Abstract
The balance between the supply and utilization of carbon (C) changes continually. It has been proposed that plants respond in an acclimatory manner, modifying C utilization to minimize harmful periods of C depletion. This hypothesis predicts that signaling events are initiated by small changes in C status. We analyzed the global transcriptional response to a gradual depletion of C during the night and an extension of the night, where C becomes severely limiting from 4 h onward. The response was interpreted using published datasets for sugar, light, and circadian responses. Hundreds of C-responsive genes respond during the night and others very early in the extended night. Pathway analysis reveals that biosynthesis and cellular growth genes are repressed during the night and genes involved in catabolism are induced during the first hours of the extended night. The C response is amplified by an antagonistic interaction with the clock. Light signaling is attenuated during the 24-h light/dark cycle. A model was developed that uses the response of 22K genes during a circadian cycle and their responses to C and light to predict global transcriptional responses during diurnal cycles of wild-type and starchless pgm mutant plants and an extended night in wild-type plants. By identifying sets of genes that respond at different speeds and times during C depletion, our extended dataset and model aid the analysis of candidates for C signaling. This is illustrated for AKIN10 and four bZIP transcription factors, and sets of genes involved in trehalose signaling, protein turnover, and starch breakdown.
Changing environmental conditions continually alter the balance between carbon (C) assimilation and utilization (Stitt, 1991; Geiger and Servaites, 1994; Stitt and Krapp, 1999; Geiger et al., 2000; Paul and Foyer, 2001). Even short periods of C starvation lead to an inhibition of growth, which is not immediately reversed when C becomes available again (Smith and Stitt, 2007; Stitt et al., 2007). For example, there is a delay before growth is reestablished when Suc is resupplied to C-depleted seedlings (Osuna et al., 2007) or wild-type plants are reilluminated after a 6-h extension of the night (Gibon et al., 2004b). A particularly dramatic example is provided by starchless pgm mutants, which deplete their sugars in the first hours of the night (Caspar et al., 1985). This is followed by an inhibition of growth, which is only slowly reversed during the next light period (Gibon et al., 2004b). Repeated periods of C depletion also have major midterm consequences. The levels of most metabolites and enzymes in pgm resemble those in wild-type plants after several days of darkness (Gibon et al., 2004a, 2006). This shows that repeated short periods clamp C depletion metabolism in a state appropriate to C starvation even if they are alternating with periods of high sugar (Gibon et al., 2006).
The diurnal cycle provides a defined experimental system to investigate how the supply and utilization of C is coordinated (Smith and Stitt, 2007; Stitt et al., 2007). Plants switch each day between a surplus of C in the light and a negative C balance at night. These diurnal changes are buffered by retaining some photosynthate in the leaves as starch in the light and remobilizing it at night to support Suc synthesis and export (Geiger and Servaites, 1994; Smith et al., 1997, 2005; Stitt et al., 2007). The importance of starch turnover is demonstrated by studies of mutants that are defective in starch synthesis or mobilization. They grow at the same rate as wild-type plants in continuous light or very long days, but show a large inhibition of growth in short days (Caspar et al., 1985, 1989; Lin et al., 1988; Schulze et al., 1991; Huber and Hanson, 1992; Zeeman et al., 1998; Zeeman and ap Rees, 1999; Gibon et al., 2004b). This inhibition is due to the transient depletion of C during the night (Gibon et al., 2004b).
Starch turnover is regulated in wild-type plants to avoid a shortfall of C at the end of the night. It is typically degraded in a near-linear manner, leaving only a small amount at the end of the night (Fondy and Geiger, 1985; Geiger and Servaites, 1994; Matt et al., 1998; Smith et al., 2004). When the C supply is decreased, for example, in short days or low irradiance, the rate of starch synthesis is increased and the rate of degradation is decreased. As a result, C reserves still last until the end of the night (Stitt et al., 1978; Chatterton and Silvius, 1979, 1980, 1981). Starch turnover starts to adjust on the first day after transfer from a long- to a short-day regime and, within 2 to 3 d, the levels of starch and sugars at the end of the night resemble those in long days (Gibon et al., 2004b). These observations raise fundamental questions about how plants sense changes in C availability over time and use this information to adjust and coordinate starch synthesis and breakdown with the use of C for growth.
Recently, Stitt et al. (2007) and Smith and Stitt (2007) proposed that plants respond to decreasing C in an acclimatory manner (i.e. where signaling triggers changes in storage, allocation, and growth before C falls so far that it exerts an acute limitation on metabolism or growth). This hypothesis provides a framework to understand how starch turnover, metabolism, and growth are coordinated to avoid C starvation. It makes several predictions. One is that sugars should inhibit starch synthesis. This prediction is counterintuitive because increased availability of C might be expected to stimulate starch synthesis. It was recently shown that starch synthesis is decreased when small amounts of Suc are included in the rooting medium of Arabidopsis (Arabidopsis thaliana) seedlings provided they are grown in short-day conditions where C is limiting (Gibon et al., 2004b; Stitt et al., 2007). Second, and crucially, changes in signaling should be initiated by relatively small changes in the C status before the onset of acute C starvation.
Transcript profiles provide the most comprehensive readout of signaling pathways that is currently available. It has been known for >20 years that sugars regulate gene expression in plants (Yu, 1999; Koch, 2004; Gibson, 2005; Rolland et al., 2006). Koch (1996) developed the concept of feast and famine genes; high sugar induces feast genes that are required for biosynthesis and growth, whereas low sugar induces famine genes that are required for C assimilation or the catabolism of alternative C sources. Prolonged C starvation or readdition of sugars to starved seedlings leads to changes of transcript levels for hundreds of genes that are involved in metabolism, signaling, and growth (Contento et al., 2004; Price et al., 2004; Thimm et al., 2004; Thum et al., 2004; Li et al., 2006; Osuna et al., 2007). However, these treatments are too drastic to reveal whether expression responds to small changes of C (Osuna et al., 2007). Thousands of genes show significant changes of their transcripts during diurnal light/dark cycles (Smith et al., 2004; Bläsing et al., 2005). Two lines of evidence indicate that C contributes to these changes. First, there is qualitative agreement between the diurnal changes of sugars and transcripts for many C-responsive genes (Bläsing et al., 2005). Second, the accentuated diurnal changes of sugars in pgm are accompanied by larger and more widespread changes of transcripts (Gibon et al., 2004b; Thimm et al., 2004; Bläsing et al., 2005). This is mainly due to depletion of C at night (Bläsing et al., 2005).
This article presents a more comprehensive analysis of the global response of transcripts to progressive depletion of C. Arabidopsis plants were darkened at the end of the night and metabolite profiling was performed to identify when C becomes severely limiting. This information was used to define times at which samples were taken for expression profiling during a transition to acute C deprivation. This dataset was combined with published datasets for diurnal changes of transcripts and analyzed to provide an overview of the global response of transcript levels during a gradual transition from high to acutely limiting C and to identify candidates for upstream components of the transcriptional response to small changes in the C status. It was also used to formulate and test a simple linear model, which predicts the global responses of gene expression in diurnal cycles from three inputs—the clock, light, and C. This allows us to validate the role of C in the diurnal regulation of a large number of genes and provides a framework in which the role of candidate genes can be quantitatively evaluated.
RESULTS
Changes of Metabolites in an Extended Night
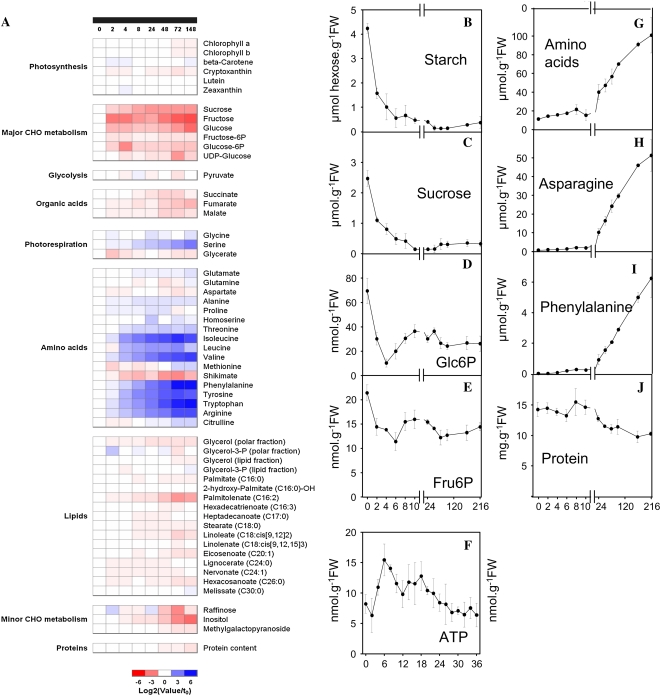
Ecotype Columbia of Arabidopsis (Col-0) was grown for 5 weeks at 20°C in a 14-h light/10-h dark cycle at a moderate light intensity in well-fertilized soil. Whole rosettes were harvested at the end of the night and at different times after transfer into continuous darkness. Figure 1A shows the response of the metabolite profile and Figure 1, B to H, the responses of selected metabolites in another experiment with more time points.
Figure 1.
Changes of metabolites measured in an extended night. Arabidopsis Col-0 was grown for 5 weeks at moderate light intensity (130 μmol m−2 s−1) in a 14-h light/10-h dark cycle at 20°C in well-fertilized soil. After 5 weeks, plants were subjected to an extended night by not illuminating them at the start of the light period and whole rosettes harvested at the end of the night and at different times into the treatment. A, Changes of metabolites summarized as a heat map; all data are normalized on the values at the end of the night, with blue and red indicating an increase and decrease, respectively (see legend for scale). Gray indicates missing values. Some of the data are recalculated from Gibon et al. (2006). B to J, Starch (B), Suc (C), Glc-6-P (D), Fru-6-P (E), ATP (F), total amino acids (G), Asn (H), Phe (I), protein (J). The results in B to I are the mean and sd of five independent samples.
Arabidopsis grown in these conditions contains about 35, 6, and 10 μmol hexose equivalents per gram fresh weight of starch, Suc, and reducing sugars in the rosette at the start of the night (Gibon et al., 2004b). At the end of the night, about 10% of the starch is left (Fig. 1B) and Suc and reducing sugars have fallen about 2-fold (Fig. 1C). Starch, Suc, and reducing sugars fell to very low levels by 8 to 12 h into the extended night. Glc-6-P (Fig. 1D) and Fru-6-P (Fig. 1E) decreased for 4 h and then rose again. Hexose-Ps are an indicator for the C status; they are produced during photosynthesis and are consumed during carbohydrate synthesis, the synthesis of structural cell components, and respiration. The marked decrease during the first 4 h of the extended night shows that carbohydrates are starting to acutely limit metabolism. The partial recovery from 4 h onward indicates that metabolism is adjusting to this shortfall. ATP rises during the first 4 to 6 h and then gradually decreases (Fig. 1F), revealing that this adjustment occurs without a major breakdown of energy metabolism.
Organic acids like fumarate and malate are present at high levels in Arabidopsis (Chia et al., 2000). They show a gradual decrease when the night is extended, which becomes more marked from 48 h onward. Other C-containing metabolites also decrease, including raffinose, inositol, and several fatty acids, especially palmitolenoate (16:2) with less marked decreases of palmitate (16:0), stearate (18:0) and linoleate (18:2), eicosenoate (20:1), and hexacosanoate (26:0). Amino acids start to rise in the first hours of the extended night (Fig. 1, A and G). It has been proposed that amino acids like Asn (Fig. 1H) and Arg increase in C starvation because they have a high nitrogen (N)-C ratio (Lam et al., 1996, 1998). However, there is an equally large increase of low-N amino acids, including aliphatic (Leu, Ile, and Val; Fig. 1A) and aromatic amino acids (Phe, Tyr, Trp; Fig. 1, I and A), and smaller increases of Thr, Ala, and Glu (Fig. 1A). The only amino acids that do not increase are Asp and Gln. This general accumulation is unlikely to be due to increased synthesis; indeed, there is a marked decrease of shikimate (Fig. 1A), an intermediate in the aromatic amino acid synthesis pathway. The amino acids are likely to be released by protein degradation (Fig. 1J).
Expression Profiling during an Extended Night
Three separate experiments provided triplicate transcript profiles for the end of the night, duplicate samples 4, 8, 24, and 48 h into an extended night, and single samples 2 and 6 h into the extended night. Robust multiarray analysis (RMA) expression measures (Bolstad et al., 2003) were calculated on all arrays used. There was close agreement between the experiments. Pairwise scatter plots between the end of the night samples yielded R2 values of 0.977, 0.975, and 0.979 (Supplemental Fig. S1a). Principal component analysis (PCA) showed samples for each time group together (Supplemental Fig. S1c). The original data are provided in Supplemental Table S1.
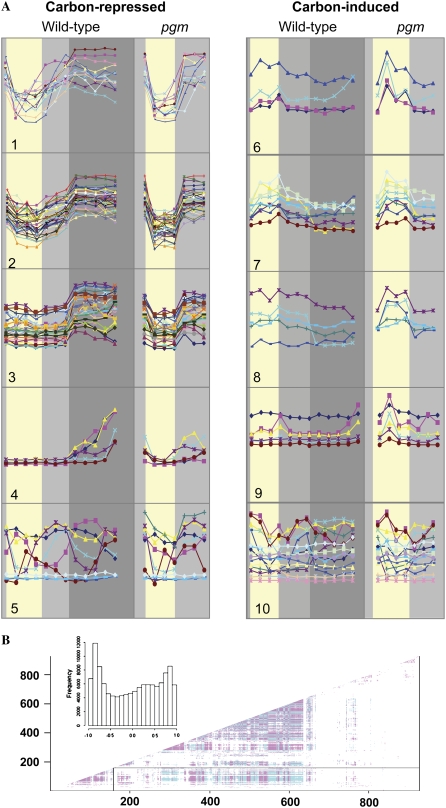
To identify genes that show significant changes of their transcript levels early in the extended night, a linear model was fitted with limma (Smyth, 2004) and corrected for multiple testing using a false discovery rate of 0.05 (Benjamini and Hochberg, 1995). Significantly changed genes are listed in Supplemental Table S2. After 4 h, 1,885 genes were induced and 2,147 were repressed, rising to 2,626 and 3,032 genes after 8 h, 3,135 and 3,388 genes after 24 h, and 3,929 and 3,888 genes after 48 h. Figure 2 shows the temporal responses of the 200 most strongly induced and the 200 most strongly repressed genes. Most show a rapid response that is near-saturated after 6 to 8 h. A smaller group shows a delayed response, with marked changes after 24 to 48 h.
Figure 2.
Temporal changes of global gene expression during an extended night. The plots show the changes of transcript levels of the 200 most strongly induced and the 200 most strongly repressed genes. Line shading is used to distinguish genes that respond early and later in the treatment.
Integration of the Extended Night Dataset and Datasets for Diurnal Changes in Gene Expression
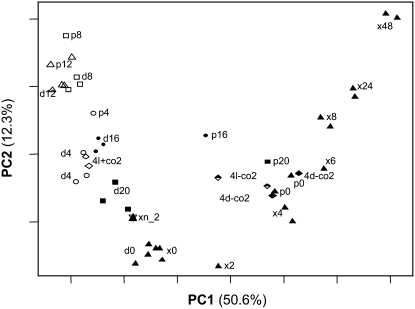
We have published replicated datasets for global changes of transcripts (1) during diurnal cycles in wild-type Col-0 (Bläsing et al., 2005); (2) after illumination of Col-0 for 4 h at the end of the night at 50 and 350 ppm [CO2] (the former prevents CO2 fixation and carbohydrate synthesis (Bläsing et al., 2005); and (3) during the diurnal cycle in the starchless pgm mutant (where there are exaggerated changes of sugars; Gibon et al., 2004b; Bläsing et al., 2005). Related treatments grouped together when the extended night dataset was combined with these datasets and subjected to hierarchical clustering (Supplemental Fig. S2) or PCA (Fig. 3).
Figure 3.
