Abstract
Habitat fragmentation commonly causes genetic problems and reduced fitness when populations become small. Stocking small populations with individuals from other populations may enrich genetic variation and alleviate inbreeding, but such artificial gene flow is not commonly used in conservation owing to potential outbreeding depression. We addressed the role of long-term population size, genetic distance between populations and test environment for the performance of two generations of offspring from between-population crosses of the locally rare plant Ranunculus reptans L. Interpopulation outbreeding positively affected an aggregate measure of fitness, and the fitness superiority of interpopulation hybrids was maintained in the second offspring (F2) generation. Small populations benefited more strongly from interpopulation outbreeding. Genetic distance between crossed populations in neutral markers or quantitative characters was not important. These results were consistent under near-natural competition-free and competitive conditions. We conclude that the benefits of interpopulation outbreeding are likely to outweigh potential drawbacks, especially for populations that suffer from inbreeding.
Keywords: interpopulation gene flow, intraspecific hybridization, fitness, heterosis, hybrid breakdown, coadapted gene complexes, genomic divergence
1. Introduction
Small and isolated populations often have genetic problems that lead to reduced fitness (Fischer et al. 2000; Reed & Frankham 2003; Spielman et al. 2004; Reed 2005). These problems include inbreeding depression, resulting from reproduction among close relatives, and increased load from fixed mutations owing to genetic drift (Willi et al. 2005). Drift may also reduce mate availability by causing unequal sex ratio or small effective number of S alleles in plants with self-incompatibility systems (e.g. Wagenius et al. 2007). Genetic problems combine with disruption of ecological interactions and demographic and environmental stochasticity to increase the extinction risk of small and isolated populations (Newman & Pilson 1997; Saccheri et al. 1998). Genetic threats are especially severe because they do not remain constant for a given population size; rather, they increase with time owing to continuous accumulation of genetic load and inbreeding, leading to a so-called mutational meltdown (Lynch & Gabriel 1990).
An obvious strategy to counteract such genetic erosion is to stock small populations with individuals from surrounding populations. Experimental studies have documented the rapid spread of immigrant genomes within inbred populations due to heterosis (Richards 2000; Ebert et al. 2002; Saccheri & Brakefield 2002) and even small amounts of artificial gene flow into natural populations can quickly reduce inbreeding depression and fitness reductions from fixed genetic load (Westemeier et al. 1998; Madsen et al. 1999; Stokstad 2005). Nevertheless, conservation biologists hold deep reservations about artificial gene flow, mostly because they fear breakdown of coadapted gene complexes and of local adaptation (Tallmon et al. 2004; Edmands 2007). These potentially negative consequences of gene flow and interpopulation outbreeding have been termed outbreeding depression (Templeton 1986). Hence, the main question that still needs to be answered is whether the net effect of heterosis and outbreeding depression is positive or negative.
Before genetic restoration can become accepted in conservation management, several fundamental issues must be resolved. First, how does population size affect the fitness consequences of gene flow in the long term? So far, long-term studies focusing on the effect of interpopulation outbreeding have not considered the size of focal populations (e.g. Edmands 1999; Fenster & Galloway 2000). These studies report outbreeding depression in some interpopulation hybrids. However, in small populations, heterosis can be so strong that outbreeding depression does not lead to a negative net effect (e.g. Heschel & Paige 1995; Willi & Fischer 2005). However, studies which account for population size have not included assessments of later generations.
Second, little is known about the impact of genetic distance between the donor and the recipient populations for genetic restoration in the long term. Increased genetic distance at neutral markers between populations may reflect differentiation in coadapted gene complexes arising from positive epistasis. According to theory, interpopulation outcrossing destroys those epistatic interactions. Although the breakdown may be masked by heterosis (dominance or overdominance) in the first (F1) generation, it becomes more obvious in the second (F2) and later generations when homozygosity increases (Lynch 1991). Therefore, it is important to test whether genetic distance to the source population influences the outcome of artificial gene flow over the long term. The best predictor of the success of interpopulation outcrossing may be the extent of genetic differentiation in potentially adaptive traits between target and partner populations, rather than the geographical or the genetic distances in neutral markers.
Finally and most importantly, for those managing small populations, one needs to know whether the net effect of artificial gene flow depends on environmental conditions. This seems possible because both heterosis and outbreeding depression might differ among environments, as occurs with inbreeding depression (Dudash 1990).
We tackled these questions by crossing plants of 14 populations of the locally endangered, tetraploid and self-incompatible Ranunculus reptans. This plant occurs in distinct populations of varying size at Lake Constance (Central Europe). In a previous study, we found that long-term small populations with low gene diversity suffered from reduced mean fitness, caused by three types of genetic problems. Pairs of plants from small populations were more often cross-incompatible than pairs of plants from large populations, indicating reduced S allele numbers in those populations. Furthermore, plants from small populations experienced higher fitness reductions due to inbreeding depression and increased fixed drift load, reducing female fertility (Willi et al. 2005). Finally, small populations benefit more than large populations in the first generation after interpopulation outbreeding (Willi & Fischer 2005).
