Abstract
Overexpression of p21-activated kinase 1 (PAK1) occurs during the progression of human breast cancer. We have investigated the role of PAK1 in the premalignant progression of the MCF10 series of human breast epithelial cell lines. Levels of PAK1 expression and activation increased with premalignant progression, and expression of dominant-negative (DN) PAK1 reduced both cell proliferation and migration/invasion. In three-dimensional (3D) overlay cultures in reconstituted basement membrane, the MCF10 series produced an in vitro model for premalignant progression. MCF10AneoT cells formed a hyperplastic morphology in which some spheroids developed abnormal lumens. The MCF10.AT1 line exhibited an atypical hyperplastic morphology of abnormal spheroid clusters that did not form lumens. The MCF10.DCIS cells exhibited dysplastic growth. Expression of DN-PAK1 promoted lumen formation in 3D-cultured MCF10A, NeoT, and AT1 structures, suggesting partial reversion of the premalignant phenotype, but did not affect the atypical budding of AT1 structures or the dysplastic growth of ductal carcinoma in situ structures. Aberrant proteolysis is another important characteristic of breast cancer progression and invasion. DN-PAK1 or knock-down of PAK1 reduced pericellular proteolysis of DQ-collagen IV in the 3D cultures. Treatment of cells with an inhibitor of Rac1 also reduced pericellular proteolysis, and this reduction was reversed by the expression of activated PAK1. Our conclusion is that overexpressed and activated PAK1 may be a key coordinator of aberrant cell survival and proteolysis in breast cancer progression.
Introduction
The malignant progression of breast cancers from normal mammary epithelia requires multiple alterations at both genetic and epigenetic levels [1]. Although deregulated expression or mutation of an oncogene or loss of function of a tumor suppressor gene may be the initiating event of cellular transformation, it is not normally sufficient [2] and so requires successive 2 or concurrent alterations in the expression of a panel of other important genes to produce or maintain a premalignant/malignant phenotype [3–5]. p21-Activated kinase 1 (PAK1) [6] is a candidate to be one of these factors, because it is required for oncogenic Ras-induced transformation of Rat-1 fibroblasts [7], for Rac3-controlled proliferation of breast cancer cells [8], and for Vav3-induced transformation, motility, and morphologic changes of NIH3T3 cells [9]. PAK1 is an immediate downstream effector of Rac/Cdc42 small GTPases, active lipids, GRB2a, PIX/Cool, and NCK adaptor proteins that receive extensive upstream cell signals from receptor kinases, G-proteins, and Ras small G proteins [10–13]. Activated PAK1 regulates many essential cell signal pathways involving the cell cycle, cytoskeleton reorganization and motility, gene expression, and survival and proliferation [14]. PAK1 expression is significantly increased in colorectal and ovarian cancers [15,16] and in primary breast cancers [17]. Amplification of the PAK1 gene occurs in bladder, ovarian, and breast cancers [16,18,19]. In breast cancer, PAK1 amplification predicts tumor recurrence and resistance to tamoxifen therapy [20], whereas transgenic overexpression of PAK1 alone is sufficient to induce breast tumorigenesis in animal models [21]. These findings could indicate that PAK1 may be a therapeutic target candidate for treatment of cancers. For example, direct inhibition of PAK1 activity by expressing a dominant-negative mutant, PAK1.K299R (DNPAK1), suppresses cellular motility and invasiveness in MDA-MB-435 and MCF-7 breast cancer cells [22–24], and increases chemotherapeutic-induced cell killing of renal carcinoma cell lines [25]. Conversely, the expression of a constitutively activated PAK1.T423E (CA-PAK1) increases cell motility, mitosis, anchorage-independent growth, and invasiveness in MCF-7 breast cancer cells [24]. Overall, there is extensive evidence for the involvement of PAK1 in metastatic human breast cancer [18].
PAK1 overexpression in human breast cancer may occur in the early stages, with marked increase during the conversion of normal epithelium to ductal carcinoma in situ (DCIS) [26]. Thus, it is important to consider whether PAK1 may contribute to the premalignant progression of the disease. In immortalized but untransformed cells, DN-PAK1 induces resistance to Ras transformation in Rat-1 fibroblasts [7] and promotes detachment-induced cell death (also termed anoikis) in MCF10A breast epithelial cells [27]. CA-PAK1 rescues MCF10A cells from undergoing anoikis [27]. A Rac/PAK pathway that is activated by the extracellular matrix through integrin α6β4 supports cell survival signaling in breast epithelial cells through NF-κB [28].
In normal breast tissues, the epithelial cells are organized both structurally and functionally in a specialized glandular architecture with polarized cell-cell and cell-basement membrane contacts [29]. Preinvasive and invasive epithelial tumors are notable for progressive disruptions of this normal morphology [30]. Human mammary tumor cell lines are useful tools for understanding breast carcinogenesis; however, it may be difficult to correlate results from studies of biologic pathways in two-dimensional (2D) cell culture to clinical aspects of breast tumors. For example, traditional cell culture techniques have limited relevance to the pathophysiological context of breast tissue [29]. In contrast, three-dimensional (3D) culture using reconstituted basement membrane (rBM) better recapitulates both structural and functional cues of breast tissue [30]. Mammary epithelial cells grown in 3D rBM overlay cultures form spheroids with lumens (commonly called acini), and thus resemble secretory alveoli in the normal breast tissue [30,31].
Long-established breast cancer cell lines isolated from clinical specimens (e.g., MCF-7, BT20, MDA-MB-231, etc.) have been routinely used as cell models in experimental cancer biology. A problem with using these cell lines for studies of breast cancer progression is that they were derived from tumor metastases, such as pleural effusions, far from the primary tumors [32]. Progression series cell models derived from nontumorigenic breast epithelial cells may provide clearer clues to the genetic and epigenetic changes that underlie tumor progression. For example, HMT-3522 human mammary epithelial cells and their tumor progression series derived by continuous cell passaging in defined medium are well-established and are used for molecular and cellular studies of cancer [33]. Another model is derived from MCF10A cells, which are a spontaneously immortalized human breast epithelial cell line that was developed from a patient who underwent a reduction mammoplasty [34]. MCF10A cells are pseudodiploid and form typical cobblestone epithelial sheets in monolayer culture and do not survive as xenografts in immunodeficient mice [34]. Stable transfection of MCF10A cells with the T24 Ha-Ras oncogene produced the MCF10AneoT (NeoT) line [35]. NeoT cells exhibit some aspects of the malignant phenotype in that they can form xenograft lesions, with characteristics of mild to moderate [grades 1 and 2] hyperplasia [36]. The validity of this model is based on the functional activation of Ras in human breast cancers due to overexpressed growth factors and their receptors [37], and the evidence that Ras-directed therapeutics may be useful in breast cancer treatment [38], although Ras mutations per se are not common. Further confirmation comes from the use of gene expression signatures to define Ras pathway activation; these studies found that Ras activation is prevalent in breast cancer and that this signature is predictive of sensitivity to Ras-directed therapeutics [39]. By serial passaging of NeoT xenograft lesions in vivo, a more advanced cell line was isolated and named MCF10.AT1 (AT1). AT1 cells produce xenograft lesions with characteristics of atypical [grade 3] hyperplasia [36]. After further serial passages in vivo, MCF10.DCIS.com (DCIS) cells were isolated. As xenografts, DCIS cells initially form lesions with the characteristics of comedo ductal carcinoma in situ, which may progress to invasive carcinoma over time [3]. Thus, the MCF10A variants that grow as orthotopic xenografts constitute an in vivo model for the analysis of progression of premalignant breast disease.
Materials and Methods
Reagents
Dulbecco's modified Eagle's medium (DMEM)/F12, sodium pyruvate solution, l-glutamine solution, horse serum, PBS, human epidermal growth factor (EGF), gelatin, Lipofectamine 2000, and DNA oligonucleotides were purchased from Invitrogen (Carlsbad, CA). DMEM, trypsin/EDTA solution, and penicillin-streptomycin were obtained from Cellgro (Herndon, VA). Fetal bovine serum (FBS) was from HyClone (Logan, UT). Protease-inhibitor cocktail tablets were obtained from Roche (Indianapolis, IN). Basement membrane matrix (Matrigel) was obtained from BD Biosciences (Bedford, MA). Quenched fluorescein-conjugated collagen type IV (DQ-collagen IV) and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) were obtained from Molecular Probes (Eugene, OR). Rac1 inhibitor (NSC23766) was obtained from Calbiochem (La Jolla, CA). Enhanced chemiluminescence Western blot analysis detection reagents were purchased from Amersham Biosciences (Piscataway, NJ). DRAQ5 vital DNA stain was purchased from Biostatus (Shepshed, UK). Bovine serum albumin (BSA), fibronectin (from human plasma), chicken ovalbumin, hexadimethrine bromide (polybrene), hydrocortisone, and other chemicals not otherwise listed were obtained from Sigma (St. Louis, MO). Rabbit anti-PAK1, rabbit anti-PAK2, rabbit anti-PAK3, rabbit anti-phospho-PAK1 (Thr423)/PAK2 (Thr402), mouse anti-myc-tag (9B11), rabbit anti-phospho-LIM kinase 1 (Thr508)/LIM kinase 2 (Thr505), and rabbit anti-cleaved caspase 3 (Asp175) antibodies were purchased from Cell Signaling Technology (Beverly, MA). Rabbit anti-phospho-cofilin (Ser3) and horseradish peroxidase-conjugated goat anti-rabbit IgG and horseradish peroxidase-conjugated goat anti-mouse IgG antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Mouse anti-tubulin antibody was obtained fromDevelopmental StudiesHybridoma Bank (Iowa City, IA).Oregon green-488-conjugated goat anti-rabbit IgG, Alexa Fluor-568 conjugated phalloidin, and 4′,6-diamidino-2-phenylindole (DAPI) were purchased from Molecular Probes.
The retrovirus expression vector pLNCX2, the enhanced green fluorescent protein (EGFP) coexpressing vector pIRES2-EGFP, and the Discosoma red fluorescent protein (RFP) coexpressing vector pIRES2-DsRed-Express were purchased from BD Clontech Laboratories (Mountain View, CA). PfuUltra high-fidelity DNA polymerase, XL10-Gold Ultracompetent E. coli, and retrovirus packaging vectors pVPack-GP and pVPack-VSV-G were purchased from Stratagene (La Jolla, CA).
Cell Lines and Culture
MCF10A, NeoT, AT1, and DCIS cell lines were obtained from the Cell Lines Resource (Karmanos Cancer Institute, Detroit, MI). Cells were maintained as monolayers in DMEM/F12 containing 5% horse serum, 20 ng/ml EGF, 0.5 µg/ml hydrocortisone, 10 µg/ml insulin, 50 U/ml penicillin, and 50 µg/ml streptomycin at 37°C and 5% CO2. For 3D culture, each 12-mm round coverslip was coated with 40 µl of ice-cold rBM (Matrigel) and incubated at 37°C for 20 minutes to allow the rBM to solidify. The coated coverslips were placed in a 24-well plate with the rBM facing up. A trypsinized single cell suspension containing 5000 cells in a 40-µl volume of the assay medium [DMEM/F12 containing 2% horse serum, 5 ng/ml EGF, 0.5 µg/ml hydrocortisone, 10 µg/ml insulin, 50 U/ml penicillin, 50 µg/ml streptomycin, and supplemented with 2% (v/v) of Matrigel] was carefully loaded on top of the rBM, and the plate was incubated at 37°C for 30 minutes to let the cells attach to the rBM. Then, 500 µl of the assay medium was added per well, and the cells were cultured at 37°Ca and 5% CO2, with the medium being changed every 4 days.
