Abstract
Iron is an essential micronutrient but is toxic if accumulated at high levels. Thus, iron uptake and distribution in plants are controlled by precise regulatory mechanisms. IRON-REGULATED TRANSPORTER1 (IRT1) is the major high affinity iron transporter responsible for iron uptake from the soil in Arabidopsis (Arabidopsis thaliana). Previously, we showed that IRT1 is subject to posttranscriptional regulation; when expressed from the constitutive cauliflower mosaic virus 35S promoter, IRT1 protein accumulates only in iron-deficient roots. IRT1 contains an intracellular loop that may be critical for posttranslational regulation by metals. Of particular interest are a histidine (His) motif (HGHGHGH) that might bind metals and two lysine residues that could serve as attachment sites for ubiquitin. We constructed a set of mutant IRT1 alleles: IRT1H154Q, IRT1H156Q, IRT1H158Q, IRT1H160Q, IRT14HQ (quadruple His mutant), IRT1K146R, IRT1K171R, and a double mutant (IRT1K146R,K171R). Mutation of the His or lysine residues did not eliminate the ability of IRT1 to transport iron or zinc. Expression of each of the IRT1 variants and an IRT1intact construct in plants from the 35S promoter revealed that either K146 or K171 is required for iron-induced protein turnover, and 35S-IRT1K146R,K171R plants contain higher levels of iron as compared to 35S-IRT1 and wild type. Furthermore, accumulation of metals in 35S-IRT1K146R,K171R plants was not associated with an increase in ferric chelate reductase activity; this result indicates that, at least under conditions when iron is abundant, reduction of ferric iron may not be the rate-limiting step in iron uptake by strategy I plants such as Arabidopsis.
Iron is an essential element for virtually all organisms, as it is a required cofactor for a number of enzymes that function in respiration and photosynthesis. Studies in many different organisms have shown that iron deficiency induces a carefully regulated set of responses that function to increase the amount of iron available for metabolism. These responses can include increased uptake of iron from the environment and release of intracellular iron stores. In plants, like other organisms, it is known that several components of the iron uptake machinery are quickly up-regulated when plants sense the onset of iron limitation (Curie et al., 2001; Connolly et al., 2002, 2003; Vert et al., 2003; Colangelo and Guerinot, 2004). Iron deficiency is a major problem in agriculture because iron, like nitrogen and phosphorus, often limits plant growth.
It is also clear that plants, like other organisms, must maintain cellular iron levels within a relatively small range. When iron accumulates inappropriately within cells, it can interact with hydrogen peroxide to form hydroxyl radicals via the Fenton reaction; hydroxyl radicals may then cause significant damage to lipids, DNA, and proteins (Halliwell and Gutteridge, 1992). It is therefore crucial for cells to maintain iron homeostasis, such that iron levels are adequate for metabolism yet do not rise inappropriately and contribute to oxidative stress. In recent years, studies in Arabidopsis (Arabidopsis thaliana) have suggested that complex regulatory circuits control the iron uptake machinery to ensure maintenance of iron homeostasis (Connolly et al., 2002, 2003; Vert et al., 2003; Colangelo and Guerinot, 2004; Jakoby et al., 2004; Yuan et al., 2005).
All plants except the grasses respond to iron limitation by induction of a set of three major activities in roots, which are collectively termed the strategy I response (Römheld, 1987). A plasma membrane H+-ATPase pumps protons into the rhizosphere to solubilize ferric iron. Additionally, a plasma membrane ferric chelate reductase enzyme serves to convert ferric iron to ferrous iron at the root surface; the Arabidopsis FERRIC REDUCTASE OXIDASE2 (FRO2) gene is known to encode the low iron-inducible root surface ferric chelate reductase (Robinson et al., 1999). Finally, an iron transporter moves ferrous iron across the plasma membrane of root epidermal cells. The Arabidopsis IRON-REGULATED TRANSPORTER1 (IRT1) gene encodes the primary high affinity ferrous iron transporter necessary for iron uptake from the soil (Eide et al., 1996; Henriques et al., 2002; Varotto et al., 2002; Vert et al., 2002).
Studies have shown that expression of the FRO2 and IRT1 genes is controlled very precisely by metal availability. In fact, FRO2 and IRT1 transcript levels coordinately rise in roots following the imposition of iron deficiency and coordinately decline following iron resupply (Connolly et al., 2003; Vert et al., 2003). Studies have shown that root iron deficiency responses are controlled both by local signals and long-distance signals from the shoot (Grusak and Pezeshgi, 1996; Schmidt et al., 1996; Schikora and Schmidt, 2001; Vert et al., 2003). However, the shoot-derived signal has not yet been identified. Finally, recent studies showed that Arabidopsis FIT (FER-like iron deficiency-induced transcription factor) is necessary for the induction of root iron deficiency responses under iron limitation (Colangelo and Guerinot, 2004; Jakoby et al., 2004; Yuan et al., 2005; Bauer et al., 2007).
In addition to metal regulation of transcript accumulation, IRT1 and FRO2 are subject to posttranscriptional regulation by iron. Overexpression of IRT1 in transgenic plants showed that while IRT1 transcript levels accumulate to high levels in roots and shoots of both iron-sufficient and iron-deficient plants, IRT1 protein only accumulates in iron-deficient roots, indicating that a posttranscriptional regulatory mechanism contributes to the control of IRT1 protein abundance (Connolly et al., 2002). Similarly, while FRO2 mRNA levels are high in both iron-sufficient and iron-deficient 35S-FRO2 plants, FRO2 activity is elevated only in iron-deficient roots (Connolly et al., 2003). Presumably, posttranscriptional regulation of IRT1 and FRO2 serves to ensure that metals do not inappropriately accumulate in plants when metals are plentiful.
Here, we describe our efforts to uncover the molecular basis for posttranscriptional regulation of IRT1 by iron. Based on reports of metal-induced ubiquitination and turnover of metal transporters in yeast (Saccharomyces cerevisiae; Gitan et al., 1998; Liu and Culotta, 1999; Gitan and Eide, 2000; Felice et al., 2005), we reasoned that iron may signal modification and turnover of the Arabidopsis IRT1 protein. We show that either of two Lys residues that reside in an intracellular loop of IRT1 is required for iron-stimulated IRT1 turnover. In contrast, we could find no evidence that a His motif proposed to function in intracellular metal sensing has a role to play in posttranslational regulation of IRT1. Our data have important implications for the nutritional enhancement of crop plants, as 35S-IRT1K146R,K171R plants accumulate more iron than wild-type plants in their roots. This is in contrast to 35S-IRT1 plants that display wild-type levels of iron (Connolly et al., 2002).
RESULTS
Mutation of Specific Lys and His Residues in the IRT1 Intracellular Loop Does Not Eliminate IRT1-Mediated Iron or Zinc Transport
Posttranscriptional regulation of IRT1 could be mediated at the level of translation and/or at the level of protein turnover. In this study, we focused on regulation of protein turnover, in part because it is known that the yeast ZIP family member ZRT1 is subject to metal-induced ubiquitination and endocytosis (Gitan et al., 1998; Gitan and Eide, 2000; Gitan et al., 2003). Indeed, regulated turnover of membrane transporters has emerged as a relatively common regulatory mechanism in yeast and mammals (Ooi et al., 1996; Liu and Culotta, 1999; Katzmann et al., 2002; Hicke and Dunn, 2003; Kim et al., 2004; Wang et al., 2004; Felice et al., 2005), and recent work in plants demonstrated boron-induced endocytosis and degradation of a boron transporter (BOR1; Takano et al., 2005).
