Abstract
The hypertrophy and hyperplasia of infected roots into clubs are the intrinsic characteristics of clubroot, one of the economically most important diseases in Brassica crops worldwide. Polyamines, arginine (Arg)-derived metabolites, have long been recognized as cell proliferation and differentiation regulators in plants and consequently are suitable candidates for potential gall development factors. Furthermore, Arg catabolism, through arginase, which is strongly connected to polyamine metabolism, would play an important role in response to wound trauma and pathogen infection. In this study, we exploited the Arabidopsis (Arabidopsis thaliana)-Plasmodiophora brassicae pathosystem to investigate the involvement of polyamine metabolism and Arg catabolism in host responses to the pathogen infection and in partial clubroot resistance mechanisms. We demonstrated at the transcriptional, enzymatic, and metabolic levels that polyamine metabolism and Arg catabolism are induced during the later stages of disease in compatible Arabidopsis-P. brassicae interactions. However, susceptible and partially resistant plants showed strikingly different Arg metabolism signatures. Susceptible plants were characterized by a transient agmatine production, a massive induction of arginase, and a strong accumulation of proline. The potential functions of this marked activation of the arginase pathway in the P. brassicae pathogenicity strategy are discussed. Partially resistant plants showed a continuous agmatine production and a weaker arginase pathway activity than the susceptible genotype. Results suggest that the symptom severity was strongly associated to the differential regulation of root polyamine metabolism and Arg catabolism. Further work using arginase transgenic plants will provide insight into the physiological function of the arginase pathway in partial clubroot resistance.
Clubroot, caused by the obligate biotrophic protist Plasmodiophora brassicae Woron., is one of the economically most important diseases of Brassica crops in the world. The life cycle of this soil-borne pathogen can be divided into two phases: a primary phase in which events are confined to the root hairs, and a secondary phase that occurs in the cortex and the stele of the hypocotyl and roots of the infected plants. During the second phase, multinucleate plasmodia cause the hypertrophy (abnormal cell enlargement) and hyperplasia (uncontrolled cell division) of infected roots into characteristic clubs (Ingram and Tommerup, 1972). These symptoms obstruct nutrient and water transport, stunt the growth of the plant, and consequently reduce crop yield and quality. Since the pathogen survives as resting spores for a long period (up to 15 years) in the soil, control of the disease by agricultural practices and/or chemical treatments is difficult and/or expensive. Thus, the development of resistant cultivars is currently the most efficient way to control clubroot among Brassica crops. Both qualitative and quantitative clubroot resistances have been identified and analyzed in different cultivated Brassicaceae species (Manzanares-Dauleux et al., 2000a; Suwabe et al., 2003; Hirai et al., 2004; Piao et al., 2004; Rocherieux et al., 2004; Hirai, 2006; Saito et al., 2006; Suwabe et al., 2006). Monogenic or oligogenic conferred clubroot resistance introduced into commercial cultivars leads to complete resistance (incompatible interaction) but is rapidly overcome. Partial resistance (compatible interaction), controlled by multiple genes, would be overcome by the pathogen more slowly and is thus an alternative for developing durable host-plant resistance. However, mechanisms associated with partial quantitative resistance remain poorly understood.
Arabidopsis (Arabidopsis thaliana), a wild Brassicaceae, also is a host species for clubroot (Koch et al., 1991). Natural variation in the responses to clubroot occurs in the model plant (Fuchs and Sacristán, 1996; Siemens et al., 2002; Alix et al., 2007) and our group discovered that the Burren-0 (Bur-0) accession is partially resistant to the eH P. brassicae isolate (Alix et al., 2007). The resistance harbored by this accession is under polygenic control (M. Jubault, C. Lariagon, M. Simon, R. Delourme, and M.J. Manzanares-Dauleux, unpublished data) and is characterized by a reduction of symptom severity. Compared to a susceptible genotype (main and secondary root systems are replaced by a big club), the partially resistant plants usually show only tiny clubs confined to the secondary root system. These findings make it possible to exploit the model plant and its panel of available biological and genomic resources to gain insight into clubroot pathogenesis and the mechanisms controlling partial resistance. Furthermore, P. brassicae does not show host specificity in Brassicaceae (i.e. the same isolate can infect different species). Consequently, knowledge acquired on Arabidopsis could then be rapidly integrated and transferred to cultivated crops. To date, research on clubroot using Arabidopsis as a model host system was mainly focused on the role of hormonal regulation by auxin (Ludwig-Muller et al., 1999; Grsic-Rausch et al., 2000; Neuhaus et al., 2000; Ando et al., 2006; Devos et al., 2006; Schuller and Ludwig-Muller, 2006) and/or cytokinins (Devos et al., 2006; Siemens et al., 2006) in the development of clubroot symptoms. However, other molecules, such as polyamines, could also be involved in cell proliferation and differentiation. These metabolites are of interest regarding the development of clubroot symptoms and were previously proposed to participate in club formation in infected roots of susceptible Brassica (Walters and Shuttleton, 1985; Cao et al., 2008).
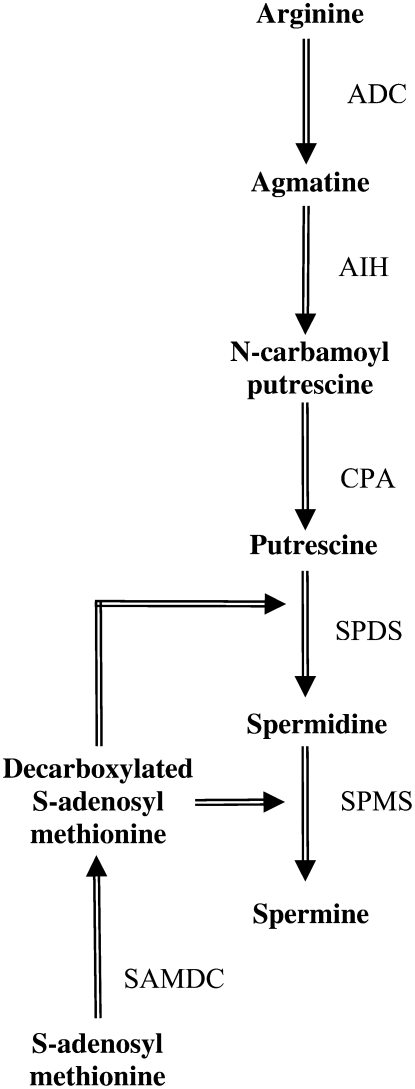
Polyamines are ubiquitous aliphatic cations that are produced by almost all living organisms, including plants, animals, fungi, and bacteria. Their biosynthesis and catabolism pathways have been fully characterized for many organisms (mammals, fungi, plants). In Arabidopsis, the amino acid Arg is the starting point for polyamine biosynthesis. Arg is decarboxylated by Arg decarboxylase to yield agmatine (Fig. 1), which then serves as the substrate for the biosynthesis of putrescine through the activities of two enzymes, agmatine iminohydrolase and N-carbamoylputrescine amidohydrolase. In higher plants, putrescine is also produced by an alternative pathway, from Orn, as the result of the action of Orn decarboxylase. However, several plant species, including the model plant Arabidopsis, show reduced or absent Orn decarboxylase activity, so that polyamine biosynthesis must be mostly dependant on the basic amino acid Arg (Hanfrey et al., 2001). Putrescine is then converted into the polyamines spermidine and spermine by addition of amino-propyl residues from decarboxylated S-adenosyl-Met, which results from decarboxylation of S-adenosyl-Met by S-adenosyl-Met decarboxylase. These reactions are sequentially catalyzed by two closely related but distinct enzymes, spermidine synthase and spermine synthase. In higher plants, in addition to their implication in fundamental cellular processes, such as chromatin organization, cell proliferation, differentiation, and programmed cell death (Thomas and Thomas, 2001; Bais and Ravishankar, 2002), polyamines were also reported to be involved in adaptive responses to abiotic (for review, see Bouchereau et al., 1999; Urano et al., 2003; Kuznetsov et al., 2006) and biotic stresses (for review, see Walters, 2003). First, they are essential components of the plant cell wall, a crucial physical barrier against pathogen invasion (Berta et al., 1997). In addition, polyamine catabolism may also contribute to defense responses through two reaction products (for review, see Cona et al., 2006): γ-aminobutyric acid, an important metabolite largely and rapidly produced in response to biotic and abiotic stresses (Bouche and Fromm, 2004), and the reactive oxygen species H2O2, which has long been recognized to play a key role in defense (Gechev et al., 2006). Last, polyamine conjugation with phenolic acids was also linked with plant defense to pathogen infection (Walters, 2000).