Plasmid Construction
Gene inserts of wild-type PAK1 (wt-PAK1) and three mutant forms, namely, a constitutively activated mutant PAK1.T423E (CA-PAK1) and two dominant-negative mutants of PAK1 (DN-PAK1), PAK1. K299R and PAK1.H83,86L,K299R, all with myc epitope tags at the N-terminus, were prepared using polymerase chain reaction from their parental plasmids. The primers were designed to introduce two new restriction enzyme digestion sites: a 5′ HindIII site and a 3′ NotI site. PfuUltra high-fidelity DNA polymerase was used to facilitate correct amplification of the 1.6-kb-long sequences. After double-enzyme digestion with HindIII and NotI, the PAK1 inserts were ligated into the pLNCX2 retroviral vector, and then their sequences were confirmed. Internal ribosomal entry site (IRES) 2-EGFP or IRES2-RFP elements were polymerase chain reaction-prepared using a pair of primers introducing a 5′ NotI site and a 3′ ClaI site. After double-enzyme digestion with NotI and ClaI, the IRES2-EGFP or IRES2-RFP elements were ligated into the pLNCX2-PAK1 plasmids to form the pLNCX2-PAK1-IRES2-EGFP and pLNCX2-PAK1-IRES2-RFP series of retrovirus constructs. The IRES2-EGFP or IRES2-RFP element was also ligated into the pLNCX2 to form control constructs.
To knock-down PAK1 expression, PAK1 siRNA hairpin oligonucleotide sequences and a nonspecific shRNA control sequence (BD Clontech) were separately cloned into RNAi-Ready pSIREN-RetroQ-DsRed-Express retrovirus vector (BD Clontech) according to the manufacturer's instructions. The first siRNA sequence, CAUCAAAUAUCACUAAGUC, which targets PAK1 326 to 344, was derived from a previous publication [40]. The second siRNA sequence CUCGAAGAAGACAUCCAAC, which targets PAK1 396 and 414, was designed using an online siRNA designer tool (http://bioinfo. clontech.com/rnaidesigner/frontpage.jsp).
Retrovirus Production and Infection of MCF10A Progression Series
HEK293T cells were maintained at 37°C in DMEM containing 10% FBS, 50 U/ml penicillin, 50 µg/ml streptomycin and supplemented with 4 mM l-glutamine, and 1 mM sodium pyruvate until 90% confluent. For transfection, the medium was changed to omit the antibiotics. Then, the cells were triply transfected with equal amounts of the retrovirus construct, pVPack-GP plasmid, and pVPack-VSV-G plasmid using Lipofectamine 2000 reagent according to the manufacturer's instructions. The medium was changed 6 hours after transfection, and the cells were cultured at 37°C for 16 hours when extensive expression of the reporter EGFP or RFP proteins could be monitored by fluorescent microscopy. The transfection efficiency was ∼100% based on fluorescence. The medium was changed and the cells were cultured at 30°C for 24 hours for the production of retroviruses. The virus broth was collected and centrifuged for 5 minutes at 200g to remove cell debris. Fresh medium was added and the HEK293 cells were continued to culture for 24 hours at 30°C to harvest a second batch of virus broth. Typically, for a 35-mm dish, 1 ml of virus broth was harvested in each batch.
The supernatant broth containing viruses was supplemented with polybrene at a final concentration of 12 µg/ml by the addition of a 4-mg/ml polybrene stock solution. Cells of the MCF10A progression series were trypsinized, centrifuged, and resuspended in growth medium to form single cell suspensions at a concentration of 2 x 106 cells/ml. The cell suspensions were mixed (3:1 v/v ratio) with virus mixture, plated onto tissue culture dishes at low starting confluence (typically ∼5%), and cultured at 37°C. The next day, the medium was changed, and the cells were subjected to a second round of infection using the second batch of viruses as described above. On the third day, the medium was changed again, and the culture continued. Virus transduction efficiency was checked using fluorescent microscopy, and the cells were used for experiments. Optimization of virus production and infection conditions/parameters in this way produced ∼95% infection efficiency.
Western Blot Assays
Cell lysates were made in a lysis buffer, composed of 50 mM Tris-HCl, pH 7.4, 100 mM NaCl, 10 mM sodium pyrophosphate, 2 mM EDTA, 1% Nonidet P-40, 1% sodium deoxycholate, 2% sodium dodecyl sulfate, 0.1% 2-mercaptoethanol, 10% glycerol, and 0.005% bromophenol blue, and supplemented with protease inhibitors (Roche) according to the manufacturer's instructions. For detecting phosphorylated proteins, this lysis buffer was further supplemented with 50 mM sodium fluoride, 0.2 mM sodium orthovanadate, and 1 µM okadaic acid. The cell lysates were subjected to brief sonication on ice, heated at 100°C for 5 minutes, and loaded onto SDS-PAGE gels for electrophoresis. The proteins were then transferred onto nitrocellulose membrane and were incubated in blocking buffer: 2% BSA, 0.5% chicken ovalbumin, 25 mM Tris-HCl, pH 7.4, 135 mM NaCl, 3 mM KCl, 50 mM sodium fluoride, 0.2 mM sodium orthovanadate, and 0.3% Tween-20. This blocking buffer was also used for the dilution of antibodies. Membranes were stripped for subsequent probing with additional antibodies as described [41].
Cell Proliferation Assay
This assay is based on the ability of living cells to convert MTT into insoluble formazan precipitate that can be dissolved in DMSO and be quantified using a spectrometer. The absorbance values are proportional to the live cell number [42]. Cells were infected with viruses, trypsinized into single cell suspensions, and inoculated into 96-well plates with 103 cells/well in 200 µl of growth medium. The plates were cultured at 37°C and 5% CO2 for the times shown. On the day of the assay, 20 µl of MTT (5 mg/ml) stock solution was added into each well, and the plate was incubated at 37°C and 5% CO2 for 4 hours for the formation of formazan. The medium was removed, and 150 µl of DMSO was added into each well to dissolve the formazan precipitate. The absorbance values at 485 nm were determined using a plate reader (SpectraFluor Plus; Tecan, Salzburg, Austria).
Cell Migration/Invasion Assay
Cell migration/invasion assays through a minimal barrier were performed using blind well chambers and polycarbonate membranes (NeuroProbe, Gaithersburg, MD). The procedure was based on the manufacturer's instructions with the modifications we previously described [43]. Briefly, polycarbonate membranes (12-µm pore, 13-mm-diameter, polyvinyl pyrrolidone-free) were soaked (both sides) in 1% gelatin solution and were air-dried to block the pores. Then, the upper side of the membrane was coated with 30 µl of 0.4 mg/ml Matrigel diluted in distilled water and then air-dried. The lower side of the membrane was further coated with fibronectin by wetting with 10 µg/ml fibronectin solution and was air-dried. The lower chamber of the blind well chamber set was filled with 214 µl of DMEM/F12 containing 10% FBS as chemoattractant, and the coated membrane was gently placed on top of the lower chamber without making air bubbles. The upper chamber was filled with 200 µl of DMEM/F12 containing 0.1% BSA. Cells were trypsinized into single cell suspensions and washed twice with DMEM/F12; 50,000 cells were loaded into the upper chamber and the cells were incubated at 37°C and 5% CO2 for 24 hours. The membrane was removed, washed briefly with PBS, and fixed with 4% paraformaldehyde for 20 minutes at room temperature. The cells were then permeabilized with 0.5% Triton X-100 in PBS for 10 minutes, quenched with 0.75% glycine in PBS, and stained with DAPI for nuclei. Finally, the membrane was mounted on a slide and sealed with a coverslip. Confocal fluorescent images of the DAPI-stained nuclei of both the cells on the upper side of the membrane (which had not migrated) and the cells on the lower side of the membrane (which had migrated) were taken in separate confocal sections at the appropriate z-axis. Cell numbers were counted based on the number of the nuclei. Migration/invasion activity was expressed as the percentage of cells that had migrated.
Immunofluorescence and Confocal Microscopy
MCF10A series cells growing on Matrigel-coated coverslips were cultured in 24-well plates. To fix structures, the coverslips were briefly washed with PBS at 37°C and incubated in 4% paraformaldehyde, pH 7.5 in PBS for 20 minutes at room temperature. The fixation was quenched by three washes with 0.75% glycine in PBS. Then, the cellular structures were permeabilized with 0.5% Triton X-100 in PBS for 10 minutes at 4°C and blocked with a 1-hour incubation in immunofluorescence (IF) buffer: 130 mM NaCl, 7 mM Na2HPO4, 3.5 mM NaH2PO4, 0.1% BSA, 0.2% Triton X-100, and 0.05% Tween-20, pH 7.5. Structures were incubated at 4°C with primary antibody in IF buffer overnight in a humidified chamber. After washing three times with IF buffer, the coverslips were further incubated with the fluorescentconjugated secondary antibodies together with other fluorescent cellular staining reagents (e.g., DAPI, Alexa Fluor-568 phalloidin) at room temperature for 2 hours. After three washes with IF buffer, the coverslips were either mounted onto slides or placed in a 35-mm dish with PBS and imaged with a confocal microscope (LSM 510; Zeiss, Göttingen, Germany) as described previously [44].
Assay of Proteolysis By Live Cells
Fluorescence-quenched DQ-collagen IV was added into the Matrigel at a concentration of 25 µg/ml. Coverslips coated with 40 µl of the supplemented Matrigel were loaded with 5000 single cells suspended in 50 µl of the assay medium. The cells were then subjected to 3D culture in 24-well plates for 2 days in 500 µl of the assay medium, in the presence or absence of treatments as indicated. Small spheroidal structures developed from single cells, and they were then stained for nuclei with DRAQ5 diluted in DMEM/F12 at 1:3000 for 30 minutes. Confocal microscopic images were taken using dipping objective lenses. Quantification of pericellular proteolysis at equatorial cross-sections of spheroids was performed using a software (MetaMorph Premiere version 7.0; Molecular Devices, Sunnyvale, CA). Red fluorescent protein expression (red channel) and DRAQ5-stained nuclei (blue channel) were used to demarcate cellular boundaries to exclude any signal from intracellular activity. For 3D reconstruction, z-axis image stacks were acquired through the spheroids at a slice thickness of 1 µm.
Statistical Analysis
For quantitative data analysis, the results were plotted as the mean ± SEM. Statistical analysis was performed using GraphPad Prism version 3.0a (GraphPad Software Inc., San Diego, CA). Statistical significance was determined using Student's t tests, or for multiple comparisons using one-way analysis of variance (ANOVA). For enumeration data analysis, the results were plotted as the frequencies. Statistical significance between groups was determined using chi-square analysis.