In the present study, we crossed plants from populations of differing allozyme genetic distance and genetic distance in quantitative traits over two generations. We reared the offspring under near-natural benign and stressful conditions in an outdoor common garden experiment. Our focus was on heterosis and gene coadaptation, independent of possible adaptation to local conditions. We addressed the following questions: (i) do F1 and F2 offspring of interpopulation crosses show outbreeding depression or outbreeding vigour? (ii) do small and inbred populations enjoy a higher fitness benefit from interpopulation outbreeding? (iii) does F1 and F2 interpopulation hybrid performance depend on genetic distance between target and partner population? (iv) is genetic distance in quantitative traits a better predictor of interpopulation hybrid performance than marker distance? and (v) how does competition modulate these relationships?
2. Material and methods
(a) Study species and plant material
Ranunculus reptans is a clonal plant with a gametophytic self-incompatibility system. The species grows in a fragmented band on the shores of Lake Constance in Central Europe. The persistence of these populations is associated with the regular occurrence of floods; R. reptans is a weak competitor but more flood tolerant than its competitors. The species probably had a fragmented distribution even before shoreline development began to affect it in the twentieth century. Our study populations covered surface areas from 40 to 2000 m2 in 2003 (Willi et al. 2005). Census population size, calculated as surface area times a measure of plant density, was positively correlated with Nei's gene diversity, Hs, assessed on eight allozyme loci. Hs is a measure of long-term population size under mutation–drift balance (Willi et al. 2005).
In spring 2002, we collected 173 plants from 14 populations of R. reptans; 12 populations at Lake Constance, 1 on the Bernina Pass and 1 South of the Alps on Lago Maggiore, Switzerland (Willi & Van Buskirk 2005; Willi et al. 2005). At each site, 14 individuals were collected at 5 m intervals along two parallel transects separated by 5 m. In six populations, the band of R. reptans was so short that we could only sample 8–12 individuals. After collection, plants were grown in separate tubs in a growth room. Five out of 173 field-collected plants died during propagation.
(b) Crossing design
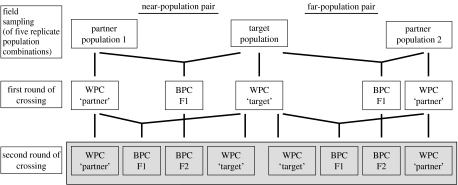
The crossing design simultaneously produced F1 and F2 interpopulation hybrid offspring between population pairs varying in the degree of genetic differentiation (figure 1). Two partner populations were crossed with the same target population. One population pair was genetically similar and the other was genetically dissimilar. The population combinations were chosen based on the knowledge of genetic marker distance at eight allozyme loci of this autotetraploid species (Willi et al. 2007). The FST values between the near-pair and between the far-pair were about 0.05 and 0.15, respectively (table 1). The experiment was replicated over five such three-population combinations, totalling 10 population pairs. All population combinations consisted of three different populations except in one case where a population was used as a partner population once in a genetically similar (near-) pair, and once in a genetically dissimilar (far-) pair. For all the population pairs, we also knew the genetic distance in seven morphological and life-history traits, measured as QST (Willi et al. 2007).
Figure 1.
Diagram illustrating the crossing design. Crosses were established over two generations. In the first round of crosses, two partner populations were crossed with a single target population (BPC F1), and within-population crosses were performed (WPC). FST was about 0.05 for the ‘near’ population pair and 0.15 for the ‘far’ population pair. In the second round, plants from the first round were crossed to produce within-population crosses (WPCtarget, WPCpartner), a new generation of F1 between-population crosses (BPC F1) and recombinant F2 between-population crosses (BPC F2). The offspring from the second round, enclosed within the grey shaded box, were reared in the outdoor competition experiment. This crossing design was applied to five replicate population combinations.
Table 1.
Genetic distance between target populations of Ranunculus reptans and their partner populations based on eight allozymes (FST), on seven quantitative traits (QST) and geographical separation by region. Gene diversity (Hs) of all target and partner populations reflects long-term population size under the assumption of mutation–drift balance.
| population combination | near-population pair | far-population pair | partner population | ||||||
|---|---|---|---|---|---|---|---|---|---|
| FST | QST | regiona | Hs | FST | QST | regiona | Hs | Hs | |
| 1 | 0.0480 | 0.0000 | LC-O | 0.4807 | 0.1525 | 0.2514 | LC | 0.3856 | 0.4444 |
| 2 | 0.0542 | 0.1528 | LC | 0.4792 | 0.1426 | 0.3028 | CH | 0.4548 | 0.4214 |
| 3 | 0.0473 | 0.2857 | LC | 0.4081 | 0.1378 | 0.1598 | LC | 0.4346 | 0.4710 |
| 4 | 0.0694 | 0.1433 | LC-B | 0.3997 | 0.1728 | 0.0983 | LC | 0.4847 | 0.4099 |
| 5 | 0.0676 | 0.1564 | LC-U | 0.4346 | 0.2391 | 0.2433 | CH | 0.4237 | 0.4909 |
Abbreviations for crosses between populations: LC, crosses between populations situated at two different basins of Lake Constance; LC-B, LC-O and LC-U, crosses between populations situated within the same lake basin (the three lake basins are Überlinger See (B), Obersee (O) and Untersee (U)); CH, crosses between populations situated in two different lake systems of Switzerland.