PCA of an extended night treatment in Col-0 (x) combined with datasets for a diurnal cycle in wild-type Col-0 (d) and pgm (p). A cluster analysis of the same datasets is shown in Supplemental Figure S2. White symbols and black symbols represent day and (extended) night time points, respectively. The star, circles (○), rectangles (□), and triangles (▵) represent time points −2, 4, 8, and 12 h into the day or night. Further, all extended night treatments are also marked with a closed triangle (▴). Treatments with ambient and compensation point [CO2] are represent by diamonds. White diamonds (⋄) represent the light treatment at ambient [CO2], black diamonds (♦) the dark treatment at compensation point [CO2], and the half-black diamond represents the light treatment at compensation point [CO2].
PC1 accounted for 50.6% of the total variation, and separated treatments according to their carbohydrate content and the duration of the dark treatment. From the Col-0 diurnal cycle dataset we find, from left to right, samples from 12 and 8 h into the light period, 4 h into the light period, 4 h into the dark period, 8 h into the dark period, the end of the night, and then 2, 4, 6, 8, 24, and 48 h into the extended night. pgm samples group with Col-0 samples in the light, but are shifted to the right in the night, mirroring the faster depletion of sugars (Gibon et al., 2004b; Bläsing et al., 2005). PC2 accounted for 12.3% of the variance and separated samples collected at the end of the night from samples collected during the next 8 to 12 h. The 24- and 48-h extended night treatments grouped with the end of the light period.
The response to an extended night was investigated in plants growing in a 14-h light/10-h dark cycle, whereas the diurnal cycle and response to [CO2] were analyzed in a 12-h light/12-h dark cycle (Bläsing et al., 2005). We checked whether this small difference in the photoperiod precludes joint analysis of the datasets. Hierarchical cluster analysis (Supplemental Fig. S2) and PCA (Fig. 3) showed that samples collected at the end of the night group together, regardless of whether they are from plants in a 12/12 (diurnal cycle) or a 14/10 cycle (extended night experiment). The dark controls from the low [CO2] experiments (12/12 cycle) group with samples collected 4 h into the extended night (14/10 cycle). Samples collected at the end of the night in a 12/12 cycle were separated from samples collected 2 h after the end of the night in a 14-h light/10-h dark cycle, even though the plants had been in the dark for 12 h in both treatments. We calculated, for all >22K genes on the ATH1 array, the log2-fold change 2, 4, 6, 8, 24, and 48 h into the extended night relative to the average level in the three end-of-the-night samples from the extended night experiments and the average level in the three end-of-the-night samples in the diurnal cycle dataset. Pairwise scatter plots of the ratios after 2, 4, 6, 8, 24, and 48 h yielded correlation coefficients of 0.88, 0.96, 0.97, 0.98, 0.98, and 0.99, respectively, which is comparable to or better than the agreement for biological replicates.
These results show that plants in 12/12 and a 14/10 light/dark cycle have similar transcript profiles at the end of the night, and show a similar response to an extension of the night. This conclusion was checked by fitting a linear model to the whole dataset and statistically analyzing the end-of-the-night samples from the 12/12 and 14/10 treatments with limma (Smyth, 2004). Only 917 genes had a P value <0.05, which is the proportion expected by chance (5% of approximately 22,000), only eight genes showed a >2-fold change, and no genes showed significant changes after correcting for multiple testing (Benjamini and Hochberg, 1995).
Contribution of Sugars to the Changes of Transcript Levels
To provide qualitative evidence that changes of C make a major contribution to the global changes of transcription in this large dataset, we compared the weighting of genes in PC1 (see Supplemental Table S3) with the changes of transcript levels in five published datasets where different treatments were used to alter C status. These were adding 15 mm Suc to C-starved seedlings for 30 min or 3 h (Osuna et al., 2007), adding 100 mm Glc to C-starved seedlings for 3 h (Bläsing et al., 2005), comparing seedlings in full nutrient medium with C-starved seedlings (Osuna et al., 2007), and illuminating rosettes for 4 h at 350 or 50 ppm [CO2] (Bläsing et al., 2005). Regression plots yielded highly significant correlation coefficients (r) of −0.44, −0.5, −0.43, −0.47, and −0.6. With 1,000 shuffled datasets, most values of r were between 0.1 and −0.1, and none were >0.25 or <−0.25 (Supplemental Fig. S3).
Temporal Kinetics of the Responses of C-Responsive Genes during Diurnal Cycles and an Extended Night
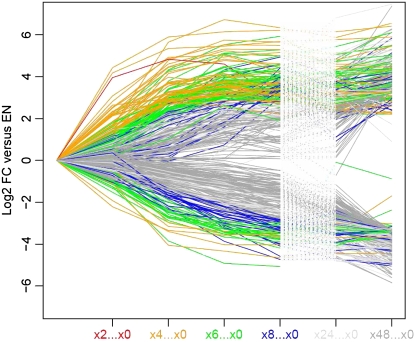
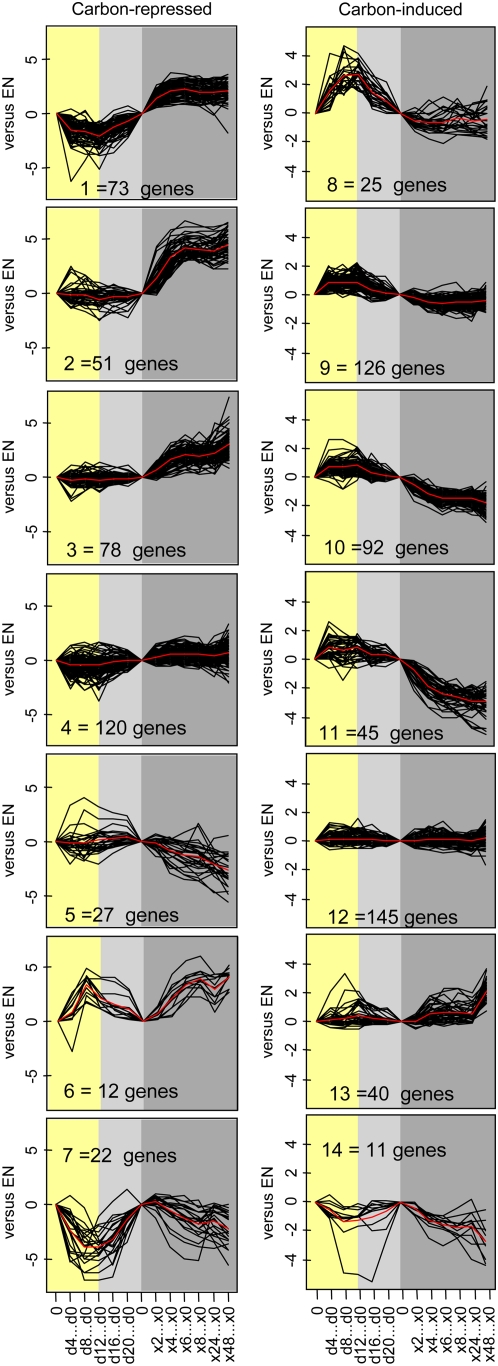
A test set for C-induced genes was generated by combining the 200 most strongly induced genes 3 h after adding 15 mm Suc to C-depleted seedlings, the 200 most strongly induced genes 3 h after adding 100 mm Glc to C-starved seedlings, and the 200 most strongly induced genes after illuminating 5-week-old plants for 4 h in 350 compared to 50 ppm [CO2] (Bläsing et al., 2005; Osuna et al., 2007). Because there was overlap, the test set contained 484 genes (Supplemental Table S4). An analogous procedure generated a test set of 383 C-repressed genes. Undirected and directed approaches were applied to test whether the temporal responses are consistent with these genes being regulated by changes of C during the diurnal cycle and the first hours of the extended night.
In the undirected approach, the C-repressed and C-induced genes were each separated into seven groups by K-means clustering (Fig. 4; genes are identified in Supplemental Table S4). Over 40% are in the clusters whose response is consistent with C regulating their expression during the night or early in the extended night. This includes cluster 8 (transcripts for 25 C-induced genes rise in the light and relax by the end of the night), clusters 9 to 11 (transcripts for 263 C-induced genes rise during the light period and relax during the night and first hours of the extended night), and cluster 1 (transcripts for 73 C-repressed genes decrease in the light and recover during the night and first 4 to 8 h of the extended night). Clusters 2 to 3 and 11 (representing 14% of the genes) are stable during the diurnal cycle, but respond strongly at the start of the extended night. A few genes did not respond until 24 to 48 h into the extended night (cluster 13). About 30% are in clusters that showed mostly <2-fold changes (clusters 4 and 12), and 10% showed qualitatively inconsistent responses.
Figure 4.
K-means clustering of the responses of sugar-responsive genes during the diurnal cycle and an extended night. The 200 genes that are induced and the 200 genes that are repressed most strongly 3 h after adding 15 mm Suc to C-depleted seedlings were downloaded from Osuna et al. (2007) and corresponding sets of genes for the responses 3 h after adding 100 mm Glc to C-starved seedlings and after illuminating 5-week-old plants for 4 h in 350 or 50 ppm [CO2] were downloaded from Bläsing et al. (2005). The genes were combined to provide a test set of 484 C-induced genes and 383 C-repressed genes. The responses of these genes in the diurnal cycle (Bläsing et al., 2005) and the extended night were extracted from Supplemental Table S4. All transcript levels were normalized on the level at the end of the night in the same experiment and K-means clustered. Clusters for C-repressed (clusters 1–7) and C-induced (clusters 8–14) genes are shown. The numbers of genes in each cluster are indicated.
In a directed Boolean approach, genes were assigned values of −1, 0, or +1 depending upon whether their transcript level decreased, remained unaltered, or increased in a given time interval. They were scored using a filter of >2-fold change. The time intervals were between the start and end of the night, between the end of the night and 8 h into the extended night, and between 8 and 48 h into the extended night. The results are summarized in Table I. Response classes that are qualitatively consistent with C induction or repression are shown in bold and italics, respectively. About 80% of C-repressed genes are in classes whose response is consistent with repression by C (transcripts rise during the night and/or the extended night). Almost one-half of the C-induced genes are in classes whose response is consistent with them being induced by C, whereas 43% do not show marked changes and the remainder show inconsistent responses. Crucially, some genes complete their response in the diurnal cycle (classes 12 and 14), larger sets start to change in a diurnal cycle and change further in the first hours of the extended night (e.g. classes 0 and 26), and others start to change early in the extended night (classes 1 and 26). A small set of C-repressed genes are not markedly induced until 24 to 48 h into the extended night (class 22). Some C-induced genes (class 22) were unexpectedly induced after 24 to 48 h in the dark.
Table I.
Boolean analysis of the responses of test sets of C- and light-responsive genes during the diurnal cycle and an extended night
A test set of 484 C-induced genes was generated by combining the 200 genes that are induced most strongly after adding 15 mm Suc for 3 h to C-depleted seedlings (Osuna et al., 2007); 200 genes that are induced most strongly after adding 100 mm Glc for 3 h to C-starved seedlings (Bläsing et al., 2005); and the 200 genes that are induced most strongly after illuminating 5-week-old plants for 4 h in 350 as compared to 50 ppm [CO2] (Bläsing et al., 2005; see gene lists). A similar approach was taken to generate a test set of 383 C-repressed genes. A set of 158 genes that respond rapidly to C was extracted from Osuna et al. (2007); these genes change by >log2 1.4-fold within 30 min of adding 15 mm Suc to C-starved seedlings. A test set of approximately 200 light-induced and 200 light-repressed genes was extracted from a treatment in AtGenExpress in which dark-grown seedlings were exposed to weak white light for 4 h (Bläsing et al., 2005; see gene list). Approximately 1,000 AKIN10-responsive genes were identified from Baena-Gonzalez et al. (2007). For each test set, genes were assigned values of −1, 0, or +1, depending upon whether they decreased, remained unaltered, or increased in a given time interval, using an arbitrary filter of 2-fold change. The time intervals were between the end of the day and end of the night, between the end of the night and 8 h into an extended night, and between 8 h and 24 or 48 h into an extended night. This gives in total 27 possible categories. Response classes that are qualitatively consistent with induction or repression by C are in bold and italic type, respectively. The table gives the numbers and the percent of the total C-repressed or C-induced genes in each class. Gene lists and assignments to the classes by the undirected and directed approach are provided in Supplemental Table S4. DC, Diurnal cycle; XN, extended night.
| Class | DC | XN | XN Late | C Repressed | C Induced | C Rapidly Repressed | C Rapidly Induced | Light Repressed | Light Induced | AKIN10 Repressed | AKIN10 Induced |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 12 | −1 | 0 | 0 | 3 | 38 | 0 | 9 | 2 | 2 | 26 | 3 |
| 3 | −1 | 0 | −1 | 1 | 10 | 0 | 0 | 0 | 0 | 8 | 0 |
| 9 | −1 | −1 | 0 | 0 | 12 | 0 | 1 | 2 | 0 | 6 | 0 |
| 0 | −1 | −1 | −1 | 1 | 43 | 0 | 1 | 0 | 6 | 14 | 1 |
| 10 | 0 | −1 | 0 | 1 | 6 | 0 | 2 | 2 | 3 | 13 | 4 |
| 1 | 0 | −1 | −1 | 16 | 100 | 0 | 2 | 4 | 67 | 55 | 24 |
| 4 | 0 | 0 | −1 | 6 | 17 | 1 | 0 | 3 | 48 | 63 | 13 |
| 2 | 1 | −1 | −1 | 13 | 5 | 5 | 4 | 2 | 24 | 5 | 12 |
| 11 | 1 | −1 | 0 | 4 | 0 | 2 | 0 | 0 | 2 | 1 | 2 |
| 5 | 1 | 0 | −1 | 7 | 0 | 2 | 0 | 1 | 11 | 0 | 7 |
| 8 | 1 | 1 | −1 | 1 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| 6 | −1 | 1 | −1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| 7 | 0 | 1 | −1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 13 | 0 | 0 | 0 | 33 | 201 | 10 | 19 | 73 | 27 | 302 | 86 |
| 14 | 1 | 0 | 0 | 22 | 0 | 12 | 0 | 2 | 1 | 3 | 26 |
| 23 | 1 | 0 | 1 | 3 | 0 | 3 | 0 | 0 | 0 | 0 | 1 |
| 17 | 1 | 1 | 0 | 9 | 0 | 8 | 0 | 1 | 0 | 0 | 9 |
| 26 | 1 | 1 | 1 | 80 | 0 | 32 | 0 | 19 | 0 | 0 | 71 |
| 16 | 0 | 1 | 0 | 11 | 2 | 2 | 0 | 2 | 0 | 1 | 19 |
| 25 | 0 | 1 | 1 | 132 | 8 | 32 | 0 | 50 | 0 | 3 | 152 |
| 22 | 0 | 0 | 1 | 24 | 30 | 4 | 2 | 28 | 2 | 13 | 55 |
| 24 | −1 | 1 | 1 | 13 | 6 | 2 | 0 | 4 | 1 | 1 | 16 |
| 15 | −1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| 21 | −1 | 0 | 1 | 1 | 5 | 0 | 3 | 1 | 1 | 1 | 3 |
| 18 | −1 | −1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 20 | 1 | −1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 19 | 0 | −1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Temporal Responses of Genes That Respond Rapidly to Added Suc
The test sets used in the last section were compiled from treatments that lasted 3 to 4 h. The different temporal responses during the diurnal cycle and extended night might reflect differences in the timing of the signal to which they are responding or differences in the speed of their response. We therefore repeated this analysis with genes that respond rapidly to added Suc.
