Abstract
Circadian rhythms are found in organisms from cyanobacteria to plants and animals. In flowering plants, the circadian clock is involved in the regulation of various physiological phenomena, including growth, leaf movement, stomata opening, and floral transitions. Molecular mechanisms underlying the circadian clock have been identified using Arabidopsis (Arabidopsis thaliana); the functions and genetic networks of a number of clock-related genes, including CIRCADIAN CLOCK ASSOCIATED1, LATE ELONGATED HYPOCOTYL (LHY), TIMING OF CAB EXPRESSION1, GIGANTEA (GI), and EARLY FLOWERING3 (ELF3), have been analyzed. The degree to which clock systems are conserved among flowering plants, however, is still unclear. We previously isolated homologs for Arabidopsis clock-related genes from monocotyledon Lemna plants. Here, we report the physiological roles of these Lemna gibba genes (LgLHYH1, LgLHYH2, LgGIH1, and LgELF3H1) in the circadian system. We studied the effects of overexpression and RNA interference (RNAi) of these genes on the rhythmic expression of morning- and evening-specific reporters. Overexpression of each gene disrupted the rhythmicity of either or both reporters, suggesting that these four homologs can be involved in the circadian system. RNAi of each of the genes except LgLHYH2 affected the bioluminescence rhythms of both reporters. These results indicated that these homologs are involved in the circadian system of Lemna plants and that the structure of the circadian clock is likely to be conserved between monocotyledons and dicotyledons. Interestingly, RNAi of LgGIH1 almost completely abolished the circadian rhythm; because this effect appeared to be much stronger than the phenotype observed in an Arabidopsis gi loss-of-function mutant, the precise role of each clock gene may have diverged in the clock systems of Lemna and Arabidopsis.
Circadian systems are important devices that allow organisms to adapt to the day/night cycle. Most organisms, including cyanobacteria, plants, insects, fish, and mammals, possess endogenous circadian clocks. Circadian clocks in plants are involved in various physiological behaviors, such as cell growth, changes in stomata aperture, metabolism, and photoperiodic flowering (Sweeney, 1987; Más, 2005). These circadian phenomena are thought to involve the regulation of gene expression. Molecular mechanisms based on circadian oscillations have been revealed using Arabidopsis (Arabidopsis thaliana), in which clock-related genes have been isolated and analyzed. CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY) encode similar Myb-related transcription factors, and their expression levels circadianly oscillate with peaks occurring around dawn (Wang and Tobin, 1998; Schaffer et al., 1998). Single mutations in cca1 or lhy shortened the period length of the circadian rhythm, and the cca1 lhy double mutant showed a damped oscillation with an extremely short period (Green and Tobin, 1999; Mizoguchi et al., 2002). Overexpression of CCA1 or LHY repressed their own gene expression and disturbed the rhythmic expression of other clock-controlled genes. Therefore, these genes play a role in a negative feedback loop that presumably forms the circadian timing machinery. LHY and CCA1 control the expression of circadian-controlled genes through direct interactions with the evening element in their promoters (Harmer and Kay, 2005). One target of LHY/CCA1 is another clock gene, TIMING OF CAB EXPRESSION1 (TOC1)/PSEUDO-RESPONSE REGULATOR1 (PRR1), for which increased mRNA levels have been observed in the early night (Makino et al., 2000; Strayer et al., 2000; Alabadí et al., 2001). Available evidence strongly suggests that LHY and CCA1 circadianly repress the gene expression of TOC1 by directly binding to an evening element in its promoter region during the morning. In contrast, TOC1 positively regulates CCA1 and LHY expression by an unknown mechanism.
GIGANTEA (GI) is another clock-related gene that plays an important role in circadian oscillations (Park et al., 1999). A gi loss-of-function mutant as well as a GI overexpressor showed short-period rhythms with lower amplitudes (Mizoguchi et al., 2005). Recent computer simulations have suggested that GI may form a feedback loop with TOC1 independent of the TOC1-LHY/CCA1 regulatory loop (Locke et al., 2006). EARLY FLOWERING3 (ELF3) encodes a clock-related component that transmits light-mediated signals to the circadian clock, possibly through an interaction with photoreceptors (Hicks et al., 2001; Liu et al., 2001). The elf3 mutant showed an arrhythmic phenotype under constant light (LL) conditions (Hicks et al., 1996). Interestingly, LHY was expressed at a lower level in the elf3 mutant than in wild-type plants (Schaffer et al., 1998), whereas TOC1 expression was maintained at a higher level in the elf3 mutant (Alabadí et al., 2001). In elf3 mutants, however, faint circadian rhythms were preserved under constant dark conditions. ELF4, LUX ARRYTHMO/PHYTOCLOCK1 (PCL1), and the PRR series of genes (PRR3/5/7/9) also function in the Arabidopsis circadian system (Matsushika et al., 2000; Doyle et al., 2002; Hazen et al., 2005; Onai and Ishiura, 2005; Mizuno and Nakamichi, 2005; McWatters et al., 2007). A mathematical model has predicted that these genes may be components of interlocking feedback loops that include LHY/CCA1, TOC1, and GI (Locke et al., 2006).
On the basis of sequence similarities to the Arabidopsis clock-related genes, homologous genes were isolated from a number of plants (Boxall et al., 2005; Ramos et al., 2005; Murakami et al., 2007). Comprehensive analysis was carried out in rice (Oryza sativa) after the complete genomic sequence of this model monocotyledonous plant was determined (Murakami et al., 2007). OsCCA1 (also called OsLHY), OsZEITLUPE (OsZTL), OsPCL1, and the OsPRR gene family were characterized by mRNA expression profiles that were similar to those of their Arabidopsis counterparts (Izawa et al., 2002; Murakami et al., 2003, 2007). It was suggested, however, that the ELF3- and ELF4-related genes found in the rice genome may not be orthologous to the Arabidopsis counterparts, and it was mentioned that those homologs may not show circadian mRNA expression profiles (Murakami et al., 2007). Because Arabidopsis ELF3 and ELF4 show robust circadian rhythms in their expression levels, a divergence in the functions of these clock-related genes may have occurred between these species. Overexpression of the rice clock-related genes in Arabidopsis demonstrated that OsPRR1 and OsZTL and their Arabidopsis homologs produced similar effects on circadian rhythms, whereas the circadian rhythm in the OsCCA1 overexpressor appeared to be almost normal (Murakami et al., 2007). As shown in this example, it is still unclear whether or not clock-related homologs are functionally conserved among flowering plants. To date, only a few reports clearly demonstrate the functions of clock-related genes in species other than Arabidopsis due to a lack of loss-of-function mutants for these genes (Hecht et al., 2007).
Lemna plants (duckweeds), a group of monocotyledonous plants with tiny, floating bodies, exemplify intragenus variability in the photoperiodic flowering response; Lemna gibba G3 is a long-day plant, whereas Lemna paucicostata 6746 is a short-day plant (Hillman, 1961a). These plants have been extensively analyzed because their close evolutionary relationship suggested they would be good model organisms for comparing the mechanisms underlying the regulation of day lengths. Recently, clock-related gene homologs have been isolated from both Lemna species (Miwa et al., 2006). Examination of their expression profiles under several light-dark conditions revealed that they were similar in these two Lemna species and were also similar to those of the Arabidopsis genes. In this report, we present functional analyses of the Lemna clock-related homologs of LHY, GI, and ELF3 using a semitransient gene expression system that allowed us to monitor the circadian expression of bioluminescent reporters in response to the overexpression or RNA interference (RNAi) of clock-related genes. Using a morning-specific and an evening-specific promoter (Nakamichi et al., 2004), we were able to observe various effects of the clock-related genes on the circadian system. We show that LgLHYH1, LgGIH1, and LgELF3H1 are involved in the circadian clock, although the effects of overexpression or knockdown of these genes are not always the same as those observed in Arabidopsis.
