Abstract
To discover genes involved in von Hippel-Lindau (VHL)-mediated carcinogenesis, we used renal cell carcinoma cell lines stably transfected with wild-type VHL-expressing transgenes. Large-scale RNA differential display technology applied to these cell lines identified several differentially expressed genes, including an alpha carbonic anhydrase gene, termed CA12. The deduced protein sequence was classified as a one-pass transmembrane CA possessing an apparently intact catalytic domain in the extracellular CA module. Reintroduced wild-type VHL strongly inhibited the overexpression of the CA12 gene in the parental renal cell carcinoma cell lines. Similar results were obtained with CA9, encoding another transmembrane CA with an intact catalytic domain. Although both domains of the VHL protein contribute to regulation of CA12 expression, the elongin binding domain alone could effectively regulate CA9 expression. We mapped CA12 and CA9 loci to chromosome bands 15q22 and 17q21.2 respectively, regions prone to amplification in some human cancers. Additional experiments are needed to define the role of CA IX and CA XII enzymes in the regulation of pH in the extracellular microenvironment and its potential impact on cancer cell growth.
Keywords: gene targets/differential display/carcinogenesis
Inactivation of the von Hippel-Lindau (VHL) tumor suppressor gene is responsible for hereditary (VHL disease) and most sporadic renal cell carcinomas (RCCs) of the clear cell type (1). The product of the gene, pVHL, was predicted to contain two protein-binding domains (E. V. Kunin, personal communication). This prediction for the second domain that maps close to the pVHL carboxyl terminus (residues 156–195) was confirmed by protein binding studies (2–5). This domain binds to elongin B and C subunits of the transcription elongation factor SIII/Elongin, which led to a hypothesis of pVHL involvement in transcription elongation regulation (3–6). The initial discovery of three pVHL target genes, VEGF, PDGF-B, and Glut1 (7, 8), stimulated more research into the molecular mechanisms of pVHL involvement in regulation of gene expression. A recent study of VEGF down-regulation by wild type (wt) VHL transgenes (9) showed that pVHL directly interacted in this system with the ubiquitous transcription factor Sp1. Thus pVHL has possible effects on initiation of transcription from Sp1-driven promoters. The other two genes, PDGF-B and Glut1 (8), and the TGF alpha gene (10) were down-regulated by pVHL at the posttranscriptional stages, most likely by reducing mRNA stability (7, 8, 10). RNA differential display (RDD) technology was used to study transcription in RCC cell lines expressing wtVHL transgenes versus parental nontransfected cells to discover target genes down-regulated by wtVHL. We report here two additional VHL target genes, encoding carbonic anhydrases (CA) CA12 and CA9 (11), which are strongly suppressed by wtVHL. Both protein binding domains of pVHL contribute to the regulation of CA12 expression whereas CA9 was regulated mostly by the elongin binding domain. High levels of CA12 expression in adult kidney, pancreas, colon, and prostate suggest an important physiological function for this enzyme.
MATERIALS AND METHODS
Molecular Techniques.
All molecular manipulations (screening cDNA libraries, Northern blot analysis, and PCR) were performed by using standard methods (12).
DNA Sequence Determination.
cDNA clones were sequenced on an Applied Biosystems 373 DNA sequencer (Stretch) using Taq Dyedeoxy Terminator Cycle Sequence kits (Applied Biosystems) with either vector or clone-specific walking primers.
mRNA Expression Analyses.
Northern blot hybridization was performed with CA12 and CA9 cDNA probes by using commercial MTN poly(A) RNA blots (CLONTECH) from a variety of adult human tissues and tumor cell lines and poly(A)+ RNA prepared from RCC and lung cancer cell lines. In addition, the presence of CA12 and CA9 transcripts was monitored in silico by blast homology searches (ref. 13 and http://www.ncbi.nlm.nih.gov/BLAST/) in public expressed sequence tag (EST) databases (http://www.ncbi.nlm.nih.gov/dbEST/index.html).
Fluorescent in Situ Hybridization.
Metaphase spreads derived from BrdUrd-synchronized normal peripheral lymphocytes were used as a template. Probes containing CA9 and CA12 cDNAs were labeled with digoxigenin 11-dUTP by nick-translation, and hybridization signals were detected with rhodamine-conjugated antidigoxigenin antibodies (Boehringer-Mannheim). The conditions of hybridization, detection of hybridization signals, and digital-image acquisition, processing, and analyses were performed as previously described (14). Chromosomes were identified by converting 4′,6-diamidino-2-phenylindole (DAPI) banding into G-simulated banding by using IP Lab Image Software (Scan Analytics, Vienna, VA). Rehybridization with alpha-satellite centromeric probes (Oncor) was performed to confirm chromosomal localization.
Sequence Analyses.
