Abstract
During Drosophila embryogenesis, both the cytoplasmic Abelson tyrosine kinase (Abl) and the membrane bound tyrosine phosphatase PTP69D are required for proper guidance of CNS and motor axons. We provide evidence that PTP69D modulates signaling by Abl and its antagonist, Ena. An Abl loss-of function mutation dominantly suppresses most Ptp69D mutant phenotypes including larval/pupal lethality and CNS and motor axon defects, while increased Abl and decreased Ena expression dramatically increase the expressivity of Ptp69D axonal defects. In contrast, Ptp69D mutations do not affect Abl mutant phenotypes. These results support the hypothesis that PTP69D antagonizes the Abl/Ena genetic pathway, perhaps as an upstream regulator. We also find that mutation of the gene encoding the cytoplasmic Src64B tyrosine kinase exacerbates Ptp69D phenotypes, suggesting that two different cytoplasmic tyrosine kinases, Abl and Src64B, modify PTP69D-mediated axon patterning in quite different ways.
Keywords: Axon guidance, RPTP, Src, Signal transduction, Tyrosine kinase, Tyrosine phosphatase
INTRODUCTION
Axon guidance establishes the nervous system by connecting neurons with their synaptic targets during development (Goodman, 1996). Many of the proteins involved in axon guidance regulate the state of tyrosine phosphorylation (Flanagan and Vanderhaeghen, 1998; Gallo and Letourneau, 1999; Stoker and Dutta, 1998). In mammals, for example, the cytoplasmic tyrosine kinases Src and Fyn mediate axonal outgrowth in the presence of cell adhesion molecules L1 and N-CAM respectively (Beggs et al., 1994; Ignelzi et al., 1994). In Drosophila, mutations in Abl tyrosine kinase cause certain axons to terminate prior to reaching peripheral muscle targets (Wills et al., 1999a). Abl interacts with two key regulators of actin polymerization, Enabled (Ena) and Chickadee, the Drosophila homologue of Profilin (Krause et al., 2002; Wills et al., 1999b). Misregulation of this pathway causes specific axonal defects. For example, both ena loss of function and Abl gain of function mutations cause CNS axons to cross the midline inappropriately and peripheral motor axons to grow past their targets (Bashaw et al., 2000; Wills et al., 1999a). These and other results strongly implicate tyrosine phosphorylation as a key regulator of actin dynamics in axon guidance.
Many tyrosine phosphatases, which reverse reactions catalyzed by tyrosine kinases, are also required by growing axons during axon guidance in both vertebrates (Stoker et al., 1995) and Drosophila (Chien, 1996; Desai et al., 1996). For example, mammalian receptor protein tyrosine phosphatases RPTPδ, RPTPκ and RPTPμ (Burden-Gulley et al., 2002; Drosopoulos et al., 1999; Wang and Bixby, 1999) and chick PTPσ (Rashid-Doubell et al., 2002) have been implicated in the growth and guidance of several populations of developing vertebrate neurons. In Drosophila, at least 5 membrane bound tyrosine phosphatases, DLAR, PTP10D, PTP52F, PTP69D and PTP99A, function in axon guidance during embryogenesis (Desai et al., 1997; Krueger et al., 1996). For example, Ptp69D is an essential gene involved in motor, central and retinal axon guidance, suggesting a pivotal role in nervous system development (Desai et al., 1996; Garrity et al., 1999).
The logic and mechanism behind the interplay of tyrosine kinases and phosphatases during axon guidance is a current area of interest. dlar mutations suppress the CNS midline crossing defect of Abl mutants (Wills et al., 2002), while Abl alleles suppress the intersegmental nerve b (ISNb) guidance defects of Dlar mutants. DLAR binds to Abl and these proteins phosphorylate/dephosphorylate one another in vitro (Wills et al., 1999a). Like DLAR, cytoplasmic domains of PTP69D can also bind to Abl in vitro (Wills et al., 1999a) and can substitute for DLAR in retinal and motor axon guidance (Desai et al., 1997; Maurel-Zaffran et al., 2001), suggesting potential involvement of PTP69D in Abl mediated signaling (Wills et al., 1999a). However, the physiological relevance of the in vitro interaction of Abl with PTP69D has not been determined. Recently identified and characterized Ptp69D alleles (Desai and Purdy, 2003; Marlo and Desai, 2006) show a broad range of developmental defects and reveal a requirement for enzymatic activity in the proximal tyrosine phosphatase domain (D1) for proper axonal guidance.
In this article, we show that the phenotypes of Ptp69D mutants are suppressed by Abl mutations and enhanced by overexpression of Abl. Furthermore, Abl gain-of-function phenotypes are supressed by PTP69D overexpression. On the other hand, Abl mutant phenotypes are not affected by loss of function or null mutation of Ptp69D. These results show that PTP69D antagonizes the effects of Abl and suggest that Abl may function downstream of PTP69D. Consistent with these conclusions, and with previous data showing that Abl acts by inhibiting Ena (Fox and Peifer, 2007; Gertler et al., 1995; Gertler et al., 1990; Grevengoed et al., 2003), ena mutations enhance Ptp69D defects. Mutations in Ptp69D also interact with mutations in another non-receptor tyrosine kinase, Src64B, a Drosophila member of the Src-family kinases (SFK). Strikingly however, Src64B mutations enhance rather than suppress most Ptp69D axonal defects. This finding raises the possibility that the relationship of tyrosine kinases and phosphatases is more complex than the evident antagonism of their biochemical activities.
RESULTS
Abl mutations suppress lethality of Ptp69D
Heteroallelic combinations of Ptp69D mutants confer partial lethality ranging from 56%- 91% (Desai and Purdy, 2003). We therefore examined the effect of Abl mutations on the lethality of Ptp69D mutants as a test for genetic interactions. Abl mutations dominantly suppressed lethality of various Ptp69D genotypes. For example, lethality of the hypomorphic genotype (Ptp69D10/Df(3L)8ex34) was decreased by reduction of Abl from 91% to 49%. Moreover, removal of one copy of Abl suppressed the embryonic/larval lethality of the null genotype (Ptp69D1/Df(3L)8ex34), allowing survival of a significant fraction of Ptp69D null animals to pupal stages (Table 1). Molecular information and severity of Ptp69D alleles are described in Materials and Methods. In brief, Ptp69D1 is null and Ptp69D10 is temperature sensitive and can be weaker or stronger than null depending on temperature and assay (Desai and Purdy, 2003).
Table 1.
