Abstract
Among several hypotheses to explain how translesion synthesis (TLS) by DNA polymerase η (pol η) suppresses ultraviolet light-induced mutagenesis in vivo despite the fact that pol η copies DNA with low fidelity, here we test whether replication accessory proteins enhance the fidelity of TLS by pol η. We first show that the single-stranded DNA binding protein RPA, the sliding clamp PCNA, and the clamp loader RFC slightly increase the processivity of yeast pol η and its ability to recycle to new template primers. However, these increases are small, and they are similar when copying an undamaged template and a template containing a cis-syn TT dimer. Consequently, the accessory proteins do not strongly stimulate the already robust TT dimer bypass efficiency of pol η. We then perform a comprehensive analysis of yeast pol η fidelity. We show that it is much less accurate than other yeast DNA polymerases and that the accessory proteins have little effect on fidelity when copying undamaged templates or when bypassing a TT dimer. Thus, although accessory proteins clearly participate in pol η functions in vivo, they do not appear to help suppress UV mutagenesis by improving pol η bypass fidelity per se.
Translesion synthesis (TLS1) is one mechanism of damage tolerance employed by cells when synthesis by the major replicative polymerases is blocked by lesions. Several polymerases in different families have been shown to bypass lesions in vitro (see ref 1 and references therein). Of these, the role of pol η in the bypass of UV photoproducts is the best understood. In humans, the loss of pol η causes the variant (XP-V) form of xeroderma pigmentosum (2, 3), a disease characterized by greatly increased susceptibility to sunlight-induced skin cancer (see refs 4 and 5 and references therein). A key property of human and yeast cells lacking pol η is increased mutagenesis following exposure to ultraviolet light (6-13). A key property of human and yeast pol η is its ability to bypass cyclobutane pyrimidine dimers (CPDs) with much higher efficiency than that of other eukaryotic DNA polymerases (14-18). These facts imply that pol η participates in the bypass of slowly repaired cyclobutane pyrimidine dimers in a manner that avoids mutations such that in its absence, other polymerases perform mutagenic bypass that ultimately results in skin cancer.
Consistent with the participation of pol η in CPD bypass that avoids mutations are seminal studies of single nucleotide insertion (17, 19, 20), demonstrating that yeast and human pol η preferentially insert dAMP opposite both template thymines of a cis-syn thymine-thymine dimer (TTD). This selectivity for inserting a correct nucleotide undoubtedly contributes to CPD bypass in cells that reduces UV-induced mutagenesis in humans, mice, and yeast. The actual rate of base substitutions generated per CPD bypass event in human cells is unknown. Fortunately, quantitative in vivo measurements in S. cerevisiae are available and indicate that bypass of TC and CC dimers generates less than one base substitution per 1000 bypass events (10). This represents a higher accuracy than that predicted by the low fidelity of TTD bypass by human pol η in vitro, which generates a T to C substitution at the 3′ T of a cis-syn TTD at an average rate of 1 in 27 (15). However, error rates for a complete TTD bypass reaction by yeast pol η are not yet available. For that reason, one goal of the present study is to determine these rates.
Several non-exclusive explanations could account for how a polymerase with intrinsically low fidelity contributes to suppressing damaged-induced mutagenesis. It is possible that the observed TTD bypass fidelity of human pol η is sufficient to suppress UV-induced mutagenesis and sunlight-induced skin cancer because CPDs are rarely encountered by the replication fork in cells that can remove most photoproducts using the nucleotide excision repair pathway. It is also possible that TLS errors made at the replication fork are corrected, for example, by extrinsic proofreading (14, 21-23) and/or by DNA mismatch repair (24). Yet another possibility is that accessory proteins may modulate the fidelity of a TLS reaction. Obvious candidates include RPA, the three subunit eukaryotic single strand DNA binding protein complex, and PCNA, the eukaryotic sliding clamp. In human cells, PCNA co-localizes with pol η after UV light irradiation (25-28), and human and yeast pol η interact with PCNA in a manner that reportedly increases DNA synthesis without altering processivity (29, 30). In addition, it has recently been shown that post-translational mono-ubiquitylation of PCNA by the Rad6/Rad18 complex is a key event in lesion bypass, triggered by a stalled replication fork (31-33). Pol η has been reported to preferentially associate with modified PCNA (28, 33, 34), and one study showed that the bypass of an AP site is stimulated by about 2-fold by monoubiquitylated PCNA in comparison to unmodified PCNA, under conditions allowing many cycles of synthesis to occur (35). The main goal of the present study is to investigate whether the accessory proteins RPA, PCNA, and the hetero-pentameric clamp loader replication factor C (RFC) affect the efficiency and/or the fidelity of TT dimer bypass by yeast pol η. We also report on the fidelity with which yeast pol η copies undamaged DNA, either alone or with the accessory proteins, and compare the results to those obtained when the same assay is applied to other S. cerevisiae polymerases.
EXPERIMENTAL PROCEDURES
Materials
Plasmid, phage, and bacterial strains and other materials for the assays described here were from previously described sources (36, 37). DNA modification enzymes and restriction enzymes were purchased from New England Biolabs (Ipswich, MA). Gel-purified DNA oligonucleotides were purchased from Oligos Etc., Inc. (Wilsonville, OR). Streptavidin was purchased from Roche Applied Science (Indianapolis, IN). Radiolabeled nucleotides and unlabeled dNTPs were purchased from Amersham Biosciences (Piscataway, NJ).
DNA Substrates
Gapped DNA substrates for the forward mutation assay are the same as those previously described (36). The template strand used in the TT dimer lesion bypass assays was a 70-mer (5′-ATGACCATGATTACGAATTCCAGCTCGGTACCGGGTTAGCCTTTGGAGTCGACCTGCAGAAATTCACTGG) with a 5′-biotin moiety, and was either undamaged or contained a cis-syn thymine-thymine dimer (underlined TT). Two primers, BP14 (5′-Bio-CCAGTGAATTTCTG) and 30Fid (5′-Bio-CCAGTGAATTTCTGCAGGTCGACTCCAAAG) also contained a 5′-biotin moiety, while primer LP18C (5′-GACTCCGACTCCAAAGGC) was unmodified. For bypass efficiency assay substrates, primer LP18C was labeled at the 5′-end using 32P-γ-ATP and T4 polynucleotide kinase. Substrates were prepared by mixing template oligonucleotide with 1.2× molar equivalents of both BP14 and LP18C in 50 mM Tris-Cl (pH 7.5) and 1× SSC and incubating at 75 °C for 5 min, followed by cooling to 25 °C over 3-4 h, protected from light. Substrates were stored at 4 °C until further use. For bypass fidelity assay substrates, the same procedure was used with unlabeled 30Fid primer. The template strand used in the AP site bypass assays was a 102-mer (5′-CCTTTGCGAATTCT25GCGGCTCCCTTCTTCTCCTCCCTCTCCCTTCCCTT29) with both 5′- and 3′-biotin moieties and was either undamaged or contained a synthetic AP site (tetrahydrofuran) residue at the underlined position. The primer used (5′-AGGGAAGGGAGAGGGAGGAGAAGAAG) pairs with the italicized region. AP site bypass substrates were prepared as described above using a 1.2× molar excess of template to the 5′-end-labeled primer.