Figure 1.
Polyamine biosynthetic pathway in Arabidopsis. ADC, Arg decarboxylase; AIH, agmatine iminohydrolase; CPA, N-carbamoylputrescine amidohydrolase; SPDS, spermidine synthase; SPMS, spermine synthase; SAMDC, S-adenosyl-Met decarboxylase.
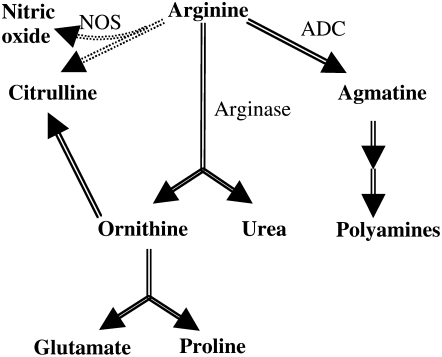
Two major metabolic pathways are closely connected to polyamine metabolism. Arg is also the precursor for the biosynthesis of nitric oxide (NO), Orn, and urea (Fig. 2). The step catalyzed by NO synthase (NOS), which allows the two-step oxidation of Arg to NO and citrulline, is intensively studied during abiotic stresses and plant-pathogen interactions (Mur et al., 2006; Arasimowicz and Floryszak-Wieczorek, 2007). The second Arg catabolism pathway is catalyzed by arginase, which hydrolyzes Arg to Orn and urea. Whereas Orn contributes to the biosynthesis of Pro and Glu, urea is further catabolized by urease to carbon dioxide and ammonium. Although research on plant arginase has mainly focused on its role in mobilizing Arg as a nitrogen source during postgermination growth (Kolloffel and van Dijke, 1975; Zonia et al., 1995; Palmieri et al., 2006), plant arginase was also reported to be involved in stress responses. Arginase activity was induced in tomato (Lycopersicon esculentum) leaves in response to wounding; treatment with jasmonic acid, a potent signal for plant defense responses; and infection with a virulent strain of Pseudomonas syringae pv tomato (Chen et al., 2004). Potential roles in protection against herbivores and in pathogen virulence were consequently proposed. However, although they are strongly interconnected because they compete for a common substrate, these three Arg catabolic pathways, i.e. biosynthesis of polyamines, biosynthesis of NO, and the urea cycle, are frequently considered independently in higher plants. In contrast to animal systems, where a switch between the different branches of Arg metabolism has long been recognized to be involved in response to wound trauma and pathogen infection (Vincendeau et al., 2003; Duleu et al., 2004; Pfaff et al., 2005), the potential role of Arg metabolism regulation in the plant-pathogen relationship is still unclear.
Figure 2.
Arg catabolism in Arabidopsis. ADC, Arg decarboxylase.
Consequently, Arg metabolism appears to be an exciting metabolic pathway potentially involved in Brassicaceae-P. brassicae interactions due to, on one hand, its central role in plant defense-responses and, on the other hand, the role of polyamines in cell proliferation and differentiation regulation. The present work aims to determine, first, whether polyamine metabolism and Arg catabolism through arginase are implicated in host responses to P. brassicae infection, and, second, whether these metabolic pathways might be involved in partial clubroot resistance mechanisms. Thus, we examined the temporal responses of polyamines and arginase to clubroot in roots of both the susceptible Columbia-0 (Col-0) accession and the partially resistant Bur-0 accession. We analyzed the expression levels of genes involved in polyamine biosynthesis and encoding arginase, and quantified arginase activity, Arg-related amino acids, and free polyamine levels. Our results show that the expression of genes involved in Arg catabolism and polyamine metabolism is induced upon inoculation with P. brassicae in both susceptible and partially resistant accessions. However, free polyamine production and Arg utilization is clearly regulated differently in partially resistant plants compared to susceptible ones.
RESULTS
Clubroot Resistance Tests
In each test, the Arabidopsis accessions Bur-0 and Col-0 were evaluated at 21 d postinoculation (dpi) for clubroot symptoms. A set of differential hosts, including susceptible and resistant genotypes of different Brassica species, was also evaluated at 49 dpi to characterize the isolate's pathogenicity. This confirmed that the selection isolate eH (Fähling et al., 2003) used in this study belongs to the most virulent P. brassicae pathotype P1 (Somé et al., 1996). ANOVA revealed no significant difference between tests but a significant phenotypic variation between genotypes (P < 0.001). The Col-0 accession, with a mean disease index (DI) of 86, was classified as significantly more susceptible than the Bur-0 accession (P < 0.05), which showed an intermediate behavior with a mean DI of 64 as previously reported (Alix et al., 2007).
Transcriptional Profiling of Genes Involved in Polyamine Metabolism and Arg Catabolism
We used quantitative real-time reverse transcription (RT)-PCR to examine the expression levels of polyamine biosynthesis and arginase-encoding genes in control and infected roots of the partially resistant Bur-0 accession and the susceptible Col-0 accession. Four independent experiments were carried out at four time points (2, 7, 14, and 21 dpi) to relate specific host responses to the life cycle of the pathogen. The first time point corresponds to the primary phase of P. brassicae infection, i.e. the first contact between primary zoospores and root hairs and development of primary plasmodia. Seven, 14, and 21 dpi correspond in a susceptible genotype to the early events of cortical cells colonization and club formation, respectively, during the secondary phase of infection (Fuchs and Sacristán, 1996). For each time point, an ANOVA was performed to evaluate inoculation and genotype effects on gene expression.
First, we could not detect any significant differences between the transcriptional profiles of genes involved in polyamine biosynthesis and Arg catabolism in control roots of the two Arabidopsis genotypes. Similarly, no significant differences were observed between control and P. brassicae-infected roots for either gene set or accession at the first two time points (data not shown). In contrast, at 14 and 21 dpi, the ANOVA showed that expression of most genes involved in polyamine biosynthesis and Arg catabolism had been significantly affected by the inoculation with transcripts accumulating in response to P. brassicae infection in both susceptible and partially resistant roots.
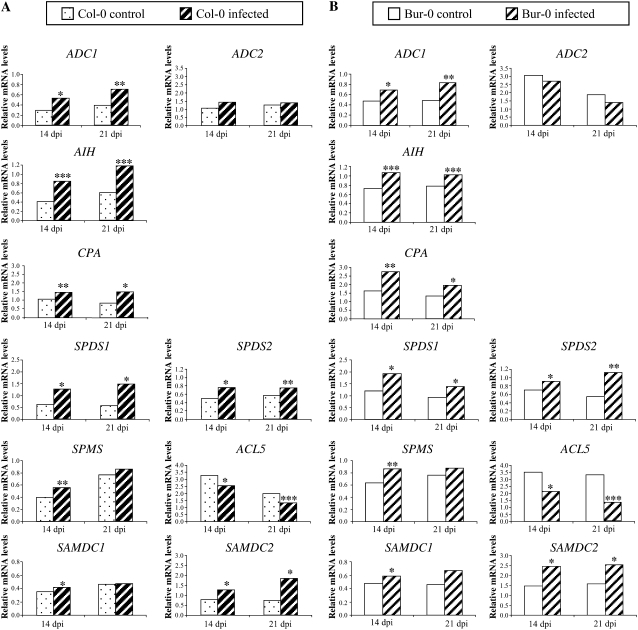
Close examination of specific gene expression profiles showed that expression of genes encoding Arg decarboxylase (ADC1), agmatine iminohydrolase (AIH), N-carbamoylputrescine amidohydrolase (CPA), spermidine synthase (SPDS1, SPDS2), and S-adenosyl-Met decarboxylase (SAMDC2) was significantly higher in P. brassicae inoculated roots than in the control at 14 and 21 dpi (P < 0.05 to P < 0.001; Fig. 3, A and B). Transcription of SAMDC1, a second gene encoding S-adenosyl-Met decarboxylase, and SPMS, encoding spermine synthase, was also induced by P. brassicae infection (P < 0.05 and P < 0.01 respectively), but only transiently at 14 dpi. In contrast, mRNA levels of ADC2, a second gene encoding Arg decarboxylase, did not change in response to infection, and expression of ACL5, a second gene encoding spermine synthase, decreased in infected plants at 14 and 21 dpi (P < 0.05 and P < 0.001). None of the genes showed significant different expression patterns between infected roots of the susceptible and partially resistant accessions.
Figure 3.
