Abstract
Unmethylated CpG dinucleotides in particular base contexts (CpG-S motifs) are relatively common in bacterial DNA but are rare in vertebrate DNA. B cells and monocytes have the ability to detect such CpG-S motifs that trigger innate immune defenses with production of Th1-like cytokines. Despite comparable levels of unmethylated CpG dinucleotides, DNA from serotype 12 adenovirus is immune-stimulatory, but serotype 2 is nonstimulatory and can even inhibit activation by bacterial DNA. In type 12 genomes, the distribution of CpG-flanking bases is similar to that predicted by chance. However, in type 2 adenoviral DNA the immune stimulatory CpG-S motifs are outnumbered by a 15- to 30-fold excess of CpG dinucleotides in clusters of direct repeats or with a C on the 5′ side or a G on the 3′ side. Synthetic oligodeoxynucleotides containing these putative neutralizing (CpG-N) motifs block immune activation by CpG-S motifs in vitro and in vivo. Eliminating 52 of the 134 CpG-N motifs present in a DNA vaccine markedly enhanced its Th1-like function in vivo, which was increased further by the addition of CpG-S motifs. Thus, depending on the CpG motif, prokaryotic DNA can be either immune-stimulatory or neutralizing. These results have important implications for understanding microbial pathogenesis and molecular evolution and for the clinical development of DNA vaccines and gene therapy vectors.
The genomic DNAs of bacteria and vertebrates differ in the frequency and methylation of CpG dinucleotides, which are relatively common in bacterial DNA (approximately 1/16 bases), but are underrepresented (“CpG suppression”; 1/50–1/60 bases) and methylated in vertebrate DNA (1). Bacterial DNA or synthetic oligodeoxynucleotides (ODN) containing unmethylated CpG dinucleotides in particular base contexts induce B cell proliferation, interleukin (IL)-6 and Ig secretion, and apoptosis resistance (2–12, 16). These immunostimulatory CpGs typically are preceded on the 5′ side by an ApA, GpA, or GpT dinucleotide and followed on the 3′ side by two pyrimidines, especially TpT (CpG-S motif) (11). Monocytes are directly activated by CpG-S motifs to secrete the Th1-like cytokine IL-12 and type I interferons (IFN), and natural killer (NK) cells respond with increased lytic activity and IFN-γ secretion, enhancing protective immune responses (4, 5, 7, 8, 13, 14). Methylated bacterial DNA or ODN in which the cytosines of CpG have been converted to 5-methyl-cytosine (the form present in vertebrate DNA) fails to induce immune activation (2). The stimulatory effects of CpG-S motifs appear to be mediated at least in part by the activation of NF-κB (8, 12, 15, 55). Thus, this simple structural difference in the frequency of CpG-S motifs between vertebrate and prokaryotic genomic DNAs appears to function as a “danger signal” to trigger innate immune defenses against infection and initiate a specific immune response (reviewed in ref. 17). Indeed, ODN containing CpG-S motifs can be mixed with antigens to promote strong Th1-like immune responses (18–23). Recent studies have suggested that effective DNA vaccines require CpG-S motifs within the plasmid backbone (24, 25).
Nearly all DNA viruses and retroviruses appear to have evolved to avoid this defense mechanism through reducing their genomic content of CpG dinucleotides by 50–94% from that expected based on random base usage (26). CpG suppression is absent from bacteriophage, indicating that it is not an inevitable result of having a small genome (26). Statistical analysis indicates that the CpG suppression in lentiviruses is an evolutionary adaptation to replication in a eukaryotic host (27). Adenoviruses, however, are an exception to this rule as they have the expected level of genomic CpG dinucleotides (26). Different groups of adenovirae can have quite different clinical characteristics. Serotype 2 and 5 adenoviruses (subgenus C) are endemic causes of upper respiratory infections and are notable for their ability to establish persistent infections in lymphocytes (28). These adenoviral serotypes are modified frequently by deletion of early genes for use in gene therapy applications, where a major clinical problem has been the frequent inflammatory immune responses to the viral particles. Serotype 12 adenovirus (subgenus A) does not establish latency, but can be oncogenic. Since viruses have evolved a broad range of sophisticated strategies for avoiding or co-opting host immune defenses, we investigated the immune effects of the DNA from these adenoviral serotypes.
METHODS
ODN and DNA.
Phosphodiester ODN were purchased from Operon Technologies (Alameda, CA), and nuclease-resistant phosphorothioate ODN were purchased from Oligos Etc. (Wilsonville, OR) or Hybridon Specialty Products (Milford, MA). All ODN had undetectable endotoxin levels (less than 1 ng/mg) by Limulus assay (BioWhittaker). Escherichia coli (strain B) DNA was purchased from Sigma, purified by repeated extraction with phenol/chloroform/isoamyl alcohol (25:24:1) and/or Triton X-114 extraction and ethanol precipitation, and made single-stranded by boiling for 10 min followed by cooling on ice for 5 min. Digestion with restriction enzymes did not reduce the stimulatory effects of the DNA (not shown). Highly purified type 2, 5, and 12 adenoviral DNA was prepared from viral preparations by using standard techniques and processed in the same manner as the E. coli DNA. Plasmids for DNA vaccination were purified by using two rounds of passage over Endo-free columns (Qiagen).
Cell Cultures and ELISA Assays for Cytokines.
ELISA assays were performed by using standard techniques and commercially available reagents as described previously (3, 11, 29). Standard deviations of the triplicate wells were <10%.
Construction of Optimized DNA Vectors.