RESULTS
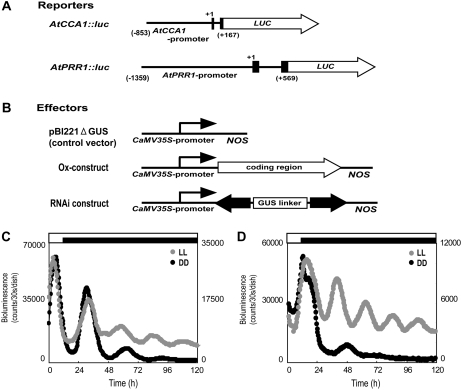
A semitransient bioluminescent reporter system with an AtCCA1 promoter was used to analyze the circadian rhythms of Lemna plants (Miwa et al., 2006). The reporter construct was introduced into plants using a particle bombardment method, and the resulting bioluminescence was continuously monitored. The reporter activity peaked during the morning phase as was observed in Arabidopsis (Nakamichi et al., 2004). The circadian rhythm (period length, approximately 25 h) continued under LL conditions but was severely damped in constant darkness (DD; Fig. 1, A and C; Table I). To examine various aspects of the circadian system of Lemna plants, we used an evening-specific promoter in the bioluminescent reporter system. Arabidopsis TOC1/PRR1, a critical component of the circadian clock, displays a rhythmic promoter activity that peaks during the evening phase (Alabadí et al., 2001). In Lemna, the AtPRR1∷luc reporter construct functioned as an evening-specific reporter as was observed in Arabidopsis (Fig. 1, A and D; Table I; Nakamichi et al., 2004). The circadian rhythmicity continued under LL conditions for more than 5 d, whereas it was damped in DD within two cycles (Fig. 1D). This similarity of the clock-controlled promoter behavior between Lemna and Arabidopsis implied that the circadian system for gene expression is likely to be conserved between these plants. Because Lemna and Arabidopsis are monocotyledonous and dicotyledonous plants, respectively, the circadian system appears to be conserved among flowering plants.
Figure 1.
The semitransient bioluminescence reporter monitoring system. A, Schemes of the reporter constructs. Structures of the AtCCA1 and AtPRR1 promoter regions that drove the firefly luciferase gene (LUC) are shown. Both reporters were constructed as translational fusion genes. Black boxes denote exons of coding regions, and +1 denotes the first base of start codon. (Nakamichi et al., 2004). B, Schemes of the effector constructs. The overexpression effector construct (Ox-construct) and RNAi-mediated knockdown construct (RNAi construct) were derived from the pBI221 vector, in which the coding region or the RNAi construct was under the control of the CaMV 35S promoter and the NOS terminator (Miwa et al., 2006). The cDNA regions used for the RNAi constructs and Ox-constructs for each gene are shown in Supplemental Figure S1. For the control experiments, we used a control vector without any insertion (pBI221ΔGUS). C, Rhythmic expression of bioluminescence following the introduction of the AtCCA1∷luc reporter into L. gibba. Plants that were cultured under 12-h-light/12-h-dark conditions were subjected to particle bombardment. They were treated with an additional 12-h-light/12-h-dark entrainment cycle and then were transferred to a bioluminescence monitoring machine under the experimental light conditions. Bioluminescence profiles of the plants in LL (gray circles) or DD (black circles) are shown (Miwa et al., 2006). The time since the last 12-h dark period is indicated. D, Rhythmic expression of bioluminescence following the introduction of the AtPRR1∷luc reporter. Measurements were performed as in C.
Table I.
Summary of circadian traits of AtCCA1∷luc and AtPRR1∷luc reporters in cotransfection assays
Averages of the period lengths, the phases, and the amplitudes ± sd are shown. Amplitudes, period lengths, and phases were estimated using the oscillation fits to sine curves (see “Materials and Methods”). NA, No samples were applicable to the fitting; ox, overexpression construct.
| Effector | Reporters
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
AtCCA1∷luc
|
AtPRR1∷luc
|
|||||||||
| No. Tested | No. Rhythmica | Amplitude | Period | Phaseb | No. Tested | No. Rhythmica | Amplitude | Period | Phasec | |
| Control vector (pBI221ΔGUS) | 12 | 12 | 0.61 ± 0.32 | 25.0 ± 1.4 | 32.3 ± 1.1 | 12 | 12 | 0.20 ± 0.10 | 22.6 ± 0.7 | 29.8 ± 0.6 |
| LgLHYH1-ox | 9 | 9 | 0.10 ± 0.03 | 23.6 ± 2.0 | 30.9 ± 0.9 | 9 | 8 | 0.10 ± 0.01 | 22.3 ± 0.3 | 29.7 ± 0.2 |
| LgLHYH1-RNAi | 9 | 8 | 0.40 ± 0.10 | 22.1 ± 1.1 | 28.7 ± 0.7 | 9 | 8 | 0.12 ± 0.02 | 22.6 ± 0.6 | 27.3 ± 0.6 |
| LgLHYH2-ox | 9 | 0 | NA | NA | NA | 9 | 9 | 0.05 ± 0.02 | 21.7 ± 0.5 | 30.6 ± 0.7 |
| LgLHYH2-RNAi | 9 | 8 | 0.57 ± 0.15 | 25.0 ± 0.5 | 32.5 ± 0.8 | 9 | 9 | 0.19 ± 0.04 | 22.9 ± 0.6 | 29.3 ± 0.8 |
| LgGIH1-ox | 9 | 9 | 0.10 ± 0.03 | 22.6 ± 2.2 | 28.1 ± 1.4 | 9 | 9 | 0.11 ± 0.03 | 22.5 ± 0.6 | 29.3 ± 0.7 |
| LgGIH1-RNAi | 9 | 0 | NA | NA | NA | 9 | 0 | NA | NA | NA |
| LgELF3H1-ox | 9 | 3 | 0.21 ± 0.02 | 32.8 ± 0.9 | 35.8 ± 0.9 | 9 | 0 | NA | NA | NA |
| LgELF3H1-RNAi | 9 | 6 | 0.14 ± 0.06 | 25.7 ± 3.3 | 34.6 ± 3.2 | 9 | 0 | NA | NA | NA |
| LgGIH1 LgELF3H1-double RNAi | 6 | 0 | NA | NA | NA | 6 | 0 | NA | NA | NA |
The sample with a bioluminescence trace properly fitted to a sine curve was counted as a rhythmic sample.
Hours in LL of the second peak of the rhythm.
Hours in LL of the second trough of the rhythm.
