Abstract
The brown planthopper (Nilaparvata lugens Stål; BPH) is a specialist herbivore on rice (Oryza sativa) that ingests phloem sap from the plant through its stylet mouthparts. Electronic penetration graphs revealed that BPH insects spent more time wandering over plants carrying the resistance genes Bph14 and Bph15, but less time ingesting phloem than they did on susceptible plants. They also showed that their feeding was frequently interrupted. Tests with [14C]sucrose showed that insects ingested much less phloem sap from the resistant than the susceptible plants. BPH feeding up-regulated callose synthase genes and induced callose deposition in the sieve tubes at the point where the stylet was inserted. The compact callose remained intact in the resistant plants, but genes encoding β-1,3-glucanases were activated, causing unplugging of the sieve tube occlusions in susceptible plants. Continuing ingestion led to a remarkable reduction in the susceptible plants' sucrose content and activation of the RAmy3D gene, leading to starch hydrolysis and ultimately carbohydrate deprivation in the plants. Our results demonstrate that BPH feeding induces the deposition of callose on sieve plates in rice and that this is an important defense mechanism that prevents insects from ingesting phloem sap. In response, however, the BPH can unplug sieve tube occlusions by activating β-1,3-glucanase genes in rice plants.
Globally there is an enormous diversity of herbivorous insects that attack and damage plants. However, long-term coevolution of herbivorous insects and plants has led to the development of an array of constitutive and induced defenses that enable plants to protect themselves from herbivore attack (Rausher, 2001; Becerra, 2007). Constitutive defense mechanisms include physical barriers (such as cuticles, thorns, trichomes, and cell walls) and preexisting metabolites that are harmful or even toxic to insects (Mithöfer et al., 2005). Induced plant defenses involve the activation of mechanisms that directly or indirectly deter herbivores (Walling, 2000). Some secondary metabolites (Baldwin et al., 2001; Benderoth et al., 2006) and induced proteins, such as lectins, chitinases, proteinase inhibitors, and enzymes (Zavala et al., 2004; Chen et al., 2005; Harfouche et al., 2006), often play direct defense roles by, for example, repelling insects, inhibiting their feeding, or damaging their digestive or neural systems. Indirect defenses include the release of volatiles that signal the location of insects on infested plants to parasitoids and predators (De Moraes et al., 2001; Schnee et al., 2006; Kempema et al., 2007). When attacked by insects, plants can also produce endogenous signal molecules, such as stress hormones, including jasmonic acid (JA), ethylene, abscisic acid (ABA), and salicylic acid (SA) that regulate signal transduction cascades in plant cells, leading to the activation and modulation of defense-related genes (Li et al., 2002; Schmelz et al., 2006).
Plants show varied responses to herbivores that are strongly correlated with the mode of herbivore feeding. Chewing insects and cell-content feeders, such as caterpillars and beetles, cause extensive tissue damage and activate wound-signaling pathways in which JA plays a central role (Kandoth et al., 2007). In contrast, attacks by phloem feeders, such as aphids, planthoppers, and whiteflies, elicit only weak wound responses, but induce the transcription of SA and pathogen-signaling pathways, which may mirror responses to virus vectors or insect-associated bacterial endosymbionts. Alternatively, this transcription signature may be due to similarities between intercellular fungal hyphae growth and stylet penetration, and hence may produce similar responses (Baldwin et al., 2001; Li et al., 2006). Limitations associated with JA-mediated defense responses may arise from antagonistic cross talk with SA and ethylene signaling or be due to stealthy feeding behavior, which minimizes the amount of tissue damage (Voelckel et al., 2004).
The brown planthopper (Nilaparvata lugens Stål; BPH) is an insect that feeds on the leaf sheath of rice (Oryza sativa) plants, ingesting nutrients specifically from the rice phloem using its piercing mouthparts (stylet), forming a stylet sheath during the feeding process. Feeding by numerous BPHs on a single plant generally results in the susceptible plants yellowing, browning, and drying. In the last decade, the BPH has frequently caused widespread destruction of rice crops and heavy losses of yields (Shi et al., 2003; Park et al., 2007).
The main methods used to control BPH pests are to apply chemical insecticides and/or develop and grow resistant varieties in an integrated pest management strategy. However, the cost of chemical control is often very high and the chemicals can destroy the natural balance of BPH predators that help to keep the BPH population in check. The misuse of chemical pesticides may also cause a resurgence of the insect. Therefore, the most economic and efficient method for controlling the BPH is to exploit the host resistance to attack (Renganayaki et al., 2002). To date, 19 BPH resistance genes in rice have been reported and several have been used in rice breeding programs (Yang et al., 2004; Jena et al., 2006; Zhang, 2007). Various molecular techniques, including suppression subtractive hybridization, northern blotting, and cDNA array analysis, have been used to study rice responses to BPH feeding (Zhang et al., 2004; Wang et al., 2005, 2008; Yuan et al., 2005; Park et al., 2007). BPH feeding is thought to result in a reorganization of the gene expression profile of rice and most of the strongly regulated genes are associated with metabolism, cell defense, cellular transport, cellular communication, or signal transduction and the biogenesis of cellular components. In contrast, the expression of genes related to the flavonoid pathway, aromatic metabolism, and the octadecanoid pathway are mostly unchanged or down-regulated. This indicates that BPH feeding induces plant responses associated with a JA-independent pathway and cross talk with responses related to abiotic stress, pathogen invasion, and phytohormone signaling pathways (Zhang et al., 2004; Wang et al., 2005, 2008; Yuan et al., 2005). Many studies have reported the effects of BPH feeding on physiological properties and metabolic changes in rice plants (Cagampang et al., 1974; Qiu et al., 2004). However, the mechanism of rice resistance to BPH attack still remains largely unknown.
The aim of this study was to further explore the interactions between the BPH insects and rice plants in an attempt to elucidate the mechanisms involved in rice resistance to the BPH. We used a susceptible rice plant variety (TN1) as a control and first studied the feeding behavior of the BPH on rice plants carrying the BPH resistance genes Bph14 and Bph15 using the electronic penetration graph (EPG) technique. We then examined the anatomical features of the punctured phloem cells, especially the induced callose, by observing and counting the number of sieve plates with callose deposition. Real-time PCR was performed to examine the expression of genes coding for callose synthases and degrading enzymes. To our knowledge, this is the first targeted callose analysis of rice resistance to BPH feeding. Our results suggest that the induced callose sealing in sieve tubes plays an important role in the inhibition of BPH feeding. However, the BPH can unplug the sieve tube occlusions by activating β-1,3-glucanase genes in rice plants.
RESULTS
EPG Monitoring the Feeding Behavior of BPH
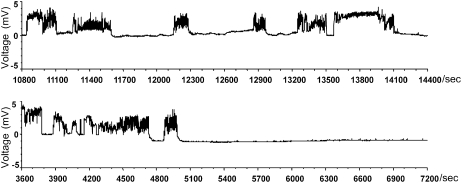
BPHs ingest phloem sap from rice plants through their narrow piercing-sucking mouthparts, which are called stylets. During the feeding process, the stylet transiently punctures the epidermis, making the first probing, and then penetrates the plant cell walls; the insect subsequently salivates into the cells and ingests the phloem sap. In this study, the feeding activities of the BPH on different varieties of rice, with varying levels of resistance, were monitored electrically using a real-time EPG technique (Tjallingii, 2006). Five types of waveform were identified, representing different types of insect feeding behavior: nonprobing (type 1), pathway (type 2), phloem puncture (type 3), xylem ingestion (type 4), and phloem sap ingestion (type 5; Fig. 1; Supplemental Fig. S1).
Figure 1.