The starting material was pUK21-A2, an expression vector containing the immediate early promoter of human cytomegalovirus (CMV IE), the bovine growth hormone (BGH) polyadenylation signal, and the kanamycin resistance gene (T.W. and H.D., unpublished data). To avoid disrupting the plasmid origin of replication, mutagenesis designed to eliminate CpG-N motifs was restricted to the kanamycin resistance gene and nonessential DNA sequences after the gene. A total of 22 point mutations were introduced to alter 15 CpG-N motifs (a “motif” refers to a hexamer containing one or more CpG dinucleotides) containing 19 CpG dinucleotides, 12 of which were eliminated and 7 of which were transformed into CpG-S motifs. Site-directed mutagenesis was performed by overlap-extension PCR as described by Ge and coworkers (30). The 1.3-kb AlwNI-EcoO109 I fragment of pUK21-A2, which contained all 22 nt to be mutated, was used as the template for PCR. The 1.3-kb fragment was regenerated by four rounds of overlap-extension PCR by using appropriate mutagenic primers and substituted for the original AlwNI-EcoO109 I fragment, resulting in pUK21-B2. All the mutations were confirmed by sequencing.
Another 37 CpG-N motifs were removed by replacing the F1 origin with a multiple cloning site. Oligonucleotides 5′-GCCCTATTTTAAATTCGAAAGTACTGGACCTGTTAACA-3′ and its complementary strand 5′-CGTGTTAACAGGTCCAGTACTTTCGAATTTAAAATAG-3′ were synthesized, and 5′-phosphorylated. Annealing of these two phosphorylated ODN resulted in a 35-bp double-stranded DNA fragment containing four unique restriction enzyme sites (DraI, ScaI, AvaII, HpaI) and two sticky ends. Replacing the 0.6-kb NarI-EcoO109 I fragment of pUK21-B2, which contained the entire F1 ori, with this double-stranded DNA fragment resulted in the master vector pMAS.
Next, different numbers of CpG-S motifs were inserted into the vector by allowing self-ligation of a 20-bp DNA fragment with the sequence 5′-GACTCCATGACGTTCCTGACGTTTCCATGACGTTCCTGACGTTG-3′ with a complementary strand and inserting different numbers of copies into the AvaII site of pMAS. Recombinant clones were screened and the two vectors were chosen for further testing with 16 and 50 CpG-S motifs, and named pMCG16 and pMCG50, respectively.
To create a DNA vaccine, the S gene encoding ay subtype of hepatitis B surface antigen (HBsAg) was amplified by PCR and cloned into the EcoRV-PstI sites of the vectors, resulting in pUK-S, pMAS-S, pMCG16-S, and pMCG50-S, respectively. Vector sequences were confirmed by sequencing and have been deposited in GenBank under accession numbers AFO53406 (pUK-S), AFO53407 (pMAS-S), AFO53408 (pMCG16-S), and AFO53409 (pMCG50-S).
Immunization of Mice Against HBsAg.
Immunization of 6- to 8-week-old female BALB/c mice (Charles River Breeding Laboratories) was done by injection into the tibialis anterior muscle (TA) of 1 μg recombinant HBsAg or 10 μg HBsAg-expressing DNA vaccine (31). Assay for antibodies against HBsAg (anti-HBs) was by endpoint dilution and for cytotoxic T lymphocytes (CTL) was by chromium release assay as described previously (19). Both the protein (±ODN) and DNA vaccines were resuspended in saline for injection.
RESULTS
Type 12 Adenoviral DNA Is Immune-Stimulatory, but Types 2 and 5 Adenoviral DNA Are Immune-Neutralizing.
To investigate possible functional differences in the immune effects of various prokaryotic DNAs, we determined their ability to induce cytokine secretion from human peripheral blood mononuclear cells (PBMC). In contrast to bacterial DNA and genomic DNA from type 12 adenovirus, DNA from types 2 and 5 adenovirus failed to induce cytokine production (Table 1). In fact, despite their similar frequency of CpG dinucleotides, type 2 or 5 adenoviral DNA severely reduced the cytokine expression induced by coadministered immunostimulatory E. coli genomic DNA (Table 2). This indicates that type 2 and 5 adenoviral DNA does not simply lack CpG-S motifs, but contains sequences that actively suppress those in E. coli DNA.
Table 1.
Genomic DNA from type 12 but not type 2 adenovirus stimulates cytokine secretion from human PBMC
| DNA added | Experiment 1
|
Experiment 2
|
||
|---|---|---|---|---|
| TNF-α | IL-6 | TNF-α | IL-6 | |
| Cells | 27 | 800 | 30 | 800 |
| E. coli (3 μg/ml) | 235 | 26,500 | 563 | 34,000 |
| CT (10 μg/ml) | 0 | 1,400 | 0 | 2,800 |
| Adv 2 (3 μg/ml) | 15.6 | 900 | 30 | 1,900 |
| Adv 12 (3 μg/ml) | 86 | 11,300 | 120 | 11,250 |
PBMC were obtained from normal human donors and cultured at 1 × 105 cells/200 μl in RPMI 1640 medium with 10% autologous serum for 4 hr (TNF-α assay) or 24 hr (IL-6 assay). The level of cytokine present in culture supernatants was determined by ELISA (pg/ml). Adv, adenovirus serotype.
Table 2.
Type 5 adenoviral DNA suppresses the human PBMC cytokine response to E. coli (EC) DNA
| DNA added | IFN-γ |
|---|---|
| EC (50 μg/ml) | 509 |
| EC (5 μg/ml) | 554 |
| EC (0.5 μg/ml) | 285 |
| EC (0.05 μg/ml) | 173 |
| Adv* (50 μg/ml) | <10 |
| Adv (5 μg/ml) | <10 |
| EC/Adv (50:50 μg/ml) | 23 |
| EC/Adv (5:50 μg/ml) | <10 |
| EC/Adv (0.5:0.5 μg/ml) | 25 |
The second column represents the level of IFN-γ production by ELISA (pg/ml) from 24-hr supernatants of human PBMC cultured at 1 × 105 cells/200 μl in RPMI 1640 medium with 10% autologous serum. Similar inhibitory effects were seen when using PBMC from several different donors and from murine spleen cells.
