Abstract
Asymmetric cell division and apoptosis (programmed cell death) are two fundamental processes that are important for the development and function of multicellular organisms. We have found that the processes of asymmetric cell division and apoptosis can be functionally linked. Specifically, we show that asymmetric cell division in the nematode Caenorhabditis elegans is mediated by a pathway involving three genes, dnj-11 MIDA1, ces-2 HLF, and ces-1 Snail, that directly control the enzymatic machinery responsible for apoptosis. Interestingly, the MIDA1-like protein GlsA of the alga Volvox carteri, as well as the Snail-related proteins Snail, Escargot, and Worniu of Drosophila melanogaster, have previously been implicated in asymmetric cell division. Therefore, C. elegans dnj-11 MIDA1, ces-2 HLF, and ces-1 Snail may be components of a pathway involved in asymmetric cell division that is conserved throughout the plant and animal kingdoms. Furthermore, based on our results, we propose that this pathway directly controls the apoptotic fate in C. elegans, and possibly other animals as well.
Author Summary
Asymmetric cell division and apoptosis (programmed cell death) are two fundamental processes that are important for the development and function of multicellular organisms. Asymmetric cell division creates daughter cells of different fates, and this is critical for the generation of cellular diversity. Apoptosis eliminates superfluous cells from the organism, which is critical for cellular homeostasis. We found that the processes of asymmetric cell division and apoptosis can be functionally linked. Specifically, we show that asymmetric cell division in the nematode Caenorhabditis elegans is mediated by a pathway involving three genes, dnj-11 MIDA1, ces-2 HLF, and ces-1 Snail, that directly control the enzymatic machinery responsible for apoptosis. Interestingly, the role of this pathway in asymmetric cell division and the control of apoptosis might be evolutionarily conserved. Furthermore, it might have an unexpected role in stem cell biology: the process of asymmetric cell division plays an essential role in the ability of stem cells to self-renew, and the mammalian counterparts of two components of the dnj-11 MIDA1, ces-2 HLF, ces-1 Snail pathway have recently been implicated in stem cell function. For this reason, we speculate that a dnj-11 MIDA1, ces-2 HLF, ces-1 Snail–like pathway might function in stem cells to coordinate self-renewal and apoptosis and, hence, the number of stem cells.
A pathway involved in asymmetric cell division in the nematode Caenorhabditis elegans, the dnj-11 MIDA1, ces-2 HLF, ces-1 Snail pathway, directly controls the enzymatic machinery responsible for apoptosis.
Introduction
Asymmetric cell division is essential for the generation of cellular diversity during animal development [1]. In certain cases, one of the cells derived from an asymmetric division is specified to undergo apoptosis (programmed cell death) [2–11]. However, the genetic and cell biological mechanisms that permit the coupling of asymmetric cell division and the adoption of the apoptotic fate are not well understood.
Previous studies have implicated members of the Snail family of transcriptional repressors in both asymmetric cell division and apoptosis, but there has been no demonstration that these processes are integrated in a given developmental context. In mammals, Snail-related proteins have been shown to have pro-survival as well as anti-apoptotic activities, and they have been causally linked to tumorigenesis and tumor progression in mammals [12,13]. In Drosophila melanogaster, the Snail-related genes snail, escargot, and worniu have been shown to function redundantly in asymmetric cell division [14,15]. Specifically, rather than dividing asymmetrically and producing two daughter cells of different sizes and fates, neuroblasts in the central nervous system of D. melanogaster mutants lacking snail, escargot, and worniu function divide symmetrically to produce two daughter cells of similar sizes and fates. The effect of snail, escargot, and worniu on asymmetric neuroblast division is mediated in part by their ability to promote the expression of the gene inscuteable, which encodes an adaptor protein that is required for the establishment and maintenance of neuroblast polarity [16]. The protein Inscuteable is localized in a polar fashion through its interaction with a complex composed of the PDZ domain–containing proteins Bazooka (also referred to as Par-3) and Par-6, which are found on the apical cell cortex of the neuroblasts [17]. Inscuteable in turn recruits the adaptor protein Pins and the alpha subunit of the heterotrimeric G protein Gi (Gαi), which initiates the displacement of the mitotic spindle along the cell division axis of the neuroblasts, resulting in their asymmetric division. Inscuteable is also at least partially required for the enrichment of cell-fate determinants such as Prospero and Staufen on the basal cell cortex of the neuroblasts and their asymmetric segregation into the basal daughter cell [17].
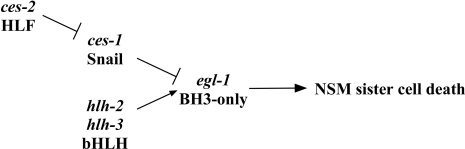
In Caenorhabditis elegans, the Snail-related protein CES-1 (cell-death specification) has been implicated in the suppression of a specific apoptotic event, the death of the neurosecretory motoneuron (NSM) sister cell [18,19]. During embryonic development, the NSM neuroblast divides to give rise to two daughter cells: the NSM, which differentiates into a serotonergic motoneuron, and the NSM sister cell, which undergoes apoptosis [3]. A gain-of-function mutation of the ces-1 gene, which most likely results in the mis- or overexpression of ces-1 in the NSM lineage, prevents the death of the NSM sister cell [18]. The death of the NSM sister cell is dependent on the transcriptional upregulation of the pro-apoptotic BH3-only gene egl-1 (egg-laying abnormal) in the NSM sister cell, a process that is at least partially dependent on a heterodimer composed of the bHLH transcription factors HLH-2 and HLH-3 (HLH-2/HLH-3) [20] (Figure 1). HLH-2/HLH-3 can bind to E-boxes/Snail-binding sites in a cis-regulatory region of the egl-1 locus referred to as Region B, which is required for the expression of egl-1 in the NSM sister cell in vivo. Therefore, it has been proposed that HLH-2/HLH-3 is a direct activator of egl-1 transcription in the NSM sister cell. The ces-1(gf) mutation prevents the death of the NSM sister cell by blocking the HLH-2/HLH-3–dependent expression of egl-1 in the NSM sister cell [18,20]. Furthermore, CES-1 can also bind to the E-boxes/Snail-binding sites in Region B of the egl-1 locus, and the ability of elevated levels of CES-1 to prevent the death of the NSM sister cell in vivo is dependent on these E-boxes/Snail-binding sites. On the basis of these observations, it has been proposed that by competing with HLH-2/HLH-3 for binding to Region B of the egl-1 locus, elevated levels of CES-1 protein in ces-1(gf) animals directly block egl-1 transcription in the NSM sister cell [20]. Finally, it has been hypothesized that the CES-1 protein might normally function in the NSM to block HLH-2/HLH-3–dependent egl-1 transcription, thereby allowing the survival of the NSM.
Figure 1. The Genetic Pathway of the NSM Sister Cell Death.
egl-1 is required for the death of the NSM sister cell. egl-1 activity in the NSM sister cell is positively regulated by the genes hlh-2 and hlh-3 and negatively regulated by the gene ces-1. The activity of ces-1 is negatively regulated by the gene ces-2. See text for details.
The function of the ces-1 gene in the NSM sister cell is negatively regulated by the gene ces-2, which is required for the death of the NSM sister cell and which encodes a bZIP transcription factor similar to the human proto-oncoprotein HLF (hepatic leukemia factor) [18,21] (Figure 1). Here we report that the previously uncharacterized protein DNJ-11 (DnaJ domain–11), a MIDA1 (mouse Id associated 1)–like chaperone, cooperates with the CES-2 protein to reduce ces-1 transcription in the NSM lineage, thereby excluding CES-1 protein from the NSM sister cell and allowing the death of the NSM sister cell. Furthermore, we show that the NSM neuroblast, which gives rise to the NSM and NSM sister cell, divides asymmetrically and that the genes dnj-11, ces-2, and ces-1 also function to cause asymmetric NSM neuroblast division. Our results reveal new developmental roles of MIDA1-like chaperones and HLF-like transcription factors. Furthermore, our results delineate a pathway involved in asymmetric cell division that directly controls the apoptotic fate.
Results
bc212 Prevents the Death of the NSM Sister Cells
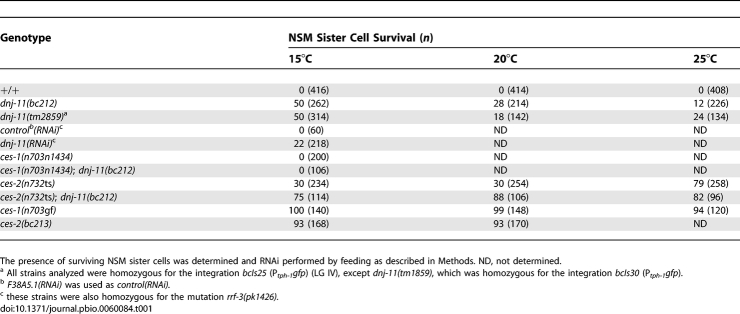
The NSM sister cells are generated about 410 min after the first cleavage of the C. elegans zygote and undergo apoptosis at about 430 min [3]. We screened for mutations that cause the NSM sister cells to survive, and we isolated the recessive mutation bc212 (J. Hatzold, B. Conradt, unpublished data). bc212 causes 12% and 50% NSM sister cell survival when raised at 25 °C or 15 °C, respectively, and hence causes a cold-sensitive NSM sister cell survival phenotype (Table 1, dnj-11(bc212)). In addition, bc212 is maternally rescued: in homozygous bc212 progeny (dnj-11(bc212)) of heterozygous bc212 hermaphrodites (dnj-11(bc212)/+), only 1% of the NSM sister cells survive (Table S1). To determine whether bc212 prevents the death of cells other than the NSM sister cells, we analyzed additional cells that undergo apoptosis during C. elegans development. We found that bc212 had no effect on their deaths, which demonstrates that bc212 does not block apoptosis in general (Table S2). However, we found that bc212 causes a variety of other defects such as morphological defects, lethality, slow growth, and reduced brood size (Figure S1 and Tables S3–S5). Therefore, the gene defined by bc212 is required for the death of the NSM sister cells and for additional processes that are important for C. elegans development and fertility.