Using this reporter system, we functionally analyzed the clock-related homologs from Lemna. Overexpression effector plasmids carrying a clock-related gene under the control of the cauliflower mosaic virus (CaMV) 35S promoter were used for a cotransfection assay in which a reporter construct was introduced together with the effector construct (Fig. 1B; Supplemental Fig. S1). We also used effector constructs for RNAi in cotransfection assays to knockdown the expression of the clock-related homologs (Fig. 1B; Supplemental Fig. S1). LgLHYH1, LgLHYH2, LgGIH1, and LgELF3H1 were subjected to overexpression and knockdown analyses using the morning-specific AtCCA1∷luc reporter and the evening-specific AtPRR1∷luc reporter. The effector constructs did not affect the luciferase activity driven by the constitutively active promoter from ZmUBIQUITIN1 (ZmUBQ1; Supplemental Fig. S2; Miwa et al., 2006).
LgLHYH1 Is a Clock Component That Functions Similarly to Arabidopsis LHY/CCA1
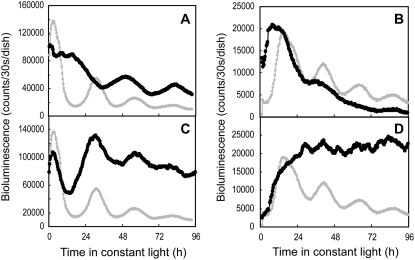
LgLHYH1 is an LHY/CCA1 homolog from L. gibba, and its expression patterns under LD and LL conditions parallel those of LHY/CCA1 (Miwa et al., 2006). Overexpression of this homolog markedly damped the bioluminescent circadian rhythm of the AtCCA1∷luc reporter (Fig. 2, A and C; Table I; Miwa et al., 2006). This phenotype resembled that of LHY/CCA1-overexpressing Arabidopsis plants (Schaffer et al., 1998; Wang and Tobin, 1998). Namely, the overexpressed genes inhibited their own expression and terminated the circadian rhythmicity. Overexpression of LHY/CCA1 in Arabidopsis also reduced the expression level of TOC1/PRR1 (Alabadí et al., 2001). Then, we examined effects of LgLHYH1 overexpression on the AtPRR1∷luc reporter in Lemna plants using the cotransfection assay, which resulted in a low-amplitude bioluminescence rhythm that was phenotypically similar to the results obtained with LHY/CCA1 overexpression in Arabidopsis (Fig. 2, B and D; Table I).
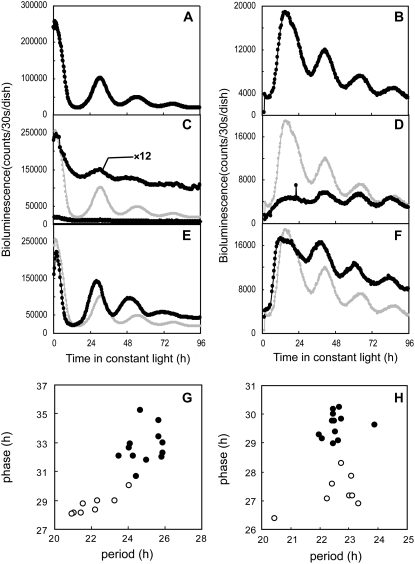
Figure 2.
Effects of overexpression and knockdown of LgLHYH1 on the bioluminescent circadian reporters. Data for the AtCCA1∷luc and AtPRR1∷luc expression patterns in LL conditions are shown in the left and right panels, respectively. The control vector (pBI221ΔGUS; A and B), the overexpression construct (LgLHYH1-ox; C and D), or the RNAi construct (LgLHYH1-RNAi; E and F) was introduced into the plants together with each reporter, and the bioluminescence profiles are shown as black circles. The traces for the control vector are also superimposed on C, D, E, and F (gray circles). Plots with a magnified scale are also shown in C. Measurements were performed as described in Figure 1. Phases and period lengths of each control (black circles) as well as RNAi-knockdown (white circles) sample are plotted in G and H. The x axis represents the period of these rhythms, and the y axis shows the phase of the second peak of AtCCA1∷luc under LL conditions (G) or the second trough of AtPRR1∷luc (H). The cotransfection assays were repeated at least nine times for each reporter. Data are representative of the independent experiments.
Previous studies showed that knockout mutants of either lhy or cca1 in Arabidopsis exhibited short-period circadian rhythms (Green and Tobin, 1999; Mizoguchi et al., 2002). We introduced an LgLHYH1-RNAi construct together with the AtCCA1∷luc reporter into plants. The bioluminescence rhythm showed a short-period phenotype (period length, approximately 22 h), suggesting that the knockdown of endogenous LgLHYH1 expression affected the circadian rhythm of Lemna cells in the same manner observed for LHY/CCA1 in Arabidopsis (Fig. 2, E and G; Table I). The bioluminescence rhythm from the AtPRR1∷luc reporter construct was also affected by cotransfection with the LgLHYH1-RNAi construct; the phase advanced by approximately 2 h compared with that observed for the control construct (Fig. 2, F and H; Table I). The average period length, however, was almost the same as that from the control sample (Table I). Although the effects of RNAi on the AtPRR1∷luc rhythm were unclear, the similarities in the gene expression patterns of Arabidopsis LHY/CCA1 and LgLHYH1 as well as the effects of gene overexpression and gene knockout/knockdown on the circadian rhythms suggest that the genes have similar functions in the clock systems of these plants.
Divergence of the Functions of LgLHYH2 and LgLHYH1
LgLHYH2 is another homolog of Arabidopsis LHY/CCA1; the encoded proteins share six conserved regions in their amino acid sequences (Miwa et al., 2006). The disrupted circadian rhythm of the AtCCA1∷luc reporter activity observed following overexpression of this gene implied that it was involved in the circadian clock (Fig. 3A; Table I; Miwa et al., 2006). We examined the effects of LgLHYH2 overexpression on the rhythmic activity of the AtPRR1∷luc reporter (Fig. 3B); overexpression markedly attenuated the rhythmicity and lowered the bioluminescence activity, indicating that overexpression of LgLHYH2 produced similar effects through the evening-specific promoter and the morning-specific promoter. This phenotype paralleled the phenotype observed in LgLHYH1-overexpressing cells (Fig. 2D).
Figure 3.
Effects of overexpression and knockdown of LgLHYH2 on the bioluminescent circadian reporters. AtCCA1∷luc and AtPRR1∷luc expression patterns under LL conditions are shown in the left and right panels, respectively. The overexpression construct (LgLHYH2-ox; A and B) and RNAi construct (LgLHYH2-RNAi; C and D) were introduced together with each reporter. The bioluminescence traces are shown as black circles. The traces for the control vector are superimposed on the panels (gray circles). Plots with a magnified scale are also shown in A. Phases and period lengths of each control (black circles) as well as RNAi-knockdown (white circles) sample are plotted in E and F. Experimental procedures and annotations are the same as those described in Figure 2. The cotransfection assays were repeated at least nine times for each reporter. Data are representative of the independent experiments.