Type 2 adenoviral DNA had indistinguishable immune effects (not shown).
Identification of Putative Immune Neutralizing CpG-N Motifs in Types 2 and 5 Adenoviral Genomes.
To identify possible nonrandom skewing of the bases flanking the CpG dinucleotides in the various adenoviral genomes, we examined their frequency of all 4,096 hexamers. The six most common hexamers in the type 2 adenoviral genome are shown in Table 3, along with their frequency in the type 12 and E. coli genomes. Remarkably, all of these overrepresented hexamers contain either direct repeats of CpG dinucleotides or CpGs that are preceded by a C and/or followed by a G. These CpG-N motifs are approximately 3- to 6-fold more common in the immune inhibitory types 2 and 5 adenoviral genomes than in those of immune-stimulatory type 12 adenoviral, E. coli, or nonstimulatory human genomic DNAs (Table 3). This hexamer analysis further revealed that the frequency of hexamers containing CpG-S motifs (e.g., GACGTT or AACGTT) in the type 2 adenoviral genome is as low as that in the human genome: only 1/3 to 1/6 of that in E. coli and type 12 adenoviral DNA (Table 3).
Table 3.
Genomic frequencies of selected hexamers
| Hexamer | Adenovirus type 2 | Adenovirus type 12 | E. coli | Human |
|---|---|---|---|---|
| GCGCGC | 1.614 | 0.498 | 0.462 | 0.153 |
| GCGGCG | 1.530 | 0.469 | 0.745 | 0.285 |
| GGCGGC | 1.419 | 0.440 | 0.674 | 0.388 |
| CGCGCG | 1.336 | 0.322 | 0.379 | 0.106 |
| GCCGCC | 1.280 | 0.410 | 0.466 | 0.377 |
| CGCCGC | 1.252 | 0.410 | 0.623 | 0.274 |
| GACGTT (CpG-S) | 0.083 | 0.234 | 0.263 | 0.068 |
| AACGTT (CpG-S) | 0.056 | 0.205 | 0.347 | 0.056 |
The frequencies of hexamers in adenoviral and E. coli genomes were kindly provided by J. Han (University of Alabama, Birmingham), who also determined those for the human genome (54). The hexamer frequencies in type 5 adenovirus are essentially identical to those in type 2 and therefore are not shown. The last two hexamers are CpG-S motifs shown for comparison and are the most stimulatory of all tested CpG-S motifs (11). Note that the expected frequency of a randomly selected hexamer is 1/4,096 = 0.244 × 10−3.
Effect of CpG-N Motifs on the Immune Stimulatory Effects of CpG-S Motifs.
To determine whether these overrepresented CpG-N motifs could explain the neutralizing properties of types 2 and 5 adenoviral DNA, we tested the in vitro immune effects of synthetic oligodeoxynucleotides bearing a CpG-S motif, one or more CpG-N motifs, or combinations of both. An ODN containing a single CpG-S motif induces spleen cell production of IL-6, IL-12, and IFN-γ (ODN 1619, Table 4). However, when the 3′ end of this ODN was modified by substituting either repeating CpG dinucleotides or a CpG dinucleotide preceded by a C, the level of cytokine production was reduced by approximately 50% (ODN 1952 and 1953, Table 4). ODN consisting exclusively of these neutralizing CpG (CpG-N) motifs induced little or no cytokine production (Table 5). Indeed, addition of ODN containing one or more CpG-N motifs to spleen cells along with the CpG-S ODN 1619 caused a substantial decrease in the induction of IL-12 expression, indicating that the neutralizing effects can be exerted in trans (Table 5).
Table 4.
Identification of neutralizing CpG motifs that reduce the induction of cytokine secretion by a CpG-S motif in the same ODN (cis-neutralization)
| ODN | Sequence 5′-3′ | ODN-induced cytokine expression
|
||
|---|---|---|---|---|
| IL-6 | IL-12 | IFN-γ | ||
| None | <5 | 206 | 898 | |
| 1619 | TCCATGTCGTTCCTGATGCT | 1,405 | 3,130 | 4,628 |
| 1952 | .............GCGCGCG | 559 | 1,615 | 2,135 |
| 1953 | ...............CC... | 577 | 1,854 | 2,000 |
Dots in the sequence of ODN 1952 and 1953 indicate identity to ODN 1619; CpG dinucleotides are underlined for clarity. ODN without CpG-N or CpG-S motifs had little or no effect on cytokine production. The data shown are representative of four experiments. All cytokines are given in pg/ml and were measured by ELISA on supernatants from DBA/2 spleen cells cultured in 96-well plates at 2 × 107 cells/ml for 24 hr with the indicated ODN at 30 μg/ml. SD of the triplicate wells was <7%. None of the ODN induced significant amounts of IL-5.
Table 5.
Inhibition of CpG-induced cytokine secretion by ODN containing CpG-N motifs
| CpG-N ODN | Sequence 5′-3′ | IL-12 secretion* | CpG-S-induced IL-12 secretion† |
|---|---|---|---|
| None | 268 | 5,453 | |
| 1895 | GCGCGCGCGCGCGCGCGCGC | 123 | 2,719 |
| 1896 | CCGGCCGGCCGGCCGGCCGG | 292 | 2,740 |
| 1955 | GCGGCGGGCGGCGCGCGCCC | 270 | 2,539 |
| 2037 | TCCATGCCGTTCCTGCCGTT | 423 | 2,847 |
To detect any effect of CpG-N ODN on basal IL-12 production, BALB/c spleen cells were cultured in 96-well plates at 2 × 107 cells/ml with the indicated ODN for 24 hr and then the supernatants were assayed for IL-12 by ELISA (pg/ml).