Table 1.
bc212 Causes a Cold-Sensitive NSM Sister Cell Survival Phenotype.
bc212 Is a Putative Null Allele of the Gene dnj-11, Which Is Widely Expressed and Encodes a Protein That Primarily Localizes to the Cytosol
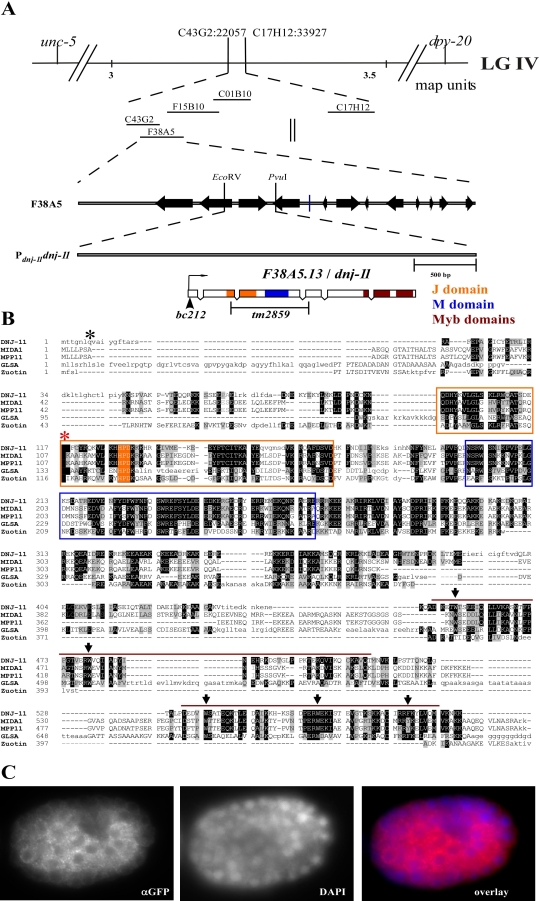
We cloned the gene defined by bc212 and found that it is identical to the previously uncharacterized gene dnj-11 (F38A5.13) (DNJ-11 accession number NP_501006) on linkage group (LG) IV (Figure 2A). dnj-11 encodes a 589–amino acid (aa) protein that is most similar to members of the family of MIDA1-like proteins, which are found in plants and animals (see below). bc212 is a C-to-T transition at position 21 of the nucleotide sequence of the dnj-11 gene and transforms codon 7 into a stop codon, which is predicted to truncate the DNJ-11 protein after aa 6 (Figure 2A and 2B). tm2859, a 641–base pair (bp) deletion of the dnj-11 gene, removes base pairs 402-1042 of the dnj-11 coding region. This deletion is predicted to result in a frameshift leading to the generation of a truncated protein composed of the first 116 aa of the wild-type protein and a 67-aa, C-terminal extension in a different reading frame (Figure 2A and 2B). Like bc212, tm2859 causes a cold-sensitive NSM sister cell survival phenotype and 50% NSM sister cell survival at 15 °C (Table 1, dnj-11(tm2859)). In addition, bc212 and tm2859 cause 24% and 26% lethality, respectively, at 15 °C (Table S3). Based on these results, we conclude that bc212 represents a strong loss-of-function mutation and putative null allele of the dnj-11 gene.
Figure 2. dnj-11 Encodes a Member of the Family of MIDA1-Like Proteins.
(A) (Top) Genes and single-nucleotide polymorphisms (SNPs) used for mapping bc212 are indicated. (Middle) Cosmids assayed for rescue of dnj-11(bc212) are shown. (Bottom) Pdnj-11dnj-11 is a subclone of F38A5 and represents the minimal dnj-11(bc212) rescue fragment. The J domain, M domain, and Myb domains of the dnj-11 ORF are indicated in orange, blue, and maroon, respectively.
(B) The alignment of the DNJ-11 sequence with the sequence of mouse MIDA1, human MPP11, V. cateri GlsA, and the S. cerevisiae Zuotin was performed using the DIALIGN algorithm. Identical amino acids and similar amino acids are shaded in black and gray, respectively. The orange box marks the J domain and the conserved tripeptide is shaded in orange. The blue box indicates the M domain. The maroon bars above the sequences represent the Myb domains. Arrows point to conserved aromatic residues in the Myb domains. bc212 results in the truncation of the DNJ-11 protein after aa 6 (asterisk). tm2859 results in a frame shift after aa 116 (red asterisk) and a stop after an additional 67 aas in a different frame.
(C) Embryos transgenic for the functional Pdnj-11dnj-11::gfp transgene were fixed as described in Materials and Methods and stained with an anti-GFP antibody (αGFP) to detect DNJ-11::GFP and with DAPI (DAPI) to detect nuclei.
Using a functional transgene that drives the expression of a DNJ-11::green fluorescent protein (GFP) fusion protein under the control of the dnj-11 promoter (Pdnj-11dnj-11::gfp), we determined the expression pattern of the dnj-11 gene and the sub-cellular localization of the DNJ-11 protein. Pdnj-11dnj-11::gfp expression was observed in most if not all somatic cells of embryos, larvae, and adult animals, including the cells of the NSM lineage (Figure 2C and unpublished data). DNJ-11::GFP protein primarily localized to the cytoplasm in a punctate pattern and could not be detected in nuclei. In addition, the dnj-11 gene is most likely also expressed in the adult germ line, because dnj-11(bc212) is maternally rescued (Table S1).
dnj-11 Cooperates with ces-2 to Negatively Regulate ces-1
The death of the NSM sister cells is dependent on the transcriptional upregulation in the NSM sister cells of the BH3-only gene egl-1 (EGL-1 accession number NP_506575) [20]. The transcriptional upregulation of egl-1 in the NSM sister cells can be blocked by the Snail-related transcription factor CES-1 (accession number NP_492338), which in turn is negatively regulated by the HLF-like transcription factor CES-2 (accession number NP_493610) [18–21]. By using an egl-1 transgene (Pegl-1his-24::gfp), we found that dnj-11(bc212) resulted in the loss of egl-1 expression in 69% of the NSM sister cells, indicating that dnj-11 acts upstream of egl-1 to promote its transcription in the NSM sister cells (Table S6). To determine where dnj-11 functions with respect to the genes ces-1 and ces-2, we analyzed the interactions of dnj-11(bc212) with a putative null mutation of ces-1— n703n1434 [19]—and a partial, temperature-sensitive loss-of-function (lf) mutation of ces-2—n732 [18]. We found that 0% of the NSM sister cells survived in ces-1(n703n1434); dnj-11(bc212) double mutants, demonstrating that the ability of dnj-11(bc212) to prevent the death of the NSM sister cells requires a functional ces-1 gene (Table 1, ces-1(n703n1434); dnj-11(bc212)). Finally, we found that at 20 °C, dnj-11(bc212) greatly enhanced the NSM sister cell survival phenotype caused by ces-2(n732) (Table 1, ces-2(n732); dnj-11(bc212)). These results indicate that, at least in the NSM lineage, dnj-11 acts upstream of and as a negative regulator of ces-1. Furthermore, these results suggest that dnj-11 and ces-2 cooperate to antagonize ces-1 function.
The J Domain and Myb Domains of the MIDA1-like DNJ-11 Protein Are Required for Its Function
MIDA1-like proteins contain three major regions: (1) an N-terminal J domain, which is a protein–protein interaction domain found in members of the J protein or Hsp40 family of chaperones [22], (2) a central M domain, which is another protein–protein interaction domain found specifically in MIDA1-like proteins [23], and (3) two C-terminal Myb domains, which are DNA-binding domains typically found in transcription factors [24,25] (Figure 1A and 1B). MIDA1-like proteins have mainly been implicated in growth control [23,26,27] and are thought to function by regulating transcription and translation [28–32]. BLAST searches [33] revealed that the DNJ-11 protein is highly similar to mouse MIDA1 (40% identical, 61% similar) (accession number NP_033610) [23], human MPP11 (M phase phosphoprotein) (38% identical, 54% similar) (accession number NP_055192) [27], Volvox carteri GlsA (gonidialess) (34% identical, 52% similar) (accession number AF_106963 GenBank) [26], and Saccharomyces cerevisiae Zuotin (37% identical, 62% similar) (accession number NP_011801) [34] (Figure 1B).
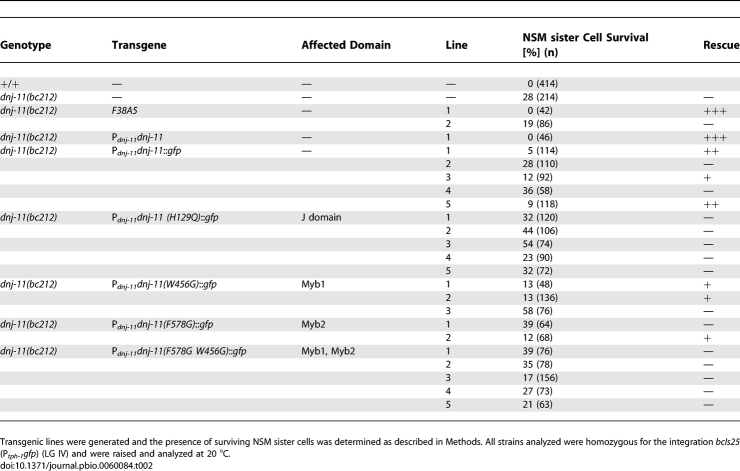
To determine whether the J domain is important for DNJ-11 activity, we replaced the histidine residue of the conserved tripeptide of the J domain with a glutamine residue (H129Q) and tested a transgene that expresses a DNJ-11(H129Q)::GFP fusion protein under the control of the dnj-11 promoter (Pdnj-11dnj-11(H129Q)::gfp) for its ability to rescue the NSM sister cell survival phenotype caused by dnj-11(bc212). In contrast to Pdnj-11dnj-11::gfp, Pdnj-11dnj-11(H129Q)::gfp failed to rescue the NSM sister cell survival phenotype of dnj-11(bc212) animals (Table 2). To determine whether the Myb domains are important for DNJ-11 activity, we replaced the tryptophan residue at position 456 in the first Myb domain (W456) and/or the phenylalanine residue at position 578 in the second Myb domain (F578) with glycine and tested the resulting transgenes (Pdnj-11dnj-11(W456G)::gfp, Pdnj-11dnj-11(F578G)::gfp, and Pdnj-11dnj-11(W456G F578G)::gfp) for their ability to rescue the NSM sister cell survival phenotype caused by dnj-11(bc212). We found that the transgenes expressing a DNJ-11::GFP fusion protein with either one of the Myb domains mutated partially rescued the NSM sister cell survival phenotype of dnj-11(bc212) animals (Table 2, dnj-11(bc212); Pdnj-11dnj-11(W456G)::gfp, dnj-11(bc212); Pdnj-11dnj-11(F578G)::gfp). However, the transgene expressing a DNJ-11::GFP fusion protein with both Myb domains mutated was no longer able to rescue (Table 2, dnj-11(bc212); Pdnj-11dnj-11(W456G F578G)::gfp). These results demonstrate that the J domain and at least one functional Myb domain are required for the ability of DNJ-11 to cause the death of the NSM sister cells.
Table 2.
The J Domain and the MYB Domains Are Required for the Ability of DNJ-11 to Cause the Death of the NSM Sister Cells
dnj-11 Is Required for Asymmetric NSM Neuroblast Division
The V. carteri ortholog of DNJ-11, GlsA, has been implicated in asymmetric cell division [35]. Specifically, during V. carteri development, an asymmetric cell division occurs that results in the generation of two daughter cells of different sizes and fates, namely a large reproductive cell and a small somatic cell. In glsA mutants, this cell division occurs symmetrically, resulting in two cells of equal size, both of which differentiate into somatic cells [26].