We then examined effects of LgLHYH2 knockdown using an LgLHYH2-RNAi construct as the effector in the cotransfection assay. Experiments using the AtCCA1∷luc reporter produced peak and trough times during the bioluminescence rhythms that were essentially the same in LgLHYH2-RNAi-expressing cells and control cells (Fig. 3, C and E; Table I). This suggested that the LgLHYH2-RNAi effector construct did not affect the rhythmic activity of this promoter. The LgLHYH2-RNAi effector also did not influence the AtPRR1∷luc reporter (Fig. 3, D and F; Table I). To confirm that this effector worked in the cotransfection assay, we examined effects of the LgLHYH2-RNAi construct on the arrhythmic phenotype induced by the overexpression of this gene. We introduced both the LgLHYH2-RNAi and the LgLHYH2-overexpression constructs into plants together with the AtCCA1∷luc reporter. The RNAi construct completely rescued the arrhythmic phenotype (Supplemental Fig. S3), suggesting that this RNAi effector suppressed the expression of genes with homologous sequences. We also introduced the LgLHYH1-RNAi and LgLHYH2-RNAi effector constructs together in the cotransfection assays to check for functional redundancies. The bioluminescence rhythms of AtCCA1∷luc and AtPRR1∷luc were similar to those observed in the assays using only the LgLHYH1-RNAi effector construct (Supplemental Fig. S4). These results implied that if LgLHYH2 plays a role in the circadian system, its function is different than that of LgLHYH1.
LgGIH1 Is a Pivotal Clock Component in Lemna
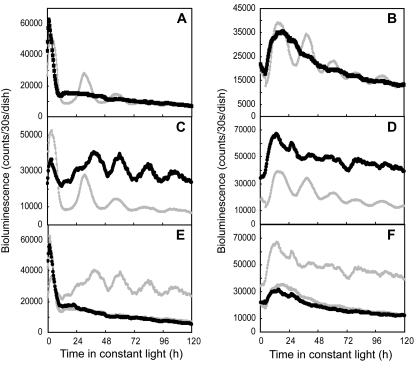
We previously reported that LgGIH1 and Arabidopsis GI showed similar expression rhythms under LD and LL conditions (Miwa et al., 2006). Overexpression of this gene damped the rhythmicity of the AtCCA1∷luc reporter (Fig. 4A; Table I; Miwa et al., 2006). We then examined the effects of LgGIH1 overexpression on the AtPRR1∷luc rhythm. The rhythmicity of the AtPRR1∷luc reporter was less affected than that of AtCCA1∷luc, suggesting that LgGIH1 may play different roles in the regulation of the morning-specific and evening-specific reporters (Fig. 4B; Table I).
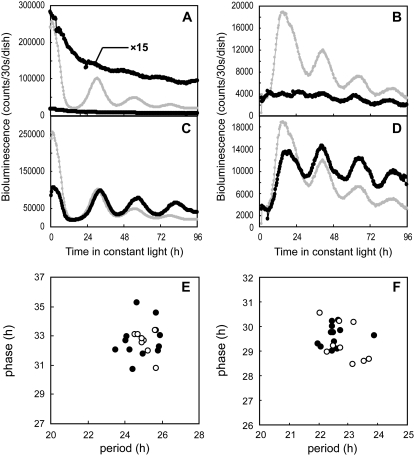
Figure 4.
Aberrant circadian rhythms of the bioluminescent reporters caused by cotransfection with the effector constructs of LgGIH1. AtCCA1∷luc and AtPRR1∷luc expression patterns under LL conditions are shown in the left (A and C) and right panels (B and D), respectively. The overexpression construct (LgGIH1-ox; A and B) and RNAi construct (LgGIH1-RNAi; C and D) were introduced together with each reporter and bioluminescence traces are shown as black circles. The traces for the control vector are superimposed on the panels (gray circles). Plots with a magnified scale are also shown in A. Measurement procedures were the same as those described in Figure 1. The cotransfection assays were repeated at least nine times for each reporter. Data are representative of the independent experiments.
We then examined the effects of LgGIH1 knockdown using an LgGIH1-RNAi effector construct in the cotransfection assay. This treatment abolished the circadian rhythmicity of both reporters (Fig. 4, C and D), which strongly suggested that LgGIH1 is essential for the circadian rhythm under LL conditions. After 12 h of LL, the expression level of AtCCA1∷luc was reduced to the lowest level observed for the control bioluminescence rhythm (Fig. 4C). A previous report demonstrated that a gi mutant in Arabidopsis sustained a robust circadian rhythm under moderate temperature conditions (17°C and 22°C), whereas the rhythm was markedly attenuated at 27°C (Gould et al., 2006). Moreover, the level of CCA1 mRNA was reduced to the lowest levels observed in wild-type plants. Thus, the results from the LgGIH1 knockdown experiments appeared to parallel the phenotypes observed in the Arabidopsis gi mutant at higher temperatures. Then, we tested whether or not the arrhythmic phenotype induced by LgGIH1 knockdown in Lemna plants was temperature dependent. In experiments performed at a lower temperature (20°C), LgGIH1 knockdown resulted in an arrhythmic phenotype as was observed with our standard conditions at 25°C (data not shown). This suggested that LgGIH1 is likely to play an essential role in the circadian system irrespective of the temperature. Although the severity of the effects of knockdown/knockout and overexpression varied between Lemna and Arabidopsis, LgGIH1 and Arabidopsis GI are likely to have similar functions in the respective clock systems of these plants.
Involvement of LgELF3H1 in the Circadian Clock
We next examined the effects of LgELF3H1 overexpression on the AtCCA1∷luc reporter. This effector construct damped the rhythmicity and lengthened the period to approximately 33 h (Fig. 5A; Table I). This phenotype was similar to that observed in Arabidopsis overexpressing ELF3 (Covington et al., 2001). We then examined the effects of LgELF3H1 overexpression on the AtPRR1∷luc reporter. This treatment markedly damped the bioluminescence rhythm (Fig. 5B). Therefore, the overexpression of LgELF3H1 disrupted the circadian regulation of both the morning-specific and evening-specific promoters.
Figure 5.
Aberrant circadian rhythms of the bioluminescent reporters caused by the effector constructs of LgELF3H1. AtCCA1∷luc and AtPRR1∷luc expression patterns under LL conditions are shown in the left (A and C) and right panels (B and D), respectively. The overexpression construct (LgELF3H1-ox; A and B) and RNAi construct (LgELF3H1-RNAi; C and D) were introduced together with each reporter and bioluminescence traces are shown as black circles. The traces for the control vector are superimposed on the panels (gray circles). Measurement procedures were the same as those described in Figure 1. The cotransfection assays were repeated at least nine times for each reporter. Data are representative of the independent experiments.
We also examined effects of LgELF3H1 knockdown on the AtCCA1∷luc reporter. Whereas the bioluminescence level of this reporter in the control experiment gradually decreased under LL conditions, treatment with the RNAi construct maintained the bioluminescence at approximately the level of the first peak of the rhythm (Fig. 5C). Although the bioluminescence level was affected by this construct, the rhythmicity was sustained with an approximately wild-type period length (Table I). This contrasted with the phenotypes observed in the Arabidopsis elf3 mutant, in which the mRNA expression level of LHY decreased without circadian rhythmicity (Schaffer et al., 1998). On the other hand, the circadian rhythm of the AtPRR1∷luc reporter was severely disrupted by the knockdown of LgELF3H1. Similar to the control bioluminescence trace, the bioluminescence level rapidly increased for approximately 12 h after the sample was exposed to light, which was followed by a more moderate increase for the next approximately 12 h (Fig. 5D). After 24 h under LL conditions, the bioluminescence remained at a high level with small fluctuations. In the Arabidopsis elf3 mutant, the expression of TOC1/PRR1 remains high without any apparent rhythmicity (Alabadí et al., 2001), suggesting that the role of LgELF3H1 in the circadian clock is similar to that of ELF3 in Arabidopsis, although knockdown/knockout of these two genes produced different effects on morning-specific gene expression.
