Abstract
The rapid loss of muscle mass that accompanies many disease states, such as cancer or sepsis, is primarily a result of increased protein breakdown in muscle, and several observations have suggested an activation of the ubiquitin–proteasome system. Accordingly, in extracts of atrophying muscles from tumor-bearing or septic rats, rates of 125I-ubiquitin conjugation to endogenous proteins were found to be higher than in control extracts. On the other hand, in extracts of muscles from hypothyroid rats, where overall proteolysis is reduced below normal, the conjugation of 125I-ubiquitin to soluble proteins decreased by 50%, and treatment with triiodothyronine (T3) restored ubiquitination to control levels. Surprisingly, the N-end rule pathway, which selectively degrades proteins with basic or large hydrophobic N-terminal residues, was found to be responsible for most of these changes in ubiquitin conjugation. Competitive inhibitors of this pathway that specifically block the ubiquitin ligase, E3α, suppressed most of the increased ubiquitin conjugation in the muscle extracts from tumor-bearing and septic rats. These inhibitors also suppressed ubiquitination in normal extracts toward levels in hypothyroid extracts, which showed little E3α-dependent ubiquitination. Thus, the inhibitors eliminated most of the differences in ubiquitination under these different pathological conditions. Moreover, 125I-lysozyme, a model N-end rule substrate, was ubiquitinated more rapidly in extracts from tumor-bearing and septic rats, and more slowly in those from hypothyroid rats, than in controls. Thus, the rate of ubiquitin conjugation increases in atrophying muscles, and these hormone- and cytokine-dependent responses are in large part due to activation of the N-end rule pathway.
The overall balance between the rates of protein synthesis and degradation in skeletal muscle determines its size and functional capacity. If the rate of degradation rises and exceeds the rate of synthesis, muscle atrophy occurs (1–3). It is now clear from studies of animal models that increased proteolysis is the major cause of the rapid muscle wasting seen in many pathological states (1–3), including sepsis (4), cancer cachexia (5, 6), metabolic acidosis (7, 8), hyperthyroidism (1, 9–12), fasting (13–16), denervation atrophy (13, 14, 16), and diabetes (17, 18).
Like other cells, skeletal muscle contains multiple intracellular proteolytic systems, including the lysosomal pathway, Ca2+-activated proteases (calpains), and the ATP-dependent system that involves ubiquitin (Ub) and the 26S proteasome. In extracts from normal muscles, the Ub-proteasome system catalyzes the degradation of the bulk of cytoplasmic proteins and individual myofibrillar components (19). A number of observations had suggested that the increased protein breakdown in muscle atrophying because of denervation (13), fasting (13), sepsis (4), acidosis (7), and cancer cachexia (5, 6) is mainly through activation of an ATP-dependent pathway with little or no change in the lysosomal or Ca2+-dependent degradative process. Several recent studies using proteasome inhibitors confirmed that the increased muscle proteolysis in sepsis (12, 20), diabetes (18), hyperthyroidism (12), and denervation atrophy (12) involves proteasomes. In these atrophying muscles, the rise in this ATP-dependent process can account for most of the enhancement of protein degradation (1–3). Furthermore, in states where muscle protein breakdown falls, as occurs in hypothyroid animals (9, 10, 11, 21, 22) or in animals on protein-deficient diets, the reduction in proteolysis appears to be primarily a result of a suppression of this ATP-dependent process (1, 11).
Stronger evidence for an activation of the Ub-proteasome system was the finding of increased levels of Ub-protein conjugates in the muscles after denervation (16), in fasting (13, 14, 16), and in tumor-bearing animals (6). In addition, in these conditions (6, 13, 14, 16), there is an increased mRNA content for critical components of this degradative pathway, including polyUb and proteasome subunits. Similar changes also occur in the muscles of rats with experimentally induced sepsis (4), diabetes (17, 18), and metabolic acidosis (7, 8). Furthermore, interventions that prevent the increased proteolysis, such as adrenalectomy (23) or administering glucocorticoid antagonists (4), blocked the rise in Ub conjugates and Ub mRNA. These findings strongly suggest that these various disease processes activate in muscle the same coordinated set of adaptations to enhance the activity of the Ub-proteasome system and thus cause muscle atrophy. However, direct evidence for changes in overall rates of Ub conjugation has been lacking. The present studies were undertaken to obtain evidence for such global regulation of ubiquitination rates and to identify enzyme system(s) that may be activated in these catabolic states.
Proteins to be hydrolyzed by the Ub-proteasome system first are modified by covalent conjugation to multiple Ub molecules, and this modification marks them for rapid ATP-dependent degradation by the 26S proteasome complex (24–27). Studies of the degradation of individual short-lived proteins indicate that the rate-limiting step in their degradation is their conjugation to Ub. In this process, the Ub first is activated by the Ub-activating enzyme, E1, which forms a Ub-thioester. The activated Ub is then transferred to one of the many Ub-carrier proteins (E2s), and, finally, in a reaction catalyzed by a Ub-protein ligase, E3, the Ub is linked to the ɛ-amino group of a lysine residue in the polypeptide substrate, or in a Ub molecule attached previously. Cells contain a large number of E2 enzymes that appear to function together with one or a few E3s in the ubiquitination of different protein substrates (24–27).
One of the best-characterized ubiquitination systems has been termed the “N-end rule pathway,” since it catalyzes the rapid degradation of proteins with certain (destabilizing) N-terminal residues in place of the usual N-terminal methionine. In mammals, this pathway involves E3α (a homolog of yeast Ubr1), which functions in concert with E214k in the selective ubiquitination of proteins that bear either basic or bulky hydrophobic residues (28–31). The recognition of such proteins is due to the presence of two distinct binding sites on E3α, one specific for basic and one for bulky hydrophobic N-terminal residues (29, 32, 33). When proteins with such N termini were produced in vivo by Varshavsky and coworkers, they were degraded very rapidly in all cells tested (28–31). However, because such proteins normally are not found in cells, the physiological importance of this ubiquitination system has remained unclear (26), especially since yeast mutants lacking this E3α (Ubr1) or E214k (Ubc2) appear to have only minor phenotypic defects (34).
