Abstract
A 9-kD proteinase inhibitor was isolated from the seeds of ginkgo (Ginkgo biloba) and purified to homogeneity. This protein was revealed to partial-noncompetitively inhibit the aspartic acid proteinase pepsin and the cysteine proteinase papain (inhibition constant = 10−5–10−4 m). The cDNA of the inhibitor was revealed to contain a 357-bp open reading frame encoding a 119-amino acid protein with a potential signal peptide (27 residues), indicating that this protein is synthesized as a preprotein and secreted outside the cells. Semiquantitative reverse transcription-polymerase chain reaction revealed that this gene expresses only in seeds, not in stems, leaves, and roots, suggesting that the protein is involved in seed development and/or germination. The inhibitor showed about 40% sequence homology with type-I nonspecific lipid transfer protein (nsLTP1) from other plant species. Actually, this inhibitor exerted both lipid transfer activity and lipid-binding activity, while the protein did not show any antifungal and antibacterial activities. Furthermore, the site-directed mutagenesis study using a recombinant ginkgo nsLTP1 revealed that proline (Pro)-79 and phenylalanine-80 are important on phospholipid transfer activity and that Pro-79 and isoleucine-82 are essential for the binding activity toward cis-unsaturated fatty acids. On the other hand, the α-helical content of P79A and F80A mutants was significantly lower than that of the wild-type protein. It was noteworthy that the papain-inhibitory activity of P79A and F80A mutants was elevated twice as much as that of the wild-type protein. In summary, we concluded that Pro-79 plays a critical role in both the lipid transfer and binding activities of ginkgo nsLTP1.
Plants are exposed to a variety of potential threatening phytopathogens and pests. As a result, they have evolved various mechanisms for self-defense such as the production of secondary metabolites and defense-related proteins. In plant life cycle, seed germination is an especially vulnerable period for phytopathogen attack, because the rupture of a seed coat allows pathogens to invade the seed-storage tissues. Many phytopathogenic fungi are known to produce extracellular proteinases (Kalashnikova et al., 2003), and recent findings suggest that proteinases play an active role in the development of diseases (Sara and Heale, 1990). In a response to proteinase attacks by phytopathogens, plants synthesize inhibitory proteins that can suppress the enzyme activities (Ryan, 1990).
Ginkgo (Ginkgo biloba) is one of the oldest gymnosperm species and is called a living fossil. This plant shows a broad spectrum of resistance or tolerance to many phytopathogens; therefore, a large number of studies on this unique plant have been carried out. So far, a chitin-binding antimicrobial peptide has been isolated, and a jasmonate-dependent defensin gene has been cloned from gingko leaves (Huang et al., 2000; Shen et al., 2005). In searching for proteinase inhibitors from various sources, the extracts from ginkgo seeds were found to contain some inhibitory activities against the Asp proteinase pepsin and the Cys proteinase papain. In a previous article, we purified a novel proteinase inhibitor toward pepsin and revealed that the inhibitor is an antifungal protein highly homologous with embryo-abundant proteins from the gymnosperm Picea abies and Picea glauca (Sawano et al., 2007).
In this article, we report the purification of the other Cys proteinase inhibitor, characterization of the inhibitory activity, and cloning of its full-length cDNA sequence. As a result of gene cloning, this inhibitor was revealed to possess high homology with type-I nonspecific lipid transfer protein (nsLTP1), especially two consensus pentapeptides that have been proposed to be important for lipid catalysis or binding (Douliez et al., 2000). Such proteins so far have been isolated from a lot of plants (e.g. Jégou et al., 2000; Douliez et al., 2001; Lin et al., 2007) and are proposed to be involved in defense, pollination, and germination, although the biological function remains to be elucidated (Kader, 1996). Here, to provide new insights into the physiological function of nsLTP1 from ginkgo, we examined the lipid transfer/binding, antifungal/antibacterial properties, and the gene expression pattern in different tissues. Furthermore, we expressed the recombinant proteins mutated in the above-mentioned pentapeptides using Escherichia coli and investigated their lipid transfer and lipid binding, proteinase inhibitory activities, and circular dichroism (CD) spectra to shed light on the structure-activity relationship of this LTP-like proteinase inhibitor. Finally, we discuss the putative physiological significance of ginkgo nsLTP1 and the essential residue(s) for the lipid transfer and lipid-binding activities.
RESULTS
Purification of a Novel Proteinase Inhibitor from Ginkgo Seeds
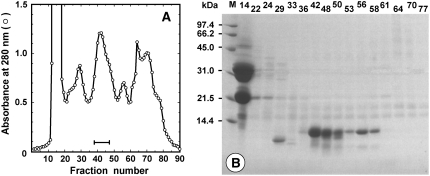
As shown in Figure 1A, the extracts from 775 g of shelled ginkgo seeds were fractionated on a Sephadex G-50 column to obtain a fraction with papain-inhibitory activity. The inhibitory fraction (0.52 g) was finally purified by ion-exchange chromatography (data not shown), and it appeared as a protein band with the molecular mass of approximately 10 kD on SDS-PAGE (Fig. 1B). The sample was estimated to be more than 95% pure and was identified with a molecular mass of 9,318.8 D by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS), and the final yield from the starting material was about 83 mg. The NH2-terminal 28 amino acids were revealed to be APGCDTVDTDLAPCISYLQTGTGNPTVQ by direct protein sequencing.
Figure 1.
Fractionation and purification of the inhibitory activity from ginkgo seeds. A, Second gel-filtration chromatography of the crude extracts (4.5 g) on a Sephadex G-50 column. The fraction size and flow rate were 5 mL and 0.33 mL min−1, respectively. The fractions with a bar were pooled. B, Gly SDS-PAGE of fractionation of the second gel-filtration chromatography. Molecular mass markers are shown in lane M.
Partial Amino Acid Sequencing of the Proteinase Inhibitor
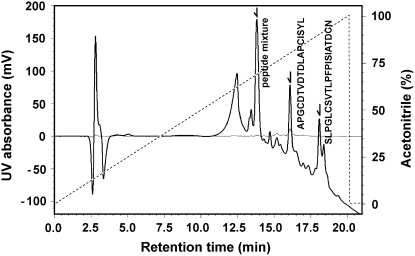
The oxidized inhibitor was digested by trypsin, and the resulting peptides were separated by HPLC. As shown in Figure 2, the elution profile of HPLC showed four major peaks at retention times of 12.5, 14.1, 16.6, and 18.8 min. To characterize all the peaks, we performed both the MALDI-TOF MS and NH2-terminal peptide sequencing. As a consequence, the peaks at retention times of 16.6 and 18.8 min were revealed as the peptides of APGCDTVDTDLAPCISYL and SLPGLCSVTLPFPISIATDCN, respectively (Fig. 2). The other peaks at retention times of 12.5 and 14.1 min exhibited a column artifact and a mixture of di- or tripeptides digested, respectively (data not shown).
Figure 2.
HPLC chromatogram of the peptides obtained by trypsin digestion. Absorbance was monitored at 215 (solid line) and 280 nm (fine line), and elution was performed with a linear gradient (dotted line) of acetonitrile. The result of sequence analysis of the fragment is shown close to the corresponding peak with an arrow.
Molecular Cloning of the Gene Encoding the Proteinase Inhibitor
We first designed degenerate primers by using the partial amino acid sequences of the inhibitor and cloned an initial 267-bp cDNA by reverse transcription (RT)-PCR with the primers. Based on the DNA sequence obtained, we synthesized the gene-specific primers of GBs1 and GBs2 for 3′ RACE and GBa3, GBa4, GBa5, GBa6, GBa7, and GBa8 for 5′ RACE. Using the RACE method, we obtained the full-length cDNA sequence of the inhibitor gene, which was 700 bp long and contained a 357-bp open reading frame encoding a 119-amino acid protein (GenBank accession no. DQ836633). An untranslated 5′ region was 46 bp upstream from the start codon, and the coding region was followed by an untranslated 3′ region that was 294 bp long downstream from the stop codon. Two possible polyadenylation signals of AATAAA were found at the 62- and 194-bp positions downstream from the stop codon. It is probable that more than one polyadenylation signal site provides a high degree of flexibility.