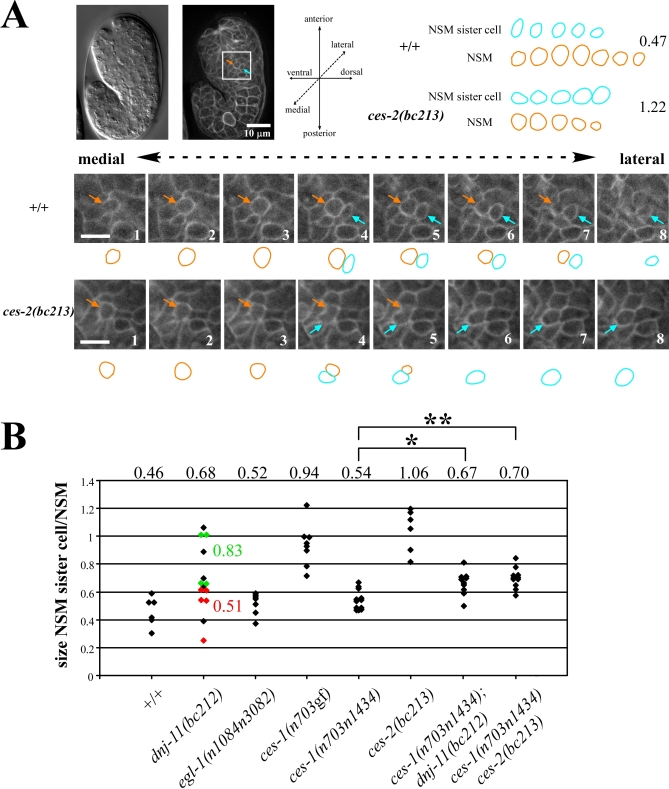
Most of the 131 cells that undergo apoptosis during C. elegans development are thought to be the result of an asymmetric cell division that gives rise to a large cell, which is programmed to survive, and a small cell, which is programmed to die [2,3]. Therefore, we determined whether the NSM neuroblast, which gives rise to the NSM and the NSM sister cell, divides asymmetrically with respect to size as well. To that end, we identified the NSM neuroblast based on its position in the embryo, observed its division at about 410 min, and determined the sizes of its daughter cells immediately after its division had been completed (Figure 3A). Using a transgene that expresses a plasma membrane–targeted GFP fusion protein (Ppie-1gfp::ph(PLC1δ1)) [36], we found that in wild-type animals, the size of the NSM sister cell on average is 0.46 times the size of the NSM (Figure 3B, +/+). Therefore, the NSM neuroblast divides asymmetrically to give rise to a large cell (the NSM) and a small cell (the NSM sister cell).
Figure 3. dnj-11, ces-1, and ces-2 Are Involved in Asymmetric NSM Neuroblast Division.
(A) (Left top) Nomarski and epifluorescence image of a wild-type embryo transgenic for the transgene Ppie-1gfp::ph(PLC1δ1) immediately after the NSM neuroblast divided. The orange arrow points to the NSM, the blue arrow points to the NSM sister cell. (Bottom) Series of eight consecutive focal planes (0.5 μm distance) (1–8) through the NSM and NSM sister cell of a wild-type (upper panel, +/+) or ces-2(bc213) (lower panel, ces-2(bc213)) embryo. (Right top) Summary of schematic representations of the outlines. Numbers on the right indicate the ratios between the sizes of the NSM sister cells and the NSMs in the Z-series shown.
(B) The ratio of the sizes of the NSM sister cell and the NSM was determined as described in Materials and Methods. The NSM sister cell was defined as the lateral cell of the two daughter cells. Each diamond represents the ratio of a single, independent NSM neuroblast division. The numbers above the diamonds represent the average ratio obtained for a given genotype. The apoptotic fate of the NSM sister cells was determined in a subset of dnj-11(bc212) embryos (red/died, green/survived). The complete genotypes of the embryos analyzed from left to right were: tIs38 (Ppie-1gfp::ph(PLC1δ1)), dnj-11(bc212) bcIs25; tIs38, egl-1(n1048n3082); tIs38, ces-1(n703gf); tIs38, ces-1(n703n1434); tIs38, ces-2(bc213); bcIs25; tIs38, ces-1(n703n1434); dnj-11(bc212) bcIs25; tIs38, and ces-1(n703n1434) ces-2(bc213); bcIs25; tIs38. p-Values were determined by the Student's t-test. *ces-1(n703n1434) compared to ces-1(n703n1434); dnj-11(bc212): p<0.002, **ces-1(n703n1434) compared to ces-1(n703n1434) ces-2(bc213): p<0.001.
Furthermore, we found that in dnj-11(bc212) animals, the difference in size between the NSM sister cell and the NSM is highly variable, ranging from a 2-fold difference as observed in wild-type animals to no difference (Figure 3B, dnj-11(bc212)). To determine whether the range observed reflects the incomplete penetrance of the NSM sister cell survival phenotype caused by dnj-11(bc212) (see Table 1), we followed the fate of NSM sister cells after the division of the NSM neuroblasts. We found that in five out of five embryos in which the division of the NSM neuroblast had occurred asymmetrically (NSM sister cell size on average 0.51 times the size of the NSM), the NSM sister cells died (Figure 3B, red diamonds). By contrast, four out of four NSM sister cells that were similar in size to the NSMs (NSM sister cell size on average 0.83 times the size of the NSM) survived (green diamonds). To rule out the possibility that the increase in NSM sister cell size observed in dnj-11(bc212) animals is a result of inappropriate NSM sister cell survival rather than a defect in asymmetric cell division, we analyzed egl-1(lf) mutants, in which many apoptotic events, including the death of the NSM sister cells, are blocked. We found that, like in wild-type animals, the NSM neuroblast divided asymmetrically in egl-1(lf) mutants (NSM sister cell size on average 0.52 times the size of the NSM) (Figure 3B, egl-1(n1084n3082)). Thus, the increase in NSM sister cell size observed in dnj-11(bc212) animals is a result of a defect in asymmetric cell division. Therefore, we conclude that dnj-11 is required for asymmetric NSM neuroblast division and, by inference, is required for the displacement of the mitotic spindle along the cell division axis. Furthermore, because in dnj-11(bc212) animals the defect in asymmetric NSM neuroblast division correlates with the defect in NSM sister cell death, asymmetric NSM neuroblast division might be critical for the specification of the apoptotic fate of the NSM sister cell.
To determine whether dnj-11 is required for asymmetric cell division in lineages other than the NSM lineage, we analyzed a subset of the other asymmetric cell divisions (including the first division of the zygote) that take place during C. elegans development and that give rise to daughter cells of different sizes and fates. We found that none of these divisions was affected by dnj-11(bc212), suggesting that dnj-11 is not required for asymmetric cell division in general (Table S7 and unpublished data).
ces-1 and ces-2 also Function in Asymmetric NSM Neuroblast Division
Like dnj-11(bc212), the ces-1(gf) mutation n703 and a putative null mutation of ces-2, bc213, prevent the death of the NSM sister cells (Table 1, ces-1(n703gf), ces-2(bc213)). Unexpectedly, we found that ces-1(n703gf) and ces-2(bc213) also disrupt the asymmetric division of the NSM neuroblast and result in the production of two cells of similar sizes (NSM sister cell sizes on average 0.94 and 1.06 times the size of the NSM) (Figure 3B, ces-1(n703gf), ces-2(bc213)). These results demonstrate that increased levels of ces-1 function can prevent asymmetric NSM neuroblast division and that, like dnj-11, ces-2 is required for asymmetric NSM neuroblast division.
To determine whether ces-1 also acts downstream of dnj-11 and ces-2 during asymmetric NSM neuroblast division, we determined whether the loss of ces-1 function can suppress the defects in asymmetric NSM neuroblast division observed in dnj-11(bc212) and ces-2(bc213) animals. We found that ces-1(n703n1434) partially suppressed the defects in asymmetric NSM neuroblast division that are observed in dnj-11(bc212) and ces-2(bc213) animals (Figure 3B). This finding suggests that ces-1 acts downstream of dnj-11 and ces-2 during asymmetric NSM neuroblast division as well.
dnj-11, ces-2, and ces-1 Also Play a Role in Establishing the Correct Orientation of the Cell Division Axis During Asymmetric NSM Neuroblast Division
Shortly after the division of the NSM neuroblast has been completed, the NSM is located at a position that is medial and ventral to the NSM sister cell (Figure 3A). The position of the NSM relative to the NSM sister cell implies that in wild-type animals, the cleavage plane of the NSM neuroblast is along the ventral/lateral to dorsal/medial axis. While determining the size of the NSM and NSM sister cell using the plasma membrane–targeted GFP fusion protein, we observed that in two out of nine ces-2(bc213) animals, the medial daughter cell was located dorsally rather than ventrally to the lateral daughter cell (Figure 3A). This observation suggests that the loss of ces-2 function not only affects the displacement of the mitotic spindle along the cell division axis in the NSM neuroblast, but also the orientation of the cell division axis and, hence, the orientation of the cleavage plane.
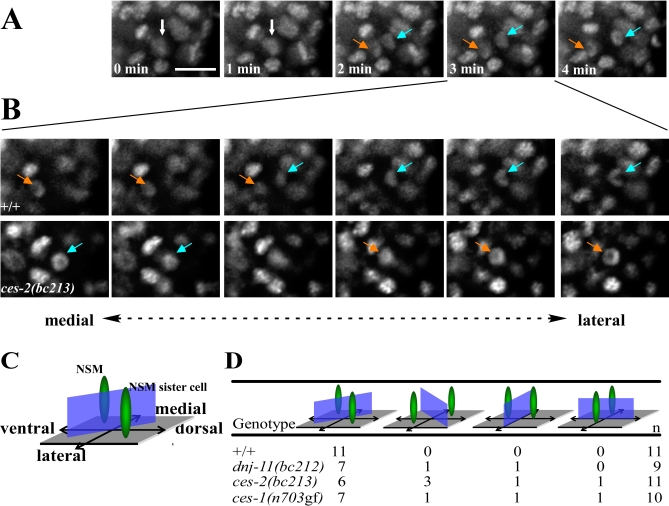
To observe more directly the division of the NSM neuroblast, we used a transgene that expresses a DNA-targeted GFP fusion protein (Phis-24his-24::gfp) (M. Dunn, G. Seydoux, J. Waddle, personal communication). This fusion protein allowed us to determine the axis along which the separated chromatids move during anaphase of the NSM neuroblast division and therefore the cleavage plane of the dividing NSM neuroblast. In wild-type embryos, the chromatids of the future NSM move to the ventral/medial side, whereas the chromatids of the future NSM sister cell move to the dorsal/lateral side, confirming that the cleavage plane of the NSM neuroblast is along the ventral/lateral to dorsal/medial axis (Figure 4 ; +/+). As in wild-type embryos, we found that in ces-2(bc213) embryos, the cleavage plane in six out of 11 NSM neuroblasts was along the ventral/lateral to dorsal/medial axis. In three cases, however, the cleavage plane was reversed, and the cells divided along the ventral/medial to dorsal/lateral axis. Furthermore, in two cases, the cleavage plane was either along the lateral to medial or ventral to dorsal axis (Figure 4B and 4D; ces-2(bc213)). We next analyzed dnj-11(bc212) and ces-1(n703gf) embryos and found that the orientation of the cleavage plane of the NSM neuroblast was disrupted in two out of nine and three out of 10 NSM neuroblasts, respectively (Figure 3D; dnj-11(bc212), ces-1(n703gf)). Based on these findings, we conclude that dnj-11, ces-2, and ces-1 not only play a role in the displacement of the mitotic spindle along the cell division axis in the NSM neuroblast but also in the orientation of the cell division axis.