World Wide Web-based servers were used to analyze the cDNAs and deduced protein sequences. Global sequence alignments were done by using blast and advanced blast programs as provided by the National Center for Biotechnology Information (ref. 13; http://www.ncbi.nlm.nih.gov/BLAST/and http://www.ncbi.nlm.nih.gov/ cgi-bin/BLAST/nph-blast?Jform = 1) and BLAST2/WU Blast, provided by the European Molecular Biology Laboratory (http://www2.ebi.ac.uk/blast2/).
Multiple sequence alignments, global and local, were done by using the clustal version w program as provided by the European Molecular Biology Laboratory (http://dot.imgen.bcm.tmc.edu:9331/seq-search/alignment.html), the Baylor Computing Center (http://gc.bcm.tmc.edu:8088/search-launcher/launcher.html), and the Wisconsin Genetics Computer Group, package 8, program (http://www.gcg.com/).
Protein domains were discovered on the Pfam (http://www.sanger.ac.uk/Pfam/) and membrane topology on the psort servers (http://www.cbs.dtu.dk/services/SignalP/). Some protein motifs were found by visually inspecting local alignments or by using the protein motifs server (http://www.mips.biochem.mpg.de/).
VHL-Expressing Constructs.
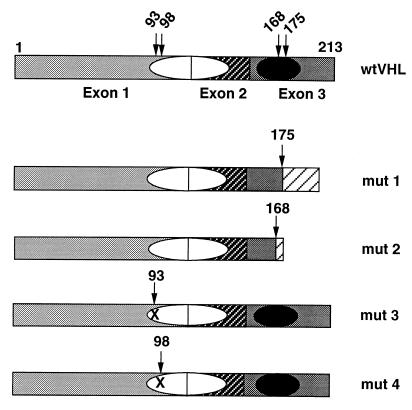
wtVHL expressing minigenes were constructed with the pRC/CMV-Htag-VHL (15) plasmid cut with HindIII–XhoI, and the insert was placed into pCEP4 (Invitrogen). H-tag was removed by partial digestion with BamHI–HindIII, followed by ligation. Another wtVHL-expressing construct used in this work was obtained by reverse transcription–PCR amplification of the wtVHL reading frame followed by insertion into the pCR 3.1 expression vector as recommended by the vendor (Invitrogen). Mutant (mut) pVHL-expressing constructs were created in a similar way by using naturally occurring mutations. Mut 1 is a 1-bp deletion creating a frameshift after residue 175; mut 2 is a 4-bp insertion creating a frameshift after residue 168; mut 3 and mut 4 are missense mutations, Gly-93–Asp and Tyr-98–His, respectively. Products of mut 1 and mut 2 are devoid of the elongin binding domain; those of mut 3 and mut 4 possess an intact functional (5) elongin binding domain (Fig. 1).
Figure 1.
Schematic representation of the domain structure of wt and naturally occurring mut pVHLs. The three exons are indicated by different shadowing; the putative protein binding domains are represented by ovals, the filled one is the elongin binding domain; numbers above the pictures are amino acid residues, arrows indicate residues involved in producing mut pVHLs. Mut 1 is a 1-bp deletion creating a frameshift after residue 175; mut 2 is a 4-bp insertion creating a frameshift after residue 168; mut 3 and mut 4 are missense mutations, Gly-93–Asp and Tyr-98–His, respectively. Mut 1 and mut 2 are devoid of the elongin binding domain; mut 3 and mut 4 possess an intact functional elongin binding domain.
RCC Cell Lines Overexpressing wtVHL and mutVHLs.
VHL transgenes were overexpressed in two clear cell type RCC cell lines. The first one, UM-RC-6 (16), contains an intragenic 9-bp deletion in the remaining single allele of the VHL gene creating a frameshift after residue 168 (17). The second one, 786–0 (18), contains a 1-bp deletion creating a frameshift in the remaining single allele after residue 104 (17). All transfections were performed by using Lipofectin (GIBCO/BRL) according to the manufacturer’s protocol. The VHL-positive clones were selected for the presence of exogenous VHL mRNA and protein by using Northern, Southern, and Western blot analyses (data not shown). VHL transgene mRNAs were amplified by reverse transcription–PCR and verified by sequencing.
Cell Lines Used in RDD.
For RDD we used two cell lines: UM-RC-6 and its wtVHL-transfected derivative harvested at a late confluent stage of growth. The following cell lines were used for subsequent Northern analyses: parental UM-RC-6 and 786–0 and the same cells stably transfected with wt and mut VHLs.
RDD Analysis.
RNA isolation for RDD, cDNA production, recovery and reamplification of differentially expressed bands, subcloning, and sequencing were performed as described previously (19). For equal loading of mRNA samples, the EagleEye Vision system (Stratagene) was used to control intensity of ethidium bromide-stained lanes on a pilot gel. Before hybridization, blots were stained with methylene blue to show equal loading of the RNA samples. Quantification of hybridization signals was done by using the PhosphorImager system (Molecular Dynamics).
RESULTS
Strategy and Experimental Design to Isolate Genes Involved in VHL-Mediated Carcinogenesis.