Protein Purification
S. cerevisiae polymerase η was purified as described previously using the plasmid obtained from Dr. Zhigang Wang (38). S. cerevisiae proliferating cell nuclear antigen, replication factor C complex, and replication protein A were purified as previously described (39-41).
Lesion Bypass Efficiency Assay
TT dimer bypass efficiency reactions (30 μL) were performed in a manner similar to those previously described (15, 42), with the following modifications. Substrate DNA was first incubated at 30 °C for 10 min with a 10-fold molar excess of streptavidin in 40 mM Tris-Cl (pH 7.8), 75 mM NaCl, 2% glycerol, and 100 μg/mL BSA. All reactions contained 1 pmol of DNA substrate and 5 fmol of polymerase (200:1 substrate to enzyme ratio). Where indicated, 1.2 pmol each of PCNA trimer, RFC complex, and RPA was added. Reactions contained 40 mM Tris-CL (pH 7.8), 75 mM NaCl, 5 mM MgCl2, 1 mM ATP, 0.1 mM each dATP, dCTP, dGTP, and dTTP, 2 mM DTT, and 100 μg/mL BSA. All components except polymerase were mixed on ice and then incubated at 30 °C for 2 min. Polymerase was added to initiate the reaction, and samples (6 μL) removed at 2, 4, and 6 min were added to 12 μL of formamide loading buffer (95% deionized formamide, 25 mM EDTA, 0.1% bromophenol blue, and 0.1% xylene cyanol). Products were heated to 95 °C for 5 min, fractionated on a 12% denaturing polyacrylamide gel, and quantified on a Molecular Dynamics Typhoon 9400 imager. Bypass parameters were calculated as described previously (42). Polymerase cycling was calculated by multiplying the percentage of primer used by the amount of starting substrate and then dividing by the amount of polymerase in the reaction. The maximum number of incorporations was determined visually from the gel images. AP site bypass reactions were performed and analyzed similarly using 0.1 pmol of substrate, 0.2 pmol polymerase, 0.3 pmol RFC complex, 0.5 pmol PCNA trimer, and 1 pmol RPA.
Lesion Bypass Fidelity Assay
The lesion bypass fidelity assay was performed essentially as previously described (37) in the presence of ∼50 μCi (3000 Ci/mmol) α-32P-dCTP. Substrates contained a single primer (30Fid) and were blocked with streptavidin as described above prior to use. Reaction conditions were as described above and contained 4 pmol of substrate and 0.8 pmol of pol η (5:1 substrate: enzyme ratio) and were incubated at 30 °C for 20 min. Where indicated, 4.8 pmol each of PCNA trimer, RFC complex, and RPA was added. All components except polymerase were mixed on ice and incubated at 30 °C for 2 min, and then polymerase was added to initiate the reaction. Reactions were brought to a total volume of 50 μL in the presence of 0.1% SDS, 25 mM EDTA, and 0.2 mg/mL proteinase K and incubated at 37 °C for 10 min, followed by the removal of the unincorporated radioactive nucleotide by G25 sephadex spin column chromatography. Reaction products were purified by phenol/chloroform extraction and ethanol precipitation. Reaction products were digested with 10 U each of EcoRI and PstI in buffer recommended by the manufacturer. Recovery of the synthesized strand, annealing to gapped M13 DNA molecules, transfection into E. coli cells, and determination of the mutation rates and spectra were performed as previously described (15, 37, 42).
Undamaged DNA Forward Mutation Assay
Gapped M13 DNA (5 fmol) was incubated with pol η (1250 fmol) either in the absence or presence of RPA, PCNA trimer, and RFC complex. When present, 7.5 fmol of PCNA trimer and RFC complex were added, and 100 fmol of RPA was added, which is approximately 1.5 times the amount needed to completely coat the 407 base gap present in the substrate, assuming a minimal binding footprint of ∼30 bases (43, 44). Reactions (25 μL) contained 40 mM Tris-CL (pH 7.8), 75 mM NaCl, 5 mM MgCl2, 1 mM ATP, 0.1 mM each dATP, dCTP, dGTP, and dTTP, 2 mM DTT, and 100 μg/mL BSA. All components except polymerase were mixed on ice and then incubated at 30 °C for 2 min. Polymerase was added to initiate the reaction and incubated for an additional 1 h at 30 °C. Details of the reaction protocol, plaque color determination, and sequence analysis are as described previously (36).
RESULTS
Processivity and Lesion Bypass Efficiency of Yeast Pol η with and without Accessory Proteins
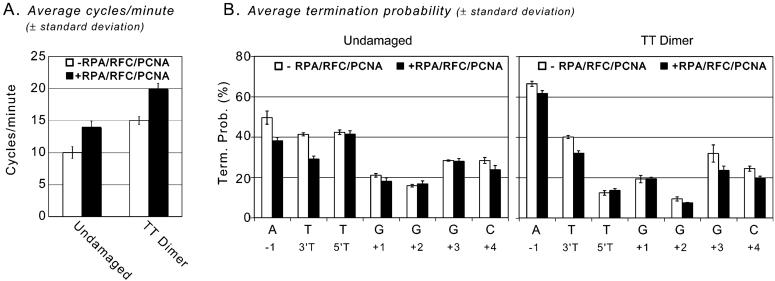
The DNA substrate depicted in Figure 1A was used to investigate whether S. cerevisiae RPA, RFC, and PCNA affect the TT dimer bypass properties of yeast pol η. This template-primer contains 38 nucleotides of single stranded template, sufficient for RPA to bind downstream of the primer terminus, and biotin-streptavidin complexes on the 5′-ends of both DNA strands to prevent PCNA from diffusing off the DNA ends after it has been loaded by RFC. Reactions were performed with a 200-fold excess of DNA over pol η, and aliquots were removed at times sufficient to extend only a small proportion of primers. Under these conditions, each polymerase molecule cycles multiple times but largely to previously unused primers such that the majority of product chains result from a single processive encounter with polymerase (42). Using such conditions, when copying undamaged DNA yeast pol η predominantly adds 1-10 nucleotides, while also generating smaller amounts of longer chains, including full-length products (Figure 1B, lanes 2-4). As in an earlier study (14), many of these products are considerably longer than those generated by human and mouse pol η during a single cycle of processive synthesis (15, 19, 20, 29, 45), confirming that yeast pol η is somewhat more processive than its mammalian homologues. Inclusion of RPA, RFC, and PCNA had a small effect on overall product distribution (Figure 1B, lanes 5-7) but slightly stimulated the production of full-length chains (compare full-length band intensities in lanes 2-4 and 5-7 in Figure 1B). The number of times the polymerase cycled was also slightly increased, from ∼10 cycles per minute in the absence of accessory proteins to ∼14 cycles per minute with accessory proteins present (Figure 2A).
Figure 1.