It has been suggested that the intracellular loop region of IRT1 might play a role in sensing intracellular metal levels and mediating protein turnover (Eide et al., 1996; Eng et al., 1998; Connolly et al., 2002). In particular, a His-rich motif found in the intracellular loop of the majority of ZIP family members is an attractive metal sensor candidate. Here, we test the hypothesis that the His motif in IRT1 (HGHGHGHG) plays a role in regulation of IRT1 protein stability. In addition, we test the hypothesis that Lys residues (that could serve as ubiquitin attachment sites) in the intracellular loop of IRT1 function in iron-induced regulation of IRT1 protein turnover.
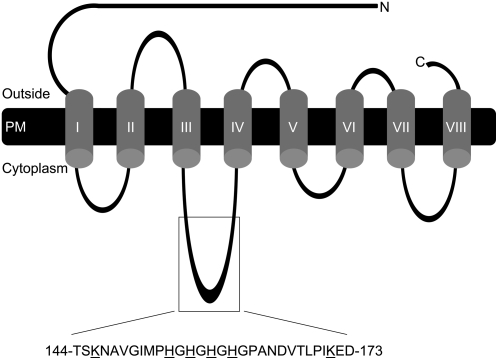
We constructed a set of IRT1 alleles: IRT1intact, IRT1H154Q, IRT1H156Q, IRT1H158Q, IRT1H160Q, IRT14HQ (quadruple His mutant), IRT1K146R, IRT1K171R, and IRT1K146R,K171R (Fig. 1). His was changed to Gln, as this is a relatively conservative change that would impair metal binding, while Lys was changed to Arg, as both amino acids are basic and this change presumably would eliminate ubiquitination but would cause the least disruption in protein structure. All constructs consisted of the IRT1 coding region and were identical except for the indicated mutations.
Figure 1.
Schematic representation of the predicted protein topology of Arabidopsis IRT1 (topology based on work of Eide et al. [1996]). Amino acids targeted for site-directed mutagenesis are underlined. PM, Plasma membrane; C, C terminus; N, N terminus.
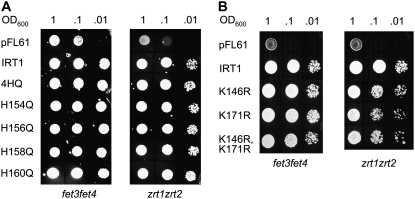
We next performed yeast complementation experiments to determine if the introduced mutations eliminated transport by IRT1. We tested whether each construct could rescue the iron-limited and zinc-limited growth defects of the fet3fet4 and zrt1zrt2 yeast strains, respectively. IRT1 was cloned by functional complementation of fet3fet4 (Eide et al., 1996) and has been shown to complement the zrt1zrt2 phenotype as well (Korshunova et al., 1999). Like the IRT1intact construct, all of the IRT1 mutant alleles were able to rescue the fet3fet4 and zrt1zrt2 mutant phenotypes (Fig. 2), showing that mutation of the intracellular loop His or Lys residues did not abolish the ability of IRT1 to transport iron or zinc. In addition, we examined growth rates of the fet3fet4 strain expressing the vector alone, the wild-type IRT1 allele, and each of the IRT1 mutant alleles, and observed that strains expressing the mutant forms of IRT1 grew at the same rate as the strain expressing wild-type IRT1 (data not shown). Further confirmation for this conclusion came from complementation of Arabidopsis irt1; expression of the various IRT1 alleles from the IRT1 promoter in irt1 showed that the mutant forms of IRT1 complemented the irt1 phenotype as well as wild-type IRT1 (Supplemental Fig. S1).
Figure 2.
Mutation of neither Lys nor His residues within the IRT1 intracellular loop region affects transport of iron or zinc by IRT1. Yeast strains defective in iron uptake (fet3fet4) and zinc uptake (zrt1ztr2) were transformed with pFL61 (vector), pFL61+ IRT1intact, pFL61+ IRT14HQ, pFL61+ IRT1H154Q, pFL61+ IRT1H156Q, pFL61+ IRT1H158Q, pFL61+ IRT1H160Q, pFL61+ IRT1K146R, pFL61+ IRT1K171R, or pFL61+ IRT1K146R,K171R and assayed for growth on SD-ura plates. The entire experiment was performed twice. A, His mutations; B, Lys mutations.
Lys-146 or Lys-171 Is Necessary for Iron-Induced Posttranslational Turnover of IRT1
Next, we tested the effect of mutation of the IRT1 intracellular loop His and Lys residues on the posttranscriptional regulation of IRT1 in transgenic plants. We transformed wild-type Arabidopsis with the IRT1intact, IRT1H154Q, IRT1H156Q, IRT1H158Q, IRT1H160Q, IRT14HQ, IRT1K146R, IRT1K171R, and IRT1K146R,K171R alleles; expression of each construct was driven by the cauliflower mosaic virus 35S promoter. Examination of the T1 plants showed that only the 35S-IRT1K146R,K171R transgenic lines displayed visible phenotypes. Out of 62 35S-IRT1K146R,K171R T1 plants, 29 were tiny, chlorotic, and produced little or no seed. The phenotype displayed by these 35S-IRT1K146R,K171R transgenic lines (reduced size, chlorosis, seedling lethality) is reminiscent of the irt1 phenotype (Henriques et al., 2002; Varotto et al., 2002; Vert et al., 2002) and thus could be due to cosuppression of both the IRT1 transgene and the IRT1 endogenous gene. Alternatively, it could be due to accumulation of elevated levels of metal, leading to metal toxicity-induced chlorosis. To test if the phenotype was due to loss of IRT1 expression, we grew a number of 35S-IRT1K146R,K171R T1 plants in the presence of high levels of a ferric iron-chelate [sequestrene; ferric ethylene-diamindei-(0-hydroxyphenylacetate)]. Previous work has shown that supplementation with sequestrene rescues the irt1 phenotype (Vert et al., 2002). Of approximately eight 35S-IRT1K146R,K171R transgenic lines that displayed the phenotype, none was rescued by supplementation with sequestrene (data not shown). This result indicates that it is unlikely that the 35S-IRT1K146R,K171R phenotype is due to cosuppression. Further support for this idea comes from the fact that overexpression of none of the other IRT1 alleles yielded plants with the phenotype. It is possible that the 35S-IRT1K146R,K171R phenotype is a result of metal toxicity-induced chlorosis. It was not possible to directly test this hypothesis because the 35S-IRT1K146R,K171R plants that displayed the phenotype were tiny and did not produce seed. Molecular and elemental analysis of the 35S-IRT1K146R,K171R transgenic lines described below lend further support for this hypothesis.