Within-population crosses (WPC) and first- and second-generation between-population crosses (BPC F1 and BPC F2) were established over two generations (figure 1). In the first round of crossing, each of the field-collected plants was crossed with two randomly chosen plants from the same population and with one plant from the other population of a population pair. Each cross was performed reciprocally; that is, each plant served as a pollen donor and a pollen recipient, leading to two maternal seed families per cross combination. We germinated the seeds, haphazardly chose one seedling per seed family and raised it in an outdoor garden. After two months, we randomly chose 24 plants per population and population pair and used them for the production of the next generation of seed families. We ensured that each field-collected genotype was represented at least once as mother and once as father (pollen donor) in a cross group so that no initial genetic variation was lost. For F1 between-population crosses, we also ensured that 12 plants had a mother from one population and 12 had a mother from the other population of a population pair.
In the second round of crossing, each of the 24 within-population cross plants was crossed reciprocally with another plant of the same group (WPC). To generate new F1 between-population crosses (BPC F1), 12 randomly chosen within-population cross plants from each of the two populations of a pair were crossed reciprocally. To produce recombinant F2 interpopulation hybrid individuals (BPC F2), first-round between-population cross plants were crossed reciprocally. This led to a total of 749 reciprocal crosses: 10 population pairs×(24 WPCpartner+24 BPC F1+24 BPC F2)+5 target populations×24 WPCtarget−24 WPCpartner (because one population was used twice)−58 crosses (some plants produced no flowers and could not be crossed)−9 crosses lost due to an error. Crosses were arranged among randomly chosen plants of a cross group. However, the random choice was restricted in the second round of crossing so that each F2 cross was the product of four different grandparental field-collected plants. Seeds were harvested one month after crossing, and the number of developed seeds and undeveloped ovules were counted.
(c) Outdoor common garden experiment
We reared two replicate seedlings of each cross in two environments, one free of competitors and one together with the natural competitor Agrostis stolonifera L. On 8 June 2004, seeds were sown into trays (3×3×5 cm) containing a 3 : 1 mixture of horticultural soil and sand. Trays were distributed in the green house and their locations were randomized weekly. Six weeks after germination began, we randomly chose four individuals of each seed family and planted each individual separately into a tub (18×13×5 cm) filled with a 1 : 2 mixture of horticultural soil and sand. Because some seed families produced less than four seedlings, the experiment included 2010 instead of 2996 plants. Directly after planting, we sowed seeds of the grass A. stolonifera into two of the four tubs per seed family. The seeds of A. stolonifera germinated within 7 days and formed a dense stand, similar to densities in the natural habitat of R. reptans. We randomly assigned each tub to a position within one of six outdoor beds covered with 50% shade cloth. The plants were watered daily, unless it rained. We checked survival after one and two weeks, and if a plant had died, it was replaced by another representative of the same seed family. A total of 41 plants were replaced because they died after heavy rainfall on the first day in the garden, and a further 34 plants were replaced during the first two weeks. We re-randomized the positions of the tubs one month after the start of the outdoor experiment.
(d) Measuring fitness
We estimated fitness of each maternal seed family in both environments with multiplicative measures of clonal and sexual performance of the progeny. Clonal growth is especially important for R. reptans in years of high lake water levels, when plants do not flower and exclusively reproduce via above-ground stolons with roots on some nodes. Eight weeks after transplantation (20–24 September 2004), we counted the number of rooted rosettes, flowers, flower buds and fruits of each plant. Clonal fitness was calculated as seed set (proportion of maternal ovules that turned into seeds)× germination rate (number of seedlings after six weeks divided by the number of sown seeds)× survival×number of rooted rosettes. Sexual fitness was calculated as seed set×germination rate×survival×number of flowers, flower buds and fruits. We log-transformed fitness measures and measures of growth. Seed set and germination rate underwent an angular transformation (Sokal & Rohlf 1995).
(e) Statistical analysis
Because multiplicative sexual and clonal fitness were positively correlated (n=2260, r=0.64, p<0.0001), our dependent variable was the first component of a principal component analysis (PCA) on sexual and clonal fitness (explaining 82% of the variation in the two measures). We applied a three-level mixed model analysis to test for differences in aggregate fitness (Singer 1998; MIXED procedure in SAS; SAS Institute 2002). The random subjects were population pair and maternal seed family nested within population pair. Fixed effects included genetic distance between pairs of populations (near, far; predictor on population pair level), cross type (WPCtarget, WPCpartner, BPC F1, BPC F2; predictor on seed family level) and interspecific competition (present, absent; predictor on plant level). Gene diversity, Hs, of target and partner populations was a predictor on the level of population pair. Denominator degrees of freedom for testing fixed effects were estimated using Satterthwaite's approximation (SAS Institute 2002). We randomly split the within-population crosses of the five partner populations into two groups to obtain independent within-population cross antagonists. Similarly, we randomly split the between-population crosses of a particular population pair into two groups and assigned them to the gene diversity of the target population, and to the gene diversity of the partner population, respectively. Two types of planned contrasts were performed. The first, a test for heterosis, compared the fitness of F1 and F2 between-population crosses with the fitness of within-population crosses of target and partner populations, the so-called midparental mean. A second contrast compared the fitness of F1 and F2 between-population crosses. This contrast tests for outbreeding depression due to a breakdown of coadapted gene complexes (Lynch 1991).
