for each pair of data sets. In all data set comparisons, R2 values were higher than 0.94 indicating high reproducibility of biological repeats. Genes were considered as differentially expressed if they showed a change by a factor of at least 2 in both biological repeats, and the change was significant at P < 0.05, according to Student's t test (Supplemental Table S1). The entire data set, including raw and normalized data from all the microarray experiments is available online from the Tomato Expression Database (http://ted.bti.cornell.edu/cgi-bin/miame/home.cgi). Genes were categorized into functional categories based on annotations of the Solanaceae Genomics Network (http://www.sgn.cornell.edu/) and of The Institute for Genomic Research (http://www.tigr.org/). The distribution in functional categories of annotated genes differentially expressed in response to Cmm was compared to that of the approximately 1,000 annotated tomato genes available from the database. P values for differences in the percentage of each category were calculated using a two-sided binomial test with false discovery rate correction for multiple testing (Benjamini and Hochberg, 1995).
qRT-PCR Analysis
Total RNA was extracted from stems of Cmm-inoculated and control plants as described elsewhere (Chang et al., 1993). RNA samples (2.5 μg) were reverse transcribed for 50 min at 42°C in a 20-μL reaction volume containing 100 units of SuperScript II RNase H− reverse transcriptase (Invitrogen), 10 mm dithiothreitol, 1× reverse transcriptase buffer (Invitrogen), 40 units of RNase inhibitor (Invitrogen), 500 μm each dNTP, and 500 ng of oligo(dT)18-24 primer. The reaction was stopped by incubation at 70°C for 15 min. Gene-specific primers for the analyzed genes were designed to have a Tm of 60°C ± 3°C, by means of the Primer 3 program of the Biology WorkBench software package (http://workbench.sdsc.edu/); they are listed in Supplemental Table S2. The PCR efficiency was determined for each set of primers by using standard curves with four cDNA dilutions. The PCR reactions were performed in triplicate and contained template cDNA, 200 nm gene-specific primers, and SYBR Green QPCR Master Mix (Stratagene) in a volume of 15 μL. Reactions were carried out with an Mx3000P QPCR system (Stratagene) with the following cycling program: 10 min at 95°C, followed by 40 cycles of 30 s at 95°C, 1 min at 60°C, and 1 min at 72°C. Fluorescence was measured at the end of each cycle. The absence of nonspecific products and primer dimers was confirmed by analysis of melting curves. For data analysis, average threshold cycle (CT) values were calculated for each gene of interest, on the basis of three or four independent biological samples, as indicated, and were normalized and used to calculate relative transcript levels of the transcript as described elsewhere (Pfaffl, 2001). The GAPDH (glyceraldehyde-3-phosphate dehydrogenase) gene from tomato (accession no. U97257) was used as an internal standard for normalization.
Semiquantitative RT-PCR Analysis
RNA samples (2.5 μg) were reverse transcribed as described above for the quantitative RT-PCR analysis. One microliter of RT reaction mixture was used for PCR in a 50-μL reaction volume containing 1 unit of Taq DNA polymerase, 200 μm of each dNTP, and 300 nm of each forward and reverse primer for each gene, as listed in Supplemental Table S2. The PCR conditions were: 2 min at 94°C, followed by 40 cycles of 30 s at 94°C, 1 min at 53°C for SlACO1 and SlACO2, 48°C for SlACO3, 55°C for SlACO4, SlACO5, and GAPDH, and 1 min at 72°C. A 10-μL aliquot of each PCR reaction mixture was removed after 25, 30, and 35 cycles, separated on a 1% gel, and visualized by staining with ethidium bromide. To ensure equal amounts of cDNA, a control reaction was performed for each PCR reaction with primers corresponding to the tomato GAPDH gene (accession no. U97257; Supplemental Table S2).
Estimation of Plant Susceptibility to Cmm
The susceptibility of the various tomato lines to Cmm was estimated by daily monitoring of the appearance of wilt symptoms in a group of at least 30 infected plants, over a period of up to 20 d. For a quantitative estimation of susceptibility to Cmm, a wilting index, defined as the number of days required until 50% of the plants showed first wilt symptoms, was determined as described elsewhere (Meletzus et al., 1993).