However, using specific inhibitors of E3α, we recently made the surprising finding (35) that the bulk of endogenous proteins degraded in extracts of rabbit muscles are primarily ubiquitinated by the N-end rule pathway. For example, several low-molecular-mass inhibitors of E3α (dipeptides or methyl esters that contain basic or hydrophobic N termini) suppressed by at least 50% the ATP-dependent degradation of endogenous soluble muscle proteins, and did so by generally inhibiting their conjugation to Ub. These reagents are competitive inhibitors of E3α; by contrast, the isomeric dipeptides and amino acid methyl esters bearing stabilizing N-terminal residues had little or no effect on muscle proteolysis. Moreover, another specific inhibitor of the N-end rule pathway, a dominant-negative mutant form of E214k [C88S, which can form an ester with Ub, but cannot transfer the Ub moiety to a substrate (36)], inhibited similarly the conjugation of Ub to proteins in muscle extracts. Furthermore, although the N-end rule pathway thus appears to play an important role in the Ub-dependent proteolysis in skeletal muscle, these inhibitors did not reduce Ub conjugation generally in HeLa cell lysate, even though these cells contain the N-end rule pathway (35).
The primary goals of the present experiments were to determine (i) whether the rates of Ub conjugation to muscle proteins generally rise in atrophying muscles when overall proteolysis increases and (ii) whether the N-end rule pathway is not only important in normal muscles but may also contribute to this increased ubiquitination. Several experimental models of muscle wasting were chosen that closely mimic major human disease states. In particular, muscle atrophy is a prominent feature of patients with systemic infections (e.g., AIDS or sepsis) and in many types of cancer cachexia. We have studied the dramatic muscle wasting in rats with sepsis, induced by cecal ligation and puncture (4), and cancer cachexia induced by transplantation of Yoshida Ascites Hepatoma (6) for different time periods. In both, there is profound muscle wasting and a large increase in proteolysis that appears to be signaled by interleukin-1 and tumor necrosis factor (1–3). In addition, to demonstrate hormonal regulation of ubiquitination rates in cells, we studied this process in muscles of animals where overall protein breakdown falls because of deficiency of thyroid hormones and rises after treatment with these hormones.
MATERIALS AND METHODS
Reagents.
Various dipeptides and methyl esters were obtained from Sigma.
Animals.
Control, hypophysectomized, and thyroidectomized CD male rats (ranging between 70 and 90 when killed) were purchased from Charles River Breeding Laboratories. To ensure the clearance of circulating hormones and proteins induced by hypophyseal or thyroid hormones, rats were used for experiments only after 15–30 days after hypophysectomy or thyroidectomy. Leg muscles were dissected after the rats were killed by CO2 asphyxiation and cervical dislocation. Muscles from each rat then were frozen separately in liquid nitrogen until use. All animal studies reported in this paper were carried out in accordance with institutional guidelines on animal care.
T3 Treatment.
To restore the function of thyroid hormones, the thyroidectomized rats received a single daily subcutaneous injection of T3 (20 μg/100 g body weight) for 7 days, and hypophysectomized animals received 100 μg/100 g body weight for 4 days. The control rats were given a similar volume of the saline solution. The body temperature and the weight of each animals were observed regularly before and during the thyroid hormone treatment. Animals from each experimental group being compared were sacrificed on the same day. Seven animals in each group generally were used for each experiment.
Induction of Sepsis.
A standard method for inducing sepsis in rats, involving cecal ligation and puncture, was used (4). The animals were sacrificed after 16 hr, when there is extensive loss of muscle mass and a large increase in the ATP-dependent proteolytic process (4).
Implantation of Yoshida Hepatoma.
For tumor-bearing studies, rats were implanted with 100 ml of ascites fluid containing Yoshida Hepatoma cells from a single animal donor, as described previously (6). The control rats were implanted with an equal volume of saline buffer. On days 3 and 5, after tumor implantation, the animals were sacrificed.
Muscle Extract Preparation.
To prepare extract, frozen muscles from three to six rats from each experimental group were combined, and 20% (wt/vol) muscle extract was prepared in the presence of various protease inhibitors, as described earlier for rabbit (19). To prevent degradation of the Ub conjugates by the 26S proteasome complex, and to remove isopeptidases associated with these particles, proteasomes were removed by centrifuging the extracts for 6–8 hr at 100,000 × g (19). The proteasome-depleted extracts were fractionated further by chromatography on DEAE-cellulose to remove endogenous ubiquitin (19). The resulting Fraction-II was dialyzed and stored in small aliquots at −70°C in a buffer containing 50 mM Tris, pH 7.6/1 mM DTT/5 mM MgCl2/20% glycerol.
Ub-Conjugation Assays.
Ub conjugation to soluble extract proteins was followed by incubating extracts (2 mg/ml) with 125I-Ub (0.15 mg/ml) at 37°C in a buffer containing 20 mM Tris, pH 7.4/1 mM DTT/5 mM MgCl2/2 mM adenosine 5′-[γ-thio]triphosphate (ATPγS). When conjugation between 125I-lysozyme and unlabeled Ub was studied, the reaction mixture contained 0.15 mg/ml labeled lysozyme and 0.25 mg/ml unlabeled ubiquitin. At indicated times, the samples were withdrawn, and 12% SDS/PAGE was performed. The gels were then dried and autoradiographed.