Overview of the EPG waveforms that occurred over 1 h. In the resistant variety B5 (top), waveform type 5 (phloem sap ingestion) was interrupted at least five times and lasted no longer than 10 min uninterrupted. In the susceptible variety TN1 (bottom), waveform type 5 occurred twice and was interrupted after 2 min (at about 4,850 s) on one occasion, whereas the other occurrence lasted for more than 35 min (from 5,000 s to 7,200 s).
Waveform type 1 appeared when the stylet was outside the plant tissue and the electronic circuit remained open. Type 1 behavior can be divided into two subtypes: (1) BPH insects resting on plants; and (2) BPH insects walking or searching for feeding sites (Supplemental Fig. S1A). In an 8-h recording period, the total duration of type 1 behavior on the resistant variety B5 (carrying Bph14 and Bph15 genes) was 168.2 min, much longer than that on the susceptible rice variety TN1 (19.7 min, carrying no BPH-resistance gene). Two moderately resistant varieties, RI35 (carrying Bph14) and YHY15 (carrying Bph15), showed similar results to B5, with total durations of type 1 behavior of 110.4 min and 120.5 min, respectively (Table I, total durations). Type 1 behavior occurred 35 times on a resistant B5 plant and only 14 times on a susceptible TN1 plant (Table I, no. of times). The mean duration for type 1 behavior per feeding event on the three resistant varieties (5.5–8.7 min) was significantly longer than on the susceptible TN1 variety (1.5 min; Table I, mean durations). These findings indicate that BPHs spent more time resting or moving about on resistant plants than on susceptible ones, probing more frequently on resistant plants; the data also show that feeding was often interrupted on resistant plants.
Table I.
Feeding behaviors of BPH recorded by EPG
| Rice Variety | Waveform Type
|
||||
|---|---|---|---|---|---|
| Nonprobing (Type 1) | Pathway (Type 2) | Pha Puncture (Type 3) | Xya Ingestion (Type 4) | Ph Ingestion (Type 5) | |
| Total duration (min) of each EPG waveform type on different host plants over an 8-h recording period | |||||
| B5 | 168.2 ± 42.7 ab | 114.6 ± 34.2 a | 88.8 ± 16.7 a | 75.5 ± 21.6 ab | 33.0 ± 6.7 c |
| RI35 | 110.4 ± 50.2 b | 97.1 ± 38.3 a | 85.1 ± 23.8 a | 123.9 ± 32.6 a | 63.6 ± 31.9 bc |
| YHY15 | 120.5 ± 62.0 ab | 127.7 ± 54.0 a | 33.1 ± 13.9 b | 125.9 ± 46.9 a | 72.9 ± 38.8 b |
| TN1 | 19.7 ± 10.2 c | 33.3 ± 7.2 b | 28.6 ± 9.4 b | 58.2 ± 23.3 b | 340.2 ± 26.8 a |
| Hc | 17.4 P < 0.001 | 15.5 P < 0.001 | 17.7 P < 0.000 | 15.2 P < 0.002 | 19.5 P < 0.000 |
| No. of times each EPG waveform type occurred on different host plants over an 8-h recording period | |||||
| B5 | 35.3 ± 11.1 ab | 58.4 ± 14.0 a | 32.9 ± 7.8 a | 10.6 ± 3.4 b | 13.1 ± 3.8 c |
| RI35 | 29.7 ± 18.2 ab | 46.3 ± 19.7 b | 28.0 ± 8.5 a | 13.3 ± 4.5 b | 10.3 ± 3.5 c |
| YHY15 | 18.0 ± 8.3 a | 29.9 ± 10.6 b | 12.1 ± 7.7 a | 8.4 ± 2.2 a | 5.1 ± 3.3 b |
| TN1 | 13.9 ± 5.3 b | 25.9 ± 5.8 b | 16.0 ± 4.0 b | 6.1 ± 2.5 b | 12.6 ± 7.1 a |
| Hc | 13.0 P < 0.005 | 15.1 P < 0.002 | 13.5 P < 0.004 | 11.6 P < 0.009 | 23.0 P < 0.000 |
| Mean duration (min) of each EPG waveform type on different host plants over an 8-h recording period | |||||
| B5 | 5.5 ± 3.3 a | 2.0 ± 0.7 a | 2.8 ± 0.7 a | 7.6 ± 3.2 ab | 2.9 ± 1.6 a |
| RI35 | 6.3 ± 6.4 ab | 2.4 ± 1.5 a | 3.1 ± 0.7 a | 9.8 ± 2.3 a | 6.5 ± 3.6 a |
| YHY15 | 8.7 ± 7.7 bc | 4.6 ± 1.7 b | 3.0 ± 1.2 b | 15.5 ± 6.8 bc | 15.0 ± 6.0 a |
| TN1 | 1.5 ± 1.0 c | 1.3 ± 0.4 b | 1.8 ± 0.2 b | 10.3 ± 4.6 c | 29.4 ± 9.8 b |
| Hc | 9.7 P < 0.022 | 14.6 P < 0.002 | 17.6 P < 0.001 | 11.3 P < 0.01 | 12.9 P < 0.005 |
Ph, Phloem; Xy, xylem.
Values (means ± sd) followed by the same letter within a column are not significantly different; Scheffe's post-hoc pairwise comparison (P < 0.05).
Kruskal-Wallis one-way ANOVA by ranks. Rice variety B5 carries both BPH resistance genes Bph14 and Bph15; RI35 and YHY15 carry genes Bph14 and Bph15, respectively; TN1 has no BPH resistance gene.
Waveform type 2 occurred when the BPH insects used their stylets to search for the target cells in plant tissues in a series of activities, including penetrating plant cells, salivating, tasting, and forming branches of the stylet sheath (Supplemental Fig. S1A). The total duration of this waveform type on the resistant B5 plant was 114.6 min, significantly longer than that on the susceptible TN1 plant (33.3 min). This suggests that the insects spend more time searching for suitable target feeding cells in the resistant plant tissue (Table I, total durations). The frequency of this behavior over the 8-h recording period showed a general tendency to increase with higher levels of plant resistance (Table I, no. of times).
Waveform type 3 occurred when the stylet penetrated the vascular bundle of the rice plant (Supplemental Fig. S1B). Overall, the total duration of waveform type 3 was correlated with the plants' level of resistance, but there was a significant difference between the two moderately resistant varieties (RI35 and YHY15) and the difference between TN1 and YHY15 was insignificant (Table I, total durations). Such differences in type 3 behavior of BPHs on these moderately resistant varieties might be attributable to the differences in resistance genes of the plants.
Waveform type 4 represented xylem ingestion by the BPHs (Supplemental Fig. S1C). We assumed that the xylem was not the resistant element within the plant because of the irregular duration of this behavior on the B5 (75.5 min), YHY15 (125.9 min), RI35 (123.9 min), and TN1 (58.2 min) varieties. There was no clear relationship between resistance level and total duration of this type of behavior (Table I, total durations).
Waveform type 5, representing phloem ingestion, gave a better indication of resistance because it reflected the relative quantity of phloem sap ingested by the BPHs (Supplemental Fig. S1D). During the 8-h recording period, the total duration of type 5 behavior on the resistant variety B5 was 33 min, approximately one-tenth of that on the susceptible variety TN1 (340.2 min; Table I, total durations). Moreover, the mean duration of each period of phloem sap ingestion was much shorter on the resistant varieties than on the susceptible control TN1 (Table I). These findings suggest that the BPHs spend less time ingesting phloem sap from resistant plants than from susceptible plants.