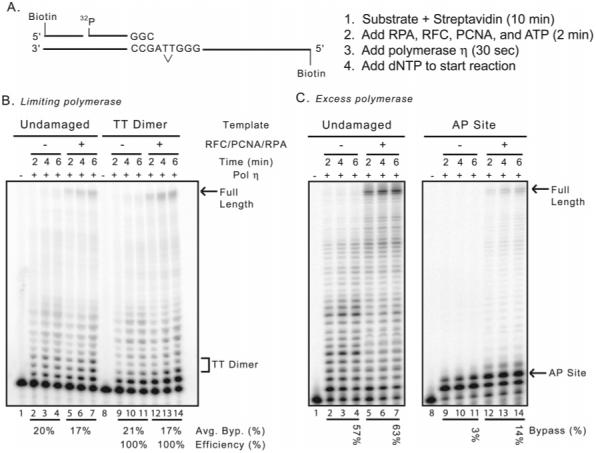
Accessory protein effects on DNA synthesis by S. cerevisiae pol η. (A) Schematic diagram of the substrate used in TLS assays. The 70-mer template oligonucleotides were either undamaged or contained a cis-syn TT dimer (designated by V). Substrate ends were first blocked by streptavidin, then RPA, RFC, and PCNA were loaded onto the DNA before the addition of the polymerase to initiate the reaction. Polymerase assays were performed at 30 °C. (B) Gel image of dPAGE showing the synthesis of the undamaged or TT dimer containing templates, in the absence of any accessory proteins, or with RPA, RFC, and PCNA. These reactions contained a 200-fold excess of substrate DNA over polymerase and are confirmed to be under single interaction conditions. The positions of full length and the TT dimer are shown on the right of the image. (C) Gel image of dPAGE showing the synthesis of the undamaged or AP site containing 102-mer templates. These reactions contain a 2-fold excess of polymerase over DNA substrate and are under multiple interaction conditions. The positions of the AP site and full length products are shown on the right.
Figure 2.
Properties of DNA synthesis reactions by pol η. (A) The number of times each polymerase molecule cycled is plotted for both undamaged and TT dimer-containing reactions, without and with the accessory proteins RPA, RFC, and PCNA. The values are the average (± standard deviation) for 2, 4, and 6 min time points of reactions verified to be under single interaction conditions. (B) Graph of the termination probability (calculated as described in ref 42) for undamaged (left) and TT dimer (right) templates in the absence and presence of the accessory proteins. The values are the average (± standard deviation) for 2, 4, and 6 min time points of reactions verified to be under single interaction conditions.
With the damaged template, yeast pol η alone efficiently bypassed the TTD, and the distribution of products (Figure 1B, lanes 9-11) was similar to that seen with the undamaged template. Inclusion of the accessory proteins did not strongly affect the overall product distribution but did slightly increase the production of full-length DNA chains (Figure 1B, compare lanes 9-11 with 12-14) and the ability of pol η to cycle to a new template primer (Figure 2A; ∼15 and ∼20 cycles per minute, without and with accessory proteins, respectively). Importantly, the termination probability patterns (Figure 2B) and the relative TLS efficiency (Figure 1B, bottom) are similar with and without RPA, RFC, and PCNA. We also performed reactions using an AP site-containing template and an excess of polymerase over substrate. These reactions, in which nearly 100% of the starting primer is used, the addition of RPA, RFC, and PCNA stimulates bypass of the more difficult to bypass AP site by ∼5-fold, similar to previous reports using this same template (35). This verifies that the proteins used here are active using these reaction conditions.
TT Dimer Bypass Fidelity of Yeast Pol η with and without Accessory Proteins
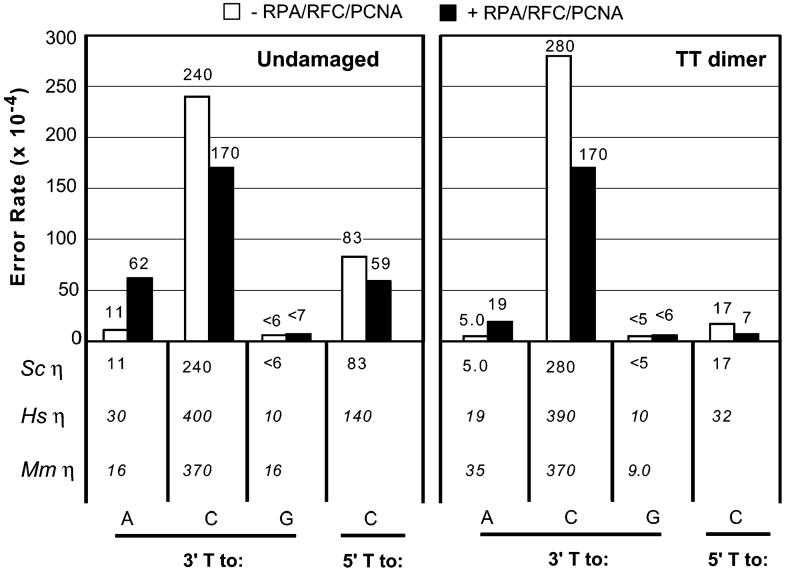
To determine if RPA, RFC, and PCNA affect the fidelity of TTD bypass by yeast pol η, we copied a substrate similar to that depicted in Figure 1A but containing a single 30-mer primer, using sufficient pol η and incubation time to achieve complete bypass of the majority of primer-template molecules. We then recovered the nascent strand, hybridized it to gapped M13mp2 DNA, and introduced these molecules into an E. coli lacZ α-complementation strain to score errors made during TLS (see Experimental Procedures). The relevant 3′ T in this substrate is in a TAG codon in the lacZ α-complementation gene of M13mp2 that when copied correctly yields a light-blue M13 plaque phenotype. Stable misincorporation (i.e., misinsertion followed by mismatch extension) of dGMP, dCMP, or dTMP opposite this 3′ T yields dark-blue plaques whose frequency is one measure of misincorporation opposite the lesion. The frequency of dark-blue plaques was similar for the products of reactions copying undamaged or TTD-containing templates by yeast pol η alone or with RPA, RFC, and PCNA (Table 1). Sequence analysis of the lacZ α-complementation gene from independent dark-blue plaques defined the substitutions at the 3′ T, while T to C substitutions (T-dGMP mismatch) at the 5′ T were monitored by plaque hybridization (15). This information was used to calculate error rates for complete bypass of a TT dimer, and these rates were compared to error rates for copying the equivalent thymines in the undamaged template (Figure 3).
Table 1.
Mutation Frequencies for TT Dimer Bypass by S. cerevisiae Pol η with and without Accessory Proteins
| Undamaged |
TT Dimer |
|||
|---|---|---|---|---|
| - RPA/RFC/PCNA | + RPA/RFC/PCNA | - RPA/RFC/PCNA | + RPA/RFC/PCNA | |
| total plaques | 6869 | 2974 | 6236 | 2884 |
| dark blue plaques | 121 | 49 | 111 | 41 |
| frequency | 1.8% | 1.7% | 1.8% | 1.4% |
| revertants sequenced | 52 | 40 | 55 | 37 |
Figure 3.
Error rates during TT dimer bypass. Lesion bypass fidelity assays were performed as described (see Experimental Procedures and refs 37 and 42). Mutant plaques were sequenced to determine the error rate at the 3′ T. Plaque hybridization (15, 37) was used to determine the rates at the 5′ T. Error rate values (×10-4) are given below the graph for yeast (this study), human (15), and mouse (45) pol η for comparison.