Double Knockdown of LgGIH1 and LgELF3H1
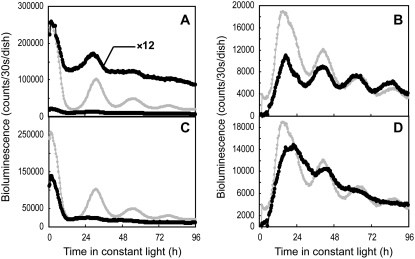
The effects of RNAi of LgGIH1 and LgELF3H1 on the reporter expression levels were markedly different (Figs. 4, C and D, and 5, C and D). Knockdown of LgGIH1 decreased the bioluminescence levels produced by both AtCCA1∷luc to the lowest levels observed during the wild-type rhythm, whereas knockdown of LgELF3H1 increased and maintained the signals at levels equivalent to the peak wild-type level (Fig. 6, A and C). Knockdown of LgELF3H1 also increased the signal of AtPRR1∷luc, but that of LgGIH1 did not (Fig. 6, B and D). To dissect the genetic relationship between these two genes, the knockdown effector constructs targeting LgGIH1 and LgELF3H1 were both cotransfected with either the AtCCA1∷luc (Fig. 6E) or AtPRR1∷luc (Fig. 6F) reporter. For both reporters, the concurrent knockdown of both genes decreased the bioluminescence levels, and the traces were essentially the same as those observed for the knockdown of LgGIH1 alone. This suggested that LgGIH1 knockdown is epistatic to LgELF3H1 and that LgGIH1 is likely to function in circadian gene regulation downstream of LgELF3H1. It was genetically demonstrated that the late flowering phenotype induced by a gi mutation was epistatic to the early flowering phenotypes induced by an elf3 mutation (Chou and Yang, 1999). Thus, the structure of the genetic relationship between GI and ELF3 appears to be conserved between Lemna and Arabidopsis.
Figure 6.
Double RNAi experiments for LgGIH1 and LgELF3H1. AtCCA1∷luc and AtPRR1∷luc expression patterns under LL conditions are shown in the left (A, C, and E) and right panels (B, D, and F), respectively. The LgGIH1-RNAi (A and B) or LgELF3H1-RNAi (C and D) construct was introduced together with each reporter and the bioluminescence traces are shown as black symbols. The traces for the control vector are superimposed on the panels (gray circles in A, B, C, and D). Both the LgGIH1-RNAi and LgELF3H1-RNAi constructs were introduced together with each reporter and the bioluminescence traces are shown as solid circles (E and F). The traces for the LgGIH1-RNAi construct alone or the LgELF3H1-RNAi construct alone are superimposed on the panels (gray symbols in E and F, respectively). Measurement procedures were the same as those described in Figure 1. The cotransfection assays were repeated at least six times for each reporter. Data are representative of the independent experiments.
DISCUSSION
In this report, we clearly demonstrated functional similarities between clock-related gene homologs from L. gibba and Arabidopsis (Table II). Our previous study showed that mRNA accumulation rhythms of LgLHYH1, LgGIH1, and LgELF3H1 were similar to those of their Arabidopsis counterparts (Miwa et al., 2006). Moreover, we have shown here that the effects of the loss-of-function of these Lemna genes on the circadian system were comparable to those observed for the corresponding Arabidopsis mutants. These similarities provide conclusive evidence that the genes are orthologs, which have similar functions as clock components. Therefore, the genetic structures of the circadian oscillators are likely conserved between monocotyledon and dicotyledon plant species; i.e. the basic clock components of circadian systems are likely conserved among flowering plants. Recently, clock-related homologs with essentially conserved expression profiles have been isolated from several plant species (Boxall et al., 2005; Ramos et al., 2005; Murakami et al., 2007). Our studies using Lemna strongly support the idea that those clock-related homologs have conserved functions in the various circadian oscillators.
Table II.
Summary of effects of Lemna clock-related genes on the circadian rhythms of two reporters
| Effector | Reporters
|
|
|---|---|---|
| AtCCA1∷luc | AtPRR1∷luc | |
| LgLHYH1-ox | Low amplitude, short period | Low amplitude |
| LgLHYH1-RNAi | Short period | Phase advance |
| LgLHYH2-ox | Arrhythmic, low level | Low amplitude |
| LgLHYH2-RNAi | Normal | Normal |
| LgGIH1-ox | Phase advance, low level | Normal |
| LgGIH1-RNAi | Arrhythmic, low level | Arrhythmic |
| LgELF3H1-ox | Low amplitude, long period | Arrhythmic |
| LgELF3H1-RNAi | Low amplitude | Arrhythmic |
| LgGIH1 LgELF3H1-double RNAi | Arrhythmic, low level | Arrhythmic |
Whereas basic clock components are conserved, their precise roles in the circadian oscillator appear to have slightly diverged. For example, the GI gene appears to have different roles in Lemna and Arabidopsis. RNAi of LgGIH1 markedly attenuated the rhythmicity of two different circadian reporters, whereas a null mutation in Arabidopsis GI resulted in a temperature-dependent phenotype that produced disordered circadian rhythms at elevated temperatures (Gould et al., 2006). We have not observed the recovery of circadian rhythmicity in cells expressing the LgGIH1-RNAi construct under various temperature conditions (data not shown). This suggests that unlike Arabidopsis GI, LgGIH1 is essential for the circadian oscillation. As predicted in a mathematical model, other genes may compensate for the function of GI in the circadian clock system of Arabidopsis; these genes may not be present in Lemna (Locke et al., 2006). Overexpression of LgGIH1 resulted in a severe damping of the morning-specific expression rhythm of AtCCA1∷luc, whereas no significant effects on the evening-specific AtPRR1∷luc reporter were observed (Fig. 4, A and B; Table I). This phenomenon is likely to parallel the low expression level of the LgLHYH1 morning clock gene, because RNAi of LgLHYH1 did not affect the circadian rhythmicity of the evening-specific AtPRR1∷luc reporter. In contrast, overexpression of GI in Arabidopsis resulted in a damping of the evening-specific expression rhythm of CCR2 but not of the morning-specific expression rhythm of LHY (Mizoguchi et al., 2005). This difference in the effects of GI overexpression on rhythmic gene expression implies a divergence of the regulatory machineries for circadian gene expression in these two species.