Results
PAK1 Expression Level Increases in Correlation with Premalignant Progression of the MCF10A Series
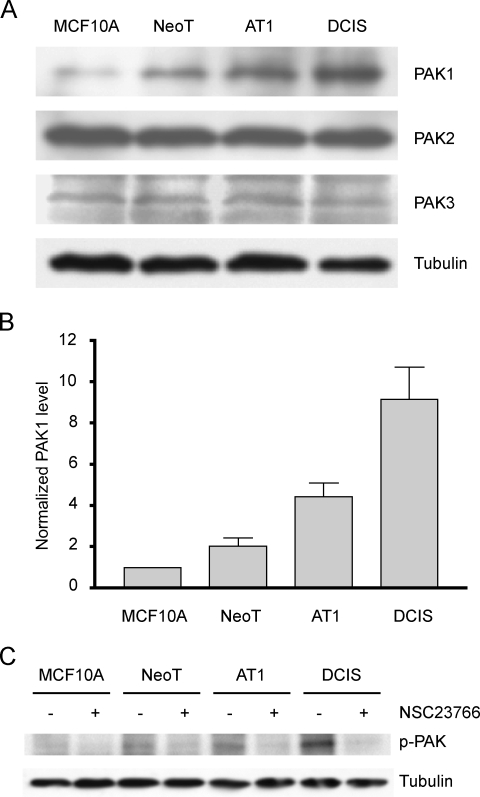
Endogenous PAKs have been reported to be elevated at both the expression level [17,45] and the activation level [46] in breast cancer cells, with evidence that PAK1 overexpression occurs during the conversion of normal epithelium to ductal carcinoma in situ [26]. We therefore tested whether there was a change in the expression levels of group I PAKs in the MCF10A breast cancer progression series. Specific PAK1, PAK2, and PAK3 antibodies were used, and the target bands were detected separately by Western blot analysis (Figure 1A). PAK1 expression levels were found to increase through the progression series of MCF10A, NeoT, AT1, and DCIS, with an approximately twofold increase at each progression step (Figure 1B). In contrast, the levels of PAK2 and PAK3 appeared constant throughout the progression series.
Figure 1.
Increase in PAK1 expression and activation in the MCF10 progression series grown in vitro. (A) Lysates of MCF10A, NeoT, AT1, and DCIS cells were subjected to immunoblot analysis for expression of group 1 PAKs [PAK1, PAK2, and PAK3]. The membranes were stripped and reprobed for tubulin to verify equal loading. (B) Densitometric analyses of PAK1 levels from four independent experiments were normalized to the level in MCF10A cells in each experiment. Data are mean ± SEM. One-way ANOVA shows that there is a significant increase in PAK1 level (P < .05) at each progression step. (C) MCF10 series cells were cultured in the presence of 75 µM NSC23766 (Rac inhibitor) or vehicle for 8 hours, followed by a further 2 hours in serum-free medium with 1 µM okadaic acid (phosphatase inhibitor) and ± NSC23766. Lysates were Western blotted first for phosphorylated PAK1 and then the membranes were stripped and reprobed for tubulin.
Although overexpression of PAK1 is likely to be significant, the activity level of PAK1 may be a more direct indicator of its function. PAK1 activation involves autophosphorylation at T423, generating an epitope that has been used to raise antibodies for detecting PAK1 activation. MCF10A cells require growth factors and hormones for cell proliferation and survival [34], and growth factors such as EGF and insulin in the growth medium can stimulate many signaling cascades that lead to the activation of PAK1 [12]. Indeed, in the regular growth culture medium for the MCF10 cell series, there is a constitutive phosphorylation of PAK1 (data not shown). To determine whether PAK1 phosphorylation results from intrinsic molecular and cellular alterations associated with progression, and not from the culture medium, we subjected the cells to serum- and hormone-starvation for 2 hours before the assay. The results showed that PAK1 phosphorylation levels increased dramatically through the series of MCF10A, NeoT, AT1, and DCIS (Figure 1C). Among the known upstream regulators, Rac1 has been delineated as a principal activator of PAK1 [6]. NSC23766, which is a commercially available, small-molecule inhibitor of the activation of Rac1 [47], significantly reduces the PAK1 phosphorylation level in bone marrow cells [48]. Addition of NSC23766 to the MCF10A series cells greatly reduced the level of PAK1 phosphorylated at T423 compared with that in vehicle-treated cells (Figure 1C).
Manipulation of PAK1 Activity By Retroviral Transduction of the MCF10A Progression Series
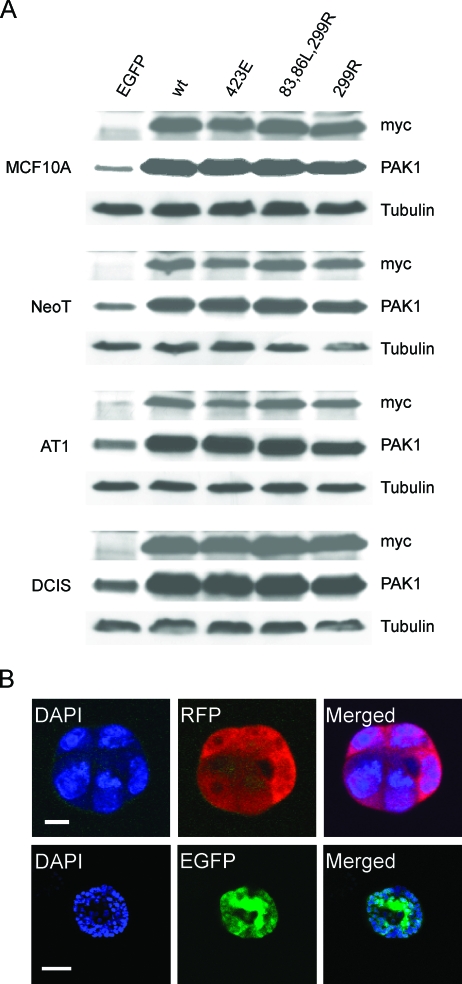
To determine the functional significance of PAK1 overexpression and activation during the progression of MCF10A cells, we developed retroviral vectors to manipulate PAK1 expression and activity. Retroviral vectors have previously been used for the stable expression of genes in MCF10A cells [29]. PAK1, CA-PAK1, and DN-PAK1 with N-terminal myc-tags were expressed from retroviral constructs that include IRES elements that allow the transcription of a single bicistronic mRNA of myc-PAK1-IRES2-FP, and so produce myc- PAK1 together with EGFP or RFP as a reporter for expression of PAK1. By optimizing infection conditions, approximately 95% infection efficiency was acquired based on fluorescent microscopic observation of the percentage of green or red fluorescent cells. By Western blot analysis, the exogenous PAK1 in the lysates of all the four cell lines was detected using the myc-tag antibody. Overexpression of PAK1 was observed by comparing to the control virus-infected cells using the PAK1 antibody. All PAK1 constructs were overexpressed to similar levels and to a relatively modest extent over that of the endogenous protein (Figure 2A). Three-dimensional rBM overlay culture of infected single cells reveals that they maintain expression of the fluorescent reporter proteins during subsequent proliferation and development of structures (Figure 2B).
Figure 2.
Expression of exogenous PAK1 constructs in MCF10 mammary epithelial progression series. (A) Cells were infected with viruses expressing exogenous myc-tagged PAK1 constructs plus EGFP, or EGFP only as the control. Cell lysates were subjected to separate Western blot assays for the myc-tag and PAK1. Overexpression of the PAK1 constructs is apparent from comparison to the signals for endogenous PAK1 in control cells exogenously expressing only EGFP. Equal loading per lane was shown by stripping and reprobing for tubulin. (B) Typical confocal fluorescent images showing equatorial slices through spheroids developed from single virus-infected cells expressing myc-tagged PAK1 constructs and coexpressing either RFP (red) or EGFP (green). Upper row: fluorescent confocal images of a spheroid cultured for 2 days; scale bar, 10 µm. Lower row: spheroid cultured for 7 days; scale bar, 50 µm. Cell nuclei were stained with DAPI (blue).
Expression of DN-PAK1 Inhibits Cell Proliferation
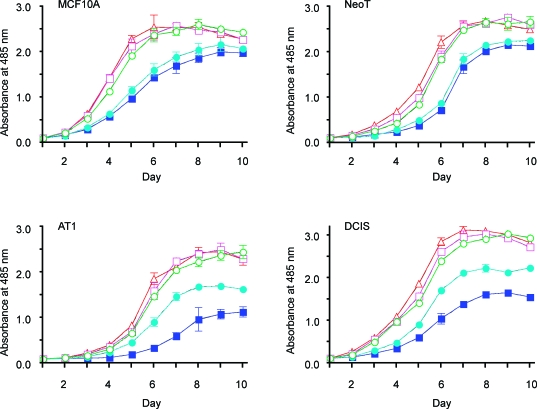
PAK1 activation has been shown to promote cell proliferation by stimulating the cell cycle [49] and inhibiting apoptosis [27]. Repression of PAK1 kinase activity correlates with cellular senescence, an irreversible cell cycle exit [50]. PAK1 activity is associated with proliferation in MCF-7 breast cancer cells [51]. To determine whether the enhanced PAK1 expression and activation that occurs in the MCF10A progression series may play a role in cell proliferation, we infected the four cell lines with the different PAK1-expressing retroviruses. An MTT assay was used to monitor cell proliferation over time. As shown in Figure 3, overexpression of wt-PAK1 or CA-PAK1 had little effect on cell proliferation, whereas expression of the two dominant-negative PAK1 mutants significantly inhibited cell proliferation, monitored at day 6, compared with EGFP controls for all four cell lines. In agreement with this result, we found that DN-PAK1 tends to increase G1 and decrease S phase populations of the cells (data not shown). By day 10, all the cultures of MCF10A and NeoT cells reached similar ∼100% confluence, as monitored by microscopy and MTT assay values. In contrast, DN-PAK1-transduced AT1 and DCIS cells remained growth-inhibited compared with EGFP-, wt-PAK1-, and CA-PAK1-transduced cells through day 10. Interestingly, the kinase-dead only mutant (PAK1.K299R) exhibited a greater growth inhibition at both days 6 and 10 than did the mutant that was both kinase-dead and disrupted for p21-binding (PAK1.H83,86L,K299R), in AT1 and DCIS cells. These findings suggest that downregulation of PAK1 activity by expression of DN-PAK1 can efficiently inhibit cell proliferation, with a much greater effect on the premalignant AT1 and DCIS cells that exhibit greater endogenous expression and activation of PAK1 (Figure 1), than on the more normal MCF10A cells.
Figure 3.
Inhibition of the proliferation of MCF10 progression series cells by DN-PAK1. Cells were passaged onto six-well plates (0.1 x 106 cells/well) and infected with viruses expressing EGFP only (○) or EGFP plus wt-PAK1 (□), CA-PAK1 (△) or DN-PAK1 [PAK1.H83,86L, K299R (●) and PAK1.K299R (■)]. The infected cells were then passaged onto 96-well plates (1 x 103 cells/well) and cultured as shown for assay of proliferation. Data are mean ± SEM from four independent experiments.
Expression of DN-PAK1 Inhibits Cell Migration/Invasion
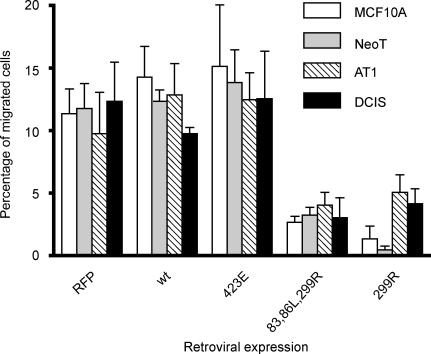
PAK1 regulates cytoskeleton reorganization and is thus likely to control cell migration and invasion [52]. We therefore tested whether alteration of PAK1 activity may affect cell migration/invasion in the MCF10 progression series. In initial experiments, we found that the invasive ability of all four cell lines was too weak to be measured using protocols that have previously been reported for the assay of the invasive phenotype of MCF10A cells that have been transformed with H-Ras.D12 [53]. Thus, we modified the method to include coating of the polycarbonate membrane and alteration of the medium compositions in the upper and lower chambers to provide a substantial and detectable control migration value for all of the cell lines. Under our modified migration/invasion assay conditions, all cell lines showed 10% to 15% cell migration. Overexpression of either wildtype or CA-PAK1 in MCF10 series cells had no significant effect on cell migration/invasion compared to the EGFP-only-infected control cells (Figure 4). The two DN-PAK1 mutants dramatically decreased the migration/invasion phenotype in all the four cell lines.