To determine whether CpG-N ODN can inhibit induced IL-12 secretion, cells were set up the same as in * except that IL-12 secretion was induced by the addition of the CpG-S ODN 1619 (TCCATGACGTTCCTGATGCT) at 30 μg/ml. The data shown are representative of five experiments.
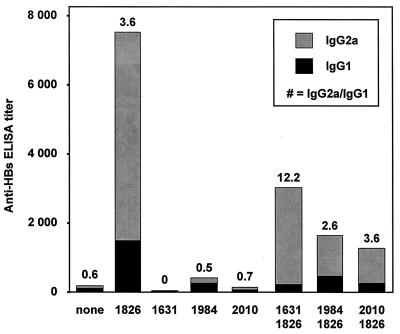
To determine whether the in vivo immune activation by ODN containing CpG-S motifs would be reversed by CpG-N motifs, we immunized mice with recombinant HBsAg, with or without nuclease-resistant phosphorothioate-modified ODN containing various types of CpG motifs. As expected, a CpG-S ODN promoted a high titer of antibodies against HBsAg (anti-HBs antibodies) that were predominantly of the IgG2a subclass, indicating a Th1-type immune response (Fig. 1; ODN 1826). The various CpG-N ODN induced either little or no production of anti-HBs antibodies (ODN 1631, 1984, and 2010) (Fig. 1). Mice immunized with combinations of CpG-S and CpG-N ODN had a reduced level of anti-HBs antibodies compared with mice immunized with CpG-S ODN alone, but these were still predominantly IgG2a (Fig. 1).
Figure 1.
Induction of a Th2-like response by a CpG-N motif, and inhibition of the Th1-like response induced by a CpG-S motif. Anti-HBs antibody titers (IgG1 and IgG2a subclasses) in BALB/c mice 12 weeks after i.m. immunization with recombinant HBsAg, which was given alone (none) or with 10 μg stimulatory ODN (1826), 10 μg of neutralizing ODN (1631, CGCGCGCGCGCGCGCGCGCG; 1984, TCCATGCCGTTCCTGCCGTT; or 2010, GCGGCGGGCGGCGCGCGCCC; CpG dinucleotides are underlined for clarity) or with 10 μg stimulatory ODN + 10 μg neutralizing ODN. To improve nuclease resistance for these in vivo experiments, all ODN were phosphorothioate-modified. Each bar represents the group mean (n = 10 for none; n = 15 for 1826 and n = 5 for all other groups) for anti-HBs antibody titers as determined by end-point dilution ELISA assay. Solid portions of bars indicate antibodies of IgG1 subclass (Th2-like), and shaded portions indicate IgG2a subclass (Th1-like). The numbers above each bar indicate the IgG2a/IgG1 ratio in which a ratio >1 than indicates a predominantly Th1-like response and a ratio <1 indicates a predominantly Th2-like response (a value of 0 indicates a complete absence of IgG2a antibodies).
Enhanced DNA Vaccination by Deletion of Plasmid CpG-N Motifs.
DNA vaccines can be highly effective inducers of Th1-like immune responses (32, 33). Based on the in vivo and in vitro effects of CpG-N motifs, we hypothesized that their presence within a DNA vaccine would decrease its immunostimulatory effects. Our starting vector, pUK21-A2, contained 254 CpG dinucleotides, of which 134 were within CpG-N motifs. To test the hypothesis that these CpG-N motifs adversely affected the efficacy of this vector for DNA-based vaccination, the number of CpG-N motifs was reduced, either by mutation or deletion. Since mutations in the plasmid origin of replication interfere with replication of the plasmid, we restricted our initial mutations to the kanamycin resistance gene and a nonessential flanking region. We were able to eliminate 19 CpG dinucleotides contained within 15 of the 20 CpG-N motifs in these regions without changing the protein sequence. The F1 origin of replication containing 37 CpG-N motifs and only 17 other CpG dinucleotides then was deleted, creating the vector pMAS. This vector was modified further by the introduction of 16 or 50 CpG-S motifs, yielding vectors pMCG16 and pMCG50, respectively. The S gene for HBsAg then was cloned into these vectors downstream from the CMV promoter, to make pUK-S, pMAS-S, pMCG16-S, and pMCG50-S, respectively.
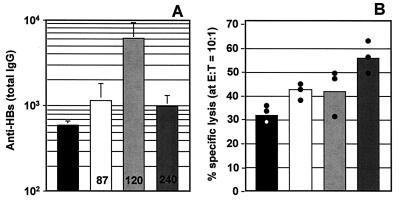
When tested for their ability to induce cytokine (IL-6 and IL-12) secretion from cultured spleen cells, we found that the pMAS-S, pMCG16-S, and pMCG50-S vectors had significantly enhanced immune stimulatory activity compared with pUK-S (not shown). When used as a DNA vaccine, the anti-HBs response at 4 and 6 weeks was substantially stronger with DNA vaccines from which CpG-N motifs had been deleted and even more so when 16 CpG-S motifs had been inserted. The vector with 50 CpG-S motifs, however, was less effective at inducing antibody production than that with 16 motifs (Fig. 2A). Removal of CpG-N motifs and addition of CpG-S motifs resulted in a more than 3-fold increase in the proportion of IgG2a relative to IgG1 anti-HBs antibodies, indicating an enhanced Th-1 response. This accentuated Th1 response also was demonstrated by the striking progressive increases in CTL responses induced by vectors from which CpG-N motifs were deleted and/or CpG-S motifs added (Fig. 2B).