At least six independent, single-insertion, homozygous lines were identified for each construct. In the case of the IRT1K146R,K171R allele, nine such lines were identified and analyzed. Previously, we examined IRT1 transcript and protein accumulation in roots over time following the transfer of plants to iron-deficient medium. We showed that IRT1 transcript and protein levels are highest 72 h after plants are transferred to iron-deficient medium (Connolly et al., 2002). For this reason, plants were grown on B5 plates for 2 weeks, then transferred to either iron-deficient or iron-sufficient medium and harvested after 3 d. Evaluation of IRT1 transcript levels in total RNA from roots and shoots was carried out by northern-blot hybridization using the full-length IRT1 cDNA as a probe. As expected, IRT1 expression was detected only in roots of iron-deprived wild-type plants (Eide et al., 1996), while IRT1 mRNA was abundant in the roots and shoots of both iron-sufficient and iron-deficient 35S-IRT1 (Fig. 3), 35S-IRT14HQ (Fig. 3A), 35S-IRT1K146R (Fig. 3B), 35S-IRT1K171R (Fig. 3C), and 35S-IRT1K146R,K171R (Fig. 3D). IRT1 transcript made from the transgene is slightly smaller than transcript made from the endogenous gene because the transgene lacks the 3′ untranslated region (UTR) present in the endogenous transcript. We tested these two versions of IRT1 (IRT1+3′UTR and IRT1-3′UTR) in both yeast and transgenic plants. We could detect no difference between the two versions in side-by-side yeast complementation assays (data not shown). In addition, we also detected no differences in IRT1 mRNA or protein levels when the two versions were expressed in plants from the 35S promoter (data not shown and compare data shown here with Connolly et al. [2002]). Finally, it is important to note that IRT1 transcript levels in the various transgenic lines are highest in iron-deficient roots because IRT1 transcript detected in iron-deficient roots corresponds to the transgene transcript plus the endogenous transcript.
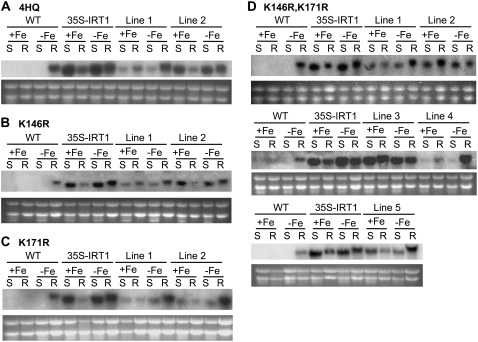
Figure 3.
IRT1 transcript levels in wild-type and transgenic plants overexpressing the intact and mutant versions of IRT1. Plants were grown in B5 medium for 2 weeks and then transferred to either iron-sufficient (+Fe) or iron-deficient (−Fe) medium for 3 d. RNA samples were prepared from shoots (S) and roots (R). The full-length IRT1 cDNA was used as a probe. Total RNA from wild-type (WT) and 35S-IRT1 plants were used as controls. Ethidium bromide-stained ribosomal RNA is shown as a loading control. A, 35S-IRT14HQ; B, 35S-IRT1K146R; C, 35S-IRT1K171R; D, 35S-IRT1K146R,K171R. Transcript levels were examined in at least four transgenic lines for each genotype. The entire experiment was performed twice.
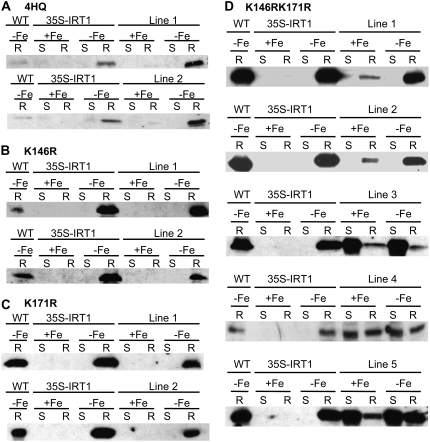
As described previously (Connolly et al., 2002), immunoblot analysis using an IRT1 peptide antibody revealed that IRT1 is detectable only in iron-deficient roots of wild-type and 35S-IRT1 plants (Fig. 4). Mutation of one or all four His residues did not affect IRT1 protein accumulation because IRT1 protein was detected only in iron-deficient roots of 35S-IRT1H154Q, 35S-IRT1H156Q, 35S-IRT1H158Q, 35S-IRT1H160Q, and 35S-IRT14HQ transgenic plants (Fig. 4A; data not shown). We further examined the abundance of IRT1 protein over time following the transfer of plants from iron-deficient medium to iron-sufficient medium and observed that IRT14HQ levels decline at a rate similar to that seen for IRT1 (Supplemental Fig. S2). These results indicate that the His motif is not required for the posttranslational regulation of IRT1 by iron. Similarly, we could not observe a role for the His motif in zinc-induced turnover of IRT1 (data not shown). Although the 35S-IRT1K146R and 35S-IRT1K171R transgenic lines displayed protein expression patterns identical to wild type and 35S-IRT1 (Fig. 4, B and C), plants overexpressing the IRT1K146R,K171R allele showed IRT1 protein accumulation in roots of both iron-deficient and iron-sufficient plants (Fig. 4D). This result was seen in all nine transgenic lines tested (Fig. 4D; data not shown). Accumulation of IRT1 protein was also observed in iron-deficient and iron-sufficient shoots in three out of the nine transgenic lines analyzed (Fig. 4D, lines 3–5). Hence, mutation of both Lys-146 and Lys-171 eliminates iron-induced turnover of IRT1, demonstrating that expression of IRT1 is controlled posttranslationally by iron status.
Figure 4.
Mutation of K146 and K171 stabilizes the IRT1 protein. Plants were grown in B5 medium for 2 weeks and then transferred to either Fe-sufficient (+Fe) or Fe-deficient (−Fe) medium for 3 d. Protein samples were prepared from shoots (S) and roots (R). IRT1 protein was detected using an IRT1 affinity-purified peptide antibody. Total protein extracts from wild-type (WT) and 35S-IRT1 were used as controls: A, 35S-IRT14HQ; B, 35S-IRT1K146R; C, 35S-IRT1K171R; and D, 35S-IRT1K146R,K171R. IRT1 protein levels were examined in at least four transgenic lines for each genotype. The entire experiment was performed twice.
35S-IRT1K146R,K171R Plants Accumulate Metals
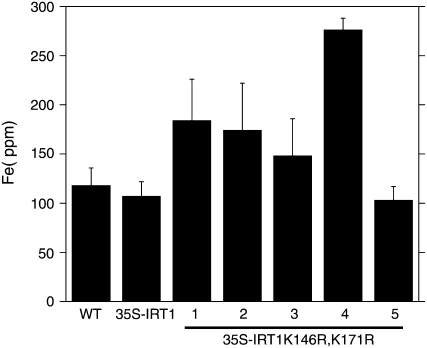
Previously, we showed that 35S-IRT1 transgenic plants do not accumulate iron or other metals when grown under iron-sufficient conditions; this result is thought to be due to the fact that IRT1 protein does not accumulate under iron-sufficient conditions (Connolly et al., 2002). We reasoned that 35S-IRT1K146R,K171R transgenic plants would accumulate metals as IRT1 protein accumulates in these lines when plants are grown on iron-sufficient medium. Elemental analysis of wild-type and transgenic seedlings grown on iron-sufficient medium revealed that 35S-IRT1K146R,K171R transgenic plants accumulate significantly more iron (1.4- to 2.6-fold) in roots than wild-type and 35S-IRT1 transgenic plants (Fig. 5).
