Abstract
Ancient septicemic plague epidemics were reported to have killed millions of people for 2 millenniums. However, confident diagnosis of ancient septicemia solely on the basis of historical clinical observations is not possible. The lack of suitable infected material has prevented direct demonstration of ancient septicemia; thus, the history of most infections such as plague remains hypothetical. The durability of dental pulp, together with its natural sterility, makes it a suitable material on which to base such research. We hypothesized that it would be a lasting refuge for Yersinia pestis, the plague agent. DNA extracts were made from the dental pulp of 12 unerupted teeth extracted from skeletons excavated from 16th and 18th century French graves of persons thought to have died of plague (“plague teeth”) and from 7 ancient negative control teeth. PCRs incorporating ancient DNA extracts and primers specific for the human β-globin gene demonstrated the absence of inhibitors in these preparations. The incorporation of primers specific for Y. pestis rpoB (the RNA polymerase β-subunit-encoding gene) and the recognized virulence-associated pla (the plasminogen activator-encoding gene) repeatedly yielded products that had a nucleotide sequence indistinguishable from that of modern day isolates of the bacterium. The specific pla sequence was obtained from 6 of 12 plague skeleton teeth but 0 of 7 negative controls (P < 0.034, Fisher exact test). A nucleic acid-based confirmation of ancient plague was achieved for historically identified victims, and we have confirmed the presence of the disease at the end of 16th century in France. Dental pulp is an attractive target in the quest to determine the etiology of septicemic illnesses detected in ancient corpses. Molecular techniques could be applied to this material to resolve historical outbreaks.
Keywords: ancient DNA/paleomicrobiology/pla gene/rpoB gene
Nucleic acid-based confirmation of ancient septicemia in which bone lesions are not a manifestation has never been achieved. Such confirmation would be useful because historical descriptions of clinical cases are difficult to interpret.
For example, the etiologies of the Great Plague in Athens during the Peloponnesian War of 430–425 B.C. described by Thucydides have been proposed as the Ebola virus (1, 2), the agent of epidemic typhus Rickettsia prowazekii (3), the agent of anthrax, Bacillus anthracis (4), or the plague bacillus Yersinia pestis (5). Although several graves of plague victims have been suspected to exist in various countries, including France, the lack of proper methodology to approach the detection of Y. pestis in ancient human remnants has prevented any microbiological or nucleic acid-based confirmation of ancient plague.
We hypothesized that the dental pulp of unerupted teeth would be a lasting refuge of Y. pestis and would be a suitable material on which to base molecular detection of the bacterium for reasons including durability, good taphonomic conservation, and encapsulation (isolating it from potentially contaminating external sources; ref. 6).
We confirmed our hypothesis by the recovery of Y. pestis-specific DNA sequences from skeletons taken from two different mass graves containing persons thought to have died of plague in Provence, southern France, and thus achieved a nucleic acid-based diagnosis of ancient plague.
MATERIALS AND METHODS
Recovery of DNA from Ancient Dental Pulp.
All manipulations of ancient teeth, including opening the teeth and DNA extraction, were performed in a laboratory located in a building different from the one where Y. pestis was cultured, its DNA was extracted, and PCR amplifications were performed. Seven negative control unerupted teeth (“negative teeth”) were extracted from seven ancient skeletons, excavated from a medieval grave in Toulon, France. These skeletons showed no macroscopic signs of infectious disease, and there was no anthropological evidence that they had plague. Historical data indicate that two mass graves, recently excavated at two different sites of future urban developments in Provence, France, were used to bury victims from ancient bubonic plague quarantine hospitals. One grave contained 133 skeletons buried between May and September 1590 in Lambesc, and the second contained approximately 200 skeletons, buried in May 1722 in Marseille. We collected four unerupted teeth from two skeletons in Lambesc and eight unerupted teeth from three skeletons in Marseille (“plague teeth”). After the skulls were radiographed, the unerupted teeth were extracted, thoroughly washed, and longitudinally fractured. Powdery remnants were scraped from the dental pulp cavities into sterile tubes for further DNA extraction. Dental pulp DNA was extracted by using a standard protocol (7).
Assessment of PCR Inhibitors in DNA Extracts.
DNA extracts derived from seven negative control teeth and four teeth collected from skeletons in the plague graves (two teeth per grave) were subjected to PCR amplification targeting a 110-bp fragment of the human β-globin gene as described (8).
Amplification and Sequence Analysis of Ancient Y. pestis DNA.