Modification of Ptp69D and Abl lethality
| Genotype of Ptp69D | Modifier | Lethality(%) | N | 1Su | p value |
|---|---|---|---|---|---|
|
*Ptp69D10/Ptp69D10 |
+/+ | 56 | 599 | Control | |
|
Abl3/+ |
1 |
662 |
++++ |
<0.0001 |
|
|
*Ptp69D1/Ptp69D10 |
+/+ | 74 | 505 | Control | |
| Abl1/+ | 42 | 270 | +++ | <0.0001 | |
|
Abl3/+ |
11 |
632 |
++++ |
<0.0001 |
|
|
*Ptp69D10/Df(3L)8ex34 |
+/+ | 91 | 643 | Control | |
|
Abl3/+ |
49 |
595 |
+++ |
<0.0001 |
|
| @Ptp69D1/Df(3L)8ex34 | +/+ | 94 | 614 | Control | |
| Abl3/+ | 73 | 777 | ++ | <0.0001 |
| Genotype of Ptp69D | Modifier | Lethality(%) | N | 2En | |
|---|---|---|---|---|---|
| Ptp69D1/Ptp69D10 | +/+ | 74 | 505 | Control | |
| enaGC5/+ | 99 | 222 | ++ | <0.0001 | |
| enaGC1/+ | 100 | 180 | ++ | <0.0001 | |
| src64Δ17/+ | 100 | 289 | ++ | <0.0001 | |
| Df(3L)10[H]/+ | 100 | 274 | ++ | <0.0001 |
| Genotype of Abl | Modifier | Lethality(%) | N | 2En | |
|---|---|---|---|---|---|
| Abl3/Abl4 | +/+ | 60 | 569 | Control | |
| Df(3L)8ex34/+ | 64 | 533 | − | NS | |
| Ptp69D10/+ | 69 | 545 | − | NS |
Su; Degree of Suppression.
En; Degree of Enhancement.
Pre-adult lethality, determined by counting adult eclosers.
Embryonic/larval lethality, determined by counting pupae. Ptp69D null animals did not survive to adulthood regardless of the Abl genotype.
NS; Difference from control not significant. p > 0.05
Abl strongly suppresses Ptp69D axon guidance defects
Ptp69D mutants display significant CNS and motor axon defects (Desai and Purdy, 2003; Marlo and Desai, 2006). For example, embryos hemizygous for Ptp69D7 (Ptp69D7/Df(3L)8ex34), which has a mutation in the proximal phosphatase domain(D1) (Desai and Purdy, 2003), displayed CNS midline crossing defects in 54% of segments (Fig.1C, Table 2), and 64% of ISNb motor nerves bypassed the ventrolateral muscle (VLM) layer (Fig. 1D, Table 2), similar to the phenotypes observed for another D1 domain mutant allele, Ptp69D21(Desai and Purdy, 2003). To test the ability of Abl to suppress Ptp69D axon defects, we introduced a heterozygous Abl mutation into embryos bearing various Ptp69D alleles. Abl mutations strongly suppressed CNS defects characteristic of several Ptp69D mutations. For example, the CNS defects of Ptp69D7/Df(3L)8ex34 dropped from 54% to 24% of segments when one copy of an Abl mutation was introduced (Fig. 1E, Table 2). Similarly, we found that Abl suppresses ISNb defects associated with Ptp69D mutations. In Ptp69D1/Ptp69D7, the ISNb bypassed its target VLMs and followed the ISN pathway towards the area of dorsal muscles in 58% of hemisegments (Fig. 1D, Table 2). In the presence of Abl1/+, the bypass rate dropped to 23% (Fig. 1F, Table 2). These results support the hypothesis that Abl antagonizes Ptp69D axon guidance function in both CNS and motor neurons.
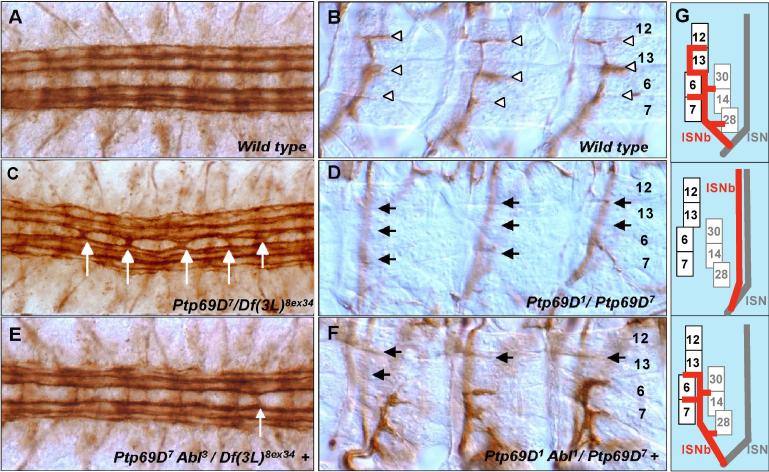
Fig.1. Abl suppresses axon defects of Ptp69D.
Late stage 16 through early stage 17 embryos were stained with anti fasciclin II (mAb 1D4) and dissected to visualize FasII positive axons in the CNS (A, C, E) and VLM(Ventro-Lateral Muscle) innervation by the ISNb (B, D,F). (A) Wild-type CNS pattern showing three pairs of longitudinal bundles flanking the midline. (C, E) CNS patterns of Ptp69D mutants in embryos that are wild type (C) or heterozygous mutant (E) for an Abl mutation. (C) Ptp69D7/Df(3L)8ex34 embryo showing a severe CNS midline defect. In contrast to wild-type, axons cross the midline in most segments (white arrows). (E) Ptp69D7Abl3/ Df(3L)8ex34 + embryo showing highly rescued CNS phenotype. Occasional segments contain a thin bundle of axon staining across the midline (white arrow) (B) Wild-type ISNb pattern showing synaptic branches in the clefts between individual VLM fibers (white arrowheads). Numbers on the right are muscle identities. (D, F) ISNbs of Ptp69D mutants in embryos that are wild type (D) or heterozygous mutant (F) for an Abl mutation. (D) Bypass phenotype in an Ptp69D1/ Ptp69D7 embryo; most ISNb axons extend past the VLM field (out of focus) and fail to innervate it (black arrows). (F) In this Ptp69D1 Abl1/ Ptp69D7 + embryo, most ISNb axons enter and innervate VLM field, although they often fail to target distal muscle fibers (black arrows). (G) Schematic of a cross section through panels B,D and F. Top : In wild type, ISN is superficial to muscles. ISNb enters muscle field. Center : Ptp69D bypasses the muscle field ; ISNb associates with ISN. Bottom: Abl rescues ISNb ability to enter muscle field, though sometimes fails to reach muscle 12. Numbered rectangles represent muscles.
Table 2.