Ethylene Measurements
For each treatment and at time points ranging from 0 to 20 dpi, 1-cm-long stem pieces were excised from nine independent plants, at 1 cm above the inoculation sites and distributed among three 25-mL flasks. Following a 3-h period during which the stem pieces were left uncovered to allow ethylene produced in response to the wounding to escape, the flasks were sealed for 4 h and 1-mL gas samples were removed with a syringe and measured for ethylene content with a Varian 3350 gas chromatograph (Varian).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Tomato genes differentially expressed in response to infection by Cmm.
Supplemental Table S2. Genes and primer sets used in quantitative and semiquantitative RT-PCR experiments.
Supplemental Table S3. Comparison between the expected and observed frequencies of functional categories for Cmm-modulated genes.
Supplemental Table S4. Induction of defense-related genes in Rio Grande and Ailsa Craig tomato lines upon Cmm infection.
Supplementary Material
Acknowledgments
We thank James Giovannoni for providing the Nr mutant; Harry Klee for the ACD transgenic plants; Yulia Soloveichik for technical assistance; and members of the Sessa Laboratory for critical reading of the manuscript.
This work was supported by the German Research Foundation program for trilateral cooperation among Israel, Palestinian Authority, and Germany (grant no. EI 535/12–1 to R.E., N.I., S.M., I.B., G.S.), and by the U.S.-Israel Binational Agricultural Research and Development Fund (grant no. IS–4047–07 to S.M., I.B., G.S.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Guido Sessa (guidos@post.tau.ac.il).
The online version of this article contains Web-only data.
References
- Abramovitch RB, Anderson JC, Martin GB (2006) Bacterial elicitation and evasion of plant innate immunity. Nat Rev Mol Cell Biol 7 601–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aderem A, Ulevitch RJ (2000) Toll-like receptors in the induction of the innate immune response. Nature 406 782–787 [DOI] [PubMed] [Google Scholar]
- Alba R, Payton P, Fei Z, McQuinn R, Debbie P, Martin GB, Tanksley SD, Giovannoni JJ (2005) Transcriptome and selected metabolite analyses reveal multiple points of ethylene control during tomato fruit development. Plant Cell 17 2954–2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloni R, Wolf A, Feigenbaum P, Avni A, Klee HJ (1998) The never ripe mutant provides evidence that tumor-induced ethylene controls the morphogenesis of Agrobacterium tumefaciens-induced crown galls on tomato stems. Plant Physiol 117 841–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415 977–983 [DOI] [PubMed] [Google Scholar]
- Barry CS, Blume B, Bouzayen M, Cooper W, Hamilton AJ, Grierson D (1996) Differential expression of the 1-aminocyclopropane-1-carboxylate oxidase gene family of tomato. Plant J 9 525–535 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc [Ser A] 57 289–300 [Google Scholar]
- Bent AF, Innes RW, Ecker JR, Staskawicz BJ (1992) Disease development in ethylene-insensitive Arabidopsis thaliana infected with virulent and avirulent Pseudomonas and Xanthomonas pathogens. Mol Plant Microbe Interact 5 372–378 [DOI] [PubMed] [Google Scholar]
- Bermpohl A, Dreier J, Bahro R, Eichenlaub R (1996) Exopolysaccharides in the pathogenic interaction of Clavibacter michiganensis subsp. michiganensis with tomato plants. Microbiol Res 151 391–399 [Google Scholar]
- Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, et al (2001) Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet 29 365–371 [DOI] [PubMed] [Google Scholar]
- Broekaert WF, Delaure SL, De Bolle MF, Cammue BP (2006) The role of ethylene in host-pathogen interactions. Annu Rev Phytopathol 44 393–416 [DOI] [PubMed] [Google Scholar]
- Browse J (2005) Jasmonate: an oxylipin signal with many roles in plants. Vitam Horm 72 431–456 [DOI] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11 113–116 [Google Scholar]
- Cohn JR, Martin GB (2005) Pseudomonas syringae pv. tomato type III effectors AvrPto and AvrPtoB promote ethylene-dependent cell death in tomato. Plant J 44 139–154 [DOI] [PubMed] [Google Scholar]
