Abstract
On Earth, it is common to employ laboratory animals such as the nematode Caenorhabditis elegans to help understand human health concerns. Similar studies in Earth orbit should help understand and address the concerns associated with spaceflight. The “International Caenorhabditis elegans Experiment FIRST” (ICE FIRST), was carried out onboard the Dutch Taxiflight in April of 2004 by an international collaboration of laboratories in France, Canada, Japan and the United States. With the exception of a slight movement defect upon return to Earth, the result of altered muscle development, no significant abnormalities were detected in spaceflown C. elegans. Work from Japan revealed apoptosis proceeds normally and work from Canada revealed no significant increase in the rate of mutation. These results suggest that C. elegans can be used to study non-lethal responses to spaceflight and can possibly be developed as a biological sensor. To further our understanding of C. elegans response to spaceflight, we examined the gene transcription response to the 10 days in space using a near full genome microarray analysis. The transcriptional response is consistent with the observed normal developmental timing, apoptosis, DNA repair, and altered muscle development. The genes identified as altered in response to spaceflight are enriched for genes known to be regulated, in C. elegans, in response to altered environmental conditions (Insulin and TGF-β regulated). These results demonstrate C. elegans can be used to study the effects of altered gravity and suggest that C. elegans responds to spaceflight by altering the expression of at least some of the same metabolic genes that are altered in response to differing terrestrial environments.
Keywords: C. elegans, Spaceflight, Microarray, Insulin, TGF-β, Dauer
1. Introduction
Human space exploration is expanding, with various space agencies planning missions to the Moon and Mars. The medical issues connected to such spaceflight will have to be addressed (Board, 1998; Buckey, 1999; Nicogossian et al., 1992). On Earth, it is common practice to employ laboratory animals to help understand human health concerns. Study of model organisms in Earth orbit should similarly help understand and address the concerns associated with spaceflight. The nematode Caenorhabditis elegans has been used as a model system, on Earth, for decades (reviewed by (Barr, 2003; Bahls et al., 2003)) and has been developed into a model organism for space life sciences research (Johnson and Nelson, 1991; Nelson et al., 1994a,b; Hartman et al., 2001; Szewczyk and McLamb, 2005; Szewczyk et al., 2005; Zhao et al., 2005, 2006; Higashitani et al., 2005; Higashibata et al., 2006).
During the Dutch Soyuz mission, DELTA, to the ISS in April of 2004, an international collaboration of laboratories carried out the “International Caenorhabditis elegans Experiment FIRST” (ICE FIRST). One of the main goals of this experiment was to validate the use of a chemically defined liquid culture for C. elegans during spaceflight (Szewczyk et al., 2003, 2006). Such culturing allows automation, which is highly desirable for experimentation during spaceflight (Board, 1998). In addition to validating such culturing, the biological response of C. elegans to the 10 day spaceflight was assessed. Animals displayed a normal rate of development in flight and returned in good apparent health. With the exception of a slight movement defect upon return to Earth, which appears to be due to altered muscle development in flight (Higashibata et al., 2006), no significant abnormalities were detected. Apoptosis proceeds normally (Higashitani et al., 2005) and the rate of mutation associated with flight was below the experiment’s limits of detection (Zhao et al., 2006). These results appear similar to what is observed for humans and suggest that C. elegans can be used to study responses to spaceflight and can possibly be developed as a biological sensor (Zhao et al., 2005; Custodia et al., 2001; Dengg and van Meel, 2004).
To further our understanding of spaceflown C. elegans, we examined the gene transcription response using a near full genome microarray analysis. We performed an analysis on three independent populations and looked for responses that were due to spaceflight across samples. These changes were confirmed in a fourth population using a different microarray platform. The transcriptional response is consistent with the observed normal rate of development, apoptosis (Higashitani et al., 2005), and DNA repair (Zhao et al., 2006). This transcriptional response is also consistent with the previously described altered muscle development in flight (Higashibata et al., 2006), with C. elegans undergoing a metabolic shift in response to spaceflight, and with the genes being regulated by the known environmental regulatory circuits in C. elegans (Insulin and TGF-β). These results demonstrate C. elegans can be used to study the effects of altered gravity and suggest that C. elegans responds to spaceflight by altering the expression of at least some of the same metabolic genes that are altered in response to differing terrestrial environments.
2. Methods
Nematode handling was performed as described (Higashibata et al., 2006; Higashitani et al., 2005; Zhao et al., 2006). Microarray analysis was carried out as described with statistics determined as described (Kim et al., 2001; Higashibata et al., 2006; Higashitani et al., 2005; Szewczyk et al., 2006). cDNA microarray analyses utilized samples hybridized versus standard laboratory C. elegans Maintenance Medium (CeMM (Szewczyk et al., 2003, 2006)) cultures. At the time of experiment conception, calculated post flight worm densities should have yielded enough RNA for one microarray hybridization per sample. Technical advances allowed replicate hybridizations to be performed for all but the first sample analyzed. With the exception of flight sample 4, all hybridizations were performed using two different prints of the microarray so as to remove any concern with print specific problems. The full dataset is available from the Stanford microarray database, www.wormbase.org, or upon request from the authors. The fold (log2) change in gene expression used as a cut off for significant change was 2, this represents p < .05 for all arrays with the p < .05 range being 1.3–1.7 fold (log2) change. When plotted against the genomic self organizing map (Kim et al., 2001) significance was accepted at p < .001. Confirmation of a subset of muscle gene microarray results by RT-PCR and western blot are reported elsewhere (Higashibata et al., 2006).
