Abstract
While conducting a search for cell cycle-regulated genes in human mammary carcinoma cells, we identified HSIX1, a recently discovered member of a new homeobox gene subfamily. HSIX1 expression was absent at the onset of and increased toward the end of S phase. Since its expression pattern is suggestive of a role after S phase, we investigated the effect of HSIX1 in the G2 cell cycle checkpoint. Overexpression of HSIX1 in MCF7 cells abrogated the G2 cell cycle checkpoint in response to x-ray irradiation. HSIX1 expression was absent or very low in normal mammary tissue, but was high in 44% of primary breast cancers and 90% of metastatic lesions. In addition, HSIX1 was expressed in a variety of cancer cell lines, suggesting an important function in multiple tumor types. These data support the role for homeobox genes in tumorigenesis/tumor progression, possibly through a cell cycle function.
Homeodomain-containing proteins act as transcription factors that regulate the coordinated expression of genes involved in both development and differentiation. They were identified initially in Drosophila, where they were found to be important in the control of segment identity (1). The genes encoding homeodomain proteins (homeobox genes) contain a common 183-nt sequence encoding a 61-aa domain that is responsible for DNA binding (2). They are postulated to act as a network of transcriptional regulators effecting cell–cell communication during normal development, alterations of which may contribute to the neoplastic phenotype. Recent studies have demonstrated homeobox gene involvement in leukemias (3) and in solid tumors such as breast, kidney, lung, and colon (4).
In this study, we cloned the HSIX1 homeobox gene from late S phase mammary carcinoma cells and demonstrated that overexpression of HSIX1 leads to an abrogation of the DNA damage-induced G2 cell cycle checkpoint. In addition, overexpression of HSIX1 occurs in a large percentage of mammary carcinomas and correlates strongly with metastatic breast disease. Preliminary tests on several cancer cell lines suggest that HSIX1 may be overexpressed in multiple types of tumors. This study links HSIX1 to the cell cycle as well as to tumor progression and lends further credence to the hypothesis that “master regulators” involved in development may contribute to tumorigenicity.
MATERIALS AND METHODS
Cell Culture and Synchronization.
21PT, NT, MT1, and MT2 breast cancer cells were derived from a patient with an infiltrating and intraductal carcinoma (5) and were obtained from the laboratory of Ruth Sager (Dana–Farber Cancer Institute). The cells were cultured at 37°C in 6.5% CO2 in α-MEM plus 10% fetal bovine serum/2 mM l-glutamine/1 mM sodium pyruvate/0.1 mM nonessential amino acids/10 mM Hepes/1 μg/ml insulin/12.5 ng/ml epidermal growth factor/1 μg/ml hydrocortisone, and antibiotics. The generated MCF7 transfectants were cultured in RPMI 1640 medium plus 10% FBS/600 μg/ml G418, and antibiotics. Normal luminal and myoepithelial cells were sorted by immunomagnetic methods from primary cultures derived from mammoplasty specimens (6). Cell synchrony with mimosine was performed as described (7); however, 150 μM mimosine was used rather than 400 μM. To monitor progression of cells through S phase, pulsed 3H-thymidine incorporation (at hourly intervals) was performed as described (8).
Differential Display (DD).
Differential display was performed with a two-step PCR and the LHA series of primers as described (9). The anchored and the arbitrary primers that led to detection of HSIX1 were LHT11C (TGC CGA AGC T11C) and LHA6 (TGC CGA AGC TTG CAG CGA). Band isolation and direct sequencing of the DD band were performed as described (9).
cDNA Construction.
Reverse transcription–PCR (RT-PCR) was performed to amplify the HSIX1 cDNA from human skeletal muscle. RT reactions were performed with 0.2 μg RNA template/25 μM dNTPs/1 mM DTT/5 μM oligo(dT)12–18, and 1× reverse transcriptase buffer (50 mM Tris⋅HCl, pH 8.3/75 mM KCl/3 mM MgCl2). The reaction conditions were as follows: 65°C, 5 min; 37°C, 60 min (5 min into this cycle, 200 units Superscript II was added to each reaction); 95°C, 5 min. After the RT reaction, PCR was performed with 2–3 μl of RT reaction, 250 μM dNTPs, 2.5 μM each primer, 1× PCR buffer (Perkin–Elmer), and a 100:1 Taq/Pfu mixture. Primers used were designed to encompass the start and stop codon of the cDNA and to incorporate BamHI and XbaI restriction sites. The PCR profile was as follows: (94°C, 45 sec; 57°C, 45 sec; 72°C) × 25, followed by an extension for 5 min at 72°C. The PCR product was then digested with BamHI and XbaI and subcloned into the BamHI-XbaI site of the pcDNA3.1(+) vector from Invitrogen. The resulting plasmid is referred to as SIXFL.
