Abstract
Understanding the conditions that favour the evolution and maintenance of antibiotic resistance is the central goal of epidemiology. A crucial feature explaining the adaptation to harsh, or ‘sink’, environments is the supply of beneficial mutations via migration from a ‘source’ population. Given that antibiotic resistance is frequently associated with antagonistic pleiotropic fitness costs, increased migration rate is predicted not only to increase the rate of resistance evolution but also to increase the probability of fixation of resistance mutations with minimal fitness costs. Here we report in vitro experiments using the nosocomial pathogenic bacterium Pseudomonas aeruginosa that support these predictions: increasing rate of migration into environments containing antibiotics increased the rate of resistance evolution and decreased the associated costs of resistance. Consistent with previous theoretical work, we found that resistance evolution arose more rapidly in the presence of a single antibiotic than two. Evolution of resistance was also more rapid when bacteria were subjected to sequential exposure with two antibiotics (cycling therapy) compared with simultaneous exposure (bi-therapy). Furthermore, pleiotropic fitness costs of resistance to two antibiotics were higher than for one antibiotic, and were also higher under bi-therapy than cycling therapy, although the cost of resistance depended on the order of the antibiotics through time. These results may be relevant to the clinical setting where immigration is known to be important between chemotherapeutically treated patients, and demonstrate the importance of ecological and evolutionary dynamics in the control of antibiotic resistance.
Keywords: evolutionary ecology, microbiology, infectious disease, antimicrobial resistance
1. Introduction
The resistance of pathogens to antimicrobials has become a worldwide problem, incurring both economic costs and loss of human lives (Evans et al. 2007). Hospitals are a hot spot for the evolution of antibiotic resistance since the high prevalence of pathogens and weakened hosts requires the use of high concentrations of antibiotics (Vincent et al. 1995; Bergmans et al. 1998). Antibiotic resistance is especially problematic in hospital-borne, or nosocomial (Garner et al. 1988), diseases where chronic infections with resistant pathogens such as Pseudomonas aeruginosa and Staphylococcus aureus are a major cause of morbidity (Vincent et al. 1995; Höffken & Niederman 2002).
The evolution and maintenance of antibiotic resistance depend on both the probability of resistance mutations and the pleiotropic fitness cost associated with resistance (Andersson 2006); the latter often manifested as slower growth rate or a reduced competitive ability in the absence of antibiotics (Andersson 2003). Although such fitness costs can be compensated for by mutation in other parts of the genome (Schrag et al. 1997; Maisnier-Patin & Andersson 2004), the probability of an antibiotic resistant strain reaching high frequency is likely to be negatively related to its fitness costs. As with the probability of resistance evolution, the fitness of a resistant mutant that goes to fixation will be a positive function of the mutation supply (Levin et al. 2000) rate, for which migration is likely to be a key determinant as higher migration brings more mutants. Under higher migration rates, many resistance mutations will arise simultaneously, allowing selection to fix the mutation with the lowest fitness cost. By contrast, if mutation supply rate is low, the less fit resistant mutation is likely to reach fixation before a better mutation appears. Given a rugged fitness landscape caused by epistasis, a costly resistance mutation is unlikely to subsequently be replaced by a less costly resistance mutation (Colegrave & Buckling 2005). It is also expected that the extent to which both the rate of evolution of resistance and fitness costs associated with resistance are affected by immigration depends on the harshness of the environment. More precisely, harsher environments reduce the rate of evolution of resistance (Gomulkiewicz et al. 1999).