RESULTS
Ubiquitin Conjugation to Proteins Increases in Atrophying Muscles.
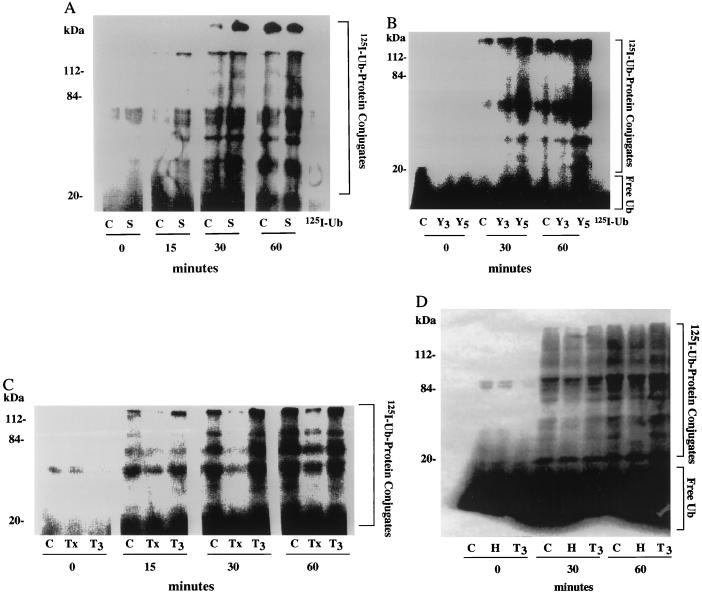
Many studies of the metabolic response to sepsis have utilized cecal puncture in rats, after which up to 25% of muscle mass is lost by 16 hr (4). When incubated, the pale muscles from such rats show a large increase in the ATP-dependent proteolytic process that can be blocked by inhibitors of proteasome function (12). To follow the overall rate of protein ubiquitination, soluble extracts were prepared from leg muscles of such septic rats and sham-operated controls, and rates of 125I-Ub conjugation to endogenous proteins were compared. The extracts were prepared as described earlier (19) by very gentle homogenization in the presence of a variety of protease inhibitors, to minimize proteolytic cleavage of muscle proteins during extract preparation (including inhibitors of lysosomal, Ca2+-dependent proteases, and the mast cell chymase). The extracts then were centrifuged at 100,000 × g for 6–8 hr to remove most of the proteasomes, to prevent the degradation of the Ub-conjugated proteins, and to remove proteasome-associated isopeptidases. Since the muscle’s Ub content can increase in atrophying muscles (2–4, 6, 14, 23) and therefore might affect the specific activity of added 125I-Ub, endogenous Ub was removed from extracts by DEAE-cellulose chromatography (19). Equal amounts of the extracts from control and septic rats (in mg total protein) then were incubated with 125I-Ub. The amount of Ub conjugated to proteins then was measured in the presence of ATPγS by the appearance of new higher-molecular-mass 125I-labeled bands (>8 kDa) detected by SDS/PAGE. ATPγS was used instead of ATP, because this nucleotide supports Ub conjugation, but not proteolysis by the 26S proteasome (37). In both extracts, Ub-conjugate formation increased at an approximately linear rate for up to 60 min. However, the overall rate of 125I-Ub conjugation to endogenous proteins in muscle extracts from the septic rats was consistently about twice that from control rats (Fig. 1A).
Figure 1.
125I-Ub conjugation to proteins rises in extracts from rat muscles when proteolysis increases and falls when proteolysis decreases. (A) Septic (S) and sham-operated control (C) rats. Muscles were removed from rats 16 hr after cecal puncture to induce sepsis, as described previously (4). (B) Rats bearing Yoshida Hepatoma for 3 (Y3) or 5 (Y5) days and pair-fed control (C) rats. Tumor implantation was performed as described previously (6). (C) Thyroidectomized (Tx), T3-treated thyroidectomized (T3), and control (C) rats. (D) Hypophysectomized rats (H), T3-treated hypophysectomized (T3), and control (C) rats. Soluble extracts were incubated with 125I-Ub for the indicated time. 125I-Ub conjugation to endogenous proteins were analyzed by SDS/PAGE as described in Materials and Methods. Free Ub runs at the dye front and may not be visible on all gels. The data in this composite figure were obtained in a single experiment by using extracts prepared from at least six animals in each group. All experiments were repeated at least three times with similar results.
Muscle wasting associated with cancer cachexia is a host response to the tumor and not simply a consequence of reduced food intake (2, 3, 6). In rats implanted with the Yoshida Ascites Hepatoma for 3–5 days, there is a rapid loss of muscle proteins mainly because of a 60–95% increase in the overall rate of protein degradation in the pale muscles. In such muscles, the levels of Ub-protein conjugates (measured immunologically) and mRNA for Ub and the proteasome subunits increase (6). Therefore, we measured the overall rate of 125I-Ub conjugation to endogenous proteins in muscle extracts (Fraction-II) from rats implanted with hepatoma for 3 or 5 days and pair-fed control rats. By 3 days after tumor implantation, the rate of 125I-Ub conjugation to endogenous proteins was 80% higher than that in controls, and by 5 days, when muscle proteolysis is increased further (6), ubiquitination of cell proteins was 2- to 3-fold higher than in control extracts (Fig. 1B). Thus, in both the septic and tumor-bearing animals, there was a general acceleration of Ub conjugation to muscle proteins that correlated with, and presumably accounts for, the increased protein degradation.
Ubiquitin Conjugation Falls When Muscle Proteolysis Is Reduced.