Generation of MCF7 Stable Transfectants.
MCF7 cells were seeded in 60-mM dishes at 5 × 105 cells per dish and transfected with SIXFL or with pcDNA3.1(CAT) by using Superfect (Qiagen). Transfections were performed according to the manufacturer’s protocol. Twenty-four hours after transfections the cells were passaged 1:15 in appropriate medium containing 600 μg/ml G418. Approximately 2 weeks later individual clones were isolated by using cloning cylinders.
X-Ray Irradiation and Subsequent Fluorescence-Activated Cell Sorter (FACS) Analysis.
MCF7 transfectants were seeded at 8 × 105 cells per 60-mM dish. Approximately 48 h later the cells were treated with x-rays (5 or 8 Gy) at a dose rate of 1.25 Gy/min using a Phillips 250-kVp x-ray machine. Sham-treated controls as well as irradiated cells were labeled with propidium iodide according to Vindelov et al. (10) at various time points after irradiation. Experiments were performed singly or in duplicate and repeated several times. FACS analysis was performed on the Becton Dickinson FACScan using cellquest (Becton Dickinson) and modfit (Verity Software, Topsham, Maine) to obtain cell cycle profiles.
RNA Isolations and Northern Blot Analysis.
All RNA extracted from cell lines (both for differential display and Northern blot analysis) and from lung tumor samples was isolated with TRIzol reagent (GIBCO/BRL) according to the manufacturer’s protocol. RNA was isolated from breast tumor specimens by the guanidinium thiocyanate/CsCl method as described (11). Multiple-tissue Northern and dot blots (both normal and cancer) were obtained from CLONTECH. All Northern blot analysis was performed as described (11).
Cloning of HSIX1 from 21PT Cells and Breast Biopsy Samples for Sequencing.
Primers were designed to the 5′ end (5′-ATG TCG ATG CTG CCG TCG TTT-3′) and 3′ end (5′-CAC TTA GGA CCC CAA GTC CAC-3′) of the HSIX1 cDNA. In some cases, BamHI and XbaI restriction sites were incorporated to assist subcloning. RT reactions were performed as above. PCR conditions were as follows: [94°C, 45 sec; 69°C or 57°C (for primers without and with restriction sites, respectively), 45 sec; 72°C, 45 sec] × 25, followed by an extension at 72°C, 5 min. The PCR products were subcloned by using the TA cloning system (Invitrogen) or by ligation into the BamHI and XbaI site of pcDNA 3.1 (Invitrogen). Sequencing was performed on multiple clones as it is known that PCR may introduce point mutations.
RESULTS AND DISCUSSION
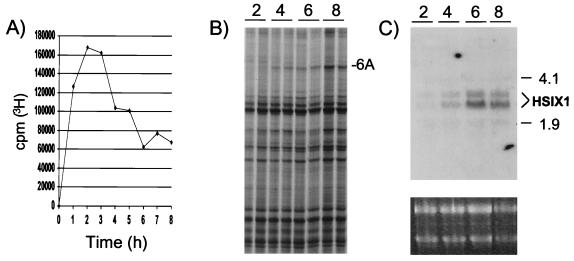
To identify genes differentially expressed in S phase of the 21PT mammary carcinoma cell line, cells synchronized with mimosine (7) were released from late G1/S phase arrest, S phase progression was monitored by 3H-thymidine incorporation (Fig. 1A), and RNA was isolated from duplicate samples for differential display analysis. Fig. 1B demonstrates increased expression of a cDNA band labeled as 6A. Direct sequencing of 6A revealed its identity as HSIX1, a homeobox gene that was cloned recently from human adult skeletal muscle (12) and whose mouse counterpart has been implicated in the development of limb tendons (13). A Northern blot probed with the HSIX1 cDNA (cloned from 21PT cells by RT-PCR) confirmed its differential expression in S phase (Fig. 1C). Levels of HSIX1 were very low in the first half of S phase and increased as cells completed S phase. This expression pattern suggests that HSIX1 may play a role at or near the end of the 21PT cell cycle.