Error rates for TTD bypass by yeast pol η without accessory proteins (Figure 3, right panel, white bars) range from <5 × 10-4 for misincorporation of dCMP opposite the 3′ T of the dimer to 280 × 10-4 for misincorporation of dGMP opposite the 3′ T of the dimer. These rates are similar to but uniformly lower than those for human and mouse pol η (Figure 3, data table below graph; human and mouse data were reproduced from refs 15 and 45). Overall, yeast pol η alone copies the 3′ T of the TT dimer with relatively low fidelity, particularly for misincorporation of dGMP that results in T to C transitions (Figure 3). The rate for this error is similar when copying the undamaged T and the 3′ T of the dimer (240 vs 280 × 10-4). Rates for misincorporation of dGMP opposite the 5′ T are 3-fold lower with the undamaged substrate (240 vs 83 × 10-4) and 16-fold lower with the dimer (280 vs 17 × 10-4), compared to that opposite the 3′ T. Thus, like mammalian pol η (15, 45), yeast pol η is relatively inaccurate for TTD bypass but does copy the damaged 5′ template T with relatively higher fidelity than it copies the corresponding undamaged T or the 3′ T of the TTD.
Most relevant to the main objective of this study are the error rates for TTD bypass with and without accessory proteins. For the major mistake made by yeast pol η, dGMP misincorporation opposite the 3′ T of the TTD, there is less than a 2-fold difference in error rate in the absence or presence of the accessory proteins (Figure 3, right panel). Similarly, dGMP misincorporation rates opposite the 5′ T of the TTD differed by less than 3-fold (17 × 10-4 and 7 × 10-4, without and with accessory proteins, respectively). Misincorporation of dTMP opposite the 3′ T of the TTD was potentially higher in the presence of the accessory proteins as compared to their absence (19 vs 5 × 10-4), although this difference is still less than 4-fold. Importantly, the bias in fidelity between the 3′ and 5′ Ts is still present when the accessory proteins are present. The bias on undamaged DNA is still ∼3 fold (170 vs 59 × 10-4), but it is 24-fold when copying the TT dimer. From this data, we conclude that the accessory proteins do not strongly alter the fidelity with which yeast pol η bypasses a TT dimer.
Yeast Pol η Fidelity when Copying Undamaged DNA with and without Accessory Proteins
Because yeast pol η was more accurate than human or mouse pol η when copying the thymines in the substrates used above (Figure 3, data table below graph), we decided to examine the fidelity of DNA synthesis by yeast pol η in a more comprehensive manner when copying an undamaged template. Synthesis of a 407-nucleotide gap template encoding the lacZ α-complementation gene within gapped M13mp2 DNA was performed using yeast pol η alone or with RPA, RFC, and PCNA. In this assay, errors are detected as light-blue and colorless lacZ mutant plaques among dark-blue plaques resulting from correct synthesis (36). Because the assay monitors the loss of a gene function that is not essential for M13 plaque formation, it scores a broad range of errors, including all 12 possible single base substitutions in a variety of sequence contexts and additions and deletions of many different template nucleotides.
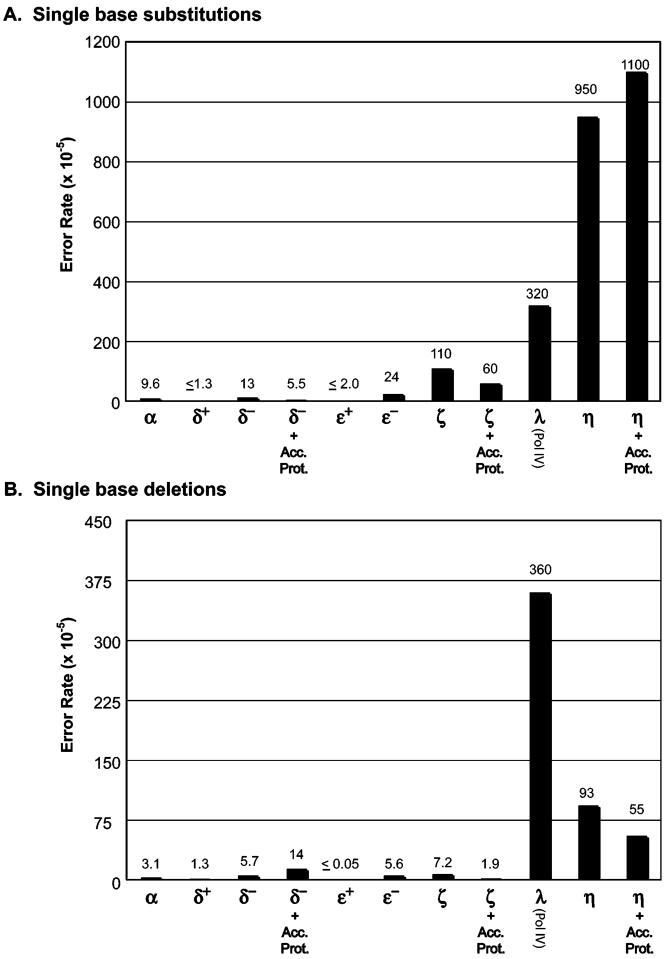
Synthesis either by yeast pol η alone or with accessory proteins generated products that yielded ∼20% lacZ mutants (Table 2). Sequence analysis of DNA from independent lacZ mutant plaques revealed that most mutants contained more than five sequence changes (Table 2), distributed nonrandomly within the 407-nucleotide template (Supporting Information, Figure S1). These changes include a large number of single base substitutions and smaller numbers of tandem double base substitutions, single base insertions, single base deletions, and various other changes involving more than one base pair (Table 2). This information was used to calculate error rates (Table 3) that were compared with human pol η (data from ref 46) as well as with data from several other yeast DNA polymerases (Figure 4) obtained using the same assay.
Table 2.
LacZα Mutant Frequencies, Sequence Changes, and Average Error Rates (× 10-4) for Gap Filling Synthesis by Yeast Pol η with and without Accessory Proteins
| - RPA/RFC/PCNA |
+ RPA/RFC/PCNA |
|||
|---|---|---|---|---|
| number | error ratea × 10-4 (human rate) | number | error ratea × 10-4 | |
| plaques counted | 889 | 704 | ||
| mutants scored | 176 | 143 | ||
| (frequency) | (20%) | (20%) | ||
| lacZ mutants sequenced | 37 | 40 | ||
| total bases sequencedb | 15030 | 16171 | ||
| total sequence changes | 188 | 222 | ||
| average changes/mutant | 5.1 | 5.6 | ||
| single base substitutions | 143 | 95 (320) | 181 | 110 |
| tandem base substitutions | 6 | 4.0 (18) | 2 | 1.2 |
| single base insertions | 13 | 8.7 (13) | 22 | 14 |
| single base deletions | 14 | 9.3 (24) | 9 | 5.5 |
| other changesc | 12 | 8 | ||
The error rates were calculated by dividing the number of a given type of change by the total number of bases sequenced. The data for human enzyme comes from ref 46.
Bases sequenced are 407 per mutant, corrected for the total number of insertions and deletions detected.
Including substitution plus insertion/deletion and the deletions of two or more bases.
Table 3.