Modification of Ptp69D axonal phenotypes
| Genotype | Modifier |
1CNS midline cross |
2ISNb bypass |
||||||
|---|---|---|---|---|---|---|---|---|---|
| % | N | 3Su | p value | % | N | 3Su | p value | ||
|
Ptp69D1/Ptp69D10 |
+/+ | 5 | 152 | Control | 25 | 223 | Control | ||
| Abl3/+ | 2 | 88 | − | NS | 7 | 132 | +++ | < 0.0001 | |
|
Abl3/Abl3 |
4 |
96 |
− |
NS |
0 |
144 |
+++ |
< 0.0001 |
|
|
Ptp69D1/Ptp69D7 |
+/+ | 56 | 144 | Control | 58 | 287 | Control | ||
|
Abl1/+ |
37 |
132 |
++ |
0.0003 |
23 |
244 |
++ |
< 0.0001 |
|
|
Ptp69D7/Df(3L)8ex34 |
+/+ | 54 | 112 | Control | 64 | 168 | Control | ||
|
Abl3/+ |
24 |
128 |
++ |
< 0.0001 |
19 |
192 |
++++ |
< 0.0001 |
|
|
Ptp69D10/Df(3L)8ex34 |
+/+ | 15 | 104 | Control | 25 | 116 | Control | ||
|
Abl3/+ |
3 |
160 |
++ |
0.0002 |
17 |
239 |
− |
NS |
|
|
UAS-Abl |
+/+ | 0 | 112 | Control | 24 | 168 | Control | ||
|
UAS-Ptp69D |
0 |
80 |
− |
NS |
0 |
120 |
++++ |
<< 0.0001 |
|
| Genotype | Modifier |
1CNS midline cross |
2ISNb bypass |
||||||
|---|---|---|---|---|---|---|---|---|---|
| % | N | 4En | p value | % | N | 4En | p value | ||
|
Ptp69D1/Ptp69D10 |
+/+ | 5 | 152 | Control | 25 | 223 | Control | ||
| UAS-Abl | 62 | 128 | +++ | < 0.0001 | 78 | 192 | +++ | < 0.0001 | |
| enaGC5/+ | 69 | 111 | +++ | < 0.0001 | 77 | 194 | +++ | < 0.0001 | |
|
src64Δ17/+ |
48 |
104 |
+++ |
< 0.0001 |
74 |
156 |
+++ |
< 0.0001 |
|
|
Ptp69D1/Ptp69D7 |
+/+ | 56 | 144 | Control | 58 | 287 | Control | ||
|
UAS-Abl |
98 |
115 |
+++ |
< 0.0001 |
99 |
166 |
+++ |
< 0.0001 |
|
|
Ptp69D1/Df(3L)8ex34 |
+/+ | 4 | 152 | Control | 6 | 227 | Control | ||
| UAS-Abl | 74 | 120 | ++++ | < 0.0001 | 59 | 170 | +++ | < 0.0001 | |
|
src64Δ17/+ |
55 |
110 |
++++ |
< 0.0001 |
59 |
132 |
+++ |
< 0.0001 |
|
|
Ptp69D7/+ |
+/+ | 1 | 120 | Control | 0 | 120 | Control | ||
|
UAS-Abl |
25 |
84 |
+++ |
< 0.0001 |
50 |
118 |
+++ |
< 0.0001 |
|
| Df(3L)8ex34/+ | +/+ | 0 | 120 | Control | 0 | 120 | Control | ||
| UAS-Abl | 32 | 88 | +++ | < 0.0001 | 38 | 132 | +++ | < 0.0001 | |
Segments (T2−3 and A1−8) harboring midline crosses were counted.
Hemisegments (A2-A7) showing that ISNb bypass the layer of target muscles were counted. Distinguishable partial bypasses were also counted.
Su; Degree of Suppression.
En; Degree of Enhancement.
NS; Difference from control not significant. p > 0.05
Ptp69D mutant phenotypes are enhanced by Abl gain-of-function
Since Abl mutations suppressed Ptp69D phenotypes, we expected that increasing Abl levels would exacerbate Ptp69D mutant phenotypes. Previous experiments demonstrated that UAS-GAL4 mediated Abl overexpression can cause mild axon defects in CNS and ISNb (Bashaw et al., 2000; Fogerty et al., 1999; Wills et al., 1999a). To test the prediction that increased Abl expression should enhance Ptp69D defects, we examined the nervous system of embryos with reduced Ptp69D expression and increased Abl expression. Under our conditions, neither loss of Ptp69D nor simple overexpression of Abl in the nervous system caused CNS defects (Fig. 2A, B). When combined, however, reduced Ptp69D together with increased Abl caused severe CNS midline defects (Fig.2C, D). We tested additional Ptp69D genotypes; in all cases, neuronal overexpression of Abl dramatically increased both the CNS and motor axon defects of Ptp69D mutants (Table 2). Conversely, Abl over-expression conferred the ISNb bypass phenotypes in about 24% of hemisegments (Fig. 2F, Table 2), and for all Ptp69D alleles tested, this phenotype was dramatically increased in expressivity by reduction of Ptp69D. For example, when a single copy of Ptp69D7 was combined with Abl overexpression, the rate at which ISNb nerves completely bypassed the VLM muscle field rose from 24% to 50%. The heterozygous Ptp69D7 by itself did not cause ISNb bypass (Fig. 2E). Moreover, heterozygosity for Ptp69D is specific in its interaction with Abl pathway and does not promiscuously interact with all mutations that affect ISNb. Thus, for example, reduction of Notch activity produces a bypass phenotype similar to that from Abl overexpression ((Crowner et al., 2003), but the Notch phenotype is not enhanced by Ptp69D heterozygosity (Supplementary Table 1).
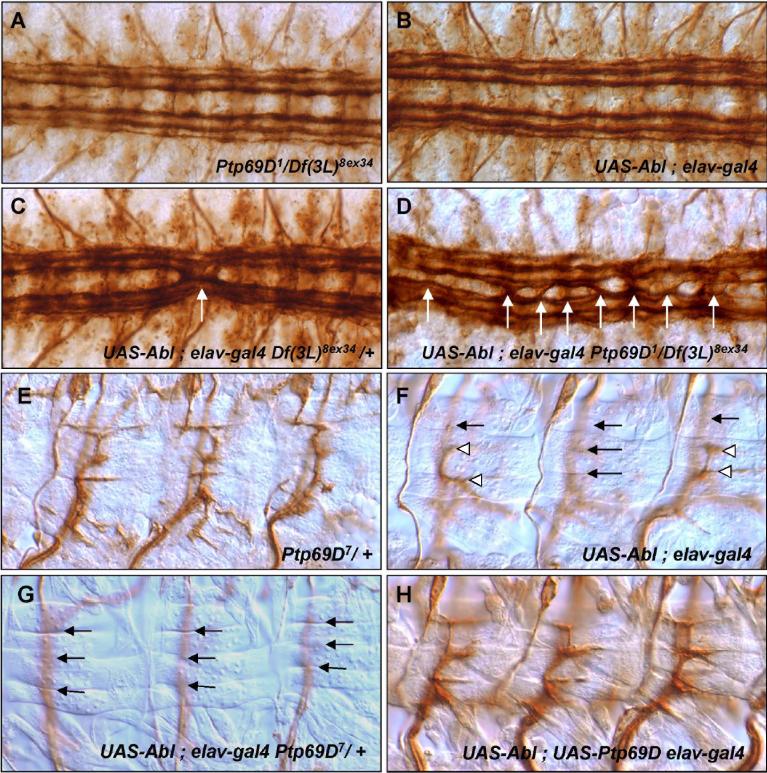
Fig. 2. Excess Abl function enhances Ptp69D axonal defects.