The changes in gene expression in RCC devoid of functional pVHL after introduction of wtVHL were studied by using differential display. This straightforward strategy was used to identify genes whose expression was up-regulated or down-regulated after introduction of wtVHL-expressing minigenes. The theoretical framework underlying the RDD technology largely depends on comparing mRNA concentrations at physiological conditions that maintain steady-state levels of gene expression. The expression of the various VHL constructs was not influenced by the growth conditions: it was similar in late logarithmic and confluent cells (data not shown). Therefore, we ran the RDD at a late confluent stage of cell growth and verified differential expression by Northern blot analyses in both growing and resting cells. Fig. 1 presents the domain structure of pVHL and the structure of mutVHL transgenes used for transfections.
Identification and Analysis of Genes Differentially Expressed by RCC Cells Overexpressing wtVHL Transgenes.
We compared steady-state RNA concentrations by RDD in late confluent UM-RC-6 cells (three independent isolates) versus the same cells stably transfected with a wtVHL minigene driven by the cytomegalovirus promoter. Eighty arbitrary 13-mer primers in combination with three anchor primers to complete 240 sets of differential display reactions were designed to scan at least 20–25% of the total gene complement expressed in these cells. Fifty-two bands were found to be differentially expressed in a reproducible fashion. All were PCR-reamplified, cloned, sequenced, and used for Northern blot analyses to confirm that they indeed represented differentially expressed RNAs. Thus far, five have been confirmed to be down-regulated by wtVHL from two to more than 30 times (data not shown). Sequence analyses revealed matches in the databases and identified three specific clones as corresponding to known human genes, namely, NOTCH 2 (ref. 20, GenBank U77493), DEC1 (ref. 21, GenBank AB004066), and an alpha CA; two clones corresponded to ESTs (GenBank D86978 and AA165698) of unknown genes.
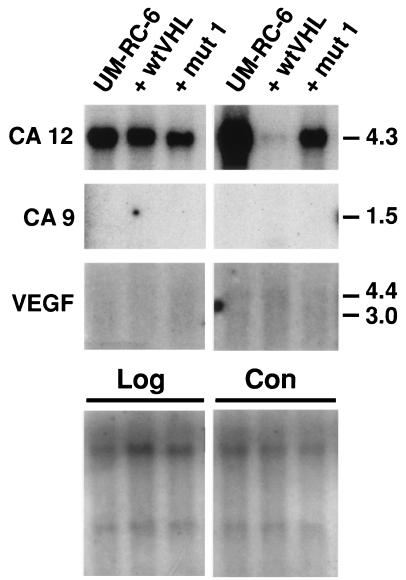
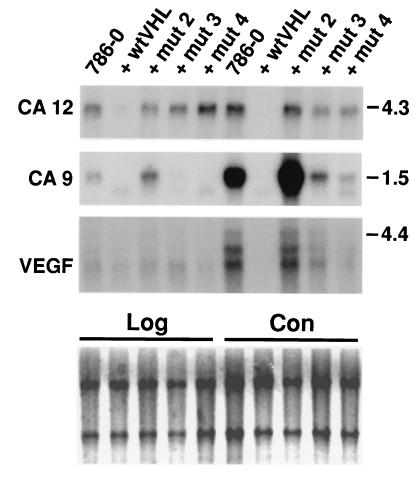
The alpha CA family (E.C. 4.2.1.1., carbonic dehydratase) (22, 23) is comprised of 12 isozymes, including the CA XII identified and cloned in this work. Here we report in detail on CA XII, and also include data on regulation of another structurally related transmembrane CA, CA IX (11) that is down-regulated by VHL (see below). First, their expression at different stages of cell growth was analyzed in another RCC cell line, 786–0, which is also devoid of a functional pVHL (17). Second, the effect of wtVHL and distinct mutVHL minigenes on the expression of CA12 and CA9 in both RCC cell lines was ascertained (Figs. 2 and 3). Two classes of naturally occurring VHL mutations (17, 24) were tested. Mut 1 and mut 2 minigenes encoded truncated proteins devoid of the elongin binding domain (Fig. 1), whereas mut 3 and mut 4 apparently encoded full-sized proteins with missense mutations in the first putative binding domain and an intact (Fig. 1) and functional (5) elongin binding domain. Mut and wt VHL proteins are highly expressed in transfected UM-RC-6 (data not shown) and 786–0 cells (3–5). The VEGF gene, repeatedly reported as a VHL gene target in 786–0 cells (7–9), was included in these analyses as a control. The results (Figs. 2 and 3) clearly show that the expression pattern of the CA9 and CA12 genes is different in the analyzed RCC lines and dramatically depends on the growth stage. The CA12 gene is highly expressed in both cell lines, whereas the CA9 and VEGF is genes are highly expressed in the 786–0 cells but not expressed at all in the UM-RC-6 line. The expression of all three genes is much higher in a late confluent stage compared with late logarithmic growth. The impact of the VHL transgenes, both wt and mut ones, on CA9, CA12, and VEGF gene expression is very striking and indeed