The deduced amino acid sequence consisted of 119 residues, including eight Cys residues (positions 4, 14, 29, 30, 50, 52, 74, and 88). A search for the full-length precursor protein in carefully prepared extracts of the fresh seeds was unsuccessful; therefore, the NH2-terminal 27 residues of MMKISWQLWLLVAFAVMVCVWTPLSTA were considered to be a propeptide. Actually, the programs SignalP-3.0 (Bendtsen et al., 2004) and PSORT (Nakai and Kanehisa, 1992) predicted that the propeptide forms a signal peptide and that the mature protein is extracellularly secreted. The Genetyx-Win program indicated that the pI and molecular mass of the mature protein are 7.45 and 9.44 kD, respectively.
A search of the National Center for Biotechnology Information (NCBI) database, using BlastP2.2.13, showed that the deduced inhibitory protein has high sequence homology with a lot of plant nsLTP. Figure 3 shows the sequence alignment of the inhibitor with gymnosperm and angiosperm nsLTP1. The plant nsLTP family can be divided into two subfamilies according to their molecular masses, namely, type-I nsLTP (nsLTP1, molecular mass about 9 kD) and type II (nsLTP2, about 7 kD; Kalla et al., 1994). According to this criterion, the inhibitory protein was named G. biloba nsLTP1 (Gb-nsLTP1). Plant nsLTP1 generally facilitate the movement of various polar lipids between membranes in vitro (Kader, 1996). However, they show an unusual lack of specificity and are proposed to be involved in a variety of biological processes such as chitin formation, embryogenesis, defense reactions against phytopathogens, symbiosis, and plant adaptation to various environmental conditions (Kader, 1996).
Figure 3.
Sequence alignment of mature Gb-nsLTP1 from ginkgo seeds with gymnosperm and angiosperm mature nsLTP1. The numbering system corresponds to that of Gb-nsLTP1 (GenBank accession no. DQ836633). Dots and asterisks below the sequences are indicated as residues conserved well and involved in ligand binding, respectively. The boxed sequences indicate two consensus pentapeptides (T/S-X-X-D-R/K and P-Y-X-I-S) that have been proposed to be important for lipid catalysis or binding (Douliez et al., 2000). Completely conserved Cys are highlighted by dark background. Sequence similarities (within parentheses) were analyzed by BLAST. The following plant nsLTP1 sequences were used for comparisons: loblolly pine (Pinus taeda), Q41073; Monterey pine (Pinus radiata), AAB80805; red pine (Pinus resinosa), AAK00625; peach (Prunus persica), P81402; tomato (Lycopersicon esculentum), CAA39512; tobacco-1, BAA03044; tobacco-2, AAA21437; tobacco-3, AAA21438; maize (Zea mays), ABA33852; rice (Oryza sativa), ABA96284; and barley, AAA32970.
Comparison of Gb-nsLTP1 with Other Homologous Proteins
The deduced sequence of Gb-nsLTP1 was 36% to 46% identical to other plant nsLTP1 sequences (Fig. 3). Gb-nsLTP1 also has two consensus pentapeptides (T/S-X-X-D-R/K and P-Y-X-I-S) that have been proposed to be important for lipid catalysis or binding (Douliez et al., 2000). The locations of four disulfide bonds in Gb-nsLTP1 could not be determined due to the complexity. However, the disulfide-bond pattern of Gb-nsLTP1 would be the same as that (Cys-4–Cys-50, Cys-14–Cys-29, Cys-30–Cys-74, and Cys-52–Cys-88) of other plant nsLTP1 (e.g. Lin et al., 2005), because the positions of the eight Cys residues were completely conserved between Gb-nsLTP1 and other plant nsLTP1 (Fig. 3). In the near future, the structural analysis of Gb-nsLTP1 by NMR or x-ray crystallography will reveal all the disulfide-bond positions.
Molecular Evolution Analysis
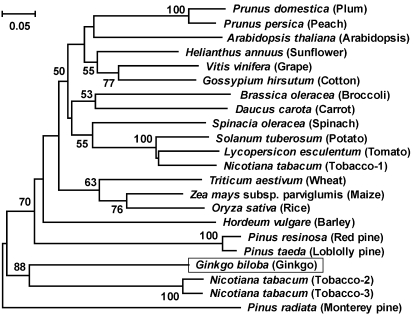
To better understand the evolutionary relationships among plant nsLTP1, we constructed an unrooted phylogenetic tree of aligned amino acid sequences by using the neighbor-joining method. As shown in Figure 4, Gb-nsLTP1 shared a common evolutionary origin and had a closer relationship with the gymnosperm species. The monocotyledonous and dicotyledonous species also formed a cluster, respectively. Tobacco (Nicotiana tabacum) plant possessed two types of nsLTP1; for instance, tobacco-2 nsLTP1 sequence was 35% and 91% identical to tobacco-1 and tobacco-3 sequences, respectively (Fig. 3). It is noteworthy that tobacco-2 and tobacco-3 nsLTP1 were grouped into a cluster with Gb-nsLTP1, indicating that ginkgo has a closer relationship with tobacco-2 and tobacco-3 than the gymnosperm species (Fig. 4). It is likely that tobacco-2, tobacco-3, and ginkgo nsLTP1 in vivo possess a unique function different from tobacco-1 and the other angiosperms.
Figure 4.
Phylogenetic tree analysis of Gb-nsLTP1 with other plant nsLTP1. This tree was constructed by the neighbor-joining method with p-distance. The number for each interior branch shows the percentage of the bootstrap value (1,000 replicates), and only the values greater than 50% are shown. The scale bar indicates the estimated number of amino acid substitutions per site. The other nsLTP1 sequences except for Figure 3 are plum (Prunus domestica), P82534; Arabidopsis, AAF76930; sunflower (Helianthus annuus), CAA63340; grape (Vitis vinifera), AAO33394; cotton (Gossypium hirsutum), AAF35186; broccoli (Brassica oleracea), AAA32995; carrot (Daucus carota), AAB96834; spinach (Spinacia oleracea), AAA34032; potato (Solanum tuberosum), AAM82607; and wheat (Triticum aestivum), ABF14725.
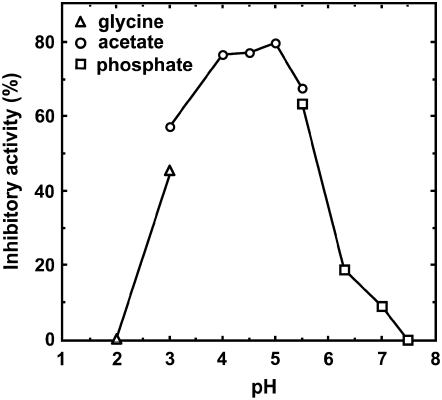
Kinetic Properties and pH Dependence of the Proteinase Inhibitory Activities
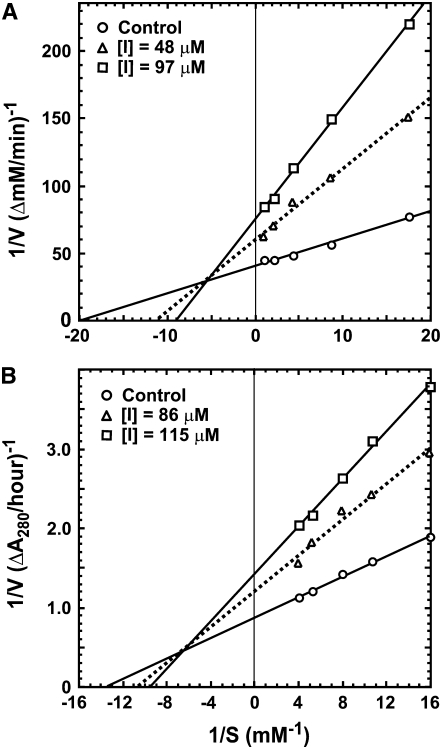
We examined whether Gb-nsLTP1 inhibits Asp-, Ser-, Cys-, or metallo-proteinases. As a consequence, this inhibitor was found to inhibit papain (74%), pepsin (43%), and the Ser proteinase trypsin (25%) under the conditions described in “Materials and Methods.” On the other hand, Gb-nsLTP1 showed no inhibitory activity against the metallo-proteinase thermolysin. The Lineweaver-Burk plots toward papain and pepsin are shown in Figure 5, A and B, respectively. These plots indicate that Gb-nsLTP1 partial-noncompetitively inhibited the activities of papain and pepsin, consistent with these Dixon plots, whose intersection points also did not exist on the y axis (data not shown). The obtained inhibition constant values against papain and pepsin were 2.40 × 10−5 and 8.50 × 10−5 M, respectively. The optimal-inhibitory pH toward papain was observed around 5 (Fig. 6). We also examined the substrate inhibition effect toward papain and trypsin by using a 60 mg mL−1 bovine serum albumin solution. As a result, the substrate inhibitions were observed to be 4% for papain and 1% for trypsin, indicating that the effect was negligible for the trypsin and papain assays.