The effects of environment quality on ecological and evolutionary dynamics have been studied using source–sink models (Pulliam 1988; Holt & Gomulkiewicz 2004). These models have been applied more recently to micro-organisms to explain the evolution of virulence (Sokurenko et al. 2006; Chattopadhyay et al. 2007). In short, a population is defined as a source if it is found in its ‘fundamental niche’, which is a set of environment conditions and resources that permit a population to persist, grow (i.e. growth rate exceeds death rate over some range of densities) and produce emigrants (Hutchinson 1978). A population outside the fundamental niche (a harsh environment) has a mean fitness lower than 1 (e.g. death rate exceeds the birth rate) and cannot be sustained without passive (Holt 1985/10) or active (Pulliam 1988) immigration and is therefore referred to as a ‘sink’ population. Recent source–sink theory predicts that given sustained immigration organisms can adapt to the sink environment and that the rate of adaptation will be faster in less harsh environments (Holt et al. 2004). Immigration facilitates adaptive evolution by exposing organisms to novel conditions and by providing a source of genetic variation that can be selected upon. Adaptation to a sink environment results in a characteristic signature of punctuated and rapid growth. Growth indicates that the new environment has been incorporated into the fundamental niche of the organism, and the new habitat now switches from the status of a sink to a source (Holt & Gaines 1992). Conversely, immigration can also constrain adaptive evolution, because gene flow can swamp locally favoured variants, and because immigrants can compete with better-adapted residents. This effect hinders adaptation by maintaining the population away from the local fitness optimum. In this study, we describe a test of the application of this theory to the evolution of resistance by experimentally investigating the effect of immigration rate and sink harshness on the evolution of antibiotic resistance and its associated pleiotropic fitness cost using the bacterial pathogen P. aeruginosa.
Using a fully factorial design, we created experimental source–sink systems in which we tested four daily immigration treatments (10, 1.0, 0.1 and 0% cells transferred from a non-evolving source population) on five antibiotic treatments. We used in the order of increasing harshness; streptomycin only, rifampicin only; two combinations of cycling therapy (alternating the two antibiotics; Raymond et al. 2001; Gruson et al. 2003); and bi-therapy (exposure to two antibiotics simultaneously; Sermet-Gaudelus et al. 2000; Châtain 2003). We replicated each treatment 12 times for a total of 240 independent sink microcosms. Each sink microcosm was initially inoculated with 104 bacterial cells and 1% was transferred daily to fresh media for 10 transfers (approx. 100 generations). Immigration from a non-evolving source was carried out at each transfer. Growth was monitored daily and recorded as absorbance reading score (OD600).
2. Material and methods
(a) Bacterial cultures
Two hundred and forty-four populations were initiated with approximately 104 cells of isogenic P. aeruginosa PA01 (Stover et al. 2000). Cultures were grown at 37°C in 150 μl of King's media B (KB) in 96-well microtitre plates. Every 24 hours, 1% of culture was transferred to a fresh microcosm for a total of 10 transfers, which would represent 106 cells from a fresh overnight culture of the ancestor. Growth was monitored as absorbance reading (600 nm).
(b) Immigration regimes
Immigration rates refer to the proportion of bacterial cells transferred from a fresh stationary phase culture of the ancestral clone grown overnight in unsupplemented KB to the selection lines. Four immigration treatments with 12 replicates each (i.e. 0%, no cells; 0.1%, 105 cells; 1.0%, 106 cells; 10%, 107 cells) were tested on each antibiotic treatment described below. The different immigration rates used in this experiment were prepared by serially diluting 15 μl of the overnight ancestral culture in 135 μl of KB. Then 15 μl of each 10-fold dilution was added to the selection lines following the initial daily transfer. We initially divided the 12 replicates into two to test an additional treatment, fluctuating immigration rates, but it proved to be non-significant and we therefore eliminated it from our statistical analyses.
(c) Antibiotic treatments
All microcosms, unless specified otherwise, were supplemented with one of four combinations of antibiotics: rifampicin (62.5 μg ml−1), streptomycin (16 μg ml−1), or rifampicin combined or alternated with streptomycin (62.5 and 16 μg ml−1, respectively). Concentrations of antibiotics creating sink conditions (population decline in the absence of immigration) were determined using a serial dilution of different concentration of antibiotics using the CLSI standards to determine minimal inhibitory concentrations (MICs). Bacterial cultures were also grown in KB for three days at the end of the experiment to test the heritability of antibiotic resistance.