3. Results
(a) Differences among cross types
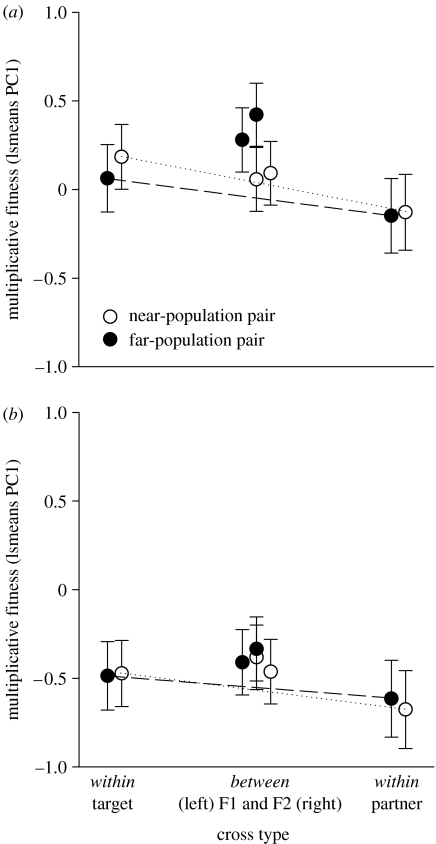
Aggregate fitness (PC1) of between-population crosses was higher than fitness of within-population crosses, as indicated by the significant contrast for within- versus between-population crosses (table 2). Figure 2 illustrates the average fitness superiority of between- over within-population crosses in the absence and presence of interspecific competitors. This fitness superiority did not appear during a particular life stage, but accumulated over several stages. The F2 between-population crosses were not significantly different from F1 between-population crosses (table 2, contrast of BPC F1 versus BPC F2). Overall, we found evidence for heterosis and no significant evidence for outbreeding depression.
Table 2.
Results of generalized linear mixed models using restricted maximum likelihood, including the fixed effects of genetic distance (near- versus far-population pair), cross type (WPCtarget, WPCpartner, BPC F1, BPC F2), competition (with and without a grass competitor) and interaction terms including gene diversity (Hs), on the first component of a PCA on sexual and clonal fitness (significant effects with p<0.05 are in italics), and on the life stage fitness components of seed set, germination rate and survival combined with growth of rooted rosettes, and production of flowers and flower buds. The denominator degrees of freedom (d.f.Den) for testing the fixed effects were estimated by Satterthwaite's approximation.
| source of variationa | PC1-multiplicative fitness | fitness componentsb | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| seed set | germination rate | survival*growth | flowers and buds | |||||||||
| d.f.Num | d.f.Den | F | p | d.f.Den | F | d.f.Den | F | d.f.Den | F | d.f.Den | F | |
| fixed effects: | ||||||||||||
| genetic distance | 1 | 8.25 | 0.12 | 0.743 | 8.13 | 0.04 | 9.29 | 0.20 | 8.46 | 0.23 | 8.25 | 0.44 |
| cross type | 3 | 21.3 | 2.66 | 0.074 | 731 | 1.40 | 666 | 2.59 | 18.1 | 2.22 | 12.2 | 1.55 |
| dist×98cross type | 3 | 21.2 | 0.74 | 0.540 | 731 | 1.22 | 666 | 2.29 | 17.6 | 2.56 | 12 | 0.83 |
| competition | 1 | 8.79 | 50.92 | <0.001 | 8.8 | 148.71*** | 8.99 | 28.25*** | ||||
| dist×comp | 1 | 8.62 | 0.17 | 0.693 | 8.37 | 0.65 | 8.66 | 0.45 | ||||
| cross type×comp | 3 | 730 | 0.30 | 0.827 | 491 | 0.23 | 15.4 | 0.19 | ||||
| dist×cross type×comp | 3 | 719 | 1.28 | 0.279 | 484 | 1.10 | 14.5 | 0.86 | ||||
| Hs×CT | 4 | 69 | 3.80 | 0.008 | 590 | 2.05 | 332 | 5.46*** | 70.4 | 1.34 | 36.6 | 1.44 |
| Hs×CT×comp | 4 | 521 | 3.16 | 0.014 | 270 | 1.25 | 67.6 | 3.19* | ||||
| contrasts | ||||||||||||
| BPC vs WPC | 1 | 26.2 | 6.55 | 0.017 | 733 | 2.36 | 669 | 0.88 | 25.7 | 2.09 | 14.8 | 4.36 |
| BPC F1 vs BPC F2 | 1 | 15.6 | 0.05 | 0.825 | 730 | 0.00 | 665 | 0.24 | 13.1 | 1.36 | 9.4 | 0.27 |
| BPC vs WPC×dist | 1 | 25.6 | 1.39 | 0.250 | 731 | 3.32 | 667 | 1.13 | 23.9 | 3.45 | 14 | 1.75 |
| BPC F1 vs BPC F2×dist | 1 | 15.5 | 0.45 | 0.512 | 730 | 0.26 | 665 | 4.71* | 12.6 | 0.00 | 9.19 | 0.51 |
| BPC vs WPC×comp | 1 | 743 | 0.43 | 0.514 | 493 | 0.39 | 22 | 0.04 | ||||
| BPC F1 vs BPC F2×comp | 1 | 715 | 0.30 | 0.583 | 490 | 0.02 | 11.1 | 0.35 | ||||
| source of variationa | d.f. | VAR | Χ2 | VAR | Χ2 | VAR | Χ2 | VAR | Χ2 | VAR | Χ2 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| random effects:b,c | |||||||||||
| population pair | 1 | 0.079 | 5.0* | 0.010 | 12.9*** | 0.003 | 2.5 | 0.016 | 2.7 | 0.008 | 2.2 |
| PP×CT | 1 | 0.008 | 0.2 | 0 | 0 | 0.002 | 0.0 | 0.004 | 0.9 | ||
| PP×com | 1 | 0.023 | 5.4* | 0.006 | 2.4 | 0.004 | 2.7 | ||||
| PP×CT×comp | 1 | 0 | 0 | 0.001 | 0.1 | ||||||
| seed family (PP) | 1 | 0.563 | 199.1*** | 0.197 | 89.7*** | 0.035 | 31.6*** | ||||
| seed family (PP)×comp | 1 | 0.034 | 1.0 | 0.005 | 0.1 | 0 | |||||
| residual | 0.836 | 0.171 | 0.158 | 0.455 | 0.182 |
Abbreviations for sources of variation: dist, genetic distance; CT, cross type; comp, competition; BPC F1/F2, between-population crosses of the first/second generation; WPC, within-population crosses of target and partner population; PP, population pair.