Figure 4.
Inhibition of migration/invasion of MCF10 progression series cells by DN-PAK1. Cellular migration/invasion was assayed over a 24-hour period through a gelatin/Matrigel barrier in response to 10% FBS as a chemoattractant in the lower chamber. Data are mean ± SEM from three independent experiments. One-way ANOVA shows that there is a significant (P < .05) inhibition of cell migration/invasion by DN-PAK1 in all the cell lines.
One defined pathway through which PAK can regulate cell motility is through phosphorylation of the LIM kinase, leading to the phosphorylation of cofilin and inhibition of its actin-severing activity [54]. In prostate carcinoma cells, for example, activation of PAK2 leading to the regulation of the LIM kinase/cofilin pathway has been proposed to increase motility in response to α2-macroglobulin, a protease inhibitor [55]. We therefore tested whether the LIM kinase/cofilin pathway was active in the MCF10 progression series and whether it was regulated by PAK. Phospho-LIM kinase levels were relatively constant through the MCF10 progression series and not affected by the overexpression of wild-type, CA-, or DN-PAK1 (Figure W1). Phosphocofilin levels, however, were virtually undetectable in the normal MCF10A cells and markedly increased through the progression series to DCIS. Again, however, overexpression of wild-type, CA-, or DNPAK1 had no significant effect on phospho-cofilin levels (Figure W1). Overall, the effect of DN-PAK1 to block migration/invasion does not correlate with the regulation of the LIM kinase/cofilin pathway.
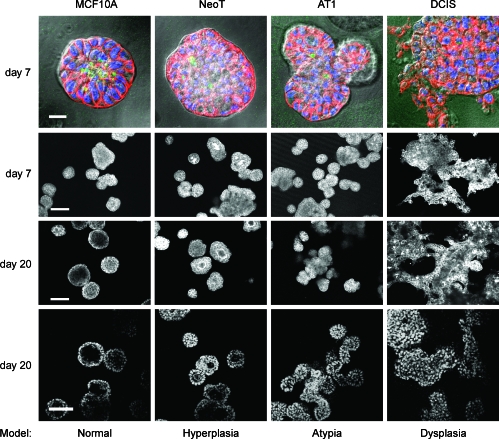
Three-Dimensional rBM Overlay Culture of the MCF10A Progression Series Provides A Model of Hyperplasia, Atypia, and Dysplasia
To develop a tractable in vitro model in which to study the role of PAK1 expression and activation as a function of breast cancer progression, we have extended an established model for analyzing morphogenesis and oncogenic transformation of MCF10A cells in a 3D laminin-rich rBM overlay culture system [29] to include the MCF10A progression series. MCF10A cells have previously been characterized in 3D rBM overlay culture as a model for normal mammary epithelia that exhibits apicobasal polarity, cell-cell junctions, strict control of cell proliferation, and apoptosis and in the formation of functional glandular structures [30,56]. Single MCF10A cells in rBM undergoes proliferation to form spheroids that develop into polarized acini (Figure 5). Hollow lumen formation in the acinar structures is because of growth arrest and detachment-induced apoptosis, also termed anoikis [57], and autophagy [56]. Strong staining for cleaved caspase 3 in the center of the MCF10A structures before the formation of the lumen supports a predominant role for apoptosis in this process (Figure 5). When the MCF10 progression series is grown in 3D rBM overlay culture, there is increasing derangement of normal acinar structures that parallels their progression to malignancy. The in vitro model thus recapitulates the stages of hyperplasia, atypical hyperplasia, and dysplasia (Figure 5) observed in vivo in xenograft studies [3,58]. By comparison to the apoptotic cells in the center of MCF10A acini, apoptotic cells were increasingly scattered and infrequent in the NeoT, AT1, and DCIS structures. After 20 days of 3D rBM overlay culture, some NeoT structures develop hollow lumens, but these acini are abnormal by several criteria: delay in formation, smaller lumens with thicker walls of multiple cell layers, and prominent staining for cortical actin [with phalloidin]. AT1 structures exhibit extensive budding of lobules from the spheroids to form clusters of spheroids. In the first few days of culture, DCIS cells proliferate to form small clumps, but the clumps do not form organized spheroids. Instead, DCIS cells form sheets and cords, with multilayers of cells that can be seen to sporadically invade into the rBM.
Figure 5.
Three-dimensional rBM overlay culture of the MCF10 progression series models hyperplastic, atypical hyperplastic, and dysplastic stages. At the times shown, the structures were fixed and processed. Top row: Differential interference contrast (DIC) images of structures with staining for: actin with phalloidin conjugated to Alexa Fluor-568 (red); DAPI as a marker of nuclei (blue); and cleaved caspase 3 as a marker of apoptosis (green); scale bar, 20 µm. The fluorescent images were captured at the equatorial plane of the structures and overlaid on a DIC image of the structure. Middle two rows: Alexa Fluor-568 phalloidin images of the actin cytoskeleton at a plane approximating the equatorial section of the majority of the structures in the field; scale bar, 100 µm. Bottom row: DAPI-stained nuclei at a plane approximating the equatorial section of the majority of the structures in the field; scale bar, 50 µm. Data are representative of results from three independent experiments.
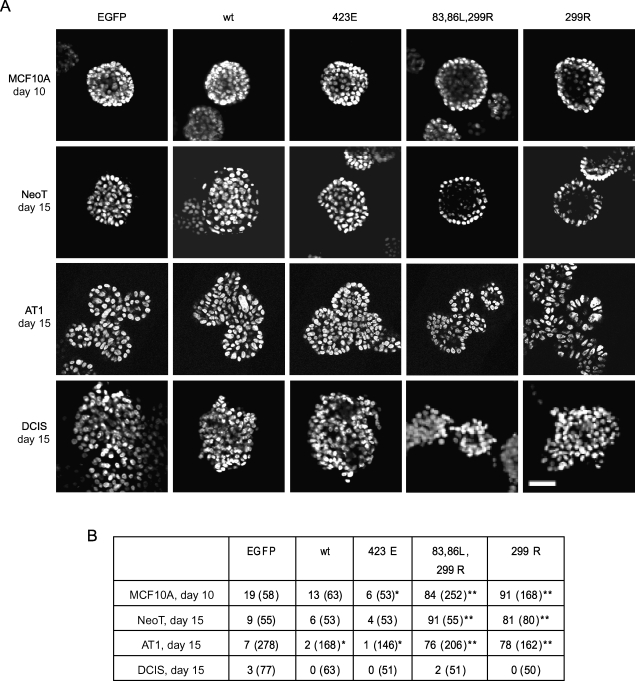
Partial Reversion of Morphology of MCF10 Progression Series By DN-PAK1
MCF10A cells form well-developed acinar structures in 3D overlay rBM culture, and so this can be used as an assay for oncogenic effects that may participate in breast cancer progression, such as cell proliferation, motility, polarity, or cell-cell adhesion that may affect the architecture [59]. PAK1 overexpression and activation correlate with inhibition of hollow lumen formation and promotion of lobular budding, which are the most typical phenotypic changes of the MCF10 progression series in 3D overlay rBM culture. Therefore, we tested whether down regulation of PAK1 activity may reverse these phenotypic changes. As shown in Figure 6, after 10 days of 3D culture, the two DN-PAK1 constructs greatly increased the formation of acini with hollow lumens (84% and 91% of spheroids were hollow, compared to 19% in control cultures). This represented an acceleration of lumen formation: EGFP-alone infected MCF10A control cells required more than 2 weeks to reach the stage where the majority of spheroids were hollow (data not shown). For NeoT cells, very few hollow lumens were formed in control structures even by day 15. Expression of DN-PAK1 induced the formation of hollow lumens in most of the structures (91% and 81% for PAK1.H83,86L,K299R and PAK1.K299R, respectively). DN-PAK1-infected AT1 cells elicited the multilobular, atypical hyperplastic phenotype, similar to the EGFP-infected AT1 controls, but with hollow lumens in most of the spheroids (76% and 78%, respectively). This was quite distinct from control AT1 structures, in which hollow lumens were rarely found. These data show a strong promotion of lumen formation by DN-PAK1, but no effect on the budding of spheroids. There was no effect of DN-PAK1 expression on the dysplastic morphology of DCIS structures, in which cells invaded into the rBM and formed amorphous patches.
Figure 6.
DN-PAK1 promotes luminal clearing in spheroids formed from cells of the MCF10 progression series in 3D rBM overlay culture. MCF10 series cells were cultured and transduced as indicated, with control retroviruses that only expressed EGFP, or bicistronic constructs expressing EGFP plus wild-type PAK1 (wt), CA-PAK1 (423E), or two forms of DN-PAK1 (83,86L,299R or 299R). The structures were fixed at the time shown, and the nuclei were stained with DAPI or DRAQ5. (A) Confocal sections at an equatorial plane through the structures are shown. Images at day 10 for MCF10A and day 15 for the other cell lines are illustrated because these were the most sensitive time points for observing the effects of PAK1 manipulation on hollow lumen formation. Data are representative of results from four independent experiments. Scale bar, 50 µm. (B) Frequencies of occurrence of hollow lumen formation (% of total structures imaged). Images of at least 50 fields were collected for each condition and analyzed for formation of hollow structures, which were defined by the absence of nuclear staining at the center of the structure. The total numbers of structures counted are listed in parentheses. To determine whether there was a significant effect of each PAK construct on the formation of hollow structures, each condition was compared to the EGFP control transduction in the same cell line by chi-square analysis (*P < .05, **P < .01).
Overexpression of wild-type or CA-PAK1 produced little observable phenotypic change in acinar development in MCF10A structures. At day 10, there was a slight effect of CA-PAK1 to reduce luminal clearing (Figure 6), but by day 20, most of the wt-PAK1- or CA-PAK1-infected MCF10A spheroids developed hollow lumens, which is similar to what occurred in the EGFP-infected controls (Figure W2). This result suggests that overexpression or activation of PAK1 alone is not sufficient to block lumen formation by MCF10A cells in 3D rBM overlay culture. It is possible that even if apoptotic clearing of the lumen by anoikis was blocked, autophagy will still be able to clear the lumen [56]. Very few hollow lumens were found in NeoT cultures infected with wt-PAK1 or CA-PAK1, even at day 20 (Figure W2). Similarly, expression of wt-PAK1 or CA-PAK1 in AT1 cells decreased formation of hollow structures. These results suggest that additional PAK1 activation in NeoT and AT1, which already have abnormal luminal clearing, can further suppress this morphology. Overexpression of PAK1 or CA-PAK1 in DCIS produced no further aberration of the observable phenotypic change compared with EGFP controls at day 15 (Figure 6).