Single Base Substitution Error Rates of Pol η
| error rate (× 10-4)a |
|||
|---|---|---|---|
| human Pol η |
S. cerevisiae Pol η |
||
| mispair | - RPA/RFC/PCNA | - RPA/RFC/PCNA | + RPA/RFC/PCNA |
| A → C A·dGMP | 130 | 52 | 25 |
| G A·dCMP | 100 | 16 | 23 |
| T A·dAMP | 160 | 36 | 51 |
| C → A C·dTMP | 26 | ≤ 2.2 | 2.0 |
| G C·dCMP | ≤ 5.0 | 4.4 | 10 |
| T C·dAMP | 100 | 4.4 | 10 |
| G → A G·dTMP | 140 | 5.7 | 7.9 |
| C G·dGMP | 27 | 14 | 11 |
| T G·dAMP | 48 | 5.7 | 2.6 |
| T → A T·dTMP | 83 | 24 | 25 |
| C T·dGMP | 560 | 180 | 220 |
| G T·dCMP | 50 | 2.2 | 10 |
The error rates for each possible base substitution were calculated from sequenced samples of mutant plaques obtained after gap-filling experiments (Table 2). As described previously (46), because of the multiple changes observed in every mutant sequenced, all sites (opposed to phenotypically detectable sites) become targets for change. Therefore, error rates are calculated by dividing the number of observed changes of a given type by the total number of sequenced bases. Comparable rates for human pol η are shown for comparison (from ref 73).
Figure 4.
Average error rates of yeast DNA polymerases when copying undamaged DNA. The average error rates for base substitution and single base deletions were calculated from sequenced samples of mutant plaques obtained after gap-filling experiments (see Tables 2 and 3). The rates given for all enzymes except pol η are for detectable sites only, whereas the rates for pol η are per incorporation (see footnote in Table 3). Pol α, 4 subunit holoenzyme (74); rates recalculated from original data based on current number of known detectable sites. Pol δ, 3 subunit holoenzyme (48, 75). Pol ε, 4 subunit holoenzyme (76). Pol ζ, dimer of Rev3p/Rev7p (49). Pol λ, also known as Pol IV (69). Pol η, this study. + Acc. Prot. refers to data obtained in the presence of RPA, RFC, and PCNA.
The results lead to three main conclusions. First, as predicted by kinetic studies of misinsertion (17, 47), yeast pol η is a highly inaccurate DNA polymerase, having much higher fidelity for single base substitutions than the other yeast DNA polymerases that have also been characterized by this approach (Figure 4). Second, yeast pol η is several-fold more accurate than human pol η. This is the case for the average rates for the five types of errors listed in Table 2 and for error rates for most of the 12 possible single base-base mismatches, where differences vary between 2- and 25-fold (Table 3). Third, the accessory proteins RPA, RFC, and PCNA have little if any affect on the fidelity of yeast pol η (Tables 2 and 3).
DISCUSSION
Although human pol η by itself synthesizes DNA with low fidelity when copying a cis-syn TT dimer (14, 15), the increase in UV light-induced mutagenesis in cells lacking functional pol η (6, 7, 45) clearly illustrates that pol η does indeed participate in a TLS pathway in vivo that suppresses UV mutagenesis. This prompted the present study to determine if accessory proteins improve the bypass properties of pol η. The observation that RPA, RFC, and PCNA slightly increase yeast pol η processivity and cycling (Figures 1 and 2) is consistent with previous studies showing that pol η is stimulated by PCNA (29, 30, 35). In the present study, using assays that allow for analysis of a single round of DNA/polymerase interaction, we report that these stimulatory effects are small and similar with damaged and undamaged templates such that the relative and high efficiency with which yeast pol η bypasses a cis-syn TT dimer is largely unchanged. Also, the previously reported switch from lower to higher termination after the lesion has been copied is still evident in the presence of the accessory proteins (e.g., compare termination probabilities at the 5′ T and +1 positions on undamaged and TT dimer templates in Figure 2B). This is consistent with a role of these accessory proteins in modulating the switch back to a major replicative polymerase following lesion bypass (15).
Our results also show that RPA, RFC, and PCNA have little effect on the fidelity with which yeast pol η bypasses a cis-syn TT dimer. The observations with undamaged DNA are consistent with recent studies showing only small effects of the accessory proteins on the fidelity of DNA synthesis by yeast pol δ and pol ζ (Figure 4) (48, 49). The small effects of yeast accessory proteins on fidelity are generally consistent with studies in other replication systems (50, 51), all indicating that replication fidelity is primarily determined by the polymerase catalytic subunits, as seen here with yeast pol η. This is not surprising, given the open active site of Y-family polymerases (52-54).
It is particularly interesting that yeast pol η, which suppresses UV-induced mutagenesis in yeast (9-13), is about 10-fold less accurate for base substitutions than yeast pol ζ (Figure 3), which promotes UV-induced mutagenesis in yeast (10, 55). This difference in the fidelity of the two polymerases is maintained in the presence of RPA, RFC, and PCNA (Figure 4). Moreover, yeast pol η fidelity remains low whether copying undamaged DNA or a TT dimer. Because accessory proteins do not improve the low fidelity of yeast pol η, we continue to entertain other possible explanations for how this (or other) intrinsically low fidelity polymerase participates in TLS that suppresses UV-induced mutagenesis. One possibility is that errors made by pol η are extrinsically proofread by the 3′ exonuclease activity of another protein. We have previously shown that errors made by pol η during in vitro TT dimer bypass can be proofread by pols δ and ε (14) and also that errors made during the replication of undamaged DNA are subject to proofreading (21). In addition, extrinsic proofreading has been shown to occur in other DNA replication pathways and systems (22, 23, 56). Another mechanism to potentially increase the in vivo fidelity of TLS is correction by DNA mismatch repair, as has been previously suggested (57). There is ample evidence that the main mismatch recognition complex hMutSα can bind to mismatched photoproduct lesions (57-59), and a recent report suggests that mismatch repair does indeed aid in controlling mutagenesis induced by UV light (60). These error correction processes seem feasible because, unlike certain other types of lesions, the template bases in CPDs retain Watson-Crick and Hoogsteen base pairing potential (61, 62). If the lesion bypass reaction takes place at the replication fork during active DNA replication, either mechanism would be expected to be available. It should be noted that recent reports (63, 64) providing support for an early model of TLS (65) that suggested that bypass occurred separate from the replication fork movement do not necessarily contradict this model. It is possible that lesions that are difficult to bypass are in fact skipped by the replication machinery and then bypassed behind the fork, whereas lesions that are easy to bypass (i.e., a TT dimer, 8-oxoguanine) could be bypassed at the fork itself. In this respect, it is important to note that both pol η and ι have been reported to be a part of the replication machinery (26, 27).