On the other hand, overall protein breakdown in muscle decreases below normal levels in certain nutritional and endocrine states, such as in hypothyroidism, induced by thyroidectomy or hypophysectomy (9–11). To test whether Ub conjugation falls under such conditions, we compared rates of conjugation of 125I-Ub to endogenous proteins in muscle extracts (Fraction-II) from thyroidectomized, hypophysectomized, and control rats (Fig. 1 C and D). In all these extracts, 125I-Ub was incorporated into high-molecular-mass 125I-Ub-protein conjugates. However, the rates of ubiquitination of muscle proteins in extracts from thyroidectomized and hypophysectomized rats were consistently about 50% lower than in those from control rats.
The decrease in muscle proteolysis observed in thyroidectomized or hypophysectomized rats can be reversed by treatment of the animals with thyroid hormones, triiodothyronine (T3), or thyroxine (T4) (1, 9–11). In fact, high levels of these hormones cause excessive protein breakdown (9) and muscle wasting, as often occurs in hyperthyroid patients (1, 11). To test whether thyroid hormones increase the rate of Ub conjugation in muscle, thyroidectomized rats were treated with T3 for 7 days, and the hypophysectomized rats were treated with T3 for 4 days before killing. These hormone treatments of the thyroidectomized animals not only restored the normal growth rate and body temperature (from 35.7°C to 36.7°C, P < 0.01), but also stimulated Ub conjugation to muscle proteins back toward levels seen in controls, which had received an equal volume of physiological saline (Fig. 1 C and D). A similar general increase in ubiquitination was seen in the muscles of hypophysectomized animals after T3 treatment. This effect is a result of the thyroid hormones and cannot be an indirect consequence of release of other pituitary or adrenal hormones. These findings further indicate that various conditions that enhance muscle proteolysis are associated with an increased capacity to ubiquitinate proteins generally, while conditions that suppress muscle protein breakdown reduce overall Ub conjugation.
Involvement of the N-End Rule Pathway.
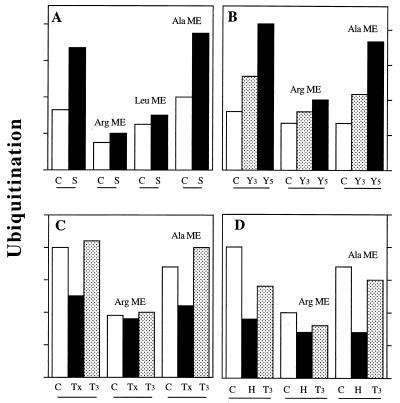
Our recent studies (35) have shown that a surprisingly large fraction, 40–70%, of the Ub-mediated degradation of soluble proteins in rabbit muscle extracts involves key components of the N-end rule pathway, E3α and E214k. We therefore investigated whether the increased ubiquitination in the atrophying muscles also involves changes in the N-end rule pathway. In the extracts (Fraction-II) from both control and septic rats, addition of the inhibitor of E3α, arginine methyl ester, reduced 125I-Ub conjugation to soluble proteins. However, ubiquitination was inhibited to a much greater extent in extracts from septic animals than in those from control animals. As a result, most of differences in rates of Ub conjugation between the two groups were eliminated by arginine methyl ester. Similarly, leucine methyl ester, which binds to a distinct site of E3α (32, 33) and also reduces proteolysis in normal rabbit muscles (35), suppressed the high rate of Ub conjugation in the extracts from septic animals much more than it affected ubiquitination in controls. Consequently, this E3α inhibitor also reduced the differences in the ubiquitination rates between these preparations. In contrast, alanine methyl ester, which does not inhibit the N-end rule pathway, had no effect on the ubiquitination of proteins in either extract (Fig. 2A).
Figure 2.
Effect of inhibitors of the E3α on increased and decreased rates of 125I-Ub conjugation to proteins in rat muscle extracts from various pathological states. (A) Septic (S) and control (C) rats. (B) Rats bearing the Yoshida Hepatoma for 3 days (Y3) or 5 days (Y5) after tumor implantation and control (C) rats. (C) Thyroidectomized (Tx), T3-treated thyroidectomized (T3), and control (C) rats. (D) Hypophysectomized (H) and T3-treated hypophysectomized rats (T3) and control (C) rats. 125I-Ub conjugates were formed and resolved by SDS/PAGE as described in Materials and Methods. Methyl esters (2 mM) were added in the presence of bestatin (20 μg/ml), and the reactions were incubated for 1 hr at 37°C. Values shown are relative rates of ubiquitination of endogenous proteins in the absence or presence of various methyl esters. Arg ME, arginine methyl ester; Leu ME, leucine methyl ester; and Ala ME, alanine methyl ester.
Similar experiments were carried out with extracts from the Hepatoma-bearing and control rats. The addition of arginine methyl ester (Fig. 2B) or the dipeptide Lys-Ala (data not shown) suppressed most of the increased ubiquitination in muscle extracts of tumor-bearing rats, but had much less of an inhibitory effect in control extracts. As a result, these agents eliminated most of the differences in Ub conjugation between control and atrophying muscle extracts. In contrast, alanine methyl ester or Ala-Lys (not shown) had no effect on protein ubiquitination in either group. Thus, the ubiquitin-protein ligase, E3α, appears to play a critical role in the increased ubiquitination of soluble proteins in septic and tumor-bearing rats.