Significance of tests for life stage fitness components and of likelihood ratio tests for random effects: * p<0.05, ** p<0.01, *** p<0.001; n=2260 for PC1-multiplicative fitness, n=749 for seed set, n=682 for germination rate, n=1996 for survival×growth of rooted rosettes, and n=1848 for production of flowers and buds.
Random effect statistics include estimates of variance components (VAR) and chi-square values of likelihood ratio tests (Χ2).
Figure 2.
Mean aggregate fitness of within-population crosses and first (F1) and second (F2) generation between-population crosses of Ranunculus reptans in (a) the absence and (b) the presence of competition. Least square means are based on crosses for 10 population pairs of two genetic distance classes, 5 near-population pairs (open circles) and 5 far-population pairs (filled circles). Dashed lines connect within-population crosses of target and partner populations and predict between-population cross performance at half-distance if all gene effects were additive. Aggregate fitness is the first principal component of a PCA on multiplicative sexual and clonal fitness. Error bars depict ±1 s.e.
(b) The role of long-term population size
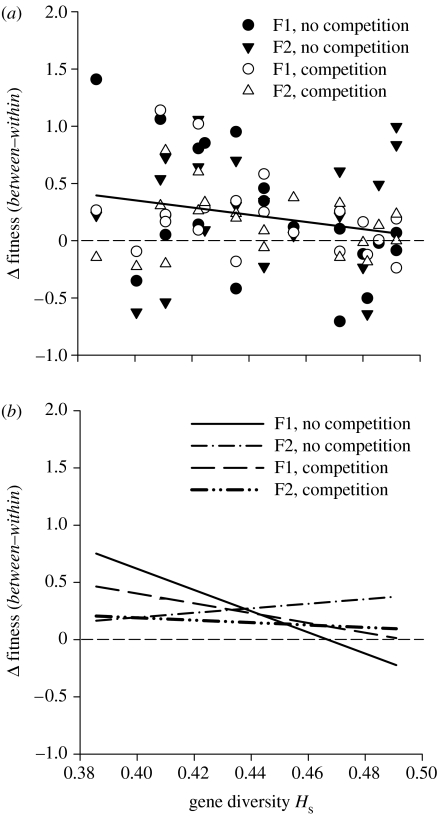
Small populations enjoyed a higher benefit from interpopulation outbreeding than large populations, in both the F1 and F2 generations, as indicated by the significant gene diversity-by-cross type interaction for aggregate fitness (table 2). Figure 3a illustrates that the fitness benefit of interpopulation outbreeding, relative to crosses within the population, depended on the population's gene diversity. The slope of the regression line is estimated to be −3.05 with lower and upper 95% confidence limits of −5.94 and −0.17. For the five populations with the lowest Hs, the fitness improvement from outbreeding (relative to within-population crosses and based on transformed data) in the F2 generation was +59% (+109%) for clonal fitness and +181% (+84%) for sexual fitness in the absence (presence) of competition. The corresponding values for the five largest populations were +18% (+3%) for clonal fitness and +123% (+18%) for sexual fitness. Analysis of the four life stages of maternal seed set, germination rate, survival and growth and flower production revealed that much of the significant interaction arose at the stage of germination (table 2).
Figure 3.
(a) Difference in aggregate fitness of between-population hybrid crosses and within-population crosses (of target or partner population) of Ranunculus reptans decreased linearly with gene diversity, a measure of long-term population size under mutation–drift balance. Long-term small populations enjoyed a greater fitness benefit from interpopulation outbreeding. (b) The fitness benefit through interpopulation outbreeding for the smallest populations occurred in the first generation of between-population crosses, and the second generation of between-population crosses when interspecific competition was present but not when there was no competition. Aggregate fitness is the first principal component from a PCA on multiplicative sexual and clonal fitness. Symbols in (a) and regression lines in (b) represent the two hybrid generations, F1 and F2, and the competition environment under which the plants were raised.