Relative transcript levels of polyamine biosynthetic genes encoding Arg decarboxylase (ADC1, ADC2), agmatine iminohydrolase (AIH), N-carbamoylputrescine amidohydrolase (CPA), spermidine synthase (SPDS1, SPDS2), spermine synthase (SPMS, ACL5), and S-adenosyl-Met decarboxylase (SAMDC1, SAMDC2) in noninfected (control) and infected roots of the susceptible accession Col-0 (A) and the partially resistant accession Bur-0 (B) at 14 and 21 dpi. Values were obtained by real-time quantitative RT-PCR and are normalized to the host Actin8 gene. Samples of control and infected roots were analyzed in duplicate in four independent experiments. Significant differences from controls are shown at *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
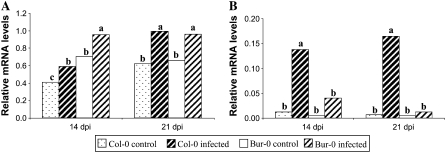
The expression of the two genes encoding arginase, ARGAH1 and ARGAH2, was also monitored throughout P. brassicae infection. ARGAH1 mRNA levels increased significantly at 14 and 21 dpi in both accessions compared to control roots (P < 0.05; Fig. 4A), but there was no significant differences in response level between the two genotypes. ARGAH2 mRNA levels were also higher in Col-0 and Bur-0 inoculated roots than in the control at 14 and 21 dpi. Interestingly, however, ARGAH2 mRNA levels were drastically induced in susceptible infected roots compared to partially resistant infected roots (Fig. 4B). This observation proved to be statistically significant with a clear interaction between genotype and inoculation factors at 14 and 21 dpi (P < 0.05 and P < 0.01). Duncan's multiple-range test (α = 0.05) performed on the four genotype × inoculation treatments also showed that ARGAH2 was expressed at significantly higher levels in infected Col-0 roots at 14 and 21 dpi. For example, at 21 dpi, the ARGAH2 expression was 25-fold higher in inoculated roots than control Col-0 roots but only 3-fold higher for the partially resistant genotype Bur-0.
Figure 4.
Relative transcript levels of the genes ARGAH1 (A) and ARGAH2 (B) encoding arginase in noninfected (control) and infected roots of the susceptible accession Col-0 and the partially resistant accession Bur-0 at 14 and 21 dpi. Values were obtained by real-time quantitative RT-PCR and are normalized to the host Actin8 gene. Samples of control and infected roots were analyzed in duplicate in four independent experiments. For each time point, same letters indicate nonsignificant difference at P = 0.05.
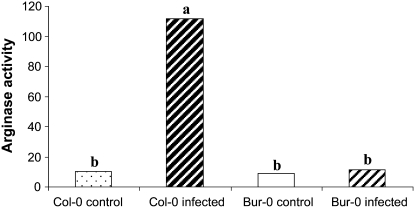
Arginase Activity
To validate our results showing induced arginase expression at the transcriptional level, arginase activity was measured in control and infected Col-0 and Bur-0 roots at 21 dpi (Fig. 5). A striking increase in arginase activity was observed in susceptible Col-0 roots in response to P. brassicae infection. Indeed, arginase activity was 10-fold higher in infected roots than in control roots. In contrast, arginase activity in infected roots of the partially resistant Bur-0 accession only increased slightly, as was observed at the transcriptional level.
Figure 5.
Levels of arginase activity in noninfected (control) and infected roots of the susceptible accession Col-0 and the partially resistant accession Bur-0 at 21 dpi. Arginase activity was expressed as nanomoles of Orn released per minute per milligram of protein. Values represent means of two replicates. Same letters indicate nonsignificant difference at P = 0.05.
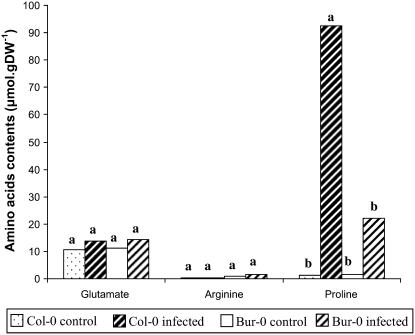
Arg-Related Amino Acids
Next, we measured the amino acid content of roots at 21 dpi, specifically looking at Arg and related amino acids, i.e. Orn, Glu, and Pro contents (Fig. 6). In noninfected roots, there was approximately 3 times more Arg in Bur-0 than Col-0. No significant change was observed for Arg contents in response to inoculation. Orn levels remained relatively low (<0.1 μmol g−1 dry weight [DW]) regardless of conditions or genotypes (data not shown). In contrast, infections had a strong impact on Pro accumulation, which reached high levels in the infected Col-0 roots. Pro also accumulated in Bur-0 roots, but in a much lower proportion. ANOVA revealed a significant interaction between genotype and inoculation factors (P < 0.05). A Duncan's multiple-range test (α = 0.05) performed on the four genotype × inoculation treatments also showed a significant higher accumulation of Pro in the infected Col-0 roots. In comparison, apparently higher levels of Glu in infected roots were very modest. ANOVA did not reveal significant changes in Glu contents in response to inoculation.
Figure 6.
Effect of P. brassicae inoculation on Arg-related amino acid contents in roots of the susceptible accession Col-0 and the partially resistant accession Bur-0 at 21 dpi. Amino acid contents were expressed as micromoles per gram of DW. Values represent means of two replicates. For each amino acid, same letters indicate nonsignificant difference at P = 0.05.
Free Polyamine Levels
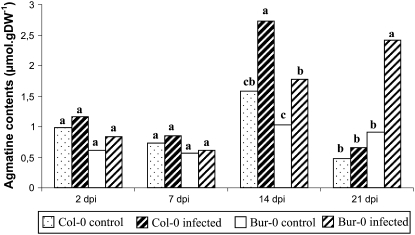
To further investigate the role of polyamine metabolism following on from the above results obtained at the transcriptional level, we quantified the levels of the precursor diamines agmatine and putrescine and the levels of polyamines spermine and spermidine at 2, 7, 14, and 21 dpi (Fig. 7; Table I). These measures were performed on the four independent experiments previously used for the transcript profiling.
Figure 7.
Contents of agmatine in noninfected (control) and infected roots of the susceptible accession Col-0 and the partially resistant accession Bur-0 at 2, 7, 14, and 21 dpi. Agmatine contents were expressed as micromoles per gram of DW. Samples of control and infected roots were analyzed in four independent experiments. For each time point, same letters indicate nonsignificant difference at P = 0.05.
Table I.
Effect of P. brassicae inoculation on free polyamine concentrations in roots of the susceptible accession Col-0 and the partially resistant accession Bur-0 at 2, 7, 14, and 21 dpi
Values represent means of four independent experiments ±se.
| Polyamine | Genotype | Inoculation | Polyamine Content
|
|||
|---|---|---|---|---|---|---|
| 2 dpi | 7 dpi | 14 dpi | 21 dpi | |||
| μmol g−1DW | ||||||
| Putrescine | Col-0 | Noninoculated | 0.37 ± 0.08 | 0.6 ± 0.16 | 0.54 ± 0.43 | 0.17 ± 0.04 |
| Inoculated | 0.32 ± 0.11 | 0.58 ± 0.3 | 0.44 ± 0.14 | 0.26 ± 0.15 | ||
| Bur-0 | Noninoculated | 0.45 ± 0.17 | 0.46 ± 0.18 | 0.25 ± 0.03 | 0.22 ± 0.10 | |
| Inoculated | 0.38 ± 0.24 | 0.45 ± 0.17 | 0.35 ± 0.10 | 0.29 ± 0.08 | ||
| Spermidine | Col-0 | Noninoculated | 0.3 ± 0.29 | 0.79 ± 0.13 | 1.8 ± 0.79 | 0.61 ± 0.82 |
| Inoculated | 0.18 ± 0.06 | 2.02 ± 1.10 | 0.44 ± 0.29 | 0.87 ± 0.84 | ||
| Bur-0 | Noninoculated | 0.18 ± 0.1 | 0.42 ± 0.39 | 0.33 ± 0.39 | 0.63 ± 0.64 | |
| Inoculated | 0.33 ± 0.33 | 0.28 ± 0.1 | 0.18 ± 0.11 | 0.69 ± 0.72 | ||
| Spermine | Col-0 | Noninoculated | 0.22 ± 0.17 | 0.35 ± 0.26 | 0.46 ± 0.55 | 0.06 ± 0.03 |
| Inoculated | 0.18 ± 0.06 | 0.49 ± 0.45 | 0.08 ± 0.01 | 0.05 ± 0.02 | ||
| Bur-0 | Noninoculated | 0.14 ± 0.1 | 0.09 ± 0.04 | 0.07 ± 0.03 | 0.06 ± 0.01 | |
| Inoculated | 0.19 ± 0.13 | 0.14 ± 0.05 | 0.06 ± 0.02 | 0.09 ± 0.04 | ||
Metabolic profiling of control Arabidopsis roots showed that agmatine and spermidine are the most abundant polyamines. An ANOVA was performed to evaluate time-point and genotype effects on each metabolite level. No significant differences in putrescine and spermine content were detected between the two genotypes. However, whereas spermine content did not change along the time course, the putrescine level decreased significantly at 21 dpi in both Col-0 and Bur-0 roots (P < 0.05). Significant increases in agmatine and spermidine were observed at 14 dpi in Col-0 roots (P < 0.05).