Because these findings indicated that the enhanced Ub conjugation in the atrophying muscles is mediated in large part by the N-end rule pathway, we tested whether the reduction in Ub conjugation that accompanies the fall in muscle proteolysis in hypothyroid animals may involve a suppression of this pathway. The addition of arginine methyl ester to the extracts of control rat muscles caused a 40–50% inhibition of Ub conjugation, but only a 10–20% inhibition of the low amount of ubiquitination in the extracts from thyroidectomized or hypophysectomized rats. As a consequence, with this inhibitor present, the rates of Ub conjugation were similar in these different extracts. In contrast, alanine methyl ester had no effect on rates of ubiquitination (Fig. 2 C and D). Thus, the fall in Ub conjugation in hypothyroid animals seems largely to be a result of a suppression of the N-end rule pathway. Furthermore, T3 treatment of the thyroidectomized or hypophysectomized rats not only restored rates of Ub conjugation (Fig. 1 C and D), but also enhanced the sensitivity of this process to the inhibitors of E3α (Fig. 2 C and D). Thus, the increase in the Ub conjugation in the T3-treated animals (as was found in the septic or tumor-bearing rats) is largely a result of activation of the N-end rule pathway, which appears to be tightly regulated by hormones and monokines.
Physiological Regulation of the Enzymes Comprising the N-End Rule Pathway.
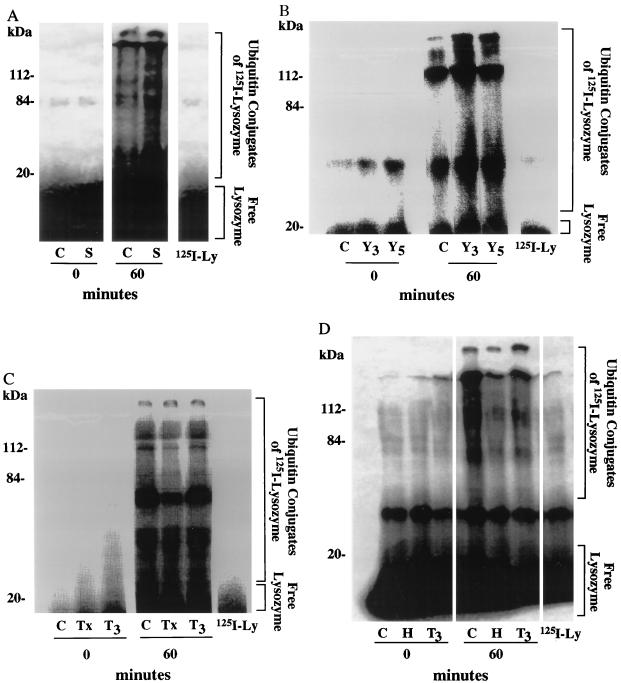
These differences in the ubiquitination of intracellular proteins may arise either through activation or inhibition of the enzymes comprising the N-end rule pathway or, alternatively, through some modification of the proteins in the muscle that makes them better (or worse) substrates of E3α. To test whether the activity of these ubiquitination enzymes is enhanced during sepsis and in tumor-bearing rats, we measured in these extracts ubiquitination of 125I-lysozyme, a model substrate of the N-end rule pathway. In muscles, as had been found in reticulocytes, Ub conjugation to 125I-lysozyme, which contains a lysine at its N terminus, was markedly inhibited by Arg methyl ester or Lys-Ala (data not shown). In extracts from septic animals, this exogenous substrate was ubiquitinated about 2-fold faster than in controls and 1.5- to 2-fold faster in rats bearing the Yoshida Hepatoma for 3 to 5 days (Fig. 3A and B). Furthermore, Ub conjugation to 125I-lysozyme was reduced to about 50% of control rates in rat muscles after thyroidectomy or hypophysectomy, and the ubiquitination of 125I-lysozyme in these extracts was restored back to levels seen in normal controls when these hypothyroid rats were treated with T3 (Fig. 3 C and D).
Figure 3.
Ub conjugation to 125I-lysozyme in extracts from muscles where proteolysis is increased or decreased. (A) Septic (S) and control (C) rats. (B) Rats bearing the Yoshida Hepatoma for 3 days (Y3) or 5 days (Y5) after tumor implantation and control (C) rats. (C) Thyroidectomized (Tx), T3-treated thyroidectomized (T3), and control (C) rats. (D) Hypophysectomized (H), T3-treated hypophysectomized (T3), and control (C) rats. 125I-lysozyme-Ub conjugation was followed by incubating 125I-lysozyme (0.15 mg/ml) with extracts (2 mg/ml) at 37°C in the presence of Ub 0.25 mg/ml. All other conditions are described in Materials and Methods. Free lysozyme runs at the dye front and may not be visible on all gels. The data in this composite figure were obtained in a single experiment by using extracts prepared from at least six animals in each group. All experiments were repeated at least three times with similar results. 125I-lysozyme, 125I-lysozyme at 60 min at 37°C.
Thus, the enzymes responsible for ubiquitination of lysozyme, E3α and E214k, are regulated in cancer cachexia and by thyroid hormones in a manner that can account for most of the changes in overall protein ubiquitination in the muscles.
DISCUSSION
Regulation of Ubiquitination in Muscle.
The present findings represent direct evidence that overall rates of protein ubiquitination in a cell can increase or decrease in response to endocrine signals or cytokines. A number of recent studies of muscle wasting in animals in diverse catabolic states (including systemic infection, cancer cachexia, and T3 treatment) all had suggested that the rapid loss of muscle mass is due to enhanced protein breakdown, primarily by the Ub-proteasome system (2–6), but the supporting evidence was of an indirect nature. The present demonstration of increased Ub conjugation in atrophying rat muscles was made at times after cecal puncture, tumor implantation, or T3 treatment, when ATP-dependent proteolysis in the leg muscles is markedly increased and when muscle proteolysis shows enhanced sensitivity to inhibitors of the proteasome (6, 12). Moreover, this finding of enhanced rates of ubiquitination of proteins in the atrophying muscles can account for the increased content of Ub-protein conjugates in muscles upon denervation, fasting, and cancer cachexia (4, 6, 16). These observations also indicate that the rate-limiting step regulating overall proteolysis in muscles is Ub conjugation, which also is the critical event determining the rate of degradation of many specific cell proteins. This general increase in ubiquitination leading to a general loss of muscle mass in these states may be a model for understanding the atrophy that occurs in other cell types during development or disease. For example, as reticulocytes develop into erythrocytes, there is a similar programmed increase in ATP-dependent proteolysis and a net loss of most cell proteins (38), which presumably also may involve a general acceleration of ubiquitination.