Use of [14C]Suc to Quantify Phloem Sap Ingestion by BPH
Suc is the main carbohydrate that is transported long distances through the phloem and ingested by the BPH. By culturing rice plants in a [14C]Suc solution, 14C can easily be introduced into the phloem. The quantity of phloem sap ingested by the BPHs can then be estimated by monitoring the radioactivity of 14C in the insects. We used the ratio of radioactivity (14C) in the insect to that in the plant, designated the I:P index, as an indicator of the relative quantity of phloem sap ingested by BPHs. The results showed that the I:P ratio for BPHs that had fed on resistant B5 plants was very low (0.02, compared with 0.85 for insects feeding on susceptible TN1 plants) over a 20-h period (Fig. 2). The I:P ratios for BPHs that fed on the moderately resistant varieties YHY15 and RI35 were 0.54 and 0.28, respectively. These results strongly indicate that the BPH insects ingested less phloem sap from the resistant rice plants than they did from the susceptible control (TN1).
Figure 2.
I:P indices for each of the rice varieties. The results suggest that the BPHs took up more phloem sap from the susceptible variety TN1 than from YHY15 or RI35, and extremely little from the resistant variety B5. Rice variety B5 carries BPH resistance genes Bph14 and Bph15; RI35 and YHY15 carry Bph14 and Bph15, respectively; TN1 has no BPH resistance gene.
Callose Deposition on the Sieve Plates of BPH-Infested Plants
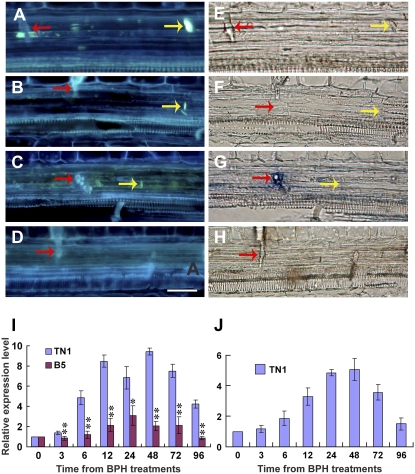
To investigate the mechanisms that prevent BPHs from continuously ingesting phloem sap from resistant rice plants, the leaf sheaths of BPH-infested and BPH-free resistant and susceptible plants were sectioned and examined histopathologically. The sections were stained with 0.1% aniline blue and examined under a fluorescence microscope. In the BPH-free untreated rice plants, there was little or no callose deposition on the sieve plates in the leaf sheaths (Fig. 3, E and F). When the plants were infested with the BPH, more callose was deposited on the sieve plates of the target sieve tubes, where the stylets had been inserted—the sieve plates were obviously thickened and emitted strong fluorescence (Fig. 3, A–D). Counts of the bright callose plugs revealed that callose deposition increased during the first 3 d of infestation in both B5 and TN1 plants, but there were more callosic sieve plates in the former (13.7 callosic sieve plates in 50 sections) than the latter (5.8). Moreover, with prolonged BPH feeding, the callose deposition decreased quickly in TN1 plants to only 2.4 callosic sieve plates in 50 sections after 4 d, but a high level of callose deposition remained in the B5 plants (12.7; Fig. 3G). Further analysis revealed that there were strong fluorescence signals from callose in almost all the sieve tubes penetrated by stylet sheaths in the resistant B5 plants, but a large proportion of the target sieve tubes did not have bright callose depositions in the susceptible TN1 plants (Fig. 4, D and H).
Figure 3.
Induced callose deposition and the expression of callose synthase genes. A to D, Induced callose deposition (yellow arrows) on the sieve plates with bright blue fluorescence in the resistant variety B5 plant infested by BPH. B and D, Light microphotographs of A and C, respectively. E and F, Longitudinal sections of an uninfested control leaf sheath showing no accumulated callose on the sieve plate (white arrows). Red arrows indicate stylet sheaths. Scale bar = 20 μm. G, Numbers of sieve plates with callose deposition in BPH-infested leaf sheaths, counted under the microscope. Standard error bars (SBs) indicate the numbers of sieve plates with callose deposition observed in 50 cross-sections, and vertical bars on the SBs indicate ses. H to J, Results of real-time PCR expression analysis of the callose synthase genes in each of the rice varieties in response to BPH feeding. H, OsGSL3. I, OsGSL1. J, OsGSL5. Total RNA was extracted from rice leaf sheaths after different BPH feeding times (h); expression of genes was quantified relative to the value obtained from 0 (h) samples (BPH-free plants). Each bar represents the mean values ± se of three replicates. Each RNA sample was extracted from approximately 200 mg of fresh leaf sheaths of 10 rice plants. Rice Actin1 gene was used as reference control. Significant differences in gene expression were indicated with * (P < 0.05) or ** (P < 0.01); Student's t test.
Figure 4.
Callose decomposition and expression of related genes. A to H, Deposited callose decomposing in longitudinal sections of leaf sheaths from susceptible TN1 plants. A and E, Deposited callose (yellow arrows) in sieve tubes on the sieve plates and embedding of the stylet sheath. B, C, F, and G, Callose in various stages of decomposition. D and H, Target sieve tubes with no bright callose deposition. Red arrows indicate stylet sheaths. Scale bar = 20 μm. I and J, Results of real-time PCR expression analysis of Gns subfamily genes: Gns5 (I) and Osg1 (J). Total RNA was extracted from rice leaf sheaths after different BPH feeding times (h). Expression of genes was quantified relative to the value obtained from 0 (h) samples (BPH-free plants). Each bar represents the mean values ± se of three replicates. Each RNA sample was extracted from approximately 200 mg of fresh leaf sheaths of 10 rice plants. Rice Actin1 gene was used as reference control. Significant differences in gene expression were indicated with * (P < 0.05) or ** (P < 0.01); Student's t test.
Expression of Callose Synthase and β-1,3-Glucanase in Rice Plants
Callose deposition is a dynamic process coordinated through the activities of callose synthase and the callose-hydrolyzing enzyme β-1,3-glucanase. To investigate the mechanisms responsible for the differential callose deposition in the resistant and susceptible rice plants, the expression of 10 callose synthase-encoding genes were investigated using semiquantitative reverse transcription (RT)-PCR. We detected transcripts of four of these genes, namely, OsGSL1, OsGSL3, OsGSL5, and OsGSL7 (Supplemental Fig. S2). Three of the detected genes, OsGSL1, OsGSL3, and OSGS5, were further analyzed by real-time PCR. These genes were clearly up-regulated after the B5 and TN1 plants were treated with BPH for 6 h, reaching high levels after 12 h (2- to 4-fold of uninfested plants). The expression levels remained high in the following 72 to 96 h, generally over 3-fold higher than those in uninfested plants (Fig. 3, H–J). These observations suggest that the callose synthase genes were up-regulated and consequently callose synthesis was enhanced in both the resistant and susceptible plants attacked by the BPH.
The expression patterns of six β-1,3-glucanase genes were also investigated. The patterns of four of them were found to differ between BPH-infested B5 and TN1 plants. Osg1 was up-regulated by 1.2-fold within 3 h in susceptible TN1 plants and kept increasing from 6 to 48 h, with a decline after 72 h. The highest expression level occurred after treatment for 48 h, about 5-fold increased in comparison to the untreated control (Fig. 4J). Gns5 showed an increase both in TN1 and B5 plants, but the increase in TN1 was much higher than in B5 plants. The greatest increase in TN1 was >9-fold relative to the uninfested control (Fig. 4I). Gns4 was constitutively expressed in B5, but was induced in TN1 plants. Gns6 shared similar expression patterns with Gns5. Little or no expression of either Gns2 or Gns3 was detected in the leaf sheath of the rice plants (Supplemental Fig. S2). The expression of β-1,3-glucanase genes, such as Osg1 and Gns5, was clearly up-regulated in the susceptible rice plants; in contrast, the expression level of these genes was up-regulated much less (Gns5) or even absent at a detectable level (Osg1) in resistant ones.
