Abstract
Root ion transport systems are regulated by light and/or sugars, but the signaling mechanisms are unknown. We showed previously that induction of the NRT2.1 NO3− transporter gene by sugars was dependent on carbon metabolism downstream hexokinase (HXK) in glycolysis. To gain further insights on this signaling pathway and to explore more systematically the mechanisms coordinating root nutrient uptake with photosynthesis, we studied the regulation of 19 light-/sugar-induced ion transporter genes. A combination of sugar, sugar analogs, light, and CO2 treatments provided evidence that these genes are not regulated by a common mechanism and unraveled at least four different signaling pathways involved: regulation by light per se, by HXK-dependent sugar sensing, and by sugar sensing upstream or downstream HXK, respectively. More specific investigation of sugar-sensing downstream HXK, using NRT2.1 and NRT1.1 NO3− transporter genes as models, highlighted a correlation between expression of these genes and the concentration of glucose-6-P in the roots. Furthermore, the phosphogluconate dehydrogenase inhibitor 6-aminonicotinamide almost completely prevented induction of NRT2.1 and NRT1.1 by sucrose, indicating that glucose-6-P metabolization within the oxidative pentose phosphate pathway is required for generating the sugar signal. Out of the 19 genes investigated, most of those belonging to the NO3−, NH4+, and SO42− transporter families were regulated like NRT2.1 and NRT1.1. These data suggest that a yet-unidentified oxidative pentose phosphate pathway-dependent sugar-sensing pathway governs the regulation of root nitrogen and sulfur acquisition by the carbon status of the plant to coordinate the availability of these three elements for amino acid synthesis.
Uptake of mineral ions by the roots is integrated in the plant to match the nutrient demand of the whole organism. This integration is ensured by regulatory mechanisms that modulate the expression and/or the activity of root ion transport systems according to the nutritional status of the plant. Specific feedback down-regulation of root ion transporters by the ions themselves, or the products of their metabolism, probably plays a central role in this context (Grignon, 1990; Clarkson and Luettge, 1991; Chrispeels et al., 1999). However, a more general control over ion uptake has also been documented that coordinates the activity of root transport systems with the photosynthetic activity of the shoot (Forde, 2002; Lejay et al., 2003). Uptake rates of many ions are dependent on light conditions and fluctuate diurnally (Clément et al., 1978; Smith and Cheema, 1985; Hatch et al., 1986; Le Bot and Kirkby, 1992; Delhon et al., 1995) or are stimulated by an increase in light intensity (Gastal and Saugier, 1989). This control over root uptake systems has often been attributed to the regulatory action of sugars produced by photosynthesis and transported downward to the roots, as shown by the positive effect of CO2 concentration in the atmosphere on NO3− uptake (Gastal and Saugier, 1989; Delhon et al., 1996) and by the stimulation of NO3− (Hänisch ten Cate and Breteler, 1981; Delhon et al., 1996; Lejay et al., 1999), NH4+ (Lejay et al., 2003), and SO42− (Smith and Cheema, 1985) uptake by the exogenous supply of sugars to the roots.
The diurnal fluctuations in root ion uptake, or its stimulation by sugars, are generally correlated with similar changes in the expression of genes encoding root ion transporters. This has been shown for iron (Vert et al., 2003), NH4+ (Gazzarrini et al., 1999; von Wiren et al., 2000; Lejay et al., 2003), NO3− (Lejay et al., 1999; Ono et al., 2000; Matt et al., 2001), K+ (Deeken et al., 2000; Ache et al., 2001; Moshelion et al., 2002; Lejay et al., 2003), phosphate (Lejay et al., 2003), and SO42− transporters (Lejay et al., 2003). Thus, it seems that the sugar regulation of ion transporter gene expression in the roots is a widespread mechanism, allowing the coordination of the transport of various ions with photosynthesis and the carbon (C) status of the plant. In a previous study, we found that six genes encoding root ion carriers in Arabidopsis (Arabidopsis thaliana), namely, NRT1.1 (NO3− transporter, formerly CHL1), NRT2.1 (NO3− transporter), AMT1.3 (NH4+ transporter), SULTR1.1 (SO42− transporter, formerly Hst1), PHT1.4 (inorganic phosphate [Pi] transporter, formerly Pt2), and KUP2 (K+ transporter), were up-regulated by light and sugars (Lejay et al., 2003). All these genes responded very similarly to the various treatments applied, suggesting the possible occurrence of a common regulatory mechanism. Further investigation on NRT2.1 indicated that its up-regulation by sugars could not be accounted for by any of the well-known sugar-sensing mechanisms (namely, specific Suc or Glc sensing, or hexokinase [HXK]-dependent sugar sensing; Sheen et al., 1999; Gibson, 2000; Smeekens, 2000; Coruzzi and Zhou, 2001; Rolland et al., 2006), but that it was dependent on C metabolism downstream of the reaction catalyzed by HXK in glycolysis (Lejay et al., 2003). NRT2.1 was chosen as a model gene because it encodes a main component of the high-affinity NO3− uptake system located at the plasma membrane of root cells (Filleur et al., 2001; Orsel et al., 2006; Chopin et al., 2007; Wirth et al., 2007) and it is a major molecular target of the regulatory mechanisms controlling root NO3− acquisition (Cerezo et al., 2001). Accordingly, its disruption results in a marked attenuation of the stimulation of root NO3− uptake by photosynthesis (Lejay et al., 2003). However, whether the control exerted by a signal originating from C metabolism downstream HXK is specific for NRT2.1 or also regulates other sugar-induced ion transporter genes has not been investigated. Furthermore, the C signal itself along with the signaling pathway involved in the sugar regulation of NRT2.1 expression is not known.
To address these questions, we combined two experimental approaches to investigate in a more systematic way the mechanisms of the sugar regulation of root ion transporters in Arabidopsis. First, we largely expanded the population of genes under study (from six to 20) to determine on a more significant basis whether a common or several different mechanisms are involved in the up-regulation of root ion transporters by sugars. Therefore, we used the microarray results of Gutierrez et al. (2007) and Price et al. (2004) to find additional sugar-induced genes encoding root ion transporters. Second, we coupled environmental and pharmacological treatments to identify more precisely which type of mechanism accounted for the sugar up-regulation of each individual gene. This was more particularly performed for NRT2.1 and NRT1.1, encoding plasma membrane transporters participating in root NO3− uptake (Tsay et al., 1993; Filleur et al., 2001) and possibly playing an additional important role as NO3− sensors modulating root development (Munos et al., 2004; Little et al., 2005; Remans et al., 2006). The two main outcomes of this work are: (1) the classification of the sugar-induced ion transporter genes according to the specific signal, signaling pathway, or step of C metabolism predominantly responsible for their regulation (light, HXK-dependent sugar sensing, sugar-sensing upstream versus downstream HXK or in upper versus lower part of glycolysis); and (2) the unraveling of a yet-unknown sugar-sensing mechanism related to the oxidative pentose phosphate pathway (OPPP) and playing a central role in the sugar regulation of NO3−, NH4+, and SO42− transporter genes in the roots.
RESULTS
Ion Transporter Genes Regulated by Light and/or Suc
A set of 20 transporter genes was selected for this work (Table I; Supplemental Table S1), including the six we investigated previously (NRT2.1, NRT1.1, AMT1.3, SULTR1.1, PHT1.4, and KUP2; Lejay et al., 2003) and 14 other ones that were induced at least 1.5-fold by Suc or Glc supply in both the Gutierrez et al. (2007) and Price et al. (2004) experiments. To determine whether all these genes were actually under the control of photosynthates, their expression was investigated in response to: (1) addition of 1% Suc in the nutrient solution during 4 h either in the dark or after transfer of the plants in the light; and (2) transfer from dark to light for 4 h in an atmosphere containing 0, 300, or 600 μL L−1 CO2.
Table I.