Figure 2.
Enhancement of in vivo immune effects with optimized DNA vaccines. Mice were injected with 10 μg of pUK-S (solid bars), pMAS-S (open bars), pMCG16-S (lightly shaded bars), or pMCG50-S (darkly shaded bars) plasmid DNA bilaterally (50 μl at 0.1 mg/ml in saline) into the TA muscle as described (53). (A) The anti-HBs antibody response at 6 weeks (detected as described in Methods). Bars represent the group means (n = 5) for ELISA end-point dilution titers (performed in triplicate), and vertical lines represent the SEM. The numbers on the bars indicate the ratio of IgG2a/IgG1 antibodies at 4 weeks, as determined in separate assays (also in triplicate) by using pooled plasma. (B) CTL activity in specifically restimulated (5 days) splenocytes taken from mice 8 weeks after DNA immunization. Bars represent the group means (n = 3) for percentage of specific lysis (performed in triplicate) at an effector-to-target (E/T) ratio of 10:1; dots represent the individual values. Nonspecific lytic activity determined with non-antigen-presenting target cells, which never exceeds 10%, has been subtracted from values with HBsAg-expressing target cells to obtain percentage of specific lysis values.
DISCUSSION
The discovery of immune-activating CpG-S motifs in bacterial DNA has led to the realization that aside from encoding genetic information, DNA can also function as a signal-transducing molecule. Our present results demonstrate that genomic DNA from type 12 adenovirus is immune-stimulatory, compatible with its relatively high content of CpG-S motifs. In contrast, genomic DNA from types 2 and 5 adenoviruses is not stimulatory, but rather is immune-neutralizing and blocks the cytokine induction of bacterial DNA (Tables 1 and 2). To identify possible differences in the CpG motifs present in these different adenoviral genomes, we analyzed the genomic frequency of all hexamer sequences. This analysis demonstrated that only the types 2 and 5 adenoviral genomes had a dramatic overrepresentation of CpG motifs containing direct repeats of CpG dinucleotides and/or CpGs preceded by a C and/or followed by a G (Table 3). Synthetic ODN containing such putative immune-neutralizing (CpG-N) motifs not only did not induce cytokine production in vitro, but also inhibited the ability of an immune-stimulatory CpG-S motif to induce cytokine expression (Tables 4 and 5). These studies reveal that there are immune-neutralizing CpG-N as well as stimulatory CpG-S motifs and that there is a surprisingly complex role for the bases flanking CpG dinucleotides in determining these immune effects. In general, CpG-N motifs oppose CpG-S motifs in cis or trans. Some of the most effective CpG-N ODN are self-complementary and/or G-rich, which may give them the capacity to form higher-ordered structures. Further studies are underway to determine the molecular mechanisms through which CpG-N and CpG-S motifs exert their respective immune effects.
Remarkably, the hexamers that contain CpG-N motifs are from 15 to 30 times more common in types 2 and 5 adenoviral genomes than those that contain immune-stimulatory CpG-S motifs. However, in type 12 adenoviral genomes the frequencies of hexamers containing CpG-N and CpG-S motifs do not differ substantially from chance. These data suggest that the immune-neutralizing effects of types 2 and 5 adenoviral DNA are not merely a result of their propagation in eukaryotic cells, but rather are due to the overall excess of CpG-N compared with CpG-S motifs. It is tempting to speculate that the marked overrepresentation of CpG-N motifs in the genomes of types 2 and 5 adenovirus may contribute to the biologic properties, such as persistent infection of lymphocytes, which distinguish them from type 12 adenovirus. The presence of large numbers of CpG-N motifs within these adenoviral genomes may have played an important role in the evolution of this virus by enabling it to avoid triggering CpG-induced immune defenses. It will be interesting to determine the general distribution of CpG-N and CpG-S motifs in different families of microbial and viral genomes, and to explore their possible roles in disease pathogenesis.
CpG-N motifs also are overrepresented in the human genome, where their hexamers are approximately 2- to 5-fold more common than CpG-S motifs. While this skewing is far less marked than that in adenoviral DNA, it would still be expected to reduce or eliminate any immune-stimulatory effect from the unmethylated CpGs present in CpG islands within vertebrate DNA. We and others have found that even when predominantly or completely unmethylated, vertebrate DNA still is not immune-stimulatory (A.M.K. and P. Jones, unpublished data; ref. 34), which is in keeping with its predominance of CpG-N motifs (Table 3). Given the overall level of CpG suppression in the human genome, the molecular mechanisms responsible for the skewing of the frequency of CpG-N to CpG-S motifs are unclear. Such a distortion from the expected random patterns would seem to require the existence of pathways that preferentially mutate the flanking bases of CpG-S motifs in vertebrate genomes, but do not affect CpG-N motifs. Indeed, statistical analyses of vertebrate genomes have provided evidence that CpGs flanked by A or T (as in CpG-S motifs) mutate at a faster rate than CpGs flanked by C or G (35).
Based on our in vitro experiments we hypothesized that the presence of CpG-N motifs in DNA vaccines interferes with the induction of the desired immune response. Indeed, the present study demonstrates that elimination of CpG-N motifs from a DNA vaccine leads to improved induction of antibodies. By removing 52 of the CpG-N motifs from a DNA vaccine (45 were deleted and 7 turned into CpG-S motifs) the serologic response was more than doubled; by then adding an additional 16 CpG-S motifs, the response was enhanced nearly 10-fold (Fig. 2A). Likewise, CTL responses were improved by removing CpG-N motifs and even more so by adding 16 or 50 CpG-S motifs (Fig. 2B). These increased responses are especially notable in view of the fact that the total number of CpG dinucleotides in the mutated vaccines is considerably below the original number.