informative (Figs. 2 and 3). The wtVHL completely suppresses the expression of CA9 and CA12 genes in both confluent and growing 786–0 cells. VEGF is completely suppressed in confluent 786–0 cells, but only partially suppressed in growing 786–0 cells. wtVHL effectively (>80%) suppresses the high level of CA12 expression in late confluent UM-RC-6 cells but not in growing cells (Fig. 2). The mutVHL transgenes differentially affect the expression of CA12, CA9, and VEGF genes (Figs. 2 and 3). Mut 1 and mut 2 transgenes, which lack the elongin binding domain, slightly suppress CA12 expression but have no effect or even slightly up-regulate the expression of CA9 and VEGF genes. In contrast, mut 3 and mut 4 transgenes, which express an intact functional elongin binding domain, suppress the expression of all three genes to varying degrees in confluent cells (Fig. 3). In growing cells, these mutant VHL proteins completely suppress CA9, show no effect on CA12, and only slightly suppress VEGF gene expression. These observations suggest that the CA12 gene discovered on the RDD screen and the previously cloned CA9 (11) gene are regulated by VHL. Both domains of the wt pVHL are required to down-regulate the expression of the CA12 gene. Naturally occurring mutants that possess an intact functional elongin binding domain alone could effectively (>90%) down-regulate the expression of the CA9 gene (Fig. 3). We next explored the possibility that VHL could regulate the expression of other members of the alpha CA gene family. wtVHL transgenes did not affect the expression of CA1, CA2, CA4, and CA5 in the UM-RC-6 and 786–0 RCC cell lines (data not shown).
Figure 2.
Northern analysis of target genes in growing (Log) and late confluent (Con) UM-RC-6 cells transfected with wt or mut 1 pVHL. Quantification of hybridization signals was done by using the PhosphorImager system (Molecular Dynamics). Before hybridization the blots were stained with methylene blue (Lower) to show approximately equal loading of the RNA samples (see also Materials and Methods). Numbers on the right represent size markers in kb.
Figure 3.
Northern analysis of target genes in growing (Log) and late confluent (Con) 786–0 cells transfected with wt or mut 2, mut 3, and mut 4 pVHL-containing plasmids. Quantification of hybridization signals was done by using the PhosphorImager system (Molecular Dynamics). Before hybridization the blots were stained with methylene blue (Lower) to show approximately equal loading of the RNA samples (see also Materials and Methods). Numbers on the right represent size markers in kb.
Cloning and Characterization of CA12, a Member of the Alpha CA Gene Family.
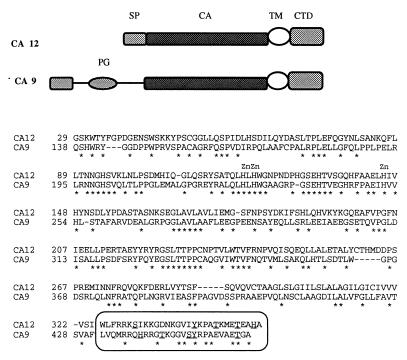
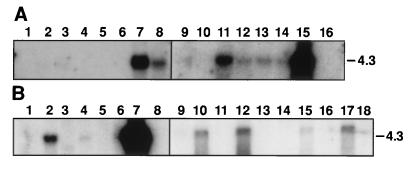
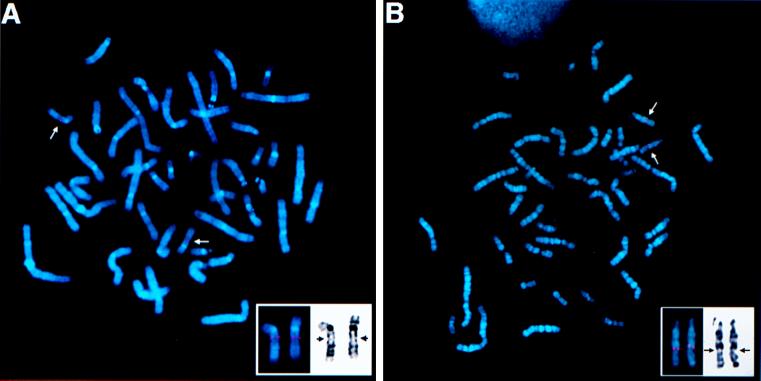
The differentially expressed cDNA clone no. 9 was used as a probe to screen a human lung cDNA library (no. 1024–018, GIBCO/BRL). As a result, several cDNA clones were obtained and then sequenced. Sequence analysis showed that all of these clones represented partial cDNA sequences of the same gene. The biggest insert, 2,771 bp in length, contained a 1,062-bp ORF (354 amino acids), a 115-bp 5′ untranslated region (UTR), and a 1,591-bp 3′ UTR, which contained the original differentially expressed cloned cDNA sequence 9. After this work was completed, Tureci et al. (25) reported the cloning of the CA12 cDNA, which contained 108 bp less in the 5′ UTR. We used a number of bioinformatics web-based servers (see Materials and Methods) to analyze the cDNA and the deduced protein sequences. Analysis of the cDNA showed high degrees of homology to members of the alpha CA gene family and established CA12 as a member of this family (data not shown). Analysis of the predicted protein sequence (354 residues) revealed a signal peptide (cleavage site between residues 24 and 25) followed by an alpha CA domain (residues 28–289), which has 55% similarity with the consensus sequence, followed by a transmembrane