In skeletal muscles, the overall rate of protein breakdown is regulated by a variety of physiological stimuli, all of which appear to influence the overall rate of Ub conjugation. The acceleration of proteolysis and presumably the increase in Ub conjugation in these models of sepsis and cancer cachexia appear to be signaled by increases in tumor necrosis factor and interleukin-1 (but other cytokines may also be involved) (1–3). In normal animals, the overall rates of muscle proteolysis are controlled by insulin, glucocorticoids, and the pituitary–thyroid axis (1–3). Thyroid hormones not only stimulate Ub conjugation and ATP-dependent proteolysis, but they also promote intralysosomal proteolysis and increase the content of lysosomal enzymes in muscle (1–3). The suppression of normal ubiquitination found here in hypothyroid animals can account for the fall in protein breakdown seen in animals on protein-deficient diets (1–3) and in long-term fasting (1–3), where thyroid status and muscle-protein breakdown fall. It is attractive to conclude that the suppression of protein ubiquitination in such animals may be an adaptive mechanism to preserve essential cell proteins.
Involvement of N-End Rule Pathway.
The most surprising aspect of the present findings was the discovery that dipeptide and methyl-ester inhibitors of the ubiquitin ligase, E3α, differentially suppress most of the Ub conjugation in the atrophying rat muscle in these different catabolic states. Using these inhibitors of E3α and a dominant-negative inhibitor of E214K, we had found previously that the N-end rule pathway was responsible for at least half the ubiquitination to soluble proteins in extracts of normal muscle (35). For example, the dipeptide and amino acid ester blockers of E3α were found to inhibit most of the breakdown of endogenous proteins in normal rabbit muscle extracts, but did not do so in HeLa cells (35). These results were quite unexpected, since E214k and E3α have been assumed to function primarily in the selective elimination of abnormal proteins with aberrant N-terminal residues, although in yeast, the breakdown of certain normal regulatory proteins (e.g., Gα and Cup9) somehow also involves the E3α homolog, Ubrl (39, 40).
The present findings were based on the use of dipeptides and amino acid esters, which are competitive inhibitors of the two substrate-binding sites of E3α. Although these inhibitors have to be used in the mM range, a variety of observations have confirmed that their large inhibitory effects on Ub conjugation in muscle extracts were in fact a result of a selective inhibition of E3α, and not a nonspecific effect. For example, the isomers of these dipeptide inhibitors (Ala-Lys or Ala-Phe) and other amino acid methyl esters (e.g., alanine methyl ester) at similar concentrations had little (<15%) or no effect either on 125I-Ub conjugation or ATP-Ub-dependent degradation of endogenous proteins in rabbit muscle extracts (35). In addition, these agents specifically inhibited Ub conjugation and ATP-dependent degradation of model substrates (lysozyme and α-lactalbumin), which are known to require E214k and E3α, but they did not affect Ub-ATP-dependent or breakdown of exogenously added 125I-troponin, 125I-myosin, or 125I-actin (35). Furthermore, these inhibitors of E3α do not nonspecifically affect protein ubiquitination; for example, they do not affect p53 ubiquitination in other cell extracts (41), and they have no effect on ubiquitination by E3L (42), another major E3 in muscles. Also, in HeLa cell lysates, these inhibitors did not reduce overall Ub conjugation to endogenous proteins (35), as they did in muscle. Thus, although not potent inhibitors, the dipeptides and methyl esters with basic or hydrophobic N termini appear to block specifically the N-end rule pathway.
A variety of observations argue against the possibility that this predominant role of the N-end rule pathway in muscle might be an in vitro artifact generated in the course of extract preparation. For example, one possible artifact might occur by endoproteolytic cleavages of the muscle proteins to generate destabilizing N-terminal residues that result in E3α-dependent ubiquitination. However, various procedures to minimize such proteolytic events (e.g., the use of a variety of protease inhibitors, preparing the homogenates at 4°C) did not reduce the extent of Ub conjugation in the muscle extracts (35). Furthermore, the present finding of increased rates of E3α-dependent ubiquitination in atrophying muscles from animals with sepsis, cancer cachexia, and after T3 treatment, as well as the decreased ubiquitination in hypothyroid states (see Results), cannot be explained by such an in vitro artifact. In all of these cases, the extracts were prepared similarly, and rates of Ub conjugation correlated with the changes in protein breakdown previously measured in vivo. Furthermore, the rates of ubiquitination of an exogenous substrate, 125I-lysozyme, increased and decreased in a similar way as did Ub conjugation to endogenous muscle proteins. Since lysozyme is a model substrate of the N-end rule pathway, the activity of this ubiquitination system must be regulated precisely under these various conditions. Although the finding of a major role for the N-end rule ubiquitination pathway in normal muscle and its activation (or suppression) in disease states were unexpected, E214k is one of the predominant E2s in this tissue, and increases in mRNA for E214k have been reported in rat muscles upon fasting (15), cancer cachexia (5, 6), glucocorticoid treatment (43), disuse (44), and sepsis (4).