(d) Statistical analyses
First, to analyse the evolution of resistance data we modelled the temporal dynamics of bacterial growth using a hierarchical linear mixed model (lme function of the nlme package using R v. 2.4 software). The response variable was optical density. Time (days since the beginning of the experiment) and treatment (drug, five levels; rate, two levels; immigration, four levels) were fixed effects. To account for nonlinear growth dynamics, we also included a quadratic effect of time as a fixed effect. Since all replicates were started under similar conditions, we constrained the model to a unique intercept. Replicates were taken to be random effects. We began by fitting the full model that included all fixed effects and their interactions, and then simplified it by sequential backward selection. We used ANOVA to compare the fit of different models. A variance function (varIdent of nlme library) that permits different variances for each level of a stratification variable (here treatment) was used to model heteroscedasticity when necessary. We also used the corAR1 function to model the autocorrelation structure in the time series. Significance of fixed effects was tested with F-tests. Differences between treatments were tested with pairwise comparisons using log likelihood ratio tests. Model parameters and confidence intervals were estimated with restricted maximum likelihood methods (Pinheiro & Bates 2000). To ensure that the rate of compensatory mutation was not significant in this analysis, we compared the optical density of bacterial populations in antibiotics just after they reached carrying capacity with the density at the 10th transfer using the non-parametric Kruskall–Wallis test. To analyse the evolution of MIC, we used a generalized linear model with relative optical densities of selection lines over five, twofold higher concentrations of antibiotics as the response variable and immigration and antibiotic treatments as fixed factors. We analysed the cost of resistance data using a generalized linear model with the relative optical density of selection line over that of the ancestral lines as response variable and immigration and antibiotic treatments as fixed factors. All analyses were performed using R v. 2.4 software, while figures were drawn using SigmaPlot 10.0.
3. Results
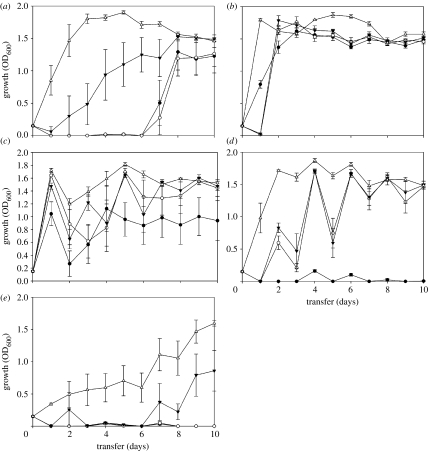
We first considered the rate of resistance evolution. The rate of evolution of resistance was determined indirectly from changes in the density of bacterial populations through time: resistant bacteria reach higher densities. The effect of immigration on the density of evolving populations was dependent on the type of antibiotic treatment and time (immigration×drug×time: F12,2335=11.231, p<0.0001). Essentially, increasing immigration rates had a significant positive effect on the density of P. aeruginosa in all environments (immigration: F3,2335=426.329, p<0.0001; figure 1). Antibiotic treatments also had an important effect on population density which varied significantly with the treatments (F4,2335=540.809, p<0.0001; figure 1). For example, rifampicin and streptomycin used in bi-therapy sustained lower bacterial densities than any other treatment despite significant positive effect of immigration and time (figure 1e). Furthermore, immigration increased significantly the MIC of antibiotics (immigration×drug: F9,160=17.82; p<0.0001). On average, MICs increased up to a 100-fold for rifampicin and more than 50-fold for streptomycin.
Figure 1.
Average density (±s.e.) of experimental lines of P. aeruginosa over 100 generations growing in a source–sink scenario where the sink environments contained: (a) rifampicin; (b) streptomycin; (c) cycling rifampicin with streptomycin; (d) cycling streptomycin with rifampicin; and (e) bi-therapy. The different migration rates used were: filled circle, 0%; open circle, 0.1%; filled down triangle, 1.0%; open up triangle, 10%.