peptide (residues 308–324) and a short cytoplasmic domain (residues 325–354). This protein is a one-pass transmembrane CA with extracellular amino and intracellular carboxy termini. The CA domain of CA12 is closely related to the CA domain of the CA6 and CA9 genes. The cytoplasmic domain of CA XII is 50% identical to the cytoplasmic domain of the CA9 gene product, which is also a transmembrane CA protein (Fig. 4). Phylogenetic reconstruction of alpha CAs (data not shown) indicates that CA XII, CA IX, and CA VI are more closely related to each other than to other alpha CAs and group together, representing a rather early branch in the evolution of the alpha CA gene family (22). The short cytoplasmic domains of these two proteins contain several conserved residues including a threonine, a motif with one tyrosine, GVXYXPA, and one or two histidine residues (Fig. 4). Both proteins possess all three zinc-binding histidines obligatory for the catalytic activity in their CA modules (Fig. 4) (22, 23). The CA IX protein was predicted (22) and reported (26) to be an active CA enzyme. Tureci et al. (25) now have shown that expressed CA XII protein is an active CA isozyme. We next analyzed the expression patterns of the CA12 gene by Northern blot hybridization to human adult tissues, some tumor cell lines, and by in silico monitoring public EST databases. Northern blot hybridization was performed with commercial multiple tissue poly(A)+ RNA blots (CLONTECH). CA12 is highly expressed in colon, kidney, and prostate, moderately in pancreas, ovary, and testis as 4.3-kb mRNA (Fig. 5A). Very low expression could be observed in lung and brain on longer exposure (data not shown). The tumor cell lines tested (Fig. 5B) showed A549 cells (adenocarcinoma of the lung) had very high level of expression, HeLa S3, and colorectal (SW480) cells moderate expression, lymphoma low expression, and no expression was detected in melanoma and several leukemias. The very high expression in the A549 cells prompted us to analyze the expression of CA12 in other small cell and nonsmall cell lung carcinoma cell lines. No expression was detected in 14 small cell lung cancer and high to moderate expression in four of 10 tested nonsmall cell lung carcinoma cell lines (Fig. 5B). The 15 CA12 ESTs were distributed in the cDNA libraries as follows: 5/15 colon, 2/15 kidney, 2/15 pancreatic islets, 2/15 ovarian carcinoma, and 1/15 in fetal brain, fetal heart, uterus, and endometrial carcinoma. These combined results suggest that CA12 is expressed in a restricted assortment of normal and tumor tissues. CA9 is far more restricted then CA12 in its tissue distribution. It is not expressed in normal kidney tissue (27), and the only normal tissue that expresses significant amounts is the gastric mucosa (ref. 27 and S. Y. Liao and E.J.S., unpublished observations) and gut enterocytes (28). The chromosomal map positions of the CA9 and CA12 genes were determined by fluorescence in situ hybridization using the corresponding cDNAs as probes. The CA9 locus was localized to 17q21.2–21.3 (Fig. 6A) and the CA12 to 15q22 (Fig. 6B). These bands are subject to amplifications in quite a number of human cancers that include prostate, lung, kidney, ovarian, breast, neuroblastoma, and head and neck tumors (29).
Figure 4.
Global amino acid alignment of CA XII and CA IX proteins obtained by using the clustalw alignment tool as provided by the ExPasy server (http://expasy.hcuge.ch/sprot/scnpsite.html). The overall identity is 35.8%, and the identical residues are marked by ∗ below the sequence. The three essential Zn-liganded histidine residues are marked by Zn symbols above them. The short cytoplasmic domains are boxed with histidine, threonine, serine, and tyrosine residues underlined. The ideograms depicting the domain structure of the genes are positioned above the alignment. SP, signal peptide; CA, carbonic anhydrase domain; TM, transmembrane peptide; CTD, cytoplasmic domain; PG, proteoglycan domain.
Figure 5.