Figure 1.
Differential expression of HSIX1 throughout S phase of 21PT cells. (A) Pulsed 3H-thymidine incorporation after release from mimosine arrest shows progression of cells through S phase. (B) Section of the differential display gel demonstrating the differential expression of 6A (subsequently identified as HSIX1) in S phase. (C) Northern blot analysis confirming the differential expression of HSIX1 throughout S phase of 21PT cells. RNA was isolated from cells after release from mimosine arrest, and Northern blot analysis was performed with the HSIX1 cDNA probe. (Lower) Ethidium bromide (EtBr) staining as a loading control. The numbers 2, 4, 6, and 8 represent time in hours after release from mimosine block.
Another clue suggesting a function of HSIX1 in cell cycle control was obtained by comparison with the Drosophila sine oculis (so) gene. The mouse Six1 gene was first cloned by virtue of its homology to so (13). It is 62% similar to the Drosophila gene and 87% similar if sequences C-terminal to the homeodomain are excluded (13). So plays a role in the development of the fly visual system. Interestingly, Drosophila eye development involves coordinate regulation of cell cycle progression and so has been suggested to play a role in the synchronization of the cell cycle because its expression precedes a burst of cell divisions (14). Additionally, complete loss of function alleles of so are embryonic lethals (14), suggesting that the gene’s expression is important for more than just eye development. These results, in conjunction with the cell cycle-regulated expression of HSIX1 in 21PT cells, suggest that HSIX1 may play a role in regulating the onset of mitosis.
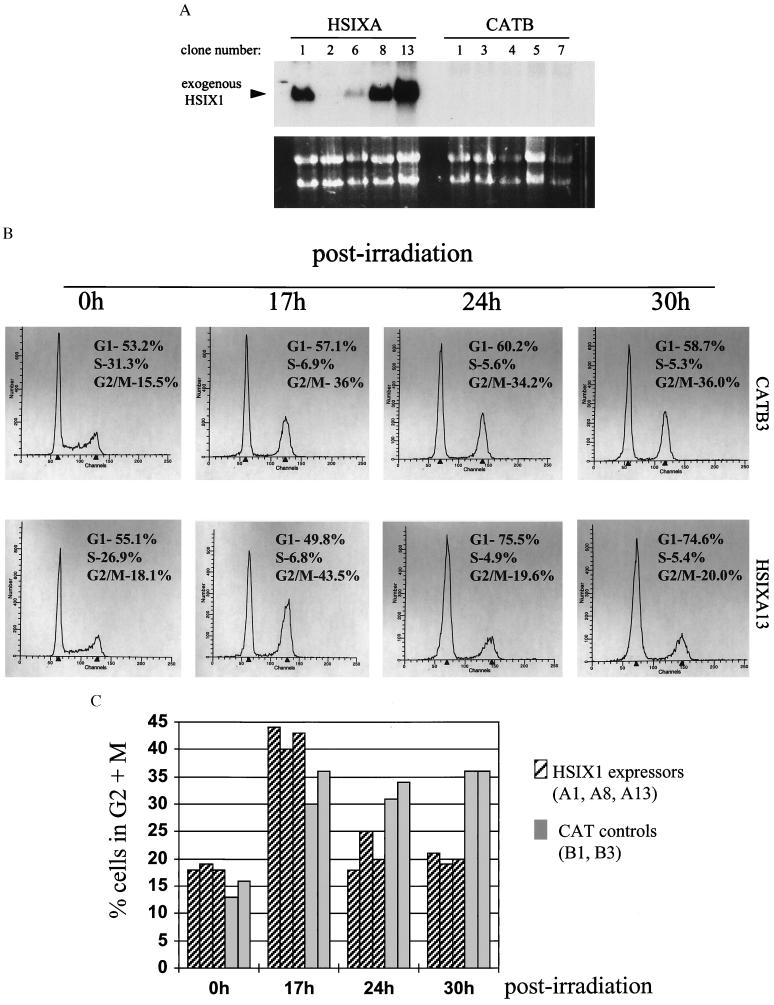
To determine whether HSIX1 plays a role in regulating the cell cycle, the MCF7 mammary carcinoma cell line was transfected with SIXFL, a construct that allows for constitutive expression of the full-length wild-type HSIX1 cDNA, or with the parent vector expressing the chloramphenicol acetyl transferase gene (CAT) as a control. MCF7 cells were chosen because they are mammary carcinoma cells with an endogenous HSIX1 level far lower than that in 21PT cells (data not shown). Stable transfectants were selected by using cloning cylinders and examined for HSIX1 expression via Northern blot analysis (Fig. 2A). For all subsequent analysis, three stable clones expressing HSIX1 (HSIXA1, A8, and A13) and two control transfectants (CATB1 and CATB3) were examined.