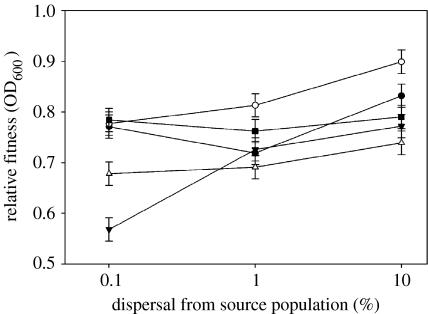
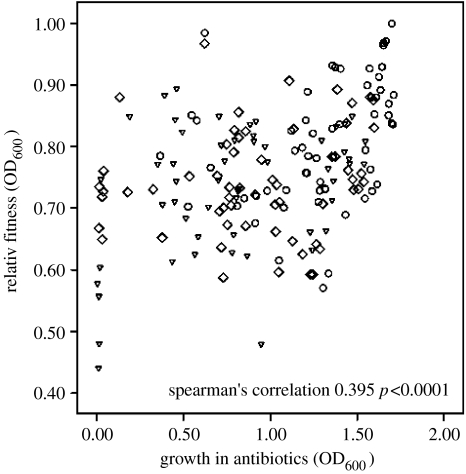
Second, we determined the antagonistic pleiotropic fitness costs associated with resistance evolution. We measured fitness costs as the mean carrying capacities through time of each population in the absence of antibiotics over approximately 30 generations. High immigration favoured the evolution of lower fitness costs in all drug treatments, although the magnitude of the change depended on the drug (cost×drug: F8,152=3.500, p<0.0001, figure 2). The harsher environments, i.e. bi-therapy and cycling therapy, imposed a greater pleiotropic cost on resistant populations. As hypothesized, pleiotropic fitness costs are negatively correlated with resistance level (mean growth rate during the course of the selection experiment), mainly due to the effect of immigration (Spearman's correlation=0.396, p<0.0001, figure 3). In other words, the cost of resistance decreased with increasing rate of resistance evolution. Since no increase in carrying capacity was detected following the evolution of resistance during the 30-generation experiment, the fixation of compensatory mutation was not important in explaining this result (Kruskall–Wallis test; H=1.032, p>0.10).
Figure 2.
The effect of immigration on experimental populations of P. aeruginosa grown in a source–sink scenario on the average antagonistic pleiotropic fitness cost (±s.e.) associated with different antibiotic treatments: open circles, streptomycin; filled circles, rifampicin; filled squares, cycling rifampicin with streptomycin; open triangles, cycling streptomycin with rifampicin; and filled triangles bi-therapy.
Figure 3.
The relative fitness of experimental lines of P. aeruginosa that have evolved resistance to antibiotics versus the average growth of each line over 300 generations of selection in antibiotics. Spearman's correlation is based on the pooled data, but data can be visualized as: triangles, 0.1%; lozenges, 1.0%; and circles, 10%.
4. Discussion
We have shown that both higher rates of migration and less harsh antibiotic sinks result in a more rapid evolution of resistance and, crucially, reduced fitness costs associated with the resistance. These data provide the first corroboration of predictions arising from recent source–sink models (Holt & Gomulkiewicz 2004). In particular we observed faster rates of adaptation in more benign environments (e.g. single versus two antibiotics) and with increasing rates of immigration (figure 1). However, theoretical models have yet to demonstrate how increasing immigration affects the costs of adaptation to novel or harsh environments.
Translated into a clinical setting where immigration is known to be important between chemotherapeutically treated patients (Bergmans et al. 1998; Massey et al. 2006), this means that in important nosocomial infections, such as those by P. aeruginosa, the immigration of susceptible bacteria established in an antibiotic-free reservoir (e.g. contaminated water; Trautmann et al. 2005) into transient secondary niches supplemented with antibiotic (e.g. respiratory tract of treated patient; Festini et al. 2006) can not only foster the rapid evolution of antibiotic resistance, but can also create resistant mutants with little or no fitness cost. It thus appears crucial to control the rate of immigration of infectious organisms in hospitals to preserve the efficacy of our treatments. The higher fitness cost fostered by harsh antibiotic treatments also highlights the importance of treatment choice. For example, the increased cost of resistance associated with bi-therapy is promising for the development of new therapy. This increased cost is probably due to the fact that two different mutations are required for resistance to this treatment as both antibiotics are affecting protein synthesis but with different target proteins (streptomycin targets the S12 protein of 30S ribosomal subunit (Carter et al. 2000), while rifampicin targets the β-subunit of RNA polymerase; Wehrli 1983). Whether the two antibiotics could have an additive qualitative effect on the mortality rate remains to be investigated. The overall effect of immigration is similar to that of mutation supply rate, which has been shown to both theoretically (Levin et al. 2000) and experimentally (Maisnier-Patin et al. 2002) impact the fitness of evolved resistant bacteria. Finally, immigration had a similar effect to that observed in clinically important populations of bacteria with elevated mutation rate, both increasing the rate of adaptation (Oliver et al. 2000; Perron et al. 2006).