The finding that the vector with 50 CpG-S motifs was inferior to that with 16 motifs for induction of humoral immunity was unexpected and may be secondary to CpG-induced production of type I interferons and subsequent reduction in the amount of antigen expressed. The decreased antibody response induced by pMCG50-S seems unlikely to be explained by vector instability since this vector gave the best CTL responses (Fig. 2B). Although the pMCG50-S vector was slightly larger than pMCG16-S, the 10-μg dose still contained 93% as many plasmid copies as it did pMCG16-S, so lower copy number is unlikely to account for the reduced antibody levels. The current generation of DNA vaccines are quite effective in mice, but much less effective in primates (36–42). Our present results suggest that attaining the full clinical potential of DNA vaccines may require using engineered vectors in which CpG-N motifs have been deleted and CpG-S motifs have been added.
On the other hand, the field of gene therapy may benefit from the discovery of CpG-N motifs through their insertion into gene transfer vectors to prevent or reduce the induction of host immune responses. Most of the CpG-N motifs in the adenoviral genome are in the left-hand (5′) side, which generally is partially or totally deleted for the preparation of gene therapy vectors, especially with the “gutless” vectors (43). This could lead to an enhanced CpG-S effect. Since nucleic acids produced in viral vectors are unmethylated, they may produce inflammatory effects if they contain a relative excess of CpG-S over CpG-N motifs and are delivered at an effective concentration (about 1 μg/ml). Gene therapy studies with adenoviral vectors have used doses up to 1010 infectious units (IU)/ml (which contains 0.4 μg of DNA/ml based on the genome size of 36 kb). Given that approximately 99% of adenoviral particles are noninfectious, this corresponds to a DNA dose of approximately 40 μg/ml, which is well within the range at which CpG DNA causes in vivo immune-stimulatory effects; just 10 μg/mouse induces IFN-γ production (5), acts as an adjuvant for immunization (19, 20–23), and causes acute pulmonary inflammation when delivered into mouse airways (44). Multiple mechanisms besides the presence of CpG-S DNA doubtless are responsible for the inflammatory responses that have limited the therapeutic development of adenoviral vectors (45, 46). Nonetheless, our present results suggest that consideration be given to the maintenance or insertion of CpG-N motifs in adenoviral vectors and to the engineering of inserts so that CpG-S motifs are mutated to reduce immune activation.
In recent years, it has become clear that effective gene expression need not require a viral delivery system. The use of plasmids for gene delivery (with or without lipids or other formulations) avoids some of the problems of viral vectors. On the other hand, much larger doses of DNA typically are required, since delivery is far less efficient than with a targeted system such as a virus. For example, effective gene expression in mice typically may require 500–1,000 μg DNA/mouse (47, 48). A recent human clinical trial using lipid/DNA complexes and naked DNA for delivery of cystic fibrosis transmembrane conductance regulator to the nasal epithelium of patients with cystic fibrosis used doses of 1.25 mg of plasmid/nostril (49). The successful application of naked DNA expression vectors for gene therapy will depend on the safety of repeatedly delivering high doses of DNA. Since the plasmids used for gene therapy typically contain several hundred unmethylated CpG dinucleotides, many of which are in CpG-S motifs, some immune activation may be expected to occur. Indeed, mice given repeated doses of just 10 μg of plasmid DNA daily develop elevated lymphocyte levels (50), and several humans who received intranasal plasmid DNA had elevated serum IL-6 levels (47). Furthermore, delivery of 4 mg of a gene therapy plasmid to cystic fibrosis patients in a recent clinical trial caused acute onset of symptoms compatible with immune activation, including fever, chills, and pulmonary congestion (J. Zabner, personal communication). Another reason to avoid the presence of CpG-S motifs in gene therapy vectors is that the cytokines that are produced because of the immune stimulation may reduce plasmid vector expression, especially when this is driven by viral promoters (32).
It is, therefore, highly desirable to develop improved gene delivery systems with reduced immune activation. It is not possible to simply methylate the CpG-S dinucleotides in gene therapy plasmids, since methylation of promoters abolishes or severely reduces their activity (29, 49). The only promoter resistant to methylation-induced silencing is the mouse mammary tumor virus promoter, which contains no essential CpGs, but is fairly weak (51). In any case, even when the promoter is unmethylated, expression is still greatly reduced if the coding sequences are methylated (50). In fact, even the strong CMV IE promoter is inactivated completely by CpG methylation (51). Deletion of all CpGs from an expression plasmid is not feasible since many of these are located in the origin of replication (approximately 1.2 kb long), where even single base changes can dramatically reduce plasmid replication. For these reasons, we propose that the addition of CpG-N motifs and/or mutation or conversion of CpG-S to CpG-N motifs may lead to the generation of less immune-stimulatory vectors for gene therapy. Studies to investigate this possibility are underway.
Acknowledgments
We thank Dr. Jian Han (University of Alabama at Birmingham) for providing the hexamer analysis of the adenoviral and E. coli genomes, Vickie McCauley for secretarial assistance, Lorraine Hamblin and Darlene Anderson for technical assistance, and the University of Iowa Gene Therapy Vector Core for providing purified adenoviral genomic DNAs. This research was supported in part by a Career Development Award from the Department of Veterans Affairs, by CpG ImmunoPharmaceuticals Inc., and by National Institutes of Health Grants R29-AR42556-01 and P01-CA66570 to A.M.K., and by the Medical Research Council (Canada) to H.L.D. Services were provided by the University of Iowa Diabetes and Endocrine Research Center (National Institutes of Health Grant DK25295). A.K.Y. was supported by a fellowship from the Arthritis Foundation, and H.L.D. was supported by a Career Scientist Award from the Ontario Ministry of Health.