The pattern of higher fitness benefit from outbreeding for populations with low gene diversity was present for the first generation of between-population crosses and in the second generation when there was interspecific competition (figure 3b). However, under no competition, populations with low gene diversity did not experience a higher fitness benefit in the F2 than populations with higher gene diversity. This is reflected by a significant Hs-by-cross type-by-competition interaction for aggregate fitness and the fitness component of flower and flower bud production (table 2).
(c) Genetic distance between target and partner population
Genetic distance between crossed populations had no significant effect on aggregate fitness of within- versus between-population crosses (table 2, contrast of BPC versus WPC×genetic distance; figure 2). Furthermore, the fitness of F1 and F2 between-population crosses did not show a different response to genetic distance (table 2, contrast of BPC F1 versus BPC F2×genetic distance).
For the fitness component of germination, there was a significant interaction between genetic distance and F1 versus F2 between-population crosses. F1 between-population crosses of near- and far-population pairs had about equally high germination rates (least square means of transformed data±SE: near 0.82±0.05, far 0.79±0.05). However, F2 between-population crosses had lower germination when populations were genetically similar (0.72±0.05) and higher rates when populations were genetically dissimilar (0.86±0.05).
Genetic distance in quantitative traits (QST) also had no impact on F1 and F2 interpopulation hybrid fitness (contrasts of cross types×genetic distance, p>0.4 for aggregate fitness).
(d) The role of the environment
Plants raised in competition with the grass A. stolonifera had significantly lower survival and growth of rooted rosettes, and lower flower production (table 2). Even though there was a strong effect of competition on aggregate fitness (figure 2), the contrasts show that competition did not significantly interact with cross type, suggesting that competitive stress did not influence the general pattern of heterosis and no significant effect of outbreeding depression (table 2, contrasts of cross type×competition).
4. Discussion
Positive effects of interpopulation outbreeding outweighed potential negative effects, especially for small and inbred populations. The results highlight the crucial role of population size in the long-term effect of interpopulation outbreeding. The fitness benefit was due to heterosis, and this effect is likely to accelerate population growth rates in small and inbred populations for several generations.
In the current study, the fitness benefit of interpopulation crosses arose from heterosis accumulating over several life stages. In an earlier study of one generation of interpopulation outbreeding, the fitness benefit through outbreeding occurred–-apart from heterosis–-also due to an increase in cross-compatibility among pairs of plants (Willi & Fischer 2005). Our experiment is based on relatively outcrossed plants originating from four different field-collected grandparents, which diminished differences in cross-compatibility between within- versus between-population crosses.
We detected little evidence for outbreeding depression in either F1 or F2 crosses, at least when the focal population had been inbred. Models suggest that heterosis due to dominance or overdominance may compensate for the negative consequences of interrupting epistatic interactions in the F1, but that further reproduction among hybrid outbred plants should diminish the effect of heterosis as heterozygosity declines (Lynch 1991). This argument is weaker for the autotetraploid R. reptans than for diploid species because the decay of heterozygosity proceeds more slowly in autotetraploids (Bever & Felber 1992). Hence, heterosis may continue to blur the potential breakdown of interacting genes in the F2. However, if divergent positive epistatic interactions were consistently strong in R. reptans populations, we should have detected at least some general pattern of decline in fitness by the F2. When R. reptans was hybridized with a closely related congener, Willi & Van Buskirk (2005) measured outbreeding depression already in the first hybrid generation.
Although there was a significant positive effect of interpopulation outbreeding on fitness, a few hybrid crosses performed less well than the midparental mean, and small populations occasionally showed no fitness benefit from outcrossing. This is the sort of variation one expects to find in comparisons of natural populations and should be anticipated by managers when artificial gene flow is applied. It may be possible to minimize the risk of a negative outcome for any particular population by using multiple individuals from several source populations for artificial gene flow.
Our results document that marker genetic distance and genetic distance in quantitative traits, FST and QST, cannot predict either hybrid vigour or outbreeding depression. It is not surprising that the two measures of genetic distance gave the same result because pairwise FST and QST values are correlated in this metapopulation (Willi et al. 2007). These findings show that source populations for artificial gene flow with allozyme FST values between 0.05 and 0.15 are unlikely to cause substantial outbreeding depression due to a breakdown of coadapted gene complexes. This also means that genetic coadaptation is likely to be similar in populations on a regional scale.
A few other studies have estimated the long-term fitness consequences of interpopulation outbreeding over genetic or geographical distances in natural populations (Edmands 1999; Fenster & Galloway 2000; Schiffer et al. 2006; Erickson & Fenster 2006), and they indicate that outbreeding depression does not follow simple theoretical predictions. Edmands (1999) found evidence for breakdown of epistatic interactions in the F2 hybrid generation of a copepod with increasing geographical and genetic distance of the partner population, but fitness rebounded in the F3 generation. Fenster & Galloway (2000) reported no outbreeding depression in F2 hybrid plants. However, cumulative fitness in the F3 showed a general decline compared with the mean of midparental crosses and F1 hybrid crosses, independent of geographical distance. Schiffer et al. (2006) detected hybrid breakdown in fitness-related traits in two Drosophila species. In one species (but not the other), the breakdown was restricted to crosses between the geographically most separated populations. Our study adds the important conclusion that the net benefit of interpopulation outbreeding depends critically on how small and inbred populations are.