For each time point, ANOVA was then performed to evaluate inoculation and genotype effects on metabolite level. At the two first time points, there was no significant difference in agmatine levels in noninfected and P. brassicae-infected roots (Fig. 7). At 14 dpi, however, the agmatine level significantly increased in response to infection in both susceptible and partially resistant roots. At 21 dpi, the effect of the interaction between accession and inoculation factors was significant (P < 0.01). Indeed, whereas agmatine level continued to rise at 21 dpi in the partially resistant roots, it stopped in the susceptible roots. A Duncan's multiple-range test (α = 0.05) performed on the four accession × inoculation treatments confirmed that there was a significantly higher level of agmatine in the Bur-0 infected roots (Fig. 7).
The level of putrescine did not change in response to clubroot infection either in susceptible or partially resistant roots (Table I). As opposed to Bur-0 roots, variations in spermidine and spermine levels were detected at 7 and 14 dpi in Col-0 roots. Upon P. brassicae infection, spermidine and spermine levels in susceptible roots tended to increase at 7 dpi and then to decrease at 14 dpi; however, these variations were not statistically significant.
DISCUSSION
This study reports the involvement of Arg metabolism in the Arabidopsis-P. brassicae interaction. Consistent results obtained at the transcriptional, enzymatic, and metabolic levels demonstrated that polyamine metabolism and Arg catabolism are induced in compatible Arabidopsis-P. brassicae interactions. Furthermore, we demonstrated that upon P. brassicae infection, susceptible and partially resistant plants exhibit striking differences in the regulation of Arg metabolism. In susceptible plants (Col-0), arginase activity was massively induced at 14 dpi and 21 dpi. This was associated with no change in Orn content but with a large accumulation of Pro. Furthermore, polyamine biosynthesis was also up-regulated with an accumulation of agmatine at 14 dpi. Partially resistant plants (Bur-0), on the other hand, exhibited a slight arginase induction and a moderate accumulation of Pro. In addition, as in susceptible plants, polyamine biosynthesis was also induced; however, agmatine accumulation, observed from 14 dpi, continued to increase at 21 dpi.
Transcriptional and Enzymatic Analyses of Arg Metabolism
We determined the transcriptional profile of genes involved in Arg metabolism in response to P. brassicae infection. In this study, we were particularly interested in the arginase and polyamine pathways. The NOS pathway was not included in this study because the nature of its coding gene remains elusive and controversial (Guo et al., 2003; Zemojtel et al., 2006). In Arabidopsis, most of the enzymes involved in Orn and polyamine production are encoded by duplicated genes. However, although they encode the same enzyme, duplicated genes encoding Arg decarboxylase (ADC1 and ADC2), spermine synthase (SPMS and ACL5), and arginase (ARGAH1 and ARGAH2) were expressed differentially following P. brassicae infection. Whereas ADC1 and SPMS transcripts accumulated at 14 and 21 dpi, like other polyamine biosynthetic genes, the expression of ADC2 and ACL5 was surprisingly opposite. While ARGAH1 transcripts accumulated in a similar fashion in both susceptible and partially resistant accessions, ARGAH2 overexpression was significantly higher in the susceptible accession than in the partially resistant accession at 14 and 21 dpi. This differential regulation of gene responsiveness was previously observed in biotic and abiotic stress. Indeed, ADC2 was found to be specifically involved in hyperosmotic stress (Soyka and Heyer, 1999), in water stress (Alcazar et al., 2006), and in response to jasmonic acid and abscisic acid applications (Perez-Amador et al., 2002; Urano et al., 2003). Exogenous abscisic acid also up-regulated SPMS expression (Hanzawa et al., 2002; Urano et al., 2003) but not ACL5, which was specifically induced upon indolacetic acid application (Hanzawa et al., 2000). Even if two genes (LeARG1 and LeARG2) encoding arginase were identified in tomato, specific induction of LeARG2 was observed in response to wounding, jasmonic acid treatment, and infection with a virulent strain of P. syringae pv tomato (Chen et al., 2004).
Arginase activity measurements do not appear to exactly reflect the expression of both ARGAH1 and ARGAH2, the two genes encoding arginase. ARGAH1 showed higher basal and P. brassicae-induced expression levels than ARGAH2, but the strong enhancement of arginase activity in susceptible infected roots appears to be more consistent with the massive increase in ARGAH2 expression in susceptible plants than with the overall higher ARGAH1 expression. Taken together, these results suggest that ARGAH2 is the predominant P. brassicae-inducible isoform in Arabidopsis roots and that the two arginase isoforms have contrasting biochemical properties or differing posttranscriptional regulations.
Polyamine Metabolism and Arginase Are Induced in Compatible Arabidopsis-P. brassicae Interactions
Arg metabolism was induced in response to clubroot infection in both susceptible and partially resistant plants. In the literature, similar induction was previously reported in biotic stress, both through arginase and polyamine pathways. Chen et al. (2004) reported an increase in arginase activity in tomato plants infected with a virulent strain of P. syringae pv tomato. Furthermore, in mammalian systems, arginase induction has long been associated with various parasite infections (Vincendeau et al., 2003; Duleu et al., 2004). The polyamine biosynthesis pathway was induced in both compatible and incompatible host-pathogen interactions (for review, see Walters, 2000, 2003). Furthermore, consistent with our results, Cao et al. (2008) also reported, at the proteome level, the up-regulation of the polyamine biosynthetic enzyme spermidine synthase in Brassica napus in response to P. brassicae infection. Previous reports on polyamine contents generally focused on the diamine putrescine and/or the polyamines spermidine and spermine; however, no information is currently available on the regulation of agmatine, the precursor to putrescine, in response to pathogen infection. The present study is thus the first report of agmatine involvement in biotic stress. Surprisingly, whereas agmatine accumulated in response to infection in Col-0 and Bur-0 roots, no significant variations in putrescine, spermidine, and spermine levels were reported. Burtin and Michael (1997) also reported that ADC overexpression in transgenic tobacco plants induced agmatine accumulation but did not affect putrescine, spermine, and spermidine levels. These results suggest that other polyamine-regulating mechanisms are involved, such as polyamine catabolism (for review, see Cona et al., 2006), conjugation, and transport (for review, see Martin-Tanguy, 2001). Hydroxycinnamic acid amide conjugates were proposed to play a role in defense mechanisms against biotic and abiotic stress, by acting as radical scavengers, antifungal agents, or in strengthening the plant cell wall against microbial degradation (Bors et al., 1989; Walters, 2000; Von Roepenack-Lahaye et al., 2003). Walters and Shuttleton (1985) measured the free polyamine levels in turnip (Brassica rapa) roots infected by P. brassicae and showed that putrescine, spermidine, and spermine concentrations were higher in “clubbed” regions of the infected turnip roots than in noninfected roots, while the concentrations were lower in regions of infected roots not exhibiting symptoms of clubroot development. These results suggest that in infected roots, homeostatic regulation, involving transport of polyamines from regions not exhibiting symptoms to “clubbed” regions, may be taking place. Because we looked at whole roots, any localized variations in polyamine levels due to this type of transport mechanism between the different parts of infected roots were not detected. Last, we cannot exclude that some of the polyamines we measured were contributed by P. brassicae. Indeed, due to its exclusive intracellular life cycle, it is difficult to distinguish between metabolites of plant or pathogen origin.