Figure 4. dnj-11, ces-1, and ces-2 Are Involved in Orienting the Mitotic Spindle of the NSM Neuroblasts.
(A) Epifluorescent images shown are maximum projections of Z-series of a wild-type embryo carrying the transgene Phis-24his-24::gfp. The scale bar represents 5 μm. Ventral is to the left and dorsal to the right. The Z-series were taken every 60 s during a 4-min time period. Chromosomal DNA in the NSM neuroblast prior to the division is indicated by white arrows. Between minute 1 and minute 2, the NSM neuroblast starts dividing. The chromatids of the NSM (indicated by orange arrows) move toward the ventral side, the chromatids of the NSM sister cell (indicated by blue arrows) move to the dorsal side.
(B) (Upper panel). Epifluorescence images of six consecutive focal planes of the Z-series at minute 3 shown in (A) as maximum projection. Medial to lateral planes are shown from left to right. The chromatids of the ventral NSM are located medially. The chromatids of the dorsal NSM sister cell are located laterally. (Lower panel). Six consecutive focal planes of a Z-series of a ces-2(bc213) embryo. Medial to lateral planes are shown from left to right. The chromatids of the ventral NSM are located laterally. The chromatids of the dorsal NSM sister cell are located medially.
(C) Schematic representation of the ventral/dorsal and medial/lateral positions of the chromatids of NSM and NSM sister cell (indicated in green) in wild-type embryos. The blue rectangle indicates the cleavage plane of the NSM neuroblast.
(D) Quantification of the orientation of the cleavage planes observed. The complete genotypes of the embryos analyzed from top to bottom were: dtIs372; dnj-11(bc212) bcls25; dtls372, ces-2(bc213); bcls25; dtls372, and ces-1(n703); bcIs25; dtIs372.
dnj-11 and ces-2 Reduce ces-1 Transcription in the NSM Lineage Thereby Restricting the Presence of CES-1 Protein to the NSM
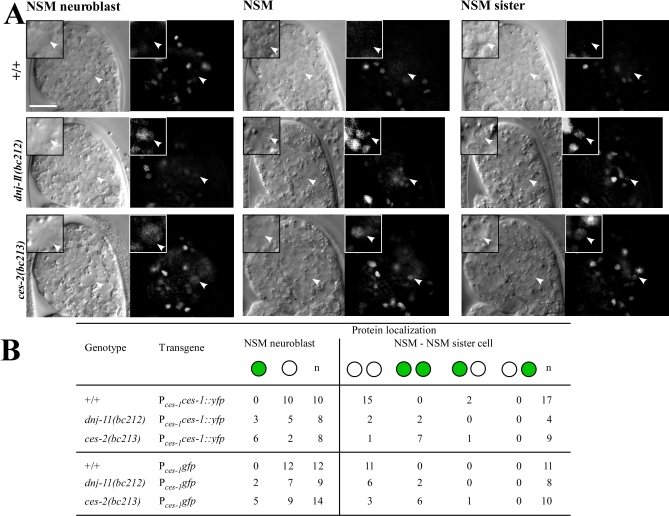
To investigate how dnj-11 and ces-2 antagonize ces-1 function in the NSM lineage, we constructed a functional transgene that drives the expression of a CES-1::yellow fluorescent protein (YFP) fusion protein under the control of the ces-1 promoter (Pces-1ces-1::yfp). In wild-type animals, we failed to detect CES-1::YFP in NSM neuroblasts or NSM sister cells. However, we observed CES-1::YFP in two out of 17 NSMs analyzed (Figure 5A and 5B, +/+). (CES-1::YFP was observed in lineages other than the NSM lineage in all animals examined.) This observation suggests that the level of CES-1::YFP and hence, most probably of endogenous CES-1 protein, is relatively low in the NSM lineage and that it is higher in NSMs than in NSM neuroblasts or NSM sister cells. In dnj-11(bc212) and ces-2(bc213) animals, we observed CES-1::YFP not only in NSMs but also in NSM sister cells and NSM neuroblasts (Figure 5A and 5B, dnj-11(bc212), ces-2(bc213)). These results demonstrate that dnj-11 and ces-2 antagonize ces-1 function in the NSM lineage by reducing the level of CES-1 protein in the NSM neuroblast, thereby restricting the presence of CES-1 protein to the NSM.
Figure 5. The Loss of dnj-11 or ces-2 Function Results in Increased Levels of CES-1 Protein and Increased ces-1 Transcription in the NSM Lineage.
(A) Nomarski and epifluorescence images of wild-type (+/+), dnj-11(bc212), and ces-2(bc213) embryos transgenic for a stable Pces-1ces-1::yfp transgene. White arrowheads indicate NSM neuroblasts (left column), NSMs (middle column), and NSM sister cells (right column). The scale bar represents 10 μm. Insets in the upper left corner of each image show the respective cells at a higher magnification.
(B) Summary of the data on the presence of CES-1 protein in the NSM lineage using the Pces-1ces-1::yfp transgene and ces-1 expression using the Pces-1gfp transgene. Green circles represent cells in which CES-1::YFP or GFP were detected. White circles represent cells in which CES-1::YFP or GFP were not detected. The complete genotypes of the embryos analyzed from top to bottom were: bcIs58 (Pces-1ces-1::yfp), dnj-11(bc212) bcIs25; bcIs58, ces-2(bc213); bcIs25; bcIs58, bcEx619 (Pces-1gfp), dnj-11(bc212) bcIs25; bcEx619, and ces-2(bc213); bcIs25; bcEx619.
To determine whether dnj-11 and ces-2 act at the transcriptional or posttranscriptional level to affect the level of CES-1 protein, we constructed a transgene that drives the expression of the GFP protein under the control of the ces-1 promoter (Pces-1gfp) and analyzed GFP expression in the NSM lineage. In wild-type animals, GFP was not detected in NSM neuroblasts, NSMs, or NSM sister cells (Figure 5B, +/+). However, in dnj-11(bc212) and ces-2(bc213) animals, GFP was detected in NSM neuroblasts, NSMs, and NSM sister cells (Figure 5B, dnj-11(bc212), ces-2(bc213)). Based on these results, we conclude that dnj-11 and ces-2 restrict the presence of CES-1 protein to the NSM by reducing ces-1 transcription in the NSM lineage.
Discussion
It is well established that asymmetric cell division can be coupled to the apoptotic fate [2–11]. However, the genetic and mechanistic basis for this relationship has not been well defined. Here we present evidence that the regulation of asymmetric cell division and apoptosis are functionally linked by a pathway that involves three evolutionarily conserved genes: dnj-11 MIDA1, ces-2 HLF and ces-1 Snail.
CES-1 Snail Levels Determine the Apoptotic Fate
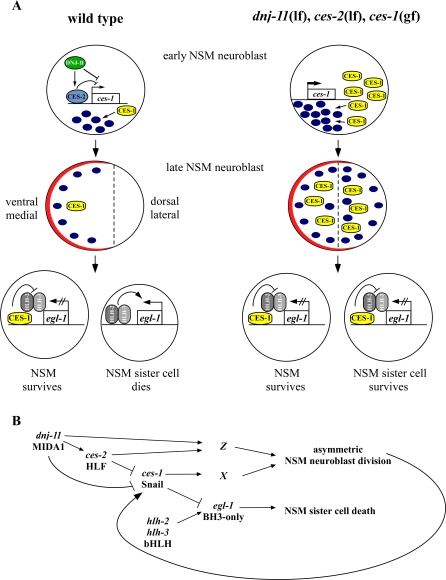
Snail family transcription factors have previously been implicated in the regulation of apoptosis in mammalian cells. In the case of C. elegans, it has been suggested that the Snail protein, CES-1, might normally function within the NSM lineage to repress transcription of the pro-apoptotic gene, egl-1, thereby allowing NSM survival. Our data support this idea, because we find that CES-1 protein is present at higher levels in the NSMs than in the NSM sister cells. Therefore, we propose that CES-1 acts as a cell-fate determinant in the NSMs to ensure the survival of the NSMs and their differentiation into serotonergic neurons. Conversely, we propose that the absence of CES-1 protein in the NSM sister cells determines the death of the NSM sister cells (Figure 6A).
Figure 6. Regulation of Asymmetric NSM Neuroblast Division and the Apoptotic Death of the NSM sister Cell.
(A) Molecular models. (Wild type) By repressing the transcription of the Snail-related gene ces-1, the proteins CES-2 HLF and DNJ-11 MIDA1 ensure that the CES-1 Snail protein is present at an appropriate, low level in the unpolarized, early NSM neuroblast. Blue oval indicates polarity factor that is required to establish polarity in the late NSM neuroblast and the synthesis of which is promoted by CES-1. Indicated in red is a complex that restricts the polarity factor to the ventral/medial side of the NSM neuroblast. Once localized to the ventral/medial side, the polarity factor is involved in restricting CES-1 to the ventral/medial side of the late NSM neuroblast. See text for details. (dnj-11(lf), ces-2(lf), ces-1(gf)) Loss-of-function mutations of dnj-11 or ces-2, or a gain-of-function mutation of ces-1 result in an increased level of CES-1 Snail protein in the early NSM neuroblast. This increased level of CES-1 protein results in a level of polarity factor too high to establish asymmetry and restrict CES-1 protein to the ventral/medial side. See text for details.
(B) Genetic pathway. dnj-11 acts upstream of or in parallel to ces-2 to negatively regulate the function of ces-1, thereby causing asymmetric NSM neuroblast division and NSM sister cell death. ces-1 function can affect asymmetric NSM neuroblast division by regulating the function of a gene or genes required for the process. By negatively regulating egl-1, ces-1 function can also prevent the hlh-2, hlh-3–dependent death of the NSM sister cells. ces-1 function in NSM sister cell death is affected by the process of asymmetric NSM neuroblast division, which causes the asymmetric distribution of the product of the ces-1 gene. See text for details.
CES-1 Snail Forges a Link between Asymmetric Cell Division and Apoptosis
Our results also indicate that the CES-1 protein acts in the NSM neuroblasts to affect the orientation of the cleavage plane and, hence, asymmetric cell division (Figure 6). Therefore, at least in the NSM lineage, the CES-1 protein represents a functional link between the cellular machinery that causes asymmetric cell division and the cellular machinery that causes the apoptotic death of specific cells during C. elegans development. At least to our knowledge, this is the first demonstration that apoptosis can directly be controlled by asymmetric cell division. Like the function of ces-1 in NSM fate determination, the function of ces-1 in asymmetric NSM neuroblast division is redundant to that of one or more unidentified genes. Similarly, the Snail-related proteins Snail, Escargot, and Worniu of D. melanogaster act redundantly to cause asymmetric cell division [14,15,17].