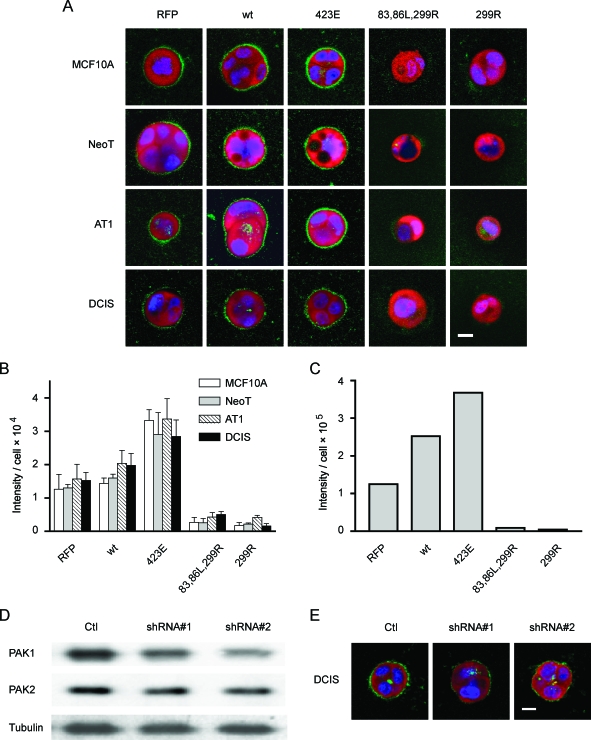
Inhibition of PAK1 Inhibits Proteolysis
Progression of premalignant breast lesions is accompanied by changes in their propensity to invade, with atypical hyperplasia classified as noninvasive and DCIS classified as preinvasive [60,61]. The conversion from preinvasive lesions to invasive cancers observed in vivo would be consistent with an increase in proteolytic activity, a characteristic that can be monitored by a number of emerging imaging techniques. The spectrum of proteases involved in pericellular proteolysis includes matrix metalloproteases, serine proteases, aspartic proteases, and cysteine proteases [62], all of which can digest collagen IV, the main component of the basement membrane of breast epithelia. We have previously investigated the proteolytic activity of BT20 human breast tumor cells in 3D [43], and this assay has also been well-documented recently for many other cancer cells [63]. Briefly, DQ-collagen IV is so heavily labeled with fluorescein that the fluorescence is quenched. On hydrolysis to single dye-labeled peptides by proteases, the quenching is relieved, and the fluorescence can be visualized by microscopy. To determine whether PAK1 expression and activity regulate pericellular proteolysis, we infected the MCF10 progression series cells with retroviral vectors that manipulate PAK1 activity and cultured them in the 3D rBM overlay system with DQ-collagen IV. The results show that at 2 days of culture, the MCF10A, NeoT, AT1, and DCIS cells all have measurable pericellular proteolytic activity (Figure 7A). Quantification of the pericellular proteolytic activity was done by pixel analysis of the equatorial section of the structures (Figure 7B). The results showed that expression of CA-PAK1 significantly enhanced pericellular degradation of DQ-collagen IV, whereas expression of the two DN-PAK1 mutants significantly inhibited pericellular proteolysis (Figure 7B). To further analyze these results, we also performed 3D reconstruction of the MCF10A structures shown in Figure 7A to quantify the pericellular proteolysis from the entire spheroid by voxel analysis of the z-stacked images. The results show that the pericellular proteolysis assayed in 3D (Figure 7C) is consistent with that measured at the equatorial plane of the structures (Figure 7B).
Figure 7.
PAK1 regulates pericellular proteolysis in the MCF10 progression series grown in 3D rBM overlay cultures. (A and B) MCF10 series cells were infected with control retroviruses that only expressed RFP, or bicistronic constructs expressing RFP plus wild-type PAK1 (wt), CA-PAK1 (423E), or two forms of DN-PAK1 (83,86L,299R or 299R) and cultured for 2 days in Matrigel plus DQ-collagen IV. The nuclei of live cells were stained with DRAQ5 for 30 minutes, and then confocal immunofluorescent images were taken. (A) Expressions of retroviral constructs (red) and nuclei (blue) are shown in equatorial sections of the cells along with the associated cleavage of DQ-collagen IV (green). Scale bar, 10 µm. (B) Quantification of the green signals from the equatorial cross-sections of three to eight structures for each condition. The data are presented as average pixel intensity per cell, with cell number determined by counting of DRAQ5-labeled nuclei. CA-PAK1 significantly increased pericellular proteolysis, and both DN-PAK1 constructs significantly inhibited pericellular proteolysis, compared to RFP control structures, in all cell lines (P < .05). (C) Representative quantification of the green signal from 3D reconstructions of the MCF10A experiments shown in (A). The data are presented as average voxel intensity per cell from the entire structure. Cell number was determined by counting the DRAQ5-labeled nuclei in z-stacks through the structures. (D and E) DCIS cells were infected with dual sequence retroviruses that expressed RFP plus a control shRNA sequence (Ctl), or plus two forms of shRNA designed to decrease PAK1 expression (shRNA #1 and shRNA #2) and cultured for 2 days. (D) DCIS Cell lysates were subjected to immunoblot analysis for PAK1 to assess knock down and were stripped and reprobed for PAK2 to assess specificity. Quantification of results from two independent experiments demonstrated that the shRNA #2 sequence decreased PAK1 expression by 55%. (E) Transduced DCIS cells were cultured in Matrigel containing DQ-collagen IV for 3 days, and images were prepared from equatorial sections of the structures for cleavage of DQ-collagen IV (green), nuclei (blue), and expression of retroviral constructs (red). Data are representative of results from two independent experiments. Scale bar, 10 µm.
To further confirm PAK1 regulation of pericellular proteolysis, we tested an alternative method to reduce PAK1 activity by knock-down of its expression. We developed dual-sequence retroviruses that encoded RFP plus shRNA sequences designed to generate RNAi constructs to target PAK1. We tested two different shRNA sequences against PAK1, one based on a published siRNA [40], and one that we designed ourselves. Both shRNA retroviruses produced a selective but modest reduction in PAK1 expression (Figure 7D) and a partial reduction in pericellular proteolysis (Figure 7E).
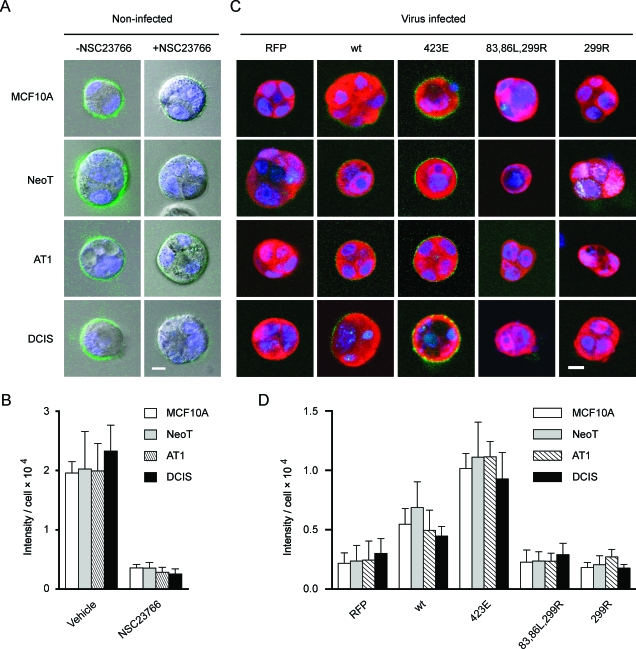
CA-PAK1 Restores Pericellular Proteolysis That Has Been Blocked By Rac Inhibition
We tested yet another alternative method to confirm the link between PAK1 activity and pericellular proteolysis. One of the most important and well-documented mechanisms for PAK1 activation is through its interaction with Rac1. The small-molecule inhibitor of Rac, which inhibits PAK1 activation in this system (Figure 1C), greatly reduced pericellular proteolysis in the 3D rBM overlay DQ-collagen IV assay (Figure 8, A and B). Overexpression of CA-PAK1 restored pericellular proteolysis to 3D rBM cultures that had been treated with NSC23766 (Figure 8, C and D). DN-PAK1 did not affect the residual pericellular proteolysis that remained in the presence of the Rac inhibitor. The rescue of inhibition by CA-PAK1 suggests that the effect of the Rac inhibitor on pericellular proteolysis is selective and not likely due to any off-target effects on other pathways. These results are consistent with a pathway in which Rac activation of PAK1 regulates pericellular proteolysis during preneoplastic progression in the MCF10 progression series.
Figure 8.
A Rac/PAK1 pathway regulates pericellular proteolysis in the MCF10 progression series grown in 3D rBM overlay culture. (A) MCF10 series cells were cultured for 2 days in Matrigel/DQ-collagen IV with or without 75 µM NSC23766. Confocal fluorescent images taken at the equatorial plane of structures show live cell nuclei (blue; DRAQ5 staining) and pericellular proteolysis (green). Scale bar, 10 µm. (B) Quantification of the green signals from the equatorial cross-sections of three to nine structures for each condition. The data are presented as average pixel intensity per cell, with cell number determined by counting of DRAQ5-labeled nuclei. Inhibition of Rac greatly inhibits pericellular proteolysis in all cell lines (P < .05). (C) MCF10 series cells were infected with control retroviruses that only expressed RFP, or bicistronic constructs expressing RFP plus wild-type PAK1 (wt), CA-PAK1 (423E), or two forms of DN-PAK1 (83,86L,299R or 299R) and were cultured for 2 days in Matrigel containing DQ-collagen IV in the presence of 75 µM NSC23766. The data are equatorial sections of the cells and show cleavage of DQ-collagen IV (green), the nuclei (blue), and the expression of retroviral constructs (red). Scale bar, 10 µm. (D) Quantification of the green signals from the equatorial cross-sections of three to eight structures for each condition. The data are presented as average pixel intensity per cell, with cell number determined by counting of DRAQ5-labeled nuclei. Expression of CAPAK1 significantly increases pericellular proteolysis in all cell lines that had been treated with NSC23766 (P < .05).
Discussion
PAK1 plays important functional roles in human breast cancer (recently reviewed in the study of [18]). For example, PAK1 is often overexpressed in breast tumors [17,64]. In some cases, there is gene amplification [19] while PAK1 mRNA also increases in a hypoxic model of the tumor microenvironment [65]. Serial analysis of gene expression demonstrates that PAK1 overexpression occurs during the premalignant conversion of normal epithelium to DCIS [26], and our data show that PAK1 expression is selectively upregulated in the MCF10A progression series in vitro (this study), where it can coordinate aberrant cell survival, through resistance to anoikis [27], and altered pericellular proteolysis (this study). The 11q13 locus that includes PAK1 is not amplified in the MCF10 series [66]. Thus, our results provide a defined premalignant progression series that recapitulates in vitro the upregulation of PAK1 that is seen in human disease and further suggest a new aspect for the potential significance of PAK1 in the regulation of pericellular proteolysis.
Disruption of polarized architecture is a prominent phenotypic change that is characteristic of carcinoma progression. Rac1 is known to control epithelial polarity [67,68]. PAK1 is an immediate downstream effector of Rac1 that regulates microtubule, actin, and microfilament organization, which are critical for maintenance of the cytoskeleton and polarity. Rac1 activity is tightly controlled to maintain normal epithelial polarity; either hyperactivity, by expression of CA-Rac1, or hypoactivity, by expression of DN-Rac1, disrupts multicellular epithelial architecture [67,68]. Similarly, physiologically relevant levels of PAK1 activity are needed for both mammary gland development and normal epithelial function [69]. Hyperactive and hypoactive PAK1 both disrupt cell polarity. DN-PAK1 promoted hollow lumen formation in the MCF10 progression series, which may be partially due to the reestablishment of cell polarity that had been suppressed by overactivated PAK1. Activated PAK1 can lead to phosphorylation of the proapoptotic proteins BAD and BIM, thus inactivating cell apoptosis [70,71], and also couples to the NF-κB pathway [28]. DN-PAK1-induced luminal cell death may be due to restored anoikis and suppressed survival in the luminal cells. We have previously shown that active PAK1 rescues MCF10A cells from undergoing anoikis [27]. Although DN-PAK1 promoted hollow lumen formation in MCF10A, NeoT, and AT1 cells, DNPAK1 was not able to reverse the multilobular morphology of AT1 cells. Further, DN-PAK1 did not reverse the dysplastic phenotype of DCIS in 3D rBM overlay culture.