Figure 2.
Overexpression of HSIX1 in MCF7 cells abrogates the G2 cell cycle checkpoint. (A) Northern blot analysis of HSIX1 transfectants and controls. Clones labeled HSIXA were stably transfected with the SIXFL plasmid; those labeled CATB were stably transfected with the parent vector containing the CAT gene [pcDNA3.1(CAT)]. Note that HSIXA2, while G418-resistant, does not express HSIX1. (Lower) EtBr staining as a loading control. (B) Representative FACS analysis on propidium iodide-stained cells before and after x-ray irradiation at a dose of 8 Gy. The panels display a time course after irradiation of one HSIX1 transfectant (HSIXA13) and one control transfectant (CATB3) cell line. The experiment was performed several times at two different doses of irradiation (5 and 8 Gy) with the same outcome. (C) Summary of the percentage of cells in G2 at various time points before and after irradiation in the transfectants and controls. The data graphed are from one experiment performed at 8 Gy and are representative of several experiments performed at 5 and 8 Gy. Note that cells expressing HSIX1 progress through the G2 arrest at a more rapid rate than transfected controls.
Exponentially growing cells overexpressing HSIX1 showed cell cycle profiles similar to transfected controls (Fig. 2B, Oh). In contrast, when the cells were irradiated at a dose of 8 Gy to examine the DNA damage-induced G2 cell cycle checkpoint, a marked difference was observed in the G2 + M population in HSIX1 transfectants vs. the CAT controls (Fig. 2 B and C). In the representative experiment, both HSIX1 expressers and CAT controls were arrested in G2 17 h after irradiation, as was expected. However, by 24 h postirradiation, all cell lines expressing HSIX1 had progressed beyond the G2 arrest, whereas the nonexpressers remained arrested in G2. The CAT control transfectants were blocked in G2 as long as 30 h postirradiation, whereas the HSIX1 transfectants had exited the G2 arrest significantly earlier. Although absolute percentages varied from experiment to experiment, the passage of HSIX1 expressers through G2 after x-ray irradiation was always more rapid than that of the controls. Note that MCF7 cells have an intact G1/S arrest in response to irradiation and that cells passing through G2 subsequently will arrest at the G1/S boundary.
In addition to the CAT controls, a cell line transfected with SIXFL (HSIXA2) that did not express HSIX1 (possibly because of silencing of the gene upon insertion into the chromosomal DNA) was tested in the x-ray irradiation assay. This cell line behaved as the CAT controls, confirming that HSIX1 expression was necessary for abrogation of the G2 cell cycle checkpoint and that the expression of CAT did not affect the checkpoint in any way (data not shown). Furthermore, the growth rates of the HSIX1 transfectants and controls in the absence of irradiation were not appreciably different (data not shown), indicating that the rapid transit of HSIX1 transfectants through the G2 arrest after DNA damage was not merely a consequence of faster growth. These data demonstrate that overexpression of HSIX1 leads to an abrogation of the DNA damage-induced G2 cell cycle checkpoint.