Lemna have at least two LHY homologues (LgLHYH1 and LgLHYH2), which show high sequence similarities to Arabidopsis LHY/CCA1 and its rice homolog OsCCA1 (Miwa et al., 2006). Although both Lemna homologs show circadian gene expression rhythms, the phase of the LgLHYH2 rhythm is delayed compared to the LgLHYH1 rhythm and also to those of LHY/CCA1 homologs in other species (Miwa et al., 2006). Because LgLHYH1 and LgLHYH2 show almost equivalent degrees of homology to LHY/CCA1 homologs in other species, the functional diversity between the two Lemna homologs was intriguing. Our RNAi assays to assess their functions in circadian rhythms suggested that only LgLHYH1 is involved in the generation of circadian oscillations. RNAi-mediated knockdown of LgLHYH1 produced a short-period length for the AtCCA1∷luc reporter, which was comparable to the phenotypes observed in cca1 and lhy Arabidopsis mutants. The period length of the AtPRR1∷luc rhythm, however, was not affected by the LgLHYH1-RNAi construct in Lemna plants. Different effects on the period lengths of these gene expression profiles mediated by morning- and evening-specific promoters were not observed in the Arabidopsis mutants (Mizoguchi et al., 2002). Thus, the circadian system in Lemna plants may contain morning- and evening-specific oscillations that are more weakly coupled than those of Arabidopsis. Although the physiological functions of LgLHYH2 are unclear, its overexpression represses circadian gene expression and suspends circadian oscillations as was observed for LgLHYH1. Because both these proteins are presumably transcription factors with similar MYB-type DNA-binding regions, they may share downstream target genes. The expression rhythm of LgLHYH2 lagged the expression of LgLHYH1 by approximately 4 h, which may be important for the functional divergence of these clock components. Knockdown of LgLHYH1 shortened the period length and altered the phase, although the effects were not as severe as those observed in the Arabidopsis lhy/cca1 double loss-of-function mutant. Another gene may compensate for the knockdown of LgLHYH1, although we have not isolated any additional LHY/CCA1 homologs from Lemna. It should be noted that rice, a model monocotyledonous plant, has only one LHY/CCA1 homolog in its genome (Murakami et al., 2007). Because Lemna is also a monocotyledon, LgLHYH1 may be the only functionally conserved ortholog in this plant genus. Overexpression analysis with LgGIH1 seemed to support this idea. The expression of the morning-specific promoter of AtCCA1 was dramatically repressed, suggesting that other morning-specific genes, such as LgLHYH1, and any potential LgLHYH1 homolog were also repressed. Even under such conditions, the circadian expression of the evening-specific promoter of AtPRR1 was robust. Therefore, normal gene expression of any potential LgLHYH1 homolog would not be required for the rhythmic expression of evening-specific genes. Thus, it is possible that LgLHYH1 is a functionally relevant ortholog of OsCCA1, and OsCCA1 and LgLHYH1 may have similar functions in the respective circadian systems.
LgELF3H1 knockdown resulted in a low-amplitude rhythmic expression of AtCCA1 and AtPRR1 under LL conditions (Fig. 5, C and D; Table I). This appears to parallel the arrhythmic phenotype induced by the elf3 mutation in Arabidopsis (Hicks et al., 1996; Schaffer et al., 1998; Alabadí et al., 2001). Thus, it is likely that the LgELF3H1 gene plays an important role in the circadian system as has been shown for Arabidopsis ELF3. In the rice genome, there are two ELF3-related genes, of which functions in the circadian system are not revealed (Murakami et al., 2007). Our functional analysis using L. gibba clearly indicated that the ELF3 homolog plays an important role in the circadian oscillations in monocotyledonous plants. Therefore, it is likely that one or both of the ELF3 homologs are involved in circadian rhythms in rice.
The functional conservation of ELF3 between L. gibba and Arabidopsis is also supported by overexpression analysis of these genes, which lengthened the period of the circadian rhythms in both plants (Fig. 5A; Covington et al., 2001). Our data also showed that the genetic relationship between ELF3 and GI is conserved in L. gibba and Arabidopsis, suggesting that the genetic networks involving these genes are also conserved (Fig. 6; Chou and Yang, 1999). Despite these broad similarities in the roles of ELF3, the effects of knockdown/knockout on the morning-specific gene expression were different in these plants. Whereas the knockdown of LgELF3H1 resulted in a higher level of AtCCA1-promoter activity, the elf3 mutation inhibited the accumulation of LHY mRNA (Fig. 5C; Schaffer et al., 1998). This contrasts with the difference in the effects of GI overexpression in these species; LgGIH1 overexpression severely lowered AtCCA1-promoter activity in L. gibba, whereas the promoter was not affected by GI overexpression in Arabidopsis (Fig. 4A; Mizoguchi et al., 2005). These phenomena imply that the genetic frameworks involving ELF3 and GI that underlie the circadian oscillations are conserved between L. gibba and Arabidopsis, but the precise roles of these genes have diverged, probably due to modifications of their molecular functions and/or networks.
A semitransient expression system using a particle bombardment method in Lemna plants has allowed us to functionally analyze clock-related genes. Moreover, a number of characteristics of Lemna plants have facilitated the use of this experimental system. The flat, tiny body of this plant allows us to keep the whole plant in small dishes under normal growth conditions throughout the experiments. The flat, smooth surface of the frond is suitable for the particle bombardment method. In these procedures, exogenous genes are introduced into mature epidermal cells. The circadian reporter activity in a single type of cell allows us to focus on the rhythmicity without variables introduced by different tissues or developmental stages (Fukuda et al., 2007). Together with these advantages, the performance of the semitransient reporter expression system is suitable for large-scale analyses of gene functions. Hence, our reporter-effector experimental system using Lemna plants can be used as a model for plant circadian systems and should allow the dynamics of the associated intracellular signal transduction systems to be examined.
It should be noted that the semitransient expression system has technical limitation when effector constructs are applied to it. The efficiency of an effector is not directly accessible in our present semitransient expression system because only tens to hundreds of epidermal cells are transfected in our experiments (data not shown). In other words, the expression levels of overexpressed or knockdown target genes in the transfected cells are unknown. This could cause difficulty in interpreting effects of overexpression or knockdown/knockout, especially when they are different between our analysis of Lemna and that of other plant species. For example, the disorder of the circadian rhythms by the LgELF3H1 overexpression appeared much more severe than that of an Arabidopsis ELF3 overexpression transgenic plant (Fig. 5; Covington et al., 2001). This might be attributed to the difference of their expression levels. As well as the expression levels of effectors, tissue specificity of circadian reporter could cause the difference of circadian behaviors between transiently transfected cells and transgenic plants. The circadian reporters expressing in different tissues show distinct features of circadian rhythms between them (Michael et al., 2003). The semitransient expression system using particle bombardment only allows expression of the reporter in the mature epidermal cells, while the luciferase reporter under the chlorophyll a/b binding protein2 promoter that is well used in Arabidopsis as circadian reporter is predominantly expressed in mesophyll cells.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Lemna gibba G3 has been maintained in our laboratory for >40 years by vegetative reproduction. L. gibba plants were kept in M medium with 1% (w/v) Suc under LL conditions (Hillman, 1961b). Approximately 10 colonies were picked from cultures and grown under 12-h-light/12-h-dark conditions for 3 weeks to use in the bombardment experiments. The growth temperature was maintained at 25°C ± 1°C and the light intensity supplied by fluorescent lamps (FLR40SW/M/36 or FL20SSW18; Mitshubishi/Osram) was approximately 25 μE m−2 s−1. Colonies were grown in 100 mL of medium in 200-mL Erlenmeyer flasks plugged with cotton. New stock cultures were made every week, and well-grown plants were used for the experiments.
Reporter and Effector Constructs
The reporter constructs pSP1-CCA1∷LUC-B and pSP1-APRR1∷LUC were kind gifts from Dr. Mizuno (Nakamichi et al., 2004). The pSP1-based ZmUBQ1 promoter-luc+ construct was described previously (Miwa et al., 2006). For overexpression constructs, coding regions for clock-related genes were amplified using PCRs and cloned into pBI221 (CLONTECH; Supplemental Fig. S1; Supplemental Table S1; Miwa et al., 2006). The pBI221 plasmid contains the GUS gene under the control of the CaMV 35S promoter; the GUS region was replaced with the coding region of the gene to be overexpressed.