List of ion transporter genes investigated and primer sequences used for real-time quantitative PCR
| Gene Family/Gene Name | Sequence Left | Sequence Right | Amplicon Size |
|---|---|---|---|
| NRT2 | |||
| NRT2.1 (At1g08090) | AACAAGGGCTAACGTGGATG | CTGCTTCTCCTGCTCATTCC | 167 |
| NRT2.4 (At5g60770) | GAACAAGGGCTGACATGGAT | GCTTCTCGGTCTCTGTCCAC | 166 |
| PTR | |||
| NRT1.1 (At1g12110) | GCACATTGGCATTAGGCTTT | CTCAATCCCCACCTCAGCTA | 181 |
| NRT1.5 (At1g32450) | ATCACATGCCTGGTTGGATT | CCTCTTCACTCTCGGTGTCA | 198 |
| At3g16180 | CCAGCTGGATCGTTTGGTAT | CCGCCATTGCTAAGAATGAT | 169 |
| At3g21670 | AGCTGGCTTAGAAGTAACCT | CGTCACTTCCTTCTCCACTG | 177 |
| At5g62680 | CATCCCTGCCGTTCTAATGT | GTTAAGCCAAGGCTGTTTCG | 166 |
| At1g59740 | GATCACGCCACAGTTCTTGA | ACGAGCACCGAGCTGAAGTA | 162 |
| AMT | |||
| AMT1.3 (At3g24300) | CCTCAAAAGGCTCAATCTGC | TAGCTGATCGAGGGAAAGGA | 152 |
| PHT | |||
| PHT3.1 (At5g14040) | CGTTTCTCATCCAGCAGACA | CAGGCCAACAAACACTTTGA | 193 |
| PHT1.4 AtPT2 (At2g38940) | CCCAATGCTACAACCTTCGT | GTATCCTGCGTCGGTCTTGT | 168 |
| SULTR | |||
| SULTR3.5 (At5g19600) | CGGAAGTGTGACCTTCTTCTT | GCCACGAAGCAATCATAGTG | 231 |
| SULTR1.1 (At4g08620) | GGAAGTGGCTGAGCAACAA | TTGTTCCCATCTCACCATTG | 202 |
| ZIP | |||
| ZIP11 (At1g55910) | GTTGCCATCGGGATAGTCAT | TCCAAACAACACAGCCAAAA | 195 |
| HAK/KUP | |||
| KUP2 (At2g40540) | GATACCTCGTGGGTCGTGTT | ACGAGCGTTGTCGTCTTCTT | 182 |
| HAK5 (At4g13420) | GTTGGTGGAGAAAGCGAGAG | AGGAATCGCAAGTGCTTTGT | 163 |
| CNGC | |||
| CNGC11 (At2g46440) | ATTGCTGGTGATTCCTGTGG | GGCGACGATACTGAGTAGCG | 160 |
| NRAMP | |||
| NRAMP4 (At5g67330) | GTACGTACGCCGGACAGTTT | AAACTGCCCATGATTTGCTC | 232 |
| YSL | |||
| YSL4 (At5g41000) | GAGCTTACTTCGCCATCGAC | CAAATGGGTGGATTGATTCTT | 181 |
| Shaker-like | |||
| AKT2 (At4g22200) | CTGTGGTGACTACAGGCAAT | GGATGTTGCAACCGTGCTTT | 162 |
| AKT1 (At2g26650) | TGACGAATGTTCTGCTGGAG | TGCCATTGTTATCCGATTCA | 155 |
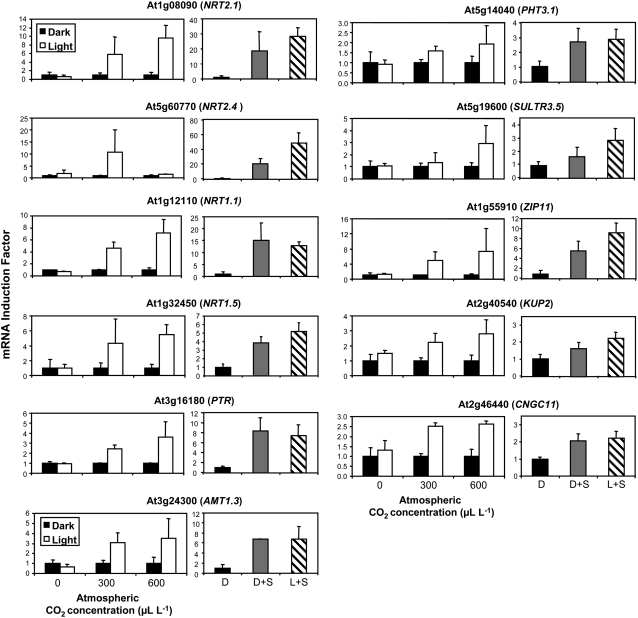
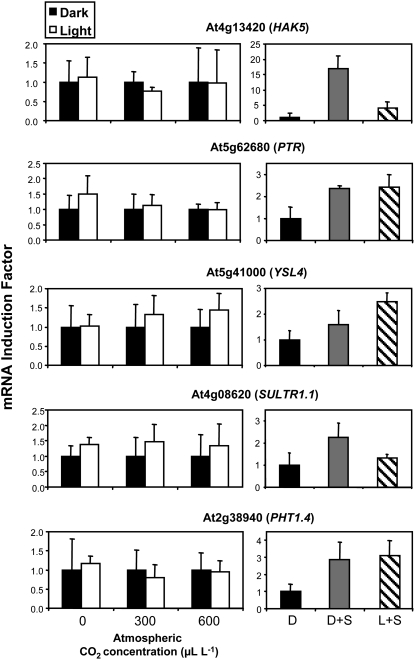
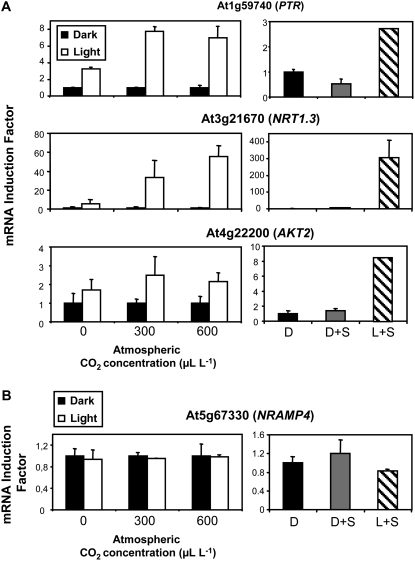
Out of the 20 genes tested, 19 were induced in the roots at various levels (but at least 2-fold) by the exogenous supply of Suc and/or after transfer in the light (Figs. 1, 2, and 3A). However, these genes did not respond similarly to the modulation of photosynthesis through CO2 concentration and could be classified into three groups (Figs. 1, 2, and 3A). The first and largest group contained 11 genes, which, like NRT2.1, NRT1.1, AMT1.3, and KUP2, were induced by Suc in the dark, by light plus Suc, and by light only in the presence of CO2 (Fig. 1). For many of these genes, the increase in transcript level resulting from illumination of the plants tended to be higher at 600 μL L−1 CO2 than at 300 μL L−1 CO2. These data strongly suggest that the light/sugar regulation of this group of genes corresponds in fact to a control exerted by photosynthesis. The second group contained five genes, which, like SULTR1.1 and PHT1.4, were induced by Suc and/or light plus Suc but that displayed very limited response, if any, to the changes in photosynthesis (Fig. 2). Indeed, their expression was not significantly increased in response to the illumination of the plants, regardless of whether CO2 was present or not in the atmosphere. The third group contained three genes, including the potassium channel gene AKT2 and two members of the PTR family (At3g21670 and At1g59740), which were clearly induced by light, even when photosynthesis was not active due to the absence of CO2 in the atmosphere (Fig. 3A). These genes were not or poorly induced by the addition of 1% Suc in the dark, but strongly responded to the light plus Suc treatment. Thus, unlike the 16 Suc-inducible genes of the two groups described above, the three genes of this third group appear to respond to light alone and not to photosynthates. Finally, only NRAMP4 did not show any regulation by light or Suc supply (Fig. 3B). This gene was therefore used as a control in further experiments (sometimes together with AKT1) to make sure that the treatments applied did not have a general effect on transporter gene expression.
Figure 1.
Ion transporter genes regulated by photosynthesis. The plants were pretreated for 40 h in darkness to repress light- or sugar-inducible transporter genes. Two experiments were performed. In the first one (left panel for each gene), the plants were transferred for 4 h in the light or left in the dark in an atmosphere containing 0, 300, or 600 μL L−1 CO2. In the second experiment (right panel for each gene), the plants were either kept for 4 additional h in the dark without Suc supply (D), kept in the dark and supplied with 1% Suc for 4 h (D+S), or transferred in the light and supplied with 1% Suc for 4 h (L+S). All transcripts were measured in the roots using real-time quantitative PCR and normalized to a putative clathrin coat-assembly protein gene (At4g24550). The data represent the mean and sd of at least three independent experiments (two replicates from each experiment).
Figure 2.
Ion transporter genes regulated by Suc. The plants were pretreated for 40 h in darkness to repress light- or sugar-inducible transporter genes. Two experiments were performed. In the first one (left panel for each gene), the plants were transferred for 4 h in the light or left in the dark in an atmosphere containing 0, 300, or 600 μL L−1 CO2. In the second experiment (right panel for each gene), the plants were either kept for 4 additional h in the dark without Suc supply (D), kept in the dark and supplied with 1% Suc for 4 h (D+S), or transferred in the light and supplied with 1% Suc for 4 h (L+S). All transcripts were measured in the roots using real-time quantitative PCR and normalized to a putative clathrin coat-assembly protein gene (At4g24550). The data represent the mean and sd of at least three independent experiments (two replicates from each experiment).
Figure 3.
Ion transporter genes regulated by light (A) or not regulated by light, Suc, and photosynthesis (B). The plants were pretreated for 40 h in darkness to repress light- or sugar-inducible transporter genes. Two experiments were performed. In the first one (left panel for each gene), the plants were transferred for 4 h in the light or left in the dark in an atmosphere containing 0, 300, or 600 μL L−1 CO2. In the second experiment (right panel for each gene), the plants were either kept for 4 additional h in the dark without Suc supply (D), kept in the dark and supplied with 1% Suc for 4 h (D+S), or transferred in the light and supplied with 1% Suc for 4 h (L+S). All transcripts were measured in the roots using real-time quantitative PCR and normalized to a putative clathrin coat-assembly protein gene (At4g24550). The data represent the mean and sd of at least three independent experiments (two replicates from each experiment).
Role of HXK in the Regulation of Root Ion Transporter Genes by Suc
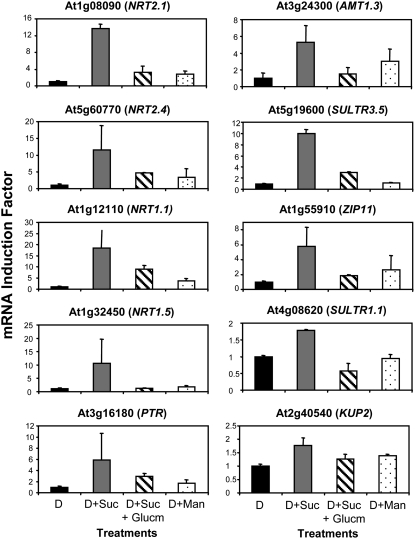
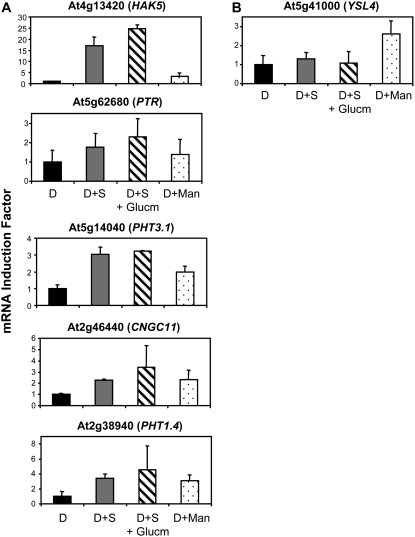
In a previous study (Lejay et al., 2003), we showed that the regulation of NRT2.1 by sugars was not related to HXK-sensing activity but was dependent on C metabolism in glycolysis downstream HXK. Part of the argument for this conclusion was that sugar induction of NRT2.1 expression was abolished in wild-type plants supplied with the HXK inhibitor glucosamine and that Man, a sugar analog that is phosphorylated by HXK (thus triggering HXK signaling) but poorly metabolized downstream in glycolysis, was not able to induce NRT2.1 expression. We thus addressed the question of whether the 16 Suc-inducible ion transporter genes identified above were all regulated the same way, and investigated the effect of glucosamine and Man on their expression. Out of the 16 genes tested, 10 genes, including two members of the NRT2 family (NRT2.1 and NRT2.4), three members of the PTR family (NRT1.1, NRT1.5, and At3g16180), AMT1.3, two members of the SULTR family (SULTR1.1 and SULTR3.5), ZIP11 (At1g55910), and KUP2, were regulated like NRT2.1 (Fig. 4). They were neither induced by Suc in the presence of glucosamine nor by Man. On the other hand, five other genes, including HAK5, a member of the PTR family (At5g62680), two members of the PHT family (PHT3.1 and PHT1.4), and CNGC11 (At2g46440), were induced by Suc even in the presence of glucosamine, but not by Man (Fig. 5A). This indicates that neither catalytic HXK activity nor HXK signaling is required for the induction of those genes by Suc. Finally, YSL4 (At5g41000) was induced by Man and thus appeared to be regulated through HXK-sensing activity (Fig. 5B).