Osuna et al. (2007) short listed >150 genes that show marked changes (>1.41, log2 scale) of their transcript levels within 30 min of adding 15 mm Suc to C-depleted seedlings. These were manually separated into groups based on the kinetics of their response during the diurnal cycle and extended night (Fig. 5A; Table II). Some show marked changes at the start of the light period that are reversed early in the night (groups 1 and 6), some show a slightly slower response that is almost completed by the end of the night (groups 2 and 7), some do not show large changes until the start of the extended night (groups 3 and 8), and some (group 4) do not respond until 24 to 48 h into the extended night. Because their transcripts respond 30 min after adding Suc to C-starved seedlings, the differing temporal kinetics during the night and extended night indicate that they respond to signals that are generated at different phases as C is depleted. The diurnal changes in pgm are summarized in the right-hand images. Responses that do not occur until the extended night in wild-type plants occur in the normal night in pgm, providing independent support that the responses in the wild type are due to the C status. A small group of genes is induced in the light in pgm, but not in wild-type plants (group 9), indicating they only respond to very high sugar.
Figure 5.
Temporal responses during a diurnal cycle and an extended night of a set of 158 genes, which respond rapidly to Suc addition. A, Clustering of rapidly responding genes. Osuna et al. (2007) short listed 158 genes as showing marked changes (>1.41, log2 scale) of their transcript levels 30 min after adding 15 mm Suc to C-depleted seedlings. The responses of these genes in the diurnal cycle (Bläsing et al., 2005) and the extended night in wild-type Col-0 and a diurnal cycle in pgm (Bläsing et al., 2005) were extracted from Supplemental Table S4. The genes were manually sorted into different response groups. The panels show groups of C-repressed (groups 1–5) and C-induced (groups 6–10) genes. The plots show absolute transcript levels. B, Heat map showing correlations between rapidly responding genes and other more slowly responding C-responsive genes. The genes in the rapid- and slow-responding groups were preordered based on internal clustering. Positive correlations are shown as pink and negative as mauve, with a cutoff filter of R2 > 0.7. The inset shows a frequency diagram of pair-pair correlations between members of a rapidly responding gene set and the other genes.
Table II.
Temporal responses during a diurnal cycle and an extended night of genes that respond rapidly after adding Suc to C-starved seedlings
A set of 158 genes that show >2-fold changes within 30 min after adding 15 mm Suc to C-starved seedlings was extracted from Osuna et al. (2007). They were manually grouped (Fig. 5) according to the time at which their transcripts respond during a diurnal cycle and an extended night in Col-0 wild type and during a diurnal cycle in pgm.
| Group | Induced | Repressed |
|---|---|---|
| Early in the light/dark cycle | At5g12110→EF1B α-subunit | At5g17300→myb family transcription factor |
| At5g48570→peptidyl-prolyl cis-trans-isomerase | At5g28770→bZIP transcription factor family protein | |
| At3g12580→HSP70 | At5g39660→Dof-type; interacts with LKP2 and FKF1 | |
| At5g64510→expressed protein| | At1g68840→RAP2.8; RAV2; DNA-binding protein | |
| At4g37610→TAZ zinc finger family protein/BTB/POZ domain protein; | ||
| At3g48360→speckle-type POZ protein | ||
| At3g23880→F-box family protein | ||
| At4g28270→zinc finger (C3HC4-type RING finger) family protein | ||
| At1g23870→TPS9 | ||
| At5g18670→β-amylase (BMY3) | ||
| At5g18630→lipase class 3 family protein | ||
| At5g61440→thioredoxin family protein | ||
| At1g10150, At3g15630, At3g52060, At3g06070, At4g39675, expressed proteins | ||
| Slightly later in the diurnal cycle | At5g20150→SPX (SYG1/Pho81/XPR1) protein | At1g13260→DNA-binding protein RAV1 |
| At5g47060→senescence-associated protein | At2g18300→basic helix-loop-helix (bHLH) | |
| At3g24500→ethylene-responsive transcriptional coactivator | At5g61590→like ethylene responsive element binding factor 5 | |
| At3g16050→ethylene-inducible protein | At5g49450→bZIP family transcription factor | |
| At3g09440→heat shock cognate 70-kD protein | At1g25560→DNA-binding protein RAV2 | |
| At3g14200→DNAJ heat shock protein | At1g69490→no apical meristem (NAM) family protein | |
| At2g22500→mitochondrial substrate carrier family protein | At1g80440→kelch repeat-containing F-box family protein | |
| At1g67360→rubber elongation factor family protein | At3g61060→F-box family protein | |
| At2g46240→IQ/BAG domain-containing protein | At1g23390→kelch repeat-containing F-box family protein | |
| At3g44450, At1g66080→expressed proteins | At2g25900→zinc finger (CCCH-type) family protein) | |
| At5g22920→zinc finger (C3HC4-type RING finger) family protein | ||
| At1g70290→TPS8 | ||
| At1g60140→TPS10 | ||
| At2g18700→TPS11 | ||
| At2g45170→autophagy 8e (APG8e) | ||
| At4g15680→glutaredoxin | ||
| At4g15700→glutaredoxin | ||
| At3g62950→glutaredoxin | ||
| At5g65660→Hyp-rich glycoprotein | ||
| At1g80920→J8-like protein similar to DnaJ | ||
| At1g13700→glucosamine/galactosamine-6-P isomerase family protein | ||
| At4g18340→glycosyl hydrolase family 17 protein | ||
| At5g08350→GRAM domain-containing protein | ||
| At2g17880→putative DnaJ protein | ||
| At3g26510→octicosapeptide/Phox/Bem1p (PB1) protein | ||
| At1g08570→thioredoxin family protein | ||
| At3g15450→ similar to auxin down-regulated protein ARG10 | ||
| At5g11070, At2g25200, At1g54740, At3g49790, At2g40000, At3g07310, At1g49500, At4g33666, At5g60680, At2g20670, At3g07350, At2g24550, At4g05070, At3g06080, At2g27830, expressed proteins | ||
| Start of the extended night | At2g01150→zinc finger (C3HC4-type RING finger) protein | At5g61600→DNA-binding protein—like DNA-binding protein EREBP-4 |
| At4g39800→myo-inositol-3-P synthase isozyme 1 | At5g39610→no apical meristem (NAM) family protein similar to CUC2 | |
| At1g61800→Glc 6-P/Pi transporter 2, GPT2 | At4g27410→no apical meristem (NAM) family protein (RD26) | |
| At3g13650→disease resistance response protein-related | At5g47390→myb family transcription factor | |
| At1g54050→17.4-kD class III heat shock protein | At1g74840→myb family transcription factor) | |
| At5g24660→expressed protein| | At4g03510→zinc finger (C3HC4-type RING finger) family protein (RMA1) | |
| At4g11360→zinc finger (C3HC4-type RING finger) family protein (RHA1b) | ||
| At5g44260→zinc finger (CCCH-type) family protein | ||
| At1g76410→zinc finger (C3HC4-type RING finger) family protein | ||
| At1g76590→zinc-binding family protein | ||
| At3g59940→kelch repeat-containing F-box family protein | ||
| At3g15500→putative jasmonic acid regulatory | ||
| At5g23350, At5g23360 _XH2→ABA-responsive GRAM domain protein | ||
| At1g03850→glutaredoxin | ||
| At5g41080→glycerophosphoryl diester phosphodiesterase family protein | ||
| At1g15040→Gln amidotransferase related | ||
| At2g29300→putative tropinone reductase | ||
| At2g32150→haloacid dehalogenase-like family protein | ||
| At1g80160→lactoylglutathione lyase family protein/glyoxalase I family protein | ||
| At5g51440→23.5-kD mitochondrial small heat shock protein (HSP23.5-M) | ||
| At1g52250→dynein light-chain type 1 family protein | ||
| At1g05575, At1g75190, At3g52070, At1g33055, At5g24890, At2g38820, At4g16000, At1g10140, At3g03870, At2g31945, At3g10120, expressed proteins | ||
| Late in the extended night | At5g63130→octicosapeptide/Phox/Bem1p (PB1) protein | At5g13080→WRKY-like protein WRKY DNA-binding protein |
| At2g20560→DNAJ heat shock family protein | At4g37010→caltractin, putative/centrin, At3g20340, At3g15440, At2g47270, At2g34600, expressed proteins | |
| At5g52640→heat shock protein 81-1 (HSP81-1) | ||
| At2g29500→17.6-kD class I small heat shock protein | ||
| At5g12020→17.6-kD class II heat shock protein | ||
| At5g12030→17.7-kD class II heat shock protein | ||
| No or unrelated to the changes of sugars | At5g15950→adenosyl-Met decarboxylase family | At1g71030→ATMYBL2 |
| At5g67420→lateral organ boundaries domain protein 37 | At5g37260→myb family transcription factor | |
| At5g49480→sodium-inducible calcium-binding (ACP1) | At1g69600→zinc finger homeobox family protein | |
| At3g21890→zinc finger (B-box type) CONSTANS family | At1g72200→zinc finger (C3HC4-type RING finger) family protein | |
| At5g35320→expressed protein| | At1g68520→zinc finger (B-box type) family protein | |
| At5g66690→UDP-glucoronosyl/UDP-glucosyl transferase | At1g49200→zinc finger (C3HC4-type RING finger) family protein | |
| At3g59220→pirin, putative | At2g34620→mitochondrial transcription termination factor related | |
| At5g51390, At5g19875, At4g39675, At3g15310, At5g59080, expressed proteins | At4g36040→DNAJ heat shock N-terminal domain-containing protein (J11) | |
| At5g43520→DC1 domain-containing protein |
This analysis identifies genes that are probably upstream components of the transcriptional response to C. Examples of C-repressed genes that respond rapidly to Suc addition and also respond early in the light period and recover during the night, include four trehalose-P synthase-like genes (TPS8, TPS9, TPS10, and TPS11), several transcription factors, members of the BTB/POZ domain family, a set of kelch domain F-box factors, ATG8e, a thioredoxin family member, and several glutaredoxins (Table II). Many rapidly responding C-induced genes are annotated to be involved in stress responses; of the genes with a functional annotation, stress-related genes account for two of three in group 1, three of 10 in group 2, three of six in group 3, and all in group 4, whereas two genes in group 7 are related to ethylene signaling (Table II).
We investigated whether subsets of genes could be identified that might lie downstream of these early responders. To do this, we first clustered the rapid responding genes and all other C-responsive genes separately, and then correlated them with each other (Fig. 5B). Using a fairly stringent filter of R2 > 0.7, about one-half of the rapidly responding genes were correlated with each other. This set of tightly clustered genes correlated with large sets of the more slowly responding genes.
Data Condensation to Identify Biological Processes
We next investigated which metabolic and cellular processes are subject to transcriptional regulation. The dataset comprising the global transcript profiles during the diurnal cycle and extended night was analyzed with the Pageman application (Usadel et al., 2006), which queries whether the response of the genes in a functional category differs from the response of other genes on the array. It uses an extensive plant-specific ontology developed in MAPMAN (Thimm et al., 2004; Usadel et al., 2005; http://gabi.rzpd.de/projects/Map-Man; TAIR6_MappingFile_ath_affy_tair6.m02), which contains >1,000 hierarchical and mainly nonredundant categories. This condenses the >22,000 features on an ATH1 array into about 1,000 features, many of which represent a defined biological process. Transcript levels were normalized on the values at the end of the night in the same experiment, the ratios averaged across the biological replicates, and imported into PAGEMAN to calculate the average change for the genes in a given category. To provide statistical support, we calculated Wilcoxon P values, which give the probability that the response of the genes in a given category is significantly different from the response of all other genes on the ATH1 array. An increasingly deep color indicates an increasingly large change of the average value or an increasingly significant P value. Blue and red distinguish between categories where expression increases or decreases, respectively. It should be noted that when a category contains a large number of genes, P values can be significant even though the average change is very small. When a category contains few genes, a large average change may not be significant. The full analysis is provided in Supplemental Table S5. Figure 6A provides a schematic representation of different types of response during the diurnal cycle and extended night.
Figure 6.
Ontology-based overview of the global responses of transcript levels during a diurnal cycle and an extended night. Transcript levels were expressed as a log2 ratio compared to the value at the end of the night in the same experiment. The response was averaged across all data points available for that time point. This procedure was repeated for each time point during the diurnal cycle and each time point during the extended night treatment. The left-hand image shows the average change of the transcript level (on a log2 scale) for all of the genes in a given BIN or subBIN. An increasingly deep blue or deep red color indicates an increasingly large average increase or decrease, respectively, of all the transcripts in a given BIN or subBIN. The right-hand image shows the P value (calculated using Wilcoxon's test) that the changes of transcript levels of genes in a given BIN or subBIN are significantly different from the response of all the other genes represented on the array. An increasingly deep blue or red color indicates an increasingly significant P value that transcript levels for genes in a given category increase or decrease compared to the level at the end of the night. The major BINS and subBINs are indicated on the left-hand side of the diagram. Selected BINS or subBINS that showed highly significant changes are indicated on the right-hand side of the diagram together with the P value. A, Schematic illustration of selected biologically relevant responses. B, Responses for selected functional categories related to metabolism and cellular growth. C, Responses for selected functional categories related to metabolism and cellular growth.
Coordinated Transcriptional Regulation of Metabolism and Cellular Growth Processes
C depletion is accompanied by transcriptional repression of biosynthetic pathways and cellular growth processes that use C and induction of processes that mobilize C from alternative sources (Fig. 6B). Some of the responses are initiated in the night and others early in the extended night.
Categories showing a coordinated increase of transcripts in the light and a decrease that starts during the night include starch synthesis, glycolysis, amino acid synthesis, nucleotide synthesis, deoxynucleotide metabolism, many aspects of protein synthesis (amino acid activation, cytosolic ribosomal proteins, translation initiation, and elongation), protein folding, cell cycle, cell division, and histone synthesis. Categories showing a coordinated decrease of transcripts in the light, a small increase during the night, and a further increase early in the extended night include invertases, autophagy, and ubiquitin-regulated protein degradation. Slightly delayed responses are shown by genes in categories related to Suc synthesis, glycolipid synthesis, and plastid and mitochondrial metabolite transport, which are repressed after extension of the night, and genes related to gluconeogenesis, lipid degradation, and amino acid degradation, which do not show marked changes during the diurnal cycle but are induced early in the extended night. As already noted, metabolite profiling reveals that catabolism of alternative sources of C, including protein, commences early in the extended night (Fig. 1).
Other functional categories show more complex responses. For example, genes in categories related to photosynthesis (light reactions, Calvin cycle, photorespiration), chlorophyll synthesis, chloroplast biogenesis (plastid ribosomal proteins), pigment synthesis (isoprenoids, flavanols, isoflavonols), and nitrate and sulfate assimilation decrease during the light period and recover during the night, but decrease during the extended night (see Supplemental Fig. S4).
Coordinated Responses of Regulatory Genes
Some gene categories involved in signaling also show marked changes during the night and early extended night (Fig. 6C). Particularly striking responses are seen for TPS-like genes and a small BTB/POZ family. These genes respond within 30 min of adding Suc to C-depleted seedlings (Osuna et al., 2007; Table III), indicating they are early components in transcriptional responses. Many genes involved in hormone metabolism, sensing, and signaling, several families of transcription factors and E3 ubiquitin ligases, and receptor kinases are also progressively induced or repressed, with the changes starting in the normal night and being completed a few hours into the extended night (a more expanded display of the response of these genes is provided in Supplemental Fig. S4).
Table III.