Saccharomyces cerevisiae has only two Y family members, REV1 and pol η, whereas mammals have four: REV1, pol η, pol κ, and pol ι. Although pol η clearly has substantially different properties from those of REV1, pol ι, and pol κ, we did not anticipate that yeast pol η would differ in its catalytic properties from human and mouse pol η. It has previously been reported that the yeast and human enzymes do differ when examined by pre steady-state kinetic analysis (66). However, although the human enzyme was shown to incorporate dNTPs at a 50-fold faster rate than the yeast enzyme, the binding affinity for dNTP was 50-fold lower, leading to similar steady-state kinetics and overall calculated fidelity for the two enzymes (17, 19, 47, 67). Nonetheless, the present results and an earlier study of processivity (14) clearly indicate that yeast pol η has somewhat higher processivity and higher fidelity than human and mouse pol η. In fact, the processivity and fidelity of yeast pol η appears to be more like those of human pol κ, which has somewhat higher processivity and higher fidelity than human and mouse pol η (68). In this sense, yeast pol η may be a hybrid that could theoretically perform multiple functions in yeast that require multiple polymerases (pol η, pol κ, and pol ι) in mammals. This idea has also been proposed for yeast pol λ (pol IV), the X family polymerase with hybrid properties and homology to four mammalian X family members, pol β, pol λ, pol μ, and TdT (69). In considering what roles pol η might have in yeast other than TLS, it is worth noting that human pol κ has recently been implicated in filling gaps during nucleotide excision repair (70). It has also recently been reported that human pol η is involved in homologous recombination (71, 72). Whether or not yeast pol η performs similar and/or different functions, function may give us further insight into the biological significance of the catalytic differences between the two enzymes.
Supplementary Material
Spectra of forward assay mutations
ACKNOWLEDGMENT
We thank Drs. Katarzyna Bebenek and Zachary Pursell for constructive comments on the manuscript.
Footnotes
This research was supported in part by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences, and in part by NIH Grant GM32431 to P.M.J.B.
- TLS
- translesion synthesis
- pol
- polymerase
- RPA
- replication protein A
- PCNA
- proliferating cell nuclear antigen
- RFC
- replication factor C
- XP-V
- Xeroderma pigmentosum-variant
- CPD
- cyclobutane pyrimidine dimer
- TTD
- thymine-thymine dimer
SUPPORTING INFORMATION AVAILABLE
Spectra of forward assay mutations. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Shcherbakova PV, Bebenek K, Kunkel TA. Functions of eukaryotic DNA polymerases. Sci. Aging Knowledge Environ. 2003;2003:RE3. doi: 10.1126/sageke.2003.8.re3. [DOI] [PubMed] [Google Scholar]
- 2.Johnson RE, Kondratick CM, Prakash S, Prakash L. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science. 1999;285:263–265. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]
- 3.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 4.Cordonnier AM, Fuchs RP. Replication of damaged DNA: molecular defect in xeroderma pigmentosum variant cells. Mutat. Res. 1999;435:111–119. doi: 10.1016/s0921-8777(99)00047-6. [DOI] [PubMed] [Google Scholar]
- 5.Cleaver JE. Common pathways for ultraviolet skin carcinogenesis in the repair and replication defective groups of xeroderma pigmentosum. J. Dermatol. Sci. 2000;23:1–11. doi: 10.1016/s0923-1811(99)00088-2. [DOI] [PubMed] [Google Scholar]
- 6.Maher VM, Ouellette LM, Curren RD, McCormick JJ. Caffeine enhancement of the cytotoxic and mutagenic effect of ultraviolet irradiation in a xeroderma pigmentosum variant strain of human cells. Biochem. Biophys. Res. Commun. 1976;71:228–234. doi: 10.1016/0006-291x(76)90272-2. [DOI] [PubMed] [Google Scholar]
- 7.Maher VM, Ouellette LM, Curren RD, McCormick JJ. Frequency of ultraviolet light-induced mutations is higher in xeroderma pigmentosum variant cells than in normal human cells. Nature. 1976;261:593–595. doi: 10.1038/261593a0. [DOI] [PubMed] [Google Scholar]
- 8.Roush AA, Suarez M, Friedberg EC, Radman M, Siede W. Deletion of the Saccharomyces cerevisiae gene RAD30 encoding an Escherichia coli DinB homolog confers UV radiation sensitivity and altered mutability. Mol. Gen. Genet. 1998;257:686–692. doi: 10.1007/s004380050698. [DOI] [PubMed] [Google Scholar]
- 9.McDonald JP, Levine AS, Woodgate R. The Saccharomyces cerevisiae RAD30 gene, a homologue of Escherichia coli dinB and umuC, is DNA damage inducible and functions in a novel error-free postreplication repair mechanism. Genetics. 1997;147:1557–1568. doi: 10.1093/genetics/147.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu SL, Johnson RE, Prakash S, Prakash L. Requirement of DNA polymerase η for error-free bypass of UV-induced CC and TC photoproducts. Mol. Cell. Biol. 2001;21:185–188. doi: 10.1128/MCB.21.1.185-188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gibbs PE, McDonald J, Woodgate R, Lawrence CW. The relative roles in vivo of Saccharomyces cerevisiae Pol eta, Pol zeta, Rev1 protein and Pol32 in the bypass and mutation induction of an abasic site, T-T (6-4) photoadduct and T-T cis-syn cyclobutane dimer. Genetics. 2005;169:575–582. doi: 10.1534/genetics.104.034611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glick E, Vigna KL, Loeb LA. Mutations in human DNA polymerase η motif II alter bypass of DNA lesions. EMBO J. 2001;20:7303–7312. doi: 10.1093/emboj/20.24.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang H, Siede W. UV-induced T→C transition at a TT photoproduct site is dependent on Saccharomyces cerevisiae polymerase eta in vivo. Nucleic Acids Res. 2002;30:1262–1267. doi: 10.1093/nar/30.5.1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCulloch SD, Kokoska RJ, Chilkova O, Welch CM, Johansson E, Burgers PM, Kunkel TA. Enzymatic switching for efficient and accurate translesion DNA replication. Nucleic Acids Res. 2004;32:4665–4675. doi: 10.1093/nar/gkh777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCulloch SD, Kokoska RJ, Masutani C, Iwai S, Hanaoka F, Kunkel TA. Preferential cis-syn thymine dimer bypass by DNA polymerase η occurs with biased fidelity. Nature. 2004;428:97–100. doi: 10.1038/nature02352. [DOI] [PubMed] [Google Scholar]