For sample 4, all transcripts which showed a greater than 4 fold change in response to spaceflight in one sample or a greater than 4 fold change in response to culturing in flight hardware in two samples are shown in Fig. 1. For sample 22, all transcripts which showed a greater than four fold change in response to spaceflight in two samples or a greater than four fold change in response to culturing in flight hardware in two samples are shown in Fig. 2. For sample 23, all transcripts which showed a greater than four fold change in response to spaceflight in two samples are shown in Fig. 3. Note that for sample 23 the transcripts which showed a greater than four fold change in response to culturing in the flight hardware in at least two samples are not displayed due to the large number of transcripts detected and the larger amount of space required to display them. For the pooled analysis across samples, the transcripts that showed a greater than four fold change in response to spaceflight, in at least three samples from at least two populations, are shown in Fig. 4. For the pooled analysis across samples, the transcripts which showed a greater than four fold change in response to spaceflight in at least two populations and not a greater than four fold change in response to culturing in the flight hardware in at least two populations are shown in Table 1.
Fig. 1.
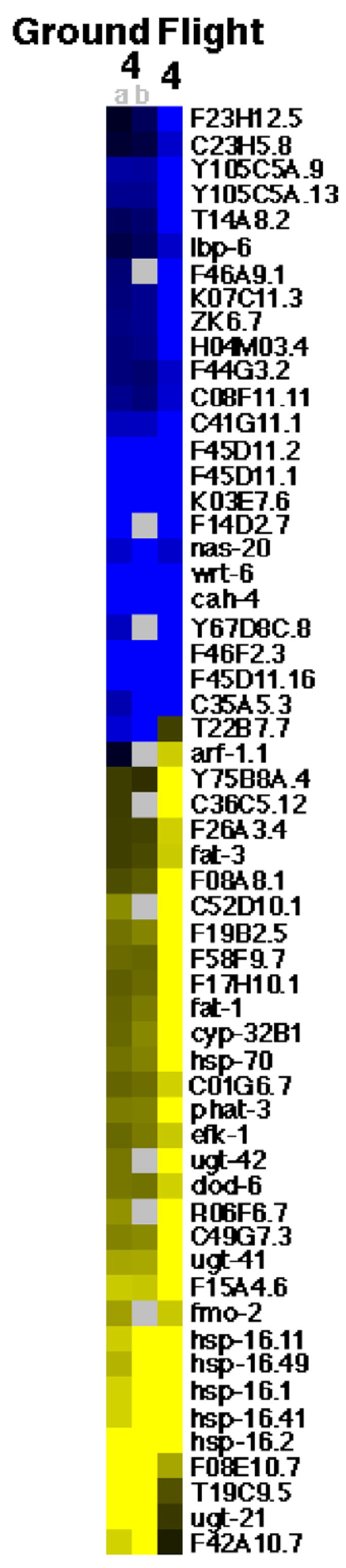
Genomic response of sample 4. Genes (black labels) that display a greater than 4 fold decrease (blue) or increase (yellow) in response to growth in flight hardware on the ground (Ground 4a and b (left)) and/or to growth on orbit (Flight 4 (right)). Both conditions are versus growth in liquid medium on Earth with grey spots indicating no data. Ground 4a and b are replicate hybridizations. (For interpretation of colour in this figure legend, the reader is referred to the web version of the article.)
Fig. 2.

Genomic response of sample 22. Genes (black labels) that display a greater than 4 fold decrease (blue) or increase (yellow) in response to growth in flight hardware on the ground (Ground 22a,b and c (left)) and/or to growth on orbit (Flight 22a,b, and c (right)). Both conditions are versus growth in liquid medium on Earth with grey spots indicating no data. Ground 22a,b, and c and Flight 22a,b, and c are replicate hybridizations. (For interpretation of colour in this figure legend, the reader is referred to the web version of the article.)
Fig. 3.