Northern blot analysis of CA12 expression in normal tissues (A) and tumor cell lines (B) using the cDNA as hybridization probe. (A) The RNA filters are from CLONTECH (nos. 7759 and 7760) and contain 2 μg of poly(A)+ mRNA per tissue indicated: lanes 1, heart; 2, brain; 3, spleen; 4, thymus; 5, prostate; 6, testis; 7, ovary; 8, small intestine; 9, colon; 10, peripheral blood leucocytes. (B) The RNA filter is from CLONTECH (no. 775) and contains 2 μg of poly(A)+ mRNA per cell line indicated: lanes 1, promyelocytic leukemia, HL-60; 2, HeLa cells S3; 3, chronic myelogenous leukemia, K-562; 4, lymphoblastic leukemia, MOLT-4; 5, Burkitt’s lymphoma, Raji; 6, colorectal adenocarcinoma, SW 480; 7, lung adenocarcinoma, A549; 8, melanoma, G361, and poly(A)+ RNA from human nonsmall cell lung carcinomas (as indicated: 9, NCI H_1373; 10, NCI H_1264; 11, NCI H_1693; 12, NCI H_1944; 13, NCI H_838; 14, NCI H_1299; 15, NCI H_157; 16, NCI H_1466; 17, NCI H_460; 18, NCI H_727).
Figure 6.
Subchromosomal localization of the human CA9 and CA12 genes. Metaphase after fluorescence in situ hybridization showing location of the (A) CA9 gene on the long arm of chromosome 17q21.2–21.3 (arrow) and (B) CA12 gene on the long arm of chromosome 15q22 (arrow). (Insets) Position of both loci, CA9 and CA12, on 4′,6-diamidino-2-phenylindole (DAPI)-banded human chromosomes, 17q21.2–21.3 and 15q22, respectively (arrows).
DISCUSSION
Analysis of VHL tumor phenotypes led to the prediction of several VHL target genes that were verified in the 786–0 cell system (7–9). Our interest in identifying more VHL gene targets was because at least some of these genes could mediate downstream events in VHL and/or general carcinogenesis. Therefore, a more global understanding of VHL-mediated changes in gene expression might direct us to new applications for diagnostic and therapeutic intervention in the VHL-caused tumors. Our study used the RDD technology applied to RCC cells overexpressing wtVHL transgenes to discover VHL gene targets. RDD analysis of 20–25% of all transcribed genes in RCC cells has allowed us to identify five additional VHL gene targets, suggesting that the total number of potential targets would not exceed 20–25 genes. RDD analysis with a complete set of primers should reveal most, if not all, of the yet-to-be-discovered VHL potential target genes. The 10 known VHL targets (six in this work and four reported previously, refs. 7–9) include only one protooncogene the NOTCH2 gene (30, 31).
The physiological functions of the numerous alpha CA enzymes (22, 23) are based on their ability to catalyze the reversible hydration of CO2, producing carbonic acid that subsequently decomposes to HCO3− and H3O+, leading to a decrease in pH. There is considerable evidence that the glycosylphosphatidylinositol-anchored plasma membrane CA IV enzyme exerts an effect on several ionic channels, which could result in shifting protons and HCO3− across the membrane, leading to a rise in cytoplasmic and decrease in extracellular pH (23, 32). The one-span transmembrane CA IX and CA XII enzymes, which were predicted and shown to be catalitically active (22, 25, 26), could serve a similar function. Their short, cytoplasmic domains containing histidine, threonine, serine, and tyrosine residues could function as sensors of cytoplasmic pH and elicit cellular responses to certain cues. The enhanced expression of CA IX (33, 34) and CA XII (35, 36) isozymes in renal and other cancers may add to our understanding of cancer development and may well explain evidence accumulated since the time of O. Warburg (37), that the extracellular pH of human tumors is on average more acidic than that of normal tissues (38). We argue that these CA isozymes acidify the immediate extracellular milieu surrounding the cancer cells and thus create a microenvironment conducive to tumor growth and spread. It has been shown that acidic pH enhances invasive behavior of tumor cells in vitro (39). The altered control of extracellular and intracellular pH caused by overexpression of the CA9 and CA12 genes could represent a pathway in the development of at least a fraction of human tumors. Cancer cells could use different mechanisms to maintain high levels of CA9 and CA12 expression such as inactivation of the VHL tumor suppressor gene and/or amplification of the corresponding loci (29). We, therefore, suggest that in addition to being used as potential diagnostic biomarkers (33, 34, 36, 40) these enzymes should be considered as targets for novel therapeutic applications.
The molecular mechanisms by which pVHL modulates the expression of target genes is not well understood. The original hypothesis based on the discovery of elongin B binding by pVHL assumed that VHL could negatively regulate transcription elongation of target genes by inhibiting the elongin/SIII function (6). Although this expectation was confirmed in vitro (2, 3), there is no compelling evidence to date that pVHL can exert the same effect in vivo. The accumulated evidence suggests that pVHL reduces the stability of mRNA (7, 8) and/or the initiation of transcription (9). Our own experiments demonstrate distinct domains of pVHL contribute unequally to the modulation of expression of different target genes (CA9, CA12, and VEGF). pVHL may operate on different levels of gene regulation depending on the particular gene and the stage of cell growth.