PCR amplifications were performed in 25-μl reaction volumes containing 5 ng of DNA in 10 mM Tris⋅HCl ( pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 200 μM each of 2′-deoxynucleoside 5′-triphosphate, 0.2 μg of BSA, and 2 μM each of the oligonucleotide primers. Sequences of the primers targeting rpoB were 5′-AACACCTTATCGTCGTGTACGT-3′ and 5′-AATCTTCTAAAAAGCGGCCTTCA-3′ (9). Sequences of the primers targeting pla (GenBank accession no. M27820) were 5′-CTTGGATGTTGAGCTTCCTA-3′ (10) and 5′-GAGATGCTGCCGGTATTTCC-3′. Taq DNA polymerase (2 units) was added, and an initial denaturation of 95°C for 90 s was followed by 40 cycles consisting of denaturation, 95°C for 20 s, primer annealing for 20 s at 58°C (rpoB primers) or 53°C (pla primers), and elongation, 72°C for 30 s. Negative controls were run in parallel with test extracts. DNA extracted from Y. pestis KIM5 strain and sterile water were also included in each run as positive and negative controls, respectively. For rpoB amplification, a second round of PCR was performed under the same conditions as the first. DNA extractions were performed in a dedicated room separate from that in which amplifications and downstream analyses were performed. Amplified products were directly sequenced by using the AmpliTaq cycle-sequencing kit and an automatic sequencer (ALF sequencer, Pharmacia). The sequences were compared with that reported for the pesticin plasmid of Y. pestis (11) and that available for Y. pestis rpoB (GenBank accession no. AF008578). Handling of and DNA extraction from ancient teeth were performed in a laboratory in which no work on Y. pestis had previously been performed. Positive control Y. pestis DNA was prepared in a building different from that in which ancient material was processed.
RESULTS
Ancient negative and plague teeth yielded powdery remnants (Fig. 1) of a white or brown color (no correlation was found between remnant color and the success rate of DNA amplifications). Amplification of a 110-bp fragment of the human β-globin gene was achieved in seven of seven ancient negative control teeth and in four of four ancient plague teeth after 40 cycles of amplification (Table 1). A single round of 40 cycle of PCR incorporating rpoB primers failed to yield an amplicon from any DNA extracts derived from ancient dental pulp. However, amplification products of the expected 133-bp size (including primers) were obtained from DNA extracts derived from four of four ancient plague teeth collected in Lambesc after the products from the first PCR were incorporated as a template in a second 40 cycles of PCR. No amplification products were obtained from the negative control teeth or PCR negative controls (Fig. 2). Direct sequence determination of each of the amplicons yielded a 100-bp sequence that was indistinguishable from the rpoB gene sequence of the 6/69 M biotype orientalis strain of Y. pestis.
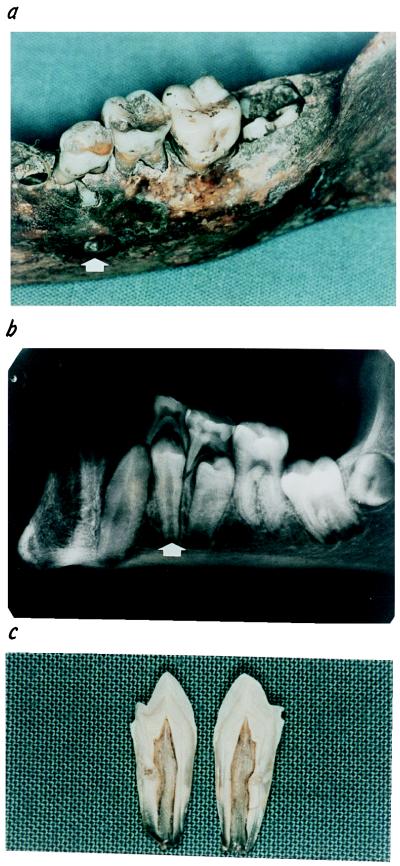
Figure 1.
Recovery of dental pulp from ancient human remains. (a) Skull remains of skeleton S61 indicating the intraosseous position of the left inferior first premolar (arrow). (b) After the skull was radiographied, the included premolar (arrow) was extracted, thoroughly washed, and fractured according to its longitudinal axis. (c) A white powder was scraped from the dental pulp cavity into a sterile tube for further DNA extraction (see Materials and Methods).
Table 1.