ABBREVIATIONS
- IL
interleukin
- IFN
interferon
- ODN
oligodeoxynucleotides
- CTL
cytotoxic T lymphocytes
- PBMC
peripheral blood mononuclear cells
- HBsAg
hepatitis B surface antigen
References
- 1. Bird A P. Trends Genet. 1987;3:342–347. [Google Scholar]
- 2.Krieg A K, Yi A-K, Matson S, Waldschmidt T J, Bishop G A, Teasdale R, Koretzky G, Klinman D. Nature (London) 1995;374:546–549. doi: 10.1038/374546a0. [DOI] [PubMed] [Google Scholar]
- 3.Klinman D, Yi A-K, Beaucage S L, Conover J, Krieg A M. Proc Natl Acad Sci USA. 1996;93:2879–2883. doi: 10.1073/pnas.93.7.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ballas Z K, Rasmussen W L, Krieg A M. J Immunol. 1996;157:1840–1845. [PubMed] [Google Scholar]
- 5.Cowdery J S, Chace J H, Yi A-K, Krieg A M. J Immunol. 1996;156:4570–4575. [PubMed] [Google Scholar]
- 6.Halpern M D, Kurlander R J, Pisetsky D S. Cell Immunol. 1996;167:72–78. doi: 10.1006/cimm.1996.0009. [DOI] [PubMed] [Google Scholar]
- 7.Yamamoto S, Kuramoto E, Shimada S, Tokunaga T. Jpn J Cancer Res. 1988;79:866–873. doi: 10.1111/j.1349-7006.1988.tb00049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stacey K J, Sweet M J, Hume D A. J Immunol. 1996;157:2116–2122. [PubMed] [Google Scholar]
- 9.Messina J P, Gilkeson G S, Pisetsky D S. J Immunol. 1991;147:1759–1764. [PubMed] [Google Scholar]
- 10.Yi A-K, Hornbeck P, Lafrenz D E, Krieg A M. J Immunol. 1996;157:4918–4925. [PubMed] [Google Scholar]
- 11.Yi A-K, Klinman D M, Martin T L, Matson S, Krieg A M. J Immunol. 1996;157:5394–5402. [PubMed] [Google Scholar]
- 12.Yi A-K, Tuetken R, Redford T, Kirsch J, Krieg A M. J Immunol. 1998;160:4755–4761. [PubMed] [Google Scholar]
- 13.Tokunaga T, Yamamoto S, Namba K. Jpn J Cancer Res. 1988;79:682–686. doi: 10.1111/j.1349-7006.1988.tb02222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yi A-K, Chace J H, Cowdery J S, Krieg A M. J Immunol. 1996;156:558–564. [PubMed] [Google Scholar]
- 15.Sparwasser T, Miethe T, Lipford G, Erdmann A, Hacker H, Heeg K, Wagner H. Eur J Immunol. 1997;27:1671–1679. doi: 10.1002/eji.1830270712. [DOI] [PubMed] [Google Scholar]
- 16.Yi A-K, Chang M, Peckham D W, Krieg A M, Ashman R F. J Immunol. 1998;160:5898–5906. [PubMed] [Google Scholar]
- 17.Krieg A M, Yi A-K, Schorr J, Davis H L. Trends Microbiol. 1998;6:23–27. doi: 10.1016/S0966-842X(97)01145-1. [DOI] [PubMed] [Google Scholar]
- 18.Roman M, Martin-Orozco E, Goodman J S, Nguyen M-D, Sato Y, Ronaghy A, Kronbluth R S, Richman D D, Carson D A, Raz E. Nat Med. 1997;3:849–854. doi: 10.1038/nm0897-849. [DOI] [PubMed] [Google Scholar]
- 19.Davis H L, Weeranta R, Waldschmidt T J, Tygrett L, Schorr J, Krieg A M. J Immunol. 1998;160:870–876. [PubMed] [Google Scholar]
- 20.Chu R S, Targoni O S, Krieg A M, Lehmann P V, Harding C V. J Exp Med. 1997;186:1623–1631. doi: 10.1084/jem.186.10.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lipford G B, Bauer M, Blank C, Reiter R, Wagner H, Heeg K. Eur J Immunol. 1997;27:2340–2344. doi: 10.1002/eji.1830270931. [DOI] [PubMed] [Google Scholar]
- 22.Weiner G J, Liu H-M, Wooldridge J E, Dahle C E, Krieg A M. Proc Natl Acad Sci USA. 1997;94:10833–10837. doi: 10.1073/pnas.94.20.10833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moldoveanu Z, Love-Homan L, Huang W Q, Krieg A M. Vaccine. 1998;16:1216–1224. doi: 10.1016/s0264-410x(98)80122-9. [DOI] [PubMed] [Google Scholar]
- 24.Sato Y, Roman M, Tighe H, Lee D, Corr M, Nguyen M-D, Silverman G J, Lotz M, Carson D A, Raz E. Science. 1996;273:352–354. doi: 10.1126/science.273.5273.352. [DOI] [PubMed] [Google Scholar]
- 25.Klinman D M, Yamshchikov G, Ishigatsubo Y. J Immunol. 1997;158:3635–3639. [PubMed] [Google Scholar]
- 26.Karlin S, Doerfler W, Cardon L R. J Virol. 1994;68:2889–2897. doi: 10.1128/jvi.68.5.2889-2897.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shpaer E G, Mullins J I. Nucleic Acids Res. 1990;18:5793–5797. doi: 10.1093/nar/18.19.5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horvath J, Palkonyay L, Weber J. J Virol. 1986;59:189–192. doi: 10.1128/jvi.59.1.189-192.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hug M, Pahl C, Novak I. FEBS Lett. 1996;379:251–254. doi: 10.1016/s0014-5793(96)01185-4. [DOI] [PubMed] [Google Scholar]