Axon patterns of CNS (A-D) and ISNb (E-H). (A) Ptp69D1/Df(3L)8ex34 null. FasII positive axons cross the midline very rarely (4% of total segment of T2−3 and A1−8). (B) P[UAS-Abl]/+; P[elav-GAL4]/+. Abl overexpressing embryos resemble the wild-type; FasII positive axons do not cross the midline. (C) In P[UAS-Abl]/+; Df(3L)8ex34 P[elav-GAL4]/++ embryos, FasII axons(white arrows) frequently cross the midline (32% of segments, Table 2). (D) P[UAS-Abl]/+; Ptp69D1 P[elav-GAL4] /Df(3L)8ex34 + embryos display severe midline crossing defects in most segments (white arrows). (E) Ptp69D7/+ embryos display normal ISNbs. (F) P[UAS-Abl+]/+; P[elav-GAL4]/+ embryos display partial ISNb bypass phenotypes, in which some axons fail to enter the VLM field while others enter and successfully innervate their target muscles (white arrowheads). (G) P[UAS-Abl+]/+; Ptp69D7 P[elav-GAL4]/+ + embryos display the complete ISNb bypass phenotype in most hemisegments (arrows). (H) Overexpression of PTP69D in P[UAS-Abl+]/+; P[UAS-Ptp69D] P[elav-GAL4]/+ + embryos fully rescue ISNb defects associated with Abl gain of function (compare to F).
These data indicate that excess Abl exacerbates defects caused by reduced PTP69D activity and vice versa, consistent with a model in which PTP69D acts in opposition to Abl function. This model is further supported by the suppression of Abl overexpression phenotypes by excess PTP69D. Specifically, the rate of ISNb bypass defects in embryos overexpressing Abl was reduced from 25% to 0% by the simultaneous overexpression of PTP69D (Fig. 2H). This result supports the hypothesis that PTP69D acts to antagonize Abl.
Is Abl downstream of Ptp69D?
Since Abl appears to modify Ptp69D mutant phenotypes, we also examined the ability of Ptp69D mutations to modify the viability and axon phenotypes of Abl mutants. Abl3/Abl4 shows 60% lethality prior to adult stage, and this rate is unchanged when Ptp69D alleles are introduced (Table 1). We obtained a similar negative result when we examined the effects of Ptp69D on Abl axon phenotypes. Stalled ISNb axons are an obvious neuronal phenotype of Abl mutants. Specifically, in both Abl1/Abl3 and Abl3/Abl3 embryos, ISNb terminates at the proximal edge of muscle 13 in 52% and 54% of hemisegments, respectively (Table 3). We introduced heterozygous or homozygous Ptp69D mutations in these Abl backgrounds, but observed no significant change in the frequency or severity of this ISNb stall phenotype, with defects ranging from 49% to 63%. Formally, these results suggest that Abl may be epistatic to Ptp69D, i.e. genetically downstream.
Table 3.
Modification of Abl axonal phenotypes
| Genotype | Modifier |
1ISNb stall |
||
|---|---|---|---|---|
| % | N | p-value | ||
|
Abl1/Abl3 |
+/+ | 52 | 188 | |
|
Ptp69D1/Ptp69D10 |
49 |
144 |
NS |
|
| Abl3/Abl3 | +/+ | 54 | 168 | |
| Ptp69D10/+ | 48 | 96 | NS | |
| Df(3L)8ex34/+ | 51 | 144 | NS | |
| Df(3L)8ex34/Df(3L)8ex34 | 63 | 142 | NS | |
Most ISNb axons reached VLM properly but arrested in the ventral region (muscle 12,13) were considered as ‘Stall’ phenotype and counted per hemisegments of A2-A7.
NS; Difference from control is not significant. p > 0.05
Ena is involved in PTP69D signaling
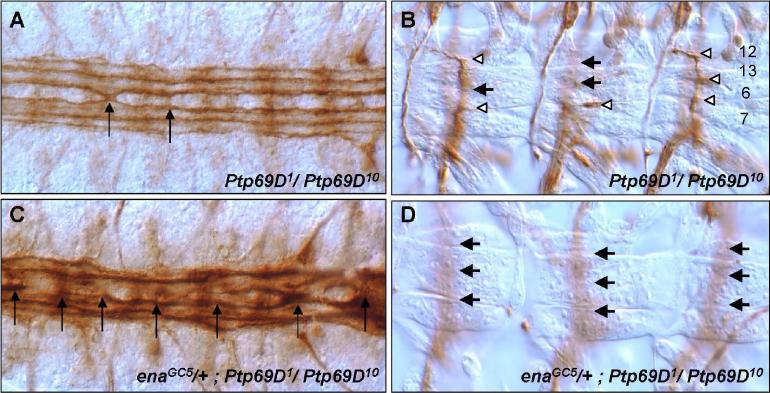
Enabled (Ena) is the sole Drosophila member of the Ena/VASP (Vasodilator-Stimulated Phosphoprotein) family of proteins that influence actin cytoskeleton architecture (Krause et al., 2003). For example, Ena/VASP proteins are involved in actin filament elongation via the recruitment of actin-profilin complexes to sites of active actin rearrangement (Reinhard et al., 1995), and also influence the activity of the Arp2/3 complex and counteract the inhibition of actin polymerization by capping proteins (Skoble et al., 2001). Ena is an antagonistic component of the Abl signaling pathway. Reduction of Ena rescues the lethality and axonal phenotypes of Abl mutations and mutant interactions (Gertler et al., 1990). It binds to the SH3 domain of Abl via a proline rich region and is an Abl substrate (Ahern-Djamali et al., 1999; Gertler et al., 1995; Gertler et al., 1990; Grevengoed et al., 2003; Grevengoed et al., 2001). We reasoned that PTP69D might function in the same biochemical pathway as Ena because both Ptp69D and ena mutants interact genetically with Abl and display ISNb bypass defects. In addition, Ena associates with the PTP69D D2 domain in vitro (Desai et al., 1997; Wills et al., 1999a). Finally, Abl binds to and phosphorylates both Ena and the D2 domain of PTP69D in vitro (Wills et al., 1999a). To test potential interaction between ena and Ptp69D, we first examined adult lethality. ena mutations dominantly enhanced the adult lethal phenotype of Ptp69D mutants (Table 1). Next, we explored the effect of ena mutation on the neural phenotypes of Ptp69D mutants. ena mutations strongly increased the axon defects of Ptp69D mutants. Specifically, the expressivity of CNS crossover defects of Ptp69D mutant embryos increased from 5% to 69% when one copy of ena was eliminated (Table 2, Figs. 3A, C). Also, enaGC5 enhanced ISNb bypass phenotypes in Ptp69D animals from 25% to 77% (Table 2, Fig. 3B, D). These results suggest that PTP69D modulates axon guidance signaling by Abl and Ena.
Fig. 3. ena enhances Ptp69D axon phenotypes.