The order in which bacteria were exposed to streptomycin and rifampicin also significantly affected the costs of resistance. Exposure to streptomycin before rifampicin resulted in a 15% higher cost than streptomycin on its own: a cost similar to that associated with bi-therapy. On the other hand, using rifampicin before streptomycin imposed a cost lower than bi-therapy and closer to that associated with rifampicin. Such considerable differences in fitness cost have important implications for the control of antibiotic resistance since a pleiotropic fitness cost of 1% could put a resistant mutant at a significant competitive disadvantage against a susceptible strain (Andersson 2006). Although, it was recently argued that the cycling of antimicrobials may actually be ineffective (Bergstrom et al. 2004), it is probable that the order in which antibiotics are used can affect the success of a therapy and the fitness cost associated with the resistance, an alternative that has not been explored yet in mathematical models.
The variation in costs of resistance observed between the immigration rates suggests that mechanisms underlying resistance are different and, therefore, likely to have different evolutionary trajectories. This study highlights the continuing importance of evolution and ecology in controlling the emergence and spread of emerging disease and antibiotic resistance in hospitals (Lipsitch et al. 2000; Antia et al. 2003; Colegrave & Buckling 2005; Sokurenko et al. 2006).
Acknowledgments
The authors would like to thank Daniel Schoen and Robert Holt for generously reviewing early drafts of the manuscript. This work was funded by the Royal Society and the Leverhulme Trust. G.G.P. is funded by the Clarendon Fund and Fonds Quebecois pour la Recherche sur la Nature et les Technologies, A.G. is funded by the National Science Engineering and Research Council of Canada and the Canada Research Chair Program and A.B. is funded by the Royal Society. The authors have no competing interests to declare. Correspondence and requests for materials should be addressed to G.G.P. (gabriel.guimond-perron@zoo.ox.ac.uk).
References
- Andersson D.I. Persistence of antibiotic resistant bacteria. Curr. Opin. Microbiol. 2003;6:452–456. doi: 10.1016/j.mib.2003.09.001. doi:10.1016/j.mib.2003.09.001 [DOI] [PubMed] [Google Scholar]
- Andersson D.I. The biological cost of mutational antibiotic resistance: any practical conclusions? Curr. Opin. Microbiol. 2006;9:461–465. doi: 10.1016/j.mib.2006.07.002. doi:10.1016/j.mib.2006.07.002 [DOI] [PubMed] [Google Scholar]
- Antia R, Regoes R.R, Koella J.C, Bergstrom C.T. The role of evolution in the emergence of infectious diseases. Nature. 2003;426:658–661. doi: 10.1038/nature02104. doi:10.1038/nature02104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmans D.C.J.J, Bonten M.J.M, Van Tiel F.H, Gaillard C.A, Van Der Geest S, Wilting R.M, De Leeuw P.W, Stobberingh E.E. Cross-colonisation with Pseudomonas aeruginosa of patients in an intensive care unit. Thorax. 1998;53:1053–1058. doi: 10.1136/thx.53.12.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstrom C.T, Lo M, Lipsitch M. Ecological theory suggests that antimicrobial cycling will not reduce antimicrobial resistance in hospitals. Proc. Natl Acad. Sci. USA. 2004;101:13 285–13 290. doi: 10.1073/pnas.0402298101. doi:10.1073/pnas.0402298101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter A.P, Clemons W.M, Brodersen D.E, Morgan-Warren R.J, Wimberly B.T, Ramakrishnan V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–348. doi: 10.1038/35030019. doi:10.1038/35030019 [DOI] [PubMed] [Google Scholar]