DNJ-11 and CES-2 Regulate ces-1 Transcription in the NSM Lineage
The HLF-like bZIP transcription factor, CES-2, is thought to act as a direct repressor of CES-1 Snail transcription [19,21]. We have found that the dnj-11 MIDA1-like gene acts in concert with CES-2 as a negative regulator of CES-1 expression. Reducing either dnj-11 or ces-2 function results in an increased level of ces-1 transcription within the NSM lineage, disrupts asymmetric NSM neuroblast division, and prevents the death of the NSM sister cells. Based on our data, dnj-11 and ces-2 could either act in parallel or in a single linear pathway to antagonize ces-1 function (Figure 6). MIDA1-like proteins have been implicated in transcriptional regulation [28–30] and are components of a ribosome-associated chaperone referred to as RAC, which co-translationally interacts with nascent polypeptides thereby affecting translational accuracy and termination as well as protein folding [31,32]. The DNJ-11 protein predominantly localizes to the cytosol in a punctate pattern, which suggests that rather than regulating ces-1 transcription directly, DNJ-11 might affect the translation and/or folding of a regulator of ces-1 transcription.
A Model for the Role of the dnj-11 MIDA1, ces-2 HLF, ces-1 Snail Pathway in Asymmetric NSM Neuroblast Division
Based on our results, we propose that by repressing the transcription of the snail-related ces-1 gene, the HLF-like bZIP transcription factor CES-2 and the MIDA1-like chaperone DNJ-11 ensure that the CES-1 protein is present at an appropriate, low level in the early NSM neuroblast (Figure 6A). A low level of CES-1 protein in the early NSM neuroblast would allow the expression at a certain level of a “polarity factor” that is required for the asymmetric division of the late NSM neuroblast. Furthermore, a complex that localizes to the ventral/medial side of the late NSM neuroblast would restrict the polarity factor to the ventral/medial side, thereby promoting the displacement of the mitotic spindle along the cell division axis and resulting in a shift of the cleavage plane and the asymmetric division of the cell. Finally, the asymmetrically localized polarity factor would also restrict CES-1 to the ventral/medial side and thereby cause its segregation predominantly into the NSM, thus resulting in the repression of egl-1/BH3-only transcription in the NSM and the survival of the NSM (Figure 6A). In this model, the CES-1 protein not only is a component of the cellular machinery that causes asymmetric NSM neuroblast division, but also one of its targets. The identity of the polarity factor as well as the signals and mechanisms that cause its asymmetric localization or activation remain to be determined. The Snail-related genes snail, escargot, and worniu of D. melanogaster function in asymmetric neuroblast division by promoting the expression of the gene inscuteable, which encodes an adaptor protein required for asymmetric neuroblast division. It will be of interest to determine whether the C. elegans ortholog of inscuteable, the gene insc-1 (F43E2.3) (INSC-1 accession number AAC71125) [37], plays a role in asymmetric cell division and in asymmetric NSM neuroblast division, in particular.
dnj-11(bc212) does not affect asymmetric cell divisions that are known to be regulated by the C. elegans genes ham-1 [4,38], pig-1 [5], dsh-2 [39], or hlh-14 [40]. Conversely, the loss of ham-1, pig-1, dsh-2, or hlh-14 function does not affect the asymmetric division of the NSM neuroblast. Therefore, the dnj-11 MIDA1, ces-2 HLF, and ces-1 Snail pathway might be independent of these genes. Furthermore, the first division of the zygote, which occurs asymmetrically and is known to be regulated by the C. elegans par genes [41], is not affected by dnj-11(bc212), ces-2(bc213), or ces-1(n703gf) (J. Hatzold, B. Conradt, unpublished data). Whether the par genes function in the asymmetric division of the NSM neuroblast remains to be determined.
The Role of the dnj-11 MIDA1, ces-2 HLF, ces-1 Snail Pathway in Asymmetric Cell Division and the Control of Apoptosis Might be Conserved
At least to our knowledge, this is the first evidence that a HLF-like bZIP transcription factor plays a role in asymmetric cell division. Furthermore, our studies provide a functional link between the known roles in asymmetric cell division of MIDA1-like chaperones and Snail-related transcription factors, and hence suggest the existence of a pathway involved in asymmetric cell division that is conserved throughout the plant and animal kingdoms [14–16,26]. Therefore, it will be of interest to determine whether HLF-like and Snail-related proteins also contribute to asymmetric cell division in V. carteri and whether MIDA1-like and HLF-like proteins also participate in asymmetric neuroblast division in D. melanogaster. Furthermore, our results hint at the possibility that MIDA1-like, HLF-like, and Snail-related proteins might play a role in asymmetric cell division in vertebrates. Specifically, it has recently been reported that HLF and the Snail-related protein Slug of mammals have functions in stem cells [12,42]. In addition, there is increasing evidence that the process of asymmetric cell division plays a crucial role in the ability of stem cells to self renew [43]. Therefore, we hypothesize that a dnj-11 MIDA1, ces-2 HLF, ces-1 Snail–like pathway might be important for stem cell renewal by allowing asymmetric stem cell division.
At least to our knowledge, this is also the first evidence that a MIDA1-like protein plays a role in the regulation of apoptosis. Furthermore, our studies have identified a new component of a conserved cell-death specification pathway composed of C. elegans ces-2 HLF and ces-1 Snail. Like CES-2 and CES-1, HLF and the Snail-related protein Slug of mammals have previously been implicated in the regulation of apoptosis [19,21,44–46]. Therefore, it will be of interest to determine whether MIDA1-like proteins also have an apoptotic role in mammals and act in a HLF-, and Slug-dependent pathway. Finally, based on our work, we consider it a possibility that the roles in vertebrates of a dnj-11 MIDA1, ces-2 HLF, ces-1 Snail–like pathway in asymmetric cell division and apoptosis might be functionally linked as well. Specifically, we speculate that such a pathway could cause stem cell renewal through asymmetric cell division and control stem cell numbers through apoptosis [47].
Implications for the Functions of the Proto-Oncoprotein HLF and the Snail-Related Protein Slug of Mammals
The oncogenic form of human HLF, the E2A-HLF fusion protein, found in patients carrying the t(17;19) (q22;p13) translocation, gives rise to pro–B cell acute lymphoblastic leukemia (ALL) in adolescents [48]. The E2A-HLF fusion protein is composed of the trans-activation domain of the bHLH protein E2A and the DNA-binding domain of HLF [49]. It has been proposed that E2A-HLF causes leukemic transformation of pro-B cells by blocking their apoptotic death. Specifically, it has been proposed that E2A-HLF inappropriately activates the transcription of the snail-related gene, slug, which encodes a direct repressor of the egl-1-like, pro-apoptotic BH3-only gene puma, thereby causing the survival of pro-B cells that are normally programmed to undergo apoptosis [44–46]. Based on our results, we speculate that like C. elegans CES-2 and CES-1, the proteins HLF and Slug might not only function to control the expression of a pro-apoptotic BH3-only gene but to cause asymmetric cell division. Hence, in patients with the t(17;19) (q22;p13) translocation, the presence of the E2A-HLF fusion protein and elevated levels of Slug protein might affect aspects of the pro-B cell fate other than their apoptotic fate and/or might alter the division of the lymphoid progenitors that produce pro-B cells. Finally, it will be of interest to determine whether the human MIDA1-like gene MPP11 [27], which is expressed in hematopoietic lineages as well as other tissues, plays a role in E2A-HLF- and Slug-mediated tumorigenesis.
Materials and Methods
Strains and genetics.
C. elegans strains were cultured as described [50]. Bristol N2 was used as the wild-type strain. The strain CB4856 (Hawaii) was used in conjunction with N2 for SNP mapping. Mutations and transgenes used in this study are listed below and are described [51], except where noted otherwise: LGI: ces-1(n703n1434), ces-1(n703gf), ces-2(n732ts), ces-2(bc213) (this study). LGII: rrf-3(pk1426) [52]. LGIII: gmIs12 (Psrb-6gfp) [53], ced-4(n1162). LGIV: unc-5(e53), dnj-11(bc212, tm2859) (this study), dpy-20(e1282), nDf41 [54], bcIs25 (Ptph-1gfp) [20]. LGV: nIs83 (Pmec-4gfp), bcIs37 (Pegl-1gfp) [20], egl-1(n1084n3083) [55]; akIs3 (Pnmr-1gfp) [56]. (LGX) nIs106 (Plin-11gfp) [57], dtIs372 (M. Dunn, G. Seydoux, J. Waddle. personal communication). Additional stable transgenes used: tIs38 (Ppie-1gfp::ph(PLC1δ1)) [36], kyIs39 (Psra-6gfp) [53], bcIs58 (Pces-1ces-1::yfp) (this study). RNA-mediated interference (RNAi) by feeding was performed as described using 6 mM IPTG [58].
Cloning of dnj-11(bc212).
Standard genetic techniques were used to map dnj-11 between unc-5 and dpy-20 on LGIV. SNP mapping was used to locate dnj-11 between the SNPs C43G2:22057 and C17H12:33927. The cosmid F38A5 as well as a 3,565-bp subclone of F38A5 contained in plasmid pBC484 (Pdnj-11dnj-11) rescued the NSM sister cell survival and brood size phenotype observed in bc212 animals (Table 2 and Table S5, dnj-11(bc212); Pdnj-11dnj-11). In addition, partially reducing dnj-11 function by RNA-mediated interference (RNAi) causes 22% NSM sister cell survival (Table 1).