Figure 5.
Elemental analysis of 35S-IRT1K146R,K171R roots. Plants were grown in B5 medium for 10 d and then transferred to 50 μm Fe-supplemented medium for 7 d. Roots were washed, harvested, dried, and analyzed by ICP-MS. Approximately 200 plants were pooled for each sample. Each value is the mean of four replicates. Error bars indicate sd. The experiment was performed twice. Results from one representative experiment are shown. Means corresponding to lines 1, 2, and 4 are statistically different as compared to 35S-IRT1 plants (P < 0.05).
It is important to note that approximately 50% of the 35S-IRT1K146R,K171R T1 transgenic plants died without setting seed; it is possible that the transgenic lines that expressed higher levels of IRT1 accumulated higher levels of iron, which would lead to metal toxicity and seedling lethality. In other words, the toxicity of iron resulted in the selection of 35S-IRT1K146R,K171R transgenic lines that express lower levels of IRT1. Only one 35S-IRT1K146R,K171R line (no. 4) displayed a significant increase (22%) in iron levels in the shoot relative to wild type and 35S-IRT1(data not shown); this line was the line that also showed the greatest accumulation of iron in roots (Fig. 5).
Manganese levels were significantly higher (1.4–2.4 times) in the shoots of four out of five 35S-IRT1K146R,K171R lines examined (data not shown), while no significant difference in zinc content was observed in the 35S-IRT1K146R,K171R transgenic plants as compared to 35S-IRT1 plants (data not shown). These observations indicate that the IRT1K146R,K171R protein is functional. Finally, elemental analysis of wild-type, 35S-IRT1, and 35S-IRT14HQ plants revealed no differences in metal accumulation (data not shown); this data is consistent with our observation that IRT14HQ protein does not accumulate in iron-sufficient plants.
Ferric Chelate Reductase Activity Is Not Altered in 35S-IRT1K146R,K171R Plants
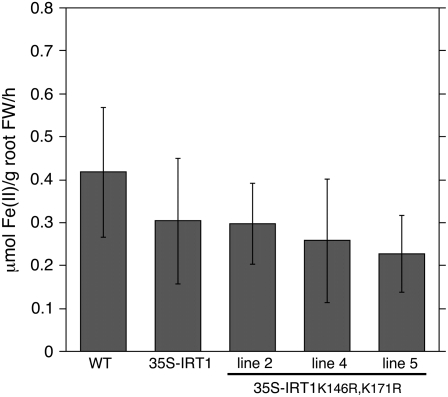
Reduction of ferric iron to ferrous iron is a prerequisite for iron uptake in Arabidopsis and is considered the rate-limiting step in iron uptake by iron-depleted roots (Grusak et al., 1990; Connolly et al., 2003). For this reason, it was of interest to determine if accumulation of elevated levels of iron by 35S-IRT1K146R,K171R plants is correlated with enhanced root ferric chelate reductase activity. We observed enhanced accumulation of iron in 35S-IRT1K146R,K171R plants only when grown on iron-sufficient medium. We therefore examined root surface ferric chelate reductase activity of wild-type, 35S-IRT1, and 35S-IRT1K146R,K171R plants grown under our typical iron-sufficient conditions (50 μm iron). No significant difference in root ferric chelate reductase activity was observed between wild-type, 35S-IRT1, and 35S-IRT1K146R,K171R plants grown under iron-sufficient conditions, indicating that the enhanced accumulation of iron in 35S-IRT1K146R,K171R roots is not associated with an enhanced ability to reduce Fe(III) (Fig. 6).
Figure 6.
Ferric chelate reductase activity is not altered in 35S-IRT1K146R,K171R plants. Plants were grown on B5 medium for 2 weeks and then transferred to Fe-sufficient medium for 3 d. Root surface ferric chelate reductase activity was measured on individual plants (n = 10) using the ferrozine assay. Error bars indicate sd. The experiment was performed four times and results from one representative experiment are shown. Means corresponding to lines 2, 4, and 5 are not statistically different as compared to wild type or the 35S-IRT1 control line (P > 0.05).
DISCUSSION
The data presented here demonstrate that the Arabidopsis IRT1 metal transporter is subject to posttranslational regulation by iron. Furthermore, either Lys-146 or Lys-171 is required for iron-induced turnover of IRT1 protein; mutation of both Lys-146 and Lys-171 stabilizes the IRT1 protein. The fact that expression of the IRT1K146R,K171R allele complements yeast mutants with defects in iron and zinc transport suggests that changing these residues does not eliminate transport. Importantly, 35S-IRT1K146R,K171R plants accumulate elevated levels of iron (in roots) and manganese (in shoots) of seedlings grown on plates. Of the five lines analyzed, one line (no. 5) does not show enhanced iron accumulation. It is not clear at this time why line 5 does not accumulate more iron, but it is noteworthy that line 5 is smaller and displays a reduced root system as compared to the four other 35S-IRT1K146R,K171R lines. Perhaps this phenotype is due to the insertion of the 35S-IRT1K146R,K171R transgene within an important gene.
IRT1 is known to transport manganese, zinc, cobalt, and cadmium in addition to iron (Eide et al., 1996; Korshunova et al., 1999; Vert et al., 2002). One might predict, then, that 35S-IRT1K146R,K171R plants would accumulate each of these elements. Zinc levels were not significantly altered in 35S-IRT1K146R,K171R plants relative to 35S-IRT1intact plants. At this time, it is not clear why this is the case, although it is known that zinc does not compete as well as iron, cadmium, and manganese for IRT1-dependent transport of iron in yeast (Eide et al., 1996). It is not clear whether or not 35S-IRT1K146R,K171R plants accumulate elevated levels of cobalt or cadmium, as elemental analysis was performed on 35S-IRT1K146R,K171R plants grown on medium that did not contain either element. However, when we examined root growth of 35S-IRT1K146R,K171R plants grown in the presence of cadmium, some lines did show enhanced sensitivity to this toxic metal (data not shown). This result suggests that 35S-IRT1K146R,K171R plants have an enhanced ability to take up cadmium.
Several studies have examined metal-induced posttranslational regulation of yeast plasma membrane metal transporters (Ooi et al., 1996; Liu and Culotta, 1999). In particular, yeast iron and zinc transport systems are known to be subject to metal-induced ubiquitination and endocytosis (Gitan et al., 1998; Gitan and Eide, 2000; Felice et al., 2005). There also are reports of posttranslational regulation of mammalian metal transporters (Sharp et al., 2002; Petris et al., 2003; Guo et al., 2004; Kim et al., 2004; Wang et al., 2004; Johnson et al., 2005). Presumably, metal-induced endocytosis serves to rapidly down-regulate metal transporters to prevent the accumulation of metals to toxic levels. Interestingly, a recent report showed that an Arabidopsis boron transporter (BOR1) is subject to boron-induced endocytosis and degradation (Takano et al., 2005).