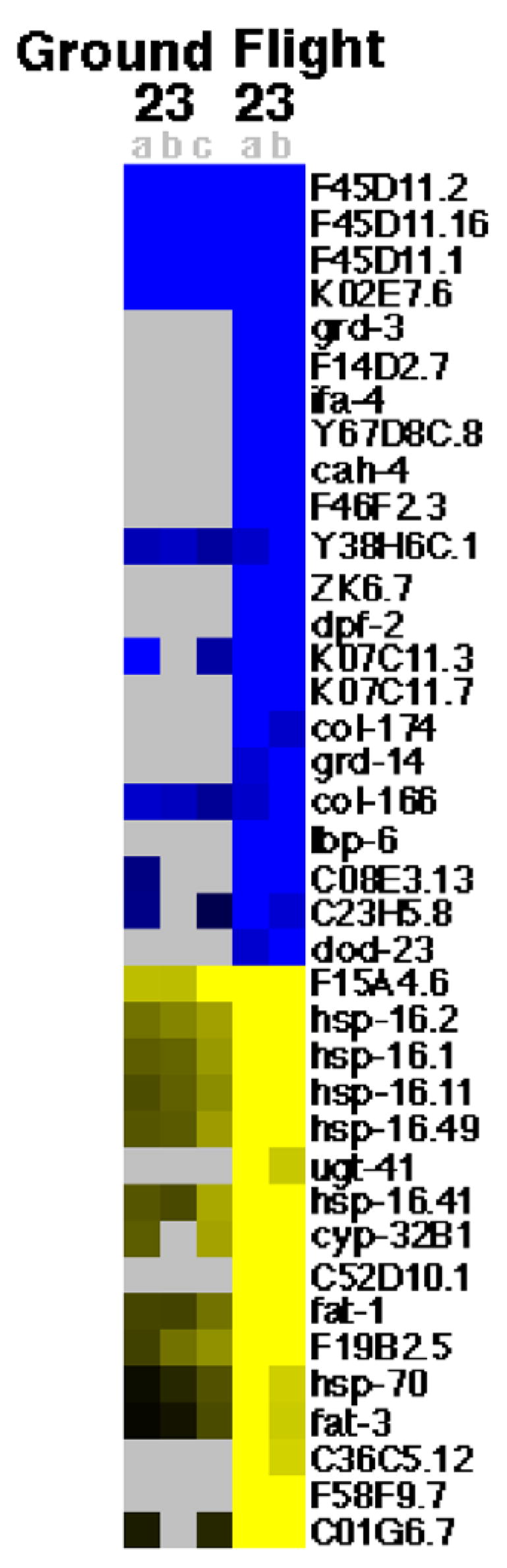
Genomic response of sample 23 to spaceflight. Genes (black labels) that display a greater than 4 fold decrease (blue) or increase (yellow) in response to growth in flight hardware on orbit (Flight 23a and b (right)) compared to growth in flight hardware on Earth (Ground 23a,b and c (left)). Both conditions are versus growth in liquid medium on Earth with grey spots indicating no data. Ground 23a,b, and c and Flight 22a and b are replicate hybridizations. Genes displayed were chosen based upon spaceflight induced reproducible changes. (For interpretation of colour in this figure legend, the reader is referred to the web version of the article.)
Fig. 4.

Genomic response of C. elegans to spaceflight. Genes (black labels) that display a greater than 4 fold decrease (blue) or increase (yellow) in response to growth in flight hardware on orbit (Flight (right)) compared with growth in flight hardware on Earth (Ground (left)). Both conditions are versus growth in liquid medium on Earth with grey spots indicating no data. Genes displayed were chosen based upon spaceflight induced reproducible changes (two independent populations based upon replicate analysis (see Figs. 1–3) and chips from multiple printings). (For interpretation of colour in this figure legend, the reader is referred to the web version of the article.)
Table 1.
Transcripts with significantly altered expression in response to spaceflight alone
| Fold Changea | Gene pairs name | Description | Regulation/mutant phenotype | |
|---|---|---|---|---|
| (a) Downregulated transcripts | ||||
| 2.05 ± .13 | Y38H6C.1 | Uncharacterized | Regulated by refeeding, TGF-β | |
| 2.09 ± .09 | C23H5.8 | Uncharacterized, isomerase? | ||
| 2.23 ± .3 | C08F11.11 | Uncharacterized | ||
| 2.3 ± .52 | C35A5.3 | Sodium-dependent phosphate transporter | Upregulated by hypoxia? | |
| 2.31 ± .28 | C41G11.1 | Uncharacterized, Metallophosphoesterase? | ||
| 2.31 ± .16 | ZK6.7 | Triacylglycerol lipase | Regulated during Dauer recovery | |
| 2.34 ± .33 | T14A8.2 | Uncharacterized, contains a GPI anchor | ||
| 2.52 ± .24 | K07C11.3 | Metalloproteinase inhibitor | Regulated during Dauer recovery | |
| 2.75 ± .52 | F14D2.7 | Uncharacterized | ||
| 3.19 ± .38 | K07C11.7 | Phosphoesterase | Regulated during Dauer recovery | |
| Fold changea | Gene pairs name | Gene name | Description | Regulation/mutant phenotype |
|
| ||||
| (b) Upregulated transcripts | ||||
| 2.00 ± .12 | W08D2.4 | fat-3 | Delta-6 fatty acid desaturase | RNAi- Altered developmental timing |
| 2.06 ± .19 | C49G7.4 | phat-3 | Secreted surface protein | Regulated during Dauer recovery |
| 2.09 ± .55 | R06F6.7 | Uncharacterized | ||
| 2.10 ± .33 | F31F4.7 | ugt-42 | UDP-glucuronosyl transferase | |
| 2.11 ± .29 | C49G7.3 | Secreted surface protein | Regulated during Dauer recovery | |
| 2.16 ± .15 | F17H10.1 | Serine active site containing 1 homologue? | PCB52 (organic toxin) induced | |
| 2.17 ± .17 | C01G6.7 | Acyl-CoA synthetase | Regulated during Dauer recovery | |
| 2.19 ± .1 | F08A8.1 | Acyl-CoA oxidase | Repressed by fasting | |
| 2.32 ± .22 | F10D2.11 | ugt-41 | UDP-glucuronosyl transferase | |
| 2.35 ± .3 | C12C8.1 | hsp-70 | Molecular chaperones HSP70 | Hypoxia induced |
| 2.38 ± .15 | Y5H2B.5 | cyp-32B1 | Cytochrome P450 | |
| 2.42 ± .06 | Y67H2B.8 | fat-1 | Omega-3 fatty acyl desaturase | Regulated by fasting |
| 2.43 ± .14 | F58F9.7 | Pristanoyl-CoA/acyl-CoA oxidase | Regulated during Dauer recovery | |
| 2.47 ± .14 | F15A4.6 | Secreted surface protein | ||
| 2.56 ± .15 | T27E4.9 | hsp-16.49 | Alpha crystallin | Insulin regulated |
| 2.56 ± .15 | Y46H3A.2 | hsp-16.41 | Alpha crystallin | Unfolded protein response induced |
| 2.59 ± .33 | C36C5.12 | Secreted cysteine rich protein | ||
| 2.84 ± .08 | F19B2.5 | Helicase-like transcription factor | Regulated by mechanosensation | |
| 3.19 ± .5 | C52D10.1 | N-myristoylated nuclear protein | Regulated during Dauer recovery | |
Value is mean ± s.d. (log2) across all flight samples (not just those >2).