Decomposition of Starch in Compensation for Sugar Losses in Susceptible Plants
To investigate the anatomical effects of BPH feeding on rice plants, leaf sheaths of plants representing the most resistant and most susceptible varieties (B5 and TN1, respectively) were sectioned, stained in 3% KI-1% I2 solution, and examined under a microscope. We found abundant starch granules in the leaf sheaths of the uninfested plants and there appeared to be more in the B5 than in the TN1 plants (Fig. 5A, top two sections). In the BPH-infested plants, starch granules were rapidly consumed after 1 d in the susceptible TN1 variety and most were exhausted after 3 d of infestation (Fig. 5A, right). In contrast, the starch granules disappeared much more slowly in the resistant variety (B5; Fig. 5A, left).
Figure 5.
Starch and Suc variation in leaf sheaths of rice varieties infested with BPH. A, Cross-sections of leaf sheaths stained with KI-I2, showing starch granules. In control plants with no BPH, leaf sheaths of both B5 and TN1 plants had large numbers of dark starch granules in their parenchyma cells (top images). After the plants had been infested with BPH for 1 d (middle images) and 3 d (bottom images), most of the starch granules still remained in the resistant B5 plants (left), but had disappeared in the susceptible TN1 plants (right). Pc, Parenchyma cells; Xy, xylem, Ph, phloem. B, Content of Suc (top) and starch (bottom) after different BPH infestation times (d) in leaf sheaths of different rice varieties. Standard bars (SBs) indicate mean content from six replicates and vertical bars on the SBs indicate ses.
The starch content of leaf sheaths was also determined. Results showed that the starch content decreased much more quickly in TN1 than in B5 under the stress of BPH infestation. However, it should be noted that there was much more starch in B5 (22.7 mg g−1 fresh weight) than in TN1 (16.7 mg g−1 fresh weight) plants that were not infested by BPHs (Fig. 5B, bottom). The Suc content varied in a similar way to that of the starch content (Fig. 5B, top). In the BPH-infested TN1 plants, Suc content fell to approximately 56% and 30% of the untreated control plant levels after 1 and 4 d, respectively (Fig. 5B), showing that susceptible TN1 plants can be rapidly and seriously deprived of carbohydrates during infestation by these insects.
The expression of nine α-amylase and three β-amylase genes in the leaf sheaths of rice plants was investigated using semiquantitative RT-PCR. We found that RAmy3D (Os08g0473900), one of the genes examined, was strongly induced in the BPH-infested TN1 plants, but was only weakly expressed in the B5 plants (Supplemental Fig. S2). This indicates that RAmy3D plays a role in the response of susceptible rice plants to BPH feeding by breaking down the starch.
DISCUSSION
Enhancing host resistance is an important component of integrated pest management. However, the mechanism of rice's resistance to BPH is still uncertain. In the past, researchers considered that it might be governed by the presence of chemicals confined to the phloem (Sogawa and Pathak, 1970). Bph1 was the first BPH resistance gene identified and is associated with the production of flavonoids, including SA; amino acids, such as Asp, Glu, Ala, Ser, Leu, Asn, and Val; and organic acids, such as succinic acid, malic acid, oxalic acid, and transaconitic acid (Sogawa and Pathak, 1970; Sogawa, 1976). There is now considerable evidence to suggest that induced defenses are effective and have low fitness costs. Several studies have revealed that gene expression profiles in rice are reorganized in response to BPH feeding (Zhang et al., 2004; Yuan et al., 2005; Park et al., 2007; Wang et al., 2008). Such reorganization has been well documented in other plant-herbivore systems, supporting the hypothesis that inducible defenses contribute to rice resistance to the BPH. The data gathered in this experiment show that BPH feeding affects the expression of genes associated with the synthesis and hydrolysis of callose and starch decomposition in rice plants, resulting in the deposition of callose plugs on the sieve plates. This prevents BPH insects from continuously ingesting phloem sap and allows normal carbohydrate levels to be maintained in resistant plants, as described below.
Phloem, the target of BPH feeding, mainly consists of sieve tubes and companion cells. The functional units of sieve tubes are series of sieve elements that have porous sieve plates at their abutting ends, allowing the phloem sap to flow continuously (Will et al., 2007). The sieve element/companion cell modules are highly sensitive to biotic and abiotic disturbance, and elaborate sealing mechanisms, such as protein plugging and callose formation, have evolved (McNairn and Currier, 1967; Will and Bel, 2006). Callose has several functions in the normal development of plants (Chen et al., 2007), and its formation and deposition can be induced by either biotic or abiotic stress (Jacobs et al., 2003; Ueki and Citovsky, 2005). We found more callose deposits on sieve plates in both the resistant and susceptible rice plants infested by BPH than in the uninfested controls (Fig. 3). However, in the resistant B5 plants, almost all the target sieve tubes showed strong fluorescence, indicating that abundant, compact callose had been deposited within them. In contrast, the callose signals were fainter in the susceptible TN1 plants, and there were no compact callose deposits in many of their sieve tubes where BPH insects had fed.
It has been reported that callose synthesis is Ca2+ dependent (King and Zeevaart, 1974), and phloem-feeding insects seem to induce the release of Ca2+ stored in the reticulum or the apoplasm, thereby activating callose synthesis (Arsanto, 1986; Volk and Franceschi, 2000). It has also been demonstrated that large amounts of callose on sieve plates reduce the rate of phloem translocation and can even block it completely (McNairn and Currier, 1967). Our results strongly suggest that the BPHs had little chance to ingest phloem sap continuously from the B5 plants. EPG data confirmed that the BPHs spent more time searching for, and tasting, cells for feeding on resistant plants than on susceptible plants, and that their feeding was often interrupted on the former (Table I, waveform types 1 and 2). The total duration of phloem sap ingestion on resistant plants was approximately one-tenth that on susceptible plants (Table I, waveform type 5). The 14C-labeling data also support the conclusions that phloem feeding was inhibited on B5 plants and that the BPHs sucked only small amounts of sap from them (Fig. 2). Therefore, we conclude that callose deposition plays an important role in preventing BPHs from ingesting phloem sap and thus contributes to the resistance of rice to these insects. Sieve tube occlusion, in response to biotic and abiotic stress, has been previously reported (Will and Bel, 2006). Cagampang et al. (1974) showed that the rate of upward sap transport in rice plants infested by BPHs was only 60% of the rate in their uninfested counterparts, and Nielson et al. (1990) found evidence that the acropetal movement of photosynthates in alfalfa (Medicago sativa) was seriously disrupted by potato leafhopper (Empoasca fabae) feeding. In cotton (Gossypium hirsutum), basipetal phloem translocation was completely inhibited by callose deposition in a 14CO2 labeling trial (McNairn and Currier, 1967). Furthermore, forisomes (spindle-like protein bodies in the sieve tubes) have been shown to inhibit aphid feeding in broad bean (Vicia faba; Will et al., 2007).