Figure 4.
Ion transporter genes regulated like NRT2.1 in response to glucosamine and Man. Roots were harvested after pretreatment of the plants for 40 h of darkness and treatment for 4 h either in the dark (D), in the dark with supply of 1% Suc (D+S), in the dark with supply of 1% Suc and 20 mm glucosamine (D+S+Glucm), or in the dark with supply of 10 mm Man (D+Man). All transcripts were measured using real-time quantitative PCR and normalized to a putative clathrin coat-assembly protein gene (At4g24550). The data represent the mean and sd of at least three independent experiments (two replicates from each experiment).
Figure 5.
Ion transporter genes differentially regulated by glucosamine and Man. Roots were harvested after pretreatment of the plants for 40 h of darkness and treatment for 4 h either in the dark (D), in the dark with supply of 1% Suc (D+S), in the dark with supply of 1% Suc and 20 mm glucosamine (D+S+Glucm), or in the dark with supply of 10 mm Man (D+Man). All transcripts were measured using real-time quantitative PCR and normalized to a putative clathrin coat-assembly protein gene (At4g24550). The data represent the mean and sd of at least three independent experiments (two replicates from each experiment).
Correlation between the Abundance of Phosphorylated Sugars and NRT2.1 or NRT1.1 Transcript Levels
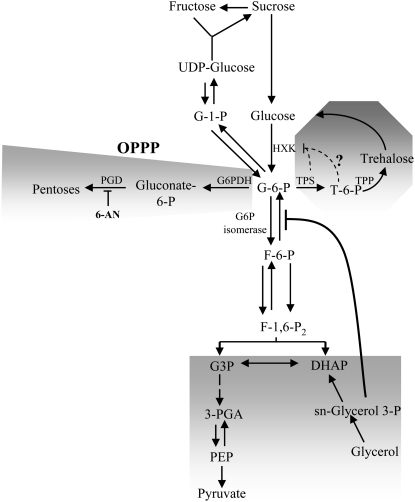
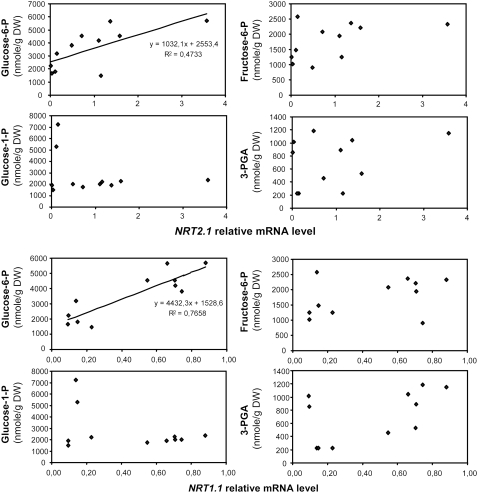
The outcome of our above results is that a majority of the ion transporter genes we found up-regulated by Suc (10 out of 16) are apparently controlled by the metabolism-dependent signaling pathway we first identified for NRT2.1 (Lejay et al., 2003). We then focused our study on the investigation of this signaling pathway, using NRT2.1 and NRT1.1 as model genes. As a first approach, we looked for correlation between the concentration of phosphorylated sugars and the expression of NRT2.1 or NRT1.1 in the roots. Transcript levels of both genes and the concentrations of Glc-6-P (G6P), Glc-1-P (G1P), Fru-6-P (F6P), and 3-phosphoglycerate (3-PGA) were monitored during a day/night cycle after 4 h of light or 16 h of dark (a normal night) plus an additional 4 h of dark with or without Suc or Man in the nutrient solution. G6P is located in the upper part of glycolysis and is the direct product of the reaction catalyzed by HXK. F6P is the product of the isomerization of G6P in the second step of the glycolysis, and 3-PGA is found in the lower part of glycolysis. G1P is not directly part of glycolysis and can be produced from G6P or from UDP-Glc (see Fig. 6). It is involved in both Suc synthesis and the first committed step of starch synthesis in the plastid. Interestingly, the results showed a correlation across the different treatments mainly between both NRT2.1 and NRT1.1 mRNA level and the concentration of G6P (Fig. 7). On the contrary, concentrations of F6P, G1P, and 3-PGA were much less or not at all correlated with the expression of both NRT2.1 and NRT1.1 (Fig. 7). These results suggest that the regulatory signal triggering induction of NRT2.1 and NRT1.1 by light and sugar is related to either G6P itself or to a product of its metabolism. G6P is at an important branched step in the upper part of glycolysis (Fig. 6) and has four main metabolic fates: (1) it is a key metabolite in Suc biosynthesis; (2) it fuels downstream glycolysis; (3) it is the starting point for the OPPP, which constitutes an alternative pathway for the oxidation of sugars in plants; and (4) it is required for the two-step process catalyzed by trehalose-6-P (T6P) synthase and T6P phosphatase to form trehalose. The two later pathways are potentially interesting for their role in nitrogen (N) metabolism and sugar signaling, respectively. The OPPP provides the reducing power for nitrite reductase and GOGAT in the roots (Oji et al., 1985; Bowsher et al., 1989, 1992), while T6P, the intermediate in trehalose biosynthesis via T6P synthase, is involved in sugar signaling (Eastmond and Graham, 2003). To further investigate the origin of the sugar signal involved in the up-regulation of NRT2.1 and NRT1.1 expression, we then used a pharmacological approach to modulate G6P concentration in the roots and to test the implication of three of the four pathways described above (glycolysis, OPPP, and T6P). The first pathway was not tested because we already showed previously that Suc itself was not involved in the regulation of NRT2.1 and NRT1.1 (Lejay et al., 2003).
Figure 6.
Scheme summarizing the fate of G6P in the OPPP, glycolysis, and trehalose synthesis, and the effect of 6-AN and glycerol on plant metabolism. F-1,6-P2, Fru-1,6-bisphosphate; DHAP, dihydroxyacetone phosphate; G6PDH, G6P dehydrogenase; PGD, 6-phosphogluconate dehydrogenase.
Figure 7.
Correlation between concentration of phosphorylated sugars in the roots and NRT2.1 or NRT1.1 transcript levels after light, dark, Suc, and Man supply. Roots were harvested after 4 h into the light period during a normal day/night cycle, or after 4 additional h in the dark after a normal night with or without supply of 1% Suc or 10 mm Man. Concentrations of G6P, F6P, G1P, and 3-PGA in root samples were determined by cycling assays. Transcript levels were measured using real-time quantitative PCR and normalized to a putative clathrin coat-assembly protein gene (At4g24550). The data are representative of at least two independent experiments (three replicates from each experiment).
Effect of Glycerol on the Level of G6P and the Regulation of NRT2.1 and NRT1.1
First, we used glycerol to inhibit G6P accumulation in roots and determine whether NRT2.1/NRT1.1 expression is dependent on G6P concentration or G6P metabolism in the upper part of glycolysis. In the absence of added sugar in the medium, the supply of glycerol to plant cells or roots leads to an accumulation of glycerol-3-P in the cytoplasm, which can be used to fuel glycolysis downstream of G3P but not as a source of C skeletons for sugar biosynthesis (Aubert et al., 1994; Brouquisse et al., 2007). Indeed, while glycerol-3-P sustains respiration, it prevents the flow back of C from triose phosphates to G6P by inhibiting G6P isomerase (Aubert et al., 1994). Furthermore, glycerol represses photosynthesis in leaves (Leegood et al., 1988; Sheen, 1990) and thus results in roots in a lowered availability of Glc originating from Suc imported from the phloem. As a consequence of both this shunt in glycolysis and diminished Glc provision, glycerol supply leads to a strong decrease of G6P concentration that impairs further metabolism of this compound, including OPPP, T6P synthesis, and the upper part of glycolysis down to F1,6P (Aubert et al., 1994; Brouquisse et al., 2007).