Figure 5.
Lineweaver-Burk plots for hemoglobin or CLN hydrolysis catalyzed by papain (A) and pepsin (B) in the presence or absence of Gb-nsLTP1.
Figure 6.
pH dependence of the inhibitory activity of Gb-nsLTP1 toward papain. The symbols used are: triangles (pH 2.0–3.0), measured under Gly-HCl buffer; circles (pH 3.0–5.5), citrate buffer; squares (pH 5.5–7.5), sodium phosphate buffer.
Assays of the Antifungal, Antibacterial, Lipid Transfer, and Lipid-Binding Activities
Gb-nsLTP1 did not exhibit antifungal activity against Fusarium oxysporum, Fusarium culmorum, Trichoderma reesei, Aspargillus fumigatus, or Mucor spinescens (data not shown). Similarly, this protein did not show antibacterial activity against E. coli (data not shown). In general, the positively-charged face of plant defensive proteins is believed to bind to the negatively-charged membrane surface of microorganisms; accordingly, Gb-nsLTP1 with the neutral pI may be unable to bind to the membrane fully.
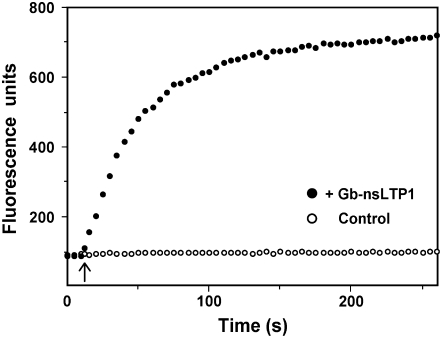
The lipid transfer activity of Gb-nsLTP1 was investigated with fluorescence spectroscopy by monitoring the increase of the fluorescence due to transferring pyrene moieties from quenched donor vesicles to unquenched acceptor vesicles. The fluorescence intensity increased as the protein catalyzed the shuffling of lipid molecules between the vesicles. The fluorescence intensity reached a plateau when the fluorescent lipid molecules were equally distributed between the vesicles (Fig. 7). The lipid transfer rate of Gb-nsLTP1 was revealed to be 2.44 nmol pyrene phospholipid min−1 mg−1.
Figure 7.
Lipid transfer assay between donor and acceptor liposomes. Five microliters of a sample solution (53 mg mL−1) was added at the time point of 10 s into a solution containing two vesicle populations (shown with an arrow). Black circles show the mean values from two independent experiments; white circles indicate the control experiment.
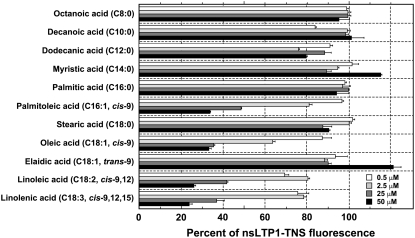
The lipid-binding assay revealed that Gb-nsLTP1 could not associate with most saturated fatty acids (C8:0–C18:0) and an unsaturated fatty acid with a trans-bond (C18:1, trans-9), because the observed fluorescence was 76% to 121% under several amounts of each fatty acid (Fig. 8). In contrast, 50 μm cis-unsaturated fatty acids, palmitoleic acid (C16:1, cis-9), oleic acid (C18:1, cis-9), linoleic acid (C18:2, cis-9,12), and linolenic acid (C18:3, cis-9,12,15) more efficiently displaced 2-p-toluidinonaphtalene-6-sulfonate (TNS), a fluorescent probe (24%–34% of the control fluorescence; Fig. 8). These results suggest that Gb-nsLTP1 has high lipid-binding activity toward cis-unsaturated fatty acids, and the substrate specificity was similar to that of tobacco nsLTP1 (Buhot et al., 2004).
Figure 8.
Effect of fatty acids on the fluorescence level of the Gb-nsLTP1-TNS complex. The amounts of each fatty acid are 0.5 (white), 2.5 (light gray), 25 (dark gray), and 50 μm (black bars). Results are expressed as the percentage of fluorescence of the complex (control fluorescence level, 100% ± 3.4%). Experiments were performed in triplicate and results are expressed as the mean values ± sds.
Expression Profile of Gb-nsLTP1
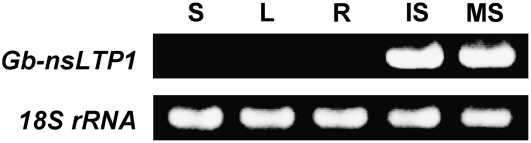
Semiquantitative RT-PCR was carried out to investigate the expression pattern of Gb-nsLTP1 in different tissues. The result showed that there was high Gb-nsLTP1 expression in the immature and mature seeds, while no expression could be detected in the stems, leaves, and roots (Fig. 9), indicating that Gb-nsLTP1 is a seed-specific expressing gene.
Figure 9.
Expression pattern of Gb-nsLTP1 in different ginkgo tissues by semiquantitative RT-PCR analysis. Each RNA sample was isolated from the stem (S), leaf (L), root (R), immature seed (IS), and mature seed (MS) and subjected to the RT-PCR analysis (top). The 18S rRNA gene was used as a control in the RT-PCR amplification (bottom).
Expression and Purification of the Recombinant Proteins
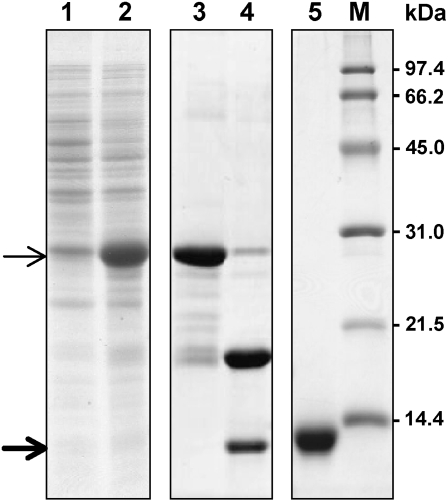
The recombinant Gb-nsLTP1 was expressed as a thioredoxin (Trx) fusion protein (Trx-Gb-nsLTP1; Mr = 25,400) in the soluble fraction of the cell lysate (Fig. 10, lane 2), and the Trx-Gb-nsLTP1 protein was purified by Ni-NTA chromatography (Fig. 10, lane 3). The fusion protein was cleaved by an HRV3C protease, resulting in the production of the recombinant protein (Mr = 9,600; Fig. 10, lane 4). Finally, the protein was purified by Mono-S chromatography to give a single band on SDS-PAGE (Fig. 10, lane 5), and the purity of the sample was estimated to be more than 95% by MALDI-TOF MS (data not shown). Almost all yields of the recombinant proteins were approximately 5 mg L−1 of bacterial culture, while the yield of the mutant P79A was less than 1 mg L−1. MS analysis gave an average molecular mass of 9,586.9 D for the wild-type recombinant (Table I) and 9,318.8 D for native Gb-nsLTP1. The difference indicates that the recombinant protein contained an additional peptide Gly-Pro derived from the cloning vector. The MS analysis of the mutant proteins revealed that they were consistent with the amino acid substitutions and properly formed four disulfide bonds (Table I).