Upon exposure to high zinc, the yeast plasma membrane-localized zinc transporter ZRT1 is rapidly ubiquitinated, undergoes endocytosis, and is degraded in the vacuole (Gitan et al., 1998; Gitan and Eide, 2000). ZRT1 is a member of the ZIP family of metal transporters and thus is related to IRT1. Like IRT1, ZRT1 contains an intracellular loop between transmembrane domains 3 and 4; a His motif (HDHTHDE) is found within this intracellular loop. In addition, it is known that a single Lys residue in the ZRT1 loop region is the site of zinc-stimulated ubiquitin attachment (Gitan and Eide, 2000). It is interesting to note that while mutation of a single Lys residue in ZRT1 is sufficient to disrupt metal-induced protein turnover, mutation of two Lys residues in IRT1 is necessary for protein stabilization; the significance of this difference is not known at this time. Our future studies are aimed at testing the hypothesis that high iron induces ubiquitination of IRT1 at Lys-146 and Lys-171. To date, we have not been able to detect ubiquitinated forms of IRT1 following immunoprecipitation of IRT1. However, this may be due to technical issues, as the IRT1 antibody recognizes a peptide (PANDVTLPIKEDDSSN) that includes Lys-171; it seems likely that the IRT1 antibody might not precipitate ubiquitinated forms of IRT1.
We have little knowledge of the regulatory circuit(s) that control IRT1 expression. Recent studies showed that the Arabidopsis bHLH transcription factor FIT is an essential gene and functions to regulate expression of both FRO2 and IRT1 (Colangelo and Guerinot, 2004; Jakoby et al., 2004; Yuan et al., 2005; Bauer et al., 2007). Another recent study suggests that ethylene may regulate the expression of FRO2 and IRT1 via FIT (Lucena et al., 2006). Surprisingly, while FRO2 mRNA is not detected in fit1, IRT1 mRNA is detected in fit1, but IRT1 protein fails to accumulate (Colangelo and Guerinot, 2004). The molecular mechanism by which FIT1 regulates IRT1 is not yet known but is of considerable interest.
Because studies have shown that reduction of ferric iron, rather than transport of ferrous iron, is the rate-limiting step for iron uptake in strategy I plants (Grusak et al., 1990; Connolly et al., 2003), it was somewhat surprising that the 35S-IRT1K146R,K171R plants accumulated elevated levels of iron. This result raised the possibility that overexpression of IRT1K146R,K171R somehow leads to coordinate overexpression of the FRO2 protein, which is responsible for reduction of ferric iron at the root surface (Robinson et al., 1999). However, we also showed that iron-replete 35S-IRT1K146R,K171R plants do not have elevated levels of root surface ferric chelate reductase activity. This result suggests that transport of ferrous iron may also be a rate-limiting step for iron uptake in strategy I plants, at least under certain conditions.
It has been suggested that the His motif found in many ZIP family members might serve as an intracellular metal-binding/-sensing domain that might mediate posttranslational regulation (Eide et al., 1996; Guerinot, 2000). We have not found any evidence that the IRT1 His motif plays a role in iron-stimulated IRT1 turnover or transport of iron or zinc. A recent study showed that the His motif (HWHD) of human ZIP1 is important for transport of zinc (Milon et al., 2006). On the other hand, the His motif (HSSHSHGGHSH) of human ZIP4 is critical for zinc-induced ubiquitination and degradation under conditions of high zinc but not for zinc-stimulated endocytosis under conditions of low zinc (Mao et al., 2007). Studies in yeast have shown that the ZRT1 His motif (HDHTHDE) is important for ZRT1 function; mutation of the ZRT1 His motif results in a 70% reduction in the Vmax, while the Km for zinc uptake is unaffected (Gitan et al., 2003). The mutant version of ZRT1 accumulated to normal levels in cells but showed reduced plasma membrane localization. Additional experiments revealed that the mutant version of ZRT1 was subject to zinc-induced inactivation, just like wild-type ZRT1 (Gitan et al., 2003). Therefore, a role for the His motif in zinc sensing and metal-induced protein turnover is not supported by data for ZRT1, and it is interesting to note that the His motif appears to play different roles in the context of different ZIPs. Further support for this conclusion comes from the work of Mao et al. (2007), who showed that the hZIP4 His motif-containing cytoplasmic loop is necessary for degradation of hZIP4 but not sufficient for zinc-induced degradation of GFP. It is interesting to note that a recent study that examined the metal-binding thermodynamics of the IRT1 His motif showed that although the His motif has a relatively low affinity for metal ions that are transported by IRT1 (Fe2+, Mn2+, Co2+, Cd2+, and Zn2+), it has a relatively high affinity for Fe3+ (Grossoehme et al., 2006).
The work described here may aid efforts to engineer crop plants with enhanced iron content. We show that 35S-IRT1K146R,K171R plants accumulate elevated levels of iron in the root. For many crops, we will want to enhance transport of iron to the aerial parts of the plant. Our knowledge of iron trafficking in plants is still limited. We do not yet have a comprehensive understanding of the molecular mechanisms that control iron distribution in plants, but it is likely that the combined modulation of IRT1 expression and specificity (Rogers et al., 2000) will be an important part of a multifaceted approach to engineer plants that contain higher levels of bioavailable iron.
MATERIALS AND METHODS
Cloning and Site-Directed Mutagenesis
The Arabidopsis (Arabidopsis thaliana) IRT1 coding sequence was amplified from pIRT-1 (Eide et al., 1996) using a high fidelity polymerase. Point mutants were constructed using overlap extension PCR. The IRT1-F and IRT1-R nonmutagenic primers were used alone (to generate an intact IRT1 construct that served as a control) and in combination with different sets of mutagenic primers (shown below). For all primers, mutagenized positions are denoted in lowercase, and restriction sites are underlined. The IRT1K171R mutant construct was used as a template to create the IRT1K146R,K171R mutant. Primers used were: IRT1-F, 5′CCCTCGAGAATTCAGCACTTCTCATGAAA3′ (XhoI); IRT1-R, 5′CCGCGGCCGCCCGCAATATCTGGAGTATTAG3′ (NotI); IRT1K146R-F, 5′CTATACACCAGCAgGAACGCAGTTGGT3′; IRT1K146R-R, 5′ACCAACTGCGTTCcTGCTGGTGTATAG3′; IRT1K171R-F, 5′ACCTTACCAATAAgAGAAGATGATTCG3′; IRT1K171R-R, 5′CGAATCATCTTCTcTTATTGGTAAGGT3′; IRT1H154Q-F, 5′GGTATCATGCCCCAaGGTCATGGTCAT3′; IRT1H154Q-R, 5′ATGACCATGACCtTGGGGCATGATACC3′; IRT1H156Q-F, 5′ATGCCCCATGGTCAaGGTCATGGTCAC3′; IRT1H156Q-R, 5′GTGACCATGACCtTGACCATGGGGCAT3′; IRT1H158Q-F, 5′CATGGTCATGGTCAaGGTCACGGCCCC3′; IRT1H158Q-R, 5′GGGGCCGTGACCtTGACCATGACCATG3′; IRT1H160Q-F, 5′CATGGTCATGGTCAaGGCCCCGCAAAT3′; IRT1H160Q-R, 5′ATTTGCGGGGCCtTGACCATGACC3′; IRT14HQ-F, 5′CAaGGTCAaGGTCAaGGTCAaGGCCCCGCAAATGATGTT3′; and IRT14HQ-R, 5′tTGACCtTGACCtTGACCtTGGGGCATGATACCAACTGC3′.