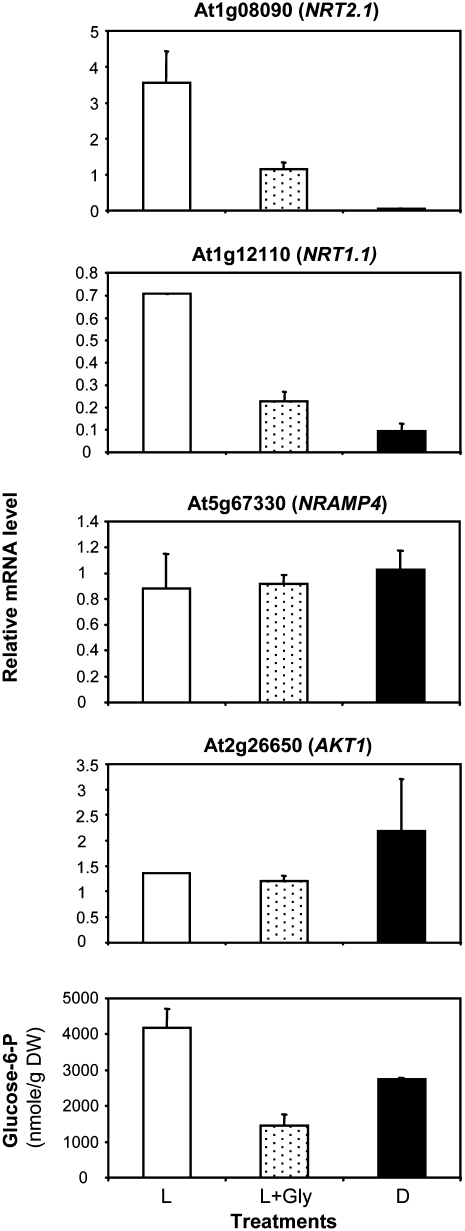
In our experiments, the exogenous supply of 30 mm glycerol for 4 h after transfer of the plants in the light resulted, as expected, in a marked decrease of G6P concentration in the roots (Fig. 8). This was associated with a strong impairment of the normal up-regulation of both NRT2.1 and NRT1.1 expression after dark/light transition (Fig. 8). In the meantime, glycerol had no effect on the expression of both NRAMP4 and AKT1 (Fig. 8), two genes not regulated by light or sugar (Fig. 3B; Lejay et al., 2003), indicating that the detrimental effect of glycerol on gene expression is not general. Altogether, these data support the hypothesis that the up-regulation of NRT2.1 and NRT1.1 by light and sugars is related to the increase in the concentration of G6P in roots or requires unaltered G6P metabolism.
Figure 8.
Effect of glycerol on NRT2.1, NRT1.1, NRAMP4, and AKT1 expression and on G6P level in roots. At the end of a normal night, the plants were either transferred for 4 h in the light (L), for 4 h in the light with supply of 30 mm glycerol (L+Gly), or kept in the dark for 4 additional h (D). All transcripts were measured using real-time quantitative PCR and normalized to a putative clathrin coat-assembly protein gene (At4g24550). Concentrations of G6P were determined by cycling assays. The data are representative of at least two independent experiments (three replicates from each experiment).
Role of T6P and the OPPP in the Regulation of NRT2.1 and NRT1.1 by Sugars
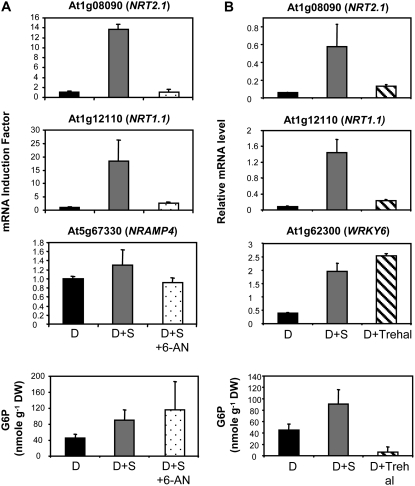
To modulate OPPP activity or T6P signaling, we treated the plants with either 6-aminonicotinamide (6-AN) or with trehalose, respectively. 6-AN impairs OPPP because it is a potent inhibitor of the phosphogluconate dehydrogenase (Kohler et al., 1970; Garlick et al., 2002), whereas trehalose supply results in an increased accumulation of T6P due to its inhibitory action on T6P phosphatase activity (Schluepmann et al., 2004).
When plants were treated in the dark with 1% Suc plus 10 mm 6-AN, sugar induction of both NRT2.1 and NRT1.1 expression was almost totally prevented, with a 95% reduction for NRT2.1 and a 75% reduction for NRT1.1, whereas NRAMP4 mRNA level was not significantly affected (Fig. 9A). These results show that an operating OPPP is important for sugar regulation of both nitrate transporter genes. On the other hand, the expression of NRT2.1 and NRT1.1 was not induced when plants were treated in the dark with 30 mm trehalose compared to 1% (30 mm) Suc (Fig. 9B). This lack of up-regulation by trehalose was not due to unsuccessful treatment, as in the same experiment, the expression of the transcription factor gene WRKY6, used as a trehalose-inducible control (Bae et al., 2005), was indeed stimulated 2- and 4-fold by 1% (30 mm) Suc and 30 mm trehalose, respectively (Fig. 9B). In both experiments with 6-AN and trehalose, the concentration of G6P was measured in the roots to determine if these compounds affected the correlation previously observed between G6P and the transcript accumulation of NRT2.1 and NRT1.1. As expected, the concentration of G6P in the roots was increased by the addition of 1% Suc, like the expression of the two nitrate transporter genes (Fig. 9, A and B). Trehalose supply did not alter the correlation between G6P accumulation and expression of the transporter genes because it led to a dramatic inhibition of both as compared to Suc supply (Fig. 9B). However, while treatment with 1% Suc plus 6-AN blocked the induction of NRT2.1 and NRT1.1, it did not reduce (or even slightly increased) the concentration of G6P compared to the treatment with 1% Suc (Fig. 9A). This lack of correlation between the changes in G6P concentration and those of the expression of NRT2.1 and NRT1.1 when plants were treated with 6-AN indicates that G6P itself is not responsible for the induction of the two genes. Rather, it suggests that the C signal triggering up-regulation of NRT2.1 and NRT1.1 is related with the activity of the OPPP.
Figure 9.
Effect of 6-AN and trehalose on NRT2.1 and NRT1.1 expression and on the root concentration of G6P. A, Roots were harvested after 40 h of darkness plus either 4 h of dark (D), 4 h of dark with supply of 1% Suc (D+S), or 4 h of dark with supply of 1% Suc and 10 mm 6-AN (D+S+6-AN). In this experiment, NRAMP4 was used as a control gene. B, Roots were harvested after 40 h of darkness plus either 4 h of dark (D), 4 h of dark with supply of 1% (30 mm) Suc (D+S), or 4 h of dark with supply of 30 mm trehalose (D+Trehal). In this experiment, WRKY6 was used as a control gene. All transcripts were measured using real-time quantitative PCR and normalized to a putative clathrin coat-assembly protein gene (At4g24550). The data represent the mean and sd of at least three independent experiments (two replicates from each experiment).
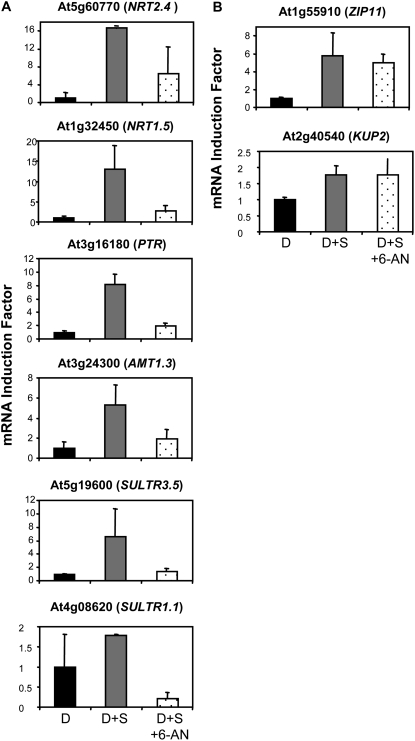
To investigate further the role of this OPPP-dependent sugar signaling, we tested the effect of 6-AN on the regulation of the other eight genes we previously found to be regulated like NRT2.1 and NRT1.1 by a metabolism-dependent signaling pathway (Fig. 4). The results show that 6-AN prevented the induction of six of them by Suc (Fig. 10A). These genes correspond to members of the NRT2 family (NRT2.4), PTR family (NRT1.5 and At3g16180), AMT family (AMT1.3), and SULTR family (SULTR3.5 and SULTR1.1), and thus appear to be also regulated by an OPPP-dependent sugar signaling like NRT2.1 and NRT1.1. The expression of the two other genes (ZIP11 and KUP2) remained induced by Suc even in the presence of 6-AN (Fig. 10B). Together with the unaltered expression of NRAMP4 (Fig. 9A), this shows that 6-AN does not have a general detrimental effect (e.g. toxic) on the expression of transporter genes, even for those regulated by sugars, and suggests that the OPPP plays only a limited role, if any, in the sugar-signaling pathway(s) governing ZIP11 and KUP2 expression.
Figure 10.
Effect of 6-AN on the group of ion transporter genes regulated like NRT2.1 and NRT1.1 in response to glucosamine and Man. Roots were harvested after 40 h of darkness plus either 4 h of dark (D), 4 h of dark with supply of 1% Suc (D+S), or 4 h of dark with supply of 1% Suc and 10 mm 6-AN (D+S+6-AN). All transcripts were measured using real-time quantitative PCR and normalized to a putative clathrin coat-assembly protein gene (At4g24550). The data represent the mean and sd of at least three independent experiments (two replicates from each experiment).
DISCUSSION
Multiple Signaling Pathways Are Involved in the Light and/or Sugar Regulation of Ion Transporter Genes in the Roots
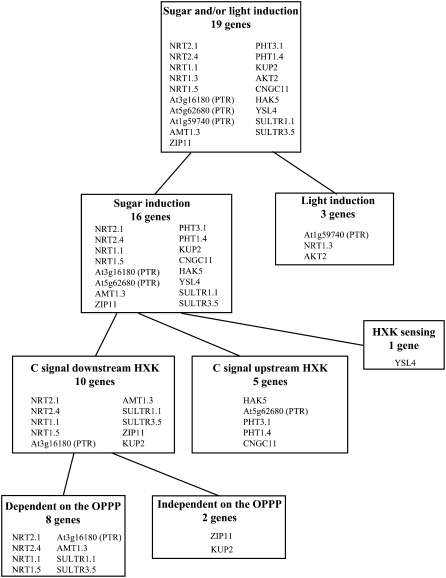
Both our previous data (Lejay et al., 2003) and those from several microarray experiments (Price et al., 2004; Gutierrez et al., 2007) indicate that genes encoding ion carriers or channels belonging to various multigenic families are strongly up-regulated by sugars in the roots. For many of these genes, this is also associated to a diurnal pattern of expression with a decay at night (Gazzarrini et al., 1999; Lejay et al., 1999, 2003; Deeken et al., 2000), suggesting dependency on photosynthesis. However, it was unclear whether these genes are coregulated by a common signaling pathway related to downward transport of photosynthates from shoot to roots. Collectively, our results indicate that root ion transporter genes do not respond to a unique sugar-signaling pathway, which would be responsible for a general control of root nutrient acquisition, but rather that they are modulated by at least four different regulatory mechanisms (Fig. 11).