DN-PAK1 inhibition of proliferation, migration, and pericellular proteolysis may be produced through suppression of multiple downstream pathways that diverge from PAK1. Some of these pathways have been well documented. For example, increased PAK1 expression in breast cancer cells stimulates the expression of cyclin D1, a cell cycle factor that promotes G1-S phase progression and cell proliferation [26], whereas PAK1-dependent activation of LIM kinase [54], Arp2/3 [72], and filamin A [73] regulates actin cytoskeleton reorganization, cell motility, and migration. In this study, we did not find that inhibition of migration/invasion by PAK1 correlated with effects on the LIM kinase/cofilin pathway, but rather tracked more closely with reduction in pericellular proteolysis. Nevertheless, the ability of DNPAK1 and the Rac inhibitor to suppress pericellular proteolysis may also be related to the regulation of actin kinetics. There is a ring of dense cortical F-actin filaments at the apical pole of epithelial cells that can form a physical barrier for exocytotic vesicle release [74,75]. PAK1 activation leads to reorganization of cortical actin and can therefore facilitate lysosomal exocytosis [76].
Although we find that inhibition of PAK1 can reduce proliferation, migration/invasion, and proteolysis, and can also promote luminal clearing in the 3D rBM overlay culture system, the major significant effect for CA-PAK1 is an increase in pericellular proteolysis. The results thus suggest that PAK1 activity may be necessary, but not sufficient, for premalignant progression in the MCF10 series. Although in vitro assays in 3D rBM overlay culture and in vivo assays as xenografts show progressive morphologic changes of the MCF10 series, our results did not suggest that overexpression of wt-PAK1 or CA-PAK1 is sufficient to drive phenotypic transformation. These results, therefore, do not directly correlate with the progressively elevated level of PAK1 expression and activation that occurs in the MCF10 series. One explanation could be that the culture media necessary for growth of the MCF10 series may cause chronic activation of PAK1 that partially suppressed the intrinsic differences in PAK1 expression and activation and their effects on phenotypic changes in the 3D rBM overlay culture conditions. Another possibility is that overactivated PAK1 may need to cooperate with other molecular alterations to drive the premalignant progression of MCF10A cells. An example of this kind of cooperation with other factors is that active Rac1-induced disruption of cell polarity must collaborate with active Akt-induced hyperproliferation to produce amorphous structures in 3D-cultured T-2 breast cancer cells [77]. It is also likely that the ability of overactivation of PAK1 to drive induction of phenotypic changes is cell type-dependent. For example, expression of CA-PAK1 in MCF-7, a breast cancer cell line that forms invasive ductal carcinoma in vivo, increases cell motility and invasiveness [24]. Different cell types may have different patterns of intracellularly expressed genes with which PAK1 could collaborate to produce transformation.
Structural and functional studies have depicted PAK1 as an interconnection point in cellular signal transduction. PAK1 kinase activity can be induced by interaction with Rac/Cdc42 through its CRIB domain, followed by autophosphorylation on T423 in its kinase domain. PAK1 also binds to PIX [10], a Rac1 guanine nucleotide exchange factor, and to NCK [78], an adaptor protein that interacts with receptor tyrosine kinases and focal adhesion kinase. These interactions localize PAK1 to focal complexes and regulate actin depolymerization. PAK1.K299R is an ATP binding-deficient mutant that is kinase-dead [79], whereas PAK1.H83,86L,K299R is a double mutant that is both Rac/Cdc42 binding-deficient and kinase-dead. Our results show that both these mutants exhibited significant inhibitory effects on cell proliferation, migration, pericellular proteolysis and anoikis. These inhibitions could be ascribed to the kinase-dead nature of the two mutants in that they can tether the upstream regulators and downstream effectors of PAK1 and knock-down the endogenous PAK1 activity. In proliferation assays, PAK1.K299R exhibited stronger inhibition than PAK1.H83,86L,299R, which might have resulted from more upstream inhibition by the former mutant than the latter, which lacks the ability to bind Rac/Cdc42. Activated Rac1, but not Cdc42, directly correlates with increased metastatic potential in a panel of cell variants derived from a single metastatic breast cancer cell line, MDA-MB-435; expression of a constitutively activated Rac1 results in a more invasive and motile phenotype whereas expressing a DN Rac1 decreases invasiveness and motility [80]. Therefore, it is possible that the elevated PAK1 activation level in the MCF10 progression series might have resulted from, in part, overactivated Rac. By inhibiting Rac1 with a selective inhibitor, NSC23766, PAK1 activation levels were significantly suppressed. This suppression of PAK1 activity by Rac inhibition greatly inhibited pericellular proteolysis and interestingly, this inhibition could be largely reversed by overexpression of CA-PAK1, which has kinase activity that is independent of Rac. Considering that Rac is upstream of PAK1 and regulates several pathways in addition to PAK1, Rac may not prove to be an effective therapeutic target for breast cancer. Conversely, and in light of the recent success of other selective inhibitors of kinase targets [81], drugs that effectively target PAK1 [82] may be a feasible approach to inhibition of breast cancer progression. In addition to the roles in premalignant progression that have been the focus of this study, PAK1 mediates the action of growth factors on the motility and invasiveness of human breast cancer cells [83], stimulates cyclin D1 expression [17,84], promotes their anchorage-independent growth [23,24], and supports epithelialto-mesenchymal transition [85]. Recently, the level of nuclear PAK1 has been strongly associated with tamoxifen resistance in breast cancer patients through phosphorylation of the estrogen receptor [45,86], which should further stimulate interest in the development of selective PAK1 inhibitors for breast cancer therapy [87].
Supplementary Material
Acknowledgments
We thank Gary M. Bokoch (Scripps Research Institute, La Jolla, CA) and Jonathan Chernoff (Fox Chase Cancer Center, Philadelphia, PA) for the original gifts of the PAK1 and mutant PAK1 constructs; Albert Chow, Andrew Jovanovski, and Mansoureh Sameni for technical assistance; and Mary Olive and Kamiar Moin for help with the imaging and quantification.
Abbreviations
- CA-PAK1
constitutively activated PAK1
- DN-PAK1
dominant-negative PAK1
- EGFP
enhanced green fluorescent protein; IRES, internal ribosomal entry site
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- PAK1
p21-activated kinase 1
- rBM
reconstituted basement membrane
- RFP
red fluorescent protein
Footnotes
This work was supported by grants from the Wilson Foundation and from the Susan G. Komen Breast Cancer Foundation and from the U.S. Public Health Service (CA56586). Confocal facilities were supported in part by Center Grants P30 ES06639 and CA22453 and a Roadmap Grant U54 RR020843.
This article refers to supplementary materials, which are designated by Figures W1 and W2 and are available online at www.neoplasia.com.
References
- 1.Beckmann MW, Niederacher D, Schnurch HG, Gusterson BA, Bender HG. Multistep carcinogenesis of breast cancer and tumour heterogeneity. J Mol Med. 1997;75:429–439. doi: 10.1007/s001090050128. [DOI] [PubMed] [Google Scholar]
- 2.Miller FR, Pauley RJ, Wang B. Activated c-Ha-ras is not sufficient to produce the preneoplastic phenotype of human breast cell line MCF10AT. Anticancer Res. 1996;16:1765–1769. [PubMed] [Google Scholar]
- 3.Miller FR, Santner SJ, Tait L, Dawson PJ. MCF10DCIS.com xenograft model of human comedo ductal carcinoma in situ. J Natl Cancer Inst. 2000;92:185–1186. doi: 10.1093/jnci/92.14.1185a. [DOI] [PubMed] [Google Scholar]
- 4.Hulit J, Di Vizio D, Pestell RG. Inducible transgenics. New lessons on events governing the induction and commitment in mammary tumorigenesis. Breast Cancer Res. 2001;3:209–212. doi: 10.1186/bcr297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ratsch SB, Gao Q, Srinivasan S, Wazer DE, Band V. Multiple genetic changes are required for efficient immortalization of different subtypes of normal human mammary epithelial cells. Radiat Res. 2001;155:143–150. doi: 10.1667/0033-7587(2001)155[0143:mgcarf]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 6.Manser E, Leung T, Salihuddin H, Zhao ZS, Lim L. A brain serine/threonine protein kinase activated by Cdc42 and Rac1. Nature. 1994;367:40–46. doi: 10.1038/367040a0. [DOI] [PubMed] [Google Scholar]
- 7.Tang Y, Chen Z, Ambrose D, Liu J, Gibbs JB, Chernoff J, Field J. Kinase-deficient Pak1 mutants inhibit Ras transformation of Rat-1 fibroblasts. Mol Cell Biol. 1997;17:4454–4464. doi: 10.1128/mcb.17.8.4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mira JP, Benard V, Groffen J, Sanders LC, Knaus UG. Endogenous, hyperactive Rac3 controls proliferation of breast cancer cells by a p21-activated kinase-dependent pathway. Proc Natl Acad Sci USA. 2000;97:185–189. doi: 10.1073/pnas.97.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sachdev P, Zeng L, Wang LH. Distinct role of phosphatidylinositol 3-kinase and Rho family GTPases in Vav3-induced cell transformation, cell motility, and morphological changes. J Biol Chem. 2002;277:17638–17648. doi: 10.1074/jbc.M111575200. [DOI] [PubMed] [Google Scholar]
- 10.Manser E, Loo TH, Koh CG, Zhao ZS, Chen XQ, Tan L, Tan I, Leung T, Lim L. PAK kinases are directly coupled to the PIX family of nucleotide exchange factors. Mol Cell. 1998;1:183–192. doi: 10.1016/s1097-2765(00)80019-2. [DOI] [PubMed] [Google Scholar]