Overlap between the C, light, and clock-regulated test sets of genes
A clock test set of 604 genes was identified from an ATH1 dataset for samples harvested 2, 6, 10, 14, 18, and 22 h into the subjective day on the second and third days after transfer from 12-h light/12-h dark into continuous light (Edwards et al., 2006). The 10,000 genes with the highest variance were subjected to a Fisher's g test to test for periodicity using the Genets package (Wichert et al., 2004), and then filtered using Benjamini-Hochberg (1995); P < 0.05. This rigorous test identified 604 circadian-regulated genes. The C-responsive tests were identified as in the legend of Table I. Sets of 196 light-induced and 198 light-repressed genes were identified from a public dataset where rosettes were darkened or illuminated for 4 h at 50 ppm [CO2] (Bläsing et al., 2005). Genes were identified that were present in the test sets of clock and C- or light-regulated genes and are given according to the subjective time at which they peaked in a free-running cycle. The numbers in parentheses give the percent of genes in that class of C- or light-regulated genes found in the corresponding class of clock-regulated genes. Time of peak describes the time in hours from subjective dawn.
| Clock Regulated
|
C Regulated
|
Light Regulated
|
||||
|---|---|---|---|---|---|---|
| Class | Time of Peak | No. | Induced | Repressed | Induced | Repressed |
| h | ||||||
| Total | 604 | 484 | 383 | 196 | 198 | |
| 1 | 2 | 135 | 6 (1%) | 13 (4%) | 13 (7%) | 2 |
| 2 | 6 | 128 | 0 | 31 (8%) | 3 | 15 (8%) |
| 3 | 10 | 101 | 3 | 10 (3%) | 0 | 6 |
| 4 | 14 | 87 | 2 | 5 | 2 | 1 |
| 5 | 18 | 73 | 2 | 2 | 3 | 0 |
| 6 | 22 | 80 | 6 (1%) | 0 | 9 (5%) | 0 |
| 19 (4%) | 61 (15%) | 30 (15%) | 24 (12%) | |||
Temporal Responses of Genes Whose Expression Is Altered by Overexpression of AKIN10
Baena-Gonzalez et al. (2007) reported that overexpression of AKIN10 leads to changes of transcripts of >1,000 genes. This included widespread induction of genes that are involved in catabolism and repression of genes that are involved in biosynthesis and cellular growth, as well as repression of TPS8 to TPS11 and APG8e. This resembles the responses that we have seen during the night and early extended night (see above). We therefore investigated how all approximately 1,000 AKIN10-responsive genes behave in our dataset. K-means clustering (Supplemental Fig. S5) and Boolean analysis (Table I) illustrated that many of them show marked changes during the night or early in the extended night. Very few respond to extreme C starvation. This provides evidence that AKIN10 contributes to the regulation of expression in response to small changes in the C status.
The diurnal cycle and extended night are complex multifactorial responses in which many other inputs, including the clock and light, may be affecting gene expression. To deepen the analysis, we investigated how C interacts with these inputs.
Interaction between C and the Clock
Using a stringent filter, we extracted 604 genes with an unambiguous timing of a circadian peak from a dataset of constant light-grown plants (free-running cycle) by Edwards et al. (2006). This is smaller than the list of 3,503 genes in Edwards et al. (2006) because our stringent filter excludes clock-regulated genes whose response for various technical or biological reasons is less clearly defined. Table III summarizes the overlap between the clock- and C-regulated genes and uncovers an unexpected antagonistic interaction. Most overlapping genes fall into two categories; some show a circadian peak in the subjective light period (i.e. 0–12 h, when the light would be on in a light/dark cycle), but are repressed by C, and the others show a circadian peak toward the end of the night, but are induced by C.
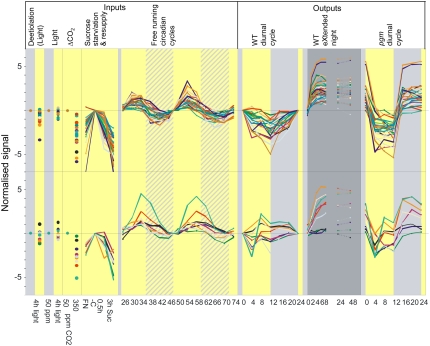
This antagonistic interaction is explored in Figures 7 to 9 (see also http://mapman.mpimp-golm.mpg.de/supplement/xn). The left-hand images show, from left to right, the response of shoots after illuminating dark-grown seedlings with weak white light for 4 h (AtGenExpress), the response after illuminating rosettes for 4 h at 50 ppm [CO2] compared to 4-h darkness (two criteria for light responsiveness), the response after illumining rosettes for 4 h at 350 ppm compared to 50 ppm [CO2], and the response 30 min or 3 h after adding Suc to C-starved seedlings (two independent criteria for C responsiveness). They also show the response in a free-running cycle, a diurnal cycle, and extended night in Col-0, and a diurnal cycle in pgm.
Figure 7.
Antagonistic interaction between C and the clock. The images show, from left to right, the response of shoots after illumination of dark-grown seedlings with weak white light for 4 h (AtGenExpress), the response after illumination of rosettes for 4 h at 50 ppm [CO2] compared to 4-h darkness (two independent inputs for light responsiveness), the response after illumination of rosettes for 4 h at 350 ppm compared to 50 ppm [CO2], and the response 30 min or 3 h after adding Suc to C-starved seedlings (two independent inputs for the C responsiveness), the response in a free-running cycle over 48 h (input for the clock) and the measured changes during a diurnal cycle and extended night in Col-0, and a diurnal cycle in the starchless pgm mutant. The original data and identities of the genes are given in Supplemental Table S4. Transcript levels in the wild-type diurnal cycle, the pgm diurnal cycle are normalized on the levels at the end of the night in wild-type plants. Treatments with light and 50 ppm or 350 ppm [CO2] are normalized on the respective dark or 50 ppm [CO2]. Transcript levels in the extended night treatment are normalized on the levels at the start of the extended night treatment (i.e. the end of the normal night in these experiments). Transcript levels in a free-running cycle are normalized on an estimated level at the end of the 24-h period. Transcript levels after illuminating etiolated seedlings are normalized on the levels in nonilluminated controls. Transcript levels in seedlings in full nutrition and 30 min and 3 h after adding 15 mm Suc to C-starved seedlings are normalized on the levels in the C-starved seedlings. The plots show genes that are regulated by C and the clock (Table III) and peak 6 h (A) and 10 h (B) into the subjective day in a free-running cycle.
Figure 8.
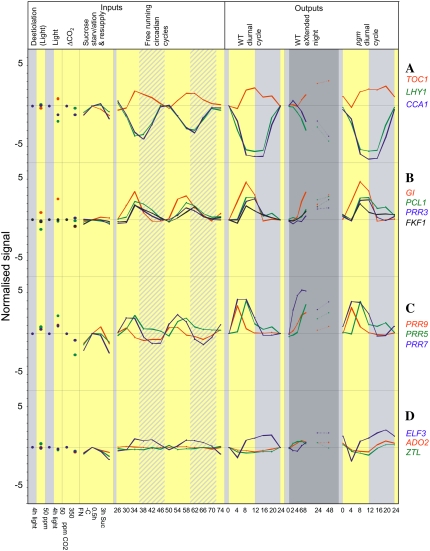
Changes of transcripts for clock genes. The display is as described in the legend of Figure 7. A, Central clock genes. B to D, Genes that entrain the clock.
Figure 9.
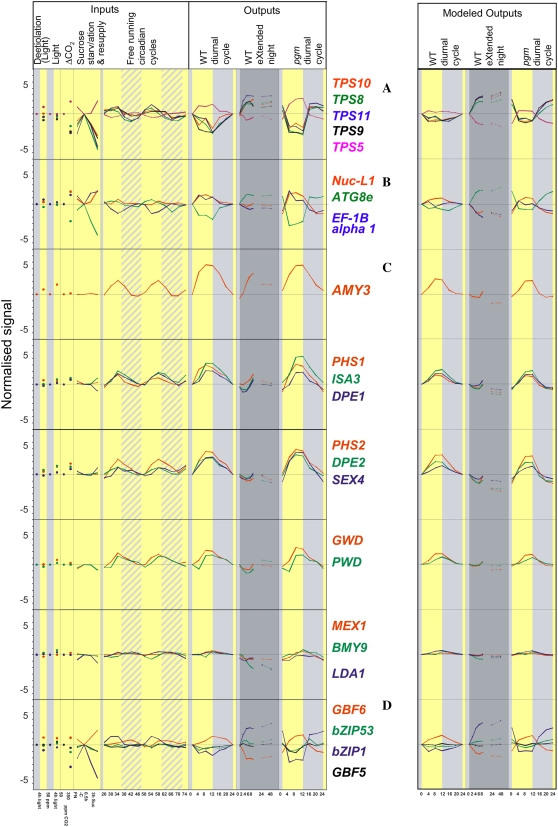
Changes of transcripts for selected genes that are antagonistically regulated by the clock output pathways and C. The left-hand display shows, from left to right, the input s for light, C, and the clock (see legend of Fig. 7 for details), and the observed responses in a diurnal cycle and extended night treatment in Col-0, and a diurnal cycle in pgm. Transcript levels were normalized as in Figure 8. The right-hand images show the response predicted by our linear model (for details, see the legend to Table IV; also “Materials and Methods” and “Results”). A, Genes involved in Tre-6-P signaling: TPS5, TPS8, TPS9, TPS10, and TPS11. B, Genes involved in the regulation of protein synthesis and degradation: EF1B α-subunit 1 (At5g12110), nucleolin organizer (NUC-L1, At1g48920), and autophagy protein 8e (ATG8e, At2g41570). C, Genes involved in starch degradation (AMY3, PHS1, ISA3, DPE2, PHS2, DPE2, SEX4, GWD, PWD, MEX1, BMY9, LDA1). D, bZIP1 (Atg5g49450), which is implicated in AKIN10 signaling (Baena-Gonzalez et al., 2007).
Figure 7 shows the responses of the genes that peak 6 or 10 h into a free-running cycle but are repressed by C. In most cases, the peak shifts to the end of the night in a light/dark cycle, presumably because sugars inhibit expression in the light. This is even more marked in pgm, where all of the genes show a clear minimum during the light period. This reflects the high levels of sugar in pgm. Figure 7 also confirms that these genes are all repressed by C; their transcripts decrease when they are illuminated at 350 compared to 50 ppm [CO2] or after adding Suc to seedlings. Although six of the genes are also light responsive (see Supplemental Table S4), the response to light is much weaker than the response to C.
Responses of Clock Genes
We next investigated the responses of central clock genes (Fig. 8A) and genes that entrain the clock (Fig. 8, B–D). The central clock genes show a strong circadian response in a free-running cycle, with TOC1 changing reciprocally to CCA1 and LHY, as typically seen (Salome and McClung, 2005; Gardner et al., 2006). The phase is unaltered and the amplitude is only slightly increased in a light/dark cycle in Col-0 or pgm or during the first 8 h of extended darkness. The clock is disrupted after a 24- to 48-h extension of the night when the transcripts resemble those at the end of the subjective light period, although these times correspond to the start of the subjective light period. A range of responses was found for genes that entrain the clock. FKF1, PCL1, and PRR3 transcripts change in parallel in a free-running cycle. These genes are not regulated by light or C and their response is not unaffected in a light/dark cycle. It should be noted that cross-hybridization of the ATH1 probe sets is to be expected between FKF1 and PCL1. GI is strongly induced by light, but comparatively unaffected by C. GI transcript rises steeply in the subjective light period in a free-running cycle. This response is retained in a light/dark cycle and is not modified in the first 8 h of an extended night. PRR5, PRR7, and PRR9 are induced by light and repressed by C. They show slightly larger amplitudes in a light/dark cycle. Potential cross-reactivity might lead to artefacts for PRR9; however, given the similar response of PRR7 and PRR5, the data probably do reflect sugar and light regulation. ELF3, ADO2, and ZTL are repressed by C. Their free-running response is modified in a light/dark cycle. ELF3 shows a stronger minimum at the start of the light period, especially in pgm. ADO2 and ZTL do not show circadian changes in a free-running cycle, but show a small diurnal change in wild-type plants with a minimum at the end of the day and an increase in the night, which is amplified in pgm. Thus, whereas C status does not modify the central clock pacemaker, it does affect some of the entraining genes.
Responses of Genes Involved in Trehalose-6-P Signaling, Protein Turnover, and Starch Degradation
There is mounting evidence that trehalose-6-P (Tre-6-P) acts as a sugar signal in plants (see “Discussion”). Arabidopsis contains a small family of genes annotated as TPS (Lunn, 2007). In a free-running cycle, TPS8 to TPS11 show a peak and TPS5 a minimum during the subjective light period (Fig. 9A). TPS8 to TPS11 are strongly repressed and TPS5 is induced by sugars. This antagonizes the circadian response, resulting in a minimum of TPS8 to TPS11 and a maximum of TPS5 during the day, and a peak of TPS8 to TPS11 and a minimum of TPS5 at the end of the night. In pgm, the diurnal changes are larger and occur even more rapidly. The clock and C act in concert in an extended night; in this case, the clock leads to an increase of TPS8 to TPS11 and decrease of TPS5 transcripts, which is reinforced by the simultaneous depletion of C.
Figure 9B shows the response of three genes that might be involved in the regulation of protein turnover. AtNUCL1 plays a key role in nucleolus organization and is required for rRNA synthesis (Kojima et al., 2007; Pontvianne et al., 2007). AtNUC-L1 transcript is subject to weak circadian regulation with a peak in the light period and is strongly C induced. This additive combination leads to a marked diurnal change with a maximum in the light and a decrease at night in Col-0, which is accentuated in the pgm. Elongation factor (EF)-α-subunit 1 is circadian regulated with a minimum during the subjective light period, but is induced by C. In this case, C antagonizes the clock. ATG8e plays a key role in the regulation of autophagy (Doelling et al., 2002; Downes and Vierstra, 2005). ATG8e shows weak circadian regulation, peaking in the subjective light period, but is repressed by C. ATG8e transcript shows a strong decrease in the light, a recovery in the night, and a further increase early in the extended night.
Figure 9C shows the response of genes involved in starch degradation (Smith et al., 2005; Zeeman et al., 2007a, 2007b). Most show a marked circadian cycle with a peak toward the end of the subjective light period and a minimum at the end of the subjective dark period (Harmer et al., 2000; Lu et al., 2005), which is retained in a light/dark cycle (see also Smith et al., 2004; Bläsing et al., 2005). Almost all show a modified response in the first hours of extended night. The circadian increase is delayed and weakened for PHS1, ISA3, and DPE1 and almost abolished for PHS2, DPE2, GWD, PWD, and SEX4. The decrease is prevented by light alone (GWD) or a combination of light and ambient [CO2]. MEX1, BMY9, and LDA1 do not show marked changes in a free-running cycle or a light/dark cycle in Col-0, but show a pronounced decrease in an extended night, and a small diurnal change in pgm. These results point to a potential mechanism to decrease the rate of starch breakdown when C is low at the beginning of the day (see “Discussion”). Surprisingly, the absolute transcript levels (data not shown) and the diurnal responses are very similar in Col-0 and pgm. This implies that expression of these genes is not regulated by events related to starch synthesis, starch accumulation, or starch degradation.
Light-Regulated Genes
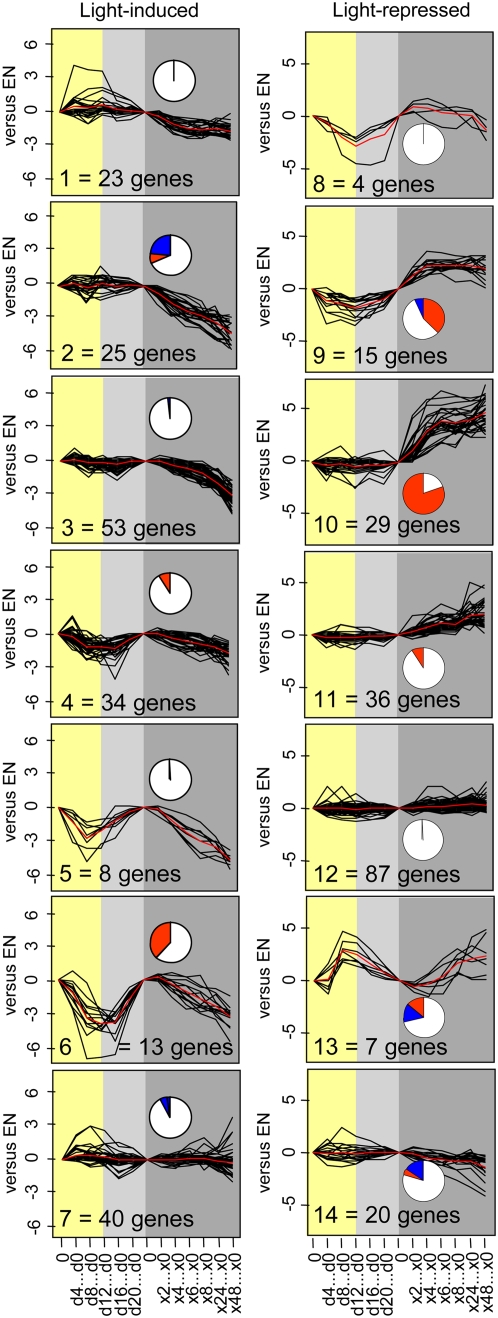
Bläsing et al. (2005) concluded that light does not play a major role in the diurnal regulation of gene expression in Arabidopsis growing in a regular light/dark cycle. This unexpected conclusion was based on the diurnal response of a test set of approximately 400 light-regulated genes, identified from a treatment in AtGenExpress in which dark-grown seedlings were exposed to weak white light for 4 h. We used K-means clustering (Fig. 10) and directed grouping (Table I) to reanalyze the response of this test set in the enlarged dataset with the extended night treatment. Lists of the genes in the different groups are provided in Supplemental Table S4.