Figure 10.
Expression and purification of the recombinant protein. In the expression of Trx-Gb-nsLTP1 fusion protein, lanes 1 and 2 indicate a soluble fraction of the cell lysate before and after induction with 0.5 mm isopropylthio-β-galactoside, respectively. Lane 3 shows a fraction with Trx-Gb-nsLTP1 fusion protein after Ni-NTA column chromatography; lane 4 is indicative of a fraction with HRV3C protease treatment. Lane 5 indicates the recombinant protein purified by Mono-S chromatography. Molecular mass markers are shown in lane M. The positions of Trx-Gb-nsLTP1 and Gb-nsLTP1 are marked by thin and thick arrows, respectively.
Table I.
MS analysis, α-helical content, lipid transfer activity, and papain inhibition of the recombinant Gb-nsLTP1 proteins
| Strain | Experimental Mass (Theoretical)a | α-Helical Contentb | Lipid Transfer Activityc | Papain Inhibitionc |
|---|---|---|---|---|
| D | % | |||
| Wild type | 9,586.9 ± 1.2 (9,587.0) | 22 | 100 ± 7 | 100 ± 12 |
| R46A mutant | 9,502.3 ± 0.5 (9,501.9) | 21 | 88 ± 5 | 94 ± 8 |
| P79A mutant | 9,561.4 ± 0.9 (9,561.0) | 11 | 58 ± 2 | 231 ± 25 |
| F80A mutant | 9,511.6 ± 0.7 (9,510.9) | 10 | 26 ± 4 | 219 ± 2 |
| I82A mutant | 9,545.6 ± 0.8 (9,544.9) | 18 | 101 ± 1 | 97 ± 12 |
Including the formation of four disulfide bonds.
Measured at pH 7.0 and estimated by the method of Chen et al. (1972).
Calculated based on the lipid transfer activity or papain inhibition of the wild-type protein. These values of the native protein were 116% and 115% under the corresponding condition, respectively.
Characterization of Gb-nsLTP1 Mutants
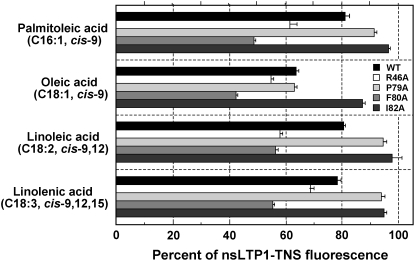
To clarify the structure-activity relationship of Gb-nsLTP1, it is very important to examine the biological activity of the mutant proteins, for instance, lipid transfer ability or papain inhibition. Especially, the biological function of plant nsLTP1 is related to the capacity of binding and/or transporting amphiphilic lipids. Therefore, we introduced four independent mutations (R46A, P79A, F80A, and I82A) into the highly conserved lipid-binding sites as known among plant nsLTP1 (Fig. 3). As a result, the phospholipid transfer activity of the mutants P79A and F80A was much lower than the wild-type protein (Table I). The activity of the R46A mutant showed 88%; on the other hand, the I82A mutant did not lose any lipid transfer activity (101%). Moreover, we measured the binding activity of the mutant proteins toward cis-unsaturated fatty acids, palmitoleic acid, oleic acid, linoleic acid, and linolenic acid. The mutants P79A and I82A showed lower binding activity against most of these fatty acids than the wild-type protein; on the other hand, the mutants R46A and F80A did not lose any lipid-binding activity (Fig. 11). It is interesting to note that the papain-inhibitory activity of the mutants P79A and F80A was elevated twice as much as the wild-type protein (Table I).
Figure 11.
Influence of cis-unsaturated fatty acids on the fluorescence level of the mutated Gb-nsLTP1-TNS complex. The percentage of fluorescence of the complex toward the wild-type, R46A, P79A, F80A, and I82A proteins indicates black, white, light gray, gray, and dark gray bars, respectively. Experiments were performed in triplicate and results are expressed as the mean values ± sds.
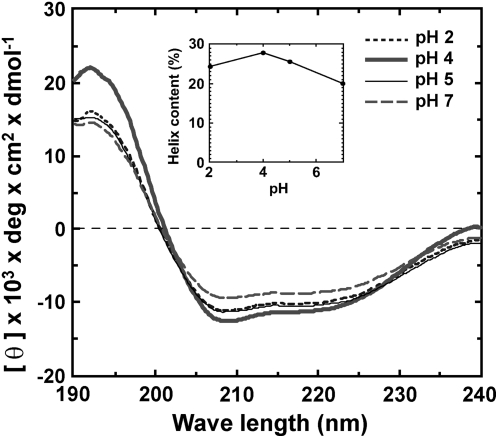
CD Measurements
Figure 12 shows CD spectra of Gb-nsLTP1 at four pH values between 2 and 7. As shown in the inset, this protein appears to be stable and rich (20%–30%) in helical structure under the above conditions. This stability is likely due to the presence of the four disulfide bonds. Analysis of the spectrum at pH 5.0 by the CONTIN program indicated that Gb-nsLTP1 is composed of α-helices (34%), β-sheets (30%), and β-turns (12%), which is roughly consistent with the estimation of helical content by the method of Chen et al. (1972). The CD spectrum of the recombinant Gb-nsLTP1 (wild type) at pH 7.0 was almost identical to that of the native one (data not shown), which indicates that the native and recombinant proteins possess a similar folding. Furthermore, the α-helical content of R46A and I82A mutants was estimated by the method of Chen et al. (1972) and was almost identical to that of the wild-type protein (Table I). In contrast, the α-helical content of P79A and F80A mutants was significantly lower than that of the wild-type protein (Table I), indicating that Pro-79 and Phe-80 are structurally important residues.
Figure 12.
Far-UV CD spectra of Gb-nsLTP1 at different pH values. The inset shows the helix content as a function of pH. The fraction of helical content was calculated according to Chen et al. (1972).
DISCUSSION
In this study, we isolated a novel proteinase inhibitor (Gb-nsLTP1) from ginkgo seeds, and its full-length cDNA was identified by RT-PCR and RACE based on the sequence information from the partial peptide sequencing. As shown in Figure 3, the deduced amino acid sequence had approximately 40% similarity to nsLTP1 from the gymnosperm and angiosperm species. Predictably, this protein had a great potential of lipid transfer activity (Fig. 7) and exhibited a unique binding ability to only unsaturated fatty acids with cis-bond(s) (Fig. 8). This indicates that linear-chain fatty acids are not easily accommodated in the hydrophobic binding site of Gb-nsLTP1 and that cis-unsaturated fatty acids exhibit more compacted structures and therefore show a higher affinity for this protein. Interestingly, P79A and F80A mutants caused a significant loss of the lipid transfer activity (Table I). Here, one may consider whether their lower transfer capabilities are due to the inability to bind either any lipid (a fatty acid as well as a phospholipid) or just a phospholipid with two acyl chains (if the peptide cavity is reduced). The binding assay using the mutant proteins revealed that the binding activity of the mutant F79A and I82A were lower than the wild-type protein (Fig. 11); therefore, it was concluded that their lower transfer capability is due to the inability to bind to any lipid.
In this article, Gb-nsLTP1 was revealed to possess inhibitory activities against the Asp and Cys proteinases. Therefore, it seems likely that Gb-nsLTP1 serves to keep inactive some proteinases that are synthesized during seed development until the plant needs them. Apparently, such proteinases are the most important enzymes involved in transforming the large ginkgo storage proteins into small nitrogenous compounds. Such proteins or peptides can easily be utilized by the growing plant in the following germination. Actually, ginkgo seeds contain several storage proteins such as a 30-kD storage glycoprotein (Kimura et al., 1999), the 11S seed storage proteins ginnacin (Arahira and Fukazawa, 1994) and legumin (Jensen and Berthold, 1989), and α-mannosidase (Woo et al., 2004). However, no proteinase activity has been detected in ginkgo seeds (data not shown); therefore, such endogenous proteinases might be expressed when the plant requires them, such as during seed germination. For instance, barley (Hordeum vulgare) plants contain several inhibitors that suppress only the Cys class proteinases, and two of these inhibitors are identified and characterized as nsLTP1 and nsLTP2 (Jones and Marinac, 2000).