3. Results
3.1. Altered gene expression for sample 4
Sample 4 was located in the fourth of eleven sample bags inside the European Space Agency (ESA) Type-1 Experiment Container (EC-1) labeled as “ICE-08”. Sample 1, which showed no movement defect post flight and a normal rate of development, was also located in this EC-1. Sufficient RNA was extracted for hybridization to one flight sample and two ground sample microarrays.
Relative to standard culturing conditions, significantly increased expression in response to spaceflight was observed for 28 genes and decreased expression was observed for 24 genes (Fig. 1). Plotted against the C. elegans genomic self organizing map (Kim et al., 2001), these data suggest decreased expression of mounts 17 (collagen), 19 (amino acid metabolism), and 22 (collagen) and increased expression of mounts 24 (fatty acid oxidation) and 36 (heat shock stress). Analysis of the sample 4 ground control revealed significantly decreased expression of 9 genes, and increased expression of 9 genes on both arrays (Fig. 1). These data suggest decreased expression of mount 17 and increased expression of mount 36. When these data for the ground sample are subtracted from flight sample data we see decreased expression of mounts 19, 21 (lipid metabolism), and 22 and increased expression of mount 24 in response to spaceflight. Taken together the data suggest both that C. elegans exhibited a stress response to culturing in the flight hardware and that C. elegans undergo a metabolic shift in response to spaceflight.
3.2. Altered gene expression for sample 22
Sample 22 was located in the eleventh of twelve sample bags inside the EC-1 labeled “ICE-05”. Sample 12, which showed a movement defect post flight and a normal rate of development, was also located in this EC-1. Sufficient RNA was extracted for hybridization to three flight sample and two ground sample microarrays.
Relative to standard culturing conditions, significantly increased expression in response to spaceflight was observed for 25 genes on at least two arrays; decreased expression of 28 genes was noted on at least two arrays (Fig. 2). Plotted against the C. elegans genomic self organizing map (Kim et al., 2001), these data suggest decreased expression of mounts 17 (collagen), 19 (amino acid metabolism), and 22 (collagen) and increased expression of mounts 21 (lipid metabolism), 24 (fatty acid oxidation) and 36 (heat shock stress). Analysis of the sample 22 ground control revealed significantly decreased expression of 27 genes, and increased expression of 13 genes on both arrays (Fig. 2). These data suggest decreased expression of mounts 8 (intestinal) and 17 and increased expression of mount 36. When these data for the ground sample are subtracted from flight sample data we see decreased expression of mounts 1 (neuromuscular), 19, and 22 and increased expression of mount 36 in response to spaceflight. Taken together the data suggest that C. elegans exhibited a stress response to culturing in the flight hardware, with spaceflight further increasing this stress response. Additionally, the data suggest that C. elegans undergo a metabolic shift in response to spaceflight and confirm the past report (Higashibata et al., 2006) of altered muscle development in response to spaceflight.
3.3. Altered gene expression for sample 23
Sample 23 was located in the last of twelve sample bags inside the EC-1 labeled “ICE-05”. This is the same EC-1 as sample 22 above. Unlike samples 4 and 22, sample 23 was immediately adjacent to the Gortex® membrane and potentially received more oxygenation. Sufficient RNA was extracted for hybridization to two flight sample and three ground sample microarrays.