Figure 10.
Temporal responses of light-regulated genes during a diurnal cycle and an extended night. Test sets of approximately 200 light-induced and 200 light-repressed genes were abstracted from a treatment in AtGenExpress in which dark-grown seedlings were exposed to weak white light for 4 h. To investigate how these genes respond during a more prolonged exposure to darkness, the responses of this test set were reanalyzed in a dataset combining the diurnal cycle (Bläsing et al., 2005) and an extended night treatment. The genes in the test sets are listed and datasets are provided in Supplemental Table S4. Clusters 1 to 7 show light-induced genes and clusters 8 to 14 show light-repressed genes. The pie chart inserts show the proportion of the genes in a given cluster that are present in the sets of C-induced (blue) or C-repressed (red) genes (white, no overlap). Lists of genes in the various clusters are provided in Supplemental Table S4.
K-means clustering (Fig. 10) showed that the vast majority of these light-regulated genes do not respond during the normal 24-h light/dark cycle, but do respond as expected in the extended night. Only approximately 10% of the light-regulated genes respond in the expected way during a light/dark cycle (23 genes in cluster 1, 19 genes in clusters 8 and 9; Fig. 10), and many of these show a larger response at the start of the extended night (especially cluster 9). A short extension of the night repressed many light-induced genes (clusters 2, 3, 4, 5, 6 = 133 genes; 66% of the set) and induced many light-repressed genes (clusters 10, 11, 13 = 78 genes; 39% of the gene set). Directed sorting (Table I) also showed that the majority of the light-regulated genes do not show a corresponding response in plants that have been grown in a light/dark cycle until the dark treatment is extended beyond the end of the normal night.
This raises the question of whether light signaling is attenuated by an interaction with the clock or C. About 13% of the light-repressed genes are present in the clock-regulated gene set. Almost all of these shared genes peak in the subjective light period (Table III), indicating that an antagonistic interaction with the clock contributes to the attenuated response of light-repressed genes. About 20% of the light-induced genes were present in the circadian set, but, in this case, the peak was at the end of the subjective night or the start of the subjective day (Table III). The pie diagrams in Figure 10 indicate the proportion of C-induced, C-repressed, and nonoverlapping genes in each cluster (blue, red, and white sectors, respectively). Many C-repressed genes in the clusters of light-repressed genes show little (9) or no (10, 11) change during the light/dark cycle, but decrease at the start of the extended night. This is at first sight surprising because additive action of light and C might be expected to generate exaggerated, rather than damped, changes during the light/dark cycle. One possible explanation is that C is still high enough during the night to repress these genes and override the effect of darkness and that a further decrease of C combined with darkness leads to strong induction when the night is extended. These clusters are enriched for genes involved in amino acid degradation (Supplemental Table S4). The situation is less clear for the light-induced genes. For example, there is no evidence that C contributes to the damped response during the light/dark cycle in the large cluster 3, which contains many (approximately 35%) genes related to photosynthesis and chlorophyll synthesis (Supplemental Table S4).
Modeling the Interaction between C, Light, and the Clock
The data analysis presented so far involved qualitative comparison of simplified experimental situations in which one input plays a dominant role and complex situations where there will be a multifactorial interaction between several inputs. It would clearly be advantageous to make the analysis more rigorous by performing it in the framework of a quantitative model. We investigated whether global transcriptional responses during the diurnal cycle in Col-0, the extended night in Col-0, and the diurnal cycle in pgm (Table IV) can be predicted by a simple linear model with three inputs: the clock, light, and C.
Table IV.
Linear model of the response of global gene expression
The model attempts to predict the response of all 22K genes from a linear combination of their response, according to the equation Yt = fcc,t * CCt + fs,t * sugar + fl,t * light. The three inputs are (1) the dataset from Edwards et al. (2006) to provide information about the response of each individual gene to the circadian clock at six times during the day (CC), normalized on its computed value at the end of the night; (2) the dataset from Bläsing et al.(2005) comparing rosettes after 4-h additional darkness or 4-h illumination at 50 ppm [CO2] to provide information about the response of every gene to light (light); and (3) the dataset from Bläsing et al. (2005) comparing rosettes after illuminating, then for 4 h in the presence of 50 or 350 ppm [CO2] to provide information about the response of every gene to C (sugar). fCC,t, fS,t, and fLt are the weighting factors for the circadian cycle, sugar, and light input datasets. We tried to predict the response of all 22K genes at each time during a diurnal cycle in wild-type Col-0, a diurnal cycle in pgm, and an extended night in wild-type Col-0. All test datasets were normalized on the wild-type end of the night (24/0 h). For this reason, no values are available for this time point. The table gives, for each time point, the R2 values for the regression against the different inputs individually and the model, and the weighting factors chosen by the model. NA, Not applicable; XN, extended night.
| Wild Type 4 h | Wild Type 8 h | Wild Type 12 h | Wild Type 16 h | Wild Type 20 h | Wild Type 24/0 h | |
|---|---|---|---|---|---|---|
| fCC,t | 0.32 | 0.88 | 0.89 | 0.84 | 0.38 | NA |
| fLt | 0.45 | 0.48 | 0.47 | 0.17 | 0.14 | NA |
| fS,t | 0.33 | 0.39 | 0.35 | 0.18 | 0.08 | NA |
| CC (R2) | 0.01 | 0.12 | 0.18 | 0.25 | 0.11 | NA |
| Sugar (R2) | 0.17 | 0.10 | 0.12 | 0.06 | 0.03 | NA |
| Light (R2) | 0.12 | 0.06 | 0.08 | 0.04 | 0.06 | NA |
| Model R2 | 0.36 | 0.39 | 0.42 | 0.35 | 0.18 | NA |
| pgm 4 h | pgm 8 h | pgm 12 h | pgm 16 h | pgm 20 h | pgm 24/0 h | |
| fCC,t | −0.03 | 0.74 | 0.80 | 0.72 | 0.34 | NA |
| fLt | 0.41 | 0.42 | 0.48 | −0.05 | −0.17 | −0.12 |
| fS,t | 0.27 | 0.47 | 0.36 | −0.36 | −0.67 | −0.75 |
| CC (R2) | 0.01 | 0.05 | 0.11 | 0.08 | <0.01 | NA |
| Sugar (R2) | 0.06 | 0.13 | 0.09 | 0.15 | 0.41 | 0.56 |
| Light (R2) | 0.06 | 0.03 | 0.07 | <0.01 | <0.01 | <0.01 |
| Model R2 | 0.14 | 0.27 | 0.32 | 0.23 | 0.43 | 0.56 |
| XN 2 h | XN 4 h | XN 6 h | XN 8 h | XN 24 h | XN 48 h | |
| fCC,t | 0.18 | 0.49 | 0.42 | 0.79 | 0.00 | 0.00 |
| fLt | −0.25 | −0.51 | −0.67 | −0.64 | −0.86 | −0.78 |
| fS,t | −0.31 | −0.57 | −0.68 | −0.65 | −0.78 | −0.99 |
| CC (R2) | 0.04 | 0.15 | 0.15 | 0.l9 | NA | NA |
| Sugar (R2) | 0.30 | 0.42 | 0.37 | 0.33 | 0.30 | 0.33 |
| Light (R2) | 0.05 | 0.08 | 0.09 | 0.08 | 0.11 | 0.05 |
| Model R2 | 0.39 | 0.60 | 0.56 | 0.56 | 0.47 | 0.42 |
In this model, the response of each individual gene to C is defined by the dataset from comparing rosettes after illuminating them for 4 h in the presence of 50 or 350 ppm [CO2] (Bläsing et al., 2005). The response to light is defined by the dataset from comparing rosettes after 4-h additional darkness or 4-h illumination at 50 ppm [CO2] in Bläsing et al. (2005). We use the dataset from Edwards et al. (2006) to define the response of each individual gene to the circadian clock six times during the 24-h cycle. The model attempts to predict the response of >22K genes from a linear combination of the response to sugar plus the response to light plus the response at a given time in the circadian cycle, each multiplied by a separate weighting factor. The weighting factor is varied to optimize the fit, but must be the same for all genes for a given input and time point (see “Materials and Methods” for details). For each time point in Col-0, signals for each gene were normalized on the signal at the end of the night in the same treatment. For pgm, the signals were normalized on the corresponding signals at the end of the night in Col-0 (as in Figs. 8–10).
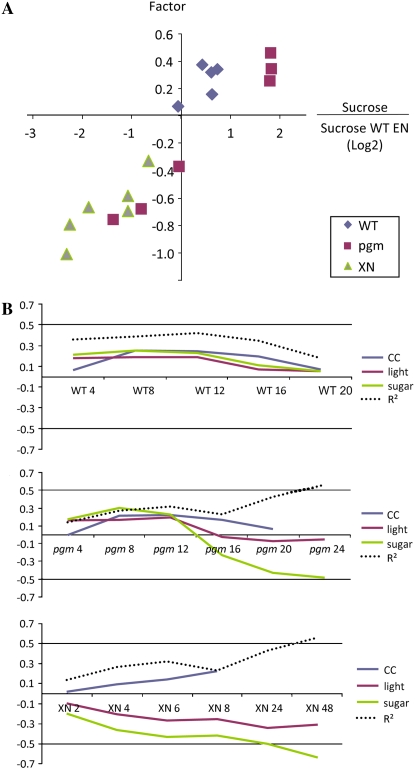
Table IV summarizes the values for the weighting factors, the fit (R2) generated by the model, and, for comparison, the fit found by regression against each individual input dataset. The model performs reasonably well. First, it provides a much better fit to the measured global response than the individual inputs. The fit was especially good in an extended night and during the night in pgm, which highlights the importance of the low sugar signal during the night in pgm (Bläsing et al., 2005; Osuna et al., 2007). Second, the weighting factors chosen by the model for C and light were qualitatively correct (i.e. daytime points were more positive and the nights lower or, in the extended night or in pgm in the dark, strongly negative). In particular, the weightings for C show very good quantitative agreement with the sugar content. When the sugar levels were extracted from Figure 1, normalized to the levels at end of the night, logged, and plotted against the weighting factors, a correlation of >0.8 was obtained for Suc (Fig. 11A) and reducing sugars (data not shown). This provides strong evidence that global transcriptional responses can be predicted from endogenous sugar levels.
Figure 11.
Modeled contribution of C. A, Comparison of the weighting factors for sugar extracted from the models and the level of Suc in the tissue at the corresponding time point. The Suc levels are normalized on the reference (wild type, end of the night) and expressed as a log ratio of Suc. B, Estimated relative contribution of the clock, light, and C to the global transcript level. For each treatment and time point, the weighting factors generated by the model were calculated by scaling the variance of the corresponding input dataset to 1 to estimate their relative contribution. The variances of the input datasets before scaling were 0.01, 0.04, 0.11, 0.08, 0.08, 0.05, and 0.04 for CC at the 2-, 4-, 6-, 8-, 12-, 16-, and 20-h time points, 0.16 for light, and 0.41 for sugar.
To estimate the relative importance of the inputs at a given time point, it is necessary to include information about the magnitude of the changes of transcript levels in each input dataset. In Figure 11B, the weightings are multiplied by the corresponding unit variances. Sugar is almost always the strongest input, especially in an extended night and in pgm in the dark. Figure 11B also summarizes how much of the total variance in the 22K datasets is explained by the model. This varies from 20% to 45% during the wild-type diurnal cycle, up to >50% in extended darkness, or at night in pgm.
Gene-Specific Assessment of the Quality of the Model
We assessed the quality of the prediction for each individual gene by plotting the measured and predicted values at all 17 modeled time points against each other and calculating the correlation coefficient (r). The average correlation coefficient was 0.41. The individual values are given in Supplemental Table S6. They allow us to investigate which sets of genes or individual genes are particularly well or badly predicted.
We first investigated model performance for genes that respond rapidly to Suc (>1.41 log2 scale within 30 min of adding 15 mm Suc to C-starved seedlings; Osuna et al., 2007; see Fig. 5). The model performed very well (r = 0.72). However, some interesting exceptions were found. GPT2, a CONSTANS-like transcription factor, several heat-responsive genes, and MBF1C (At3g24500, a transcriptional coactivator) performed particularly badly. Interestingly, MBF1C responded reciprocally to the CONSTANS-like transcription factor (Supplemental Table S6). Noise can be excluded as an explanation for the discrepancies because the signals and replicates were good (data not shown). Possible explanations include strong nonlinear interactions or that these genes are regulated by an input that is not included by our model.
The model performed well with the test set of light-regulated genes that was identified from AtGenexpress (see Fig. 10; Supplemental Table S4; r = 0.62). This is interesting because most of these genes do not respond to light during the 24-h light/dark cycle (Fig. 10). The good performance of our model confirms that this is at least partly due to an interaction between light signaling and circadian and C signaling. We also tested whether clock-regulated genes performed qualitatively well in the model and whether this could partially be attributed to the presence of cis-elements known to be involved in driving circadian response. To this end, we investigated whether CCA1-containing genes were generally better predicted than non-CCA1-containing genes. Genes containing CCA1 showed only a small, but significantly better, overall correlation between modeled and observed values (r = 0.47 versus r = 0.41; P < 0.01).
We also assessed the model using a quantitative measure of performance for each individual model (i.e. treatment). To do this, we extracted the model residual for each gene and time point (i.e. the difference between modeled and observed values at each time point). Comparing these across the 17 time points again showed that many genes are well predicted, but uncovered some that are badly predicted (data not shown). One example is DOMAIN OF UNKNOWN FUNCTION26 (DUF26)-containing genes. These lacked a significant response in the input datasets, but showed strong responses in pgm. This might indicate a higher order of long-term C integration only apparent in pgm or that these genes react to an input that is not included in the model. Several DUF26-containing genes respond to biotic stress (Nielsen et al., 2007).
As already mentioned, the model performs well with genes that respond to the model inputs. We formalized this relation by using a weighted sum of inputs (weighting each input set by how often it was used in modeling). Higher input did generally lead to better qualitative agreement. However, even though binning data into different variance sets and regressing performance versus weighted input showed dependence (data not shown), there were clearly genes that were not well explained despite the apparent strong inputs (see above for examples). Thus, we were not able to identify a clear-cut threshold for the inputs or gene classes that always performed well in the model. Imposing a threshold would lead to potentially important discrepancies not being found. In general, the model is strongly underdetermined because it has only three inputs and makes no assumptions about the dependency between these inputs.
Examples of Genes and Functional Classes That Are Well and Poorly Predicted
The right-hand image of Figure 9 shows the predicted responses of genes that may be involved in signaling or starch turnover. In almost all cases, there is very good qualitative agreement. The model correctly predicts that the response of the free-running cycle is reversed in the light for the TPS genes (Fig. 9A), ATG8e AT-NUC1-L1 and EF1Bα (Fig. 9B) and in the first part of the extended night for genes involved in starch degradation (Fig. 9C). There is also very good quantitative agreement in the night and extended night in Col-0, and in the dark in pgm. The only exception was AMY3, where the model predicted a decrease during the first hours of the extended night, whereas we observed an increase like that in a free-running cycle. The model predicts a decrease in an extended night because AMY3 is light induced (see Fig. 9). Possible reasons for the discrepancy between the predicted and observed response might be that weighting factors selected on the basis of all 22K genes are not appropriate for AMY3, that the clock and light do not act additively on AMY3 expression, or that there is a further unknown input.