- Davis MJ, Gillespie AG Jr, Vidaver AK, Harris RW (1984) Clavibacter: a new genus containing some phytopathogenic coryneform bacteria, including Clavibacter xyli subsp. xyli sp. nov., subsp. nov. and Clavibacter xyli subsp. cynodontis subsp. nov., pathogens that cause ratoon stunting disease of sugarcane and bermudagrass stunting disease. Int J Syst Bacteriol 34 107–117 [Google Scholar]
- del Pozo O, Pedley KF, Martin GB (2004) MAPKKKα is a positive regulator of cell death associated with both plant immunity and disease. EMBO J 23 3072–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreier J, Meletzus D, Eichenlaub R (1997) Characterization of the plasmid encoded virulence region pat-1 of phytopathogenic Clavibacter michiganensis subsp. michiganensis. Mol Plant Microbe Interact 10 195–206 [DOI] [PubMed] [Google Scholar]
- Eulgem T (2005) Regulation of the Arabidopsis defense transcriptome. Trends Plant Sci 10 71–78 [DOI] [PubMed] [Google Scholar]
- Feys B, Benedetti CE, Penfold CN, Turner JG (1994) Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6 751–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys BJ, Parker JE (2000) Interplay of signaling pathways in plant disease resistance. Trends Genet 16 449–455 [DOI] [PubMed] [Google Scholar]
- Gartemann KH, Kirchner O, Engemann J, Grafen I, Eichenlaub R, Burger A (2003) Clavibacter michiganensis subsp. michiganensis: first steps in the understanding of virulence of a Gram-positive phytopathogenic bacterium. J Biotechnol 106 179–191 [DOI] [PubMed] [Google Scholar]
- Gibly A, Bonshtien A, Balaji V, Debbie P, Martin GB, Sessa G (2004) Identification and expression profiling of tomato genes differentially regulated during a resistance response to Xanthomonas campestris pv. vesicatoria. Mol Plant Microbe Interact 17 1212–1222 [DOI] [PubMed] [Google Scholar]
- Groppa MD, Benavides MP (2008) Polyamines and abiotic stress: recent advances. Amino Acids 34 35–45 [DOI] [PubMed] [Google Scholar]
- Gu YQ, Wildermuth MC, Chakravarthy S, Loh YT, Yang C, He X, Han Y, Martin GB (2002) Tomato transcription factors Pti4, Pti5, and Pti6 activate defense responses when expressed in Arabidopsis. Plant Cell 14 817–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutterson N, Reuber TL (2004) Regulation of disease resistance pathways by AP2/ERF transcription factors. Curr Opin Plant Biol 7 465–471 [DOI] [PubMed] [Google Scholar]
- Ingle RA, Carstens M, Denby KJ (2006) PAMP recognition and the plant-pathogen arms race. Bioessays 28 880–889 [DOI] [PubMed] [Google Scholar]
- Jahr H, Bahro R, Burger A, Ahlemeyer J, Eichenlaub R (1999) Interactions between Clavibacter michiganensis and its host plants. Environ Microbiol 1 113–118 [DOI] [PubMed] [Google Scholar]
- Jahr H, Dreier J, Meletzus D, Bahro R, Eichenlaub R (2000) The endo-β-1,4-glucanase CelA of Clavibacter michiganensis subsp. michiganensis is a pathogenicity determinant required for induction of bacterial wilt of tomato. Mol Plant Microbe Interact 13 703–714 [DOI] [PubMed] [Google Scholar]
- Jones JD, Dangl JL (2006) The plant immune system. Nature 444 323–329 [DOI] [PubMed] [Google Scholar]
- Kawchuk L, Hachey J, Lynch DR, Klcsar F, van Rooijen G, Waterer DR, Robertson A, Kokko E, Byers R, Howard RJ, et al (2001) Tomato Ve disease resistance genes encode cell surface-like receptors. Proc Natl Acad Sci USA 98 6511–6515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klee HJ, Hayford MB, Kretzmer KA, Barry GF, Kishore GM (1991) Control of ethylene synthesis by expression of a bacterial enzyme in transgenic tomato plants. Plant Cell 3 1187–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel BN, Brooks DM (2002) Cross talk between signaling pathways in pathogen defense. Curr Opin Plant Biol 5 325–331 [DOI] [PubMed] [Google Scholar]