Because callose synthase genes were up-regulated and callose deposition occurred in both the resistant and susceptible rice plants a short time after BPH feeding commenced (Fig. 3), the insects had to overcome the physical barriers imposed by callose to obtain sufficient food even from susceptible varieties. In the aphid/broad bean system, aphid saliva can prevent sieve tube plugging by forisomes (Will et al., 2007), which have only been found in the Fabaceae. Our results, in contrast, indicate that BPH feeding induced the expression of genes encoding β-1,3-glucanases, causing decomposition of the callose barriers in the susceptible rice plants (Fig. 4, A–H). These enzymes play important roles in plant defense and development. β-Glucanase-encoding genes have been classified into four subfamilies, based on their structure and function. Two tandem gene clusters, Gns2–Gns3–Gns4 and Gns5–Gns6, have been identified in the defense-related subfamily A (Romero et al., 1998). In addition, the novel Osg1 gene of rice has also been assigned to this subfamily (Tomoya et al., 2002). Isozymes belonging to the Gns subfamily A differ widely in their requirements, needing 1,6-β-glucan branch linkages nearby in the polymer chain for activity. Polymers of β-1,3-glucan are found in both plants and fungi, but polymers of 1,3; 1,6-β-glucans are found only in fungi. Thus, GNS4 (which specifically hydrolyzes 1,3; 1,6-β-glucans) is likely to play an important role in antifungal defense rather than callose decomposition (Akiyama et al., 1997; Tomoya et al., 2002); thus, high levels of β-1,3-glucanase may not be directly correlated with reductions in callose levels. However, isozymes that mainly hydrolyze 1,3-β-glucans, such as OSG1 and GNS5, have been presumed to play important functions in the breakdown of callose, as well as in defending against pathogen attack (Akiyama et al., 1997). The Osg1 and Gns5 genes were clearly induced by BPH attack and are likely to play important roles in callose decomposition, which ultimately facilitates ingestion of phloem sap by BPHs from susceptible rice plants. Therefore, the absence of expression (or limited expression) of these genes allows the sieve tube occlusions to be maintained in the resistant plants, and this is probably the key reason for their resistance. This conclusion is further supported by the findings that the expression of Osg1 is up-regulated in Hejiang19, another variety that is susceptible to the BPH, but cannot be detected in the moderately resistant varieties RI35 and YHY15, either in the control or BPH-treated plants (Supplemental Fig. S3).
Starch is a major end product of photosynthesis; it is produced in chloroplasts and is the main energy storage substance in cereal grains and leaf sheaths. In the chloroplast and amyloplast, starch metabolism is closely related to other metabolic processes in the cytosol, such as Suc metabolism, glycolysis, and glyconeogenesis, so starch content is indicative of the dynamic transformation of carbohydrates. In treated susceptible TN1 plants, a large amount of phloem sap was ingested by BPHs and plant Suc content rapidly declined. In contrast, photosynthesis was suppressed in the BPH-infested susceptible plants (Yuan et al., 2005). Consequently, these plants had to up-regulate genes such as RAmy3D to decompose the starch stored in leaf sheaths to meet their carbohydrate and energy requirements (Supplemental Fig. S2). This decomposition process may continue until the stored starch has been exhausted. In this aspect, starch is also important in resisting (or tolerating) the BPH, by providing sugars for the plants. Greater starch storage would allow the plant to tolerate BPH feeding for a longer period of time. In this study, we found there was more starch in the uninfested plants of resistant variety B5 than in the uninfested susceptible TN1 plants, indicating that B5 plants were capable of surviving feeding stress for a longer period than TN1. Under the stress imposed by BPH, the resistant B5 plants also faced the threat of losing phloem sap, but ingestion was inhibited by the synthesis of callose, which effectively blocked the target sieve tubes. In such cases, the plant only needs to provide carbohydrate for itself and, consequently, less Suc loss and less starch decomposition will occur. Therefore, the resistant plants can survive a long period of stress from BPH, even when the phloem transportation is affected to some extent; eventually, the BPH will die of starvation. Competition for sugar, as well as other nutrients, plays an important role in the interaction between the herbivore and the plant. However, mechanisms allowing plants to resist (or tolerate) herbivore attack may differ widely between phloem-sucking insects and chewing insects. For example, in response to foliar herbivores, the allocation of sugars to roots increased in the annual Nicotiana attenuata so that plants better tolerate herbivores (Schwachtje et al., 2006).
Our understanding of the resistance mechanism can be encapsulated in the following model of interactions between the BPH and the rice plant (Fig. 6). First, the BPH acts on the plant by penetrating its tissues, ejecting saliva into its cells, and sucking up phloem sap. In response to BPH feeding, the plant up-regulates expression of its callose synthase and β-1,3-glucanase genes. Consequently, callose deposition occludes the sieve tubes and prevents the BPH from ingesting the phloem sap. However, β-1,3-glucanases that decompose the deposited callose and thereby facilitate the BPH's continued feeding from the phloem are strongly induced in susceptible plants, but much more weakly induced in resistant plants. Thus, differential expression of β-1,3-glucanases can account for between-plant differences in resistance levels.
Figure 6.
Model of the interaction between the BPH and rice plants. The BPH first acts on a plant by penetrating its tissues, ejecting saliva into its cells, and sucking up phloem sap. In response to feeding by the BPH, the plant up-regulates genes encoding callose synthases and β-1,3-glucanases. Consequently, callose deposition occludes the sieve tubes and prevents the BPH from ingesting the phloem sap. Then, specific β-1,3-glucanases decompose the deposited callose in susceptible plants (but little in resistant plants), allowing the BPH to resume feeding from the phloem. Thus, differential expression of β-1,3-glucanases accounts for the differences in their resistance levels. Arrows indicate promotion or positive modulation of the process; vertical bars indicate inhibition or negative modulation of the process.
CONCLUSION
We have demonstrated that feeding by the BPH can induce callose synthesis and deposition on the sieve plates of rice plants. Callose deposition affects phloem transportation and plays an important role in preventing the BPH from ingesting the phloem sap. Our results show not only that callose deposition is sufficient for resistant plants to defend themselves against the BPH, but also that some specific β-1,3-glucanases are active callose-decomposing enzymes, induced by BPH activity and responsible for the susceptibility of TN1 plants. The differential expression of these enzymes may result in different resistance levels in rice plants.
MATERIALS AND METHODS
Plants and Insects
Five rice (Oryza sativa) varieties were used in this study. B5 is a line carrying BPH resistance genes Bph14 and Bph15 from wild rice (Oryza officinalis) and exhibits high resistance to BPH with the severity score below 3.0 in the seedling bulk test (Huang et al., 2001). RI35 (carrying Bph14) and YHY15 (carrying Bph15) are progeny lines of B5 and are moderately resistant to BPH with the severity score about 5. TN1 and Hejiang 19 are conventional varieties susceptible to BPH and their severity score is 9.0. All experiments were carried out on rice plants at the three- to five-leaf stage.
Unless otherwise stated, the brown planthopper (Nilaparvata lugens Stål; BPH) insects were three- to four-instar nymphae, and the insects were maintained on TN1 plants in the Genetics Institute, Wuhan University.
EPG Waveform Characterization and Quantification
To link the EPG waveforms with the feeding behavior patterns of the BPH, a microscope was coupled to EPG equipment, as follows. Special plastic slides, each with a 1-cm-diameter hole in the center, were prepared and covered with stretched parafilm. Suc solution or tap water (each with a small amount of active carbon powder to trace the water flow) was dropped onto the parafilm to serve as an artificial food source and mounted under a coverslip. BPH insects with a gold wire (length 3–5 cm, diameter 20 μm) attached to the dorsum by conductive silver glue were then allowed to probe the food through the parafilm. The gold wire from each insect and a copper wire (diameter 0.1 mm) immersed in the food were linked to a Giga 4 model DC-EPG amplifier (Wageningen University). The EPG setup was housed in a climate-controlled room (25°C ± 2°C) and shielded from electrical noise by an earthed Faraday cage. The EPG was also linked to a computer running PROBE 3.1 software (attached to the EPG equipment). The electronic signals from the different channels were converted into digital data using a DI-710 data logger (DATAQ) and transformed into waveforms displayed on the computer screen in real time. By relating the feeding behavior of BPH insects under the microscope with the real-time EPG waveforms displayed on the screen, we were able to categorize the waveform types (Supplemental Fig. S1).