Interestingly, another homeobox gene, HOX11, recently was found to disrupt the G2 cell cycle checkpoint by interacting with protein phosphatase 2A (PP2A) (15). HOX11 has been implicated in cancer (15), as it was isolated from a chromosomal breakpoint in human T cell leukemia (16–18). In addition, transgenic mice expressing HOX11 in the thymus demonstrated cell cycle alterations and progression to malignancy (19). Since we originally cloned HSIX1 from a mammary carcinoma cell line (21PT), and since overexpression of this gene leads to altered cell cycle control similar to that seen with HOX11, we reasoned that HSIX1 may be differentially expressed in cancer.
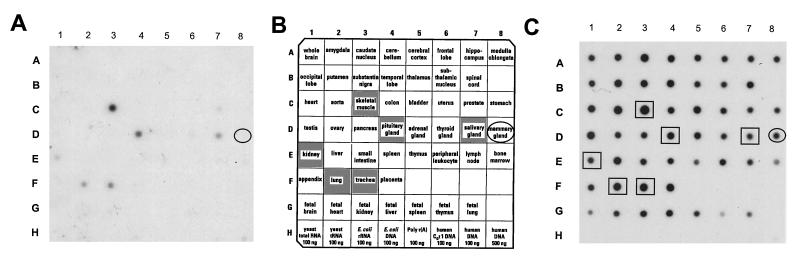
For comparison with the 21PT mammary carcinoma cells, a Human RNA Master Blot (CLONTECH) was probed to determine the pattern of HSIX1 expression in normal human adult tissue (Fig. 3). Normal mammary tissue, pooled from 20 women ages 24–40 who died of trauma, does not express detectable levels of HSIX1, suggesting that its expression in cultured lines of mammary carcinoma cells is aberrant. Expression was confirmed in normal adult skeletal muscle and was also observed in pituitary gland, salivary gland, and trachea, with low levels in the lung and kidney.
Figure 3.
HSIX1 is not expressed in normal breast, but is expressed in normal skeletal muscle, salivary gland, lung, trachea, and kidney. A Human RNA Master Blot from CLONTECH was probed with the HSIX1 cDNA. (A) HSIX1 expression pattern in normal tissues. (B) Identities of all the samples included on the blot. (C) The blot was stripped and reprobed with ubiquitin to ensure equal loading.
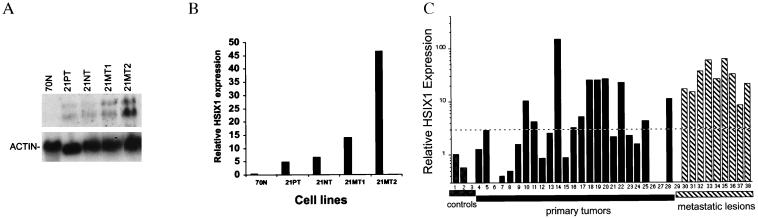
21PT cells, the source of the original HSIX1 clone, were derived from the primary tumor of a patient who had an infiltrating and intraductal mammary carcinoma (5). Other cell lines derived from the same patient include 21NT, also derived from the primary tumor, and 21MT-1 and 21MT-2, which were established from a metastatic pleural effusion. HSIX1 expression was detected in all the tumor lines examined, but was not detected in a normal breast cell line, 70N (20) (Fig. 4 A and B). When normalized to actin, levels of HSIX1 expression in 21MT-1 and 21MT-2 cells were 2- to 10-fold higher than levels in 21PT and 21NT cells (Fig. 4B). This result suggests that HSIX1 expression increases with metastasis. It should be noted that the 70N normal mammary cell line is mainly myoepithelial-like (21), thus requiring additional controls in subsequent analyses since breast tumors generally are considered more like luminal epithelial cells.
Figure 4.
Expression of HSIX1 correlates with metastatic disease. (A) Northern blot analysis of normal mammary epithelial cells (70N) and the 21T series of mammary carcinoma cells. 21PT and 21NT cells were derived from a primary tumor, whereas 21MT1 and 21MT2 were established from a pleural effusion from the same patient (7). (B) Quantitative representation of HSIX1 expression in the cell lines after normalization to actin. (C) Quantitative representation of a Northern blot analysis of 3 control tissues (normal adjacent breast, normal luminal cells, and normal myoepithelial cells; lanes 1, 2, and 3, respectively), 25 primary breast tumor biopsy samples (lanes 4–28), and 10 metastatic lesions (lanes 29–38). The blot was stripped and reprobed with 36B4 (16) for normalization, and relative HSIX1 expression was plotted. A 3-fold increase over normal adjacent breast was considered positive for HSIX1 and is marked by a dashed line. Forty-four percent of primary and 90% of metastatic lesions express greater than a 3-fold increase in HSIX1 mRNA over the normal adjacent breast control. It is not known whether the patients that expressed HSIX1 in the primary tumor went on to develop metastatic disease.