- Châtain P. Strategy of antibiotic therapy by various routes of administration in the initial colonization by Pseudomonas aeruginosa. Rev. Mal. Respir. 2003;20:S105–S112. [PubMed] [Google Scholar]
- Chattopadhyay S, Feldgarden M, Weissman S.J, Dykhuizen D.E, Van Belle G, Sokurenko E.V. Haplotype diversity in “source–sink” dynamics of Escherichia coli urovirulence. J. Mol. Evol. 2007;64:204–214. doi: 10.1007/s00239-006-0063-5. doi:10.1007/s00239-006-0063-5 [DOI] [PubMed] [Google Scholar]
- Colegrave N, Buckling A. Microbial experiments on adaptive landscapes. BioEssays. 2005;27:1167–1173. doi: 10.1002/bies.20292. doi:10.1002/bies.20292 [DOI] [PubMed] [Google Scholar]
- Evans H.L, et al. Cost of gram-negative resistance. Crit. Care Med. 2007;35:89–95. doi: 10.1097/01.CCM.0000251496.61520.75. doi:10.1097/01.CCM.0000251496.61520.75 [DOI] [PubMed] [Google Scholar]
- Festini F, Buzzetti R, Bassi C, Braggion C, Salvatore D, Taccetti G, Mastella G. Isolation measures for prevention of infection with respiratory pathogens in cystic fibrosis: a systematic review. J. Hosp. Infect. 2006;64:1–6. doi: 10.1016/j.jhin.2006.02.021. doi:10.1016/j.jhin.2006.02.021 [DOI] [PubMed] [Google Scholar]
- Garner J.S, Jarvis W.R, Emori T.G, Horan T.C, Hughes J.M. CDC Definitions for nosocomial infections, 1988. Am. J. Infect. Control. 1988;16:128–140. doi: 10.1016/0196-6553(88)90053-3. doi:10.1016/0196-6553(88)90053-3 [DOI] [PubMed] [Google Scholar]
- Gomulkiewicz R, Holt R.D, Barfield M. The effects of density dependence and immigration on local adaptation and niche evolution in a black-hole sink environment. Theor. Popul. Biol. 1999;55:283–296. doi: 10.1006/tpbi.1998.1405. doi:10.1006/tpbi.1998.1405 [DOI] [PubMed] [Google Scholar]
- Gruson D, Hilbert G, Vargas F, Valentino R, Bui N, Pereyre S, Bebear C, Bebear C.M, Gbikpi-Benissan G. Strategy of antibiotic rotation: long-term effect on incidence and susceptibilities of gram-negative bacilli responsible for ventilator-associated pneumonia. Crit. Care Med. 2003;31:1908–1914. doi: 10.1097/01.CCM.0000069729.06687.DE. doi:10.1097/01.CCM.0000069729.06687.DE [DOI] [PubMed] [Google Scholar]
- Höffken G, Niederman M.S. Nosocomial pneumonia: the importance of a de-escalating strategy for antibiotic treatment of pneumonia in the ICU. Chest. 2002;122:2183–2196. doi: 10.1378/chest.122.6.2183. doi:10.1378/chest.122.6.2183 [DOI] [PubMed] [Google Scholar]
- Holt R.D. Population dynamics in two-patch environments: some anomalous consequences of an optimal habitat distribution. Theor. Popul. Biol. 1985/10;28:181–208. doi:10.1016/0040-5809(85)90027-9 [Google Scholar]
- Holt R.D, Gaines M.S. Analysis of adaptation in heterogeneous landscapes: implications for the evolution of fundamental niches. Evol. Ecol. 1992;6:433–447. doi:10.1007/BF02270702 [Google Scholar]
- Holt R.D, Gomulkiewicz R. Conservation implications of niche conservatism and evolution in heterogeneous environments. In: Ferrière U, Dieckmann D, Couvet D, editors. Evolutionary conservation biology. Cambridge University Press; Cambridge, UK: 2004. pp. 244–264. [Google Scholar]
- Holt R.D, Barfield M, Gomulkiewicz R. Temporal variation can facilitate niche evolution in harsh sink environments. Am. Nat. 2004;164:187–200. doi: 10.1086/422343. doi:10.1086/422343 [DOI] [PubMed] [Google Scholar]
- Hutchinson G.E. Yale University Press; New York, NY: 1978. An introduction to population ecology. [Google Scholar]