For EPG recordings of BPH insects feeding on rice plants, adult brachypterous females (2 d after the final molt) were collected at 9 am, and attached to a gold wire, as described above. After being starved (but provided with water) for 1 h, each insect was placed on the leaf sheath of the plant to be tested and the gold wire from its dorsum was connected to the EPG. Before acquiring and processing data, WINDAQ Waveform Browser software (DATAQ) was run for 30 min to pretest the activity of the insect. Data were acquired at 100-Hz sample frequency, stored on the computer's hard disc, and simultaneously displayed on a screen. The data were analyzed using ANA3.0 software (Wageningen University). EPG recordings were carried out for 8 h/insect/plant, with at least seven replicates for each variety, using fresh seedlings and insects in each case. In a single experiment, all four genotypes were examined simultaneously, one channel for each genotype, and then the experiment was repeated. Data were compared using Kruskal-Wallis one-way ANOVA ranking and Scheffe's post-hoc pairwise comparisons (P < 0.05).
Isotope 14C-Labeling and Determination
For 14C-labeling, rice seedlings at the three-leaf stage were transferred to a vial containing a 1-mL solution of [14C]Suc from Sigma and nonlabeled Suc (4 μCi and 15 mg/vial, respectively). Each seedling was held in place with a sponge and pushed into the neck of the vial so that the root was immersed in the solution. Each plant, together with 10 insects, was placed in a test tube (30 mm × 200 mm) and covered with gauze to prevent the insects from escaping. Seedlings were allowed to take up the Suc solution at 25°C ± 2°C in darkness. Twenty hours later, the seedlings were removed from the tubes, their roots were discarded, and their remaining parts were cut into 1-cm-long segments. Insects and plant segments were then plunged into 5 mL of 80% ethanol-water solution (v/v), boiled for 10 min, and then centrifuged at 4,000g for 5 min at 4°C. The supernatants containing the extracted soluble fractions were collected and concentrated to 500 μL, 100 μL of which was used to determine their 14C content, using a Beckman LS6500 liquid scintillation spectrometer (Beckman). The I:P index was used to evaluate the distribution of 14C between the insects and plants, reflecting the proportion of soluble 14C ingested by the insects from the phloem.
Histochemistry and Microscopy
Rice plants were each infested with 10 BPHs. Leaf sheaths were collected, fixed in FAE (formaldehyde:acetic acid:70% ethanol, 5:5:90 [v/v/v]), dehydrated, embedded in paraffin, and cut into 10-μm-thick sections using a microtome. The sections were mounted on microscope slides, dewaxed, and rehydrated for staining at room temperature.
To highlight starch and saliva sheaths, the rehydrated sections were stained in 3% (w/v) KI-1% I2 solution for 1 min, then examined under a light microscope. For callose observations, 10-μm-thick sections were mounted on glass slides (50 sections/slide). Callose staining was performed as described by Dietrich et al. (1994) with some modifications. Rehydrated sections were stained with 0.1% (w/v) aniline blue in 0.15 m K2HPO4 for 5 min and examined under a UV epifluorescence microscope (Olympus BX51). Callose on individual sieve plates was classified as either faint or bright: Faint types included clearly visible plates with a thin, green-yellow appearance, whereas bright was used to describe all thickly callosed sieve plates with bright blue fluorescence (McNairn and Currier, 1967). The amount of callose deposition in each plant examined was evaluated by counting the number of sieve plates that had bright callose. At least 400 sections were examined for each treatment. Photographs were taken with a Coolpix 995 digital camera (Nikon).
Determination of Suc and Starch Content
Fresh leaf sheaths (approximately 1 g) were powdered in liquid nitrogen, homogenized in 4 mL of 80% (v/v) ethanol, heated in a water bath at 80°C for 40 min, and centrifuged at 4,000g for 10 min. The supernatant fraction was collected and the solid fraction was washed with 80% ethanol and centrifuged; this procedure was repeated twice. The supernatants collected from each sample were combined and then active carbon was added and filtered for Suc analysis, whereas the pellet was dried for starch determination.
Suc was measured using the anthrone-sulfuric acid method (Trevelyan and Harrison, 1952) with modifications. For colorimetric determination, 0.5 mL of the 80% ethanol extract was added to 1 mL of water and digested with 1 mL 10% (w/v) aqueous KOH in a water bath at 100°C for 3 min. The cooled reaction mixture was placed immediately in an ice-water bath, and then 4 mL of anthrone reagent (1 g of anthrone dissolved in 500 mL of 98% H2SO4) was added to the cooled mixture. The mixture was again incubated in a 100°C water bath for 1 min, and then placed for 90 s in an ice-water bath. The mixture was transferred by pipette into a 1-cm-diameter spectrophotometer cup, and its A625 was measured using a UV-1601 spectrophotometer (Shimadzu).
The dried pellet was added to 5 mL of 80% Ca(NO3)2 (w/v), placed in a 100°C water bath for 10 min, and then centrifuged at 4,000g for 4 min. The supernatant fraction was collected and the solid fraction was washed with 80% Ca(NO3)2 and centrifuged; this procedure was repeated twice. All the supernatants from each sample collected were combined and added to 20 mL of water to prepare the starch solution. One milliliter of starch solution was mixed with 2 mL of 80% Ca(NO3)2 and 100-μL solution of 0.01 n I2-KI (1.3 g I2 and 4.0 g KI in water, final volume 1 L). The mixture was transferred by pipette into a 1-cm-diameter spectrophotometer cup, and its A620 was measured using a UV-1601 spectrophotometer.
Semiquantitative RT-PCR
Total RNA was extracted from the leaf sheaths (approximately 200 mg fresh weight) with TRIzol reagent (Invitrogen), and the remaining DNA was degraded using a Turbo DNA-free kit (Ambion). cDNA was synthesized from the total RNA (4 μg) using a Thermoscript RT-PCR system (Invitrogen), with oligo(dT)20 primers, following the manufacturer's instructions. RT-PCR was performed using Taq DNA polymerase (MBI Fermentas) in 10-μL reaction mixtures with the gene-specific primers listed in Supplemental Table S1, which were either directly synthesized according to previously published information (Tomoya et al., 2002, 2006; Fukao et al., 2006) or designed using the PCR primer design tool primer3 (http://frodo.wi.mit.edu) according to cDNA sequences obtained from the National Center for Biotechnology Information (NCBI) GenBank (http://www.ncbi.nlm.nih.gov). Actin1 control primers were used as standards for mRNA expression; all the templates for RT-PCR of different genes were from the same individual cDNA samples. The amplification program consisted of an initial denaturation step at 94°C for 5 min, followed by 28 to 40 cycles of 94°C for 30 s, 58°C for 30 s, 72°C for 1 min, and a final extension at 72°C for 5 min. This procedure was repeated at least three times.