- Lanahan MB, Yen HC, Giovannoni JJ, Klee HJ (1994) The never ripe mutation blocks ethylene perception in tomato. Plant Cell 6 521–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln JE, Richael C, Overduin B, Smith K, Bostock R, Gilchrist DG (2002) Expression of the antiapoptotic baculovirus p35 gene in tomato blocks programmed cell death and provides broad-spectrum resistance to disease. Proc Natl Acad Sci USA 99 15217–15221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhang S (2004) Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 16 3386–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund ST, Stall RE, Klee HJ (1998) Ethylene regulates the susceptible response to pathogen infection in tomato. Plant Cell 10 371–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meletzus D, Bermphol A, Dreier J, Eichenlaub R (1993) Evidence for plasmid-encoded virulence factors in the phytopathogenic bacterium Clavibacter michiganensis subsp. michiganensis NCPPB382. J Bacteriol 175 2131–2136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeder W, Barry CS, Tauriainen AA, Betz C, Tuomainen J, Utriainen M, Grierson D, Sandermann H, Langebartels C, Kangasjarvi J (2002) Ethylene synthesis regulated by biphasic induction of 1-aminocyclopropane-1-carboxylic acid synthase and 1-aminocyclopropane-1-carboxylic acid oxidase genes is required for hydrogen peroxide accumulation and cell death in ozone-exposed tomato. Plant Physiol 130 1918–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mysore KS, Crasta OR, Tuori RP, Folkerts O, Swirsky PB, Martin GB (2002) Comprehensive transcript profiling of Pto- and Prf-mediated host defense responses to infection by Pseudomonas syringae pv. tomato. Plant J 32 299–315 [DOI] [PubMed] [Google Scholar]
- Navarro L, Zipfel C, Rowland O, Keller I, Robatzek S, Boller T, Jones JD (2004) The transcriptional innate immune response to flg22. Interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiol 135 1113–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman-Setterblad C, Vidal S, Palva ET (2000) Interacting signal pathways control defense gene expression in Arabidopsis in response to cell wall-degrading enzymes from Erwinia carotovora. Mol Plant Microbe Interact 13 430–438 [DOI] [PubMed] [Google Scholar]
- Nurnberger T, Brunner F, Kemmerling B, Piater L (2004) Innate immunity in plants and animals: striking similarities and obvious differences. Immunol Rev 198 249–266 [DOI] [PubMed] [Google Scholar]
- O'Donnell PJ, Jones JB, Antoine FR, Ciardi J, Klee HJ (2001) Ethylene-dependent salicylic acid regulates an expanded cell death response to a plant pathogen. Plant J 25 315–323 [DOI] [PubMed] [Google Scholar]
- O'Donnell PJ, Schmelz E, Block A, Miersch O, Wasternack C, Jones JB, Klee HJ (2003) Multiple hormones act sequentially to mediate a susceptible tomato pathogen defense response. Plant Physiol 133 1181–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedley KF, Martin GB (2003) Molecular basis of Pto-mediated resistance to bacterial speck disease in tomato. Annu Rev Phytopathol 41 215–243 [DOI] [PubMed] [Google Scholar]
- Perez-Donoso AG, Greve LC, Walton JH, Shackel KA, Labavitch JM (2007) Xylella fastidiosa infection and ethylene exposure result in xylem and water movement disruption in grapevine shoots. Plant Physiol 143 1024–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29 e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilloff RK, Devadas SK, Enyedi A, Raina R (2002) The Arabidopsis gain-of-function mutant dll1 spontaneously develops lesions mimicking cell death associated with disease. Plant J 30 61–70 [DOI] [PubMed] [Google Scholar]
- Restrepo S, Myers KL, del Pozo O, Martin GB, Hart AL, Buell CR, Fry WE, Smart CD (2005) Gene profiling of a compatible interaction between Phytophthora infestans and Solanum tuberosum suggests a role for carbonic anhydrase. Mol Plant Microbe Interact 18 913–922 [DOI] [PubMed] [Google Scholar]
- Robison MM, Shah S, Tamot B, Pauls KP, Moffat BA, Glick BR (2001) Reduced symptoms of Verticillium wilt in transgenic tomato expressing bacterial ACC deaminase. Mol Plant Pathol 2 135–145 [DOI] [PubMed] [Google Scholar]