CNS (A, C) and ISNb (B, D) Ptp69D mutant phenotypes in the absence (A,B) and presence (C,D) of a heterozygous ena mutation. (A) CNS in Ptp69D1/Ptp69D10 embryo shows occasional midline crossovers (arrow). (C) CNS in enaGC5/+; Ptp69D1/Ptp69D10 embryos display severe defects, including roundabouts, fusions and crossovers (arrows). (B) ISNbs in Ptp69D1/Ptp69D10 embryos often display partial bypass phenotypes (arrows), while still innervating some muscles(white arrowheads) (D) ISNbs in enaGC5/+;Ptp69D1/Ptp69D10 embryos show complete bypass phenotypes (arrows).
Src64B acts positively in PTP69D signaling
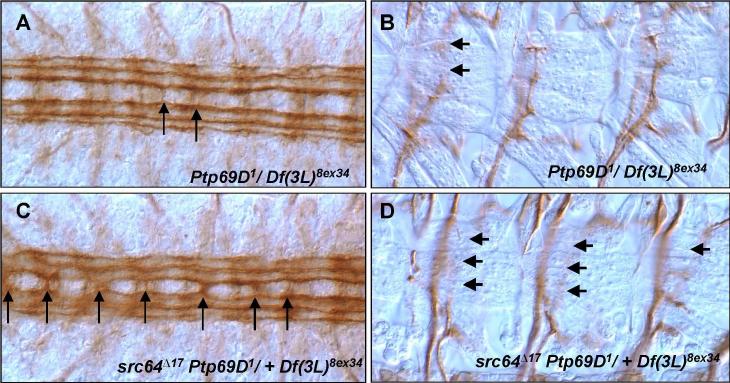
Our results so far suggest that PTP69D negatively regulates Abl, an interpretation consistent with their opposite biochemical functions and reminiscent of the antagonism between DLAR and Abl. On the other hand, it has been reported that RPTPs are positively regulated by Src family kinases (SFK). For example, neurotrophin signaling through SFK in mammalian neurons is promoted by LAR (Yang et al., 2006). We wondered whether the opposite nature of the reported interactions of Abl and Src with RPTPs reflect differences in the two biological systems assayed or different specificities of SFK vs Abl. To address this question, we tested genetic interactions of Ptp69D with a Drosophila Src gene, Src64B. Interestingly, heterozygosity for the hypomorphic Src64B allele, Src64BΔ17 enhanced the adult lethality of a Ptp69D mutant, increasing it from 74% to 100% , and a heterozygous deficiency of the Src64B region gave similar results (Table 1). Furthermore, this allele dramatically increased the axonal defects of Ptp69D embryos. Removing one copy of Src64B converted the mild, low penetrance Ptp69D midline crossing phenotype to severe, highly penetrant robo-like defects (Table 2, Figs 4A, B). Src64BΔ17 also both quantitatively increased the expressivity of the ISNb phenotype of Ptp69D mutants and qualitatively enhanced the phenotype from a partial to a complete bypass defect (Table 2, Figs. 4C, D). A heterozygous Src mutant alone does not produce axonal defects (not shown). These results reveal that Src64B acts cooperatively with PTP69D in embryonic axon guidance and are in striking contrast to the antagonistic interaction between PTP69D and Abl.
Fig. 4. Src64B enhances axonal defects of Ptp69D mutants.
CNS (A, C) and ISNb (B, D) axon patterns of Ptp69D mutants in the absence (A, B) and presence(C, D) of a heterozygous Src64B mutation. (A) A few FasII positive axons occasionally cross the midline in Ptp69D1/Df(3L)8ex34 embryos (arrows). (C) Severe midline crossing phenotypes (arrows) characterize Src64BΔ17Ptp69D1/+ Df(3L)8ex34 embryos. (B) ISNbs in Ptp69D1/Df(3L)8ex34 embryos occasionally display partial bypass and early termination phenotypes (arrows). (D) ISNbs in Src64BΔ17Ptp69D1/+ Df(3L)8ex34 embryo frequently show the complete bypass phenotype. Most axons fail to enter VLM field.
DISCUSSION
Two enzyme classes, tyrosine kinases and tyrosine phosphatases, dynamically maintain protein phosphotyrosine modifications that are critical for axon guidance. Studies that revealed physical interactions between members of these families (Wills et al., 1999a; Wills et al., 2002) led us to investigate the relationship between the membrane bound tyrosine phosphatase, PTP69D and cytoplasmic tyrosine kinases. Here we provide evidence that PTP69D modulates signaling by the tyrosine kinase, Abl, and its substrate Ena. First, Ptp69D mutant phenotypes, including adult lethality, embryonic CNS and ISNb motor axon defects, are significantly suppressed by loss of Abl function, and dramatically enhanced by gain of Abl function (Table 1, Fig. 1 and 2). Second, Ptp69D does not suppress Abl, suggesting that their interaction is asymmetric(Table 1 and 3). Third, Ena, a strong suppressor and a downstream substrate of Abl, (Gertler et al., 1995; Grevengoed et al., 2003) dominantly exacerbates the defects of Ptp69D (Fig. 3).
Abl mutants display evident phenotypes such as adult lethality and ISNb arrest defects (Gertler et al., 1990; Wills et al., 1999b). We expected to observe bi-directional suppression between Ptp69D and Abl by analogy to Dlar mutations, which interact symmetrically with Abl for such phenotypes as midline crossing defects in the CNS (Wills et al., 2002). However, although Abl mutants modify Ptp69D phenotypes, we failed to observe any evidence of reciprocal suppression by introducing Ptp69D mutations into Abl embryos despite trying various combinations of alleles (Table 1 and 3). While several models might explain this, one simple interpretation is that Abl is epistatic to Ptp69D, i.e. Ptp69D acts through Abl.
What could be downstream targets of PTP69D and Abl? As a substrate of Abl and antagonistic genetic component of the Abl pathway, Ena is an excellent candidate (Gertler et al., 1995; Gertler et al., 1990; Grevengoed et al., 2003; Grevengoed et al., 2001), and indeed, we found that ena mutations enhanced the lethality and axonal defects of Ptp69D mutants. Ena is known to play a role in cell motility (Goh et al., 2002; Krause et al., 2002), and likely supports F-actin assembly within cells by antagonizing capping protein at the barbed ends of actin and reducing filament branching (Bear et al., 2000). In Drosophila, Ena associates with the PTP69D D2 domain and is phosphorylated by Abl in vitro (Wills et al., 1999a), and its specific cellular localization is regulated by Abl (Grevengoed et al., 2003). The consistent pattern of interactions of Ptp69D with Abl and ena – suppressed by Abl mutations and enhanced by the Abl antagonist, ena – supports the idea that the Abl effector, Ena is also a key to signaling by PTP69D.