Intact and mutant IRT1 cDNAs were cloned into the XhoI and NotI sites of pBluescript SK+. After the correct sequence was verified for each construct, the various IRT1 cassettes were blunt-end cloned into the NotI site of the yeast (Saccharomyces cerevisiae) expression vector pFL61 behind the phosphoglycerate kinase promoter (Minet et al., 1992) and were subcloned into the XhoI and NotI sites of the 35SpBARN vector (a kind gift from Bonnie Bartel, Rice University; LeClere and Bartel, 2001). Yeast expression constructs made for this study include: IRT1intact (pELC220), IRT14HQ (pIC9), IRT1K146R (pELC221), IRT1K171R (pELC222), and IRT1K146R,K171R (pELC223). Plant overexpression constructs made for this study include: IRT1intact (pELC224), IRT14HQ (pIC3), IRT1K146R (pELC225), IRT1K171R (pELC226), and IRT1K146R,K171R (pELC227). In addition, IRT1H154Q, IRT1H156Q, IRT1H158Q, and IRT1H160Q constructs were tested in both yeast and transgenic Arabidopsis plants. Constructs in which the IRT1 promoter is fused upstream of the IRT1K146R,K171R and IRT14HQ cDNAs also were created for this study. Plasmid A5 (containing a 1,762-bp BamHI/SacI fragment with approximately 1.5 kb of IRT1 promoter in pCAMBIA1300) and plasmid A12 (containing the IRT1 promoter fused upstream of the entire IRT1 cDNA) were the kind gift of Aaron Atkinson and Mary Lou Guerinot (Dartmouth College). The IRT1K146R,K171R and IRT14HQ full-length cDNAs were cloned into the SacI site of A5.
Yeast Complementation
IRT1 constructs and empty vector (pFL61) were transformed into yeast strains fet3fet4 (DEY1453; MATa/MATa ade2/+ can1 his3 leu2 trp1 ura fet3-2∷HIS3 fet4-1∷LEU2; Dix et al., 1994) and zrt1zrt2 (ZHY3; MATa ade6 can1 his3 leu2 trp1 ura3 zrt1∷LEU2 zrt2∷HIS3; Zhao and Eide, 1996b) using the lithium acetate transformation method (Elble, 1992). fet3fet4 transformants were selected on SD medium lacking uracil (SD − ura) and supplemented with 10 μm FeCl3. zrt1zrt2 transformants were selected on SD − ura and supplemented with 50 μm ZnSO4. fet3fet4 and zrt1zrt2 transformants were grown overnight in liquid SD − ura and supplemented with 10 μm FeCl3 or 50 μm ZnSO4, respectively. Yeast cells were recovered by centrifugation, resuspended in SD − ura (without added Fe or Zn) at an OD600 of 1, 0.1, and 0.01. Then 20 μL of each culture was spotted on SD − ura plates; plates were incubated at 30°C for 2 d and then photographed.
Plant Growth Conditions
Wild-type (ecotype Columbia gl-1) and transgenic Arabidopsis seeds were surface sterilized in 25% bleach and 0.2% SDS for 15 min. After five washes with deionized water, seeds were suspended in sterile 0.15% agar solution and stored in the dark at 4°C for 2 to 4 d to synchronize germination and then sown on plates containing Gamborg's B5 medium (Phytotechnology Laboratories) supplemented with 2% Suc, 1 mm MES, and 0.6% agar, final pH 5.8. Transgenic plants were grown on Gamborg's B5 medium supplemented with 50 μm Basta. Plants were grown in a controlled environment plant growth chamber (Percival Scientific) under constant illumination (100 μE m−2 s−1) at 22°C for 2 weeks. The seedlings were placed beneath a yellow filter (acrylic yellow-2208; Cadillac Plastic and Chemical) to prevent the photochemical degradation of Fe(III)-EDTA (Hangarter and Stasinopoulos, 1991). Seedlings were transferred to either iron-sufficient [50 μm Fe(III)-EDTA] or iron-deficient (300 μm FerroZine [3-(2-pyridyl)-5,6-diphenyl-1,2,4-triazine sulfonate]; HACH Chemical) medium containing macro- and micronutrients [2 mm CaNO3, 750 μm K2SO4, 650 μm MgSO4, 100 μm KH2PO4, 10 μm H3BO3, 0.1 μm MnSO4, 0.05 μm CuSO4, 0.05 μm ZnSO4, 0.005 μm (NH4)6Mo24] as described (Marschner et al., 1982) and 1 mm MES, 0.6% agar, pH 6.0. Plants were grown for 3 d in a growth chamber as described above.
Plant Transformation
The 35S-IRT1intact and each of the 35S-IRT1 mutant constructs described above were used to transform Agrobacterium tumefaciens strain GV3101 (Koncz and Schell, 1986). Agrobacterium-mediated transformation of wild-type Arabidopsis plants was accomplished using the floral dip method (Clough and Bent, 1998). T1 seeds obtained from self-fertilization of the primary transformants were sown on Gamborg's B5 medium supplemented with 50 μm Basta (glufosinate ammonium; Crescent Chemical), and Basta-resistant seedlings were transferred to soil for propagation. Transgenic lines that displayed a 3:1 segregation ratio for Basta resistance to Basta sensitivity in the T2 generation and that were 100% Basta resistant in the T3 generation were selected for further analysis. At least six independent, single-insertion lines were identified for each construct; all experiments were performed with T4 seed.
Analysis of IRT1 Expression
Total RNA was prepared (Verwoerd et al., 1989) from shoots and roots of plants that were grown axenically on B5 medium and subsequently transferred to iron-sufficient or iron-deficient plates. RNA samples (10 μg) were covalently modified by treatment with glyoxal (McMaster and Carmichael, 1977), separated on a 1.2% agarose gel containing 10 mm NaPO4, pH 6.5, transferred to a nylon membrane (Nytran SuPerCharge; Schleicher and Schuell), and attached to the membrane by UV cross-linking. Hybridizations were performed in 50% formamide at 42°C using standard procedures (Ausubel et al., 2005). The IRT1 cDNA was radiolabeled according to the random primer method (Feinberg and Vogelstein, 1984) and used to probe northern blots. Following overnight hybridizations, blots were washed twice for 15 min at 42°C in 1× SSC/0.1% SDS, followed by two 15-min washes in 0.1× SSC/0.1% SDS at 65°C and subsequently exposed to x-ray film at −80°C and later to a phosphor imager screen.
Western analysis was carried out essentially as previously described (Connolly et al., 2002). Total protein was prepared from the roots and shoots of plants that were grown on B5 medium and then transferred to plates that were either iron sufficient or iron deficient. Extracts were prepared by grinding tissue (3 mL buffer/1 g fresh weight) on ice in extraction buffer containing 50 mm Tris, pH 8.0, 5% glycerol, 4% SDS, 1% polyvinylpolypyrrolidone, 1 mm phenylmethylsulfonyl fluoride, and 2 mm Pefabloc (Roche), followed by centrifugation at 4°C for 15 min at 13,000 rpm. The supernatant was recovered, and protein concentration was estimated using the bicinchoninic acid protein assay (Pierce). Samples for SDS-PAGE were diluted with an equal volume of 2× sample loading buffer (Ausubel et al., 2005) and boiled for 2 min. Total protein (15 μg) was separated by SDS-PAGE (Laemmli, 1970) and transferred to polyvinylidene difluoride membrane by electroblotting (Towbin et al., 1979). Western blots were processed as described previously using an IRT1-specific polyclonal antibody (Connolly et al., 2002).