Susceptible and Partially Resistant Plants Showed Differences in Arg Catabolism Regulation following P. brassicae Infection
Arginase catabolism was strongly induced in the susceptible plants. ARGAH1 and particularly ARGAH2 expression and arginase activity markedly increased upon P. brassicae infection. Induction of arginase may represent a pathogenicity strategy by P. brassicae. Indeed, because arginase competes with NOS for a common substrate, its induction could play an important role in pathogenesis by attenuating the production of NO-mediated host defenses. This hypothesis is supported by increasing evidence from mammalian systems (Vincendeau et al., 2003). For example, trypanosomes can evade host defenses by stimulating the expression of macrophage arginase, which effectively inhibits NO production and NO-mediated trypanosome killing (Duleu et al., 2004). Arginase induction could also be a way of diverting nitrogen metabolism in favor of the pathogen. Induction occurred at 14 d and 21 dpi, corresponding to the second phase of the P. brassicae life cycle (Fuchs and Sacristán, 1996). During this phase, multinucleate plasmodia grow by mitotic division and consequently cause the hypertrophy and hyperplasia of host cells. By analogy to the proposed role of arginase in nitrogen metabolism during postgerminative growth (Kolloffel and van Dijke, 1975; Zonia et al., 1995), P. brassicae-induced degradation of Arg to ammonium and Orn may provide a mechanism to divert plant nitrogen into the production of amino acids indispensable for pathogen multiplication. Plasmodia would thus redirect host nutrients to their own benefits, thereby acting as a metabolic sink. P. brassicae was previously proposed to interfere in host carbon metabolism following observations that carbohydrates accumulate in infected tissues (Evans and Scholes, 1995). We also observed an acute accumulation of Pro in Col-0 infected roots, which strengthens the idea that susceptibility is associated with an enhancement of the metabolic flux from Arg to Pro. Free Pro biosynthesis and accumulation at high levels is very common in plants subjected to osmotic, drought, or saline strains (Delauney and Verma, 1993). Of note, although reasonable, the amounts of Pro detected in infected roots in this experiment were of surprising magnitude (92 μmol g DW−1), and this may warrant further attention. Pro, γ-aminobutyric acid, and α-aminoadipic acid were recently observed to strongly accumulate in T-DNA-induced Arabidopsis tumors cells and were viewed by the authors as stress metabolites (Deeken et al., 2006). Our findings with this pathosystem, however, raise several issues. Was all the neosynthetized Pro pool actually derived from Orn through an Orn aminotransferase activity or was a more classically described stress-induced Glu to Pro biosynthetic pathway involved? In which cellular and subcellular compartments, including intracellular pathogen, do Pro biosynthesis and accumulation really take place? Without answers to these questions, it is difficult to conclude if, on one hand, Pro accumulated inside the pathogen favoring its growth through, for instance, osmotic or oxidative protection (Chen and Dickman, 2005), or if, on the other hand, Pro accumulation in the plant tissue simply reflects attempts by plant metabolism to manage the cellular stress caused by pathogen growth. In the partially resistant accession, agmatine accumulation occurred at 14 and 21 dpi and slight arginase induction and Pro accumulation were observed upon inoculation. This weaker response in partially resistant plants may suggest that the pathogen influence on host-metabolism was attenuated or delayed compared to the situation in susceptible plants. However, as yet we cannot conclude whether this regulation of Arg metabolism is the cause or the result of the partial clubroot resistance.
Further investigations using various sets of mutants and overexpressors are planned in our laboratory to test some of these hypotheses. For instance, genetic manipulation of arginase expression in Arabidopsis transgenic plants or quantification of arginase activity in a range of Brassicaceae, showing extreme and intermediate levels of resistance to clubroot, will provide insights into the physiological function of arginase in host-pathogen interactions and in partial clubroot resistance. Moreover, mutant genotypes for genes encoding enzymes involved in polyamine metabolism will be assessed for responsiveness to P. brassicae. However that may be, the coordination of the multiple routes from Arg directing metabolites toward nitrogen and carbon trophic channels, defense systems, growth regulators, or signaling compounds remains undoubtedly a major challenge for stressed plants and may be considered as a target of prime interest within the scope of quantitative resistance to P. brassicae.
MATERIALS AND METHODS
Pathogen
The selection isolate eH (Fähling et al., 2003), used in this study, belongs to the most virulent Plasmodiophora brassicae pathotype P1, according to the host differential set established by Somé et al. (1996). It was kindly provided by J. Siemens (University of Dresden, Germany).
Plant Materials
Seeds of the Arabidopsis (Arabidopsis thaliana) accessions Bur-0 (172AV) and Col-0 (186AV) were obtained from the Versailles Resource Center. These accessions are partially resistant and susceptible, respectively, to the P. brassicae isolate eH (Alix et al., 2007). Brassica napus subsp. oleifera ‘Nevin’ (ECD6), B. napus subsp. rapifera ‘Wilhelmsburger’ (ECD10), and B. napus subsp. oleifera (Brutor), which constitute the host differential set established by Somé et al. (1996), and the highly clubroot susceptible Brassica rapa subsp. pekiniensis ‘Granaat’ (ECD5) were included as controls in each clubroot test.
Growth Conditions and Inoculation Procedure
Five independent experiments were performed using a two-block design. Arabidopsis seeds were placed on wet blotting paper in petri dishes at 4°C for 3 d to synchronize germination. Seeds were then individually sown in 4-cm-diameter pots containing a [2/3 compost, 1/3 vermiculite] mix sterilized by autoclaving. Arabidopsis plants were grown under controlled environmental conditions (16 h light at 22°C and 8 h dark at 19°C) and inoculated 7 d after germination (stage 1.04; Boyes et al., 2001). The inoculum was prepared according to Manzanares-Dauleux et al. (2000a), and inoculation was performed by applying 1 mL of resting spore suspension (107 spores mL−1) to the crown of each seedling. The resting spore suspension was replaced by distilled water for control plants. To relate specific host responses to the life cycle of the pathogen, infected and control plants were then harvested at four time points: 2, 7, 14, and 21 dpi (respectively, stages 1.04, 1.08, 1.12, and 3.90–6.50; Boyes et al., 2001). In the last experiment, infected and control plants were only harvested at 21 dpi (stages 3.90–6.50; Boyes et al., 2001). Depending on the kinetic point, between 84 and 600 individual plants were harvested per analysis point. Roots of the whole plants were thoroughly rinsed in different baths of distilled water. Roots and leaves dissected from whole plants were frozen in liquid nitrogen and stored at −80°C. To check that the inoculation was successful, in each test, six to 12 plants of each Arabidopsis accession were evaluated for clubroot resistance at 21 dpi and symptoms were recorded using the scale previously described for Brassica oleracea (Manzanares-Dauleux et al., 2000b): 0, no visible swelling; 1, very slight swelling usually confined to lateral roots; 2, moderate swelling on lateral roots and taproot; 2+, severe clubs on all roots but some roots remain; and 3, no root left, only one big gall. DI was calculated: DI = (n1 × 25 + n2 × 50 + n2+ × 75 + n3 × 100)/N, where “ni” is the number of plants in the symptom class “i” and N is the total number of plants tested; an accession displaying a DI of zero is completely resistant and develops no clubroot symptoms while an accession with a DI of 100 is highly susceptible (Manzanares-Dauleux et al., 2000b).
Real-Time RT-PCR
Total RNA was extracted from approximately 30 mg of frozen root samples using the SV Total RNA Isolation kit (Promega). Any remaining genomic DNA was removed by digestion with DNase I (DNA-free; Ambion) according to the manufacturer's protocol. To check for genomic DNA contamination, a PCR reaction was carried out on the RNA samples using Actin8 primers. The total RNA was quantified with a spectrophotometer and electrophoresed on a 2% agarose gel to check the concentration and integrity. First-strand complementary DNA (cDNA) synthesis was performed in a 20-μL total reaction volume using 250 ng of DNAse-digested total RNA, 1 μm oligo(dT) primer, 1 mm dNTPs, 1× first-strand buffer (Invitrogen), 20 mm dithiothreitol (Invitrogen), 40 units RNaseOUT recombinant ribonuclease inhibitor (Invitrogen), and 200 units SuperScript II reverse transcriptase (Invitrogen) by incubating for 2 h at 42°C. The reaction was terminated by incubation for 15 min at 70°C. The cDNAs were diluted by 1/40 and their quality was confirmed by conventional RT-PCR with Actin8 primers (Table II).
Table II.