Figure 11.
Scheme summarizing the different sugar-/light-signaling pathways found for the 19 genes coding for root ion transporters.
The first and most important distinction that can be established relates to the role of C metabolites or light as the main signal. Most ion transporter genes investigated (16 out of 19) were clearly induced by Suc supply in the dark (Figs. 1 and 2). Among these, a majority (11 genes) was also responsive to light in the absence of Suc supply but only when CO2 was present in the atmosphere (Fig. 1). Thus, these 11 genes appear to be mainly regulated in the roots by downward transport of photosynthates from the shoot. This was already suggested for four of them (NRT2.1, NRT1.1, AMT1.3, and KUP2) by the strong correlation found between their responses to illumination of the plant on the one hand and to Suc supply to the roots on the other hand (Lejay et al., 2003). For most of these 11 genes, induction by light was also more pronounced at high than at low CO2 concentration (Fig. 1), suggesting a quantitative dependence on photosynthesis. This is consistent with the earlier observation that root transcript level of three of these genes (NRT1.1, NRT2.1, and AMT1.3) increased with light intensity (Lejay et al., 2003). Concerning NRT2.1 and NRT1.1, these results are also in agreement with earlier studies with CO2-free air, showing that the diurnal changes of root NO3− uptake are caused by photosynthesis and not by light per se (Delhon et al., 1996). For the other five Suc-inducible genes (HAK5, At5g62680, YSL4, SULTR1.1, and PHT1.4), light had no significant effect on their expression in the roots, independently of whether photosynthesis is allowed or not (Fig. 2). No definite conclusion can be drawn from this unexpected result, but at least three hypotheses may be proposed: (1) these genes are quantitatively less sensitive to sugars compared to the 11 genes of the former group, and the low light intensity used in this experiment (80 μmol m−2 s−1) did not allow us to reach the threshold level of sugar transport to the roots required to induce their expression; (2) their induction by 1% Suc in the nutrient solution is related to an osmotic effect and not to the specific action of sugars as signaling molecules; and (3) some of these genes are responding to more complex interactions between light and C signaling (Thum et al., 2003). For instance, HAK5 and SULTR1.1 were strongly induced by the addition of Suc in the dark but much less in the light (Fig. 2), suggesting that light may counteract their induction by photosynthates. Finally, three genes, including two members of the PTR family (NRT1.3 and At1g59740) and AKT2 encoding a potassium channel, were found to be induced by light even in CO2-free air (Fig. 3A). Interestingly, these genes responded to the light and Suc treatment but not to Suc supply in the dark (Fig. 3A). This indicates that light per se, and not Suc, is the predominant signal involved (Fig. 11). However, the level of induction by light is always higher in the presence of CO2 for the three genes, suggesting that sugars can also have an additive effect to that of light. For AKT2, these data closely parallel those previously reported for expression in leaves (Deeken et al., 2000). The pattern of expression of these three genes thus suggest that light could be perceived by the roots and could act directly to regulate gene expression in these organs. This kind of regulation has already been described in pea for light repression of AS1, a gene coding for Asn synthetase in roots (Tsai and Coruzzi, 1991). The authors showed that the small amount of light that passes through the soil is sufficient to repress AS1 expression in roots. Furthermore, recent microarray experiments identified several genes differentially expressed in roots of dark-grown Arabidopsis seedlings exposed for 1 h to red light (Molas et al., 2006). In agreement with the hypothesis of a direct action of light in roots, three classes of light receptors have been found in Arabidopsis roots: the phytochromes, the cryptochromes, and phototropin (Neff et al., 2000; Quail, 2002). Alternatively, light perception by the shoots could also act indirectly on roots through changes in long-distance auxin transport, for instance, as shown recently for the effect of light on Arabidopsis root development (Salisbury et al., 2007).
Following this distinction between 16 Suc-inducible and three light-inducible genes, further experiments indicated that at least three different signaling pathways are involved in the regulation of the 16 Suc-inducible genes (Fig. 11). In our previous study (Lejay et al., 2003), we found that the stimulation of NRT2.1 expression by Suc or Glc required HXK catabolic activity but not HXK signaling function. Indeed, underexpression of HXK in transgenic lines or inhibition of its activity by glucosamine prevented this stimulation, while exogenous supply of Man (triggering HXK signaling) failed to mimic it. This conclusion appears to hold true for the majority (10 out of 16) of Suc-inducible ion transporter genes investigated in this study (Fig. 4). This suggests that these 10 genes are regulated by the metabolic signaling pathway (Rolland et al., 2006) dependent on sugar metabolites or metabolism downstream HXK in glycolysis (Fig. 11). Interestingly, with the exception of PHT3.1 and CNGC11, all the genes regulated by photosynthesis fall into this major category (compare Fig. 1 and Fig. 4). However, it cannot be ruled out that the effect of Man on the expression of some of the genes is due to Pi sequestration and subsequent imbalance of metabolism due to a decreased synthesis of ATP (Herold and Lewis, 1977; Brouquisse et al., 2001). Nevertheless two arguments do not support this hypothesis: (1) our previous study showed that the effect of Man on the expression of NRT2.1 was not due to a problem of toxicity as confirmed by transgenic plants underexpressing HXK or transformed with yeast HXK (Lejay et al., 2003); and (2) glucosamine that is not involved in Pi sequestration has the same effect as Man on the expression of all the genes. Our data also reveal that a second important regulatory pathway is involved. Indeed, five genes show no diminution of Suc response upon glucosamine supply (Fig. 5A), indicating that their up-regulation seems to be dependent either on sugar transport or metabolism upstream the HXK step or on Suc itself (Fig. 11). The role of Suc as a signal molecule has already been proposed for the regulation of several transporter genes, including those encoding the proton Suc symporter of Beta vulgaris (Chiou and Bush, 1998), the VvHT1 Glc transporter of Vitis vinifera (Atanassova et al., 2003), and the CitAMT1 ammonium transporter of Citrus (Camanes et al., 2007). Alternatively, Glc transport may also be a key step of sugar metabolism involved in signaling (Lalonde et al., 1999; Chen and Jones, 2004). Finally, only one gene encoding the metal transporter YSL4 was strongly induced by Man (Fig. 5B), indicating a role of the HXK signaling function in its regulation (Fig. 11). Collectively, these results suggest that genes of root ion transporters respond to three of the main Glc signal transduction pathways defined in plants, namely glycolysis-related metabolic signaling pathway, Suc and/or Glc sensing, and HXK sensing (Rolland et al., 2006).
OPPP as a Major Pathway Involved in the Sugar Induction of NO3−, NH4+, and SO42− Transporters in Roots
Our data extend to nine other ion transporter genes our previous conclusion that NRT2.1 expression is modulated by a signal originating from C metabolism downstream HXK (Figs. 4 and 11). There are very few genes reported to be regulated this way (e.g. two PR genes in Arabidopsis; Xiao et al., 2000). To gain further insight on this yet-uncharacterized sugar-signaling pathway, NRT2.1 and NRT1.1 were used as model genes to look for possible correlations between the level of C metabolites downstream of the HXK step in glycolysis and gene expression across different treatments. The best correlation was obtained with G6P, a metabolite involved in the upper part of glycolysis (Fig. 7). The strong repression of both NRT1.1 and NRT2.1 expression by glycerol (Fig. 8) further pinpoints a tight relationship between NRT1.1 or NRT2.1 regulation and G6P or at least C metabolism in upper glycolysis. Indeed, glycerol leads to a decrease of G6P concentration while channeling glycerol-3-P into the lower part of the glycolytic pathway downstream of G3P dehydrogenase (Aubert et al., 1994; see Fig. 6). In sycamore (Acer pseudoplatanus) cells and in maize (Zea mays) root tips, glycerol has thus been successfully used to discriminate between the respective roles of the lower and upper parts of glycolysis in autophagy in response to C starvation and in the regulation of proteolysis by sugars (Aubert et al., 1994, 1996; Brouquisse et al., 2007). Interestingly, the hypothesis that the signal regulating NRT1.1 and NRT2.1 may originate from upper glycolysis and not from lower glycolysis downstream the G3P dehydrogenase step is in agreement with our previous finding that carboxylic acids, such as malate and 2-oxoglutarate, are unable to mimic the inductive effect of sugars on the expression of both genes (Lejay et al., 2003).
The correlation between the concentration of G6P in root cells and NRT1.1 or NRT2.1 expression suggested three hypotheses concerning the signaling pathway involved (Fig. 6). First, G6P itself could be the signal molecule, as suggested by its role in the regulation of phosphoenolpyruvate carboxylase and of Suc phosphate synthase (Matsumura et al., 2002; Takahashi-Terada et al., 2005). Second, G6P metabolization down to F6BP, or within OPPP, may result in the synthesis of the signal molecule or may sustain a specific reaction from which the signal originates. Third, G6P as a component of trehalose synthesis may trigger trehalose signaling (Fig. 6; Bae et al., 2005). Our results do not support the hypotheses that either G6P or trehalose signaling are directly involved in the regulation of NRT1.1 and NRT2.1 expression by sugars. On the one hand, treatment with the phosphogluconate dehydrogenase inhibitor 6-AN broke down the correlation between G6P concentration in roots and NRT1.1 or NRT2.1 transcript accumulation (Fig. 9A). On the other hand, the addition of trehalose in the nutrient solution did not mimic the inductive effect of Suc in the dark (Fig. 9B).