On the other hand, it was predicted that the mature protein is secreted extracellularly due to the NH2-terminal propeptide, which conflicts with the above hypothesis that the inhibitor localizes intracellularly with the target proteinases. Plant nsLTP1 are found primarily on the cell walls of aerial organs and seeds; therefore, nsLTP1 could be involved indirectly in plant defense by creating a mechanical barrier of cutin and might be involved directly due to their intrinsic antibiotic properties (Douliez et al., 2000). In a previous article, it was revealed that ginkgo seeds contain a novel antifungal protein (Sawano et al., 2007), indicating that Gb-nsLTP1 might possess antifungal or antibacterial activity. However, in this study, the protein exerted neither antifungal activity against several fungi nor antibacterial activity against E. coli. Furthermore, Gb-nsLTP1 was revealed to be a tissue-specific expressing gene and expressed only in both the immature and mature seeds, not in the stems, leaves, and roots (Fig. 9). Presumably, this protein could be involved in the formation of hydrophobic cutin or suberin layers that prevent water diffusion into the seeds and fungal attacks. In short, Gb-nsLTP1 would be engaged in early embryogenesis by participating in the formation of a protective layer around the young embryo, although the polymerization mechanism of cutin and suberin monomers is still unknown.
Several three-dimensional structures of nsLTP1-lipid complexes have revealed that the hydrophobic ligands are inserted into the internal cavity of nsLTP1 (e.g. Shin et al., 1995; Lerche and Poulsen, 1998; Han et al., 2001). All the structures possess the common folding of four α-helices and a long CO2H-terminal tail region, which forms a tunnel-like hydrophobic cavity for lipid binding. This tail region undergoes significant conformational changes upon ligand binding; Tyr-80 and Ile-82 are thought to be crucial for lipid-binding or lipid transfer activities of plant nsLTP1 (Han et al., 2001; Cheng et al., 2004). In this article, mutagenesis experiments were performed on Arg-46, Pro-79, Phe-80, and Ile-82 in the two conserved pentapeptides, and Pro-79 and Phe-80 in the latter conserved peptide were revealed to be important for phospholipid transfer activity (Table I). Arg-46 in the former peptide was slightly involved in lipid transfer activity, while the mutation of Ile-82 to Ala did not have any influence on the activity (Table I). On the other hand, the lipid-binding experiments indicate that Pro-79 and Ile-82 are essential for the binding activity toward cis-unsaturated fatty acids (Fig. 11). It is worth noting that Phe-80 was not involved in the recognition of the chain length and the geometry of double bonds, because the F80A mutant did not lose the activities toward these fatty acids (Fig. 11). In regards to the papain-inhibitory activity, the mutations R46A and I82A had very little influence on the papain inhibition, but the inhibitory activity of P79A and F80A mutants was elevated twice as much as the wild-type protein (Table I).
In the solution structure of nsLTP1 from mung bean (Vigna radiata), Mb-nsLTP1, Arg-44 and Tyr-79 are located near the entrance of the tunnel-like hydrophobic cavity and interact with the polar headgroup of lipid molecules (Lin et al., 2005). Ile-81 locates near the bottom of the lipid-binding cavity, in which it may interact with the aliphatic chain of the lipid. Therefore, our results suggest that Phe-80 and Ile-82 are involved in the entrance and bottom conformations of the lipid-binding cavity, respectively. It is particularly noteworthy that Pro-79 is essential for both the lipid transfer and binding activities of Gb-nsLTP1, while the ionic interaction between Arg-46 and lipid molecules was not so important for these activities. The key residues Pro-79 and Phe-80 (Pro-78 and Tyr-79 in Mb-nsLTP1) is located at the CO2H-terminal tail loop, which is reported as a flexible region with much larger root mean squared deviation values than other regions (Lin et al., 2005). These mutations might have a great influence on the entrance conformation of the hydrophobic cavity, especially the CO2H-terminal α-helix and tail loop. In the case of the papain inhibition, by the mutations of Pro-79 or Phe-80 to Ala, the conformation of the tail region might be changed to a more desirable shape that allows the putative inhibitory loop to fit well into the active site cleft of the target enzyme. Further structural analysis of Gb-nsLTP1 by NMR or x-ray crystallography will provide new insights into our understanding of structure-activity relationship of the lipid binding/transfer and proteinase inhibitory activities.
MATERIALS AND METHODS
Materials
Mature ginkgo (Ginkgo biloba) seeds were obtained from the garden of our university in September, 2004. The immature seeds, leaves, and stems were collected there in May. The roots were obtained from the seedlings kindly gifted by Japan Ginkgo Farm (Fukushima, Japan). All the tissues were immediately frozen in liquid nitrogen and were stored at −80°C. Purified proteinases and the synthetic substrates were from Sigma-Aldrich. Oligonucleotide primers were synthesized by Sigma Genosys Japan KK and ThermoElectron GmbH Molecular Biology. All other reagents were of the highest grade commercially available.
Purification of Proteinase Inhibitors from Ginkgo Seeds
Unless otherwise stated, all purification steps were carried out at 4°C. The mature ginkgo seeds were shelled and fully homogenized with 2 to 3 volumes of an extraction buffer containing 50 mm Tris-HCl buffer at pH 7.5, 10 mm KCl, 1 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, and 5 mm iodoacetic acid. After stirring overnight, the homogenate was centrifuged at 33,570g for 10 min at 4°C, and the clear supernatant was freeze-dried. The resulting powder was resuspended in distilled water and dialyzed against the extraction buffer by using a 1,000-Mr cutoff membrane. The dialyzed solution was then applied onto a Sephadex G-50 gel-filtration column (2.2 × 96 cm; GE Healthcare Life Sciences) equilibrated with 50 mm Tris-HCl buffer, pH 7.5, containing 0.1 m NaCl.
The resulting sample was finally purified on an SP-Sepharose high-performance column (1.8 × 15 cm; GE Healthcare Life Sciences) equilibrated with 10 mm ammonium acetate buffer at pH 4.3. The adsorbed protein with the papain-inhibitory activity was eluted using a linear NaCl gradient (0–1 m). The identification and purity of the sample were confirmed by both MALDI-TOF MS and SDS-PAGE (15% gels) without dithiothreitol (DTT; Laemmli, 1970). MS experiments were performed in the linear mode on an AXIMA-CFR Plus MALDI-TOF mass spectrometer (Shimadzu). 3,5-Dimethoxy-4-hydroxycinnamic acid was used for the matrix, which was prepared as a saturated solution in a 1:2 mixture of acetonitrile and 0.1% trifluoroacetic acid (TFA).
Partial Amino Acid Sequencing
The sample was oxidized with performic acid (Hirs, 1967), and the resulting peptide was digested by trypsin in 50 mm Tris-HCl buffer, pH 6.8, at 37°C for 20 h (enzyme:substrate mass ratio, 1:100). The digested products were purified on a RESOURCE RPC analytical column (3 mL; GE Healthcare Life Sciences) equilibrated with 1% (v/v) acetonitrile and 0.1% (v/v) TFA by using a Shimadzu LC10-Vp HPLC system. Elution was achieved at a flow rate of 1.0 mL min−1 with a linear gradient of 1% to 99% (v/v) acetonitrile in 0.1% TFA. The samples were blotted on a polyvinylidene fluoride membrane by using a ProSorb sample preparation cartridge (Applied Biosystems), and the amino acid sequences were determined by an Applied Biosystems model 491 sequencer.