Relative to standard culturing conditions, significantly increased expression in response to spaceflight was observed for 16 genes on both arrays; decreased expression of 23 genes was noted on both arrays (Fig. 3). Plotted against the C. elegans genomic self organizing map (Kim et al., 2001), these data suggest decreased expression of mounts 8 (intestinal), 17 (collagen), 19 (amino acid metabolism), and 22 (collagen) and increased expression of mount 36 (heat shock). Analysis of the sample 23 ground control revealed significantly decreased expression of 14 genes, and increased expression of 173 genes on at least two arrays. These data suggest increased expression of mount 1 (neuromuscular). When these data for the ground sample are subtracted from flight sample data we see decreased expression of mounts 1, 8, 19, and 22 and increased expression of mounts 16 (muscle) and 36 in response to spaceflight. Taken together the data suggest that C. elegans exhibited no stress response to culturing in the flight hardware with spaceflight alone causing a stress response. Additionally, the data suggest that C. elegans undergo a metabolic shift, largely in the intestines, in response to spaceflight and confirm the past report (Higashibata et al., 2006) of altered muscle development in response to spaceflight is not limited to a single population of spaceflown worms.
3.4. Altered gene expression across samples
To examine common responses of the C. elegans genome to spaceflight we compared the data obtained for the three independent populations described above. Several approaches were taken, the results of three methods are described below with two displayed as graphics.
When the data from all three populations are pooled and analyzed for genes that show greater than a four fold change in expression for at least three samples from at least two of the populations, 22 genes show increased expression while 17 genes show decreased expression in response to spaceflight (Fig. 4). Plotted against the C. elegans genomic self organizing map (Kim et al., 2001), these data suggest decreased expression of mounts 17 (collagen), 19 (amino acid metabolism), and 22 (collagen) and increased expression of mount 36 (heat shock stress). Subtraction of the pooled results from the similarly pooled ground controls indicates decreased expression of mounts 1 (neuromuscular), 19 (amino acid metabolism), and 22 (collagen) and increased expression of mounts 8 (intestinal) and 36 (heat shock stress) in response to spaceflight.
When the data from all three populations are pooled and analyzed for genes that show greater than a four fold change in expression for at least two populations in response to spaceflight and not greater than a four fold change in expression for at least two ground populations, 19 genes show increased expression while 10 genes show decreased expression in response to spaceflight (Table 1). Plotted against the C. elegans genomic self organizing map (Kim et al., 2001), these data suggest decreased expression of mounts 19 (amino acid metabolism), and 22 (collagen) and increased expression of mount 36 (heat shock stress). Of note, 13 of the 29 identified genes are genes known to be regulated in response to changing environmental conditions in C. elegans (regulated by Insulin and TGF-β signaling in C. elegans (Hu, 2007)).
The combined data suggest a transcriptional response consistent with the observed normal rate of development, apoptosis (Higashitani et al., 2005), and DNA repair (Zhao et al., 2006). The transcriptional response is also consistent with the previously described altered muscle development in flight (Higashibata et al., 2006) and with C. elegans undergoing a metabolic shift in response to spaceflight. The results of our pooled data analysis were confirmed by determining that changes also occurred in a fourth population for which data were validated by RT-PCR and western blot (Higashibata et al., 2006). Of note, this fourth population also utilized a different microarray platform, indicating that the expression changes we report here are highly reproducible across populations from this flight regardless of procedural differences in microarray printing, hybridization, and analysis.
3.5. Gene expression in response to form of medium
Increased mechanical load is commonly employed as a countermeasure to spaceflight induced biological changes, although it is also well recognized that exercise results in only partial amelioration of the negative effects of spaceflight (Board, 1998; Buckey, 1999; Nicogossian et al., 1992). The on-flight centrifuge failed, precluding us from directly looking at the effects of increased mechanical load on the transcriptional response. We therefore investigated the transcriptional response of C. elegans grown on solid or in liquid chemically defined medium (Szewczyk et al., 2003). We find 78 genes show decreased expression and 173 genes show increased expression across three replicates. As this response is distinct from that seen in response to changing both diet and form of the medium (Szewczyk et al., 2006) it may be, at least partially due to the difference in externally-applied surface tension, a force roughly equivalent to 10,000× unit Gravity (Szewczyk et al., 2005). Plotting this response against the C. elegans gene expression self organizing map reveals mounts 8 (intestinal), 12 (neuronal), 15, 22 (collagen), and 36 (heat shock stress) are down regulated and 4 (sperm), 19 (amino acid metabolism), 21 (lipid metabolism), and 24 (fatty acid oxidation) are up regulated in response to increased mechanical load. When compared to the response to spaceflight (Fig. 5) these results suggest that 70% of the genes induced and 40% of the genes repressed by spaceflight could, in C. elegans, be blocked by changing the form of the medium from liquid to solid. While this suggestion remains to be confirmed in flight, these observations suggest that future missions utilizing increased mechanical load might be able to block the increased expression of stress response but not the decreased expression of muscle or skeletal (collagen) genes. Alternatively, our results may suggest that the majority of gene expression changes observed in flight are due to culture conditions (e.g engineered environmental conditions) present on board the International Space Station. As examples, increased hypoxia or vibration may occur both in culturing on board the International Space Station (for a variety of reasons) and in response to liquid culture vs. culturing on solid medium.
Fig. 5.

Increased mechanical load appears to partially reverse the genomic response to spaceflight. Genes (black labels) that display a greater than 4 fold decrease (blue) or increase (yellow) in response to growth in liquid during spaceflight (μG (left)) compared with growth on solid medium (10,000× G (right)). Both conditions are versus growth in liquid medium on Earth with grey spots indicating no data. Genes displayed were chosen based upon spaceflight induced reproducible changes (four independent populations based upon replicate analysis and utilizing two different microarray platforms (Stanford cDNA (see Figs. 1–4) and Affymetrix C. elegans) and chips from multiple printings). (For interpretation of colour in this figure legend, the reader is referred to the web version of the article.)