- 16.Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, pol η. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 17.Washington MT, Johnson RE, Prakash S, Prakash L. Accuracy of thymine-thymine dimer bypass by Saccharomyces cerevisiae DNA polymerase η. Proc. Natl. Acad. Sci. U.S.A. 2000;97:3094–3099. doi: 10.1073/pnas.050491997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masutani C, Araki M, Yamada A, Kusumoto R, Nogimori T, Maekawa T, Iwai S, Hanaoka F. Xeroderma pigmentosum variant (XP-V) correcting protein from HeLa cells has a thymine dimer bypass DNA polymerase activity. EMBO J. 1999;18:3491–3501. doi: 10.1093/emboj/18.12.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson RE, Washington MT, Prakash S, Prakash L. Fidelity of human DNA polymerase η. J. Biol. Chem. 2000;275:7447–7450. doi: 10.1074/jbc.275.11.7447. [DOI] [PubMed] [Google Scholar]
- 20.Masutani C, Kusumoto R, Iwai S, Hanaoka F. Mechanisms of accurate translesion synthesis by human DNA polymerase η. EMBO J. 2000;19:3100–3109. doi: 10.1093/emboj/19.12.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bebenek K, Matsuda T, Masutani C, Hanaoka F, Kunkel TA. Proofreading of DNA polymerase η-dependent replication errors. J. Biol. Chem. 2001;276:2317–2320. doi: 10.1074/jbc.C000690200. [DOI] [PubMed] [Google Scholar]
- 22.Pavlov YI, Frahm C, McElhinny SA, Niimi A, Suzuki M, Kunkel TA. Evidence that errors made by DNA polymerase α are corrected by DNA polymerase δ. Curr. Biol. 2006;16:202–207. doi: 10.1016/j.cub.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 23.McElhinny SA, Pavlov YI, Kunkel TA. Evidence for extrinsic exonucleolytic proofreading. Cell Cycle. 2006;5:958–962. doi: 10.4161/cc.5.9.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Young LC, Hays JB, Tron VA, Andrew SE. DNA mismatch repair proteins: potential guardians against genomic instability and tumorigenesis induced by ultraviolet photoproducts. J. Invest. Dermatol. 2003;121:435–440. doi: 10.1046/j.1523-1747.2003.12450.x. [DOI] [PubMed] [Google Scholar]
- 25.Haracska L, Acharya N, Unk I, Johnson RE, Hurwitz J, Prakash L, Prakash S. A single domain in human DNA polymerase ι mediates interaction with PCNA: implications for translesion DNA synthesis. Mol. Cell. Biol. 2005;25:1183–1190. doi: 10.1128/MCB.25.3.1183-1190.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kannouche P, Fernandez de Henestrosa AR, Coull B, Vidal AE, Gray C, Zicha D, Woodgate R, Lehmann AR. Localization of DNA polymerases η and ι to the replication machinery is tightly co-ordinated in human cells. EMBO J. 2003;22:1223–1233. doi: 10.1093/emboj/cdf618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kannouche P, Broughton BC, Volker M, Hanaoka F, Mullenders LH, Lehmann AR. Domain structure, localization, and function of DNA polymerase η, defective in xeroderma pigmentosum variant cells. Genes Dev. 2001;15:158–172. doi: 10.1101/gad.187501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase η with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell. 2004;14:491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- 29.Haracska L, Johnson RE, Unk I, Phillips B, Hurwitz J, Prakash L, Prakash S. Physical and functional interactions of human DNA polymerase η with PCNA. Mol. Cell. Biol. 2001;21:7199–7206. doi: 10.1128/MCB.21.21.7199-7206.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haracska L, Kondratick CM, Unk I, Prakash S, Prakash L. Interaction with PCNA is essential for yeast DNA polymerase η function. Mol. Cell. 2001;8:407–415. doi: 10.1016/s1097-2765(01)00319-7. [DOI] [PubMed] [Google Scholar]
- 31.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 32.Stelter P, Ulrich HD. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003;425:188–191. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- 33.Watanabe K, Tateishi S, Kawasuji M, Tsurimoto T, Inoue H, Yamaizumi M. Rad18 guides poleta to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 2004;23:3886–3896. doi: 10.1038/sj.emboj.7600383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bienko M, Green CM, Crosetto N, Rudolf F, Zapart G, Coull B, Kannouche P, Wider G, Peter M, Lehmann AR, Hofmann K, Dikic I. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 2005;310:1821–1824. doi: 10.1126/science.1120615. [DOI] [PubMed] [Google Scholar]
- 35.Garg P, Burgers PM. Ubiquitinated proliferating cell nuclear antigen activates translesion DNA polymerases η and REV1. Proc. Natl. Acad. Sci. U.S.A. 2005;102:18361–18366. doi: 10.1073/pnas.0505949102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bebenek K, Kunkel TA. Analyzing fidelity of DNA polymerases. Methods Enzymol. 1995;262:217–232. doi: 10.1016/0076-6879(95)62020-6. [DOI] [PubMed] [Google Scholar]
- 37.McCulloch SD, Kunkel TA. Measuring the fidelity of translesion DNA synthesis. Methods Enzymol. 2006;408:341–355. doi: 10.1016/S0076-6879(06)08021-9. [DOI] [PubMed] [Google Scholar]
- 38.Yuan F, Zhang Y, Rajpal DK, Wu X, Guo D, Wang M, Taylor JS, Wang Z. Specificity of DNA lesion bypass by the yeast DNA polymerase η. J. Biol. Chem. 2000;275:8233–8239. doi: 10.1074/jbc.275.11.8233. [DOI] [PubMed] [Google Scholar]
- 39.Henricksen LA, Umbricht CB, Wold MS. Recombinant replication protein A: expression, complex formation, and functional characterization. J. Biol. Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- 40.Ayyagari R, Gomes XV, Gordenin DA, Burgers PM. Okazaki fragment maturation in yeast. I. Distribution of functions between FEN1 and DNA2. J. Biol. Chem. 2003;278:1618–1625. doi: 10.1074/jbc.M209801200. [DOI] [PubMed] [Google Scholar]
- 41.Fien K, Stillman B. Identification of replication factor C from Saccharomyces cerevisiae: a component of the leadingstrand DNA replication complex. Mol. Cell. Biol. 1992;12:155–163. doi: 10.1128/mcb.12.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kokoska RJ, McCulloch SD, Kunkel TA. The efficiency and specificity of apurinic/apyrimidinic site bypass by human DNA polymerase η and Sulfolobus solfataricus Dpo4. J. Biol. Chem. 2003;278:50537–50545. doi: 10.1074/jbc.M308515200. [DOI] [PubMed] [Google Scholar]
- 43.Sibenaller ZA, Sorensen BR, Wold MS. The 32- and 14-kilodalton subunits of replication protein A are responsible for species-specific interactions with single-stranded DNA. Biochemistry. 1998;37:12496–12506. doi: 10.1021/bi981110+. [DOI] [PubMed] [Google Scholar]
- 44.Sugiyama T, Zaitseva EM, Kowalczykowski SC. A single-stranded DNA-binding protein is needed for efficient presynaptic complex formation by the Saccharomyces cerevisiae Rad51 protein. J. Biol. Chem. 1997;272:7940–7945. doi: 10.1074/jbc.272.12.7940. [DOI] [PubMed] [Google Scholar]
- 45.Lin Q, Clark AB, McCulloch SD, Yuan T, Bronson RT, Kunkel TA, Kucherlapati R. Increased susceptibility to UV-induced skin carcinogenesis in polymerase η-deficient mice. Cancer Res. 2006;66:87–94. doi: 10.1158/0008-5472.CAN-05-1862. [DOI] [PubMed] [Google Scholar]
- 46.Matsuda T, Bebenek K, Masutani C, Rogozin IB, Hanaoka F, Kunkel TA. Error rate and specificity of human and murine DNA polymerase η. J. Mol. Biol. 2001;312:335–346. doi: 10.1006/jmbi.2001.4937. [DOI] [PubMed] [Google Scholar]