To determine whether HSIX1 expression is increased in primary and metastatic breast cancer, 35 human breast tumor samples were obtained and examined for HSIX1 expression by Northern blot analysis. Normalization to 36B4 was performed on these samples, as it has been shown to be a good control for breast cancer samples (22). The results were quantitated and plotted as relative HSIX1 expression (Fig. 4C). While normal adjacent breast, normal breast luminal epithelial cells, and normal breast myoepithelial cells demonstrated almost no HSIX1 expression (lanes 1–3, respectively), 44% of the primary tumors (lanes 4–27) and 90% of the metastatic lesions (lanes 28–37) expressed greater than a 3-fold increase in HSIX1 mRNA over normal adjacent breast. Some of the women with HSIX1 expression in their primary tumors already had metastasis at the time of biopsy; however, whether all the women who expressed HSIX1 in the primary tumor went on to develop metastatic disease is not known.
As the metastatic lesions came from a secondary site, it was necessary to consider the expression levels of the tissue at this site to confirm that the expression observed is from the lesion and not from contaminating adjacent tissue. The 10 metastatic lesions utilized in our analysis came from either the lymph nodes (six samples), bone/soft tissue (two samples), the lung (one sample), or the pleural wall (one sample). While no information regarding the expression of HSIX1 in normal bone/soft tissue or pleura was available, the Human RNA Master blot allowed us to examine its expression in normal lymph nodes and lung. Five of the six lymph node metastases expressed HSIX1; however, HSIX1 expression was not observed in normal lymph nodes. This suggests that the high expression levels in lymph node lesions came from the metastatic tumor itself. Normal lung does express the gene at low levels, but densitometric scanning and subsequent normalization demonstrated that expression in the metastatic lesion from the lung was equal to that in normal adult skeletal muscle (data not shown), which expresses four times more HSIX1 than normal lung (Fig. 3). This suggests that HSIX1 expression in the lung metastases cannot be explained by normal tissue contaminating the sampled metastasis.
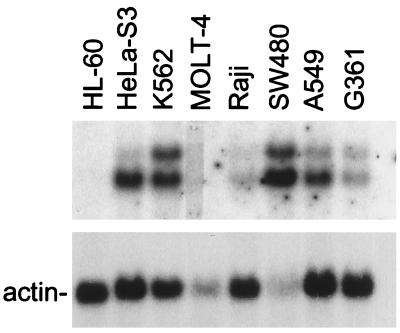
To investigate whether HSIX1 expression is observed in cancers other than breast, several additional cancer cell lines were examined. A multiple-tissue Northern blot containing 2 μg poly(A)+ RNA isolated from eight different human cancer cell lines (CLONTECH) was probed with the HSIX1 cDNA. Fig. 5 demonstrates expression of HSIX1 in cancer cell lines including HeLa S3, chronic myelogenous leukemia K562, colorectal adenocarcinoma SW480, and lung carcinoma A549. Low levels of expression also were observed in melanoma G361 and Burkitt’s lymphoma Raji. No expression was observed in promyelocytic leukemia HL-60 or lymphoblastic leukemia MOLT-4, although inadequate RNA loading may be responsible for the absence of HSIX1 expression in the MOLT-4 lane. In addition, preliminary data indicate that HSIX1 is overexpressed in non-small-cell lung carcinoma as contrasted to normal adjacent lung tissue (data not shown). Therefore, these studies suggest that HSIX1 is overexpressed in several types of cancer in addition to breast.
Figure 5.
HSIX1 is expressed in other cancers. Northern blot analysis of multiple cancer cell lines. A human cancer cell line Northern blot containing 2 μg of poly(A)+ RNA from each cell line was probed with HSIX1 cDNA. HL-60, promyelocytic leukemia; HeLa S3, cervical cancer; K-562, chronic myelogenous leukemia; MOLT-4, lymphoblastic leukemia; Raji, Burkitt’s lymphoma; SW480, colorectal carcinoma; A549, lung carcinoma; G361, melanoma. Actin mRNA is shown as a loading control.