RNAi effector constructs were constructed using a MultiSite Gateway Three-Fragment Vector Construction kit (Invitrogen; Supplemental Fig. S5). A fragment of each clock-related gene was amplified in a PCR using two sets of primers. The amplified regions, the direction of the RNAi region, and the primer sequences are shown in Supplemental Figure S1 and Supplemental Table S1. 5′-f and 3′-rv primers contained the same target sequence at an end of the amplicon for each target, and 5′-rv and 3′-f primers contained the same target at the other end of the amplicon. 5′-f, 5′-rv, 3′-f, and 3′-rv primers contained attB4, attB1, attB2, and attB3 sequences next to the target sequences, respectively. An amplicon produced with 5′-f and 5′-rv primers was cloned into the pDONR P4-P1R vector to make a pENTR-5′ vector. An amplicon produced with 3′-f and 3′-rv primers was cloned into the pDONR P2R-P3 to make a pENTR-3′ vector. Between the attL1 and attL2 regions, the pENTR-GUS vector contains a GUS intron sequence (Ohta et al., 1990) and a spectinomycin-resistant gene (Omega fragment) inside the intron. This drug-resistant gene was used in the selection process after the LR-plus reaction to increase the efficiency. These three vectors were integrated into a pBI221-based destination vector (pBI221+DEST) using the LR-plus reaction. Between the CaMV 35S promoter and the NOS terminator of pBI221, pBI221+DEST contains recombination sequences used during the LR reaction. The RNAi expression vector expresses the RNAi region under the control of the CaMV 35S promoter. As a control for the effector construct, the control vector (pBI221ΔGUS) was used.
Particle Bombardment
pSP1-CCA1∷LUC-B and pSP1-APRR1∷LUC were used as bioluminescent reporter constructs (Nakamichi et al., 2004). These plasmid vectors were introduced using particle bombardment. A 25-μL aliquot of prewashed gold particle suspension (1-μm diameter; Bio-Rad) in 50% glycerol (60 mg mL−1) was mixed with the plasmid DNA mixture, in which a 3-μg aliquot of the effector plasmid and 1 μg of pBI221 were mixed with 6 μL of reporter plasmid DNA solution (0.5 mg mL−1), 25 μL of CaCl2 (2.5 m), and 1 μL of spermidine (1 m). For the LgGIH1-LgELF3H1 double RNAi knockdown, the gold particle suspension was mixed with 3 μL of reporter plasmid DNA solution, 2 μL of the LgGIH1-RNAi plasmid DNA solution (1 mg mL−1), and 2 μL of the LgELF3H1-RNAi plasmid DNA solution (1 mg mL−1). After vortexing for 3 min, the tube was briefly centrifuged. The supernatant was discarded, 200 μL of 70% ethanol was added, and the samples were mixed well. The suspension was briefly centrifuged and the supernatant was discarded. The DNA-coated particles were washed again with 100% ethanol and resuspended in 30 μL 100% ethanol. A helium gun device (GIE-III IDERA; Tanaka) was used for particle bombardment according to the manufacturer's instructions (vacuum, 800 hectoPa; helium gauge pressure, 5.0 hectoPa). Approximately 10 Lemna colonies were set on a 35-mm polystyrene dish (Asahi Techno Glass) and covered with a small piece of plastic mesh. The dish was set underneath the muzzle of the gun, and 8 μL of the DNA-coated particle suspension was fired into the sample. After the bombardment, 3 mL of medium containing firefly luciferin (1 mm potassium salt; Biosynth) was added to the dish. The samples were cultured under light-dark entrainment conditions for at least 1 d before bioluminescence measurements began.
Bioluminescence Monitoring
Monitoring the bioluminescence of Lemna plants was basically done as described previously (Miwa et al., 2006). The luminescence dish-monitoring system used photomultiplier tubes (R329P; Hamamatsu Photonics K.K.) for bioluminescence detection. To reduce the fluorescence signals from chlorophyll, a short-pass filter (SVO630; Asahi Spectra) was set at the detection site of the photomultiplier tubes. Each dish was subjected to 30-s measurements of bioluminescence every 30 min.
Time series of bioluminescence data were fitted with sine curves using nonlinear least-squares fitting analysis. The following fitting function was used:
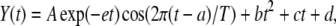 |
where T is circadian period, A is initial amplitude value, e is a coefficient for exponential decay of amplitude, a is the phase offset, and bt2 + ct + d is the trend component for quadratic function. Time series data from 24 to 72 h were applied to the fitting for AtCCA1∷luc, and those from 24 to 96 h for AtPRR1∷luc. Optimal values for parameters (A, T, a, b, c, d, and e) were estimated using the Gauss-Newton algorithm and the open-source statistics software R (version 2.4.1). The representative amplitude for each time series was calculated as A exp(−et)/(bt2 + ct + d) at t = 36 h.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AB210848 to AB210851.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Structure of the effector constructs.
Supplemental Figure S2. Effects of the knockdown of the four Lemna clock-related genes and of LgELF3H1 overexpression on ZmUBQ-promoter activity.
Supplemental Figure S3. Suppression of the effects of LgLHYH2 overexpression by LgLHYH2-RNAi knockdown.
Supplemental Figure S4. Double RNAi experiments for LgLHYH1 and LgLHYH2.
Supplemental Figure S5. Procedures for the construction of RNAi vectors using MultiSite Gateway technology.
Supplemental Table S1. Primer sequences for the RNAi constructs.
Supplementary Material
Acknowledgments
We thank Drs. Norihito Nakamichi and Takeshi Mizuno for their generous gifts of plasmids.
This work was supported by the Ministry of Education, Culture, Sports, Science and Technology of Japan (Grants-in-Aid nos. 15GS0308 and COE 13CE2005 to T.K. and T.O. and nos. 15031215 and 17370088 to T.O.) and by JSPS (fellowship for young scientists no. 1500948 to K.M.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Tokitaka Oyama (oyama@bio.nagoya-u.ac.jp).
The online version of this article contains Web-only data.