4. Discussion
4.1. Altered gene expression in response to spaceflight
Analysis of four independent populations of C. elegans, utilizing two different microarray platforms, indicates a reproducible genomic response to spaceflight. This transcriptional response is consistent with the observed biology of spaceflown animals. Specifically, spaceflown animals undergo normal developmental timing, apoptosis (Higashitani et al., 2005), and DNA repair (Zhao et al., 2006), but altered muscle development (Higashibata et al., 2006). At least one additional flight is required to confirm that this response is not unique to ICE-FIRST. At present our results suggest that C. elegans can be used to study both the genomic and physiologic effects of spaceflight with an eye toward human spaceflight countermeasure development.
The other principle finding of our microarray analysis is that spaceflown C. elegans undergo a metabolic shift, which may occur largely in the intestines, and exhibit a stress response. Our analysis identified a set of genes that is significantly enriched for genes regulated by Insulin and/or TGF-β signalling. These observations suggest that altered Insulin and/or TGF-β signalling underlie all or part of the metabolic changes observed. Altered lipid metabolism can lead to altered endocrine signalling (Dowell et al., 2005), so future experiments will be required to determine if altered lipid metabolism leads to transcriptional responses like those controlled by Insulin and TGF-β, or conversely if altered Insulin and TGF-β signalling lead to altered lipid metabolism.
Full genome analysis for other whole organisms also needs to be completed (Nichols et al., 2006) to determine how much of our findings are C. elegans specific. A comparison of our results with those obtained from spaceflown Drosophila melanogaster suggests that while an altered profile of metabolic genes is conserved, the specific genes affected are mostly organism specific (Leandro et al., 2007). In contrast, our suggestion that altered insulin like signalling is responsible for the bulk of the gene changes observed is consistent with the observation of altered insulin like signalling in astronauts (Macho et al., 2003; Tobin et al., 2002). Additionally, given the fact that some observed muscle gene expression changes are identical in spaceflown chick, rodent, and human muscle (Nichols et al., 2006; Higashibata et al., 2006) it may be the case that decreased mechanical load alone does not account for the altered muscle biology observed in these species; this has previously been suggested to be true for humans (Nicogossian et al., 1992). An attractive preliminary model, that our data appear to support, is that altered metabolism, perhaps under the control of insulin like signaling, underlies at least part of the altered muscle biology (Tobin et al., 2002). If this turns out to be the case, it raises the hope that nutritional and/or small molecule countermeasures that are developed for Earth based metabolic muscle pathologies can be successfully used to counter spaceflight induced muscle alterations.
4.2. C. elegans as a model for space life research
With an increasing number of people spending an increasing amount of time in space, and with discussions of new missions for human planetary exploration, it seems sensible to develop existing Earth based life sciences models into models for space life sciences. The nematode C. elegans is a commonly employed genetic model organism on Earth. A number of laboratories are already studying processes relevant to astronaut health, including radiation and muscle biology and immune and stress responses.
ICE-FIRST was the fifth spaceflight for C. elegans, and the first on-board the International Space Station. Data from STS-42 (Nelson et al., 1994a,b), STS-107 (Szewczyk et al., 2005), and ICE-FIRST, demonstrate that C. elegans can successfully complete at least two continuous full life cycles in space without developmental abnormalities that impair survival. It also has been demonstrated that animals will exhibit an increased rate of mutation as the direct effect of any increased cosmic radiation, independent of microgravity (Hartman et al., 2001). Evidence from ICE-FIRST suggests that when animals are protected from cosmic radiation, rates of mutation are comparable to those on Earth (Zhao et al., 2006), and there are no obvious differences in expression of known radiation or DNA repair genes. Together these results suggest there are no difficulties in employing C. elegans as a model organism for space life sciences. However, results from STS-107 (Szewczyk et al., 2005), STS-95, and ICE-FIRST all suggest that bio-incompatibility of the flight hardware can pose a significant challenge to successful experimentation in space. Therefore, careful testing of flight hardware bio-compatibility should be viewed as an essential step of any future experimentation with C. elegans in space.