PCR amplification results in negative control (Toulon), and plague-infected ancient teeth (Marseille, Lambesc)
| Origin of tooth
|
Targeted sequence
|
||||
|---|---|---|---|---|---|
| Location | Individual | Tooth | β-Globulin | rpoB | pla |
| Marseille | S61 | 46 | + | NT | + |
| 34 | NT | NT | + | ||
| 16 | NT | NT | + | ||
| S122 | 16 | NT | NT | + | |
| 75 | + | NT | − | ||
| 36 | NT | NT | − | ||
| 46 | NT | NT | − | ||
| S117 | 46 | NT | NT | − | |
| 34 | NT | NT | |||
| Lambesc | S19 | 33 | + | +* | + |
| 35 | NT | +* | − | ||
| S87 | 36 | + | +* | + | |
| 85 | NT | +* | − | ||
| Toulon | 1 | 35 | + | − | − |
| 2 | 47 | + | − | − | |
| 8 | 23 | + | NT | − | |
| 4 | 48 | + | − | − | |
| 5 | 35 | + | − | − | |
| 3 | 24 | + | NT | − | |
| 7 | 36 | + | NT | − | |
Teeth are numbered according to the international nomenclature. +, Positive PCR with first amplification; +*, PCR amplicon only visible following second round PCR; −, no amplicon; NT, not tested.
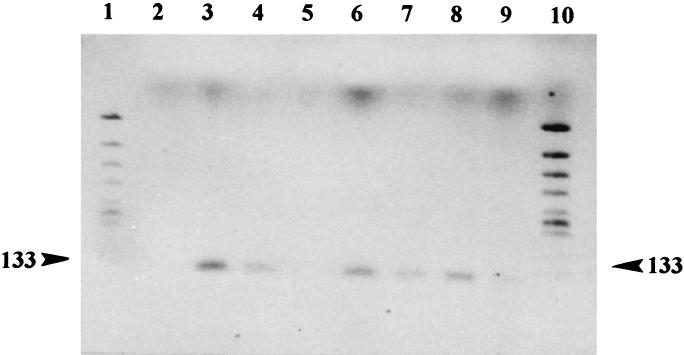
Figure 2.
Agarose gel stained with ethidium bromide showing the 133-bp amplified Y. pestis rpoB gene fragment obtained from ancient DNA after two PCR rounds. Products resulting from DNA extracted from Y. pestis strain 6/69 M biotype orientalis positive controls (lanes 3 and 4), mock extraction controls (lanes 2 and 5), and dental pulp tissues collected from specimen S19, Lambesc (lanes 6 and 7) and specimen S87, Lambesc (lanes 8 and 9) are shown. Lanes 1 and 10, molecular weight marker fragments. The measured molecular weight of the amplicons is indicated in the margin.
Incorporation of primers targeting the pla gene of Y. pestis yielded an amplification product of the expected 300-bp size (including primers) in the positive control, whereas an extraction blank processed in parallel and seven negative control teeth did not yield any detectable products. Two of four teeth collected in Lambesc and four of eight teeth collected in Marseille yielded a PCR product of the expected size after a single round of PCR amplifications (P < 0.034, Fisher’s exact test; Fig. 3). Direct sequence determination of each of the amplicons yielded a 160-bp sequence identical with that of the pla gene sequence of the KIM5 strain of Y. pestis (12).
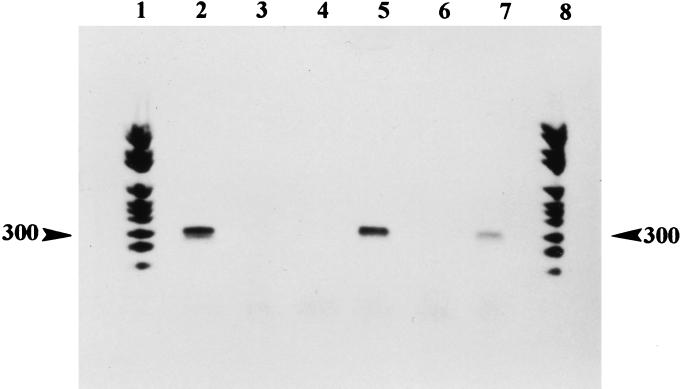
Figure 3.
Agarose gel stained with ethidium bromide showing the 300-bp amplified Y. pestis pla gene fragment obtained from ancient DNA. Products resulting from DNA extracted from Y. pestis strain 6/69 M biotype orientalis positive control (a gift from G. Baranton, Institut Pasteur, Paris; lane 2), mock extraction control (lane 3), and dental pulp tissues collected from specimen S61, Marseille (lanes 4 and 5) and specimen S122 (lanes 6 and 7) are shown. Lanes 1 and 8, molecular weight marker fragments. The measured molecular weight of the amplicons is indicated in the margin.