We have demonstrated overexpression of the HSIX1 homeobox gene in approximately half of primary and nine-tenths of metastatic breast cancer lesions. In addition, smaller-scale analysis of several different tumor cell lines suggests that HSIX1 may be expressed in a wide variety of tumors in addition to breast. Recently, numerous other homeobox genes, including members of the Hox and Pax families, have been identified as oncogenic transcription factors (3, 23). Homeobox genes often are translocated to produce a chimeric protein with a new function, particularly in leukemias (4). However, others retain their wild-type function and are overexpressed (3, 4, 23). Gross genetic alterations do not exist in HSIX1, since the molecular weight of HSIX1 mRNA is unchanged in 21PT cells and breast tumors as compared with normal skeletal muscle (data not shown). However, a translocation occurring upstream of the transcription start site might have led to aberrant expression, or a point mutation or small deletions/insertions may exist in the gene. We have determined that wild-type HSIX1 mRNA is overexpressed in at least some of these cancers by cloning and sequencing the HSIX1 cDNA by RT-PCR from 21PT cells and from one of the primary breast tumor samples (data not shown). The sequences obtained represent wild-type HSIX1 cDNA when compared with the GenBank database. This result fits a model summarized by Sager (24), who suggests that tumorigenesis is not only the result of genetic mutations, but also of overexpression of wild-type genes. Whether HSIX1 utilizes its wild-type promoter and whether its expression is cell cycle-regulated in malignant tumors remain to be determined.
In conclusion, HSIX1 is a homeobox gene that is differentially expressed in the cell cycle and whose overexpression leads to an abrogation of the DNA damage-induced G2 cell cycle checkpoint. Diseases associated with G2 checkpoint control, such as Ataxia telangiectasia (25, 26), Li Fraumeni (26), Bloom’s Syndrome (27), and Fanconi Anemia (28), all demonstrate cancer susceptibility. This lends credence to the hypothesis that overexpression of HSIX1 may be involved in tumorigenesis/tumor progression. Indeed, HSIX1 is overexpressed in a large proportion of breast cancers, and preliminary data suggest that it may be overexpressed in a variety of cancers. The significance of these findings and of future research in this area is that HSIX1 provides a potential diagnostic/prognostic marker for metastatic disease as well as a potential target for therapeutic intervention. In addition, it also suggests that important links remain to be discovered between the processes of development, cell cycle, and cancer.
Acknowledgments
We thank Drs. K. J. Martin and Krishnarao Appasani as well as other members of the Pardee laboratory for critical reading of the manuscript, helpful discussions, and support. The research was funded by an R0-1 Grant CA61253 from the National Institutes of Health. H.L.F. was supported by the Dana–Farber Cancer Institute Tumor Biology Grant T32-CA09361-16,17.