The data we report define a functional relationship among PTP69D, Abl and Ena, but what could be their physical relationship? Extrapolating from genetic interactions to molecular mechanism is not straightforward, however, an attractive speculation that could provide one framework for further thinking is the idea that PTP69D, Abl and Ena may coexist in a complex, where PTP69D inhibits Abl, which in turn inhibits Ena. Such a model would be consistent with the available biochemical evidence (Wills et al., 1999a) , as well as with the genetic interactions we observed. Many other models are equally possible, however, and a great deal of additional experimentation would be required to establish this hypothesis. For example, we have not investigated whether these three proteins co-immunoprecipitate, nor have we demonstrated that they act simultaneously in the same cell. Moreover, although the kinase activity of Abl is required for its axonal function (Wills et al., 2002), it is thought that tyrosine phosphorylation of Ena is not the sole function of Abl (Comer et al., 1998). Axon guidance by Abl seems also, for example, to be associated with the action of small Rho family GTPases, particularly Rac (Hakeda-Suzuki et al., 2002; Luo, 2002; Maurel-Zaffran et al., 2001), and any model for the mechanism of axon guidance by Abl and its partners will have to take these data into account.
The patterns of genetic interactions of Ptp69D with Abl and ena described in our paper bear some similarities to those of Dlar (Wills et al., 1999a; Wills et al., 2002). Does either RPTP substitute for each other? Previous studies demonstrated that Ptp69D and DLAR cooperate at growth cone choice points along one nerve, ISNb, while along another nerve, ISN, they do not act together (Desai et al., 1997). In the adult eye, moreover, a Dlar transgene rescues Ptp69D R7 axon phenotype, but not vice versa (Maurel-Zaffran et al., 2001). Thus, the relationship between PTP69D and DLAR is complex and depends on cellular context.
We extended our analysis of PTP69D by investigating the functional interaction of PTP69D with a Drosophila Src gene, Src64B. Mammalian Src protein (c-src) has been shown to regulate the stability and remodeling of actin structures (Boschek et al., 1981; Boyce et al., 1992). In Drosophila, Src64B has been shown to function in nervous system development in the embryo (unpublished data described in Wills et al., 1999a), the mushroom body of the adult brain and in the adult eye (Kussick et al., 1993; Nicolai et al., 2003), making it a plausible candidate for interacting with PTP69D in axon guidance. Moreover, in mouse the RPTP CD45 functions to positively regulate SFK in T cells (Hermiston et al., 2003). Indeed, our data show that Ptp69D does interact with Src64B, but in a sense opposite to that with Abl: Ptp69D and Src64B interact synergistically rather than antagonistically (Fig. 4 and Table 2).
Hints as to a possible mechanism that could underlie the interaction of PTP69D with Src64B are suggested by experiments in mammals. The SFK Fyn binds to LAR and phosphorylates the LAR D2 domain. In turn, LAR dephosphorylates a c-terminal inhibitory motif of Fyn (Tsujikawa et al., 2002), increasing Fyn activity. Our data as well could potentially be explained by an analogous model whereby PTP69D derepresses Src activity by removing an inhibitory phosphate, though other models are clearly possible and more study is required to test this speculation. It is interesting that the biochemical association of LAR with Fyn in mammals is reminiscent of that observed for DLAR with Abl in Drosophila (Wills et al., 1999a), but the biological consequences in the two cases are quite different, and in fact opposite : activation of Fyn activity, but suppression of Abl.
Superficially, it seemed surprising that two cytoplasmic kinases had opposite interactions with PTP69D, antagonizing Abl but cooperating with Src64B. To test this further, we examined the genetic interaction between Src64B and Abl. Although Src64BΔ17 did not show any significant effect on Abl lethality, it dramatically suppressed the ISNb stall defect of Abl mutants (Supplementary Table 2), further supporting the hypothesis that Src64B and Abl kinases may have opposing functions in axon guidance.
In summary, the receptor protein tyrosine phosphatase PTP69D interacts both with the Abl-Ena tyrosine kinase pathway and with Src64B to control axon patterning in Drosophila. PTP69D antagonizes Abl, perhaps as an upstream regulator, but functions synergistically with Src64B, thus revealing previously unrecognized specificity in the action of these tyrosine kinase pathways.
MATERIALS AND METHODS
Fly stocks
All experiments were done at 25°C except analysis of interaction with Notchts (Crowner et al., 2003). Ptp69D alleles were described previously (Marlo and Desai, 2006; Desai and Purdy, 2003; Desai et al., 1996). Ptp69D1 is a null allele, which lacks all but the first 115bp of the 5'UTR and sequences that encode most of the extracellular domain. Ptp69D7 has a 3 amino acid deletion in the D1 domain. Ptp69D7 has a phenotype stronger than that of a null allele, as is true of other D1 domain mutants such as Ptp69D21. However, as Ptp69D7 is recessive, and a Ptp69D7 homozygote phenotype is suppressed by duplication of the wild type gene (Desai and Purdy, 2003), it is unlikely that Ptp69D7and Ptp69D21 are neomorphic, but rather that they act negatively within the wild type PTP69D genetic pathway. Ptp69D10 is a temperature sensitive allele and has a V to D mutation at the junction between the first and second IgG domains. At the temperature used in this study (25°C), Ptp69D10 shows substantial axonal defects but is viable. Severity of alleles may be ordered as Ptp69D7=Ptp69D1 > Ptp69D10 for animal lethality and Ptp69D7 > Ptp69D10 ≥ Ptp69D1 for axon patterning. Abl, Src64B and ena stocks were obtained from Van Vactor (Harvard University), Liebl (Danison University) and the Bloomington Drosophila Stock Center. Gal4 expression was driven by neuronal specific P[elav-GAL4] driver chromosomes (obtained from C.S. Goodman). Stocks were balanced over TM6B, Tb or TM6B-T8-lacZ, Tb (gift of Dr. Peter Kolodziej).
Viability test / Scoring
10 males and 25−30 virgin females were placed in a food vial at room temperature and parents were transferred to a fresh food vial every 12 hours for 7 days. For hypomorphic combinations (Ptp69D1/ Ptp69D10 and Ptp69D10/Df(3L)8ex34), pre-adult lethality was assayed by daily counting of hatched F1 adults. The null combination,Ptp69D1/Df(3L)8ex34 produced almost no viable pupae and was considered to be embryonic/larval lethal. For this genotype, lethality was determined by counting pupal cases, including both hatched pupal cases and dead pharate adults, after no new adults eclosed for at least 24hr. Ptp69D null animals did not survive to adulthood regardless of the Abl genotype.
Immunohistochemistry
Embryos were fixed, collected, stained, analyzed and photographed as described (Desai et al., 1996). Mutant embryos from stocks balanced by TM6B T8-lacZ, Tb were identified by their failure to stain with anti-ß-galactosidase sera (Jackson Laboratories, Bar Harbor, ME).
Statistics
All data were analyzed with Microsoft excel 11.0 software. Statistically-significant differences were determined by CHITEST. Difference with a p value greater than 0.05 were considered to be not significant (NS).