Molecular analysis.
dnj-11 plasmids: pBC484 (Pdnj-11dnj-11) was generated by inserting a EcoRV-PvuI subclone of cosmid F38A5 into the EcoRV site of pBluescript II KS + (Stratagene). The gfp sequence was amplified from pPD95.02 (gift of A. Fire, Stanford Scool of Medicine, Stanford, California) with appropriate primers (sequence of these and all other primers are available on request) and inserted into the BsmI site of pBC484 to create a C terminal in frame fusion of dnj-11 to gfp (Pdnj-11dnj-11::gfp). The Pdnj-11dnj-11::gfp transgene rescued the NSM sister cell survival phenotype of dnj-11(bc212) mutants, demonstrating that it is functional (Table 2, dnj-11(bc212); Pdnj-11dnj-11::gfp). To generate Pdnj-11dnj-11::gfp mutant plasmids, site-directed PCR mutagenesis was performed to mutate CAC to CAA (H129Q), TGG to GGG (W456G), and TTC to GGC (F578G). The expression levels of the transgenes Pdnj-11dnj-11(H129Q)::gfp, Pdnj-11dnj-11(W456G)::gfp, Pdnj-11dnj-11(F578G)::gfp, and Pdnj-11dnj-11(W456G F578G)::gfp were similar to that of the wild-type transgene (Pdnj-11dnj-11::gfp), and the subcellular localization of the resulting fusion proteins was indistinguishable from that of DNJ-11::GFP (unpublished data).
ces-1 plasmids: The plasmid pBC510 (Pces-1ces-1::yfp) was generated by cloning an AflII-SpeI fragment from cosmid F43G9 containing the ces-1 rescuing fragment [19] into the EcoRV site of pBluescript II KS + (pBC482A), and inserting yfp amplified from pvdB#3 [59] into the SwaI site to generate a C terminal in frame fusion. The Pces-1ces-1::yfp transgene was able to block the ability of ces-1(n703n1434) to suppress the NSM sister cell survival phenotype of dnj-11(bc212) and ces-2(n732) animals, demonstrating that it is functional (unpublished data). To generate plasmid pBC664 (Pces-1gfp), first ces-1 upstream and downstream regulatory regions were amplified by PCR, a PmeI site was introduced 3′ of upstream and 5′ of downstream regulatory regions, and PCR products were cloned into XmaI/NcoI digested pBC482A to obtain the ces-1 locus without coding region, 5′ and 3′ untranslated regions (UTRs), and introns (pBC656). Because intron 4 of ces-1 is highly conserved between C. elegans and C. briggsae, intron 1 of the gfp sequence of plasmid pPD95.77 (gift of A. Fire) was replaced with intron 4 of ces-1 by PCR fusion. The SmaI/SpeI fragment of pPD95.77_ces-1 intron containing the gfp sequence and the unc-54 3′ UTR was inserted into the PmeI site of pBC656 to generate pBC664.
Transgenic animals.
Germline transformation was performed as described [60]. Cosmids were injected at a concentration of 10 ng/μl with pPD93.97 (Pmyo-3gfp) at 50 ng/μl as coinjection marker. Plasmids were injected at a concentration of 10 ng/μl with pRF4 (rol-6(su1006)) at 50 ng/μl as coinjection marker. pBC510 (Pces-1ces-1::yfp) was injected into N2 to create an extrachromosomal array and integrated using EMS mutagenesis [50] to generate bcIs58. The strain carrying bcIs58 was backcrossed three times to N2.
Phenotypic analysis and microscopy.
The NSM sister cell survival was scored as previously described [20]. Microscopy of living embryos was performed by mounting embryos on 2%–5% agar pads in M9 buffer, sealing them with petroleum jelly, and using a Zeiss Axioskop2 equipped with epifluorescence, a Micromax CCD camera (Princeton Instruments), and Metamorph software. NSM neuroblasts, NSMs, and NSM sister cells were identified based on the position of their nuclei using Nomarski optics. Z-series were taken with a Z-distance of 0.5 μm (analysis of Pces-1ces-1::yfp, Pces-1gfp, and Phis-24his-24::gfp expression) and 0.25 μm (determination of cell size). Epifluorescence Z-series were deconvolved using the AutoDeblur Gold WF AutoVisualize software (Media Cybernetics). The cell size of NSMs and NSM sister cells was determined 10 to 15 min after the NSM neuroblast had started to divide, as indicated by the breakdown of the nuclear envelop, which was observed by Nomarski optics. To visualize the outline of a cell, a plasma membrane–targeted GFP fusion protein (Ppie-1gfp::ph(PLC1δ1)) was used. The area of the cross section of a cell was measured in each section of a Z-series using Metamorph software. To estimate the difference in volume between NSM and NSM sister cell, the values of each section were added, and the sum obtained for the NSM sister cell was divided by the sum obtained for the NSM. To determine the orientation of the cleavage plane of the NSM neuroblast, chromatids were visualized using a His24-GFP fusion protein (Phis-24his-24::gfp, dtIs372) (gift of M. Dunn and G. Seydoux, Johns Hopkins University, Baltimore, Maryland; and J. Waddle, Southern Methodist University, Dallas, TX). The NSM neuroblast was identified by Nomarski optics before the start of the division and consecutive Z series were taken in 1 min time intervals until the completion of the NSM neuroblast division.
Immunohistochemistry.
Embryos were prepared in 10 μl on poly L-lysine coated slides and fixed and stained as described [61]. Slides were mounted in 1 μg/ml DAPI in PBS 1:1 diluted with VectaShield (Vector Laboratories). GFP was detected using the anti-AFP antibody monoclonal antibody 3E6 (Qbiogene).
Supporting Information
(A) Nomarski images of wild-type (+/+) and dnj-11(bc212) embryos at the 1.5- and 2-fold stage, respectively. Note that dnj-11(bc212) embryos show morphological defects.
(B and C) Nomarski images of wild-type (+/+) and dnj-11(bc212) larvae. Arrows point to mis-shaped structures.
(D) Nomarski images of the vulva region of wild-type (+/+) and dnj-11(bc212) adults. The arrow points to a defective vulva, the arrowhead points to a distorted body area. The complete genotypes of the analyzed animals were: bcIs25 and dnj-11(bc212) bcIs25.
(796.79 KB).
The NSM sister cell survival was scored as described in Materials and Methods. Strains were grown and analyzed at 20 °C. The complete genotype of the strains analyzed was bcIs25, dnj-11(bc212) bcIs25, unc-5(e53) dpy-20(e1282) bcIs25 / dnj-11(bc212) bcIs25, unc-5(e53) dnj-11(bc212) dpy-20(e1282) bcIs25, dnj-11(bc212) bcIs25; bcEx513 (Pdnj-11dnj-11), and dnj-11(bc212) bcIs25. The complete maternal genotype of the strains analyzed was bcIs25, dnj-11(bc212) bcIs25, unc-5(e53) dpy-20(e1282) bcIs25, unc-5(e53) dnj-11(bc212) dpy-20(e1282) bcIs25 / bcIs25, dnj-11(bc212) bcIs25; bcEx513 (Pdnj-11dnj-11), and dnj-11(bc212) bcIs25; bcEx513 (Pdnj-11dnj-11).
(33.50 KB ).
The presence of surviving cells in the anterior pharynx was scored by Nomarski Optics as described [55,62]. The presence of surviving cells in the ventral cord was scored using the reporter Plin-11gfp [57]. The presence of surviving PVQ sister cells was determined using Psra-6gfp [53]. The presence of extra PHB cells was scored using Psrb-6gfp [53]. All strains were raised and analyzed at 15 °C. The complete genotype of the strains analyzed was bcIs25 (Ptph-1gfp) (+/+) and dnj-11(bc212) bcIs25 (dnj-11(bc212)), nIs106 (Plin-11gfp) (+/+) and dnj-11(bc212) bcIs25; nIs106 (dnj-11(bc212)), kyIs39 (Psra-6gfp) (+/+) and dnj-11(bc212) bcIs25; kyIs39 (dnj-11(bc212)), and gmIs12 (Psrb-6gfp) (+/+) and dnj-11(bc212) bcIs25; gmIs12 (dnj-11(bc212)).
(56.50 KB).
Embryonic lethality was analyzed by allowing adults of genotype bcIs25 (+/+) or dnj-11(bc212) bcIs25 (dnj-11(bc212)) or bcIs30 and dnj-11(tm2859)*; bcIs30 to lay eggs for 3 h. Adults were removed and eggs laid during the 3-h period were counted. Unhatched eggs were scored after 1, 2, and 3 d. The animals were raised and the experiment performed at 15 °C.
(25.50 KB).
Strains were grown and analyzed at 15 °C. The growth rate was analyzed by allowing five adults of genotype bcIs25 (+/+) or dnj-11(bc212) bcIs25 (dnj-11(bc212)) to lay eggs for 3 h. Adults were removed and the number of eggs laid during the 3-h period was determined. The presence of L4 larvae was monitored daily starting at day 5.
(27.00 KB ).
Animals were grown and analyzed at 15 °C. Individual L4 larvae were plated onto plates and transferred to fresh plates daily for 4–5 d. The total number of progeny laid during the 4–5-d period was determined by counting the presence of L4 larvae. The standard deviation (STDEV) of the brood size of five different animals is indicated. All animals analyzed were homozygous for the integration bcIs25 (Ptph-1gfp).
(28.00 KB).
The presence of GFP in the NSM sister cells was scored in L1 larvae using Nomarski optics and the integrated egl-1 reporter bcIs37 (Pegl-1his24-gfp). Animals were grown at 15 °C.
(29.00 KB ).
The asymmetric division of cells indicated in the left column was assayed by scoring missing or extra cells generated by their respective lineage. Missing or extra Psrb-6gfp–expressing PHB neurons were scored to analyze the asymmetric division of ABpl/rapppap (the disruption of the asymmetry of this division by mutations in ham-1 results in missing or extra Psrb-6gfp expressing PHB neurons [38)]. Missing or extra Pmec-4gfp–expressing PLM neurons were scored to analyze the asymmetric division of ABpl/rapapppp (the disruption of the asymmetry of this division by mutations in ham-1 or pig-1 results in missing or extra Pmec-4gfp expressing PLM neurons [5,38]). Missing or extra I2 neurons were scored by Nomarski optics to analyze the asymmetric division of ABalpappaa/ABarapapaa (the disruption of the asymmetry of this division by mutations in ham-1 or pig-1 results in extra I2 neurons [4,5,38]). Missing or extra M4 neurons were scored by Nomarski optics to analyze the asymmetric division of MSpaaaaa (the disruption of the asymmetry of this division by mutations in pig-1 results in extra I2 neurons [5]). Missing or extra Pmec-4gfp–expressing A/PVM neurons were scored to analyze the asymmetric division of QR/L.p (the disruption of the asymmetry of this division by mutations in pig-1 results in extra A/PVM neurons [5]). Missing or extra Pnmr-1gfp–expressing PVC neurons were scored to analyze the asymmetric division of ABpl/rpppa (the disruption of the asymmetry of this division by mutations in dsh-2 results in extra PVC neurons [39]). Missing or extra Psra-6gfp–expressing PVQ neurons were scored to analyze the asymmetric division of ABpl/rappp (the disruption of the asymmetry of this division by mutations in hlh-14 results in missing PVQ neurons [40]).
(86.00 KB).
Accession Numbers
The NCBI Entrez Protein (http://www.ncbi.nlm.nih.gov/sites/entrez?db=protein) accession numbers for genes and gene products discussed in this paper are: NP_501006 (DNJ-11), NP_506575 (EGL-1), NP_492338 (CES-1), NP_493610 (CES-2), NP_033610 (mouse MIDA1), NP_055192 (human MPP11), NP_011801 (S. cerevisiae Zuotin).
The GenBank (http://www.ncbi.nlm.nih.gov/) accession number for V. carteri GlsA is AF_106963.
Acknowledgments
We thank V. Ambros, E. Lambie, and S. Rolland for comments on the manuscript; D. Mayka and E. Stenvers for excellent technical support; E. Lambie for use of the micro-injection set-up; J. Audhya for plasmid Ppie-1gfp::ph(PLC1δ1); M. Dunn, G. Seydoux, and J. Waddle for plasmid pJH2.19 and integation dtIs372; A. Fire for plasmids; the Sanger Centre (Hinxton, UK) for cosmids, S. Mitani at the National BioResource Project (Tokyo, Japan) for dnj-11(tm2859); and the C. elegans Genetics Center (CGC, supported by the NIH National Center for Research Resources) for strains.