Ferric Chelate Reductase Assays
Plants (wild type and transgenic) were grown for 2 weeks on B5 medium and then transferred to iron-sufficient medium for 3 d before analysis. Root surface ferric chelate reductase activity assays were performed as described previously (Yi and Guerinot, 1996). Assays were performed on 10 individual plants; statistical analysis was performed using the Student's t test. The experiment was performed four times.
Elemental Analysis
Wild-type and transgenic plants were grown on Gamborg's B5 medium for 10 d and then transferred to iron-sufficient medium for seven additional days. The roots and shoots of plants were separated, washed several times in deionized water, and dried at 65°C for 48 h. Elemental analysis was carried out on pools of approximately 200 plants using inductively coupled plasma mass spectrometry. Values are the means of four replicates; statistical analysis was performed using the student's t test. The entire experiment was performed two times.
Accession Number
The accession number for the Arabidopsis IRT1 gene is At4g19690.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Complementation of irt1 by IRT1promoter-IRT1K146R,K171R and IRT1promoter-IRT14HQ constructs.
Supplemental Figure S2. Mutation of the IRT1 His motif does not affect iron-induced IRT1 protein turnover.
Supplementary Material
Acknowledgments
We thank Joshua Ash for assistance in the preparation of IRT1 constructs. We are grateful to Aaron Atkinson and Mary Lou Guerinot for the gift of A5 and AI2 plasmids as well as helpful discussion throughout the course of this study and for critical reading of the manuscript. We also thank Liz Colangelo and members of the Connolly Lab for critical reading of the manuscript.
This work was supported by the U.S. Department of Agriculture (NRICGP grant nos. 2001–35100–10752 and 2004–35100–14934 to E.L.C.) and by the National Science Foundation (grant no. IOB–0419695 to D.E.S.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Erin L. Connolly (erinc@biol.sc.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (2005) Current Protocols in Molecular Biology. John Wiley & Sons, New York
- Bauer P, Ling HQ, Guerinot ML (2007) FIT, the FER-LIKE IRON DEFICIENCY INDUCED TRANSCRIPTION FACTOR in Arabidopsis. Plant Physiol Biochem 45 260–261 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Colangelo EP, Guerinot ML (2004) The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. Plant Cell 16 3400–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly EL, Campbell N, Grotz N, Prichard CL, Guerinot ML (2003) Overexpression of the FRO2 iron reductase confers tolerance to growth on low iron and uncovers post-transcriptional control. Plant Physiol 133 1102–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly EL, Fett JP, Guerinot ML (2002) Expression of the IRT1 metal transporter is controlled by metals at the levels of transcript and protein accumulation. Plant Cell 14 1347–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curie C, Panaviene Z, Loulergue C, Dellaporta SL, Briat JF, Walker EL (2001) Maize yellow stripe1 encodes a membrane protein directly involved in Fe(III) uptake. Nature 409 346–349 [DOI] [PubMed] [Google Scholar]
- Dix DR, Bridgham JT, Broderius MA, Byersdorfer CA, Eide DJ (1994) The FET4 gene encodes the low affinity Fe(II) transport protein of Saccharomyces cerevisiae. J Biol Chem 269 26092–26099 [PubMed] [Google Scholar]
- Eide D, Broderius M, Fett J, Guerinot ML (1996) A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc Natl Acad Sci USA 93 5624–5628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elble R (1992) A simple and efficient procedure for transformation of yeasts. Biotechniques 13 18–22 [PubMed] [Google Scholar]
- Eng BH, Guerinot ML, Eide D, Saier MHJ (1998) Sequence analyses and phylogenetic characterization of the ZIP family of metal ion transport proteins. J Membr Biol 166 1–7 [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B (1984) A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem 137 266–267 [DOI] [PubMed] [Google Scholar]
- Felice MR, De Domenico I, Li L, McVey Ward D, Bartok B, Musci G, Kaplan J (2005) Post-transcriptional regulation of the yeast high affinity iron transport system. J Biol Chem 280 22181–22190 [DOI] [PubMed] [Google Scholar]
- Gitan RS, Eide DJ (2000) Zinc-regulated ubiquitin conjugation signals endocytosis of the yeast ZRT1 zinc transporter. Biochem J 346 329–336 [PMC free article] [PubMed] [Google Scholar]
- Gitan RS, Luo H, Rodgers J, Broderius M, Eide D (1998) Zinc-induced inactivation of the yeast ZRT1 zinc transporter occurs through endocytosis and vacuolar degradation. J Biol Chem 273 28617–28624 [DOI] [PubMed] [Google Scholar]
- Gitan RS, Shababi M, Kramer M, Eide DJ (2003) A cytosolic domain of the yeast Zrt1 zinc transporter is required for its post-translational inactivation in response to zinc and cadmium. J Biol Chem 278 39558–39564 [DOI] [PubMed] [Google Scholar]
- Grossoehme NE, Akilesh S, Guerinot ML, Wilcox DE (2006) Metal-binding thermodynamics of the histidine-rich sequence from the metal-transport protein IRT1 of Arabidopsis thaliana. Inorg Chem 45 8500–8508 [DOI] [PubMed] [Google Scholar]
- Grusak MA, Pezeshgi S (1996) Shoot-to-root signal transmission regulates root Fe(III) reductase activity in the dgl mutant of pea. Plant Physiol 110 329–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grusak MA, Welch RM, Kochian LV (1990) Does iron deficiency in Pisum sativum enhance the activity of the root plasmalemma iron transport protein? Plant Physiol 94 1353–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerinot ML (2000) The ZIP family of metal transporters. Biochim Biophys Acta 1465 190–198 [DOI] [PubMed] [Google Scholar]
- Guo Y, Smith K, Lee J, Thiele DJ, Petris MJ (2004) Identification of methionine-rich clusters that regulate copper-stimulated endocytosis of the human Ctr1 copper transporter. J Biol Chem 279 17428–17433 [DOI] [PubMed] [Google Scholar]
- Halliwell B, Gutteridge JMC (1992) Biologically relevant metal ion-dependent hydroxyl radical generation. FEBS Lett 307 108–112 [DOI] [PubMed] [Google Scholar]
- Hangarter RP, Stasinopoulos TC (1991) Effect of Fe-catalyzed photooxidation of EDTA on root growth in plant culture media. Plant Physiol 96 843–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques R, Jasik J, Klein M, Martinoia E, Feller U, Schell J, Pais MS, Koncz C (2002) Knock-out of Arabidopsis metal transporter gene IRT1 results in iron deficiency accompanied by cell differentiation defects. Plant Mol Biol 50 587–597 [DOI] [PubMed] [Google Scholar]
- Hicke L, Dunn R (2003) Regulation of membrane protein transport by ubiquitination and ubiquitin-binding proteins. Annu Rev Cell Dev Biol 19 141–172 [DOI] [PubMed] [Google Scholar]