We have already short listed some genes as good candidates for upstream components because they respond rapidly to Suc (Osuna et al., 2007). We used a filter of r > 0.9 to identify genes in this set whose diurnal response is qualitatively extremely well predicted by our model (see above). Among them were TPS8 to TPS11, several transcription factors, and some genes of unknown function. The second best-predicted transcription factor was At5g49450 (bZIP1). This transcription factor is repressed by Suc and induced by the clock in the subjective light period (Fig. 9D). The model correctly predicts that an antagonistic interaction leads to an accentuated increase of bZIP1 transcript at the start of the extended night (Fig. 9D). Intriguingly, this bZIP1 factor has been shown to have significant synergism with AKIN10/At3g01090), implicating it in integrating sugar and energy metabolism (Baena-Gonzalez et al., 2007). We also inspected the measured and predicted responses for three further members of this family, which also modify AKIN10 signaling (Baena-Gonzalez et al., 2007). Intriguingly, each showed a different response, which was qualitatively predicted by the model. GBF6/At4g34590 was induced by Suc and repressed by the clock in the subjective light period, leading to an increase in light and strong repression in an extended night. bZIP53/At3g62420 and bZIP2/GBF5/At2g18160 were repressed by C, but were not circadian regulated, resulting in weak repression in the light period but no strong response in an extended night. Incidentally, this set of genes illustrates the consequences of the presence or absence of an antagonistic interaction of C with the clock.
We also extracted all the genes regulated by AKIN10 as determined by Baena-Gonzalez et al. (2007) and investigated their performance in the model. The performance was very good (r = 0.73, compared to an average of 0.41; see above). This confirms the qualitative conclusion (see above) that AKIN10 is deeply involved in the sugar-dependent regulation of transcriptional responses during the diurnal cycle and the first hours of the extended night.
Finally, we investigated which functional classes of genes are particularly well or poorly predicted by the model. To do this, we extracted all of the genes that showed r > 0.7 or r < 0 between the modeled and predicted values and subjected them to an over- or underrepresentation analysis using the online version of Page-Man (http://mpimp-golm.mpg.de/ora; Table V; see also Supplemental Table S6). In the well-predicted genes (r > 0.7), we found high enrichment of genes involved in amino acid and nucleotide metabolism, protein synthesis, and histones, and an underrepresentation of transposases, biotic stress genes, and MADS-box transcription factors. Genes involved in autophagy were overrepresented, although at a slightly higher P value (data not shown). In the badly predicted genes (r < 0), we found an enrichment of retrotransposons, MADS-box transcription factors, and F-box proteins, and an underrepresentation of genes that are involved in proteins synthesis, the cell cycle, and structural integrity. This quantitative analysis strengthens our earlier conclusion (Fig. 6) that small changes of C status regulate the expression of large numbers of genes that are required for biosynthesis and cellular growth processes. Poor prediction of transposases is to be expected because the expression of these is most likely to reflect noise. The poor performance of biotic stress genes might suggest a biotic stress component that could not be modeled using the inputs.
Table V.
Well-predicted categories of genes
All genes that showed an r > 0.7 between the modeled and predicted values in the linear model (see Table IV; Supplemental Table S6) were extracted and subjected to an over/underrepresentation analysis. The enrichment/depletion value is the relative enrichment of the given class when compared to the ATH1 background. Thus an enrichment/depletion value above 1 indicates an enrichment of this class compared to all ATH1 genes, and a value below 1 a depletion.
| Bin | Name | P Value | Enrichment/Depletion |
|---|---|---|---|
| 28.1.1 | Retrotransposon/transposase | 7.2E-38 | 0.08 |
| 29.2.2 | Protein synthesis miscellaneous ribososomal protein | 3.0E-29 | 1.98 |
| 29.2 | Protein synthesis | 1.6E-20 | 1.61 |
| 13 | Amino acid metabolism | 4.6E-16 | 1.73 |
| 28.1 | DNA synthesis/chromatin structure | 2.8E-14 | 0.61 |
| 28 | DNA | 7.1E-14 | 0.66 |
| 28.1.1.5 | CACTA-like transposase | 5.8E-12 | 0.07 |
| 29 | Protein | 7.0E-12 | 1.17 |
| 28.1.1.3 | Copia-like retrotransposon | 2.6E-11 | 0.04 |
| 20.1 | Stress biotic | 3.7E-11 | 0.57 |
| 27.3.24 | MADS-box transcription factor family | 2.8E-10 | 0.14 |
| 13.1 | Amino acid metabolism synthesis | 1.0E-09 | 1.69 |
| 23 | Nucleotide metabolism | 2.4E-07 | 1.63 |
| 31 | Cell | 3.5E-07 | 1.30 |
| 29.5.11.4.3 | Protein degradation ubiquitin E3 SCF | 4.6E-07 | 0.66 |
| 29.5.11.4.3.2 | Protein degradation ubiquitin E3 SCF F-box | 1.2E-06 | 0.66 |
| 13.2 | Amino acid metabolism degradation | 1.4E-06 | 1.68 |
| 28.1.3 | DNA synthesis/chromatin structure histone | 2.1E-06 | 2.00 |
DISCUSSION
Plants Retain Only a Small Reserve of C at the End of the Night
Stitt et al. (2007) and Smith and Stitt (2007) proposed that plants respond to decreasing C in an acclimatory manner, with falling C triggering changes in storage, allocation, and growth before there is an acute limitation of metabolism or growth. The experiments in this article were carried out to test a central prediction of this hypothesis, namely, that changes in signaling are initiated by small changes in C status. Transcript profiles were used to test this prediction because they provide a comprehensive readout of many signaling pathways.
Typically, only a small amount of starch remains at the end of the night (see introduction). Comparison with Gibon et al. (2004b) indicates that the amount of starch and sugars retained at the end of the night is equivalent to approximately 10% of the C assimilated in a 24-h cycle. Extending the night for just 2 to 4 h leads to an acute C limitation and catabolism of protein, lipids, and other sources of C. Whereas this resembles the response in prolonged starvation (Brouquisse et al., 1991; Dieuaide et al., 1993; Aubert et al., 1996), it is striking that these changes occur so rapidly. Holding C in reserve will decrease investment in growth and lead to an incremental decrease of biomass. Indeed, there is a strong negative correlation between the starch content at the end of the night and the biomass of 20 Arabidopsis accessions in short-day conditions (Cross et al., 2006). These results indicate that Arabidopsis grows almost as fast as the available C will permit.
Small Changes of C Status Trigger Major Changes in Gene Expression during the Night
The progressive decrease of C during the night and extended night is accompanied by widespread changes of transcripts for thousands of genes. Three lines of evidence show that C contributes to these changes. First, the response during the diurnal cycle and an extended night resembles that expected from the endogenous changes of sugars. Second, many of the changes of transcript levels occur earlier and are accentuated in the starchless pgm mutant (Figs. 5 and 8–10; Bläsing et al., 2005). Third, the global response can be predicted by a simple linear model in which C plays a major role (see later for further discussion). Analysis of the temporal kinetics reveals several waves of transcriptional responses. Some changes start early in the night and are completed or nearly completed by the end of the night; some are initiated in the first 2 to 4 h of the extended night; and some do not start until much later in the extended night.
Pathway analysis revealed that there is a coordinated global transcriptional reprogramming of biosynthesis and cellular growth during the night and first hours of the extended night. Genes that are involved in the use of C for respiration, the biosynthesis of small structural metabolites, nucleic acid and protein synthesis, and cell division are repressed in parallel with the gradual decrease of carbohydrates during the night. Pathways for lipid and protein degradation and gluconeogenesis are induced during the night or early in the extended night in parallel with the final stages of carbohydrate depletion. Earlier studies of C depletion (Contento et al., 2004; Price et al., 2004; Thimm et al., 2004) showed that many of these processes respond to sugar depletion, but they lacked the density of time points and the evaluative tools to characterize the breadth of the response and show that it occurs so early in the transition to low C.
The biological impact of changes of transcript levels depends upon whether they are followed by changes of the encoded proteins and how quickly this occurs. Our results show there is often agreement between the changes of transcripts and metabolites; for example, repression of genes involved in protein synthesis and induction of genes for protein degradation correlate with a marked decrease of polysome loading (M. Piques and M. Stitt, unpublished data), a decrease of protein, and an increase of amino acids (Fig. 1). However, this does not mean that there is a direct causal link. Gibon et al., (2004a, 2006) showed for many enzymes in central C and N metabolism that there is a lag of days until the encoded enzymes are affected. In such cases, the shifts in the diurnal responses of transcripts are integrated over time, allowing gradual adaptation to a sustained change in the conditions. Rapid adjustment within a single diurnal cycle will require that some proteins change rapidly and/or that the changes of transcription are accompanied by rapid posttranslational regulation of existing proteins.
Genes Involved in Signaling Small Changes in C Status
To identify upstream components that orchestrate this set of broad and coordinated transcriptional responses, we focused on the approximately 150 genes whose transcripts respond rapidly after adding Suc to C-starved seedlings, highlighting them as early components of the transcriptional response. We identified a small set of genes whose transcripts increase rapidly after adding Suc to seedlings and also respond during a diurnal cycle (Table II). Examples include EF1B α-subunit 1 (At5g12110), a SPX (SYG1/Pho81/XPR1) domain-containing protein (At5g20150), INOSITOL-3-PHOSPHATE SYNTHASE ISOZYME1 (IPS1; At4g39800), a Glc-6-P/Pi transporter (GPT2; At1g61800), a zinc finger (C3HC4-type RING finger) family protein (At2g01150), MBF1c (At3g24500), which makes plants more resistant to stress, but also increases trehalose levels in planta (Suzuki et al., 2005), and some further genes that are annotated as involved in stress responses. Apparently, high C generates a stress response, regardless of whether sugars are added exogenously (Price et al., 2004; Osuna et al., 2007) or synthesized in the tissue. Some C-induced genes induced after 24- to 48-h extension of the night, again indicating their involvement in stress, rather than a specific role in C signaling.
Another set of genes was identified whose transcripts fall rapidly after adding Suc to starved seedlings and are also decreased early in the light period and recover during the night (Table II). Examples include four TPS-like genes, several transcription factors, members of the BTB/POZ domain family, a set of kelch domain F-box factors, ATG8e, a thioredoxin family member, and several glutaredoxins. Several of these genes, including TPS8 to TPS11, APG8e, and two bZIPs, are regulated by an antagonistic interaction between sugars and the clock. This generates a complex response, which is well-predicted by our model. Taken together, this provides strong evidence that they are upstream components in the transcriptional response to small changes of sugars and highlight potentially important cross-talk between the sugar signaling and the clock, which (see below) may serve to sensitize sugar signaling to a shortage of C at dawn (see below).
There is mounting evidence that Tre-6-P acts as a sugar signal in plants (Schluepmann et al., 2003; Kolbe et al., 2005; Lunn et al., 2006). Tre-6-P is synthesized by TPS. TPS is encoded by a small gene family (Leyman et al., 2001; Lunn, 2007). Whereas TPS1 encodes a functional TPS, the precise function of the TPS-like members, TPS5 to TPS11, is still unclear. Our results show that TPS8 to TPS11are induced in response to small changes in C status early in the night. The function of these proteins is unclear. Sequence analysis and the absence of direct evidence for enzymatic activity indicate that they may not catalyze the synthesis of Tre-6-P (Lunn, 2007). Chary et al. (2007) recently showed that TPS6 complements TPS-deficient yeast (Saccharomyces cerevisiae). BTB/POZ domain proteins are thought to interact with cullin3 (Figueroa et al., 2005) or bromodomain proteins (Du and Pooviah, 2004), implicating them in targeted protein degradation or chromatin modeling. They have also been shown to interact with Ca2+ (Du and Pooviah, 2004) and to be involved in the regulation of telomerase activity (Ren et al., 2007). Their strong and early response to changes of C indicates a rather fundamental role in controlling metabolism or growth in response to C status. ATG8e is involved in the regulation of autophagy (Doelling et al., 2002; Downes and Vierstra, 2005). Its early induction during the night mirrors the widespread changes in genes for protein synthesis and indicates that one of the earliest responses as C falls is a coordinated switch from protein synthesis to protein degradation. A potential interaction of C with redox signaling is revealed by the repression of thioredoxin and several glutaredoxins. It has recently been shown that redox signaling affects a wide spectrum of biosynthesis pathways (Kolbe et al., 2006), including starch synthesis (Tiessen et al., 2002; Hendriks et al., 2003; Lunn et al., 2006). For starch synthesis, there is already evidence for close interaction between redox signaling and C signaling mediated by Tre-6-P (Kolbe et al., 2005; Lunn et al., 2006).
Our results, by themselves, do not provide any information about pretranscriptional signaling. Baena-Gonzalez et al. (2007) noted a striking overlap between the transcriptional response to overexpression of the protein kinase AKIN10 and various treatments that lead to C depletion. This included induction of genes that are involved in catabolism, repression of genes that are involved in biosynthesis and cellular growth, and responses of several genes (e.g. TPS8–TPS11, APG8e, bZIP1), which we highlighted as possible upstream transcriptional components in the previous paragraph. Baena-Gonzalez et al. (2007) identified >1,000 genes whose expression is increased or decreased by constitutive overexpression of AKIN10. When we analyzed the responses of their transcripts in our dataset, we found that many started to respond during the normal night or early in the extended night. This provides strong evidence that AKIN10 is involved in the orchestration of acclimatory changes in gene expression in response to small changes in C status. Intriguingly, some of the genes that are downstream of AKIN10 show changes in their transcripts early in the night, whereas others do not respond until after the start of the extended night. This implies that additional factors are modulating the downstream output.
Interaction with Light
The response of gene expression to light is strongly attenuated during the 24-h light/dark cycle. This is seen regardless of whether the test set contains genes that respond to light after illumination of dark-grown seedlings or genes that respond when plants that have grown in a light/dark cycle are illuminated at 50 ppm [CO2] for 4 h at the start of the day. Strikingly, many light-regulated genes do show marked changes in an extended night, some in the first hours, and others after 1 to 2 d (Fig. 10). The contrast between the responses of light- and C-regulated genes is striking. Whereas C-regulated genes respond strongly to changes of C during the diurnal cycle, light signaling is desensitized to the changes of illumination during the diurnal light/dark cycle and only responds strongly when they fall outside the range experienced in the recent past.
Kim and von Arnim (2006) reported changes of transcript levels for approximately 800 genes after transferring seedlings growing in continuous light on 1% Suc to the dark for 1 to 8 h. This contrasts with our study, where plants were grown in a light/dark cycle, and most light-regulated genes did not respond strongly until the dark treatment was extended beyond the end of the normal night. This indicates that light signaling is desensitized to changes of light during the diurnal light/dark cycle. The response of light-regulated genes is reasonably well predicted by our model. Qualitative inspection of the overlap with the clock output pathways and C signaling could contribute to the damped response of light-repressed genes. Further, it is known that, although ELF3 is required for circadian regulation of CAB expression in constant light, it also attenuates gating of the acute response of CAB expression to light (McWaters et al., 2000; Schultz and Kay, 2003). It is possible that ELF3 attenuates a wider set of the light-induced changes in expression during the diurnal cycle. ELF3 interacts with PHYB, indicating a mechanistic model that is consistent with it affecting a large sector of the light-signaling response (Schultz and Kay, 2003).