Real-Time PCR Analysis
Genes for real-time PCR analysis were screened based on the results of semiquantitative RT-PCR; the genes that showed obvious variation were chosen for real-time PCR, whereas the genes that could not be detected or showed no obvious variation were not chosen for further study. The primers (Table II) for real-time PCR were redesigned using primer premier 5.0 software according to cDNA sequences obtained from NCBI GenBank (see above). Reactions were carried out on the ABI PRISM 7300 real-time PCR system (Applied Biosystems) using three-step cycling conditions of 95°C for 1 min, followed by 40 cycles of 95°C for 15 s, 55°C to 60°C for 15 s, and 72°C for 28 s. After the amplification steps, the melting curve was determined for each primer pair at a final stage of 15 s at 95°C, 15 s at 60°C, and 15 s at 95°C to verify the presence of only one specific product. The reaction mixture (20 μL) contained 2 μL of cDNA solution, 10 μL SYBR Green real-time PCR master mix (QPK-201; TOYOBO), and about 5 pmol each primer. The reactions were performed in triplicate and the results were averaged. A standard curve was prepared using 5 μL of cDNA solutions in which serially diluted samples (original, 5-, 25-, 125-diluted) were included. The slopes of Ct and ΔCt (target gene) − (reference gene) and R2 values of each sample were calculated by the ABI PRISM 7300 SDS vl.X and Microsoft Excel 2003. Relative quantification was performed with the 2−ΔΔCt method (Livak and Schmittgen, 2001). Actin1 was used as reference for mRNA expression.
Table II.
Sequences of primers for real-time PCR
Gene-specific primers were designed with the PCR primer design tool primer premier 5.0 according to cDNA sequences obtained from NCBI GenBank (http://www.ncbi.nlm.nih.gov). Genes for real-time PCR were screened based on the results of semiquantitative RT-PCR.
| Gene | Specific Primers Used for Real-Time PCR
|
Expected Size | Accession No. | |
|---|---|---|---|---|
| Forward Primer (5′-3′) | Reverse Primer (5′-3′) | |||
| bp | ||||
| OsGSL1 | TGAGGACCTGCCACGATT | CACGCTGATTGCGAACAT | 119 | AP001389 |
| OsGSL3 | TGGCAAGCGACCACATAG | AGACCTTAGCACGGACTG | 283 | AP003268 |
| OsGSL5 | GTGGTGTCCCTGCTATGA | GTTGTTTGCTATTCTCCC | 185 | AP008212 |
| Osg1 | GGCGTATGGGACAAAGGA | TTCAGAGGCGAAGGATGG | 237 | AB070742 |
| Gns5 | TTGCGGCCATTCCTACAGT | TGGTGAGGGCGATGCTTG | 183 | U72251 |
| Actin1 | CAGCACATTCCAGCAGAT | GGCTTAGCATTCTTGGGT | 108 | AB047313 |
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Details of the EPG waveforms shown in Figure 1.
Supplemental Figure S2. Results of semiquantitative RT-PCR for callose synthases, β-1,3-glucanases, and amylases.
Supplemental Figure S3. Results of RT-PCR analysis of Osg1.
Supplemental Table S1. Sequences of primers for semiquantitative RT-PCR.
Supplementary Material
Acknowledgments
We thank Freddy Tjallingii (Wageningen University) for valuable technical advice about EPG, and Jie Zhao and Yingtang Lu (Whan University) for valuable help with the epifluorescence microscope and liquid scintillation spectrometer. We also thank Hongyu Yuan (Xinyang Normal University) for valuable advice and assistance with the real-time PCR system. We acknowledge Yanchang Wang (Wuhan Institute of Botany, Chinese Academy of Sciences) and members of our laboratory for their assistance with this project.
This work was supported by the National Natural Science Foundation of China (grant nos. 30730062 and 30671287) and the National Special Key Project on Functional Genomics and Biochips (grant no. 2006AA10A103).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Guangcun He (gche@whu.edu.cn).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Akiyama T, Shibuya N, Hrmova M, Fincher GB (1997) Purification and characterization of a (1→3)-β-d-glucan endohydrolase from rice (Oryza sativa) bran. Carbohydr Res 297 365–374 [DOI] [PubMed] [Google Scholar]
- Arsanto JP (1986) Ca2+-binding sites and phosphatase activities in sieve element reticulum and P-protein of chick-pea phloem. A cytochemical and X-ray microanalysis survey. Protoplasma 132 160–171 [Google Scholar]
- Baldwin IT, Halitschke R, Kessler A, Schittko U (2001) Merging molecular and ecological approaches in plant-insect interactions. Curr Opin Plant Biol 4 351–358 [DOI] [PubMed] [Google Scholar]
- Becerra JX (2007) The impact of herbivore-plant coevolution on plant community structure. Proc Natl Acad Sci USA 104 7483–7488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benderoth M, Textor S, Windsor AJ, Mitchell-Olds T, Gershenzon J, Kroymann J (2006) Positive selection driving diversification in plant secondary metabolism. Proc Natl Acad Sci USA 103 9118–9123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagampang GB, Pathak MD, Juliana OB (1974) Metabolic changes in the rice plant during infestation by the brown planthopper, Nilaparvata lugens Stål (Atemiptera: Delphacidae). Appl Entomol Zool (Jpn) 9 174–184 [Google Scholar]
- Chen H, Wilkerson CG, Kuchar JA, Phinney BS, Howe GA (2005) Jasmonate-inducible plant enzymes degrade essential amino acids in the herbivore midgut. Proc Natl Acad Sci USA 102 19237–19242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RZ, Zhao X, Shao Z, Wei Z, Wang YY, Zhu LL, Zhao J, Sun MX, He RF, He GC (2007) Rice UDP-glucose pyrophosphorylase1 is essential for pollen callose deposition and its cosuppression results in a new type of thermosensitive genic male sterility. Plant Cell 19 847–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Moraes CM, Mescher MC, Tumlinson JH (2001) Caterpillar-induced nocturnal plant volatiles repel conspecific females. Nature 410 577–580 [DOI] [PubMed] [Google Scholar]
- Dietrich RA, Delaney TP, Uknes SJ, Ward ER, Ryals JA, Dangl JL (1994) Arabidopsis mutants simulating disease resistance response. Cell 77 565–577 [DOI] [PubMed] [Google Scholar]
- Fukao T, Xu K, Ronald PC, Julia BS (2006) A variable cluster of ethylene response factor-like genes regulates metabolic and developmental acclimation responses to submergence in rice. Plant Cell 18 2021–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfouche AL, Shivaji R, Stocker R, Williams PW, Luthe DS (2006) Ethylene signaling mediates a maize defense response to insect herbivory. Mol Plant Microbe Interact 19 189–199 [DOI] [PubMed] [Google Scholar]
- Huang Z, He GC, Shu LH, Li XH, Zhang QF (2001) Identification and mapping of two brown planthopper resistance genes in rice. Theor Appl Genet 102 929–934 [Google Scholar]
- Jacobs AK, Lipka V, Burton RA, Panstruga R, Strizhov N, Paul SL, Fincher GB (2003) An Arabidopsis callose synthase, GSL5, is required for wound and papillary callose formation. Plant Cell 15 2503–2513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jena KK, Jeung JU, Lee JH, Choi HC, Brar DS (2006) High-resolution mapping of a new brown planthopper (BPH) resistance gene, Bph18(t), and marker-assisted selection for BPH resistance in rice (Oryza sativa L.). Theor Appl Genet 112 288–297 [DOI] [PubMed] [Google Scholar]