On the contrary, the marked inhibitory effect of 6-AN on NRT1.1 and NRT2.1 expression strongly suggests that sustained C flow through OPPP is required for sugar induction of both NRT genes. Moreover, this hypothesis could be generalized to most of the transporter genes we found regulated by the metabolic signaling pathway because eight (out of 10) of these genes responded to 6-AN in a similar way as NRT1.1 and NRT2.1 (Figs. 9A and 10A). Only ZIP11 and KUP2 were insensitive to 6-AN, suggesting that an OPPP-independent signaling operates to regulate these genes. It is noteworthy that the genes we found to be dependent on OPPP for their sugar induction belong to NO3−/peptide, NH4+, and SO42− transporter families (Figs. 9A and 10A; Supplemental Table S1). This certainly has a strong physiological significance for at least two main reasons. First, N and sulfur (S) are two elements entering, along with C, in the composition of amino acids. As a consequence, S and N assimilatory pathways are well coordinated, so that the availability of one element regulates the other pathway. For instance, SO42− transporters are repressed by N deprivation (Ehira et al., 2003) and induced by NO3− (Wang et al., 2003). Thus, it is not surprising to find that C availability also plays a role and coregulates transporters involved in N or S acquisition or utilization in the plant. Second, there is a strong link in roots between N and S metabolism and the OPPP because it provides the reducing power for nitrite reductase, GOGAT, and the assimilation of SO42− into Cys (Oji et al., 1985; Bowsher et al., 1989, 1992; Neuhaus and Emes, 2000; Yonekura-Sakakibara et al., 2000; Kopriva and Rennenberg, 2004). Furthermore, 3-PGA generated through the OPPP could also serve as the precursor of Ser and O-acetyl-l-Ser biosynthesis, the amino acid skeleton for SO42− assimilation in the plastids of root tissues (Ho and Saito, 2001). As a consequence, OPPP and N or S assimilation are tightly coordinated processes. In particular, N availability exerts a strong influence on the regulation of the OPPP. For instance, genes encoding OPPP enzymes are among those most affected by NO3− signaling in Arabidopsis roots (Wang et al., 2000, 2003), and NH4+ can induce an isoform of G6P dehydrogenase in barley (Hordeum vulgare) roots (Esposito et al., 2001). Thus, the reverse control of N acquisition and metabolism by C signaling originating from OPPP is highly conceivable. A strong effect of SO42− on regulation of the OPPP genes has not been reported (Hirai et al., 2003; Maruyama-Nakashita et al., 2003; Nikiforova et al., 2003). However, this can be easily explained by the low level of SO42− uptake and assimilation fluxes as compared to those of NO3− and NH4+ (the S:N molar ratio is 1:25; Rennenberg, 1984), suggesting that OPPP regulation by N largely prevails and masks any effect of S nutrition. Altogether, these results and our findings support the existence of a common OPPP-dependent sugar signaling mechanism for regulation of N and S acquisition in roots, which would coordinate the availability of all three amino acid components (C, N, and S) for adequate amino acid synthesis. Such a signaling pathway has not been described to date, even for the sugar regulation of the few N or S assimilatory enzymes investigated, e.g. nitrate reductase (Jang et al., 1997), Asn synthetase (Xiao et al., 2000), and adenosine 5′-phosphosulfate reductase (Hesse et al., 2003). Concerning the mechanism involved in this new signaling pathway, three hypotheses can be made: (1) one of the C metabolites generated through the OPPP could play the role of a signal molecule; (2) an enzyme of the OPPP could generate a signal like HXK in glycolysis; and (3) the reducing power produced by the OPPP could be involved in redox regulation of root ion transporters via, for example, an NADPH-dependent signaling pathway. This kind of regulation has been found in animals for the redox regulation of fertilization in the mouse (Urner and Sakkas, 2005), and in plants, reactive oxygen species produced by NADPH oxidase are involved in the regulation of root cell growth (Foreman et al., 2003; Jones et al., 2007).
In conclusion, in addition to the observation that the sugar regulation of root ion transporters involves multiple signaling mechanisms (Fig. 11), our study reveals for the first time, to our knowledge, the occurrence of an OPPP-dependent sugar signaling pathway in plants. We propose that this signaling pathway participates in the integration of N and S uptake by ensuring their coordination with the production of reducing equivalents required for assimilating these mineral nutrients into amino acids.
MATERIALS AND METHODS
Plant Material
Plants of Arabidopsis (Arabidopsis thaliana) ecotype Columbia were grown hydroponically under nonsterile conditions as described by Lejay et al. (1999). Briefly, the seeds were germinated directly on top of modified Eppendorf tubes filled with prewetted sand. The tubes were then positioned on floating rafts and transferred to tap water in a growth chamber under the following environmental conditions: 8-/16-h photoperiod at 250 μmol m−2 s−1, temperature of 22°C/20°C, and relative humidity of 70%. After 1 week, the tap water was replaced with a complete nutrient solution. The experiments were performed on plants grown on 1 mm NO3− as an N source. The other nutrients were added as described by Lejay et al. (1999). The plants were allowed to grow for five additional weeks before the experiments. Nutrient solutions were renewed weekly and on the day before the experiments. pH was adjusted to 5.8. All experiments were repeated two or three times.
Supply of C Metabolites, Sugar Analogs, or Inhibitor
The dependence of the expression of root ion transporters on photosynthesis was investigated by modifying the CO2 concentration in the atmosphere. After a pretreatment of 40 h in the dark, plants were placed for 4 h in the light or in the dark in a 240-L, airtight plexiglass chamber connected to a computerized device for controlling temperature, humidity, and CO2 concentration in the atmosphere (Atelliance Instruments; see Delhon et al. [1996] for details). The CO2 concentration in the atmosphere was held constant during the treatments at 0, 300, or 600 μL L−1.
The treatments involving the supply of sugars or inhibitor into the nutrient solution were performed on plants pretreated during 40 h in the dark except in the experiment testing the effect of glycerol (Figs. 7 and 8) where plants were treated immediately after a normal night. The plants were transferred during 4 h to fresh nutrient solution, pH 5.8, supplemented with the various compounds investigated at the concentration indicated in the figures. After harvest, the roots were frozen at −80°C.
RNA Extraction and Reverse Transcription
RNA extraction was performed on roots as described previously (Lobreaux et al., 1992) using guanidine hydrochloride and lithium chloride. Subsequently 40 μg of RNA were treated with DNase (RNase-Free DNase kit; Qiagen) and purified (RNeasy MinElute Cleanup kit; Qiagen) following the manufacturer's instructions. The absence of genomic DNA was verified by PCR using specific primers spanning an intron in the gene APTR (At1g27450; APTR FW, CGCTTCTTCTCGACACTGAG and APTR REV, CAGGTAGCTTCTTGGGCTTC). Reverse transcription was performed with 4 μg of purified RNA and oligo(dT)18 primers. The mix was heated for 5 min at 72°C and progressively (−1°C/10 s) cooled down to allow hybridization of the primers. The reaction was carried out in a volume of 20 μL in the presence of 200 units of Moloney murine leukemia virus reverse transcriptase (Promega) at 42°C during 90 min. The quality of the cDNA was verified by PCR using the primers for the gene APTR.
Quantitative PCR
Real-time amplification was performed in a LightCycler (Roche Diagnostics) with the kit SyberGreen (LightCycler FastStart DNA Master Syber Green1; Roche Diagnostics) according to the manufacturer's instructions with 1 μL of cDNA in a total volume of 10 μL. The following conditions of amplifications were applied: 10 min at 95°C; 45 cycles of 5 s at 95°C, 7 s at 65°C, and 8 s at 72°C. A melting curve was then performed to verify the specificity of the amplification. Successive dilutions of one sample were used as a standard curve. Amplification efficiency was around 1. All the results presented were standardized using the housekeeping gene Clathrin (At4g24550) with the following primers: Clath. FW, AGCATACACTGCGTGCAAAG and Clath. REV, TCGCCTGTGTCACATATCTC. The primers used for the genes coding for root ion transporters are described in Table I.
Metabolite Measurements
In the experiment described in Figure 7, two different kinds of extractions from lyophilized root samples were performed.
Ethanolic Extraction
A volume of 250 μL of 80% ethanol (v/v) was added to 10 mg of root sample. The mixture was vortex shaken and incubated for 20 min at 80°C. After centrifugation at 16,000g for 5 min, the supernatant (S1) was collected and put on ice. The extraction procedure was repeated twice as described above, first with 150 μL of 80% ethanol and then with 250 μL of 50% ethanol (v/v). The supernatants (S2 and S3) collected after the second and third extractions were added to S1 and kept at −20°C. The extract was used for the determination of G6P, G1P, and F6P.
TCA-Ether Extraction
A volume of 400 μL of cold 16% TCA in diethylether (v/v) was added to 5 mg of root sample, mixed by vortex-shaking, and put on ice for 20 min. Then 250 μL of 16% TCA in water (v/v) containing 5 mm EGTA was added, vortex-shaken, and left for 2.5 h on ice. After centrifugation at 16,000g for 5 min at 4°C, the aqueous (lower) phase was transferred into a new Eppendorf tube. It was washed three times with 500 μL of water-saturated ether by centrifugation for 10 min at 4°C. The upper phase (ether) was discarded. The final aqueous phase was neutralized (pH 6–7) with 5 m KOH/1 m triethanolamine. The pH was determined with narrow-range pH paper. The extract was used for the determination of 3-PGA.
The level of hexose phosphate (G6P, F6P, and G1P) and 3-PGA was determined using, respectively, NADP+ and 3-PGA cycling assays as described by Gibon et al. (2002). Absorbance was monitored at 570 nm in an Anthos htII microplate reader.
In the rest of the experiments, the level of G6P was measured enzymatically by the method of Lowry and Passonneau (1972). Frozen root tissue was ground to a fine powder in a mortar precooled with liquid N2 and extracted with ethanol as described above. Absorbance was measured at 570 nm using a WallacVictor 2 spectrofluorimeter (Perkin Elmer).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Function of the root ion transporter genes investigated.
Supplementary Material
Acknowledgments
We thank Mark Stitt for his support with C metabolites measurements and Caroline Pey for technical assistance.
This work was supported by the European Union program PLUSN (HPRN–CT–2002–00247 to L.L., J.W., J.M.F.C., and A.G.) and by the ANR project CASAH-BI (ANR–05–JCJC–0052–01 to L.L., M.P., P.T., and A.G.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Laurence Lejay (lejay@supagro.inra.fr).