Isolation of Total RNA and Cloning of a Partial Fragment of cDNA
The cDNA encoding the proteinase inhibitor was isolated based on a PCR cloning strategy. Total RNA was extracted from the mature ginkgo seeds by using Plant RNA Isolation Reagent (Invitrogen). To isolate the cDNA, RT-PCR was performed after DNase treatment using a TURBO DNA-free kit (Ambion). The first-strand cDNA was synthesized from total RNA with AccuScript reverse transcriptase (Stratagene) by using oligo(dT)12-18 primer (Invitrogen). PCR was carried out using AmpliTaq Gold DNA polymerase (Applied Biosystems). The degenerate primers for amplification were 5′-GCICCIGGITGYGAYACIGT-3′ (GBf1) and 5′-RTTRCARTCIGTIGCDAT-3′ (GBr2; R = A/G, Y = C/T, D = G/A/T, I = inosine), which were respectively designed based on the NH2-terminal (APGCDTV) and internal amino acid sequences (IATDCN). PCR was performed using the first-strand cDNA as templates and with GBf1 and GBr4 as primers. The amplified DNA fragments were cloned into a pCR2.1-TOPO vector by using the TOPO TA Cloning kit (Invitrogen) and sequenced using a BigDye Terminator kit on an ABI Prism3700 DNA Analyzer (Applied Biosystems).
Cloning of the Full-Length cDNA
To obtain the full-length cDNA, 3′ RACE and 5′ RACE were performed (Frohman et al., 1988). The gene-specific primers were designed according to the partial cDNA sequence. To obtain the 3′ end of the cDNA, the first-strand cDNA was synthesized with the adaptor-combined poly(dT) primer of 5′-CTGTGAATGCTGCGACTACGA(T)17V-3′ (V = A/C/G). After RNase-H treatment, the cDNA was amplified by PCR using PrimeSTAR HS DNA polymerase (TaKaRa), the gene-specific sense primer of 5′-TGGCACCTTGCATTTCCTATCTTCAGACC-3′ (GBs1), and an adaptor primer as the antisense primer of 5′-CTGTGAATGCTGCGACTACGA-3′. Nested PCR was performed using the resulting PCR product as a template with the nested gene-specific sense primer of 5′-CCGGCAACCCAACTGTACAATGCTG-3′ (GBs2) and the adaptor primer as above.
For 5′ RACE, first-strand cDNA synthesis was performed with the antisense primer of 5′-GAAAGGCAGAGTAACGCTACACAGTCCCG-3′ (GBa3). A poly(A) tail was added to the 3′ ends of the first-strand cDNA with terminal transferase (Roche Diagnostics) after RNase-H treatment. The poly(A)-tailed cDNA was used as a template for the first round of PCR with the adaptor primer and the nested antisense primer of 5′-AGGCTTTTGACGGCGTTCGCAACC-3′ (GBa4). The resulting product was subjected to nested PCR with the adaptor primer and another nested primer located at the 5′ end of GBa4, 5′-ACTCGAATAGCGGCCGTCTTTATACATTCAC-3′ (GBa5). The fragments amplified by 3′ RACE and 5′ RACE were cloned into HincII-digested pUC19 vector via blunt-end ligation and were sequenced. To obtain further 5′ sequences, a second round of 5′ RACE was performed as follows: the first-strand cDNA synthesis was done with the antisense primer of 5′-GCAGTTCCGGCCAGCGTTTTGACACC-3′ (GBa6) and was tailed at the 3′ end. Primary PCR was performed using the poly(A)-tailed cDNA as a template with the adaptor primer and the nested antisense primer of 5′-AGCATTGTACAGTTGGGTTGCCGGTTCC-3′ (GBa7), followed by nested PCR with the adaptor primer and another nested primer specific for the new 5′ end, 5′-GTGTCTACAGTGTCACATCCGGGTGCTGC-3′ (GBa8).
Bioinformatics and Molecular Evolution Analyses
The obtained sequences were analyzed using bioinformatics tools at Web sites (www.ncbi.nlm.nih.gov, www.expasy.org). Before analysis, the putative and experimentally verified signal sequences were added. The phylogenetic analysis of several plant nsLTP1 were aligned with ClustalW (Thompson et al., 1994), and a phylogenetic tree was subsequently constructed by the neighbor-joining method (Saitou and Nei, 1987) with MEGA-3 software (Kumar et al., 2004). The reliability of the tree was measured by bootstrap analysis with 1,000 replicates (Felsenstein, 1985). Sequence similarity searches were performed at the NCBI using BLAST services.
Characterization of Proteinase Inhibitory Activities
Inhibitory activity toward pepsin was determined by the method of Anson (1938) with some modifications. Seventy-two microliters of a sample solution (final concentration of 172 μm) was mixed with 286 μL of 2.5% acid-denatured hemoglobin in 50 mm sodium phosphate buffer at pH 4.0 and was preincubated at 37°C for 2 min. About 1.0 mg mL−1 enzyme solution (72 μL) was added to the mixture and incubated at 37°C for 60 min. To this was added 570 μL of 5% (w/v) TCA; the mixture was incubated at 4°C for 10 min and centrifuged at 5,800g for 10 min; the A280 of the supernatant was measured. Blank and control experiments were carried out in the same way, except that 5% TCA was added prior to addition of the enzyme solution and that the sample solution was replaced with water, respectively. The percent inhibition was calculated by the following equation: Inhibition (%) = [A280 (control) − A280 (sample) + A280 (blank)]/A280 (control) × 100.
In the case of papain, we thoroughly mixed 16 μL of about 0.1 mg mL−1 enzyme solution, 16 μL of a sample solution (final concentration of 147 μm), and 652 μL of 50 mm sodium acetate buffer, pH 5.0, containing 2 mm EDTA and 1 mm DTT in a 1-cm path length quartz cuvette. In the case of trypsin, we used about 0.01 mg mL−1 enzyme solution and 20 mm sodium phosphate buffer, pH 6.3, containing 0.6 mm CaCl2. The cuvette was placed in the thermostatted cell compartment (25°C) of a Beckman DU-640 spectrophotometer. After at least 2 min of preincubation, 16 μL of the substrate p-nitrophenyl benzyloxycarbonyl-l-lysinate (1.1 mg mL−1) was added to the tip of a flattened glass rod; the solution was then stirred for 10 s. The initial velocity (v0) was calculated from an increase in A340 of the released p-nitrophenol. The percent inhibition was calculated by the following equation: Inhibition (%) = [1 − (v0 with sample)/(v0 without sample)] × 100. The inhibition constant values for pepsin and papain were calculated by a Dixon plot analysis (Dixon, 1953).
In the case of thermolysin, the buffer used was 50 mm Tris-HCl buffer, pH 7.5, containing 10 mm CaCl2 and 100 mm NaCl; N-(3-[2-furyl]acryloyl)-Gly-Leu amide (1.0 mg mL−1) was used as a substrate for the enzyme. Fifty microliters of about 0.01 mg mL−1 enzyme solution, 16 μL of a sample solution (final concentration of 49 μm), and 584 μL of the buffer were mixed thoroughly in the quartz cuvette. After at least 2 min of preincubation at 37°C, 50 μL of the substrate was added to the tip of a flattened glass rod; the solution was then stirred for 10 s. Hydrolysis of the substrate by thermolysin was measured by a decrease in A345.
The pH dependence of the inhibitory activity toward papain was determined using a solution of 50 mm buffer, 2 mm EDTA, and 1 mm DTT over the pH range of 2.0 to 7.5. The buffers used were Gly-HCl, pH 2.0 to 3.0; sodium citrate, pH 3.0 to 5.5; and sodium phosphate, pH 5.5 to 7.5. A sample solution (final concentration of 145 μm) and about 0.1 mg mL−1 enzyme solution were mixed in the appropriate buffer. The other methods were the same as described above.
Assays of Antifungal and Antibacterial Activities
We carried out antifungal activity assay against several fungi, including plant and human pathogens Fusarium oxysporum, Fusarium culmorum, Trichoderma reesei, Aspargillus fumigatus, and Mucor spinescens, in 100- × 15-mm petri plates containing 20 mL of potato dextrose agar. Sterile blank paper discs (0.6 cm in diameter) were placed at a distance of 0.5 cm from the rim of mycelial colony. Thirty microliters of a sample solution (25 mg mL−1) was added to a disc. The plates were incubated at 24°C for 72 h until mycelial growth had enveloped discs containing the control and formed crescents of inhibition around discs containing antifungal samples.