At present, four lines of future research with C. elegans in space seem obvious. First, it seems logical to determine if factors that increase C. elegans radiation resistance on Earth (for example oxidative stress pretreatment (Yanase et al., 1999)) also do so in-flight. These clearly represent avenues for countermeasures development. Second, the observations of altered muscle development (Higashibata et al., 2006) and increased telomere length (Zhao et al., 2006) should be examined for reproducibility. The microarray analysis presented here suggests that while altered muscle development is seen across four populations of spaceflown C. elegans, it is not seen to the same extent. For example, sample 4 showed less of a depression in muscle gene expression than samples 22 and 23, and altered movement was only noted for animals housed with the latter samples. Understanding the mechanisms underlying altered muscle biology in space is an important issue for countermeasure development. Longer telomeres were noted in spaceflown animals (Zhao et al., 2006) and our microarray data suggest altered expression of PIF1 (Schulz and Zakian, 1994), which is reported to regulate telomere length. Given hypotheses of altered telomere length leading to premature death and/or development of cancer (Bertuch and Lundblad, 2006) it seems prudent to follow up on the response of C. elegans telomere length to spaceflight. Third, spaceflown C. elegans should be examined for altered pathogen resistance. The genes identified as altered in response to spaceflight are enriched for those regulated by TGF-β and insulin signaling, pathways that regulate innate immunity in C. elegans (Millet and Ewbank, 2004). Given the altered immunity of astronauts (Board, 1998; Buckey, 1999; Nicogossian et al., 1992), a genetic small animal model for spaceflight induced alterations in immunity would be of great value. Lastly, with ICE-FIRST we have demonstrated that it is possible to culture C. elegans in chemically defined liquid medium on-orbit. It now seems reasonable to begin automated culturing of C. elegans on-orbit. Such culturing raises the possibility of examining long term responses of C. elegans to spaceflight and the development of C. elegans as a bio-monitor for radiation (Zhao et al., 2005), environmental toxicity (Custodia et al., 2001; Dengg and van Meel, 2004), and/or other factors of medical concern that are associated with spaceflight. Given the suitability of C. elegans, such future automated experimentation could be done on unmanned satellites or interplanetary missions.
Acknowledgments
The Dutch Soyuz mission, DELTA, was facilitated by the Dutch Government. Funding for the experiment was provided by NASA, CNES, JAXA and CSA. Thanks to Professor Eberhard Horn for use of the modified EC1s. Thanks to Dr. Andre Kuipers, Cdr. Gannady Padalka, Flt. Eng. Michael Fincke, Cdr. Michael Foale, and Flt. Eng. Alexander Kaleri for in flight payload operations and support. Thanks to Comat, ESA, Roscosmos and Energia for payload support. Thanks to Gilbert Gasset, Brigitte Eche, Didier Chaput, Michel Viso, and Stuart Kim for technical assistance. Gene assignments in Table 1 were facilitated by use of www.wormbase.org and Worm-PD™ (www.biobase-international.com).
References
- Bahls C, Weitzman J, Gallagher R. Biology’s Models. The Scientist. 2003;17:S5. [Google Scholar]
- Barr MM. Super models. Physiol Genomics. 2003;13:15–24. doi: 10.1152/physiolgenomics.00075.2002. [DOI] [PubMed] [Google Scholar]
- Bertuch AA, Lundblad V. The maintenance and masking of chromosome termini. Curr Opin Cell Biol. 2006;18:247–253. doi: 10.1016/j.ceb.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Board SS. A Strategy for Research in Space Biology and Medicine in the New Century. National Research Council; Washington, D.C: 1998. [Google Scholar]
- Buckey JC., Jr Preparing for Mars: the physiologic and medical challenges. Eur J Med Res. 1999;4:353–356. [PubMed] [Google Scholar]
- Custodia N, Won SJ, Novillo A, Wieland M, Li C, Callard IP. Caenorhabditis elegans as an environmental monitor using DNA microarray analysis. Ann NY Acad Sci. 2001;948:32–42. doi: 10.1111/j.1749-6632.2001.tb03984.x. [DOI] [PubMed] [Google Scholar]
- Dengg M, van Meel JC. Caenorhabditis elegans as model system for rapid toxicity assessment of pharmaceutical compounds. J Pharmacol Toxicol Methods. 2004;50:209–214. doi: 10.1016/j.vascn.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Dowell P, Hu Z, Lane MD. Monitoring energy balance: metabolites of fatty acid synthesis as hypothalamic sensors. Annu Rev Biochem. 2005;74:515–534. doi: 10.1146/annurev.biochem.73.011303.074027. [DOI] [PubMed] [Google Scholar]
- Hartman PS, Hlavacek A, Wilde H, Lewicki D, Schubert W, Kern RG, Kazarians GA, Benton EV, Benton ER, Nelson GA. A comparison of mutations induced by accelerated iron particles versus those induced by low earth orbit space radiation in the FEM-3 gene of Caenorhabditis elegans. Mutat Res. 2001;474:47–55. doi: 10.1016/s0027-5107(00)00154-8. [DOI] [PubMed] [Google Scholar]