The online version of this article contains Web-only data.
References
- Ache P, Becker D, Deeken R, Dreyer I, Weber H, Fromm J, Hedrich R (2001) VFK1, a Vicia faba K+ channel involved in phloem unloading. Plant J 27 571–580 [PubMed] [Google Scholar]
- Atanassova R, Leterrier M, Gaillard C, Agasse A, Sagot E, Coutos-Thevenot P, Delrot S (2003) Sugar-regulated expression of a putative hexose transport gene in grape. Plant Physiol 131 326–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert S, Gout E, Bligny R, Douce R (1994) Multiple effects of glycerol on plant cell metabolism. Phosphorus-31 nuclear magnetic resonance studies. J Biol Chem 269 21420–21427 [PubMed] [Google Scholar]
- Aubert S, Gout E, Bligny R, Marty-Mazars D, Barrieu F, Alabouvette J, Marty F, Douce R (1996) Ultrastructural and biochemical characterization of autophagy in higher plant cells subjected to carbon deprivation: control by the supply of mitochondria with respiratory substrates. J Cell Biol 133 1251–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae H, Herman E, Bailey B, Bae HJ, Sicher R (2005) Exogenous trehalose alters Arabidopsis transcripts involved in cell wall modification, abiotic stress, nitrogen metabolism, and plant defense. Physiol Plant 125 114–126 [Google Scholar]
- Bowsher CG, Boulton EL, Rose J, Nayagam S, Emes MJ (1992) Reductant for glutamate synthase is generated by the oxidative pentose phosphate pathway in non-photosynthetic root plastids. Plant J 2 893–898 [Google Scholar]
- Bowsher CG, Hucklesby DP, Emes MJ (1989) Nitrite reduction and carbohydrate metabolism in plastids purified from roots of Pisum sativum L. Planta 177 359–366 [DOI] [PubMed] [Google Scholar]
- Brouquisse R, Evrard A, Rolin D, Raymond P, Roby C (2001) Regulation of protein degradation and protease expression by mannose in maize root tips. Pi sequestration by mannose may hinder the study of its signaling properties. Plant Physiol 125 1485–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouquisse R, Rolin D, Cortes S, Gaudillere M, Evrard A, Roby C (2007) A metabolic study of the regulation of proteolysis by sugars in maize root tips: effects of glycerol and dihydroxyacetone. Planta 225 693–709 [DOI] [PubMed] [Google Scholar]
- Camanes G, Cerezo M, Primo-Millo E, Gojon A, Garcia-Agustin P (2007) Ammonium transport and CitAMT1 expression are regulated by light and sucrose in citrus plants. J Exp Bot 58 2811–2825 [DOI] [PubMed] [Google Scholar]
- Cerezo M, Tillard P, Filleur S, Munos S, Daniel-Vedele F, Gojon A (2001) Major alterations of the regulation of root NO(3)(-) uptake are associated with the mutation of Nrt2.1 and Nrt2.2 genes in Arabidopsis. Plant Physiol 127 262–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JG, Jones AM (2004) AtRGS1 function in Arabidopsis thaliana. Methods Enzymol 389 338–350 [DOI] [PubMed] [Google Scholar]
- Chiou TJ, Bush DR (1998) Sucrose is a signal molecule in assimilate partitioning. Proc Natl Acad Sci USA 95 4784–4788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopin F, Wirth J, Dorbe MF, Lejay L, Krapp A, Gojon A, Daniel-Vedele F (2007) The Arabidopsis nitrate transporter AtNRT2.1 is targeted to the root plasma membrane. Plant Physiol Biochem 45 630–635 [DOI] [PubMed] [Google Scholar]
- Chrispeels MJ, Crawford NM, Schroeder JI (1999) Proteins for transport of water and mineral nutrients across the membranes of plant cells. Plant Cell 11 661–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson DT, Luettge U (1991) Inducible and repressible nutrient transport systems. Prog Bot 52 61–83 [Google Scholar]
- Clément CR, Hopper MJ, Jones LHP, Leafe EL (1978) The uptake of nitrate by Lolium perenne from flowing nutrient solution. Effect of light, defoliation, and relationship to CO2 flux. J Exp Bot 29 1173–1183 [Google Scholar]
- Coruzzi GM, Zhou L (2001) Carbon and nitrogen sensing and signaling in plants: emerging “matrix effects”. Curr Opin Plant Biol 4 247–253 [DOI] [PubMed] [Google Scholar]
- Deeken R, Sanders C, Ache P, Hedrich R (2000) Developmental and light-dependent regulation of a phloem-localised K+ channel of Arabidopsis thaliana. Plant J 23 285–290 [DOI] [PubMed] [Google Scholar]
- Delhon P, Gojon A, Tillard P, Passama L (1995) Diurnal regulation of NO3− uptake in soybean plants. I. Changes in NO3− influx, efflux, and N utilization in the plant during the day/night cycle. J Exp Bot 46 1585–1594 [Google Scholar]
- Delhon P, Gojon A, Tillard P, Passama L (1996) Diurnal regulation of NO3- uptake in soybean plants IV. Dependence on current photosynthesis and sugar availability to the roots. J Exp Bot 47 893–900 [Google Scholar]
- Eastmond PJ, Graham IA (2003) Trehalose metabolism: a regulatory role for trehalose-6-phosphate? Curr Opin Plant Biol 6 231–235 [DOI] [PubMed] [Google Scholar]
- Ehira S, Ohmori M, Sato N (2003) Genome-wide expression analysis of the responses to nitrogen deprivation in the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. DNA Res 10 97–113 [DOI] [PubMed] [Google Scholar]
- Esposito S, Massaro G, Vona V, Di Martino Rigano V, Carfagna S, Rigano C (2001) Ammonium induction of a novel isoform of glucose-6P dehydrogenase in barley roots. Physiol Plant 113 469–476 [DOI] [PubMed] [Google Scholar]
- Filleur S, Dorbe MF, Cerezo M, Orsel M, Granier F, Gojon A, Daniel-Vedele F (2001) An Arabidopsis T-DNA mutant affected in Nrt2 genes is impaired in nitrate uptake. FEBS Lett 489 220–224 [DOI] [PubMed] [Google Scholar]
- Forde BG (2002) Local and long-range signaling pathways regulating plant responses to nitrate. Annu Rev Plant Biol 53 203–224 [DOI] [PubMed] [Google Scholar]
- Foreman J, Demidchik V, Bothwell JH, Mylona P, Miedema H, Torres MA, Linstead P, Costa S, Brownlee C, Jones JD, et al (2003) Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422 442–446 [DOI] [PubMed] [Google Scholar]
- Garlick AP, Moore C, Kruger NJ (2002) Monitoring flux through the oxidative pentose pathway using [1-14C]gluconate. Planta 216 265–272 [DOI] [PubMed] [Google Scholar]
- Gastal F, Saugier B (1989) Relationships between nitrogen uptake and carbon assimilation in whole plants of tall fescue. Plant Cell Environ 12 407–418 [Google Scholar]
- Gazzarrini S, Lejay L, Gojon A, Ninnemann O, Frommer WB, von Wiren N (1999) Three functional transporters for constitutive, diurnally regulated, and starvation-induced uptake of ammonium into Arabidopsis roots. Plant Cell 11 937–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon Y, Vigeolas H, Tiessen A, Geigenberger P, Stitt M (2002) Sensitive and high throughput metabolite assays for inorganic pyrophosphate, ADPGlc, nucleotide phosphates, and glycolytic intermediates based on a novel enzymic cycling system. Plant J 30 221–235 [DOI] [PubMed] [Google Scholar]
- Gibson SI (2000) Plant sugar response pathways: part of a complex regulatory web. Plant Physiol 124 1532–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grignon C (1990) Transports in roots. Symbiosis 9 3–17 [Google Scholar]
- Gutierrez RA, Lejay LV, Dean A, Chiaromonte F, Shasha DE, Coruzzi GM (2007) Qualitative network models and genome-wide expression data define carbon/nitrogen-responsive molecular machines in Arabidopsis. Genome Biol 8 R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänisch ten Cate CH, Breteler H (1981) Role of sugars in nitrate utilization by roots of dwarf bean. Physiol Plant 52 129–135 [Google Scholar]
- Hatch DJ, Hopper MJ, Dhanoas MS (1986) Measurements of ammonium ions in flowing solution culture and diurnal variation in uptake by Lolium perenne L. J Exp Bot 37 589–596 [Google Scholar]
- Herold A, Lewis DH (1977) Mannose and green plants: occurence, physiology and metabolism, and use as a tool to study the role of orthophosphate. New Phytol 79 1–40 [Google Scholar]
- Hesse H, Trachsel N, Suter M, Kopriva S, von Ballmoos P, Rennenberg H, Brunold C (2003) Effect of glucose on assimilatory sulphate reduction in Arabidopsis thaliana roots. J Exp Bot 54 1701–1709 [DOI] [PubMed] [Google Scholar]
- Hirai MY, Fujiwara T, Awazuhara M, Kimura T, Noji M, Saito K (2003) Global expression profiling of sulfur-starved Arabidopsis by DNA macroarray reveals the role of O-acetyl-l-serine as a general regulator of gene expression in response to sulfur nutrition. Plant J 33 651–663 [DOI] [PubMed] [Google Scholar]
- Ho CL, Saito K (2001) Molecular biology of the plastidic phosphorylated serine biosynthetic pathway in Arabidopsis thaliana. Amino Acids 20 243–259 [DOI] [PubMed] [Google Scholar]
- Jang JC, Leon P, Zhou L, Sheen J (1997) Hexokinase as a sugar sensor in higher plants. Plant Cell 9 5–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MA, Raymond MJ, Yang Z, Smirnoff N (2007) NADPH oxidase-dependent reactive oxygen species formation required for root hair growth depends on ROP GTPase. J Exp Bot 58 1261–1270 [DOI] [PubMed] [Google Scholar]
- Kohler E, Barrach H, Neubert D (1970) Inhibition of NADP dependent oxidoreductases by the 6-aminonicotinamide analogue of NADP. FEBS Lett 6 225–228 [DOI] [PubMed] [Google Scholar]
- Kopriva S, Rennenberg H (2004) Control of sulphate assimilation and glutathione synthesis: interaction with N and C metabolism. J Exp Bot 55 1831–1842 [DOI] [PubMed] [Google Scholar]