Acknowledgments
We thank William Kaelin (Dana Farber Cancer Institute, Boston) for kindly providing the 786–0 RCC cell line and the pRC/CMV-Htag-VHL expressing plasmid and David Hewett-Emmett (University of Texas Health Science Center, Houston) for helpful discussions on the phylogenetics of carbonic anhydrases. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. NO1-CO-56000. It also was supported, in part, by a grant (CA19401) from the National Cancer Institute (to E.J.S.).
ABBREVIATIONS
- CA
carbonic anhydrase (CA genes are denoted by Arabic numerals and the corresponding isozymes by Roman numerals)
- EST
expressed sequence tag
- mut
mutant
- RCC
renal cell carcinoma
- RDD
RNA differential display
- VHL
von Hippel-Lindau
- wt
wild type
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Data deposition: The CA12 cDNA sequence reported in this paper has been deposited in the GenBank database (accession no. AF037335).
References
- 1. Latif F, Tory K, Gnarra J, Yao M, Duh F M, Orcutt M L, Stackhouse T, Kuzmin I, Modi W, Geil L, et al. Science. 1993;260:1317–1320. doi: 10.1126/science.8493574. [DOI] [PubMed] [Google Scholar]
- 2.Kishida T, Stackhouse T M, Chen F, Lerman M I, Zbar B. Cancer Res. 1995;55:4544–4548. [PubMed] [Google Scholar]
- 3.Duan D R, Pause A, Burgess W H, Aso T, Chen D Y, Garrett K P, Conaway R C, Conaway J W, Linehan W M, Klausner R D. Science. 1995;269:1402–1406. doi: 10.1126/science.7660122. [DOI] [PubMed] [Google Scholar]
- 4.Kibel A, Iliopoulos O, DeCaprio J A, Kaelin W G., Jr Science. 1995;269:1444–1446. doi: 10.1126/science.7660130. [DOI] [PubMed] [Google Scholar]
- 5.Duan D R, Humphrey J S, Chen D Y, Weng Y, Sukegawa J, Lee S, Gnarra J R, Linehan W M, Klausner R D. Proc Natl Acad Sci USA. 1995;92:6459–6463. doi: 10.1073/pnas.92.14.6459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krumm A, Groudine M. Science. 1995;269:1400–1401. doi: 10.1126/science.7660121. [DOI] [PubMed] [Google Scholar]
- 7.Gnarra J R, Zhou S, Merrill M J, Wagner J R, Krumm A, Papavassiliou E, Oldfield E H, Klausner R D, Linehan W M. Proc Natl Acad Sci USA. 1996;93:10589–10594. doi: 10.1073/pnas.93.20.10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iliopoulos O, Levy A P, Jiang C, Kaelin W G, Jr, Goldberg M A. Proc Natl Acad Sci USA. 1996;93:10595–10599. doi: 10.1073/pnas.93.20.10595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mukhopadhyay D, Knebelmann B, Cohen H T, Ananth S, Sukhatme V P. Mol Cell Biol. 1997;9:5629–5639. doi: 10.1128/mcb.17.9.5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knebelmann B, Ananth S, Cohen H T, Sukhatme V P. Cancer Res. 1998;58:226–231. [PubMed] [Google Scholar]
- 11.Opavsky R, Pastorekova S, Zelnik V, Gibadulinova A, Stanbridge E J, Zavada J, Kettmann R, Pastorek J. Genomics. 1996;33:480–487. doi: 10.1006/geno.1996.0223. [DOI] [PubMed] [Google Scholar]
- 12.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 13.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pack S D, Tanigami A, Ledbetter D H, Sato T, Fukuda M N. Cytogenet Cell Genet. 1997;79:123–124. doi: 10.1159/000134698. [DOI] [PubMed] [Google Scholar]
- 15.Iliopoulos O, Kibel A, Gray S, Kaelin W G., Jr Nat Med. 1995;8:822–826. doi: 10.1038/nm0895-822. [DOI] [PubMed] [Google Scholar]
- 16.Grossman H B, Wedemeyer G, Ren L. J Surg Oncol. 1985;28:237–244. doi: 10.1002/jso.2930280320. [DOI] [PubMed] [Google Scholar]
- 17.Gnarra J R, Tory K, Weng Y, Schmidt L, Wei M H, Li H, Latif F, Liu S, Chen F, Duh F-M, et al. Nat Genet. 1994;7:85–90. doi: 10.1038/ng0594-85. [DOI] [PubMed] [Google Scholar]
- 18.Williams R D, Elliott A Y, Stein N, Fraley E E. In Vitro. 1978;14:779–786. doi: 10.1007/BF02617972. [DOI] [PubMed] [Google Scholar]