- 11.Knaus UG, Bokoch GM. The p21Rac/Cdc42-activated kinases (PAKs) Int J Biochem Cell Biol. 1998;30:857–862. doi: 10.1016/s1357-2725(98)00059-4. [DOI] [PubMed] [Google Scholar]
- 12.Menard RE, Mattingly RR. Cell surface receptors activate p21-activated kinase 1 via multiple Ras and PI3-kinase-dependent pathways. Cell Signal. 2003;15:1099–1109. doi: 10.1016/s0898-6568(03)00087-1. [DOI] [PubMed] [Google Scholar]
- 13.Menard RE, Mattingly RR. Gbetagamma subunits stimulate p21-activated kinase 1 (PAK1) through activation of PI3-kinase and Akt but act independently of Rac1/Cdc42. FEBS Lett. 2004;556:187–192. doi: 10.1016/s0014-5793(03)01406-6. [DOI] [PubMed] [Google Scholar]
- 14.Manser E, Lim L. Roles of PAK family kinases. Prog Mol Subcell Biol. 1999;22:115–133. doi: 10.1007/978-3-642-58591-3_6. [DOI] [PubMed] [Google Scholar]
- 15.Carter JH, Douglass LE, Deddens JA, Colligan BM, Bhatt TR, Pemberton JO, Konicek S, Hom J, Marshall M, Graff JR. Pak-1 expression increases with progression of colorectal carcinomas to metastasis. Clin Cancer Res. 2004;10:3448–3456. doi: 10.1158/1078-0432.CCR-03-0210. [DOI] [PubMed] [Google Scholar]
- 16.Schraml P, Schwerdtfeger G, Burkhalter F, Raggi A, Schmidt D, Ruffalo T, King W, Wilber K, Mihatsch MJ, Moch H. Combined array comparative genomic hybridization and tissue microarray analysis suggest PAK1 at 11q13.5-q14 as a critical oncogene target in ovarian carcinoma. Am J Pathol. 2003;163(985) doi: 10.1016/S0002-9440(10)63458-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Balasenthil S, Sahin AA, Barnes CJ, Wang RA, Pestell RG, Vadlamudi RK, Kumar R. p21-Activated kinase-1 signaling mediates cyclin D1 expression in mammary epithelial and cancer cells. J Biol Chem. 2004;279:1422–1428. doi: 10.1074/jbc.M309937200. [DOI] [PubMed] [Google Scholar]
- 18.Kumar R, Gururaj AE, Barnes CJ. p21-Activated kinases in cancer. Nat Rev Cancer. 2006;6:459–471. doi: 10.1038/nrc1892. [DOI] [PubMed] [Google Scholar]
- 19.Bekri S, Adelaide J, Merscher S, Grosgeorge J, Caroli-Bosc F, Perucca-Lostanlen D, Kelley PM, Pebusque MJ, Theillet C, Birnbaum D, et al. Detailed map of a region commonly amplified at 11q13→q14 in human breast carcinoma. Cytogenet Cell Genet. 1997;79:125–131. doi: 10.1159/000134699. [DOI] [PubMed] [Google Scholar]
- 20.Bostner J, Ahnstrom Waltersson M, Fornander T, Skoog L, Nordenskjold B, Stal O. Amplification of CCND1 and PAK1 as predictors of recurrence and tamoxifen resistance in postmenopausal breast cancer. Oncogene. 2007;26:6997–7005. doi: 10.1038/sj.onc.1210506. [DOI] [PubMed] [Google Scholar]
- 21.Wang RA, Zhang H, Balasenthil S, Medina D, Kumar R. PAK1 hyperactivation is sufficient for mammary gland tumor formation. Oncogene. 2006;25:2931–2936. doi: 10.1038/sj.onc.1209309. [DOI] [PubMed] [Google Scholar]
- 22.Adam L, Vadlamudi R, Kondapaka SB, Chernoff J, Mendelsohn J, Kumar R. Heregulin regulates cytoskeletal reorganization and cell migration through the p21-activated kinase-1 via phosphatidylinositol-3 kinase. J Biol Chem. 1998;273:28238–28246. doi: 10.1074/jbc.273.43.28238. [DOI] [PubMed] [Google Scholar]
- 23.Adam L, Vadlamudi R, Mandal M, Chernoff J, Kumar R. Regulation of microfilament reorganization and invasiveness of breast cancer cells by kinase dead p21-activated kinase-1. J Biol Chem. 2000;275:12041–12050. doi: 10.1074/jbc.275.16.12041. [DOI] [PubMed] [Google Scholar]
- 24.Vadlamudi RK, Adam L, Wang RA, Mandal M, Nguyen D, Sahin A, Chernoff J, Hung MC, Kumar R. Regulatable expression of p21-activated kinase-1 promotes anchorage-independent growth and abnormal organization of mitotic spindles in human epithelial breast cancer cells. J Biol Chem. 2000;275:36238–36244. doi: 10.1074/jbc.M002138200. [DOI] [PubMed] [Google Scholar]
- 25.O'Sullivan GC, Tangney M, Casey G, Ambrose M, Houston A, Barry OP. Modulation of p21-activated kinase 1 alters the behavior of renal cell carcinoma. Int J Cancer. 2007;121:1930–1940. doi: 10.1002/ijc.22893. [DOI] [PubMed] [Google Scholar]
- 26.Abba MC, Drake JA, Hawkins KA, Hu Y, Sun H, Notcovich C, Gaddis S, Sahin A, Baggerly K, Aldaz CM. Transcriptomic changes in human breast cancer progression as determined by serial analysis of gene expression. Breast Cancer Res. 2004;6:R499–R513. doi: 10.1186/bcr899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Menard, RE, Jovanovski AP, Mattingly RR. Active p21-activated kinase 1 rescues MCF10A breast epithelial cells from undergoing anoikis. Neoplasia. 2005;7:638–645. doi: 10.1593/neo.04736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friedland JC, Lakins JN, Kazanietz MG, Chernoff J, Boettiger D, Weaver VM. α6β4 integrin activates Rac-dependent p21-activated kinase 1 to drive NF-κB-dependent resistance to apoptosis in 3D mammary acini. J Cell Sci. 2007;120:3700–3712. doi: 10.1242/jcs.03484. [DOI] [PubMed] [Google Scholar]
- 29.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 30.Debnath J, Brugge JS. Modelling glandular epithelial cancers in three-dimensional cultures. Nat Rev Cancer. 2005;5:675–688. doi: 10.1038/nrc1695. [DOI] [PubMed] [Google Scholar]
- 31.Li ML, Aggeler J, Farson DA, Hatier C, Hassell J, Bissell MJ. Influence of a reconstituted basement membrane and its components on casein gene expression and secretion in mouse mammary epithelial cells. Proc Natl Acad Sci USA. 1987;84:136–140. doi: 10.1073/pnas.84.1.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burdall SE, Hanby AM, Lansdown MR, Speirs V. Breast cancer cell lines: friend or foe? Breast Cancer Res. 2003;5:89–95. doi: 10.1186/bcr577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang F, Weaver VM, Petersen OW, Larabell CA, Dedhar S, Briand P, Lupu R, Bissell MJ. Reciprocal interactions between β1-integrin and epidermal growth factor receptor in three-dimensional basement membrane breast cultures: a different perspective in epithelial biology. Proc Natl Acad Sci USA. 1998;95:14821–14826. doi: 10.1073/pnas.95.25.14821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soule HD, Maloney TM, Wolman SR, Peterson WD Jr, Brenz R, McGrath CM, Russo J, Pauley RJ, Jones RF, Brooks SC. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 1990;50:6075–6086. [PubMed] [Google Scholar]
- 35.Basolo F, Elliott J, Tait L, Chen XQ, Maloney T, Russo IH, Pauley R, Momiki S, Caamano J, Klein-Szanto AJ, et al. Transformation of human breast epithelial cells by c-Ha-ras oncogene. Mol Carcinog. 1991;4:25–35. doi: 10.1002/mc.2940040106. [DOI] [PubMed] [Google Scholar]
- 36.Dawson PJ, Wolman SR, Tait L, Heppner GH, Miller FR. MCF10AT: a model for the evolution of cancer from proliferative breast disease. Am J Pathol. 1996;148:313–319. [PMC free article] [PubMed] [Google Scholar]
- 37.Eckert LB, Repasky GA, Ulku AS, McFall A, Zhou H, Sartor CI, Der CJ. Involvement of Ras activation in human breast cancer cell signaling, invasion, and anoikis. Cancer Res. 2004;64:4585–4592. doi: 10.1158/0008-5472.CAN-04-0396. [DOI] [PubMed] [Google Scholar]
- 38.Li T, Sparano JA. Inhibiting Ras signaling in the therapy of breast cancer. Clin Breast Cancer. 2003;3:405–416. doi: 10.3816/CBC.2003.n.005. discussion 417–420. [DOI] [PubMed] [Google Scholar]
- 39.Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, Joshi MB, Harpole D, Lancaster JM, Berchuck A, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 40.Tang DD, Bai Y, Gunst SJ. Silencing of p21-activated kinase attenuates vimentin phosphorylation on Ser-56 and reorientation of the vimentin network during stimulation of smooth muscle cells by 5-hydroxytryptamine. Biochem J. 2005;388:773–783. doi: 10.1042/BJ20050065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mattingly RR, Milstein ML, Mirkin BL. Down-regulation of growth factor-stimulated MAP kinase signaling in cytotoxic drug-resistant human neuroblastoma cells. Cell Signal. 2001;13:499–505. doi: 10.1016/s0898-6568(01)00173-5. [DOI] [PubMed] [Google Scholar]
- 42.Alley MC, Scudiero DA, Monks A, Hursey ML, Czerwinski MJ, Fine DL, Abbott BJ, Mayo JG, Shoemaker RH, Boyd MR. Feasibility of drug screening with panels of human tumor cell lines using a microculture tetrazolium assay. Cancer Res. 1988;48:589–601. [PubMed] [Google Scholar]
- 43.Sameni M, Dosescu J, Moin K, Sloane BF. Functional imaging of proteolysis: stromal and inflammatory cells increase tumor proteolysis. Mol Imaging. 2003;2:159–175. doi: 10.1162/15353500200303136. [DOI] [PubMed] [Google Scholar]
- 44.Yang H, Mattingly RR. The Ras-GRF1 exchange factor coordinates activation of H-Ras and Rac1 to control neuronal morphology. Mol Biol Cell. 2006;17:2177–2189. doi: 10.1091/mbc.E05-10-0913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Holm C, Rayala S, Jirstrom K, Stal O, Kumar R, Landberg G. Association between Pak1 expression and subcellular localization and tamoxifen resistance in breast cancer patients. J Natl Cancer Inst. 2006;98:671–680. doi: 10.1093/jnci/djj185. [DOI] [PubMed] [Google Scholar]
- 46.Stofega MR, Sanders LC, Gardiner EM, Bokoch GM. Constitutive p21-activated kinase (PAK) activation in breast cancer cells as a result of mislocalization of PAK to focal adhesions. Mol Biol Cell. 2004;15:2965–2977. doi: 10.1091/mbc.E03-08-0604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao Y, Dickerson JB, Guo F, Zheng J, Zheng Y. Rational design and characterization of a Rac GTPase-specific small molecule inhibitor. Proc Natl Acad Sci USA. 2004;101:7618–7623. doi: 10.1073/pnas.0307512101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cancelas JA, Lee AW, Prabhakar R, Stringer KF, Zheng Y, Williams DA. Rac GTPases differentially integrate signals regulating hematopoietic stem cell localization. Nat Med. 2005;11:886–891. doi: 10.1038/nm1274. [DOI] [PubMed] [Google Scholar]