ABBREVIATIONS
- RT-PCR
reverse transcription–PCR
- FACS
fluorescence-activated cell sorter
- CAT
chloramphenicol acetyltransferase
References
- 1. Lewis E B. Nature (London) 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- 2.McGinnis W, Krumlauff R. Cell. 1992;68:283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- 3.Lawrence H J, Sauvageau G, Humphries R K, Largman C. Stem Cells. 1996;14:281–291. doi: 10.1002/stem.140281. [DOI] [PubMed] [Google Scholar]
- 4.Cillo C. Invasion Metastasis. 1994;14:38–49. [PubMed] [Google Scholar]
- 5.Band V, Zajchowski D, Swisshelm K, Trask D, Kulesa V, Cohen C, Connolly J, Sager R. Cancer Res. 1990;50:7351–7357. [PubMed] [Google Scholar]
- 6.Clarke C, Titley J, Davies S, O’Hare M J. Epithelial Cell Biol. 1994;3:38–46. [PubMed] [Google Scholar]
- 7.Alpan R S, Pardee A B. Cell Growth Differ. 1996;7:893–901. [PubMed] [Google Scholar]
- 8.Keyomarsi K, Sandoval L, Band V, Pardee A B. Cancer Res. 1991;51:3602–3609. [PubMed] [Google Scholar]
- 9.Martin K J, Kwan C P, Sager R. In: Methods in Molecular Biology–Differential Display Methods and Protocols. Pardee A B, Liang P, editors. Vol. 85. Totowa, NJ: Humana; 1996. pp. 77–85. [DOI] [PubMed] [Google Scholar]
- 10.Vindelov L L, Christensen I J, Nissen N I. Cytometry. 1983;3:323–327. doi: 10.1002/cyto.990030503. [DOI] [PubMed] [Google Scholar]
- 11.Maniatis T, Fritsch E F, Sambrook J. Molecular Cloning. 2nd Ed. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 12.Boucher C A, Carey N, Edwards Y H, Siciliano M J, Johnson K J. Genomics. 1996;33:140–142. doi: 10.1006/geno.1996.0172. [DOI] [PubMed] [Google Scholar]
- 13.Oliver G, Wehr R, Jenkins N A, Copeland B G, Cheyette B N R, Hartenstein V, Zipursky S L, Gruss P. Development. 1995;121:693–705. doi: 10.1242/dev.121.3.693. [DOI] [PubMed] [Google Scholar]
- 14.Cheyette B N R, Green P J, Martin K, Garren H, Hartenstein V, Zipursky S L. Neuron. 1994;12:977–996. doi: 10.1016/0896-6273(94)90308-5. [DOI] [PubMed] [Google Scholar]
- 15.Kawabe T, Muslin A J, Korsmeyer S. Nature (London) 1997;385:454–458. doi: 10.1038/385454a0. [DOI] [PubMed] [Google Scholar]
- 16.Hatano M, Roberts C W, Minden M, Crist W M, Korsemeyer S J. Science. 1991;253:79–82. doi: 10.1126/science.1676542. [DOI] [PubMed] [Google Scholar]
- 17.Kennedy M A, Gonzales-Sarmiento R, Kees U R, Lampert F, Dear N, Boehm T, Rabbitts T H. Proc Natl Acad Sci USA. 1991;88:8900–8904. doi: 10.1073/pnas.88.20.8900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dube I D, Kamel-Reid S, Yuan C C, Lu M, Wu X, Corpus G, Raimondi S C, Crist W M, Carroll A J, Minowada J, et al. Blood. 1991;78:2996–3002. [PubMed] [Google Scholar]
- 19.Hatano M, McGuire E A, Roberts C, Kawabe T, Korsmeyer S J. Curr Opin Oncol. 1992;4:24–26. [Google Scholar]
- 20.Band V, Sager R. Proc Natl Acad Sci USA. 1989;86:1249–1253. doi: 10.1073/pnas.86.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trask D K, Band V, Zajchowski D A, Yaswen P, Suh T, Sager R. Proc Natl Acad Sci USA. 1990;87:2319–2323. doi: 10.1073/pnas.87.6.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laborda J. Nucleic Acids Res. 1991;19:3998. doi: 10.1093/nar/19.14.3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stuart E T, Gruss P. Human Mol Genet. 1995;4:1717–1720. doi: 10.1093/hmg/4.suppl_1.1717. [DOI] [PubMed] [Google Scholar]
- 24.Sager R. Proc Natl Acad Sci USA. 1997;94:952–955. doi: 10.1073/pnas.94.3.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scott, D., Spreadborough, A. R. & Roberts, S. A. (1994) Int. J. Radiat. Biol. 66, Suppl., 157s–163s. [PubMed]
- 26.Paules R S, Levendakou E N, Wilson S J, Innes C L, Rhodes N, Tlsty T D, Galloway D A, Donehower L A, Tainsky M A, Kaufmann W K. Cancer Res. 1995;55:1763–1773. [PubMed] [Google Scholar]
- 27.Davey S, Han C S, Ramer S A, Klassen J C, Jacobsen A, Eisenberger A, Hopkins K M, Lieberman H B, Freyer G A. Mol Cell Biol. 1998;18:2721–2728. doi: 10.1128/mcb.18.5.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D’Andrea A D, Kupfer G M. Blood. 1996;88:1019–1025. [PubMed] [Google Scholar]