Acknowledgement
We thank Drs. David Van Vactor, Chi-Hon Lee and Pamela Bradley for comments; Drs David Van Vactor, Eric Liebl, Corey Goodman and Peter Kolodziej for Src, Abl, elav-Gal4 and T8-lacZ balancer lines; the Bloomington Drosophila Stock Center for ena alleles; Drs. David Miller, Liliana Solnika-Krezel, Bruce Carter and Peter Kolodziej for the general advice. This work was supported by awards to C.J.D. from the NIH grant (NSHD38141) and also supported by NINDS Intramural Basic Neuroscience Program (Z01 NS003013) to E.G.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary Material
Reference
- Ahern-Djamali SM, Bachmann C, Hua P, Reddy SK, Kastenmeier AS, Walter U, Hoffmann FM. Identification of profilin and src homology 3 domains as binding partners for Drosophila enabled. Proc Natl Acad Sci U S A. 1999;96:4977–82. doi: 10.1073/pnas.96.9.4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashaw GJ, Kidd T, Murray D, Pawson T, Goodman CS. Repulsive axon guidance: Abelson and Enabled play opposing roles downstream of the roundabout receptor. Cell. 2000;101:703–15. doi: 10.1016/s0092-8674(00)80883-1. [DOI] [PubMed] [Google Scholar]
- Bear JE, Loureiro JJ, Libova I, Fassler R, Wehland J, Gertler FB. Negative regulation of fibroblast motility by Ena/VASP proteins. Cell. 2000;101:717–28. doi: 10.1016/s0092-8674(00)80884-3. [DOI] [PubMed] [Google Scholar]
- Beggs HE, Soriano P, Maness PF. NCAM-dependent neurite outgrowth is inhibited in neurons from Fyn-minus mice. J Cell Biol. 1994;127:825–33. doi: 10.1083/jcb.127.3.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boschek CB, Jockusch BM, Friis RR, Back R, Grundmann E, Bauer H. Early changes in the distribution and organization of microfilament proteins during cell transformation. Cell. 1981;24:175–84. doi: 10.1016/0092-8674(81)90513-4. [DOI] [PubMed] [Google Scholar]
- Boyce BF, Yoneda T, Lowe C, Soriano P, Mundy GR. Requirement of pp60c-src expression for osteoclasts to form ruffled borders and resorb bone in mice. J Clin Invest. 1992;90:1622–7. doi: 10.1172/JCI116032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burden-Gulley SM, Ensslen SE, Brady-Kalnay SM. Protein tyrosine phosphatase-mu differentially regulates neurite outgrowth of nasal and temporal neurons in the retina. J Neurosci. 2002;22:3615–27. doi: 10.1523/JNEUROSCI.22-09-03615.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien CB. PY in the fly receptor-like tyrosine phosphatases in axonal pathfinding. Neuron. 1996;16:1065–8. doi: 10.1016/s0896-6273(00)80131-2. [DOI] [PubMed] [Google Scholar]
- Comer AR, Ahern-Djamali SM, Juang JL, Jackson PD, Hoffmann FM. Phosphorylation of Enabled by the Drosophila Abelson tyrosine kinase regulates the in vivo function and protein-protein interactions of Enabled. Mol Cell Biol. 1998;18:152–60. doi: 10.1128/mcb.18.1.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowner D, Le Gall M, Gates MA, Giniger E. Notch steers Drosophila ISNb motor axons by regulating the Abl signaling pathway. Curr Biol. 2003;13:967–72. doi: 10.1016/s0960-9822(03)00325-7. [DOI] [PubMed] [Google Scholar]
- Desai C, Purdy J. The neural receptor protein tyrosine phosphatase DPTP69D is required during periods of axon outgrowth in Drosophila. Genetics. 2003;164:575–88. doi: 10.1093/genetics/164.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai CJ, Gindhart JG, Jr., Goldstein LS, Zinn K. Receptor tyrosine phosphatases are required for motor axon guidance in the Drosophila embryo. Cell. 1996;84:599–609. doi: 10.1016/s0092-8674(00)81035-1. [DOI] [PubMed] [Google Scholar]
- Desai CJ, Krueger NX, Saito H, Zinn K. Competition and cooperation among receptor tyrosine phosphatases control motoneuron growth cone guidance in Drosophila. Development. 1997;124:1941–52. doi: 10.1242/dev.124.10.1941. [DOI] [PubMed] [Google Scholar]
- Drosopoulos NE, Walsh FS, Doherty P. A soluble version of the receptor-like protein tyrosine phosphatase kappa stimulates neurite outgrowth via a Grb2/MEK1-dependent signaling cascade. Mol Cell Neurosci. 1999;13:441–9. doi: 10.1006/mcne.1999.0758. [DOI] [PubMed] [Google Scholar]
- Flanagan JG, Vanderhaeghen P. The ephrins and Eph receptors in neural development. Annu Rev Neurosci. 1998;21:309–45. doi: 10.1146/annurev.neuro.21.1.309. [DOI] [PubMed] [Google Scholar]
- Fogerty FJ, Juang JL, Petersen J, Clark MJ, Hoffmann FM, Mosher DF. Dominant effects of the bcr-abl oncogene on Drosophila morphogenesis. Oncogene. 1999;18:219–32. doi: 10.1038/sj.onc.1202239. [DOI] [PubMed] [Google Scholar]
- Fox DT, Peifer M. Abelson kinase (Abl) and RhoGEF2 regulate actin organization during cell constriction in Drosophila. Development. 2007;134:567–78. doi: 10.1242/dev.02748. [DOI] [PubMed] [Google Scholar]
- Gallo G, Letourneau PC. Axon guidance: A balance of signals sets axons on the right track. Curr Biol. 1999;9:R490–2. doi: 10.1016/s0960-9822(99)80304-2. [DOI] [PubMed] [Google Scholar]
- Garrity PA, Lee CH, Salecker I, Robertson HC, Desai CJ, Zinn K, Zipursky SL. Retinal axon target selection in Drosophila is regulated by a receptor protein tyrosine phosphatase. Neuron. 1999;22:707–17. doi: 10.1016/s0896-6273(00)80730-8. [DOI] [PubMed] [Google Scholar]
- Gertler FB, Comer AR, Juang JL, Ahern SM, Clark MJ, Liebl EC, Hoffmann FM. enabled, a dosage-sensitive suppressor of mutations in the Drosophila Abl tyrosine kinase, encodes an Abl substrate with SH3 domain-binding properties. Genes Dev. 1995;9:521–33. doi: 10.1101/gad.9.5.521. [DOI] [PubMed] [Google Scholar]
- Gertler FB, Doctor JS, Hoffmann FM. Genetic suppression of mutations in the Drosophila abl proto-oncogene homolog. Science. 1990;248:857–60. doi: 10.1126/science.2188361. [DOI] [PubMed] [Google Scholar]