- 49.Xiao GH, Gallagher R, Shetler J, Skele K, Altomare DA, Pestell RG, Jhanwar S, Testa JR. The NF2 tumor suppressor gene product, merlin, inhibits cell proliferation and cell cycle progression by repressing cyclin D1 expression. Mol Cell Biol. 2005;25:2384–2394. doi: 10.1128/MCB.25.6.2384-2394.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alexander K, Yang HS, Hinds PW. Cellular senescence requires CDK5 repression of Rac1 activity. Mol Cell Biol. 2004;24:2808–2819. doi: 10.1128/MCB.24.7.2808-2819.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bagheri-Yarmand R, Mandal M, Taludker AH, Wang RA, Vadlamudi RK, Kung HJ, Kumar R. Etk/Bmx tyrosine kinase activates Pak1 and regulates tumorigenicity of breast cancer cells. J Biol Chem. 2001;276:29403–29409. doi: 10.1074/jbc.M103129200. [DOI] [PubMed] [Google Scholar]
- 52.Royal I, Lamarche-Vane N, Lamorte L, Kaibuchi K, Park M. Activation of cdc42, rac, PAK, and rho-kinase in response to hepatocyte growth factor differentially regulates epithelial cell colony spreading and dissociation. Mol Biol Cell. 2000;11:1709–1725. doi: 10.1091/mbc.11.5.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moon A, Kim MS, Kim TG, Kim SH, Kim HE, Chen YQ, Kim HR. H-ras, but not N-ras, induces an invasive phenotype in human breast epithelial cells: a role for MMP-2 in the H-ras-induced invasive phenotype. Int J Cancer. 2000;85:176–181. doi: 10.1002/(sici)1097-0215(20000115)85:2<176::aid-ijc5>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 54.Edwards DC, Sanders LC, Bokoch GM, Gill GN. Activation of LIM-kinase by Pak1 couples Rac/Cdc42 GTPase signalling to actin cytoskeletal dynamics. Nat Cell Biol. 1999;1:253–259. doi: 10.1038/12963. [DOI] [PubMed] [Google Scholar]
- 55.Misra UK, Deedwania R, Pizzo SV. Binding of activated alpha2-macroglobulin to its cell surface receptor GRP78 in 1-LN prostate cancer cells regulates PAK-2-dependent activation of LIMK. J Biol Chem. 2005;280:26278–26286. doi: 10.1074/jbc.M414467200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Debnath J, Mills KR, Collins NL, Reginato MJ, Muthuswamy SK, Brugge JS. The role of apoptosis in creating and maintaining luminal space within normal and oncogene-expressing mammary acini. Cell. 2002;111:29–40. doi: 10.1016/s0092-8674(02)01001-2. [DOI] [PubMed] [Google Scholar]
- 57.Reginato MJ, Mills KR, Paulus JK, Lynch DK, Sgroi DC, Debnath J, Muthuswamy SK, Brugge JS. Integrins and EGFR coordinately regulate the proapoptotic protein Bim to prevent anoikis. Nat Cell Biol. 2003;5:733–740. doi: 10.1038/ncb1026. [DOI] [PubMed] [Google Scholar]
- 58.Miller FR. Xenograft models of premalignant breast disease. J Mammary Gland Biol Neoplasia. 2000;5:379–391. doi: 10.1023/a:1009577811584. [DOI] [PubMed] [Google Scholar]
- 59.Debnath J, Walker SJ, Brugge JS. Akt activation disrupts mammary acinar architecture and enhances proliferation in an mTOR-dependent manner. J Cell Biol. 2003;163:315–326. doi: 10.1083/jcb.200304159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Burstein HJ, Polyak K, Wong JS, Lester SC, Kaelin CM. Ductal carcinoma in situ of the breast. N Engl J Med. 2004;350:1430–1441. doi: 10.1056/NEJMra031301. [DOI] [PubMed] [Google Scholar]
- 61.Simpson PT, Reis-Filho JS, Gale T, Lakhani SR. Molecular evolution of breast cancer. J Pathol. 2005;205:248–254. doi: 10.1002/path.1691. [DOI] [PubMed] [Google Scholar]
- 62.Podgorski I, Linebaugh BE, Sameni M, Jedeszko C, Bhagat S, Cher ML, Sloane BF. Bone microenvironment modulates expression and activity of cathepsin B in prostate cancer. Neoplasia. 2005;7:207–223. doi: 10.1593/neo.04349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sloane BF, Sameni M, Podgorski I, Cavallo-Medved D, Moin K. Functional imaging of tumor proteolysis. Annu Rev Pharmacol Toxicol. 2006;46:301–315. doi: 10.1146/annurev.pharmtox.45.120403.095853. [DOI] [PubMed] [Google Scholar]
- 64.Salh B, Marotta A, Wagey R, Sayed M, Pelech S. Dysregulation of phosphatidylinositol 3-kinase and downstream effectors in human breast cancer. Int J Cancer. 2002;98:148–154. doi: 10.1002/ijc.10147. [DOI] [PubMed] [Google Scholar]
- 65.Knowles HJ, Phillips RM. Identification of differentially expressed genes in experimental models of the tumor microenvironment using differential display. Anticancer Res. 2001;21:2305–2311. [PubMed] [Google Scholar]
- 66.Worsham MJ, Pals G, Schouten JP, Miller F, Tiwari N, van Spaendonk R, Wolman SR. High-resolution mapping of molecular events associated with immortalization, transformation, and progression to breast cancer in the MCF10 model. Breast Cancer Res Treat. 2006;96:177–186. doi: 10.1007/s10549-005-9077-8. [DOI] [PubMed] [Google Scholar]
- 67.Liu H, Radisky DC, Bissell MJ. Proliferation and polarity in breast cancer: untying the Gordian knot. Cell Cycle. 2005;4:646–649. doi: 10.4161/cc.4.5.1674. [DOI] [PubMed] [Google Scholar]
- 68.Mostov K, Brakeman P, Datta A, Gassama A, Katz L, Kim M, Leroy P, Levin M, Liu K, Martin F, et al. Formation of multicellular epithelial structures. Novartis Found Symp. 2005;269:193–200. discussion 200–205, 223–230. [PubMed] [Google Scholar]
- 69.Wang RA, Vadlamudi RK, Bagheri-Yarmand R, Beuvink I, Hynes NE, Kumar R. Essential functions of p21-activated kinase 1 in morphogenesis and differentiation of mammary glands. J Cell Biol. 2003;161:583–592. doi: 10.1083/jcb.200212066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schurmann A, Mooney AF, Sanders LC, Sells MA, Wang HG, Reed JC, Bokoch GM. p21-Activated kinase 1 phosphorylates the death agonist bad and protects cells from apoptosis. Mol Cell Biol. 2000;20:453–461. doi: 10.1128/mcb.20.2.453-461.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bouillet P, Purton JF, Godfrey DI, Zhang LC, Coultas L, Puthalakath H, Pellegrini M, Cory S, Adams JM, Strasser A. BH3-only Bcl-2 family member Bim is required for apoptosis of autoreactive thymocytes. Nature. 2002;415:922–926. doi: 10.1038/415922a. [DOI] [PubMed] [Google Scholar]
- 72.Vadlamudi RK, Li F, Barnes CJ, Bagheri-Yarmand R, Kumar R. p41-Arc subunit of human Arp2/3 complex is a p21-activated kinase-1-interacting substrate. EMBO Rep. 2004;5:154–160. doi: 10.1038/sj.embor.7400079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vadlamudi RK, Li F, Adam L, Nguyen D, Ohta Y, Stossel TP, Kumar R. Filamin is essential in actin cytoskeletal assembly mediated by p21-activated kinase 1. Nat Cell Biol. 2002;4:681–690. doi: 10.1038/ncb838. [DOI] [PubMed] [Google Scholar]
- 74.Wu K, Jerdeva GV, da Costa SR, Sou E, Schechter JE, Hamm-Alvarez SF. Molecular mechanisms of lacrimal acinar secretory vesicle exocytosis. Exp Eye Res. 2006;83:84–96. doi: 10.1016/j.exer.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 75.Rodriguez A, Martinez I, Chung A, Berlot CH, Andrews NW. cAMP regulates Ca2+-dependent exocytosis of lysosomes and lysosome-mediated cell invasion by trypanosomes. J Biol Chem. 1999;274:16754–16759. doi: 10.1074/jbc.274.24.16754. [DOI] [PubMed] [Google Scholar]
- 76.Li Q, Ho CS, Marinescu V, Bhatti H, Bokoch GM, Ernst SA, Holz RW, Stuenkel EL. Facilitation of Ca(2+)-dependent exocytosis by Rac1-GTPase in bovine chromaffin cells. J Physiol. 2003;550:431–445. doi: 10.1113/jphysiol.2003.039073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu H, Radisky DC, Wang F, Bissell MJ. Polarity and proliferation are controlled by distinct signaling pathways downstream of PI3-kinase in breast epithelial tumor cells. J Cell Biol. 2004;164:603–612. doi: 10.1083/jcb.200306090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bokoch GM, Wang Y, Bohl BP, Sells MA, Quilliam LA, Knaus UG. Interaction of the Nck adapter protein with p21-activated kinase (PAK1) J Biol Chem. 1996;271:25746–25749. doi: 10.1074/jbc.271.42.25746. [DOI] [PubMed] [Google Scholar]
- 79.Lei M, Lu W, Meng W, Parrini MC, Eck MJ, Mayer BJ, Harrison SC. Structure of PAK1 in an autoinhibited conformation reveals a multistage activation switch. Cell. 2000;102:387–397. doi: 10.1016/s0092-8674(00)00043-x. [DOI] [PubMed] [Google Scholar]
- 80.Baugher PJ, Krishnamoorthy L, Price JE, Dharmawardhane SF. Rac1 and Rac3 isoform activation is involved in the invasive and metastatic phenotype of human breast cancer cells. Breast Cancer Res. 2005;7:R965–R974. doi: 10.1186/bcr1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cohen P. Protein kinases—he major drug targets of the twenty-first century? Nat Rev Drug Discov. 2002;1:309–315. doi: 10.1038/nrd773. [DOI] [PubMed] [Google Scholar]
- 82.Porchia LM, Guerra M, Wang YC, Zhang Y, Espinosa AV, Shinohara M, Kulp SK, Kirschner LS, Saji M, Chen CS, et al. 2-Amino-N-{4-[5-(2-phenanthrenyl)-3-(trifluoromethyl)-1H-pyrazol-1-yl]-phenyl} acetamide (OSU-03012), a celecoxib derivative, directly targets p21-activated kinase. Mol Pharmacol. 2007;72:1124–1131. doi: 10.1124/mol.107.037556. [DOI] [PubMed] [Google Scholar]
- 83.Yang Z, Bagheri-Yarmand R, Wang RA, Adam L, Papadimitrakopoulou VV, Clayman GL, El-Naggar A, Lotan R, Barnes CJ, Hong WK, et al. The epidermal growth factor receptor tyrosine kinase inhibitor ZD1839 (Iressa) suppresses c-Src and Pak1 pathways and invasiveness of human cancer cells. Clin Cancer Res. 2004;10:658–667. doi: 10.1158/1078-0432.ccr-0382-03. [DOI] [PubMed] [Google Scholar]
- 84.Cai D, Iyer A, Felekkis KN, Near RI, Luo Z, Chernoff J, Albanese C, Pestell RG, Lerner A. AND-34/BCAR3, a GDP exchange factor whose overexpression confers antiestrogen resistance, activates Rac, PAK1, and the cyclin D1 promoter. Cancer Res. 2003;63:6802–6808. [PubMed] [Google Scholar]
- 85.Yang Z, Rayala S, Nguyen D, Vadlamudi RK, Chen S, Kumar R. Pak1 phosphorylation of snail, a master regulator of epithelial-to-mesenchyme transition, modulates snail's subcellular localization and functions. Cancer Res. 2005;65:3179–3184. doi: 10.1158/0008-5472.CAN-04-3480. [DOI] [PubMed] [Google Scholar]
- 86.Rayala SK, Talukder AH, Balasenthil S, Tharakan R, Barnes CJ, Wang RA, Aldaz M, Khan S, Kumar R. P21-Activated kinase 1 regulation of estrogen receptor-alpha activation involves serine 305 activation linked with serine 118 phosphorylation. Cancer Res. 2006;66:1694–1701. doi: 10.1158/0008-5472.CAN-05-2922. [DOI] [PubMed] [Google Scholar]
- 87.Rayala SK, Molli PR, Kumar R. Nuclear p21-activated kinase 1 in breast cancer packs off tamoxifen sensitivity. Cancer Res. 2006;66:5985–5988. doi: 10.1158/0008-5472.CAN-06-0978. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.