Our finding that the muscle’s capacity to ubiquitinate a model N-end rule substrate rises or falls under different conditions is strong evidence that the activity or the amounts of E3α (and perhaps E214k) are regulated precisely by hormones and cytokines. The present findings with the inhibitors of the substrate-binding sites of E3α suggest that the N-end rule pathway is responsible for most of the increased Ub conjugation to soluble proteins in the muscles of septic, tumor-bearing, and hyperthyroid animals. Moreover, in related unpublished studies, we have found that in other pathological states characterized by muscle wasting, there is a similar increase in the activity of the N-end rule pathway, including rats with renal failure, diabetes, and disuse atrophy (S. H. Lecker, V.S., W. E. Mitch, and A.L.G., unpublished data). Thus, the overall increase in ubiquitination in muscle through the activation of the N-end rule pathway seems to represent a general mechanism for enhancing muscle proteolysis in disease states, although additional adaptations may also be occurring under these different conditions, e.g., other E3s or E2s may also be activated, or additional mechanisms may be promoting myofibrillar disassembly.
The most fundamental question raised by these findings is why such a large fraction of intracellular proteins are substrates of E3α in normal muscles and why these proteins comprise an even larger fraction in atrophying muscles. Clearly, the suppression of the N-end rule pathway in the hypothyroid rats indicates that cells can function with lower rates of E214k-E3α-dependent process than are seen normally. One intriguing possibility is that in muscles, in addition to the regulation of the ubiquitinating enzymes, E214k or E3α, there may be a mechanism to activate protein degradation generally by altering the N termini of cellular proteins, for example, through endoproteolytic cleavages or some other N-terminal modification. A tRNA-dependent enzymatic reaction that conjugates arginine to the N termini of proteins bearing N-terminal aspartate or glutamate is known to direct substrates to E3α-E214k (28, 31, 45–48), and we have obtained evidence suggesting that a fraction of the proteins in normal rabbit muscle extracts undergoes such a tRNA-dependent modification leading to their degradation by the N-end rule pathway (35). Perhaps in catabolic states, an endoproteolytic clipping or a novel chemical modification of the N termini of the bulk of muscle proteins also could be a critical regulated step that triggers the general acceleration of muscle proteolysis. However, it is also possible that E3α, while preferentially acting on substrates with basic or bulky hydrophobic N termini, also may mediate ubiquitination of other cell proteins generally in a less selective, but still dipeptide-sensitive, manner. Although these findings demonstrate a crucial role for the N-end rule pathway in pathological states in which Ub-dependent proteolysis is either enhanced or suppressed, further studies clearly will be necessary to clarify specifically which endogenous proteins are ubiquitinated in these extracts, how these proteins are recognized by E3α, and how hormones and cytokines alter the activities of this ubiquitination pathway.
Acknowledgments
We are grateful to Mrs. Aurora Scott for assistance in preparing this manuscript. This work was supported by grants to A.L.G. from the National Space Biomedical Research Institute, the National Institute of General Medical Sciences, and the Muscular Dystrophy Association.
ABBREVIATION
- Ub
ubiquitin
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1. Tawa N E, Goldberg A L. In: Myology. Engel A G, Franzini Armstrong C, editors. New York: McGraw–Hill; 1994. pp. 683–707. [Google Scholar]
- 2.Attaix D, Taillandier D, Temparis S, Larbaud D, Aurousseau E, Combaret L, Voisin L. Nutr Dev. 1994;34:583–597. doi: 10.1051/rnd:19940605. [DOI] [PubMed] [Google Scholar]
- 3.Mitch W E, Goldberg A L. N Engl J Med. 1996;335:1897–1905. doi: 10.1056/NEJM199612193352507. [DOI] [PubMed] [Google Scholar]
- 4.Tiao G, Fagan J, Roegner V, Lieberman M, Wang J J, Fischer J E, Hasselgren P O. J Clin Invest. 1996;97:339–348. doi: 10.1172/JCI118421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Temparis S, Asensi M, Taillandier D, Aurousseau E, Larbaud D, Obled A, Bechet D, Ferrara M, Estrela J M, Attaix D. Can Res. 1994;54:5568–5573. [PubMed] [Google Scholar]
- 6.Baracos V E, Devivo C, Hoyle D H, Goldberg A L. Am J Physiol. 1995;268:E996–E1006. doi: 10.1152/ajpendo.1995.268.5.E996. [DOI] [PubMed] [Google Scholar]
- 7.Mitch W E, Medina R, Grieber S, May R C, England B K, Price S R, Bailey J L, Goldberg A L. J Clin Invest. 1994;93:2127–2133. doi: 10.1172/JCI117208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bailey J L, Wang X, England B K, Price S R, Ding X, Mitch W E. J Clin Invest. 1996;97:1447–1453. doi: 10.1172/JCI118566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldberg A L, Griffin G E. J Physiol. 1977;270:51P–52P. [PubMed] [Google Scholar]