- Higashibata A, Szewczyk NJ, Conley CA, Imamizo-Sato M, Higashitani A, Ishioka N. Decreased expression of myogenic transcription factors and myosin heavy chains in Caenorhabditis elegans muscles developed during spaceflight. J Exp Biol. 2006;209:3209–3218. doi: 10.1242/jeb.02365. [DOI] [PubMed] [Google Scholar]
- Higashitani A, Higashibata A, Sasagawa Y, Sugimoto T, Miyazawa Y, Szewcyk NJ, Viso M, Gasset G, Eche B, Fukui K, Shimazu T, Fujimoto N, Kuriyama K, Ishioka N. Checkpoint and physiological apoptosis in germ cells proceeds normally in spaceflown Caenorhabditis elegans. Apoptosis. 2005;10:949–954. doi: 10.1007/s10495-005-1323-3. [DOI] [PubMed] [Google Scholar]
- Hu PJ Dauer. Riddle DL, editor. The C. elegans Research Community. Wormbook. 2007 August 08;:1–13. doi: 10.1895/wormbook.1.144.1. http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Johnson TE, Nelson GA. Caenorhabditis elegans: a model system for space biology studies. Exp Gerontol. 1991;26:299–309. doi: 10.1016/0531-5565(91)90024-g. [DOI] [PubMed] [Google Scholar]
- Kim SK, Lund J, Kiraly M, Duke K, Jiang M, Stuart JM, Eizinger A, Wylie BN, Davidson GS. A gene expression map for Caenorhabditis elegans. Science. 2001;293:2087–2092. doi: 10.1126/science.1061603. [DOI] [PubMed] [Google Scholar]
- Leandro LJ, Szewcyk NJ, Benguria A, Herranz R, Lavan D, Medina FJ, Gasset G, van Loon J, Conley CA, Marco R. Comparative analysis of Drosophila melanogaster and Caenorhabditis elegans gene expression experiments in the European Soyuz flights to the International Space Station. Adv Space Res. 2007;40:506–512. doi: 10.1016/j.asr.2007.05.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho L, Koska J, Ksinantova L, Pacak K, Hoff T, Noskov VB, Grigoriev AI, Vigas M, Kvetnansky R. The response of endocrine system to stress loads during space flight in human subject. Adv Space Res. 2003;31:1605–1610. doi: 10.1016/s0273-1177(03)00097-8. [DOI] [PubMed] [Google Scholar]
- Millet AC, Ewbank JJ. Immunity in Caenorhabditis elegans. Curr Opin Immunol. 2004;16:4–9. doi: 10.1016/j.coi.2003.11.005. [DOI] [PubMed] [Google Scholar]
- Nelson GA, Schubert WW, Kazarians GA, Richards GF. Development and chromosome mechanics in nematodes: results from IML-1. Adv Space Res. 1994a;14:209–214. doi: 10.1016/0273-1177(94)90405-7. [DOI] [PubMed] [Google Scholar]
- Nelson GA, Schubert WW, Kazarians GA, Richards GF, Benton EV, Benton ER, Henke R. Radiation effects in nematodes: results from IML-1 experiments. Adv Space Res. 1994b;14:87–91. doi: 10.1016/0273-1177(94)90455-3. [DOI] [PubMed] [Google Scholar]
- Nichols HL, Zhang N, Wen X. Proteomics and genomics of microgravity. Physiol Genomics. 2006;26:163–171. doi: 10.1152/physiolgenomics.00323.2005. [DOI] [PubMed] [Google Scholar]
- Nicogossian AE, Rummel JD, Leveton L, Teeter R. Development of countermeasures for medical problems encountered in space flight. Adv Space Res. 1992;12:329–337. doi: 10.1016/0273-1177(92)90301-d. [DOI] [PubMed] [Google Scholar]
- Schulz VP, Zakian VA. The saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell. 1994;76:145–155. doi: 10.1016/0092-8674(94)90179-1. [DOI] [PubMed] [Google Scholar]
- Szewczyk NJ, Kozak E, Conley CA. Chemically defined medium and Caenorhabditis elegans. BMC Biotechnology 3. 2003;19(1–7) doi: 10.1186/1472-6750-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szewczyk NJ, Mancinelli RL, McLamb W, Reed D, Blumberg BS, Conley CA. Caenorhabditis elegans survives atmospheric breakup of STS-107, space shuttle Columbia. Astrobiology. 2005;5:690–705. doi: 10.1089/ast.2005.5.690. [DOI] [PubMed] [Google Scholar]
- Szewczyk NJ, McLamb W. Surviving atmospheric spacecraft breakup. Wilderness Environ Med. 2005;16:27–32. doi: 10.1580/pr21-03.1. [DOI] [PubMed] [Google Scholar]
- Szewczyk NJ, Udranszky IA, Kozak E, Sunga J, Kim SK, Jacobson LA, Conley CA. Delayed development and lifespan extension as features of metabolic lifestyle alteration in C. elegans under dietary restriction. J Exp Biol. 2006;209:4129–4139. doi: 10.1242/jeb.02492. [DOI] [PubMed] [Google Scholar]
- Tobin BW, Uchakin PN, Leeper-Woodford SK. Insulin secretion and sensitivity in space flight: diabetogenic effects. Nutrition. 2002;18:842–848. doi: 10.1016/s0899-9007(02)00940-1. [DOI] [PubMed] [Google Scholar]
- Yanase S, Hartman PS, Ito A, Ishii N. Oxidative stress pretreatment increases the X-radiation resistance of the nematode Caenorhabditis elegans. Mutat Res. 1999;426:31–39. doi: 10.1016/s0027-5107(99)00079-2. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Johnsen R, Baillie D, Rose A. Worms in space? A model biological dosimeter. Gravit Space Biol Bull. 2005;18:11–16. [PubMed] [Google Scholar]
- Zhao Y, Lai K, Cheung I, Youds J, Tarailo M, Tarailo S, Rose A. A mutational analysis of Caenorhabditis elegans in space. Mutat Res. 2006;601:19–29. doi: 10.1016/j.mrfmmm.2006.05.001. [DOI] [PubMed] [Google Scholar]