The fact that consequences of outcrossing were broadly similar in treatments with and without competitors makes our results especially convincing. Unlike inbreeding, for which the impact can depend on the environment (Dudash 1990; Armbruster & Reed 2005), outbreeding seems to be beneficial for small and inbred populations regardless of the environment. The environments chosen for this study reflect natural conditions during a good part of the year. When the lake water level drops after snow melt finishes during mid-summer, plants close to the water live in an environment with little competition, whereas those a few metres inland occur in relatively thick wetland vegetation.
Our results show that the scepticism towards artificial gene flow in conservation is not justified in those cases where populations are small and show significant fitness reductions due to inbreeding depression, fixed mutational load or reduced cross-compatibility. If source material comes from populations within the same region, negative fitness consequences from a breakdown of coadapted gene complexes are unlikely. Furthermore, dilution of local adaptation can be minimized by the appropriate choice of number of immigrants. Tufto (2001) demonstrated that even importing individuals that deviate from the local adaptive optimum can positively affect local population size as long as the number of immigrants is kept below some threshold value. Even if the benefit of outcrossing and heterosis declines over a couple of generations, the temporary increase in fitness could prove crucial for small populations in the longer term by reducing ecological Allee effects and extinction probability from demographic stochasticity.
Acknowledgments
We thank L. Blischke, J. Kaufmann, F. Kloß, V. Pasqualetto and D. Raudnitschka for their technical assistance and J. Van Buskirk for constructive comments on the manuscript. H.-R. Roth and R. Liesch of the Seminar für Statistik, ETH Zürich helped with the data analysis. Y.W. was supported by the Swiss National Science Foundation.
References
- Armbruster P, Reed D.H. Inbreeding depression in benign and stressful environments. Heredity. 2005;95:235–242. doi: 10.1038/sj.hdy.6800721. doi:10.1038/sj.hdy.6800721 [DOI] [PubMed] [Google Scholar]
- Bever J.D, Felber F. The theoretical population genetics of autopolyploidy. Oxford Surv. Evol. Biol. 1992;8:185–217. [Google Scholar]
- Dudash M.R. Relative fitness of selfed and outcrossed progeny in a self-compatible, protandrous species, Sabatia angularis L. (Gentianaceae): a comparison in three environments. Evolution. 1990;44:1129–1139. doi: 10.1111/j.1558-5646.1990.tb05220.x. doi:10.2307/2409277 [DOI] [PubMed] [Google Scholar]
- Ebert D, Haag C, Kirkpatrick M, Riek M, Hottinger J.W, Pajunen V.I. A selective advantage to immigrant genes in a Daphnia metapopulation. Science. 2002;295:485–488. doi: 10.1126/science.1067485. doi:10.1126/science.1067485 [DOI] [PubMed] [Google Scholar]
- Edmands S. Heterosis and outbreeding depression in interpopulation crosses spanning a wide range of divergence. Evolution. 1999;53:1757–1768. doi: 10.1111/j.1558-5646.1999.tb04560.x. doi:10.2307/2640438 [DOI] [PubMed] [Google Scholar]
- Edmands S. Between a rock and a hard place: evaluating the relative risks of inbreeding and outbreeding for conservation and management. Mol. Ecol. 2007;16:463–475. doi: 10.1111/j.1365-294X.2006.03148.x. doi:10.1111/j.1365-294X.2006.03148.x [DOI] [PubMed] [Google Scholar]
- Erickson D.L, Fenster C.B. Intraspecific hybridization and the recovery of fitness in the native legume Chamaecrista fasciculata. Evolution. 2006;60:225–233. [PubMed] [Google Scholar]
- Fenster C.B, Galloway L.F. Population differentiation in an annual legume: genetic architecture. Evolution. 2000;54:1157–1172. doi: 10.1111/j.0014-3820.2000.tb00551.x. [DOI] [PubMed] [Google Scholar]
- Fischer M, van Kleunen M, Schmid B. Genetic allee effects on performance, plasticity and developmental stability in a clonal plant. Ecol. Lett. 2000;3:530–539. doi:10.1046/j.1461-0248.2000.00188.x [Google Scholar]
- Heschel M.S, Paige K.N. Inbreeding depression, environmental stress, and population size variation in scarlet gilia (Ipomopsis aggregata) Conserv. Biol. 1995;9:126–133. doi:10.1046/j.1523-1739.1995.09010126.x [Google Scholar]
- Lynch M. The genetic interpretation of inbreeding depression and outbreeding depression. Evolution. 1991;45:622–629. doi: 10.1111/j.1558-5646.1991.tb04333.x. doi:10.2307/2409915 [DOI] [PubMed] [Google Scholar]
- Lynch M, Gabriel W. Mutation load and the survival of small populations. Evolution. 1990;44:1725–1737. doi: 10.1111/j.1558-5646.1990.tb05244.x. doi:10.2307/2409502 [DOI] [PubMed] [Google Scholar]
- Madsen T, Shine R, Olsson M, Wittzell H. Conservation biology: restoration of an inbred adder population. Nature. 1999;402:34–35. doi:10.1038/46941 [Google Scholar]
- Newman D, Pilson D. Increased probability of extinction due to decreased genetic effective population size: experimental populations of Clarkia pulchella. Evolution. 1997;51:354–362. doi: 10.1111/j.1558-5646.1997.tb02422.x. doi:10.2307/2411107 [DOI] [PubMed] [Google Scholar]
- Reed D.H. Relationship between population size and fitness. Conserv. Biol. 2005;19:563–568. doi:10.1111/j.1523-1739.2005.00444.x [Google Scholar]
- Reed D.H, Frankham R. Correlation between fitness and genetic diversity. Conserv. Biol. 2003;17:230–237. doi:10.1046/j.1523-1739.2003.01236.x [Google Scholar]
- Richards C.M. Inbreeding depression and genetic rescue in a plant metapopulation. Am. Nat. 2000;155:383–394. doi: 10.1086/303324. doi:10.1086/303324 [DOI] [PubMed] [Google Scholar]
- Saccheri I.J, Brakefield P.M. Rapid spread of immigrant genomes into inbred populations. Proc. R. Soc. B. 2002;269:1073–1078. doi: 10.1098/rspb.2002.1963. doi:10.1098/rspb.2002.1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccheri I, Kuussaari M, Kankare M, Vikman P, Fortelius W, Hanski I. Inbreeding and extinction in a butterfly metapopulation. Nature. 1998;392:491–494. doi:10.1038/33136 [Google Scholar]
- SAS Institute Inc. 2002 SAS OnlineDoc. Version 9.1 SAS Institute Inc., Cary, NC.