- 10.Brown J G, Bates P C, Holliday M A, Millward D. J Biochem J. 1981;194:771–782. doi: 10.1042/bj1940771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kettelhut I C, Wing S S, Goldberg A L. Diabetes Metab Rev. 1988;4:751–772. doi: 10.1002/dmr.5610040805. [DOI] [PubMed] [Google Scholar]
- 12.Tawa N E, Jr, Odessey R, Goldberg A L. J Clin Invest. 1997;100:197–203. doi: 10.1172/JCI119513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Medina R, Wing S S, Haas A, Goldberg A L. Biomed Biochim Acta. 1991;50:347–356. [PubMed] [Google Scholar]
- 14.Medina R, Wing S S, Goldberg A L. Biochem J. 1995;307:631–637. doi: 10.1042/bj3070631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wing S S, Banville D. Am J Physiol. 1994;267:E39–E48. doi: 10.1152/ajpendo.1994.267.1.E39. [DOI] [PubMed] [Google Scholar]
- 16.Wing S S, Haas A, Goldberg A L. Biochem J. 1995;307:639–645. doi: 10.1042/bj3070639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pepato M T, Migliorini R H, Goldberg A L, Kettelhut I C. Am J Physiol. 1996;271:E340–E347. doi: 10.1152/ajpendo.1996.271.2.E340. [DOI] [PubMed] [Google Scholar]
- 18.Price S R, Bailey J L, Wang X, Jurkovitz C, England B K, Ding X, Phillips L S, Mitch W E. J Clin Invest. 1996;98:1703–1708. doi: 10.1172/JCI118968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Solomon V, Goldberg A L. J Biol Chem. 1996;271:26690–26697. doi: 10.1074/jbc.271.43.26690. [DOI] [PubMed] [Google Scholar]
- 20.Hobler S C, Tiao G, Fischer J E, Monaco J, Hasselgren P O. Am J Physiol. 1998;274:R30–R37. doi: 10.1152/ajpregu.1998.274.1.R30. [DOI] [PubMed] [Google Scholar]
- 21.Tawa N E, Jr, Goldberg A L. Surg Forum. 1991;42:25–28. [Google Scholar]
- 22.Tawa N E, Kettelhut I C, Goldberg A L. Am J Physiol. 1992;263:E326–E334. doi: 10.1152/ajpendo.1992.263.2.E326. [DOI] [PubMed] [Google Scholar]
- 23.Wing S S, Goldberg A L. Am J Physiol. 1993;264:E668–E676. doi: 10.1152/ajpendo.1993.264.4.E668. [DOI] [PubMed] [Google Scholar]
- 24.Hershko A, Ciechanover A. Annu Rev Biochem. 1992;61:761–807. doi: 10.1146/annurev.bi.61.070192.003553. [DOI] [PubMed] [Google Scholar]
- 25.Jentsch S. Annu Rev Gene. 1992;26:179–207. doi: 10.1146/annurev.ge.26.120192.001143. [DOI] [PubMed] [Google Scholar]
- 26.Ciechanover A. Cell. 1994;79:13–21. doi: 10.1016/0092-8674(94)90396-4. [DOI] [PubMed] [Google Scholar]
- 27.Coux O, Tanaka K, Goldberg A L. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 28.Bachmair A, Finley D, Varshavsky A. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 29.Gonda D K, Bachmair A, Wunning I, Tobias J W, Lane W S, Varshavsky A. J Biol Chem. 1989;264:16700–16712. [PubMed] [Google Scholar]
- 30.Baker R T, Varshavsky A. Proc Natl Acad Sci USA. 1991;88:1090–1094. doi: 10.1073/pnas.88.4.1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Varshavsky A. Proc Natl Acad Sci USA. 1996;93:12142–12149. doi: 10.1073/pnas.93.22.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reiss Y, Kaim D, Hershko A. J Biol Chem. 1988;263:2693–2698. [PubMed] [Google Scholar]
- 33.Reiss Y, Hershko A. J Biol Chem. 1990;265:3685–3690. [PubMed] [Google Scholar]
- 34.Bartel B, Wunning I, Varshavsky A. EMBO. 1990;9:3179–3189. doi: 10.1002/j.1460-2075.1990.tb07516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Solomon, V., Lecker, S. H. & Goldberg, A. L. (1998) J. Biol. Chem., in press. [DOI] [PubMed]
- 36.Sung P, Prakash S, Prakash L. J Mol Biol. 1991;221:745–749. doi: 10.1016/0022-2836(91)80169-u. [DOI] [PubMed] [Google Scholar]
- 37.Johnston N L, Cohen R E. Biochemistry. 1991;30:7514–7522. doi: 10.1021/bi00244a021. [DOI] [PubMed] [Google Scholar]
- 38.Boches F S, Goldberg A L. Science. 1982;215:978–980. doi: 10.1126/science.7156977. [DOI] [PubMed] [Google Scholar]
- 39.Madura K, Varshavsky A. Science. 1994;265:1454–1458. doi: 10.1126/science.8073290. [DOI] [PubMed] [Google Scholar]
- 40.Byrd C, Turner G C, Varshavsky A. EMBO. 1998;17:269–277. doi: 10.1093/emboj/17.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scheffner M, Takahashi T, Huibregtse J M, Minna J D, Howley P M. J Virol. 1992;66:5100–5105. doi: 10.1128/jvi.66.8.5100-5105.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gonen H, Stancovski I, Shkedy D, Hadari T, Bercovich B, Bengal E, Mesilati S, Abu-Hatoum O, Schwartz A L, Ciechanover A. J Biol Chem. 1996;271:302–310. doi: 10.1074/jbc.271.1.302. [DOI] [PubMed] [Google Scholar]
- 43.Auclair D, Garrel D R, Chaouki Zerouala A, Ferland L H. Am J Physiol. 1997;272:C1007–C1016. doi: 10.1152/ajpcell.1997.272.3.C1007. [DOI] [PubMed] [Google Scholar]
- 44.Taillandier D, Aurousseau E, Meynial-Denis D, Bechet D, Ferrara M, Cottin P, Ducastaing A, Bigard X, Guezennec C Y, Schmid H P. Biochem J. 1996;316:65–72. doi: 10.1042/bj3160065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ferber S, Ciechanover A. Nature (London) 1987;326:808–811. doi: 10.1038/326808a0. [DOI] [PubMed] [Google Scholar]
- 46.Balzi E, Choder M, Chen W, Varshavsky A, Goffeau A. J Biol Chem. 1990;265:7464–7471. [PubMed] [Google Scholar]
- 47.Ciechanover A, Ferber S, Ganosh D, Elias S, Hershko A, Arfin S. J Biol Chem. 1988;263:11155–11167. [PubMed] [Google Scholar]
- 48.Elias S, Ciechanover A. J Biol Chem. 1990;265:15511–15517. [PubMed] [Google Scholar]