DISCUSSION
The lack of suitable ancient remains has prevented direct confirmation of ancient septicemia. DNA has been recovered from naturally preserved (13–15) and mummified soft tissues (16–21); however, such tissues are only preserved under exceptional circumstances. The fixation and embedding of soft tissues are relatively modern practices, although molecular-based analysis of such samples has allowed confirmation of the identity of the 1918 “Spanish” influenza virus (22) and of the Bacillus anthracis 1979 Sverdlovsk outbreak (23). Mycobacterial DNA has been recovered from medieval tuberculous bones (24, 25) and 600 A.D. lepromatous bones (26), but because buried bones are contaminated by their environment and require decalcification before DNA extraction (27), their diagnostic use may be restricted to rare diseases that appear as osseous lesions. The exploitation of dental pulp offers a practical alternative, allowing the recovery of DNA from a naturally enclosed cavity without the necessity of decalcification. Whole teeth, but not the dental pulp, have previously been used as templates for PCR-based amplifications of human (28) and animal (29) DNA. The use of unerupted teeth, still within the jawbone, further ensures the isolation of pulp from the environment.
Contamination of the samples during handling is highly improbable. A recent study conducted in urban rats in Marseille found no evidence of Y. pestis, there is no modern case of plague in southern France, and no Y. pestis isolate has been used in the laboratory where ancient teeth samples were prepared and amplified. All precautions were taken during the experimental phase to avoid cross-contamination with Y. pestis, its DNA, and its amplicons. The validity of our conclusions is supported by independent amplification of two different genes in Y. pestis, one of them being located in the pesticin gene region. This region has been shown to be specific for Y. pestis among several Yersinia spp. and to be homologous in several Y. pestis isolates from different sources and different countries (10, 30). At last, we sequenced the amplicons we obtained to confirm their specificity.
Specific amplification products were obtained from repeated testing of the original DNA extracts. Although reproducibility is a standard in experimental methods, it has often been overlooked in previous studies of ancient pathogens. Either the minute quantities of extracted ancient DNA did not allow reproducibility to be tested or reproducibility was not achieved. That the pla gene fragment was amplified in only 6 of 12 dental pulp samples may be related to its 300-bp size; ancient DNA may have degradated to such an extent that larger fragments no longer exist.
Our protocol could potentially be applied to virtually all blood-borne pathogens lacking a specific tissue signature and should elucidate current questions about the antiquity and the geographical origin of systemic infectious diseases such as typhus and leprosy. Also, our protocol could be applied to resolve more recent historical outbreaks and to understand the progression of epidemics. The findings from this study confirm that an accurate diagnosis of bubonic plague can be made by the end of 16th century in France, probably through specific buboes recognition. Whether septicemic plague was specifically diagnosed before that period remains to be confirmed by the study of teeth collected from victims of, for example, medieval Black Death or the Athenian Great Plague. Such epidemics killed as many as one-third of the population, and to confirm the diagnosis of plague or alternative viral hypotheses is a challenging task in the perspective of reemergence of infectious diseases.
Acknowledgments
The authors acknowledge Dr. R. Birtles for expert review of the manuscript. The osteoarcheological material was excavated under the supervision of the Service Régional de l’Archéologie, French Ministry for Culture (B. Bizot) by AFAN (excavation 210134062, F. Conche, Marseille; excavation 13180032 AH, P. Reynaud, Lambesc).