- Levin B.R, Perrot V, Walker N. Compensatory mutations, antibiotic resistance and the population genetics of adaptive evolution in bacteria. Genetics. 2000;154:985–997. doi: 10.1093/genetics/154.3.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsitch M, Bergstrom C.T, Levin B.R. The epidemiology of antibiotic resistance in hospitals: paradoxes and prescriptions. Proc. Natl Acad. Sci. USA. 2000;97:1938–1943. doi: 10.1073/pnas.97.4.1938. doi:10.1073/pnas.97.4.1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisnier-Patin S, Andersson D.I. Adaptation to the deleterious effects of antimicrobial drug resistance mutations by compensatory evolution. Res. Microbiol. 2004;155:360–369. doi: 10.1016/j.resmic.2004.01.019. doi:10.1016/j.resmic.2004.01.019 [DOI] [PubMed] [Google Scholar]
- Maisnier-Patin S, Berg O.G, Liljas L, Andersson D.I. Compensatory adaptation to the deleterious effect of antibiotic resistance in Salmonella typhimurium. Mol. Microbiol. 2002;46:355–366. doi: 10.1046/j.1365-2958.2002.03173.x. doi:10.1046/j.1365-2958.2002.03173.x [DOI] [PubMed] [Google Scholar]
- Massey R.C, Horsburgh M.J, Lina G, Höök M, Recker M. The evolution and maintenance of virulence in Staphylococcus aureus: a role for host-to-host transmission? Nat. Rev. Microbiol. 2006;4:953–958. doi: 10.1038/nrmicro1551. doi:10.1038/nrmicro1551 [DOI] [PubMed] [Google Scholar]
- Oliver A, Cantón R, Campo P, Baquero F, Blázquez J. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science. 2000;288:1251–1253. doi: 10.1126/science.288.5469.1251. doi:10.1126/science.288.5469.1251 [DOI] [PubMed] [Google Scholar]
- Perron G.G, Zasloff M, Bell G. Experimental evolution of resistance to an antimicrobial peptide. Proc. R. Soc. B. 2006;273:251–256. doi: 10.1098/rspb.2005.3301. doi:10.1098/rspb.2005.3301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro J.C, Bates D.M. Springer; New York, NY: 2000. Mixed-effects models in S and S-PLUS. [Google Scholar]
- Pulliam H.R. Sources, sinks and population regulation. Am. Nat. 1988;132:652–661. doi:10.1086/284880 [Google Scholar]
- Raymond D.P, Pelletier S.J, Crabtree T.D, Gleason T.G, Hamm L.L, Pruett T.L, Sawyer R.G. Impact of a rotating empiric antibiotic schedule on infectious mortality in an intensive care unit. Crit. Care Med. 2001;29:1101–1108. doi: 10.1097/00003246-200106000-00001. doi:10.1097/00003246-200106000-00001 [DOI] [PubMed] [Google Scholar]
- Schrag S.J, Perrot V, Levin B.R. Adaptation to the fitness costs of antibiotic resistance in Escherichia coli. Proc. R. Soc. B. 1997;264:1287–1291. doi: 10.1098/rspb.1997.0178. doi:10.1098/rspb.1997.0178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sermet-Gaudelus I, Ferroni A, Gaillard J.L, Silly C, Chretiennot C, Lenoir G, Berche P. Antibiotic therapy in cystic fibrosis. Arch. Pediatr. 2000;7:645–656. doi: 10.1016/s0929-693x(00)80134-7. doi:10.1016/S0929-693X(00)80134-7 [DOI] [PubMed] [Google Scholar]
- Sokurenko E.V, Gomulkiewicz R, Dykhuizen D.E. Source–sink dynamics of virulence evolution. Nat. Rev. Microbiol. 2006;4:548–555. doi: 10.1038/nrmicro1446. doi:10.1038/nrmicro1446 [DOI] [PubMed] [Google Scholar]
- Stover C.K, et al. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. doi:10.1038/35023079 [DOI] [PubMed] [Google Scholar]
- Trautmann M, Lepper P.M, Haller M. Ecology of Pseudomonas aeruginosa in the intensive care unit and the evolving role of water outlets as a reservoir of the organism. Am. J. Infect. Control. 2005;33:S41–S49. doi: 10.1016/j.ajic.2005.03.006. doi:10.1016/j.ajic.2005.03.006 [DOI] [PubMed] [Google Scholar]
- Vincent J.L, Bihari D.J, Suter P.M, Bruining H.A, White J, Nicolas-Chanoin M.H, Wolff M, Spencer R.C, Hemmer M. The prevalence of nosocomial infection in intensive care units in Europe: results of the European Prevalence of Infection in Intensive Care (EPIC) study. J. Am. Med. Assoc. 1995;274:639–644. doi:10.1001/jama.274.8.639 [PubMed] [Google Scholar]
- Wehrli W. Rifampin: mechanisms of action and resistance. Rev. Infect. Dis. 1983;5:S407–S411. doi: 10.1093/clinids/5.supplement_3.s407. [DOI] [PubMed] [Google Scholar]