Genes and oligonucleotides used in the real-time RT-PCR experiments
| Genes | Loci | Encoded Proteins | Primers (5′–3′) |
|---|---|---|---|
| Actin8 | At1g49240 | Actin | TTACCCGACGGACAAGTGATC |
| ATGATGGCTGGAAAAGGACTTC | |||
| ADC1 | At2g16500 | Arg decarboxylase | CCAAGGTGTGTATCCTGTGAAAT |
| AGCTTCTAAACCGAATCGAAAAC | |||
| ADC2 | At4g34710 | Arg decarboxylase | GCGATGGACCACACAGCTTT |
| AGAACATCCGCTGAGGACTGA | |||
| AIH | At5g08170 | Agmatine iminohydrolase | TCGAGAATGCAAGAGAGATCGTT |
| CATTTTCGGCGACGGAAGTA | |||
| CPA | At2g27450 | N-carbamoylputrescine amidohydrolase | GATCAAGTCGAAAAGGCAAAGCT |
| CCATCCATAGTAAGAAGCACCTTGT | |||
| SPDS1 | At1g23820 | Spermidine synthase | CTAATCTGGAGGATCGTTGAATTT |
| ATAGTCGCTCTTGCATTGTGTTAC | |||
| SPDS2 | At1g70310 | Spermidine synthase | TTGCCCGTGAAGAGACCTAGA |
| TCCACCGTTCTCTGTTTCCAT | |||
| SPMS | At5g53120 | Spermine synthase | TGGCTCCATACTCATCTTATTGAA |
| CGCATAGTGAACACTTTTGAATG | |||
| ACL5 | At5g19530 | Spermine synthase | CCATCATTTGCGGACACATG |
| GAGACGAAAGAAGGAGCGTTTAGA | |||
| SAMDC1 | At3g02470 | S-adenosyl-Met decarboxylase | TCTTTGAGCCAAGCATCTTTCA |
| GCAGCAGGTGTAAGAATTTCATCA | |||
| SAMDC2 | At5g15950 | S-adenosyl-Met decarboxylase | TCTCCGAGATCTACCTTGAAATG |
| GATTCCCTATTCCTTCTCGTCCT | |||
| ARGAH1 | At4g08900 | Arginase/agmatinase | GTCGAGGATTATTGGTAGAAAAGG |
| GAACGACGCAGAATTTAGTCTATG | |||
| ARGAH2 | At4g08870 | Arginase/agmatinase | TGGTACTTTGGAGTTTAATCGTTG |
| TGTCTATGAGACCACACTTATTGC |
For each gene, primers for real-time RT-PCR were designed on gene sequence tags (Hilson et al., 2004) with Primer Express v1.5 software (Applied Biosystems) and synthesized by Eurogentec. The genes, as well as the sequence of their specific oligonucleotides, are presented in Table I. Three microliters of cDNA was used for PCR amplification using the SYBR Green PCR Master kit containing a Hot Start Taq polymerase (Applied Biosystems) and 0.9 μm of each specific primer, in the ABI PRISM 7700 sequence detection system (Applied Biosystems). Ten-microliter reactions were run in duplicate in a 96-well thin-wall PCR plate (MicroAmp Optical 96-wells reaction plate; Applied Biosystems). The PCR amplification protocol consisted of a denaturation step at 95°C for 10 min, followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. The SYBR Green I fluorescence signal was measured during the 60°C annealing step. To check the annealing specificity of each oligonucleotide, melting-curve analysis (55°C–94°C) was carried out at the end of amplification. For calculations, a standard curve was determined for each gene using different dilutions of cDNA products. Expression levels for each target gene were then quantified following normalization to Actin8, the endogenous reference.
Arginase Assays
A known weight of frozen powdered root tissues (usually 0.5 g) was ground and homogenized in 1 mL of extraction buffer for 3 min. The extraction medium consisted of 100 mm Tris-HCl, pH 7.5, 1% (v/v) 2-mercaptoethanol, and 0.1 mm phenylmethylsulfonyl fluoride (Chen et al., 2004). Homogenates were centrifuged at 10,000g for 15 min at 4°C and the supernatants were used as the enzyme source. Protein concentrations were determined by the Bradford method (Bradford, 1976). Before the arginase activity was assayed, the enzyme extract was made up to 5 mm in MnCl2 and left for 15 min at room temperature to activate arginase (Goldraij and Polacco, 1999). A standard reaction mixture contained 450 μL of 125 mm Tris-HCl, pH 9.5, 50 μL of 550 mm l-Arg (adjusted to pH 9.5), 25 μL of 44 mm MnCl2, and 25 μL of enzyme source. Control assays were concurrently performed by removing l-Arg or replacing native enzyme extract by boiled enzyme extract or increasing enzyme dose in the assay. The reaction mixture was incubated with continuous agitation (500 rpm) for 45 min at 30°C, and the reaction was stopped at 0, 15, 30, and 45 min by adding 500 μL of 10% (v/v) perchloric acid. Precipitated proteins were removed by centrifugation at 12,000g for 15 min, and Orn released in the medium was spectrophotometrically measured by the Chinard method (Chinard, 1952) modified according to Roubelakis and Kliewer (1978). Absorbances were read at 515 nm, and standard l-Orn solutions (0–250 μm) were used for calibration. To verify the results obtained by this colorimetric method, the amino acid composition of the reaction mixture was determined by UPLC with the Waters AccQ-Tag amino acid analysis system as described below. Arginase activity was expressed as the average of triplicates assays as nanomoles of Orn released per minute per milligram of protein.
Arg-Related Amino Acid Contents
Samples were ground in liquid nitrogen and freeze-dried. Methanol-chloroform-water-based extractions were made on 10 mg of the resulting dry powder using the following procedure: the powder was suspended in 400 μL of a 100 μm dl-3-aminobutyric acid solution in methanol, followed by 15 min of agitation at room temperature. Two hundred microliters of chloroform was then added, followed by a 5-min agitation step. Finally, 400 μL of water was added, and samples were then vortexed vigorously to induce phase separation and centrifuged at 13,000g for 5 min. The upper phase, which contains amino acids, was transferred to a clean vial and dried under vacuum. Dry residues were resuspended in 50 μL of ultra-pure water and 10 μL was used for the derivatization using the AccQ-Tag Ultra derivitization kit (Waters). Derivatized amino acids were analyzed using an Acquity UPLC system (Waters). One microliter of the reaction mixture was injected onto an Acquity UPLC BEH C18 1.7-μm 2.1- × 100-mm column heated at 55°C. Amino acids were eluted with a mixture of 10-fold diluted AccQ-Tag Eluent A (Waters) and pure acetonitrile at a flow rate of 0.7 mL min−1 according to the following gradient: initial, 99.9% A; 0.54 min, 99.9% A; 6.50 min, 90.9% A, curve 7; 8.50 min, 78.8% A, curve 6; 8.90 min, 40.4% A, curve 6; 9.50 min, 40.4% A, curve 6; 9.60 min, 99.9% A, curve 6; and 10.10 min, 99.9% A. Derivatized amino acids were detected at 260 nm using a photo diode array detector. Amino acids were characterized by cochromatography of individual standards and quantified by comparison of individual external calibration curves.
Isolation of Free Polyamines
Extraction
DWs were determined after freeze-drying. Approximately 5 to 30 mg of powdered root samples were thoroughly mixed with 800 μL of 1 m HCl supplemented with 10 μm heptanediamine, as internal standard, on a magnetic stirring plate (2,000 rpm) for 1 h at 4°C. The homogenates were then centrifuged for 15 min at 10,000g at 4°C and the supernatants collected. The pellets were further extracted twice with 600 μL of 1 m HCl and 10 μm heptanediamine and stirred for 30 min at 4°C. The homogenates were centrifuged for 15 min at 10,000g at 4°C. The combined supernatants were used as the crude extract for characterization and determination of free polyamines.
Chromatographic Analysis
Extracted polyamines were derivatized with dansyl chloride (5-dimethylamino-1-naphthalenesulfonyl chloride) according to the method of Smith and Davies (1985). Two hundred microliters of supernatant was added to 100 mg of sodium carbonate and 600 μL of dansyl chloride in acetone (10 mg mL−1). The mixture was incubated overnight at room temperature. Three hundred microliters of aqueous Pro (150 mg mL−1) was added to the mixture to remove the excess dansyl chloride. The dansylated polyamines were further extracted with ethyl acetate. The organic phase containing polyamines was dried under nitrogen stream and the residue solubilized with 800 μL of methanol and stored at −20°C until analysis. Free polyamines were separated and quantified through HPLC after yield correction with the internal standard and calibration with external standards (Hayman et al., 1985; Duhaze et al., 2002). The mobile phase consisted of a solution of 17.5 mm potassium acetate (pH 7.17) as eluant A and acetonitrile as eluant B. The solvent gradient was modified according to Hayman et al. (1985) as follows: t = 0 min, 70% A; t = 2 min, 58% A; t = 24 min, 58% A; t = 25 min, 55% A; t = 32 min, 55% A; t = 52 min, 45% A; t = 53 min, 35% A; t = 64 min, 35% A; t = 70 min, 20% A; t = 75 min, 100% B; and t = 80 min, 70% A. The flow rate of the mobile phase was 1.5 mL min−1. The column was then rinsed for 8 min with 70% A before the next injection. Fluorescence was measured with an excitation wavelength of 366 nm and an emission wavelength of 490 nm. The HPLC design consisted of an HP series 1050 system, an autosampler with a 20-μL injection loop, a Shimadzu RF-10AXL fluorometer, and a Waters Spherisorb 5-μm ODS2 column (4.6 × 250 mm). Signals were computed and analyzed using AZUR software (Datalys).