Interaction with the Clock
There is marked interaction between the clock and C responses (Table III; Figs. 7–9). This was analyzed by investigating the responses of genes that are shared between stringently filtered sets of 604 clock-regulated genes and 867 C-repressed or C-induced genes. Transcripts for the C-repressed genes usually peak in the subjective light period in a free-running cycle. In a diurnal light/dark cycle, the clock is antagonized by rising sugars and, depending on the gene, the increase of the transcript is attenuated or even reversed. Transcripts for the C-induced genes often peak toward the end of the night in a free-running cycle. This rise is attenuated or reversed in a light/dark cycle because sugars are at a minimum at this time. The antagonism between the clock and C signaling is especially clear in pgm, where large changes of sugar levels completely override the clock.
In the first hours of an extended night, the clock and C signaling act in the same direction, increasing transcripts for C-repressed genes and decreasing transcripts for C-induced genes. This may sensitize these genes to changes in the C status during the dark-light transition and early in the day. Examples where it leads to a particularly strong increase of the transcript levels in the first hours of the extended night include TPS8 to TPS11 (see above), three genes involved in protein synthesis or degradation: EF-α-subunit 1, AtNUCL1, and ATG8e and two bZIP transcription factors. AtNUC-L1 plays a key role in nucleolus organization and rRNA synthesis (Pontvianne et al., 2007; Kojima et al., 2007) and EF-α-subunit 1β could contribute to the regulation of translation, whereas APG8e plays a key role in the regulation of autophagy (Doelling et al., 2002; Downes and Vierstra, 2005). Baena-Gonzalez et al. (2007) provided evidence that at least some of the responses to AKIN10 are mediated by bZIP transcription factors. All four bZIP members identified in their study show different responses in the diurnal cycle and perturbations of it, which are predicted by our model. Their varying responses are at least partly due to the differences in the direction and strength of their response to C and its interaction with the clock.
A contrasting type of interaction between the clock and C is seen for PHS1, ISA3, DPE1, PHS2, DPE2, GWD, PWD, and SEX4, which are involved in starch degradation (Zeeman et al., 2007a, 2007b). Their transcripts rise during the subjective light period in a free-running cycle. Most of these genes are induced by light and/or C. In agreement, the circadian response is retained almost unaltered during a light/dark cycle. However, this increase is weakened or abolished early in an extended night when the clock is antagonized by falling C or darkness. This provides a potential mechanism to decrease the rate of starch breakdown when C is in short supply in the light period, although the impact will depend upon the speed and extent to which the encoded proteins change.
Modeling Global Transcriptional Responses
The global response of transcripts during diurnal cycles and an extended night are generated by a complex interplay between the clock, C, light, and many other inputs. The main features of this complex multifactorial response can nevertheless be predicted by a simple linear model. The input data are experimental datasets that provide empirical values for the response of >22K genes to the clock in a free-running cycle and to changes of C or light. The model searches for the best fit to the measured data, allowing the relative importance of the three inputs to vary independently of each other at each time point. This simple linear model allowed good qualitative prediction of the response of >22K genes during the diurnal cycle and extended night in Col-0 and a diurnal cycle in pgm, and also allowed qualitative modeling of the responses of 23 of 24 individual candidates that were chosen because they are subject to interactive regulation by the clock, light, and C. Crucially, the weightings the model selected for C and light varied in a biologically meaningful manner. Light was given a stronger and negative weighting in the dark, especially the extended night, than in the light. For C, the weighting chosen by the model correlated remarkably well with the rosette sugar content. This model provided a good qualitative prediction of the global responses of C and light-regulated genes. It also accurately predicted the responses of the genes that we have highlighted as potential upstream components in the transcriptional response to small changes of sugar (see above), including bZIP transcription factors that interact with AKIN10 and also accurately predicted the responses of >1,000 genes, which Baena-Gonzalez et al. (2007) identified as being regulated by AKIN10. Taken together, this provides good evidence that C, light, and the clock play a prominent role in regulating global gene expression in these complex physiological situations.
The agreement is remarkable because the model has several limitations. First, the input datasets have obvious limitations. For example, the input set for the clock is derived from seedlings growing on 3% Suc (Edwards et al., 2006), whereas our dataset was for 5-week-old plants on soil. The general applicability is nevertheless shown by the similar absolute levels and responses of individual genes, including central clock components and strongly clock-regulated genes in starch degradation (Figs. 7–9). Second, the model assumes a simple linear response of genes to an input (e.g. that all C-responsive genes show a linear response to increasing C across the whole range found in the test treatments, which (Figs. 4 and 5) is probably not the case. Part of this successive response can be explained by interactions with the clock and light. However, the quality of the prediction might be further improved by extending the input dataset for C to include several treatments that generate different levels of sugars at the same time during the 24-h cycle. Third, the model assumes linear interactions between the different inputs. It is likely that complex interactions between the signaling networks sometimes modify their output in a nonadditive manner. Indeed, specific examples were found of genes that were poorly predicted. The model, in combination with gene-wise and treatment-wise quality criteria, provides a starting point to search for further important inputs and, following this, nonlinear interactions. This will be aided by the simple structure of the model and the absence of any threshold filters.
In conclusion, global transcript profiling reveals major changes in C signaling during the night and the first hours of the extended night before acute C depletion develops. As a result, transcripts decrease for genes involved in biosynthesis and cellular growth and increase for genes involved in the remobilization of alternative C sources. Different groups of genes respond at different times during this transition, indicating that progressive changes in C status trigger a sequence of responses. These responses can be modeled by a simple linear model, which captures interactions between C, light, and the clock. Plants possess many different mechanisms to sense changes in sugars and other C sources (Koch, 2004; Gibson, 2005; Rolland et al., 2006; Smith and Stitt, 2007; Stitt et al., 2007). By identifying sets of genes that respond at different speeds and times during C depletion, our extended dataset and model will aid the analysis of specific candidates for C signaling, as illustrated here for AKIN10/bZIP proteins, a set of genes in trehalose signaling, genes controlling protein turnover, and genes involved in starch breakdown.
MATERIALS AND METHODS
Plant Growth
For the extended night experiments, Arabidopsis (Arabidopsis thaliana) ecotype Col-0 was grown on soil in a 14-h light/10-h dark day/night cycle at a light intensity of 130 μmol/m2 s, as in Thimm et al. (2004). Plants were harvested in the vegetative state with three rosettes per sample and typically at least five replicate samples. Rosettes were transferred into liquid N under ambient growth irradiance or darkness. Data for responses during diurnal cycles were obtained from Col-0 and the pgm mutant (in a Col-0 background) grown on soil in the same conditions, except that a 12-h light/12-h dark cycle was used (Bläsing et al., 2005).
Reagents
Chemicals were purchased from Sigma, except NADH (Roche). Enzymes for analysis were purchased from Roche, except invertase (Sigma) and UMP kinase, which was overexpressed in Escherichia coli and purified as in Serina et al. (1995).
Extraction and Assay of Metabolites
Suc, Glc, and Fru were determined in ethanol extracts as described in Geigenberger et al. (1996), starch was determined as in Hendriks et al. (2003), and Glc-6-P as in Gibon et al. (2002). Assays were prepared in 96-well microplates using a Multiprobe II pipetting robot (Perkin-Elmer). The absorbances were red at 340 nm or at 570 nm in a Synergy or an ELX-800-UV microplate reader (Bio-Tek). Mass spectrometry-based metabolite profiling was performed as in Gibon et al. (2006) and Osuna et al. (2007).
RNA Isolation and Expression Analysis with 22K Affymetrix Arrays
Isolation of total RNA, cDNA synthesis, cRNA labeling, and the hybridization on the GeneChip Arabidopsis ATH1 genome array was done exactly as described by Thimm et al. (2004) and as recommended by the manufacturer (part no. 900385; Affymetrix UK Ltd.) packages in R (R Development Core Team, 2006) were used. In brief, RMA expression measures (Bolstad et al., 2003) were calculated on all arrays used and a linear model was fitted to the data using the limma package. Afterward, contrasts of interest were extracted and P values were corrected first across genes and then across contrasts. All R code is available upon request. Microarray data have been deposited with the National Center for Biotechnology Information Gene Expression Omnibus data repository (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE10016 and data can be inspected at http://mapman.mpimp-golm.mpg.de/supplement/xn.
Statistical Methods
All calculations were performed in R (R Development Core Team, 2006). PCA was performed using the prcomp procedure and K-means clustering of genes was performed using the Hartigan and Wong (1979) algorithm on the normalized (logged) RMA expression measures produced as described above. To cluster experiments, complete linkage hierarchical clustering was performed on the RMA-normalized values, using one minus the correlation between two experiments as the distance measure.
Normalization of the Test and Input Datasets
After obtaining RMA expression values, these were converted into response ratios as follows. For all data points from diurnal cycles of wild type and the pgm mutant, the RMA values at the end of the night of the wild type was subtracted. Thus, by definition, the wild-type response at the end of the night is zero for all genes. For the circadian cycle data, for each individual time point, the average of the RMA expression values at the time points 46 and 50 was subtracted because the end of the night would be at the 48-h time point. For the response to sugar, plants grown at ambient or compensation point [CO2] were sampled and the values subtracted from each other. For the response to light, from values of plants that experienced an extended night for 4 h at compensation point [CO2], the RMA measures at plants 4 h in the light at compensation point [CO2] were subtracted (see Bläsing et al., 2005).
To obtain values from an independent sample, seedlings grown in tissue culture were subjected to sugar starvation and were sampled before and during sugar starvation as well as after 0.5 and 3 h of sugar readdition. For all samples, the RMA measures at sugar starvation were subtracted.
As a further independent dataset for the response to light, the AtGenExpress light treatment was taken, where 7-d-old etiolated seedlings were either left further in the dark or illuminated for 4 h with weak white light. Here, the dark RMA expression measures were subtracted from the respective light values.
Modeling
Modeling was performed by fitting a different linear model to each time point in the diurnal sets for the wild-type and the pgm mutant as well as for time points in the extended night. For each individual time point and treatment, we tried to explain the response by a combination of the response of the circadian cycle plus sugar plus light. Thus, the model can be formulated as follows:
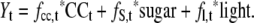 |
Here, t indicates the time within the diurnal (or circadian) cycle. The input datasets are: CC, the response of the 22K transcripts on the ATH1 array at a given time in a free-running circadian cycle, normed on the estimated value at the subjective end of the night (Edwards et al., 2006); sugar, the response of the 22K transcripts after illumination with 350 compared to 50 ppm [CO2] (Bläsing et al., 2005); and light, the response of the 22K genes after illumination for 4 h to 50 ppm [CO2] compared to darkness (Bläsing et al., 2005). fCC,t, fS,t, and fLt are fitting factors for the input datasets CC, sugar, and light, respectively. The fitting factor by definition has the same value for all genes in an input dataset (i.e. it is a common value by which the response ratio for each of the 22K genes in multiplied) at a given time and treatment. The fitting factors vary freely between the different inputs and between different times and treatments. The model was fitted to optimize the fit for all 22K genes simultaneously using least-square fitting and the factors fi,t were extracted. For each time point in Col-0, the signal for each of the 22K genes was normalized on the corresponding signal at the end of the night in the same treatment. For pgm, the signals were normalized on the corresponding signals at the end of the night in Col-0. There were 17 models (five wild type, six pgm, and six for the extended night) and thus 45 factors in total.
The factors describing the influence of sugar at a given time t (f3,t) were regressed against the logarithm of the sugar level, normed to sugar level at the respective end of the night. However, when comparing the importance of different inputs, it should be noted that given the difference variance in the input datasets (ranging from lower variance in circadian cycle time points close to the end of the night through medium variance in the circadian time points close to the middle of the day to high variance in the sugar input), the model might artificially scale small variance by a higher factor.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Additional PCA, scatter plots, and bar chart of changing genes.
Supplemental Figure S2. Hierarchical cluster analysis of the dataset.
Supplemental Figure S3. Scatter plots of PCA loadings versus various experiments and shuffled data.
Supplemental Figure S4. Further ontological analysis of selected categories.
Supplemental Figure S5. Clustering of the AKIN10 dataset from Baena-Gonzalez et al. (2007)
Supplemental Table S1. Absolute values for the Affymetrix array analyses for extended night samples.
Supplemental Table S2. LIMMA analysis of the array data.
Supplemental Table S3. Weightings of genes in the principle components of a PCA.
Supplemental Table S4. Lists, groupings, and responses of genes in the test sets for clock-, light-, and C-responsive genes.
Supplemental Table S5. Dataset to load into PageMan.
Supplemental Table S6. Model errors.
Supplementary Material
Acknowledgments
We thank Dr. Florian Wagner at the RZPD Company (Berlin) for carrying out the microarray hybridizations.