- Schiffer M, Gilchrist A.S, Hoffmann A.A. The contrasting genetic architecture of wing size, viability, and development time in a rainforest species and its more widely distributed relative. Evolution. 2006;60:106–114. [PubMed] [Google Scholar]
- Singer J.D. Using SAS PROC MIXED to fit multilevel models, hierarchical models, and individual growth models. J. Ed. Behav. Stat. 1998;24:323–355. doi:10.2307/1165280 [Google Scholar]
- Sokal R.R, Rohlf F.J. Freeman; New York, NY: 1995. Biometry: the principles and practice of statistics in biological research. [Google Scholar]
- Spielman D, Brook B.W, Frankham R. Most species are not driven to extinction before genetic factors impact them. Proc. Natl Acad. Sci. USA. 2004;101:15 261–15 264. doi: 10.1073/pnas.0403809101. doi:10.1073/pnas.0403809101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokstad E. ‘Genetic rescue’ helps panthers but puts researchers on the spot. Science. 2005;309:1162–1162. doi: 10.1126/science.309.5738.1162. doi:10.1126/science.309.5738.1162 [DOI] [PubMed] [Google Scholar]
- Tallmon D.A, Luikart G, Waples R.S. The alluring simplicity and complex reality of genetic rescue. Trends Ecol. Evol. 2004;19:489–496. doi: 10.1016/j.tree.2004.07.003. doi:10.1016/j.tree.2004.07.003 [DOI] [PubMed] [Google Scholar]
- Templeton A.R. Coadaptation and outbreeding depression. In: Soulé M.E, editor. Conservation biology: the science of scarcity and diversity. Sinauer Associates; Sunderland, MA: 1986. pp. 105–116. [Google Scholar]
- Tufto J. Effects of releasing maladapted individuals: a demographic-evolutionary model. Am. Nat. 2001;158:331–340. doi: 10.1086/321987. doi:10.1086/321987 [DOI] [PubMed] [Google Scholar]
- Wagenius S, Lonsdorf E, Neuhauser C. Patch aging and the S-allee effect: breeding system effects on the demographic response of plants to habitat fragmentation. Am. Nat. 2007;169:383–397. doi: 10.1086/511313. doi:10.1086/511313 [DOI] [PubMed] [Google Scholar]
- Westemeier R.L, Brawn J.D, Simpson S.A, Esker T.L, Jansen R.W, Walk J.W, Kershner E.L, Bouzat J.L, Paige K.N. Tracking the long-term decline and recovery of an isolated population. Science. 1998;282:1695–1698. doi: 10.1126/science.282.5394.1695. doi:10.1126/science.282.5394.1695 [DOI] [PubMed] [Google Scholar]
- Willi Y, Van Buskirk J. Genomic compatibility occurs over a wide range of parental genetic similarity in an outcrossing plant. Proc. R. Soc. B. 2005;272:1333–1338. doi: 10.1098/rspb.2005.3077. doi:10.1098/rspb.2005.3077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willi Y, Fischer M. Genetic rescue in interconnected populations of small and large size of the self-incompatible Ranunculus reptans. Heredity. 2005;95:437–443. doi: 10.1038/sj.hdy.6800732. doi:10.1038/sj.hdy.6800732 [DOI] [PubMed] [Google Scholar]
- Willi Y, Van Buskirk J, Fischer M. A threefold genetic allee effect: population size affects cross-compatibility, inbreeding depression and drift load in the self-incompatible Ranunculus reptans. Genetics. 2005;169:2255–2265. doi: 10.1534/genetics.104.034553. doi:10.1534/genetics.104.034553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willi Y, Van Buskirk J, Schmid B, Fischer M. Genetic isolation of fragmented populations is exacerbated by drift and selection. J. Evol. Biol. 2007;20:534–542. doi: 10.1111/j.1420-9101.2006.01263.x. doi:10.1111/j.1420-9101.2006.01263.x [DOI] [PubMed] [Google Scholar]