Statistical Analysis
The data were statistically analyzed using a generalized linear model (PROC GLM of Statistical Analysis System software; SAS Institute Inc., 2000). Multiple comparisons of means were performed using Duncan's multiple-range test (α = 0.05).
Acknowledgments
We acknowledge Henri Bellis, Pascal Glory, Marcellin Deschamps, and our colleagues of OUEST-Génopole for technical assistance, and Dr. Françoise Hennion for her help in polyamine investigations. Mélanie Jubault is a Ph.D. student funded by the French Ministry of Research.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Maria J. Manzanares-Dauleux (maria.manzanares@agrocampus-rennes.fr).
References
- Alcazar R, Cuevas JC, Patron M, Altabella T, Tiburcio AF (2006) Abscisic acid modulates polyamine metabolism under water stress in Arabidopsis thaliana. Physiol Plant 128 448–455 [Google Scholar]
- Alix K, Lariagon C, Delourme R, Manzanares-Dauleux MJ (2007) Exploiting natural genetic diversity and mutant resources of Arabidopsis thaliana to study the A. thaliana-Plasmodiophora brassicae interaction. Plant Breed 126 218–221 [Google Scholar]
- Ando S, Tsushima S, Tagiri A, Kamachi S, Konagaya KI, Hagio T, Tabei Y (2006) Increase in BrAO1 gene expression and aldehyde oxidase activity during clubroot development in Chinese cabbage (Brassica rapa L.). Mol Plant Pathol 7 223–234 [DOI] [PubMed] [Google Scholar]
- Arasimowicz M, Floryszak-Wieczorek J (2007) Nitric oxide as a bioactive signalling molecule in plant stress responses. Plant Sci 172 876–887 [Google Scholar]
- Bais HP, Ravishankar GA (2002) Role of polyamines in the ontogeny of plants and their biotechnological applications. Plant Cell Tissue Organ Cult 69 1–34 [Google Scholar]
- Berta G, Altamura MM, Fusconi A, Cerruti F, Capitani F, Bagni N (1997) The plant cell wall is altered by inhibition of polyamine biosynthesis. New Phytol 137 569–577 [Google Scholar]
- Bors W, Langebartels C, Michel C, Sandermann H Jr (1989) Polyamines as radical scavengers and protectants against ozone damage. Phytochemistry 28 1589–1595 [Google Scholar]
- Bouche N, Fromm H (2004) GABA in plants: just a metabolite? Trends Plant Sci 9 110–115 [DOI] [PubMed] [Google Scholar]
- Bouchereau A, Aziz A, Larher F, Martin-Tanguy J (1999) Polyamines and environmental challenges: recent development. Plant Sci 140 103–125 [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Gorlach J (2001) Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72 248–254 [DOI] [PubMed] [Google Scholar]
- Burtin D, Michael AJ (1997) Overexpression of arginine decarboxylase in transgenic plants. Biochem J 325 331–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao T, Srivastava S, Rahman MH, Kav NNV, Hotte N, Deyholos MK, Strelkov SE (2008) Proteome-level changes in the roots of Brassica napus as a result of Plasmodiophora brassicae infection. Plant Sci 174 97–115 [Google Scholar]
- Chen CB, Dickman MB (2005) Proline suppresses apoptosis in the fungal pathogen Colletotrichum trifolii. Proc Natl Acad Sci USA 102 3459–3464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, McCaig BC, Melotto M, He SY, Howe GA (2004) Regulation of plant arginase by wounding, jasmonate, and the phytotoxin coronatine. J Biol Chem 279 45998–46007 [DOI] [PubMed] [Google Scholar]
- Chinard FP (1952) Photometric estimation of proline and ornithine. J Biol Chem 199 91–95 [PubMed] [Google Scholar]
- Cona A, Rea G, Angelini R, Federico R, Tavladoraki P (2006) Functions of amine oxidases in plant development and defence. Trends Plant Sci 11 80–88 [DOI] [PubMed] [Google Scholar]
- Deeken R, Engelmann JC, Efetova M, Czirjak T, Muller T, Kaiser WM, Tietz O, Krischke M, Mueller MJ, Palme K, et al (2006) An integrated view of gene expression and solute profiles of Arabidopsis tumors: a genome-wide approach. Plant Cell 18 3617–3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delauney AJ, Verma DPS (1993) Proline synthesis and osmoregulation in plants. Plant J 4 215–223 [Google Scholar]
- Devos S, Laukens K, Deckers P, Van der Straeten D, Beeckman T, Inzé D, Van Onckelen H, Witters E, Prinsen E (2006) A hormone and proteome approach to picturing the initial metabolic events during Plasmodiophora brassicae infection on Arabidopsis. Mol Plant Microbe Interact 19 1431–1443 [DOI] [PubMed] [Google Scholar]
- Duhaze C, Gouzerh G, Gagneul D, Larher F, Bouchereau A (2002) The conversion of spermidine to putrescine and 1,3-diaminopropane in the roots of Limonium tataricum. Plant Sci 163 639–646 [Google Scholar]
- Duleu S, Vincendeau P, Courtois P, Semballa S, Lagroye I, Daulouede S, Boucher JL, Wilson KT, Veyret B, Gobert AP (2004) Mouse strain susceptibility to trypanosome infection: an arginase-dependent effect. J Immunol 172 6298–6303 [DOI] [PubMed] [Google Scholar]
- Evans JL, Scholes JD (1995) How does clubroot alter the regulation of carbon metabolism in its host? Asp Appl Biol 42 125–132 [Google Scholar]
- Fähling M, Graf H, Siemens J (2003) Pathotype separation of Plasmodiophora brassicae by the host plant. J Phytopathol 151 425–430 [Google Scholar]
- Fuchs H, Sacristán MD (1996) Identification of a gene in Arabidopsis thaliana controlling resistance to clubroot (Plasmodiophora brassicae) and characterization of the resistance response. Mol Plant Microbe Interact 9 91–97 [Google Scholar]
- Gechev TS, Van Breusegem F, Stone JM, Denev I, Laloi C (2006) Reactive oxygen species as signals that modulate plant stress responses and programmed cell death. Bioessays 28 1091–1101 [DOI] [PubMed] [Google Scholar]
- Goldraij A, Polacco JC (1999) Arginase is inoperative in developing soybean embryos. Plant Physiol 119 297–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grsic-Rausch S, Kobelt P, Siemens JM, Bischoff M, Ludwig-Muller J (2000) Expression and localization of nitrilase during symptom development of the clubroot disease in Arabidopsis. Plant Physiol 122 369–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F-Q, Okamoto M, Crawford NM (2003) Identification of a plant nitric oxide synthase gene involved in hormonal signaling. Science 302 100–103 [DOI] [PubMed] [Google Scholar]
- Hanfrey C, Sommer S, Mayer MJ, Burtin D, Michael AJ (2001) Arabidopsis polyamine biosynthesis: absence of ornithine decarboxylase and the mechanism of arginine decarboxylase activity. Plant J 27 551–560 [DOI] [PubMed] [Google Scholar]
- Hanzawa Y, Imai A, Michael AJ, Komeda Y, Takahashi T (2002) Characterization of the spermidine synthase-related gene family in Arabidopsis thaliana. FEBS Lett 527 176–180 [DOI] [PubMed] [Google Scholar]
- Hanzawa Y, Takahashi T, Michael AJ, Burtin D, Long D, Pineiro M, Coupland G, Komeda Y (2000) ACAULIS5, an Arabidopsis gene required for stem elongation, encodes a spermine synthase. EMBO J 19 4248–4256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayman AR, Gray DO, Evans SV (1985) New high-performance liquid chromatography system for the separation of biogenic amines as their Dns derivatives. J Chromatogr 325 462–466 [DOI] [PubMed] [Google Scholar]
- Hilson P, Allemeersch J, Altmann T, Aubourg S, Avon A, Beynon J, Bhalerao RP, Bitton F, Caboche M, Cannoot B, et al (2004) Versatile gene-specific sequence tags for Arabidopsis functional genomics: transcript profiling and reverse genetics applications. Genome Res 14 2176–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai M (2006) Genetic analysis of clubroot resistance in Brassica crops. Breed Sci 56 223–229 [Google Scholar]