This work was supported by the Max Planck Society, the Bildungsministerium für Bildung und Forschung-funded Genomanalyse im biologischen System Pflanze project Arabidopsis III Gauntlets, Carbon and Nutrient Signaling: Test Systems and Metabolite and Transcript Profiles (grant no. 0312277A), and the AGRON-OMICS project within the sixth framework program of the European Union.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Björn Usadel (usadel@mpimp-golm.mpg.de).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Aubert S, Gout E, Bligny R, Marty-Mazars D, Barrieu F, Alabouvette J, Marty F, Douce R (1996) Ultrastructural and biochemical characterization of autophagy in higher plant cells subjected to C deprivation: control by the supply of mitochondria with respiratory substrates. J Cell Biol 133 1251–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-Gonzalez E, Rolland F, Thevelein JM, Sheen J (2007) A central integrator of transcription networks in plant stress and energy signalling. Nature 448 938–942 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Statist Soc Ser B Methodological 57 289–300 [Google Scholar]
- Bläsing O, Gibon Y, Günther M, Höhne M, Osuna D, Thimm T, Scheible W-R, Morcuende R, Stitt M (2005) Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell 17 3257–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP (2003) A comparison of normalization methods for high density oligonucleotide array data based on bias and variance. Bioinformatics 19 185–193 [DOI] [PubMed] [Google Scholar]
- Brouquisse R, James F, Raymond P, Pradet A (1991) Study of glucose starvation in excised maize root tips. Plant Physiol 96 619–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspar T, Huber SC, Somerville CR (1985) Alterations in growth, photosynthesis and respiration in a starch deficient mutant of Arabidopsis thaliana (L.) Heynh deficient in chloroplast phosphoglucomutase. Plant Physiol 79 11–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspar T, Lin TP, Kakefuda G, Bernhard W, Spilatro S, Preiss J, Somerville CR (1989) Altered regulation of β-amylase activity in mutants of Arabidopsis with lesions in starch metabolism. Proc Natl Acad Sci USA 86 5830–5833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chary SN, Hicks GR, Choi YG, Carter D, Raikhel NV (2007) Trehalose-6-phosphate synthase/phosphatase regulates cell shape and plant architecture in Arabidopsis. Plant Physiol 146 97–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterton NJ, Silvius JE (1979) Photosynthate partitioning into starch in soybean leaves: effects of photoperiod versus photosynthetic period duration. Plant Physiol 64 749–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterton NJ, Silvius JE (1980) Photosynthate partitioning as affected by daily photosynthetic period duration in six species. Physiol Plant 49 141–144 [Google Scholar]
- Chatterton NJ, Silvius JE (1981) Photosynthate partitioning into starch in soybean leaves: irradiance level and daily photsynthetic period duration effects. Plant Physiol 67 257–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia DW, Yoder TJ, Reiter WD, Gibson SI (2000) Fumaric acid: an overlooked form of fixed carbon in Arabidopsis and other plant species. Planta 211 743–751 [DOI] [PubMed] [Google Scholar]
- Contento AL, Kim SJ, Bassham DC (2004) Transcriptome profiling of the response of Arabidopsis suspension culture cells to sucrose starvation. Plant Physiol 135 2330–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross JM, von Korff M, Altmann T, Bartzenko L, Sulpice R, Gibon Y, Palacios N, Stitt M (2006) Natural variation in carbon-nitrogen interactions: changes of metabolite levels and enzyme activities across 24 Arabidopsis thaliana accessions. Plant Physiol 142 1574–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieuaide M, Couée I, Pradet A, Raymond P (1993) Effects of glucose starvation on the oxidation of fatty acids by maize root tip mitochondria and peroxisomes: evidence for mitochondrial fatty acid beta-oxidation and acyl-CoA dehydrogenase activity in a higher plant. Biochem J 296 199–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doelling JH, Walker JM, Friedman EM, Thompson AR, Vierstra RD (2002) The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J Biol Chem 277 33105–33114 [DOI] [PubMed] [Google Scholar]
- Downes B, Vierstra RD (2005) Post-translational regulation in plants employing a diverse set of polypeptide tags. Biochem Soc Trans 33 394–398 [DOI] [PubMed] [Google Scholar]
- Du L, Pooviah PW (2004) A novel family of C22+/calmodulin-binding proteins involved in transcriptional regulation: interaction with fsh/Ring3 class transcription factors. Plant Mol Biol 54 549–569 [DOI] [PubMed] [Google Scholar]
- Edwards KD, Anderson PE, Hall A, Salathia NS, Locke JC, Lynn JR, Straume M, Smith JQ, Millar AJ (2006) FLOWERING LOCUS C mediates natural variation in the high-temperature response of the Arabidopsis circadian clock. Plant Cell 18 639–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa P, Gusmaroli G, Serino G, Habashi J, Ma L, Shen Y, Feng S, Bostick M, Callis J, Hellmann H, et al (2005) Arabidopsis has two redundant Cullin3 proteins that are essential for embryo development and that interact with RBX1 and BTB proteins to form multisubunit E3 ubiquitin ligase complexes in vivo. Plant Cell 17 1180–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fondy BR, Geiger DR (1985) Diurnal changes of allocation of newly fixed carbon in exporting sugar beet leaves. Plant Physiol 78 753–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MJ, Hubbard KE, Hotta CT, Dodd AN, Webb AAR (2006) Circadian clocks. How plants tell the time. Biochem J 397 15–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geigenberger P, Lerchl J, Stitt M, Sonnewald U (1996) Phloem-specific expression of pyrophosphatase inhibits long distance transport of carbohydrates and amino acids in tobacco plants. Plant Cell Environ 19 43–55 [Google Scholar]
- Geiger DR, Servaites JC (1994) Diurnal regulation of photosynthetic carbon metabolism in C3 plants. Annu Rev Plant Biol 45 235–256 [Google Scholar]
- Geiger DR, Servaites JC, Fuchs MA (2000) Role of starch in carbon translocation and partitioning at the plant level. Aust J Plant Physiol 27 571–582 [Google Scholar]
- Gibon Y, Blaesing OE, Hannemann J, Carillo P, Hohne M, Hendriks JH, Palacios N, Cross J, Selbig J, Stitt M (2004. a) A robot-based platform to measure multiple enzyme activities in Arabidopsis using a set of cycling assays, comparison of changes of enzyme activities and transcript levels during diurnal cycles and in prolonged darkness. Plant Cell 16 3304–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon Y, Blaesing OE, Palacios-Rojas N, Pankovic D, Hendriks JH, Fisahn J, Hohne M, Gunther M, Stitt M (2004. b) Adjustment of diurnal starch turnover to short days, depletion of sugar during the night leads to a temporary inhibition of carbohydrate utilization, accumulation of sugars and post-translational activation of ADP-glucose pyrophosphorylase in the following light period. Plant J 39 847–862 [DOI] [PubMed] [Google Scholar]
- Gibon Y, Usadel B, Blaesing OE, Kamlage K, Höhne M, Trethewey R, Stitt M (2006) Integration of metabolite with transcript and enzyme activity profiling during diurnal cycles in Arabidopsis rosettes. Genome Biol 7 R76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon Y, Vigeolas H, Tiessen A, Geigenberger P, Stitt M (2002) Sensitive and high throughput metabolic assays fro inorganic pyrophosphate, ADPglc, nucleotide phosphates, and glycolytic intermediates based on a novel enzymic cycling system. Plant J 30 221–235 [DOI] [PubMed] [Google Scholar]
- Gibson SI (2005) Control of plant development and gene expression by sugar signalling. Curr Opin Plant Biol 8 93–102 [DOI] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch JB, Straume M, Chang HS, Han B, Zhu T, Wang X, Kreps JA, Kay SA (2000) Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290 2110–2113 [DOI] [PubMed] [Google Scholar]
- Hartigan JA, Wong MA (1979) A K-means clustering algorithm. Appl Stat 28 100–108 [Google Scholar]
- Hendriks JHM, Kolbe A, Gibon Y, Stitt M, Geigenberger P (2003) ADP-glucose pyrophosphorylase is activated by posttranslational redox modification in response to light and to sugars in leaves of Arabidopsis and other plant species. Plant Physiol 133 838–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber SC, Hanson KR (1992) Carbon partitioning and growth of a starchless mutant of Nicotiana sylvestris. Plant Physiol 99 1449–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B-H, von Arnim AG (2006) The early dark-response in Arabidopsis thaliana revealed by cDNA microarray analysis. Plant Mol Biol 60 321–342 [DOI] [PubMed] [Google Scholar]
- Koch KE (1996) Carbohydrate-modulated gene expression in plants. Annu Rev Plant Biol 47 509–540 [DOI] [PubMed] [Google Scholar]
- Koch KE (2004) Sucrose metabolism, regulatory mechanisms and pivotal roles in sugar sensing and plant development. Curr Opin Plant Biol 7 235–246 [DOI] [PubMed] [Google Scholar]
- Kojima H, Suzuki T, Kato T, Enomoto KI, Sato S, Tabate S, Saez-Vasqzez J, Echeverria M, Nakagawa T, Ishiguro S, et al (2007) Sugar-inducible expression of the nucleolin-1 gene of Arabidopsis thaliana and its role in ribosome synthesis, growth and development. Plant J 49 1053–1063 [DOI] [PubMed] [Google Scholar]
- Kolbe A, Oliver S, Fernie AR, Stitt M, van Doongen JT, Geigenberger P (2006) Combined transcript and metabolite profiling of Arabidopsis leaves reveals fundamental effects of the thiol-disulfide status on plant metabolism. Plant Physiol 141 412–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolbe A, Tiessen A, Schluepmann H, Paul M, Ulrich S, Geigenberger P (2005) Trehalose 6-phosphate regulates starch synthesis via posttranslational redox activation of ADP-glucose pyrophosphorylase. Proc Natl Acad Sci USA 102 11118–11123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam HM, Coshigano K, Oliveira I, Melo-Oliveira R, Coruzzi G (1996) The molecular genetics of N assimilation into amino acids in higher plants. Annu Rev Plant Physiol Plant Mol Biol 47 569–593 [DOI] [PubMed] [Google Scholar]
- Lam HM, Hsieh MH, Coruzzi G (1998) Reciprocal regulation of distinct asparagine synthetase genes by light and metabolites in Arabidopsis thaliana. Plant J 16 345–353 [DOI] [PubMed] [Google Scholar]
- Leyman B, Van Dijck P, Thevelein JM (2001) An unexpected plethora of trehalose biosynthesis genes in Arabidopsis thaliana. Trends Plant Sci 6 510–513 [DOI] [PubMed] [Google Scholar]
- Li Y, Lee KK, Walsh S, Smith C, Hadingham S, Sorefan K, Cawley G, Bevan MW (2006) Establishing glucose- and ABA-regulated transcription networks in Arabidopsis by microarray analysis and promotor classification using a relevance vector machine. Genome Res 16 414–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin TP, Caspar T, Somerville CR, Preiss J (1988) A starch deficient mutant of Arabidopsis thaliana with low ADP-glucose pyrophosphorylase activity lacks one of the 2 subunits of the enzyme. Plant Physiol 88 1175–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Gehan JP, Sharkey TD (2005) Daylength and circadian effects on starch degradation and maltose metabolism. Plant Physiol 138 2280–2291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn J, Feil R, Hendriks JH, Gibon Y, Mocuende R, Scheible WR, Osuna D, Carillo P, Hajirezaei M, Stitt M (2006) Sugars lead to large changes of trehalose-6-phosphate in Arabidopsis, which are correlated with changes in the post-translational activation of AGPase and the rate of starch synthesis. Biochem J 397 139–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn JE (2007) Gene families and evolution of trehalose metabolism in plants. Funct Plant Biol 34 550–563 [DOI] [PubMed] [Google Scholar]
- Matt P, Schurr U, Krapp A, Stitt M (1998) Growth of tobacco in short day conditions leads to high starch, low sugars, altered diurnal changes of the Nia transcript and low nitrate reductase activity, and an inhibition of amino acid synthesis. Planta 207 27–41 [DOI] [PubMed] [Google Scholar]
- McWaters HG, Bastow R, Hall A, Millar AR (2000) The ELF3 Zeitnehmer regulates light signalling to the circadian clock. Nature 408 716–720 [DOI] [PubMed] [Google Scholar]
- Nielsen HB, Mundy J, Willenbrock H (2007) Functional associations by response overlap (FARO), a functional genomics approach matching gene expression phenotypes. PLoS ONE 2 e676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osuna D, Usadel B, Morcuende R, Gibon Y, Blasing OE, Hohne M, Gunter M, Kamlage B, Trethewey R, Scheible W-R, et al (2007) Temporal responses of transcripts, enzyme activities and metabolites after adding sucrose to carbon-deprived Arabidopsis seedlings. Plant J 49 463–491 [DOI] [PubMed] [Google Scholar]
- Paul MJ, Foyer CH (2001) Sink regulation of photosynthesis. J Exp Bot 52 1383–1400 [DOI] [PubMed] [Google Scholar]
- Price J, Laxmi A, Martin SK, Jang JC (2004) Global transcription profiling reveals multiple sugar signal transduction mechanisms in Arabidopsis. Plant Cell 16 2128–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontvianne F, Matia I, Douet J, Toumente S, Medina FJ, Echeverria M, Sáez-Vásques J (2007) Characterization of AtNUC-L1 reveals a central role of nucleolin in nucleolar organization and silencing of AtNUC-L2 gene in Arabidopsis. Mol Biol Cell 18 369–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2006) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria
- Ren S, Mandradi KK, Boedeker AL, Rathore KS, McKnight TD (2007) Regulation of telomerase in Arabidopsis by BT2, an apparent target of TELOMERASE ACTIVATOR1. Plant Cell 19 23–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland F, Baena-Gonzalez E, Sheen J (2006) Sugar sensing and signalling in plants, conserved and novel mechanisms. Annu Rev Plant Biol 57 676–709 [DOI] [PubMed] [Google Scholar]
- Salome PA, McClung CR (2005) What makes the Arabidopsis clock tick on time. A review on entrainment. Plant Cell Environ 28 21–38 [Google Scholar]
- Schluepmann H, Pellny T, van Dijken A, Smeekens S, Paul M (2003) Trehalose 6-phosphate is indispensable for carbohydrate utilization and growth in Arabidopsis thaliana. Proc Natl Acad Sci USA 100 6849–6854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz TF, Kay SE (2003) Circadian clocks in daily and seasonal control of development. Science 301 326–328 [DOI] [PubMed] [Google Scholar]
- Schulze W, Stitt M, Schulze ED, Neuhaus HE, Fichtner K (1991) A quantification of the significance of assimilatory starch for growth of Arabidopsis thaliana L. Heynh. Plant Physiol 95 890–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serina L, Blondin C, Krin E, Sismeiro O, Danchin A, Sakamoto H, Gilles AM, Barzu O (1995) Escherichia coli UMP-kinase, a member of the aspartokinase family, is a hexamer regulated by guanine-nucleotides and UTP. Biochemistry 34 5066–5074 [DOI] [PubMed] [Google Scholar]
- Smith AM, Denyer K, Martin C (1997) The synthesis of the starch granule. Annu Rev Plant Physiol Plant Mol Biol 48 67–87 [DOI] [PubMed] [Google Scholar]
- Smith AM, Zeeman SC, Smith SM (2005) Starch degradation. Annu Rev Plant Biol 56 73–98 [DOI] [PubMed] [Google Scholar]
- Smith SM, Fulton DC, Chia T, Thorneycroft D, Chapple A, Dunstan H, Hylton C, Zeeman SC, Smith AM (2004) Diurnal changes in the transcriptome encoding enzymes of starch metabolism provide evidence for both transcriptional and post-transcriptional regulation of starch metabolism in Arabidopsis leaves. Plant Physiol 136 2687–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Stitt M (2007) Coordination of carbon supply and plant growth. Plant Cell Environ 30 1126–1149 [DOI] [PubMed] [Google Scholar]
- Smyth GK (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3 Article 3 [DOI] [PubMed]
- Stitt M (1991) Rising CO2 levels and their potential significance for carbon flow in photosynthetic cells. Plant Cell Environ 14 741–762 [Google Scholar]
- Stitt M, Bulpin PV, ap Rees T (1978) Pathway of starch breakdown in photosynthetic tissue of Pisum sativum. Biochim Biophys Acta 544 200–214 [DOI] [PubMed] [Google Scholar]
- Stitt M, Gibon Y, Lunn J, Piques M (2007) Multilevel genomics analysis of carbon signaling during low carbon availability: coordination of the supply and of utilization carbon in a fluctuating environment. Funct Plant Biol 34 526–549 [DOI] [PubMed] [Google Scholar]
- Stitt M, Krapp A (1999) The molecular physiological basis for the interaction between elevated carbon dioxide and nutrients. Plant Cell Environ 22 583–622 [Google Scholar]
- Suzuki N, Rizhsky L, Liang H, Shuman J, Shulaev V, Mittler R (2005) Enhanced tolerance to environmental stress in transgenic plants expressing the transcriptional coactivator multiprotein bridging factor 1c. Plant Physiol 139 1313–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Blaesing OE, Gibon Y, Nagel A, Meyer S, Krueger P, Selbig J, Mueller LA, Rhee SY, Stitt M (2004) Mapman: a user-driven tool to display genomics datasets onto diagrams of metabolic pathways and other biological processes. Plant J 37 914–939 [DOI] [PubMed] [Google Scholar]
- Thum KE, Shin MJ, Palenchar PM, Kouranov A, Coruzzi GM (2004) Genome wide investigation of light- and C-signaling interactions in Arabidopsis. Genome Biol 5 R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiessen A, Hendriks JHM, Stitt M, Branscheid A, Gibon Y, Farre EM, Geigenberger P (2002) Starch synthesis in potato tubers is regulated by post-translational redox modification of ADP-glucose pyrophosphorylase: a novel regulatory mechanism linking starch synthesis to the sucrose supply. Plant Cell 14 2191–2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Steinhauser D, Gibon Y, Blasing OE, Redestig H, Sreenivasulu N, Krall L, Hannah MA, Poree F, et al (2006) PageMan: an interactive ontology tool to generate, display, and annotate overview graphs for profiling experiments. BMC Bioinformatics 7 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Thimm O, Redestig H, Blaesing OE, Palacios-Rojas N, Selbig J, Hannemann J, Piques MC, Steinhauser D, et al (2005) Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes and comparison with known responses. Plant Physiol 138 1195–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichert S, Fokianos K, Strimmer K (2004) Identifying periodically expressed transcripts in microarray time series data. Bioinformatics 20 5–20 [DOI] [PubMed] [Google Scholar]
- Yu S (1999) Cellular and genetic responses of plants to sugar starvation. Plant Physiol 121 687–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeman SC, ap Rees T (1999) Changes in carbohydrate metabolism and assimilate export in starch-excess mutants of Arabidopsis. Plant Cell Environ 22 1445–1453 [Google Scholar]
- Zeeman SC, Northrop F, Smith AM, ap Rees T (1998) A starch-accumulating mutant of Arabidopsis thaliana deficient in a chloroplastic starch-hydrolysing enzyme. Plant J 15 357–365 [DOI] [PubMed] [Google Scholar]
- Zeeman SC, Delatte T, Messerli G, Umhang M, Stettler M, Mettler T, Streb S, Reinhold H, Kötting O (2007. a) Starch breakdown: recent discoveries suggest distinct pathways and novel mechanisms. Funct Plant Biol 34 465–473 [DOI] [PubMed] [Google Scholar]
- Zeeman SC, Smith SM, Smith AM (2007. b) The diurnal metabolism of leaf starch. Biochem J 401 13–28 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.