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1. Olson P E, Hames C S, Benenson A S, Genovese E N. Emerging Infect Dis. 1996;2:155–156. doi: 10.3201/eid0202.960220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holden C. Science. 1996;272:1591. [Google Scholar]
- 3.Weiss E. In: Encyclopedia of Microbiology. 1st Ed. Lederberg J, editor. Vol. 3. San Diego: Academic; 1992. pp. 585–610. [Google Scholar]
- 4.McSherry J, Kilpatrick R. J R Soc Med. 1992;85:713. [PMC free article] [PubMed] [Google Scholar]
- 5.Perry R D, Fetherston J D. Clin Microbiol Rev. 1997;10:35–66. doi: 10.1128/cmr.10.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bercovier H, Mollaret H H. In: Bergey’s Manual of Systematic Bacteriology. 9th Ed. Krieg N R, Holt J G, editors. Baltimore: Williams & Wilkins; 1984. pp. 498–506. [Google Scholar]
- 7.Golenberg E M. In: Ancient DNA. Herrmann B, Hummel S, editors. New York: Springer; 1994. pp. 237–256. [Google Scholar]
- 8.Greer C E, Peterson S L, Kiviat N B, Manos M M. Am J Clin Pathol. 1991;95:117–124. doi: 10.1093/ajcp/95.2.117. [DOI] [PubMed] [Google Scholar]
- 9.Mollet C, Drancourt M, Raoult D. Mol Microbiol. 1997;26:1005–1011. doi: 10.1046/j.1365-2958.1997.6382009.x. [DOI] [PubMed] [Google Scholar]
- 10.Hinnebusch J, Schwan T G. J Clin Microbiol. 1993;31:1511–1514. doi: 10.1128/jcm.31.6.1511-1514.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McDonough K A, Schwan T G, Thomas R E, Falkow S. J Clin Microbiol. 1988;26:2515–2519. doi: 10.1128/jcm.26.12.2515-2519.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sodeinde O A, Goguen J D. Infect Immun. 1989;57:1517–1523. doi: 10.1128/iai.57.5.1517-1523.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Higuchi R, Bownan B, Freiberger M, Ryder O, Wilson A C. Nature (London) 1984;312:282–284. doi: 10.1038/312282a0. [DOI] [PubMed] [Google Scholar]
- 14.Golenberg E M, Giannasi D E, Clegg M T, Smiley C J, Durbin M, Henderson D, Zurawski G. Nature (London) 1990;344:656–658. doi: 10.1038/344656a0. [DOI] [PubMed] [Google Scholar]
- 15.Lawlor D A, Dickel C D, Hauswirth W W, Parham P. Nature (London) 1991;349:786–788. doi: 10.1038/349785a0. [DOI] [PubMed] [Google Scholar]
- 16.Salo W L, Aufderheide A C, Buikstra J, Holcomb T A. Proc Natl Acad Sci USA. 1994;91:2091–2094. doi: 10.1073/pnas.91.6.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pääbo S. Nature (London) 1985;314:644–645. doi: 10.1038/314644a0. [DOI] [PubMed] [Google Scholar]
- 18.Doran G H, Dickel D N, Ballinger W E, Jr, Agee O F, Laipis P J, Hauswirth W W. Nature (London) 1986;323:803–806. doi: 10.1038/323803a0. [DOI] [PubMed] [Google Scholar]
- 19.Del Pozzo G, Guardiola J. Nature (London) 1989;339:431–432. doi: 10.1038/339431b0. [DOI] [PubMed] [Google Scholar]
- 20.Thomas R H, Schaffner W, Wilson A C, Pääbo S. Nature (London) 1989;340:465–467. doi: 10.1038/340465a0. [DOI] [PubMed] [Google Scholar]
- 21.Guhl F, Jaramillo C, Yockteng R, Vallejo G A, Cardenas-Arroyo F. Lancet. 1997;349:1370. doi: 10.1016/s0140-6736(05)63207-2. [DOI] [PubMed] [Google Scholar]
- 22.Taubenberger J K, Reid A H, Krafft A E, Bijwaard K E, Fanning T G. Science. 1997;275:1793–1796. doi: 10.1126/science.275.5307.1793. [DOI] [PubMed] [Google Scholar]
- 23.Jackson P J, Hugh-Jones M E, Adair D M, Green G, Hill K K, Kuske C R, Grinberg L M, Abramova F A, Keim P. Proc Natl Acad Sci USA. 1998;95:1224–1229. doi: 10.1073/pnas.95.3.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spigelman M, Lemma E. Int J Osteoarchaeol. 1993;3:137–143. [Google Scholar]
- 25.Baron H, Hummel S, Herrmann B. J Archeol Sci. 1996;23:667–671. [Google Scholar]
- 26.Rafi A, Spigelman M, Standford J, Lemma E, Donoghue H, Zias J. Lancet. 1994;343:1360. [PubMed] [Google Scholar]
- 27.Hagelberg E, Sykes B, Hedges R. Nature (London) 1989;342:485. doi: 10.1038/342485a0. [DOI] [PubMed] [Google Scholar]
- 28.Hänni C, Laudet V, Sakka M, Bègue A, Stéhelin D. C R Acad Sci. 1990;310:365–370. [PubMed] [Google Scholar]
- 29.Hoöss M, Dilling A, Currant A, Pääbo S. Proc Natl Acad Sci USA. 1996;93:181–185. doi: 10.1073/pnas.93.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campbell J, Lowe J, Walz S, Ezzell J. J Clin Microbiol. 1993;31:758–759. doi: 10.1128/jcm.31.3.758-759.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]