- Ron M, Avni A (2004) The receptor for the fungal elicitor ethylene-inducing xylanase is a member of a resistance-like gene family in tomato. Plant Cell 16 1604–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk PM, Kazan K, Wilson I, Anderson JP, Richmond T, Somerville SC, Manners JM (2000) Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc Natl Acad Sci USA 97 11655–11660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Xie Z, Chen W, Glazebrook J, Chang HS, Han B, Zhu T, Zou G, Katagiri F (2003) Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. Plant Cell 15 317–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thilmony R, Underwood W, He SY (2006) Genome-wide transcriptional analysis of the Arabidopsis thaliana interaction with the plant pathogen Pseudomonas syringae pv. tomato DC3000 and the human pathogen Escherichia coli O157:H7. Plant J 46 34–53 [DOI] [PubMed] [Google Scholar]
- Thomma BP, Eggermont K, Tierens KF, Broekaert WF (1999) Requirement of functional ethylene-insensitive 2 gene for efficient resistance of Arabidopsis to infection by Botrytis cinerea. Plant Physiol 121 1093–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman W, de Zabala MT, Grant M (2006) Type III effectors orchestrate a complex interplay between transcriptional networks to modify basal defence responses during pathogenesis and resistance. Plant J 46 14–33 [DOI] [PubMed] [Google Scholar]
- Tuomainen J, Betz C, Kangasjarvi J, Ernst D, Yin ZH, Langebartels C, Sandermann H (1997) Ozone induction of ethylene emission in tomato plants: regulation by differential accumulation of transcripts for the biosynthetic enzymes. Plant J 12 1151–1162 [Google Scholar]
- Van Alfen NK, McMillan BD, Wang Y (1987) Properties of the extracellular polysaccharides of Clavibacter michiganense subsp. insidiosum that may affect pathogenesis. Phytopathology 77 501–505 [Google Scholar]
- Van den Bulk RW, Loffler HJM, Dons JJM (1989) Effect of phytotoxic compounds produced by Clavibacter michiganensis subsp. michiganensis on resistant and susceptible tomato plants. Neth J Plant Pathol 95 107–117 [Google Scholar]
- Van der Hoeven R, Ronning C, Giovannoni J, Martin G, Tanksley S (2002) Deductions about the number, organization, and evolution of genes in the tomato genome based on analysis of a large expressed sequence tag collection and selective genomic sequencing. Plant Cell 14 1441–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Molen GE, Labavitch JM, Strand LL, DeVay JE (1983) Pathogen-induced vascular gels: ethylene as a host intermediate. Physiol Plant 59 573–580 [Google Scholar]
- Walters DR (2003) Polyamines and plant disease. Phytochemistry 64 97–107 [DOI] [PubMed] [Google Scholar]
- Wei G, Shirsat AH (2006) Extensin over-expression in Arabidopsis limits pathogen invasiveness. Mol Plant Pathol 7 579–592 [DOI] [PubMed] [Google Scholar]
- Wiermer M, Feys BJ, Parker JE (2005) Plant immunity: the EDS1 regulatory node. Curr Opin Plant Biol 8 383–389 [DOI] [PubMed] [Google Scholar]
- Wise RP, Moscou MJ, Bogdanove AJ, Whitham SA (2007) Transcript profiling in host-pathogen interactions. Annu Rev Phytopathol 45 329–369 [DOI] [PubMed] [Google Scholar]
- Yao N, Greenberg JT (2006) Arabidopsis ACCELERATED CELL DEATH2 modulates programmed cell death. Plant Cell 18 397–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Thilmony R, Bender CL, Schaller A, He SY, Howe GA (2003) Virulence systems of Pseudomonas syringae pv. tomato promote bacterial speck disease in tomato by targeting the jasmonate signaling pathway. Plant J 36 485–499 [DOI] [PubMed] [Google Scholar]
- Zhong GY, Burns JK (2003) Profiling ethylene-regulated gene expression in Arabidopsis thaliana by microarray analysis. Plant Mol Biol 53 117–131 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Felix G (2005) Plants and animals: a different taste for microbes? Curr Opin Plant Biol 8 353–360 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, Felix G (2006) Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125 749–760 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, Felix G, Boller T (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428 764–767 [DOI] [PubMed] [Google Scholar]
- Zou J, Rodriguez-Zas S, Aldea M, Li M, Zhu J, Gonzalez DO, Vodkin LO, DeLucia E, Clough SJ (2005) Expression profiling soybean response to Pseudomonas syringae reveals new defense-related genes and rapid HR-specific downregulation of photosynthesis. Mol Plant Microbe Interact 18 1161–1174 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.