- Jakoby M, Wang HY, Reidt W, Weisshaar B, Bauer P (2004) FRU (BHLH029) is required for induction of iron mobilization genes in Arabidopsis thaliana. FEBS Lett 577 528–534 [DOI] [PubMed] [Google Scholar]
- Johnson DM, Yamaji S, Tennant J, Srai SK, Sharp PA (2005) Regulation of divalent metal transporter expression in human intestinal epithelial cells following exposure to non-haem iron. FEBS Lett 579 1923–1929 [DOI] [PubMed] [Google Scholar]
- Katzmann DJ, Odorizzi G, Emr SD (2002) Receptor downregulation and multivesicular-body sorting. Nat Rev Mol Cell Biol 3 893–905 [DOI] [PubMed] [Google Scholar]
- Kim BE, Wang F, Dufner-Beattie J, Andrew GK, Eide DJ, Petris MJ (2004) Zn2+-stimulated endocytosis of the mZIP4 zinc transporter regulates its location at the plasma membrane. J Biol Chem 279 4523–4530 [DOI] [PubMed] [Google Scholar]
- Koncz C, Schell J (1986) The promoter of the TL-DNA gene5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vectors. Mol Gen Genet 204 383–396 [Google Scholar]
- Korshunova YO, Eide D, Clark WG, Guerinot ML, Pakrasi HB (1999) The IRT1 protein from Arabidopsis thaliana is a metal transporter with broad specificity. Plant Mol Biol 40 37–44 [DOI] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685 [DOI] [PubMed] [Google Scholar]
- LeClere S, Bartel B (2001) A library of Arabidopsis 35S-cDNA lines for identifying novel mutants. Plant Mol Biol 46 695–703 [DOI] [PubMed] [Google Scholar]
- Liu XF, Culotta VC (1999) Post-translation control of Nramp metal transport in yeast. J Biol Chem 274 4863–4868 [DOI] [PubMed] [Google Scholar]
- Lucena C, Waters BM, Romera FJ, Garcia MJ, Morales M, Alcantara E, Perez-Vincente R (2006) Ethylene could influence ferric chelate reductase, iron transporter, and H+-ATPase gene expression by affecting FER (or FER-like) gene activity. J Exp Bot 57 4145–4154 [DOI] [PubMed] [Google Scholar]
- Mao X, Kim BE, Wang F, Eide DJ, Petris MJ (2007) A histidine-rich cluster mediates the ubiquitination and degradation of the human zinc transporter, hZIP4, and protects against zinc cytotoxicity. J Biol Chem 282 6992–7000 [DOI] [PubMed] [Google Scholar]
- Marschner H, Römheld V, Ossenberg-Neuhaus H (1982) Rapid method for measuring changes in pH and reducing processes along roots of intact plants. Z Pflanzenphysiol 105 407–416 [Google Scholar]
- McMaster GK, Carmichael GG (1977) Analysis of single- and double-stranded nucleic acids on polyacrylamide and agarose gels using glyoxal and acridine orange. Proc Natl Acad Sci USA 74 4835–4839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milon B, Wu Q, Zou J, Costello LC, Franklin RB (2006) Histidine residues in the region between transmembrane domains III and IV of hZip1 are required for zinc transport across the plasma membrane in PC-3 cells. Biochim Biophys Acta 1758 1696–1701 [DOI] [PubMed] [Google Scholar]
- Minet M, Dufour ME, Lacroute F (1992) Complementation of Saccharomyces cerevisiae auxotrophic mutants by Arabidopsis thaliana cDNAs. Plant J 2 417–422 [DOI] [PubMed] [Google Scholar]
- Ooi CE, Rabinovich E, Dancis A, Bonifacino JS, Klausner RD (1996) Copper-dependent degradation of the Saccharomyces cerevisiae plasma membrane copper transporter Ctr1p in the apparent absence of endocytosis. EMBO J 15 3515–3523 [PMC free article] [PubMed] [Google Scholar]
- Petris MJ, Smith K, Lee J, Thiele DJ (2003) Copper-stimulated endocytosis and degradation of the human copper transporter, hCtr1. J Biol Chem 278 9639–9646 [DOI] [PubMed] [Google Scholar]
- Robinson NJ, Proctor CM, Connolly EL, Guerinot ML (1999) A ferric-chelate reductase for iron uptake from soils. Nature 397 694–697 [DOI] [PubMed] [Google Scholar]
- Rogers EE, Eide DJ, Guerinot ML (2000) Altered selectivity in an Arabidopsis metal transporter. Proc Natl Acad Sci USA 97 12356–12360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römheld V (1987) Different strategies for iron acquisition in higher plants. Physiol Plant 70 231–234 [Google Scholar]
- Schikora A, Schmidt W (2001) Iron stress-induced changes in root epidermal cell fate are regulated independently from physiological responses to low iron availability. Plant Physiol 125 1679–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt W, Boomgaarden B, Ahrens V (1996) Reduction of root iron in Plantago lanceolata during recovery from Fe deficiency. Physiol Plant 98 587–593 [Google Scholar]
- Sharp PA, Tandy S, Yamaji S, Tennant J, Williams M, Singh Srai SK (2002) Rapid regulation of divalent metal transporter (DMT1) protein but not mRNA expression by non-haem iron in human intestinal Caco-2 cells. FEBS Lett 510 71–76 [DOI] [PubMed] [Google Scholar]
- Takano J, Miwa K, Yuan L, von Wiren N, Fujiwara T (2005) Endocytosis and degradation of BOR1, a boron transporter of Arabidopsis thaliana, regulated by boron availability. Proc Natl Acad Sci USA 102 12276–12281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA 76 4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varotto C, Maiwald D, Pesaresi P, Jahns P, Francesco S, Leister D (2002) The metal ion transporter IRT1 is necessary for iron homeostasis and efficient photosynthesis in Arabidopsis thaliana. Plant J 31 589–599 [DOI] [PubMed] [Google Scholar]
- Vert G, Briat JF, Curie C (2003) Dual regulation of the Arabidopsis high affinity root iron uptake system by local and long-distance signals. Plant Physiol 132 796–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G, Grotz N, Dedaldechamp F, Gaymard F, Guerinot ML, Briat JF, Curie C (2002) IRT1, an Arabidopsis transporter essential for iron uptake from the soil and plant growth. Plant Cell 14 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verwoerd TC, Dekker BM, Hoekema A (1989) A small scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res 17 2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Dufner-Beattie J, Kim BE, Petris MJ, Andrews G, Eide DJ (2004) Zinc-stimulated endocytosis controls activity of the mouse ZIP1 and ZIP3 zinc uptake transporters. J Biol Chem 279 24631–24639 [DOI] [PubMed] [Google Scholar]
- Yi Y, Guerinot ML (1996) Genetic evidence that induction of root Fe(III) chelate reductase activity is necessary for iron uptake under iron deficiency. Plant J 10 835–844 [DOI] [PubMed] [Google Scholar]
- Yuan YX, Zhang J, Wang DW, Ling HQ (2005) AtbHLH29 of Arabidopsis thaliana is a functional ortholog of tomato FER involved in controlling iron acquisition in strategy I plants. Cell Res 15 613–621 [DOI] [PubMed] [Google Scholar]
- Zhao H, Eide D (1996. b) The ZRT2 gene encodes the low affinity zinc transporter in Saccharomyces cerevisiae. J Biol Chem 271 23203–23210 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.