The assay for antibacterial activity was conducted using sterile petri plates (100 × 15 mm) containing 10 mL of Luria-Bertani agar (1.5% agar). Three milliliters of warm nutrient agar (0.7%) containing Escherichia coli was poured into the plates. The sterile blank paper discs were placed on the agar; to one of the discs was then added a sample solution (20 μL) in 10 mm Tris-HCl buffer at pH 7.4. Only the same buffer was added to the control disc. The plate was incubated at 30°C for 20 to 24 h. A transparent ring around the paper disc signifies antibacterial activity.
Lipid Transfer and Lipid-Binding Assays
Lipid transfer activity was assayed using fluorescence spectroscopy as described previously (Samuel et al., 2002) with minor modifications. 1,2-Dimyristoyl-sn-glycerol-3-phospho-rac-1-glycerol (Myr2PtdGro) multilamellar vesicles were prepared vortexing a Myr2PtdGro solution (1 mg mL−1) above its transition temperature (23°C) in 10 mm sodium phosphate buffer at pH 7.0 and by freeze-thawing. Homogeneously sized small unilamellar vesicles were obtained by sonicating Myr2PtdGro multilamellar vesicles at 4°C for 30 min. Four microliters of 1-palmitoyl-2-pyrenyl-decanoyl-sn-phosphorylcholine (0.1 mg mL−1) in ethanol and 10 μL of the Myr2PtdGro small unilamellar vesicle solution were mixed in a cuvette in 2 mL of 50 mm phosphate buffer at pH 7.0. After 10 s of incubation, 5 μL of the sample solution (53 mg mL−1) was added to the mixed solution. Fluorescence intensities were monitored at 396 nm with excitation at 346 nm by using a Shimadzu RF-5300PC fluorescence spectrophotometer at 25°C. The experimental values were obtained from two independent experiments. The lipid transfer rate between donor and acceptor vesicles was estimated by the method described by Van Paridon et al. (1988).
The lipid-binding assay of Gb-nsLTP1 was carried out using TNS. Fluorescence experiments were performed at 25°C in the fluorescence spectrophotometer as described previously (Buhot et al., 2004) with minor modifications. The excitation and emission wavelengths were set at 355 and 430 nm, respectively. Before the initial fluorescence (F0) was recorded, 20 μm TNS with or without 0.5, 2.5, 25, and 50 μm (2.5 μm for the mutant proteins) of each fatty acid was incubated for 1 min in 2 mL of a measurement buffer (175 mm mannitol, 0.5 mm K2SO4, 0.5 mm CaCl2, and 5 mm MES at pH 7.0). The equilibrated fluorescence (F) was recorded after 2-min incubation with Gb-nsLTP1 (final concentration of 2.8 μm). The lipid-binding activity was represented as a percentage of (F − F0)/FC × 100, where FC is the fluorescence of an nsLTP1-TNS complex without any fatty acid.
Expression Profile of Gb-nsLTP1 in Different Tissues
Total RNA was extracted separately from different tissues (stem, leaf, root, immature seed, and mature seed). An aliquot of 1 μg of total RNA was used as a template in a single-step RT-PCR analysis using PrimeScript One Step RT-PCR kit version 2 (TaKaRa) with the primers of 5′-ATGATGAAGATATCGTGGCAGCTC-3′ and 5′-TTAAACGATCTTGTTACAGTCGGTGG-3′. The template was reversely transcribed at 50°C for 30 min and denatured at 94°C for 2 min, followed by 25 cycles of amplification (94°C for 30 s, 60°C for 30 s, and 72°C for 45 s) and by extension at 72°C for 10 min. As a control, RT-PCR reaction for the housekeeping gene (18S rRNA gene) was performed using the specific primers of 5′-ATGATAACTCGACGGATCGC-3′ and 5′-CTTGGATGTGGTAGCCGTTT-3′. The PCR products were separated on 2% (w/v) agarose gel stained with SYBR Safe DNA gel stain (Invitrogen).
Construction of Expression Vector of the Recombinant Proteins
The DNA sequence encoding Gb-nsLTP1 was amplified from the first-strand cDNA mixture using PfuUltra Hotstart DNA polymerase (Stratagene). The forward primer was 5′-GCACCCGGATGTGACACTGTAGAC-3′, and the reverse was 5′-CGGGGATCCTTATTAAACGATCTTGTTACAGTCGG-3′, designed to contain an additional stop codon and a BamHI site. The PCR product was phosphorylated by T4 polynucleotide kinase (TaKaRa), digested with BamHI, and ligated into a SmaI/BamHI-digested expression vector of pET48 (Novagen) driven by a T7 promoter. The resulting vector of pET48-Gb-nsLTP1 encodes a Trx-tag, a (His)6-tag, an HRV3C protease recognition site, and Gb-nsLTP1 in this order. The expression vectors for the mutants were constructed from a pET48-Gb-nsLTP1 vector by using the QuikChange mutagenesis kit (Stratagene).
Expression and Purification of the Recombinant Proteins
E. coli Origami2(DE3) cells (Novagen) were transformed with pET48-Gb-nsLTP1. The cells were cultured at 37°C by shaking in a Luria-Bertani medium containing kanamycin (30 μg mL−1) and tetracycline (10 μg mL−1). The production of the recombinant proteins was induced at an optical density at 600 nm of 0.5 by addition of isopropyl-β-d-thiogalactopyranoside at a final concentration of 0.5 mm. After induction at 25°C for 16 h, the cells were harvested by centrifugation, washed once with 0.5% (w/v) NaCl, and stored at −80°C. The frozen cells were resuspended in a binding buffer (50 mm sodium phosphate buffer at pH 8.0, 300 mm NaCl, and 10 mm imidazole) containing 0.1 mm 4-(2-aminoethyl)-benzenesulfonyl fluoride hydrochloride, 80 nm aprotinin, 2 μm leupeptin, 4 μm bestatin, 1.5 μm pepstatin A, and 1.4 μm E-64; the resulting suspension was lysed by ultrasonication. The soluble fraction was separated from the insoluble by centrifugation (40,000g at 4°C for 30 min), and the supernatant was added onto a Ni-NTA-agarose column (Qiagen) equilibrated with the binding buffer. After washing with the buffer, the sample was eluted with 50 mm sodium phosphate buffer, pH 8.0, containing 300 mm NaCl and 300 mm imidazole and was dialyzed against a 20 mm Tris-HCl buffer at pH 7.5. The regions of Trx-tag and (His)6-tag were digested with HRV3C protease (Novagen) according to the manufacturer's instructions. The sample was applied onto a Mono-S 10/10 column (GE Healthcare Life Sciences) equilibrated with 20 mm MES buffer at pH 5.5 and was eluted with a linear gradient of 0 to 0.2 m NaCl in the MES buffer. The resulting sample was pooled, dialyzed against 10 mm MES buffer at pH 6.0, and concentrated by ultrafiltration using a 3,000-Mr cutoff Vivaspin (Sartorius). The purity and identification of the sample were checked by SDS-PAGE and MALDI-TOF MS as described above. The mutant proteins were expressed and purified as described above.
CD Measurements
All CD spectra were recorded at room temperature with a Jasco J-600 spectropolarimeter (JASCO International). Gb-nsLTP1 was adjusted to pH 2.0, 4.0, 5.0, or 7.0 with protein concentrations of approximately 70 μm, while the recombinant Gb-nsLTP1 proteins were only to pH 7.0. Individual CD spectra were averaged over two scans with 0.2-nm increments using a 0.5-mm path length cuvette. The spectra thus obtained were submitted to a smoothing procedure and expressed in terms of mean residue ellipticity [θ] in degree cm2 dmol−1. The fraction of helical content (fH) was calculated using the following equation (Chen et al., 1972): fH = ([θ]222 + 2,340)/30,300 × 100, where [θ]222 is the observed ellipticity value at 222 nm. The CD spectrum of the native protein at pH 5.0 was analyzed with the program CONTIN (Provencher and Glöckner, 1981) to estimate the relative amount of each secondary structure element. The protein quantitation was carried out using a bicinchoninic acid assay with bovine serum albumin as a standard (Smith et al., 1985).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number DQ836633.