Abbreviations
- aa
amino acid
- CES
cell-death specification
- CED
cell-death abnormal
- DNJ
DnaJ domain
- Egl
egg-laying abnormal
- GFP
green fluorescent protein
- Gls
gonidialess
- HLF
hepatic leukemia factor
- Insc
Inscuteable (Drosophila asymmetric cell division protein) homolog
- LG
linkage group
- MIDA
mouse Id associated
- MPP
M phase phosphoprotein
- NSM
neurosecretory motoneuron
Footnotes
Author contributions. JH and BC conceived and designed the experiments, analyzed the data, and wrote the paper. JH performed the experiments.
Funding. This work was supported by funding from the Howard Hughes Medical Institute Award 76200–560801 to Dartmouth Medical School under the Biomedical Research Support Program for Medical Schools and National Institutes of Health grant R01-GM069950.
Competing interests. The authors have declared that no competing interest exists.
References
- Horvitz HR, Herskowitz I. Mechanisms of asymmetric cell division: two Bs or not two Bs, that is the question. Cell. 1992;68:237–255. doi: 10.1016/0092-8674(92)90468-r. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans . Dev Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans . Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Frank CA, Hawkins NC, Guenther C, Horvitz HR, Garriga G. C. elegans HAM-1 positions the cleavage plane and regulates apoptosis in asymmetric neuroblast divisions. Dev Biol. 2005;284:301–310. doi: 10.1016/j.ydbio.2005.05.026. [DOI] [PubMed] [Google Scholar]
- Cordes S, Frank CA, Garriga G. The C. elegans MELK ortholog PIG-1 regulates cell size asymmetry and daughter cell fate in asymmetric neuroblast divisions. Development. 2006;133:2747–2756. doi: 10.1242/dev.02447. [DOI] [PubMed] [Google Scholar]
- Bossing T, Udolph G, Doe CQ, Technau GM. The embryonic central nervous system lineages of Drosophila melanogaster. I. Neuroblast lineages derived from the ventral half of the neuroectoderm. Dev Biol. 1996;179:41–64. doi: 10.1006/dbio.1996.0240. [DOI] [PubMed] [Google Scholar]
- Orgogozo V, Schweisguth F, Bellaiche Y. Binary cell death decision regulated by unequal partitioning of Numb at mitosis. Development. 2002;129:4677–4684. doi: 10.1242/dev.129.20.4677. [DOI] [PubMed] [Google Scholar]
- Schmidt H, Rickert C, Bossing T, Vef O, Urban J, et al. The embryonic central nervous system lineages of Drosophila melanogaster. II. Neuroblast lineages derived from the dorsal part of the neuroectoderm. Dev Biol. 1997;189:186–204. doi: 10.1006/dbio.1997.8660. [DOI] [PubMed] [Google Scholar]
- Shen Q, Zhong W, Jan YN, Temple S. Asymmetric Numb distribution is critical for asymmetric cell division of mouse cerebral cortical stem cells and neuroblasts. Development. 2002;129:4843–4853. doi: 10.1242/dev.129.20.4843. [DOI] [PubMed] [Google Scholar]
- Roach HI, Erenpreisa J, Aigner T. Osteogenic differentiation of hypertrophic chondrocytes involves asymmetric cell divisions and apoptosis. J Cell Biol. 1995;131:483–494. doi: 10.1083/jcb.131.2.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieberich E, MacKinnon S, Silva J, Noggle S, Condie BG. Regulation of cell death in mitotic neural progenitor cells by asymmetric distribution of prostate apoptosis response 4 (PAR-4) and simultaneous elevation of endogenous ceramide. J Cell Biol. 2003;162:469–479. doi: 10.1083/jcb.200212067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobaleda C, Perez-Caro M, Vicente-Duenas C, Sanchez-Garcia I. Function of zinc-finger transcription factor SNA12 in cancer and development. Annu Rev Genet. 2007;41:41–61. doi: 10.1146/annurev.genet.41.110306.130146. [DOI] [PubMed] [Google Scholar]
- Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype. Nat Rev Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- Cai Y, Chia W, Yang X. A family of snail-related zinc finger proteins regulates two distinct and parallel mechanisms that mediate Drosophila neuroblast asymmetric divisions. Embo J. 2001;20:1704–1714. doi: 10.1093/emboj/20.7.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf SI, Ip YT. The Snail protein family regulates neuroblast expression of inscuteable and string, genes involved in asymmetry and cell division in Drosophila . Development. 2001;128:4757–4767. doi: 10.1242/dev.128.23.4757. [DOI] [PubMed] [Google Scholar]
- Kraut R, Chia W, Jan LY, Jan YN, Knoblich JA. Role of inscuteable in orienting asymmetric cell divisions in Drosophila . Nature. 1996;383:50–55. doi: 10.1038/383050a0. [DOI] [PubMed] [Google Scholar]
- Betschinger J, Knoblich JA. Dare to be different: asymmetric cell division in Drosophila, C. elegans and vertebrates. Curr Biol. 2004;14:R674–685. doi: 10.1016/j.cub.2004.08.017. [DOI] [PubMed] [Google Scholar]
- Ellis RE, Horvitz HR. Two C. elegans genes control the programmed deaths of specific cells in the pharynx. Development. 1991;112:591–603. doi: 10.1242/dev.112.2.591. [DOI] [PubMed] [Google Scholar]
- Metzstein MM, Horvitz HR. The C. elegans cell death specification gene ces-1 encodes a snail family zinc finger protein. Mol Cell. 1999;4:309–319. doi: 10.1016/s1097-2765(00)80333-0. [DOI] [PubMed] [Google Scholar]
- Thellmann M, Hatzold J, Conradt B. The Snail-like CES-1 protein of C. elegans can block the expression of the BH3-only cell-death activator gene egl-1 by antagonizing the function of bHLH proteins. Development. 2003;130:4057–4071. doi: 10.1242/dev.00597. [DOI] [PubMed] [Google Scholar]
- Metzstein MM, Hengartner MO, Tsung N, Ellis RE, Horvitz HR. Transcriptional regulator of programmed cell death encoded by Caenorhabditis elegans gene ces-2 . Nature. 1996;382:545–547. doi: 10.1038/382545a0. [DOI] [PubMed] [Google Scholar]
- Walsh P, Bursac D, Law YC, Cyr D, Lithgow T. The J-protein family: modulating protein assembly, disassembly and translocation. EMBO Rep. 2004;5:567–571. doi: 10.1038/sj.embor.7400172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji W, Inoue T, Yamamoto T, Obinata M. MIDA1, a protein associated with Id, regulates cell growth. J Biol Chem. 1995;270:24818–24825. doi: 10.1074/jbc.270.42.24818. [DOI] [PubMed] [Google Scholar]
- Oh IH, Reddy EP. The myb gene family in cell growth, differentiation and apoptosis. Oncogene. 1999;18:3017–3033. doi: 10.1038/sj.onc.1202839. [DOI] [PubMed] [Google Scholar]
- Aasland R, Stewart AF, Gibson T. The SANT domain: a putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. Trends Biochem Sci. 1996;21:87–88. [PubMed] [Google Scholar]
- Miller SM, Kirk DL. glsA, a Volvox gene required for asymmetric division and germ cell specification, encodes a chaperone-like protein. Development. 1999;126:649–658. doi: 10.1242/dev.126.4.649. [DOI] [PubMed] [Google Scholar]
- Matsumoto-Taniura N, Pirollet F, Monroe R, Gerace L, Westendorf JM. Identification of novel M phase phosphoproteins by expression cloning. Mol Biol Cell. 1996;7:1455–1469. doi: 10.1091/mbc.7.9.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Shoji W, Obinata M. MIDA1 is a sequence specific DNA binding protein with novel DNA binding properties. Genes Cells. 2000;5:699–709. doi: 10.1046/j.1365-2443.2000.00362.x. [DOI] [PubMed] [Google Scholar]
- Inoue T, Shoji W, Obinata M. MIDA1, an Id-associating protein, has two distinct DNA binding activities that are converted by the association with Id1: a novel function of Id protein. Biochem Biophys Res Commun. 1999;266:147–151. doi: 10.1006/bbrc.1999.1779. [DOI] [PubMed] [Google Scholar]
- Yoshida M, Inoue T, Shoji W, Ikawa S, Obinata M. Reporter gene stimulation by MIDA1 through its DnaJ homology region. Biochem Biophys Res Commun. 2004;324:326–332. doi: 10.1016/j.bbrc.2004.09.059. [DOI] [PubMed] [Google Scholar]
- Otto H, Conz C, Maier P, Wolfle T, Suzuki CK, et al. The chaperones MPP11 and Hsp70L1 form the mammalian ribosome-associated complex. Proc Natl Acad Sci U S A. 2005;102:10064–10069. doi: 10.1073/pnas.0504400102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundley HA, Walter W, Bairstow S, Craig EA. Human Mpp11 J protein: ribosome-tethered molecular chaperones are ubiquitous. Science. 2005;308:1032–1034. doi: 10.1126/science.1109247. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Lockshin C, Herbert A, Winter E, Rich A. Zuotin, a putative Z-DNA binding protein in Saccharomyces cerevisiae. Embo J. 1992;11:3787–3796. doi: 10.1002/j.1460-2075.1992.tb05464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Q, Fowler R, Tam LW, Edwards L, Miller SM. The role of GlsA in the evolution of asymmetric cell division in the green alga Volvox carteri . Dev Genes Evol. 2003;213:328–335. doi: 10.1007/s00427-003-0332-x. [DOI] [PubMed] [Google Scholar]
- Audhya A, Hyndman F, McLeod IX, Maddox AS, Yates JR, 3rd, et al. A complex containing the Sm protein CAR-1 and the RNA helicase CGH-1 is required for embryonic cytokinesis in Caenorhabditis elegans . J Cell Biol. 2005;171:267–279. doi: 10.1083/jcb.200506124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh M. Identification and characterization of human Inscuteable gene in silico. Int J Mol Med. 2003;11:111–116. [PubMed] [Google Scholar]
- Guenther C, Garriga G. Asymmetric distribution of the C. elegans HAM-1 protein in neuroblasts enables daughter cells to adopt distinct fates. Development. 1996;122:3509–3518. doi: 10.1242/dev.122.11.3509. [DOI] [PubMed] [Google Scholar]
- Hawkins NC, Ellis GC, Bowerman B, Garriga G. MOM-5 frizzled regulates the distribution of DSH-2 to control C. elegans asymmetric neuroblast divisions. Dev Biol. 2005;284:246–259. doi: 10.1016/j.ydbio.2005.05.024. [DOI] [PubMed] [Google Scholar]
- Frank CA, Baum PD, Garriga G. HLH-14 is a C. elegans Achaete-Scute protein that promotes neurogenesis through asymmetric cell division. Development. 2003;130:6507–6518. doi: 10.1242/dev.00894. [DOI] [PubMed] [Google Scholar]