References
- Alabadí D, Oyama T, Yanovsky MJ, Harmon FG, Más P, Kay SA (2001) Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293 880–883 [DOI] [PubMed] [Google Scholar]
- Boxall SF, Foster JM, Bohnert HJ, Cushman JC, Nimmo HG, Hartwell J (2005) Conservation and divergence of circadian clock operation in a stress-inducible Crassulacean acid metabolism species reveals clock compensation against stress. Plant Physiol 137 969–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou ML, Yang CH (1999) Late-flowering genes interact with early-flowering genes to regulate flowering time in Arabidopsis thaliana. Plant Cell Physiol 40 702–708 [DOI] [PubMed] [Google Scholar]
- Covington MF, Panda S, Liu XL, Strayer CA, Wagner DR, Kay SA (2001) ELF3 modulates resetting of the circadian clock in Arabidopsis. Plant Cell 13 1305–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle MR, Davis SJ, Bastow RM, McWatters HG, Kozma-Bognár L, Nagy F, Millar AJ, Amasino RM (2002) The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 419 74–77 [DOI] [PubMed] [Google Scholar]
- Fukuda H, Nakamichi N, Hisatsune M, Murase H, Mizuno T (2007) Synchronization of plant circadian oscillators with phase delay effect of the vein network. Phys Rev Lett 99 098102. [DOI] [PubMed] [Google Scholar]
- Gould PD, Locke JC, Larue C, Southern MM, Davis SJ, Hanano S, Moyle R, Milich R, Putterill J, Millar AJ, et al (2006) The molecular basis of temperature compensation in the Arabidopsis circadian clock. Plant Cell 18 1177–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RM, Tobin EM (1999) Loss of circadian clock-associated protein 1 in Arabidopsis results in altered clock-regulated gene expression. Proc Natl Acad Sci USA 96 4176–4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer SL, Kay SA (2005) Positive and negative factors confer phase-specific circadian regulation of transcription in Arabidopsis. Plant Cell 17 1926–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen SP, Schultz TF, Pruneda-Paz JL, Borevitz JO, Ecker JR, Kay SA (2005) LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc Natl Acad Sci USA 102 10387–10392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht V, Knowles CL, Vander Schoor JK, Liew LC, Jones SE, Lambert MJ, Weller JL (2007) Pea LATE BLOOMER1 is a GIGANTEA ortholog with roles in photoperiodic flowering, deetiolation, and transcriptional regulation of circadian clock gene homologs. Plant Physiol 144 648–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks KA, Albertson TM, Wagner DR (2001) EARLY FLOWERING3 encodes a novel protein that regulates circadian clock function and flowering in Arabidopsis. Plant Cell 13 1281–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks KA, Millar AJ, Carré IA, Somers DE, Staume M, Straume M, Meeks-Wagner DR, Kay SA (1996) Conditional circadian dysfunction of the Arabidopsis early-flowering 3 mutant. Science 274 790–792 [DOI] [PubMed] [Google Scholar]
- Hillman WS (1961. a) The Lemnaceae, or duckweeds. A review of the descriptive and experimental literature. Bot Rev 21 221–287 [Google Scholar]
- Hillman WS (1961. b) Experimental control of flowering in Lemna. III. A relationship between medium composition and the opposite photoperiodic responses of L. perpusilla 6746 and L. gibba G3. Am J Bot 48 413–419 [Google Scholar]
- Izawa T, Oikawa T, Sugiyama N, Tanisaka T, Yano M, Shimamoto K (2002) Phytochrome mediates the external light signal to repress FT orthologs in photoperiodic flowering of rice. Genes Dev 16 2006–2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XL, Covington MF, Fankhauser C, Chory J, Wagner DR (2001) ELF3 encodes a circadian clock-regulated nuclear protein that functions in an Arabidopsis PHYB signal transduction pathway. Plant Cell 13 1293–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke JC, Kozma-Bognár L, Gould PD, Fehér B, Kevel É, Nagy F, Turner MS, Hall A, Millar AJ (2006) Experimental validation of a predicted feedback loop in the multi-oscillator clock of Arabidopsis thaliana. Mol Syst Biol 2 59 [DOI] [PMC free article] [PubMed]
- Makino S, Kiba T, Imamura A, Hanaki N, Nakamura A, Suzuki T, Taniguchi M, Ueguchi C, Sugiyama T, Mizuno T (2000) Genes encoding pseudo-response regulators: insight into His-to-Asp phosphorelay and circadian rhythm in Arabidopsis thaliana. Plant Cell Physiol 41 791–803 [DOI] [PubMed] [Google Scholar]
- Más P (2005) Circadian clock signaling in Arabidopsis thaliana: from gene expression to physiology and development. Int J Dev Biol 49 491–500 [DOI] [PubMed] [Google Scholar]
- Matsushika A, Makino S, Kojima M, Mizuno T (2000) Circadian waves of expression of the APRR1/TOC1 family of pseudo response regulators in Arabidopsis thaliana: insight into the plant circadian clock. Plant Cell Physiol 41 1002–1012 [DOI] [PubMed] [Google Scholar]
- McWatters HG, Kolmos E, Hall A, Doyle MR, Amasino RM, Gyula P, Nagy F, Millar AJ, Davis SJ (2007) ELF4 is required for oscillatory properties of the circadian clock. Plant Physiol 144 391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael TP, Salomé PA, McClung CR (2003) Two Arabidopsis circadian oscillators can be distinguished by differential temperature sensitivity. Proc Natl Acad Sci USA 100 6878–6883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa K, Serikawa M, Suzuki S, Kondo T, Oyama T (2006) Conserved expression profiles of circadian clock-related genes in two Lemna species showing long-day and short-day photoperiodic flowering responses. Plant Cell Physiol 47 601–612 [DOI] [PubMed] [Google Scholar]
- Mizoguchi T, Wheatley K, Hanzawa Y, Wright L, Mizoguchi M, Song H-R, Carré IA, Coupland G (2002) LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev Cell 2 629–641 [DOI] [PubMed] [Google Scholar]
- Mizoguchi T, Wright L, Fujiwara S, Cremer F, Lee K, Onouchi H, Mouradov A, Fowler S, Kamada H, Putterill J, et al (2005) Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. Plant Cell 17 2255–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T, Nakamichi N (2005) Pseudo-response regulators (PRRs) or True oscillator components (TOCs). Plant Cell Physiol 46 677–685 [DOI] [PubMed] [Google Scholar]
- Murakami M, Ashikari M, Miura K, Yamashino T, Mizuno T (2003) The evolutionarily conserved OsPRR quintet: rice pseudo-response regulators implicated in circadian rhythm. Plant Cell Physiol 44 1229–1236 [DOI] [PubMed] [Google Scholar]
- Murakami M, Tago Y, Yamashino T, Mizuno T (2007) Comparative overviews of clock-associated genes of Arabidopsis thaliana and Oryza sativa. Plant Cell Physiol 48 110–121 [DOI] [PubMed] [Google Scholar]
- Nakamichi N, Ito S, Oyama T, Yamashino T, Kondo T, Mizuno T (2004) Characterization of plant circadian rhythms by employing Arabidopsis cultured cells with bioluminescence reporters. Plant Cell Physiol 45 57–67 [DOI] [PubMed] [Google Scholar]
- Ohta S, Mita S, Hattori T, Nakamura K (1990) Construction and expression in tobacco of a β-glucuronidase (GUS) reporter gene containing an intron within the coding sequence. Plant Cell Physiol 31 805–813 [Google Scholar]
- Onai K, Ishiura M (2005) PHYTOCLOCK 1 encoding a novel GARP protein essential for the Arabidopsis circadian clock. Genes Cells 10 963–972 [DOI] [PubMed] [Google Scholar]
- Park DH, Somers DE, Kim YS, Choy YH, Lim HK, Soh MS, Kim HJ, Kay SA, Nam HG (1999) Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285 1579–1582 [DOI] [PubMed] [Google Scholar]
- Ramos A, Pérez-Solís E, Ibáñez C, Casado R, Collada C, Gómez L, Aragoncillo C, Allona I (2005) Winter disruption of the circadian clock in chestnut. Proc Natl Acad Sci USA 102 7037–7042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carré IA, Coupland G (1998) The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93 1219–1229 [DOI] [PubMed] [Google Scholar]
- Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Más P, Panda S, Kreps JA, Kay SA (2000) Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289 768–771 [DOI] [PubMed] [Google Scholar]
- Sweeney BM (1987) Rhythmic Phenomena in Plants, Ed 2. Academic Press, San Diego
- Wang ZY, Tobin EM (1998) Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93 1207–1217 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.