This work was supported by three national projects from the Ministry of Education, Culture, Sports, Science and Technology of Japan: Protein Structure and Functional Analyses, the Targeted Proteins Research Program, and the 21st Century Center of Excellence Program.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Ken-ichi Hatano (hatano@chem-bio.gunma-u.ac.jp).
References
- Anson ML (1938) The estimation of pepsin, trypsin, papain, and cathepsin with hemoglobin. J Gen Physiol 22 79–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arahira M, Fukazawa C (1994) Ginkgo 11S seed storage protein family mRNA: unusual Asn-Asn linkage as post-translational cleavage site. Plant Mol Biol 25 597–605 [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S (2004) Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340 783–795 [DOI] [PubMed] [Google Scholar]
- Buhot N, Gomès E, Milat ML, Ponchet M, Marion D, Lequeu J, Delrot S, Coutos-Thévenot P, Blein JP (2004) Modulation of the biological activity of a tobacco LTP1 by lipid complexation. Mol Biol Cell 15 5047–5052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, Yang JT, Martinez HM (1972) Determination of the secondary structures of proteins by circular dichroism and optical rotatory dispersion. Biochemistry 11 4120–4131 [DOI] [PubMed] [Google Scholar]
- Cheng HC, Cheng PT, Peng P, Lyu PC, Sun YJ (2004) Lipid binding in rice nonspecific lipid transfer protein-1 complexes from Oryza sativa. Protein Sci 13 2304–2315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon M (1953) The determination of enzyme inhibitor constants. Biochem J 55 170–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douliez JP, Jégou S, Pato C, Larré C, Mollé D, Marion D (2001) Identification of a new form of lipid transfer protein in wheat seeds. J Agric Food Chem 49 1805–1808 [DOI] [PubMed] [Google Scholar]
- Douliez JP, Michon T, Elmorjani K, Marion D (2000) Structure, biological and technological functions of lipid transfer proteins and indolines, the major lipid binding proteins from cereal kernels. J Cereal Sci 32 1–20 [Google Scholar]
- Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution Int J Org Evolution 39 783–791 [DOI] [PubMed] [Google Scholar]
- Frohman MA, Dush MK, Martin GR (1988) Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA 85 8998–9002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han GW, Lee JY, Song HK, Chang C, Min K, Moon J, Shin DH, Kopka ML, Sawaya MR, Yuan HS, et al (2001) Structure basis of non-specific lipid binding in maize lipid transfer protein complexes revealed by high-resolution x-ray crystallography. J Mol Biol 308 263–278 [DOI] [PubMed] [Google Scholar]
- Hirs CHW (1967) Performic acid oxidation. Methods Enzymol 11 197–199 [Google Scholar]
- Huang X, Xie WJ, Gong ZZ (2000) Characteristics and antifungal activity of a chitin binding protein from Ginkgo biloba. FEBS Lett 478 123–126 [DOI] [PubMed] [Google Scholar]
- Jégou S, Douliez JP, Mollé D, Boivin P, Marion D (2000) Purification and structural characterization of LTP1 polypeptides from beer. J Agric Food Chem 48 5023–5029 [DOI] [PubMed] [Google Scholar]
- Jensen U, Berthold H (1989) Legumin-like proteins in gymnosperms. Phytochemistry 28 1389–1394 [Google Scholar]
- Jones BL, Marinac LA (2000) Purification and partial characterization of a second cysteine proteinase inhibitor from ungerminated barley (Hordeum vulgare L.). J Agric Food Chem 48 257–264 [DOI] [PubMed] [Google Scholar]
- Kader JC (1996) Lipid-transfer proteins in plants. Annu Rev Plant Physiol Plant Mol Biol 47 627–654 [DOI] [PubMed] [Google Scholar]
- Kalashnikova EE, Chernyshova MP, Ignatov VV (2003) The extracellular proteases of the phytopathogenic bacterium Xanthomonas campestris. Mikrobiologiia 72 498–502 [PubMed] [Google Scholar]
- Kalla R, Shimamoto K, Potter R, Nielsen PS, Linnestad C, Olsen OA (1994) The promotor of the barley aleurone-specific gene encoding a putative 7 kDa lipid transfer protein confers aleurone cell-specific expression in transgenic rice. Plant J 6 849–860 [DOI] [PubMed] [Google Scholar]
- Kimura Y, Harada T, Matsuo S, Yonekura M (1999) Purification and some chemical properties of 30 kDa Ginkgo biloba glycoprotein, which reacts with antiserum against β1→xylose-containing N-glycans. Biosci Biotechnol Biochem 63 463–467 [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5 150–163 [DOI] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685 [DOI] [PubMed] [Google Scholar]
- Lerche MH, Poulsen FM (1998) Solution structure of barley lipid transfer protein complexed with palmitate. Two different binding modes of palmitate in the homologous maize and barley nonspecific lipid transfer proteins. Protein Sci 7 2490–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin KF, Liu YN, Hsu ST, Samuel D, Cheng CS, Bonvin AMJJ, Lyu PC (2005) Characterization and structural analyses of nonspecific lipid transfer protein 1 from mung bean. Biochemistry 44 5703–5712 [DOI] [PubMed] [Google Scholar]
- Lin P, Xia L, Wong JH, Ng TB, Ye X, Wang S, Shi X (2007) Lipid transfer proteins from Brassica campestris and mung bean surpass mung bean chitinase in exploitability. J Pept Sci 13 642–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K, Kanehisa M (1992) A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics 14 897–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provencher SW, Glöckner J (1981) Estimation of globular protein secondary structure from circular dichroism. Biochemistry 20 33–37 [DOI] [PubMed] [Google Scholar]
- Ryan CA (1990) Protease inhibitors in plants: genes for improving defenses against insects and pathogens. Annu Rev Phytopathol 28 425–449 [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4 406–418 [DOI] [PubMed] [Google Scholar]
- Samuel D, Lin YJ, Cheng CS, Lyu PC (2002) Solution structure of plant nonspecific lipid transfer protein-2 from rice (Oryza sativa). J Biol Chem 277 35267–35273 [DOI] [PubMed] [Google Scholar]
- Sara M, Heale JB (1990) The roles of aspartic proteinase and endo-pectin lyase enzymes in the primary stages of infection and pathogenesis of various host tissues by different isolates of Botrytis cinerea Pers ex. Pers. Physiol Mol Plant Pathol 36 303–324 [Google Scholar]
- Sawano Y, Miyakawa T, Yamazaki H, Tanokura M, Hatano K (2007) Purification, characterization, and molecular cloning of an antifungal protein gene from Ginkgo biloba seeds. Biol Chem 388 273–280 [DOI] [PubMed] [Google Scholar]
- Shen G, Pang Y, Wu W, Deng Z, Liu X, Lin J, Zhao L, Sun X, Tang K (2005) Molecular cloning, characterization and expression of a novel jasmonate-dependent defensin gene from Ginkgo biloba. Plant Physiol Biochem 43 836–843 [DOI] [PubMed] [Google Scholar]
- Shin DH, Lee JY, Hwang KY, Kim KK, Suh SW (1995) High-resolution crystal structure of the non-specific lipid-transfer protein from maize seedlings. Structure 3 189–199 [DOI] [PubMed] [Google Scholar]
- Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC (1985) Measurement of protein using bicinchonic acid. Anal Biochem 150 76–85 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Paridon PA, Gadella TWJ, Wirtz KWA (1988) The effect of polyphosphoinositides and phosphatidic acid on the phosphatidylinositol transfer protein from bovine brain: a kinetic study. Biochim Biophys Acta 943 76–86 [DOI] [PubMed] [Google Scholar]
- Woo KK, Miyazaki M, Hara S, Kimura M, Kimura Y (2004) Purification and characterization of a Co(II)-sensitive alpha-mannosidase from Ginkgo biloba seeds. Biosci Biotechnol Biochem 68 2547–2556 [DOI] [PubMed] [Google Scholar]