- Gönczy P, Rose LS. Asymmetric cell division and axis formation in the embryo. WormBook, The C. elegans Research Community. 2005. doi/ 10.1895/wormbook.1.30.1. Available at: http://www.wormbook.org/. Accessed 10 March 2008. [DOI] [PMC free article] [PubMed]
- Shojaei F, Trowbridge J, Gallacher L, Yuefei L, Goodale D, et al. Hierarchical and ontogenic positions serve to define the molecular basis of human hematopoietic stem cell behavior. Dev Cell. 2005;8:651–663. doi: 10.1016/j.devcel.2005.03.004. [DOI] [PubMed] [Google Scholar]
- Lin H. Cell biology of stem cells: an enigma of asymmetry and self-renewal. J Cell Biol. 2008;180:257–260. doi: 10.1083/jcb.200712159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba T, Inukai T, Yoshihara T, Seyschab H, Ashmun RA, et al. Reversal of apoptosis by the leukaemia-associated E2A-HLF chimaeric transcription factor. Nature. 1996;382:541–544. doi: 10.1038/382541a0. [DOI] [PubMed] [Google Scholar]
- Inukai T, Inoue A, Kurosawa H, Goi K, Shinjyo T, et al. SLUG, a ces-1-related zinc finger transcription factor gene with antiapoptotic activity, is a downstream target of the E2A-HLF oncoprotein. Mol Cell. 1999;4:343–352. doi: 10.1016/s1097-2765(00)80336-6. [DOI] [PubMed] [Google Scholar]
- Wu WS, Heinrichs S, Xu D, Garrison SP, Zambetti GP, et al. Slug antagonizes p53-mediated apoptosis of hematopoietic progenitors by repressing puma. Cell. 2005;123:641–653. doi: 10.1016/j.cell.2005.09.029. [DOI] [PubMed] [Google Scholar]
- Sommer L, Rao M. Neural stem cells and regulation of cell number. Prog Neurobiol. 2002;66:1–18. doi: 10.1016/s0301-0082(01)00022-3. [DOI] [PubMed] [Google Scholar]
- Seidel MG, Look AT. E2A-HLF usurps control of evolutionarily conserved survival pathways. Oncogene. 2001;20:5718–5725. doi: 10.1038/sj.onc.1204591. [DOI] [PubMed] [Google Scholar]
- Inaba T, Roberts WM, Shapiro LH, Jolly KW, Raimondi SC, et al. Fusion of the leucine zipper gene HLF to the E2A gene in human acute B-lineage leukemia. Science. 1992;257:531–534. doi: 10.1126/science.1386162. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans . Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle DL, Blumenthal T, Meyer B, Priess JR, editors. C. elegans II. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- Simmer F, Tijsterman M, Parrish S, Koushika S, Nonet M, et al. Loss of the Putative RNA-Directed RNA Polymerase RRF-3 Makes C. elegans Hypersensitive to RNAi. Curr Biol. 2002;12:1317. doi: 10.1016/s0960-9822(02)01041-2. [DOI] [PubMed] [Google Scholar]
- Troemel ER, Chou JH, Dwyer NE, Colbert HA, Bargmann CI. Divergent seven transmembrane receptors are candidate chemosensory receptors in C. elegans . Cell. 1995;83:207–218. doi: 10.1016/0092-8674(95)90162-0. [DOI] [PubMed] [Google Scholar]
- Williams BD, Waterston RH. Genes critical for muscle development and function in Caenorhabditis elegans identified through lethal mutations. J Cell Biol. 1994;124:475–490. doi: 10.1083/jcb.124.4.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conradt B, Horvitz HR. The C. elegans protein EGL-1 is required for programmed cell death and interacts with the Bcl-2-like protein CED-9. Cell. 1998;93:519–529. doi: 10.1016/s0092-8674(00)81182-4. [DOI] [PubMed] [Google Scholar]
- Brockie PJ, Madsen DM, Zheng Y, Mellem J, Maricq AV. Differential expression of glutamate receptor subunits in the nervous system of Caenorhabditis elegans and their regulation by the homeodomain protein UNC-42. J Neurosci. 2001;21:1510–1522. doi: 10.1523/JNEUROSCI.21-05-01510.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien PW, Cameron S, Horvitz HR. Phagocytosis promotes programmed cell death in C. elegans . Nature. 2001;412:198–202. doi: 10.1038/35084096. [DOI] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans . Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Labrousse AM, Zappaterra MD, Rube DA, van der Bliek AM. C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol Cell. 1999;4:815–826. doi: 10.1016/s1097-2765(00)80391-3. [DOI] [PubMed] [Google Scholar]
- Mello C, Fire A. DNA transformation. Methods Cell Biol. 1995;48:451–482. [PubMed] [Google Scholar]
- Van Furden D, Johnson K, Segbert C, Bossinger O. The C. elegans ezrin-radixin-moesin protein ERM-1 is necessary for apical junction remodelling and tubulogenesis in the intestine. Dev Biol. 2004;272:262–276. doi: 10.1016/j.ydbio.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Hengartner MO, Ellis RE, Horvitz HR. Caenorhabditis elegans gene ced-9 protects cells from programmed cell death. Nature. 1992;356:494–499. doi: 10.1038/356494a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Nomarski images of wild-type (+/+) and dnj-11(bc212) embryos at the 1.5- and 2-fold stage, respectively. Note that dnj-11(bc212) embryos show morphological defects.
(B and C) Nomarski images of wild-type (+/+) and dnj-11(bc212) larvae. Arrows point to mis-shaped structures.
(D) Nomarski images of the vulva region of wild-type (+/+) and dnj-11(bc212) adults. The arrow points to a defective vulva, the arrowhead points to a distorted body area. The complete genotypes of the analyzed animals were: bcIs25 and dnj-11(bc212) bcIs25.
(796.79 KB).
The NSM sister cell survival was scored as described in Materials and Methods. Strains were grown and analyzed at 20 °C. The complete genotype of the strains analyzed was bcIs25, dnj-11(bc212) bcIs25, unc-5(e53) dpy-20(e1282) bcIs25 / dnj-11(bc212) bcIs25, unc-5(e53) dnj-11(bc212) dpy-20(e1282) bcIs25, dnj-11(bc212) bcIs25; bcEx513 (Pdnj-11dnj-11), and dnj-11(bc212) bcIs25. The complete maternal genotype of the strains analyzed was bcIs25, dnj-11(bc212) bcIs25, unc-5(e53) dpy-20(e1282) bcIs25, unc-5(e53) dnj-11(bc212) dpy-20(e1282) bcIs25 / bcIs25, dnj-11(bc212) bcIs25; bcEx513 (Pdnj-11dnj-11), and dnj-11(bc212) bcIs25; bcEx513 (Pdnj-11dnj-11).
(33.50 KB ).
The presence of surviving cells in the anterior pharynx was scored by Nomarski Optics as described [55,62]. The presence of surviving cells in the ventral cord was scored using the reporter Plin-11gfp [57]. The presence of surviving PVQ sister cells was determined using Psra-6gfp [53]. The presence of extra PHB cells was scored using Psrb-6gfp [53]. All strains were raised and analyzed at 15 °C. The complete genotype of the strains analyzed was bcIs25 (Ptph-1gfp) (+/+) and dnj-11(bc212) bcIs25 (dnj-11(bc212)), nIs106 (Plin-11gfp) (+/+) and dnj-11(bc212) bcIs25; nIs106 (dnj-11(bc212)), kyIs39 (Psra-6gfp) (+/+) and dnj-11(bc212) bcIs25; kyIs39 (dnj-11(bc212)), and gmIs12 (Psrb-6gfp) (+/+) and dnj-11(bc212) bcIs25; gmIs12 (dnj-11(bc212)).
(56.50 KB).
Embryonic lethality was analyzed by allowing adults of genotype bcIs25 (+/+) or dnj-11(bc212) bcIs25 (dnj-11(bc212)) or bcIs30 and dnj-11(tm2859)*; bcIs30 to lay eggs for 3 h. Adults were removed and eggs laid during the 3-h period were counted. Unhatched eggs were scored after 1, 2, and 3 d. The animals were raised and the experiment performed at 15 °C.
(25.50 KB).
Strains were grown and analyzed at 15 °C. The growth rate was analyzed by allowing five adults of genotype bcIs25 (+/+) or dnj-11(bc212) bcIs25 (dnj-11(bc212)) to lay eggs for 3 h. Adults were removed and the number of eggs laid during the 3-h period was determined. The presence of L4 larvae was monitored daily starting at day 5.
(27.00 KB ).
Animals were grown and analyzed at 15 °C. Individual L4 larvae were plated onto plates and transferred to fresh plates daily for 4–5 d. The total number of progeny laid during the 4–5-d period was determined by counting the presence of L4 larvae. The standard deviation (STDEV) of the brood size of five different animals is indicated. All animals analyzed were homozygous for the integration bcIs25 (Ptph-1gfp).
(28.00 KB).
The presence of GFP in the NSM sister cells was scored in L1 larvae using Nomarski optics and the integrated egl-1 reporter bcIs37 (Pegl-1his24-gfp). Animals were grown at 15 °C.
(29.00 KB ).
The asymmetric division of cells indicated in the left column was assayed by scoring missing or extra cells generated by their respective lineage. Missing or extra Psrb-6gfp–expressing PHB neurons were scored to analyze the asymmetric division of ABpl/rapppap (the disruption of the asymmetry of this division by mutations in ham-1 results in missing or extra Psrb-6gfp expressing PHB neurons [38)]. Missing or extra Pmec-4gfp–expressing PLM neurons were scored to analyze the asymmetric division of ABpl/rapapppp (the disruption of the asymmetry of this division by mutations in ham-1 or pig-1 results in missing or extra Pmec-4gfp expressing PLM neurons [5,38]). Missing or extra I2 neurons were scored by Nomarski optics to analyze the asymmetric division of ABalpappaa/ABarapapaa (the disruption of the asymmetry of this division by mutations in ham-1 or pig-1 results in extra I2 neurons [4,5,38]). Missing or extra M4 neurons were scored by Nomarski optics to analyze the asymmetric division of MSpaaaaa (the disruption of the asymmetry of this division by mutations in pig-1 results in extra I2 neurons [5]). Missing or extra Pmec-4gfp–expressing A/PVM neurons were scored to analyze the asymmetric division of QR/L.p (the disruption of the asymmetry of this division by mutations in pig-1 results in extra A/PVM neurons [5]). Missing or extra Pnmr-1gfp–expressing PVC neurons were scored to analyze the asymmetric division of ABpl/rpppa (the disruption of the asymmetry of this division by mutations in dsh-2 results in extra PVC neurons [39]). Missing or extra Psra-6gfp–expressing PVQ neurons were scored to analyze the asymmetric division of ABpl/rappp (the disruption of the asymmetry of this division by mutations in hlh-14 results in missing PVQ neurons [40]).
(86.00 KB).