- 19.Ivanova A V, Bonaduce M J, Ivanov S V, Klar A J S. Nat Genet. 1998;19:192–195. doi: 10.1038/566. [DOI] [PubMed] [Google Scholar]
- 20.Weinmaster G, Roberts V J, Lemke G. Development (Cambridge, UK) 1992;116:931–941. doi: 10.1242/dev.116.4.931. [DOI] [PubMed] [Google Scholar]
- 21.Shen M, Kawamoto T, Yan W, Nakamasu K, Tamagami M, Koyano Y, Noshiro M, Kato Y. Biochem Biophys Res Commun. 1997;236:294–298. doi: 10.1006/bbrc.1997.6960. [DOI] [PubMed] [Google Scholar]
- 22.Hewett-Emmett D, Tashian R E. Mol Phylogenet Evol. 1996;5:50–77. doi: 10.1006/mpev.1996.0006. [DOI] [PubMed] [Google Scholar]
- 23.Sly W S, Hu P Y. Annu Rev Biochem. 1995;64:375–401. doi: 10.1146/annurev.bi.64.070195.002111. [DOI] [PubMed] [Google Scholar]
- 24.Zbar B, Kishida T, Chen F, Schmidt L, Maher E R, Richards F M, Crossey P A, Webster A R, Affara N A, Ferguson-Smith M A, et al. Hum Mutat. 1996;8:348–357. doi: 10.1002/(SICI)1098-1004(1996)8:4<348::AID-HUMU8>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 25.Tureci O, Sahin U, Vollmar E, Siemer S, Gottert E, Seitz G, Parkkila A K, Shah G N, Grubb J H, Pfreundschuh M, Sly W S. Proc Natl Acad Sci USA. 1998;95:7608–7613. doi: 10.1073/pnas.95.13.7608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pastorek J, Pastorekova S, Callebaut I, Mornon J P, Zelnik V, Opavsky R, Zat’ovicova M, Liao S, Portetelle D, Stanbridge E J, et al. Oncogene. 1994;9:2877–2888. [PubMed] [Google Scholar]
- 27.Liao S Y, Brewer C, Zavada J, Pastorek S, Pastorekova S, Manetta A, Berman M L, DiSaia P J, Stanbridge E J. Am J Pathol. 1994;145:598–609. [PMC free article] [PubMed] [Google Scholar]
- 28.Saarnio J, Parkkila S, Parkkila A K, Waheed A, Casey M C, Zhou X Y, Pastorekova S, Pastorek J, Karttunen T, Haukipuro K, et al. J Histochem Cytochem. 1998;46:497–504. doi: 10.1177/002215549804600409. [DOI] [PubMed] [Google Scholar]
- 29.Mitelman F. In: Catalog of Chromosome Aberrations in Cancer. 5th Ed. Mitelman F, Johansson B, Mertens F, editors. Vol. 2. New York: Wiley; 1994. pp. 2485–2619. and 2748–2930. [Google Scholar]
- 30.Capobianco A J, Zagouras P, Blaumueller C M, Artavanis-Tsakonas S, Bishop J M. Mol Cell Biol. 1997;17:6265–6273. doi: 10.1128/mcb.17.11.6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Egan S E, St-Pierre B, Leow C C. Curr Top Microbiol Immunol. 1998;228:273–324. doi: 10.1007/978-3-642-80481-6_11. [DOI] [PubMed] [Google Scholar]
- 32.Wu Q, Pierce W M, Jr, Delamere N A. J Membr Biol. 1998;162:31–38. doi: 10.1007/s002329900339. [DOI] [PubMed] [Google Scholar]
- 33.Oosterwijk E, Ruiter D J, Hoedemaeker P J, Pauwels E K, Jonas U, Zwartendijk J, Warnaar S O. Int J Cancer. 1986;38:489–494. doi: 10.1002/ijc.2910380406. [DOI] [PubMed] [Google Scholar]
- 34.Liao S Y, Aurelio O N, Jan K, Zavada J, Stanbridge E J. Cancer Res. 1997;57:2827–2831. [PubMed] [Google Scholar]
- 35.Sahin U, Tureci O, Schmitt H, Cochlovius B, Johannes T, Schmits R, Stenner F, Luo G, Schobert I, Pfreundschuh M. Proc Natl Acad Sci USA. 1995;92:11810–11813. doi: 10.1073/pnas.92.25.11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Torczynski, R. M. & Bollon, A. P. (1996) U.S. Patent 5,589,579.
- 37.Warburg O. Uber den Stoffwechsel der Tumoren. Cambridge, U.K.: Cambridge Univ. Press; 1926. [Google Scholar]
- 38.Griffiths J R. Br J Cancer. 1991;64:425–437. doi: 10.1038/bjc.1991.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martinez-Zaguilan R, Seftor E A, Seftor R E, Chu Y W, Gillies R J, Hendrix M J. Clin Exp Metastasis. 1996;14:176–186. doi: 10.1007/BF00121214. [DOI] [PubMed] [Google Scholar]
- 40.Liao S Y, Stanbridge E J. Cancer Epidemiol Biomarkers. 1996;5:549–557. [PubMed] [Google Scholar]