- Hirai M, Harada T, Kubo N, Tsukada M, Suwabe K, Matsumoto S (2004) A novel locus for clubroot resistance in Brassica rapa and its linkage markers. Theor Appl Genet 108 639–643 [DOI] [PubMed] [Google Scholar]
- Ingram DS, Tommerup IC (1972) The life history of Plasmodiophora brassicae Woron. Proc R Soc Lond B Biol Sci 180 103–112 [Google Scholar]
- Koch E, Cox R, Williams PH (1991) Infection of Arabidopsis thaliana by Plasmodiophora brassicae. J Phytopathol 132 99–104 [Google Scholar]
- Kolloffel C, van Dijke HD (1975) Mitochondrial arginase activity from cotyledons of developing and germinating seeds of Vicia faba L. Plant Physiol 55 507–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov VV, Radyukina NL, Shevyakova NI (2006) Polyamines and stress: biological role, metabolism, and regulation. Russ J Plant Physiol 53 583–604 [Google Scholar]
- Ludwig-Muller J, Pieper K, Ruppel M, Cohen JD, Epstein E, Kiddle G, Bennett R (1999) Indole glucosinolate and auxin biosynthesis in Arabidopsis thaliana (L.) Heynh. glucosinolate mutants and the development of clubroot disease. Planta 208 409–419 [DOI] [PubMed] [Google Scholar]
- Manzanares-Dauleux MJ, Delourme R, Baron F, Thomas G (2000. a) Mapping of one major gene and of QTLs involved in resistance to clubroot in Brassica napus. Theor Appl Genet 101 885–891 [Google Scholar]
- Manzanares-Dauleux MJ, Divaret I, Baron F, Thomas G (2000. b) Evaluation of French Brassica oleracea landraces for resistance to Plasmodiophora brassicae. Euphytica 113 211–218 [Google Scholar]
- Martin-Tanguy J (2001) Metabolism and function of polyamines in plants: recent development (new approaches). Plant Growth Regul 34 135–148 [Google Scholar]
- Mur LAJ, Carver TLW, Prats E (2006) NO way to live; the various roles of nitric oxide in plant-pathogen interactions. J Exp Bot 57 489–505 [DOI] [PubMed] [Google Scholar]
- Neuhaus K, Grsic-Rausch S, Sauerteig S, Ludwig-Muller J (2000) Arabidopsis plants transformed with nitrilase 1 or 2 in antisense direction are delayed in clubroot development. J Plant Physiol 156 756–761 [Google Scholar]
- Palmieri L, Todd CD, Arrigoni R, Hoyos ME, Santoro A, Polacco JC, Palmieri F (2006) Arabidopsis mitochondria have two basic amino acid transporters with partially overlapping specificities and differential expression in seedling development. Biochim Biophys Acta 1757 1277–1283 [DOI] [PubMed] [Google Scholar]
- Perez-Amador MA, Leon J, Green PJ, Carbonell J (2002) Induction of the arginine decarboxylase ADC2 gene provides evidence for the involvement of polyamines in the wound response in Arabidopsis. Plant Physiol 130 1454–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaff AW, Villard O, Klein JP, Mousli M, Candolfi E (2005) Regulation of Toxoplasma gondii multiplication in BeWo trophoblast cells: cross-regulation of nitric oxide production and polyamine biosynthesis. Int J Parasitol 35 1569–1576 [DOI] [PubMed] [Google Scholar]
- Piao ZY, Deng YQ, Choi SR, Park YJ, Lim YP (2004) SCAR and CAPS mapping of CRb, a gene conferring resistance to Plasmodiophora brassicae in Chinese cabbage (Brassica rapa ssp. pekinensis). Theor Appl Genet 108 1458–1465 [DOI] [PubMed] [Google Scholar]
- Rocherieux J, Glory P, Giboulot A, Boury S, Barbeyron G, Thomas G, Manzanares-Dauleux MJ (2004) Isolate-specific and broad-spectrum QTLs are involved in the control of clubroot in Brassica oleracea. Theor Appl Genet 108 1555–1563 [DOI] [PubMed] [Google Scholar]
- Roubelakis KA, Kliewer WM (1978) Enzymes of Krebs-Henseleit cycle in Vitis vinifera L. III. In vivo and in vitro studies of arginase. Plant Physiol 62 344–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito M, Kubo N, Matsumoto S, Suwabe K, Tsukada M, Hirai M (2006) Fine mapping of the clubroot resistance gene, Crr3, in Brassica rapa. Theor Appl Genet 114 81–91 [DOI] [PubMed] [Google Scholar]
- Schuller A, Ludwig-Muller J (2006) A family of auxin conjugate hydrolases from Brassica rapa: characterization and expression during clubroot disease. New Phytol 171 145–158 [DOI] [PubMed] [Google Scholar]
- Siemens J, Keller I, Sarx J, Kunz S, Schuller A, Nagel W, Schmulling T, Parniske M, Ludwig-Muller J (2006) Transcriptome analysis of Arabidopsis clubroots indicate a key role for cytokinins in disease development. Mol Plant Microbe Interact 19 480–494 [DOI] [PubMed] [Google Scholar]
- Siemens J, Nagel M, Ludwig-Muller J, Sacristán MD (2002) The interaction of Plasmodiophora brassicae and Arabidopsis thaliana: parameters for disease quantification and screening of mutant lines. J Phytopathol 150 592–605 [Google Scholar]
- Smith MA, Davies PJ (1985) Separation and quantitation of polyamines in plant tissue by high performance liquid chromatography of their dansyl derivatives. Plant Physiol 78 89–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somé A, Manzanares MJ, Laurens F, Baron F, Thomas G, Rouxel F (1996) Variation for virulence on Brassica napus L. amongst Plasmodiophora brassicae collections from France and derived single-spore isolates. Plant Pathol 45 432–439 [Google Scholar]
- Soyka S, Heyer AG (1999) Arabidopsis knockout mutation of ADC2 gene reveals inducibility by osmotic stress. FEBS Lett 458 219–223 [DOI] [PubMed] [Google Scholar]
- Suwabe K, Tsukazaki H, Iketani H, Hatakeyama K, Fujimura M, Nunome T, Fukuoka H, Matsumoto S, Hirai M (2003) Identification of two loci for resistance to clubroot (Plasmodiophora brassicae Woronin) in Brassica rapa L. Theor Appl Genet 107 997–1002 [DOI] [PubMed] [Google Scholar]
- Suwabe K, Tsukazaki H, Iketani H, Hatakeyama K, Kondo M, Fujimura M, Nunome T, Fukuoka H, Hirai M, Matsumoto S (2006) Simple sequence repeat-based comparative genomics between Brassica rapa and Arabidopsis thaliana: the genetic origin of clubroot resistance. Genetics 173 309–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas T, Thomas TJ (2001) Polyamines in cell growth and cell death: molecular mechanisms and therapeutic applications. Cell Mol Life Sci 58 244–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano K, Yoshiba Y, Nanjo T, Igarashi Y, Seki M, Sekiguchi F, Yamaguchi-Shinozaki K, Shinozaki K (2003) Characterization of Arabidopsis genes involved in biosynthesis of polyamines in abiotic stress responses and developmental stages. Plant Cell Environ 26 1917–1926 [Google Scholar]
- Vincendeau P, Gobert AP, Daulouede S, Moynet D, Mossalayi MD (2003) Arginases in parasitic diseases. Trends Parasitol 19 9–12 [DOI] [PubMed] [Google Scholar]
- Von Roepenack-Lahaye E, Newman M-A, Schornack S, Hammond-Kosack KE, Lahaye T, Jones JDG, Daniels MJ, Dow JM (2003) p-Coumaroylnoradrenaline, a novel plant metabolite implicated in tomato defense against pathogens. J Biol Chem 278 43373–43383 [DOI] [PubMed] [Google Scholar]
- Walters DR (2000) Polyamines in plant-microbe interactions. Physiol Mol Plant Pathol 57 137–146 [Google Scholar]
- Walters DR (2003) Polyamines and plant disease. Phytochemistry 64 97–107 [DOI] [PubMed] [Google Scholar]
- Walters DR, Shuttleton MA (1985) Polyamines in the roots of turnip infected with Plasmodiophora brassicae Wor. New Phytol 100 209–214 [Google Scholar]
- Zemojtel T, Frohlich A, Palmieri MC, Kolanczyk M, Mikula I, Wyrwicz LS, Wanker EE, Mundlos S, Vingron M, Martasek P, et al (2006) Plant nitric oxide synthase: a never-ending story? Trends Plant Sci 11 524–525 [DOI] [PubMed] [Google Scholar]
- Zonia LE, Stebbins NE, Polacco JC (1995) Essential role of urease in germination of nitrogen-limited Arabidopsis thaliana seeds. Plant Physiol 107 1097–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]