- Goh KL, Cai L, Cepko CL, Gertler FB. Ena/VASP proteins regulate cortical neuronal positioning. Curr Biol. 2002;12:565–9. doi: 10.1016/s0960-9822(02)00725-x. [DOI] [PubMed] [Google Scholar]
- Goodman CS. Mechanisms and molecules that control growth cone guidance. Annu Rev Neurosci. 1996;19:341–77. doi: 10.1146/annurev.ne.19.030196.002013. [DOI] [PubMed] [Google Scholar]
- Grevengoed EE, Fox DT, Gates J, Peifer M. Balancing different types of actin polymerization at distinct sites: roles for Abelson kinase and Enabled. J Cell Biol. 2003;163:1267–79. doi: 10.1083/jcb.200307026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grevengoed EE, Loureiro JJ, Jesse TL, Peifer M. Abelson kinase regulates epithelial morphogenesis in Drosophila. J Cell Biol. 2001;155:1185–98. doi: 10.1083/jcb.200105102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakeda-Suzuki S, Ng J, Tzu J, Dietzl G, Sun Y, Harms M, Nardine T, Luo L, Dickson BJ. Rac function and regulation during Drosophila development. Nature. 2002;416:438–42. doi: 10.1038/416438a. [DOI] [PubMed] [Google Scholar]
- Hermiston ML, Xu Z, Weiss A. CD45: a critical regulator of signaling thresholds in immune cells. Annu Rev Immunol. 2003;21:107–37. doi: 10.1146/annurev.immunol.21.120601.140946. [DOI] [PubMed] [Google Scholar]
- Ignelzi MA, Jr., Miller DR, Soriano P, Maness PF. Impaired neurite outgrowth of src-minus cerebellar neurons on the cell adhesion molecule L1. Neuron. 1994;12:873–84. doi: 10.1016/0896-6273(94)90339-5. [DOI] [PubMed] [Google Scholar]
- Krause M, Bear JE, Loureiro JJ, Gertler FB. The Ena/VASP enigma. J Cell Sci. 2002;115:4721–6. doi: 10.1242/jcs.00218. [DOI] [PubMed] [Google Scholar]
- Krause M, Dent EW, Bear JE, Loureiro JJ, Gertler FB. Ena/VASP proteins: regulators of the actin cytoskeleton and cell migration. Annu Rev Cell Dev Biol. 2003;19:541–64. doi: 10.1146/annurev.cellbio.19.050103.103356. [DOI] [PubMed] [Google Scholar]
- Krueger NX, Van Vactor D, Wan HI, Gelbart WM, Goodman CS, Saito H. The transmembrane tyrosine phosphatase DLAR controls motor axon guidance in Drosophila. Cell. 1996;84:611–22. doi: 10.1016/s0092-8674(00)81036-3. [DOI] [PubMed] [Google Scholar]
- Kussick SJ, Basler K, Cooper JA. Ras1-dependent signaling by ectopically-expressed Drosophila src gene product in the embryo and developing eye. Oncogene. 1993;8:2791–803. [PubMed] [Google Scholar]
- Luo L. Actin cytoskeleton regulation in neuronal morphogenesis and structural plasticity. Annu Rev Cell Dev Biol. 2002;18:601–35. doi: 10.1146/annurev.cellbio.18.031802.150501. [DOI] [PubMed] [Google Scholar]
- Marlo JE, Desai CJ. Loss of phosphatase activity in Ptp69D alleles supporting axon guidance defects. J Cell Biochem. 2006;98:1296–307. doi: 10.1002/jcb.20862. [DOI] [PubMed] [Google Scholar]
- Maurel-Zaffran C, Suzuki T, Gahmon G, Treisman JE, Dickson BJ. Cell-autonomous and -nonautonomous functions of LAR in R7 photoreceptor axon targeting. Neuron. 2001;32:225–35. doi: 10.1016/s0896-6273(01)00471-8. [DOI] [PubMed] [Google Scholar]
- Nicolai M, Lasbleiz C, Dura JM. Gain-of-function screen identifies a role of the Src64 oncogene in Drosophila mushroom body development. J Neurobiol. 2003;57:291–302. doi: 10.1002/neu.10277. [DOI] [PubMed] [Google Scholar]
- Rashid-Doubell F, McKinnell I, Aricescu AR, Sajnani G, Stoker A. Chick PTPsigma regulates the targeting of retinal axons within the optic tectum. J Neurosci. 2002;22:5024–33. doi: 10.1523/JNEUROSCI.22-12-05024.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard M, Giehl K, Abel K, Haffner C, Jarchau T, Hoppe V, Jockusch BM, Walter U. The proline-rich focal adhesion and microfilament protein VASP is a ligand for profilins. Embo J. 1995;14:1583–9. doi: 10.1002/j.1460-2075.1995.tb07146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoble J, Auerbuch V, Goley ED, Welch MD, Portnoy DA. Pivotal role of VASP in Arp2/3 complex-mediated actin nucleation, actin branch-formation, and Listeria monocytogenes motility. J Cell Biol. 2001;155:89–100. doi: 10.1083/jcb.200106061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoker A, Dutta R. Protein tyrosine phosphatases and neural development. Bioessays. 1998;20:463–72. doi: 10.1002/(SICI)1521-1878(199806)20:6<463::AID-BIES4>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Stoker AW, Gehrig B, Haj F, Bay BH. Axonal localisation of the CAM-like tyrosine phosphatase CRYP alpha: a signalling molecule of embryonic growth cones. Development. 1995;121:1833–44. doi: 10.1242/dev.121.6.1833. [DOI] [PubMed] [Google Scholar]
- Wang J, Bixby JL. Receptor tyrosine phosphatase-delta is a homophilic, neurite-promoting cell adhesion molecular for CNS neurons. Mol Cell Neurosci. 1999;14:370–84. doi: 10.1006/mcne.1999.0789. [DOI] [PubMed] [Google Scholar]
- Wills Z, Bateman J, Korey CA, Comer A, Van Vactor D. The tyrosine kinase Abl and its substrate enabled collaborate with the receptor phosphatase Dlar to control motor axon guidance. Neuron. 1999a;22:301–12. doi: 10.1016/s0896-6273(00)81091-0. [DOI] [PubMed] [Google Scholar]
- Wills Z, Emerson M, Rusch J, Bikoff J, Baum B, Perrimon N, Van Vactor D. A Drosophila homolog of cyclase-associated proteins collaborates with the Abl tyrosine kinase to control midline axon pathfinding. Neuron. 2002;36:611–22. doi: 10.1016/s0896-6273(02)01022-x. [DOI] [PubMed] [Google Scholar]
- Wills Z, Marr L, Zinn K, Goodman CS, Van Vactor D. Profilin and the Abl tyrosine kinase are required for motor axon outgrowth in the Drosophila embryo. Neuron. 1999b;22:291–9. doi: 10.1016/s0896-6273(00)81090-9. [DOI] [PubMed] [Google Scholar]
- Yang T, Massa SM, Longo FM. LAR protein tyrosine phosphatase receptor associates with TrkB and modulates neurotrophic signaling pathways. J Neurobiol. 2006;66:1420–36. doi: 10.1002/neu.20291. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.