- 30.Prosch S, Stein J, Staak K, Liebenthal C, Volk H D, Kruger D H. Biol Chem. 1996;377:195–201. doi: 10.1515/bchm3.1996.377.3.195. [DOI] [PubMed] [Google Scholar]
- 31.Chace J H, Hooker N A, Mildenstein K L, Krieg A M, Cowdery J S. Clin Immunol Immunopathol. 1997;84:185–193. doi: 10.1006/clin.1997.4380. [DOI] [PubMed] [Google Scholar]
- 32.Raz E, Tighe H, Sato Y, Corr M, Dudler J A, Roman M, Swain S L, Spiegelberg H L, Carson D A. Proc Natl Acad Sci USA. 1996;93:5141–5145. doi: 10.1073/pnas.93.10.5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Donnelly J J, Ulmer J B, Shiver J W, Liu M A. Annu Rev Immunol. 1997;15:617–648. doi: 10.1146/annurev.immunol.15.1.617. [DOI] [PubMed] [Google Scholar]
- 34.Sun S, Beard C, Jaenisch R, Jones P, Sprent J. J Immunol. 1997;159:3119–3125. [PubMed] [Google Scholar]
- 35.Bains W. Mutation Res. 1992;267:43–54. doi: 10.1016/0027-5107(92)90109-f. [DOI] [PubMed] [Google Scholar]
- 36.Davis H L, McCluskie M J, Gerin J L, Purcell R H. Proc Natl Acad Sci USA. 1996;93:7213–7218. doi: 10.1073/pnas.93.14.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Letvin N L, Montefiori D C, Yasutomi Y, Perry H C, Davies M E, Lekutis C, Alroy M, Freed D C, Lord C I, Handt L K, et al. Proc Natl Acad Sci USA. 1997;94:9378–9383. doi: 10.1073/pnas.94.17.9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fuller D H, Murphey-Corb M, Clements J, Barnett S, Haynes J R. J Med Primatol. 1996;25:236–241. doi: 10.1111/j.1600-0684.1996.tb00021.x. [DOI] [PubMed] [Google Scholar]
- 39.Lu S, Arthos J, Monteflori D C, Yasutomi Y, Manson K, Mustafa F, Johnson E, Santoro J C, Wissink J, Mullins J I, et al. J Virol. 1996;70:3978–3991. doi: 10.1128/jvi.70.6.3978-3991.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu M A, McClements W, Ulmer J B, Shiver J, Donnelly J. Vaccine. 1997;15:909–912. doi: 10.1016/s0264-410x(96)00280-0. [DOI] [PubMed] [Google Scholar]
- 41.Prince A M, Whalen R, Brotman B. Vaccine. 1997;15:916–919. doi: 10.1016/s0264-410x(96)00248-4. [DOI] [PubMed] [Google Scholar]
- 42.Gramzinski R A, Brazolot Millan C L, Obaldia N, Hoffman S L, Davis H L. Mol Med. 1998;4:109–119. [PMC free article] [PubMed] [Google Scholar]
- 43.Kochanek S, Clemens P R, Mitani K, Chen H H, Chan S, Caskey C T. Proc Natl Acad Sci USA. 1996;93:5731–5736. doi: 10.1073/pnas.93.12.5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwartz D, Quinn T J, Thorne P S, Sayeed S, Yi A-K, Krieg A M. J Clin Invest. 1997;100:68–73. doi: 10.1172/JCI119523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Newman K D, Dunn P F, Owens J W, Schulick A H, Virmani R, Sukhova G, Libby P, Dichek D A. J Clin Invest. 1995;96:2955–2965. doi: 10.1172/JCI118367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zabner J, Ramsey B W, Meeker D P, Aitken M L, Balfour R P, Gibson R L, Launspach J, Moscicki R A, Richards S M, Standaert T A, et al. J Clin Invest. 1996;97:1504–1511. doi: 10.1172/JCI118573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Philip R, Liggitt D, Philip M, Dzain P, Debs R. J Biol Chem. 1993;268:16087–16090. [PubMed] [Google Scholar]
- 48.Wang C, Chao L, Chao J. J Clin Invest. 1995;95:1710–1715. doi: 10.1172/JCI117847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zabner J, Cheng S H, Meeker D, Launspach J, Balfour R, Perricone M A, Morris J E, Marshall J, Fasbender A, Smith A E, et al. J Clin Invest. 1997;100:1529–1537. doi: 10.1172/JCI119676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parker S E, Vahlsing H L, Serfilippi L M, Franklin C L, Doh S G, Gromkowski S H, Lew D, Manthorpe M, Norman J. Hum Gene Ther. 1995;6:575–590. doi: 10.1089/hum.1995.6.5-575. [DOI] [PubMed] [Google Scholar]
- 51.Muiznieks I, Doerfler W. FEBS Lett. 1994;344:251–254. doi: 10.1016/0014-5793(94)00394-7. [DOI] [PubMed] [Google Scholar]
- 52.Ge L, Rudolph P. BioTechniques. 1997;22:28–30. doi: 10.2144/97221bm03. [DOI] [PubMed] [Google Scholar]
- 53.Davis H L, Whalen R G, Demeneix B A. Hum Gene Ther. 1993;4:151–159. doi: 10.1089/hum.1993.4.2-151. [DOI] [PubMed] [Google Scholar]
- 54.Han J, Zhu Z, Hsu C, Finley W H. Antisense Res Dev. 1994;4:53–65. doi: 10.1089/ard.1994.4.53. [DOI] [PubMed] [Google Scholar]
- 55.Yi A-K, Krieg A M. J Immunol. 1998;160:1240–1245. [PubMed] [Google Scholar]