- Lalonde S, Boles E, Hellmann H, Barker L, Patrick JW, Frommer WB, Ward JM (1999) The dual function of sugar carriers. Transport and sugar sensing. Plant Cell 11 707–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bot J, Kirkby EA (1992) Diurnal uptake of nitrate and potassium during the vegetative growth of tomato plants. J Plant Nutr 15 247–264 [Google Scholar]
- Leegood RC, Labate CA, Huber SC, Neuhaus HE, Stitt M (1988) Phosphate sequestration by glycerol and its effects on photosynthetic carbon assimilation by leaves. Planta 176 117–126 [DOI] [PubMed] [Google Scholar]
- Lejay L, Gansel X, Cerezo M, Tillard P, Muller C, Krapp A, von Wiren N, Daniel-Vedele F, Gojon A (2003) Regulation of root ion transporters by photosynthesis: functional importance and relation with hexokinase. Plant Cell 15 2218–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejay L, Tillard P, Lepetit M, Olive F, Filleur S, Daniel-Vedele F, Gojon A (1999) Molecular and functional regulation of two NO3- uptake systems by N- and C-status of Arabidopsis plants. Plant J 18 509–519 [DOI] [PubMed] [Google Scholar]
- Little DY, Rao H, Oliva S, Daniel-Vedele F, Krapp A, Malamy JE (2005) The putative high-affinity nitrate transporter NRT2.1 represses lateral root initiation in response to nutritional cues. Proc Natl Acad Sci USA 102 13693–13698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobreaux S, Massenet O, Briat JF (1992) Iron induces ferritin synthesis in maize plantlets. Plant Mol Biol 19 563–575 [DOI] [PubMed] [Google Scholar]
- Lowry OH, Passonneau JV (1972) A Flexible System of Enzymatic Analysis. Academic Press, New York
- Maruyama-Nakashita A, Inoue E, Watanabe-Takahashi A, Yamaya T, Takahashi H (2003) Transcriptome profiling of sulfur-responsive genes in Arabidopsis reveals global effects of sulfur nutrition on multiple metabolic pathways. Plant Physiol 132 597–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura H, Xie Y, Shirakata S, Inoue T, Yoshinaga T, Ueno Y, Izui K, Kai Y (2002) Crystal structures of C4 form maize and quaternary complex of E. coli phosphoenolpyruvate carboxylases. Structure 10 1721–1730 [DOI] [PubMed] [Google Scholar]
- Matt P, Geiger M, Walch-Liu P, Engels C, Krapp A, Stitt M (2001) Elevated carbon dioxide increases nitrate uptake and nitrate reductase activity when tobacco is growing on nitrate, but increases ammonium uptake and inhibits nitrate reductase activity when tobacco is growing on ammonium nitrate. Plant Cell Environ 24 1119–1137 [Google Scholar]
- Molas ML, Kiss JZ, Correll MJ (2006) Gene profiling of the red light signalling pathways in roots. J Exp Bot 57 3217–3229 [DOI] [PubMed] [Google Scholar]
- Moshelion M, Becker D, Czempinski K, Mueller-Roeber B, Attali B, Hedrich R, Moran N (2002) Diurnal and circadian regulation of putative potassium channels in a leaf moving organ. Plant Physiol 128 634–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munos S, Cazettes C, Fizames C, Gaymard F, Tillard P, Lepetit M, Lejay L, Gojon A (2004) Transcript profiling in the chl1-5 mutant of Arabidopsis reveals a role of the nitrate transporter NRT1.1 in the regulation of another nitrate transporter, NRT2.1. Plant Cell 16 2433–2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff MM, Fankhauser C, Chory J (2000) Light: an indicator of time and place. Genes Dev 14 257–271 [PubMed] [Google Scholar]
- Neuhaus HE, Emes MJ (2000) Nonphotosynthetic metabolism in plastids. Annu Rev Plant Physiol Plant Mol Biol 51 111–140 [DOI] [PubMed] [Google Scholar]
- Nikiforova V, Freitag J, Kempa S, Adamik M, Hesse H, Hoefgen R (2003) Transcriptome analysis of sulfur depletion in Arabidopsis thaliana: interlacing of biosynthetic pathways provides response specificity. Plant J 33 633–650 [DOI] [PubMed] [Google Scholar]
- Oji Y, Watanabe M, Wakiuchi N, Okamoto S (1985) Nitrite reduction in barley-root plastids: dependence on NADPH coupled with glucose-6-phoshate and 6-phosphogluconate dehydrogenases, and possible involvement of an electron carrier and a diaphorase. Planta 165 85–90 [DOI] [PubMed] [Google Scholar]
- Ono F, Frommer WB, von Wiren N (2000) Coordinated diurnal regulation of low- and high-affinity nitrate transporters in tomato. Plant Biol 2 17–23 [Google Scholar]
- Orsel M, Chopin F, Leleu O, Smith SJ, Krapp A, Daniel-Vedele F, Miller AJ (2006) Characterization of a two-component high-affinity nitrate uptake system in Arabidopsis. Physiology and protein-protein interaction. Plant Physiol 142 1304–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J, Laxmi A, St Martin SK, Jang JC (2004) Global transcription profiling reveals multiple sugar signal transduction mechanisms in Arabidopsis. Plant Cell 16 2128–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail PH (2002) Photosensory perception and signalling in plant cells: new paradigms? Curr Opin Cell Biol 14 180–188 [DOI] [PubMed] [Google Scholar]
- Remans T, Nacry P, Pervent M, Girin T, Tillard P, Lepetit M, Gojon A (2006) A central role for the nitrate transporter NRT2.1 in the integrated morphological and physiological responses of the root system to nitrogen limitation in Arabidopsis. Plant Physiol 140 909–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennenberg H (1984) The fate of excess sulfur in higher plants. Annu Rev Plant Physiol 35 121–153 [Google Scholar]
- Rolland F, Baena-Gonzalez E, Sheen J (2006) Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu Rev Plant Biol 57 675–709 [DOI] [PubMed] [Google Scholar]
- Salisbury FJ, Hall A, Grierson CS, Halliday KJ (2007) Phytochrome coordinates Arabidopsis shoot and root development. Plant J 50 429–438 [DOI] [PubMed] [Google Scholar]
- Schluepmann H, van Dijken A, Aghdasi M, Wobbes B, Paul M, Smeekens S (2004) Trehalose mediated growth inhibition of Arabidopsis seedlings is due to trehalose-6-phosphate accumulation. Plant Physiol 135 879–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J (1990) Metabolic repression of transcription in higher plants. Plant Cell 2 1027–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J, Zhou L, Jang JC (1999) Sugars as signaling molecules. Curr Opin Plant Biol 2 410–418 [DOI] [PubMed] [Google Scholar]
- Smeekens S (2000) Sugar-induced signal transduction in plants. Plant Mol Biol 51 49–81 [DOI] [PubMed] [Google Scholar]
- Smith IK, Cheema HK (1985) Sulphate transport into plants and excised roots of soybean (Glycine max L.). Ann Bot (Lond) 56 219–224 [Google Scholar]
- Takahashi-Terada A, Kotera M, Ohshima K, Furumoto T, Matsumura H, Kai Y, Izui K (2005) Maize phosphoenolpyruvate carboxylase. Mutations at the putative binding site for glucose 6-phosphate caused desensitization and abolished responsiveness to regulatory phosphorylation. J Biol Chem 280 11798–11806 [DOI] [PubMed] [Google Scholar]
- Thum KE, Shasha DE, Lejay LV, Coruzzi GM (2003) Light- and carbon-signaling pathways. Modeling circuits of interactions. Plant Physiol 132 440–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai FY, Coruzzi GM (1991) Light represses the transcription of asparagine synthetase genes in photosynthetic and non-photosynthetic organs of plants. Mol Cell Biol 11 4966–4972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsay YF, Schroeder JI, Feldmann KA, Crawford NM (1993) The herbicide sensitivity gene CHL1 of Arabidopsis encodes a nitrate-inducible nitrate transporter. Cell 72 705–713 [DOI] [PubMed] [Google Scholar]
- Urner F, Sakkas D (2005) Involvement of the pentose phosphate pathway and redox regulation in fertilization in the mouse. Mol Reprod Dev 70 494–503 [DOI] [PubMed] [Google Scholar]
- Vert GA, Briat JF, Curie C (2003) Dual regulation of the Arabidopsis high-affinity root iron uptake system by local and long-distance signals. Plant Physiol 132 796–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wiren N, Lauter FR, Ninnemann O, Gillissen B, Walch-Liu P, Engels C, Jost W, Frommer WB (2000) Differential regulation of three functional ammonium transporter genes by nitrogen in root hairs and by light in leaves of tomato. Plant J 21 167–175 [DOI] [PubMed] [Google Scholar]
- Wang R, Guegler K, LaBrie ST, Crawford NM (2000) Genomic analysis of a nutrient response in Arabidopsis reveals diverse expression patterns and novel metabolic and potential regulatory genes induced by nitrate. Plant Cell 12 1491–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Okamoto M, Xing X, Crawford NM (2003) Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1,000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiol 132 556–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth J, Chopin F, Santoni V, Viennois G, Tillard P, Krapp A, Lejay L, Daniel-Vedele F, Gojon A (2007) Regulation of root nitrate uptake at the NRT2.1 protein level in Arabidopsis thaliana. J Biol Chem 282 23541–23552 [DOI] [PubMed] [Google Scholar]
- Xiao W, Sheen J, Jang JC (2000) The role of hexokinase in plant sugar signal transduction and growth and development. Plant Mol Biol 44 451–461 [DOI] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Onda Y, Ashikari T, Tanaka Y, Kusumi T, Hase T (2000) Analysis of reductant supply systems for ferredoxin-dependent sulfite reductase in photosynthetic and nonphotosynthetic organs of maize. Plant Physiol 122 887–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.