- Kandoth PK, Ranf S, Pancholi SS, Jayanty S, Walla MD, Miller W, Howe GA, Lincoln DE, Stratmann JW (2007) Tomato MAPKs LeMPK1, LeMPK2, and LeMPK3 function in the systemin-mediated defense response against herbivorous insects. Proc Natl Acad Sci USA 104 12205–12210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempema LA, Cui X, Holzer FM, Walling LL (2007) Arabidopsis transcriptome changes in response to phloem-feeding silverleaf whitefly nymphs. Similarities and distinctions in responses to aphids. Plant Physiol 143 849–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RW, Zeevaart JAD (1974) Enhancement of phloem exudation from cut petioles by chelating agents. Plant Physiol 53 96–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Xie QG, Smith-Becker J, Navarre DA, Kaloshian I (2006) Mi-1-mediated aphid resistance involves salicylic acid and mitogen-activated protein kinase signaling cascades. Mol Plant Microbe Interact 19 655–664 [DOI] [PubMed] [Google Scholar]
- Li X, Schuler MA, Berenbaum MR (2002) Jasmonate and salicylate induce expression of herbivore cytochrome P450 genes. Nature 419 712–715 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25 402–408 [DOI] [PubMed] [Google Scholar]
- McNairn RB, Currier HB (1967) Sieve plate callose. A factor in blockage of axial phloem transport. Naturwissenschaften 54 591. [DOI] [PubMed] [Google Scholar]
- Mithöfer A, Wanner G, Boland W (2005) Plant resistance towards insect herbivores: a dynamic interaction. Plant Physiol 137 1160–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielson GR, Lamp WO, Stutte GW (1990) Potato leafhopper (Homoptera: Cicadellidae) feeding disruption of phloem translocation in alfalfa. J Econ Entomol 83 807–813 [Google Scholar]
- Park DS, Lee SK, Lee JH, Song MY, Song SY, Kwak DY, Yeo US, Jeon NS, Park SK, Yi G, et al (2007) The identification of candidate rice genes that confer resistance to the brown planthopper (Nilaparvata lugens) through representational difference analysis. Theor Appl Genet 115 537–547 [DOI] [PubMed] [Google Scholar]
- Qiu HM, Wu JC, Yang GQ, Dong B, Li DH (2004) Changes in the uptake function of the rice root to nitrogen, phosphorus and potassium under brown planthopper, Nilaparvata lugens (Stål) (Homoptera: Delphacidae) and pesticide stresses, and effect of pesticides on rice-grain filling in field. Crop Prot 23 1041–1048 [Google Scholar]
- Rausher MD (2001) Co-evolution and plant resistance to natural enemies. Nature 411 857–864 [DOI] [PubMed] [Google Scholar]
- Renganayaki K, Fritz AK, Sadasivam S, Pammi S, Harrington SE, McCouch SR, Kumar SM, Reddy AS (2002) Mapping and progress toward map-based cloning of brown planthopper biotype-4 resistance gene introgressed from Oryza officinalis into cultivated rice, O. sativa. Crop Sci 42 2112–2117 [Google Scholar]
- Romero GO, Simmons C, Yaneshita M, Doan M, Thomas BR, Rodriguez RL (1998) Characterization of rice endo-β-glucanase genes (Gns2–Gns14) defines a new subgroup within the gene family. Gene 223 311–320 [DOI] [PubMed] [Google Scholar]
- Schmelz EA, Carroll MJ, LeClere S, Phipps SM, Meredith J, Chourey PS, Alborn HT, Teal PEA (2006) Fragments of ATP synthase mediate plant perception of insect attack. Proc Natl Acad Sci USA 103 8894–8899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnee C, Köllner TG, Held M, Turlings TCJ, Gershenzon J, Degenhardt J (2006) The products of a single maize sesquiterpene synthase form a volatile defense signal that attracts natural enemies of maize herbivores. Proc Natl Acad Sci USA 103 1129–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwachtje J, Minchin PEH, Jahnke S, van Dongen JT, Schittko U, Baldwin IT (2006) SNF1-related kinases allow plants to tolerate herbivory by allocating carbon to roots. Proc Natl Acad Sci USA 103 12935–12940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi ZY, Ren X, Weng QM, Li XH, He GC (2003) Construction of a genomic library from a brown planthopper resistant rice line using a transformation-competent vector and identification of clones spanning the Qbp1 locus. Plant Sci 165 879–885 [Google Scholar]
- Sogawa K (1976) Studies on the feeding habits of the brown planthopper, Nilaparvata lugens (Stål) (Hemiptera: Delphacidae). V. Probing stimulatory effect of rice flavonoid. Appl Entomol Zool (Jpn) 11 160–164 [Google Scholar]
- Sogawa K, Pathak MD (1970) Mechanisms of brown planthopper resistance in Mudgo variety of rice (Hemiptera: Delphacidae). Appl Entomol Zool (Jpn) 5 145–158 [Google Scholar]
- Tjallingii WF (2006) Salivary secretions by aphids interacting with proteins of phloem wound responses. J Exp Bot 57 739–745 [DOI] [PubMed] [Google Scholar]
- Tomoya Y, Katsuhiro N, Takami H, Yoshiyuki Y, Setsuo K (2002) Molecular cloning and characterization of a novel β-1,3-glucanase gene from rice. Biosci Biotechnol Biochem 66 1403–1406 [DOI] [PubMed] [Google Scholar]
- Tomoya Y, Takami H, Katsuhiro N, Setsuo K (2006) Expression analysis of genes for callose synthases and rho-type small GTP-binding proteins that are related to callose synthesis in rice anther. Biosci Biotechnol Biochem 70 639–645 [DOI] [PubMed] [Google Scholar]
- Trevelyan WE, Harrison JS (1952) Studies on yeast metabolism 1. Fractionation and microdetermination of cell carbohydrates. Biochem J 50 298–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki S, Citovsky V (2005) Identification of an interactor of cadmium ion-induced glycine-rich protein involved in regulation of callose levels in plant vasculature. Proc Natl Acad Sci USA 102 12089–12094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelckel C, Weisser WW, Baldwin IT (2004) An analysis of plant-aphid interactions by different microarray hybridization strategies. Mol Ecol 13 3187–3195 [DOI] [PubMed] [Google Scholar]
- Volk GM, Franceschi FR (2000) Localization of a calcium-like in the sieve element plasma membrane. Aust J Plant Physiol 27 779–786 [Google Scholar]
- Walling LL (2000) The myriad plant responses to herbivores. J Plant Growth Regul 19 195–216 [DOI] [PubMed] [Google Scholar]
- Wang XL, He RF, He GC (2005) Construction of suppression subtractive hybridization libraries and identification of brown planthopper-induced genes. J Plant Physiol 162 1254–1262 [DOI] [PubMed] [Google Scholar]
- Wang YY, Wang XL, Yuan HY, Chen RZ, Zhu LL, He RF, He GC (2008) Responses of two contrasting genotypes of rice to brown planthopper. Mol Plant Microbe Interact 21 122–132 [DOI] [PubMed] [Google Scholar]
- Will T, Bel A (2006) Physical and chemical interactions between aphids and plants. J Exp Bot 57 729–737 [DOI] [PubMed] [Google Scholar]
- Will T, Tjallingii WF, Thönnessen A, Bel A (2007) Molecular sabotage of plant defense by aphid saliva. Proc Natl Acad Sci USA 104 10536–10541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HY, You AQ, Yang ZF, Zhang FT, He RF, Zhu LL, He GC (2004) High-resolution genetic mapping at the Bph15 locus for brown planthopper resistance in rice (Oryza sativa L.). Theor Appl Genet 110 182–191 [DOI] [PubMed] [Google Scholar]
- Yuan HY, Chen XP, Zhu LL, He GC (2005) Identification of genes responsive to brown planthopper Nilaparvata lugens Stål (Homoptera: Delphacidae) feeding in rice. Planta 221 105–112 [DOI] [PubMed] [Google Scholar]
- Zavala JA, Patankar AG, Gase K, Baldwin IT (2004) Constitutive and inducible trypsin proteinase inhibitor production incurs large fitness costs in Nicotiana attenuata. Proc Natl Acad Sci USA 101 1607–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang FT, Zhu LL, He GC (2004) Differential gene expression in response to brown planthopper feeding in rice. J Plant Physiol 161 53–62 [DOI] [PubMed] [Google Scholar]
- Zhang QF (2007) Strategies for developing Green Super Rice. Proc Natl Acad Sci USA 104 16402–16409 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.