- 47.Washington MT, Johnson RE, Prakash S, Prakash L. Fidelity and processivity of Saccharomyces cerevisiae DNA polymerase η. J. Biol. Chem. 1999;274:36835–36838. doi: 10.1074/jbc.274.52.36835. [DOI] [PubMed] [Google Scholar]
- 48.Fortune JM, Stith CM, Kissling GE, Burgers PM, Kunkel TA. RPA and PCNA suppress formation of large deletion errors by yeast DNA polymerase delta. Nucleic Acids Res. 2006;34:4335–4341. doi: 10.1093/nar/gkl403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhong X, Garg P, Stith CM, McElhinny SA, Kissling GE, Burgers PM, Kunkel TA. The fidelity of DNA synthesis by yeast DNA polymerase zeta alone and with accessory proteins. Nucleic Acids Res. 2006;34:4731–4742. doi: 10.1093/nar/gkl465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bebenek A, Carver GT, Dressman HK, Kadyrov FA, Haseman JK, Petrov V, Konigsberg WH, Karam JD, Drake JW. Dissecting the fidelity of bacteriophage RB69 DNA polymerase: site-specific modulation of fidelity by polymerase accessory proteins. Genetics. 2002;162:1003–1018. doi: 10.1093/genetics/162.3.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bebenek A, Carver GT, Kadyrov FA, Kissling GE, Drake JW. Processivity clamp gp45 and ssDNA-binding-protein gp32 modulate the fidelity of bacteriophage RB69 DNA polymerase in a sequence-specific manner, sometimes enhancing and sometimes compromising accuracy. Genetics. 2005;169:1815–1824. doi: 10.1534/genetics.104.037630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ling H, Boudsocq F, Woodgate R, Yang W. Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell. 2001;107:91–102. doi: 10.1016/s0092-8674(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 53.Silvian LF, Toth EA, Pham P, Goodman MF, Ellenberger T. Crystal structure of a DinB family error-prone DNA polymerase from Sulfolobus solfataricus. Nat. Struct. Biol. 2001;8:984–989. doi: 10.1038/nsb1101-984. [DOI] [PubMed] [Google Scholar]
- 54.Trincao J, Johnson RE, Escalante CR, Prakash S, Prakash L, Aggarwal AK. Structure of the catalytic core of S. cerevisiae DNA polymerase η: implications for translesion DNA synthesis. Mol. Cell. 2001;8:417–426. doi: 10.1016/s1097-2765(01)00306-9. [DOI] [PubMed] [Google Scholar]
- 55.Lawrence CW, Maher VM. Mutagenesis in eukaryotes dependent on DNA polymerase ζ and Rev1p. Philos. Trans. R. Soc. London, Ser. B. 2001;356:41–46. doi: 10.1098/rstb.2000.0746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Perrino FW, Loeb LA. Proofreading by the ε subunit of Escherichia coli DNA polymerase III increases the fidelity of calf thymus DNA polymerase α. Proc. Natl. Acad. Sci. U.S.A. 1989;86:3085–3088. doi: 10.1073/pnas.86.9.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang H, Lawrence CW, Li G-M, Hays JB. Specific binding of human MSH2ΣMSH6 mismatch-repair protein heterodimers to DNA incorporating thymine- or uracil-containing UV light photoproducts opposite mismatched bases. J. Biol. Chem. 1999;274:16894–16900. doi: 10.1074/jbc.274.24.16894. [DOI] [PubMed] [Google Scholar]
- 58.Hoffman PD, Wang H, Lawrence CW, Iwai S, Hanaoka F, Hays JB. Binding of MutS mismatch repair protein to DNA containing UV photoproducts, “mismatched” opposite Watson-Crick and novel nucleotides, in different DNA sequence contexts. DNA Repair. 2005;4:983–993. doi: 10.1016/j.dnarep.2005.04.018. [DOI] [PubMed] [Google Scholar]
- 59.Mu D, Tursun M, Duckett DR, Drummond JT, Modrich P, Sancar A. Recognition and repair of compound DNA lesions (base damage and mismatch) by human mismatch repair and excision repair systems. Mol. Cell. Biol. 1997;17:760–769. doi: 10.1128/mcb.17.2.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Borgdorff V, Pauw B, van Hees-Stuivenberg S, de Wind N. DNA mismatch repair mediates protection from mutagenesis induced by short-wave ultraviolet light. DNA Repair. 2006;5:1364–1372. doi: 10.1016/j.dnarep.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 61.Park H, Zhang K, Ren Y, Nadji S, Sinha N, Taylor JS, Kang C. Crystal structure of a DNA decamer containing a cis-syn thymine dimer. Proc. Natl. Acad. Sci. U.S.A. 2002;99:15965–15970. doi: 10.1073/pnas.242422699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ling H, Boudsocq F, Plosky BS, Woodgate R, Yang W. Replication of a cis-syn thymine dimer at atomic resolution. Nature. 2003;424:1083–1087. doi: 10.1038/nature01919. [DOI] [PubMed] [Google Scholar]
- 63.Lopes M, Foiani M, Sogo JM. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol. Cell. 2006;21:15–27. doi: 10.1016/j.molcel.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 64.Heller RC, Marians KJ. Replication fork reactivation downstream of a blocked nascent leading strand. Nature. 2006;439:557–562. doi: 10.1038/nature04329. [DOI] [PubMed] [Google Scholar]
- 65.Rupp WD, Howard-Flanders P. Discontinuities in the DNA synthesized in an excision-defective strain of Escherichia coli following ultraviolet irradiation. J. Mol. Biol. 1968;31:291–304. doi: 10.1016/0022-2836(68)90445-2. [DOI] [PubMed] [Google Scholar]
- 66.Washington MT, Johnson RE, Prakash L, Prakash S. The mechanism of nucleotide incorporation by human DNA polymerase η differs from that of the yeast enzyme. Mol. Cell. Biol. 2003;23:8316–8322. doi: 10.1128/MCB.23.22.8316-8322.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Washington MT, Johnson RE, Prakash L, Prakash S. Accuracy of lesion bypass by yeast and human DNA polymerase η. Proc. Natl. Acad. Sci. U.S.A. 2001;98:8355–8360. doi: 10.1073/pnas.121007298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ohashi E, Bebenek K, Matsuda T, Feaver WJ, Gerlach VL, Friedberg EC, Ohmori H, Kunkel TA. Fidelity and processivity of DNA synthesis by DNA polymerase κ, the product of the human DINB1 gene. J. Biol. Chem. 2000;275:39678–39684. doi: 10.1074/jbc.M005309200. [DOI] [PubMed] [Google Scholar]
- 69.Bebenek K, Garcia-Diaz M, Patishall SR, Kunkel TA. Biochemical properties of Saccharomyces cerevisiae DNA polymerase IV. J. Biol. Chem. 2005;280:20051–20058. doi: 10.1074/jbc.M501981200. [DOI] [PubMed] [Google Scholar]
- 70.Ogi T, Lehmann AR. The Y-family DNA polymerase kappa (pol kappa) functions in mammalian nucleotide-excision repair. Nat. Cell Biol. 2006;8:640–642. doi: 10.1038/ncb1417. [DOI] [PubMed] [Google Scholar]
- 71.Kawamoto T, Araki K, Sonoda E, Yamashita YM, Harada K, Kikuchi K, Masutani C, Hanaoka F, Nozaki K, Hashimoto N, Takeda S. Dual roles for DNA polymerase eta in homologous DNA recombination and translesion DNA synthesis. Mol. Cell. 2005;20:793–799. doi: 10.1016/j.molcel.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 72.McIlwraith MJ, Vaisman A, Liu Y, Fanning E, Woodgate R, West SC. Human DNA polymerase eta promotes DNA synthesis from strand invasion intermediates of homologous recombination. Mol. Cell. 2005;20:783–792. doi: 10.1016/j.molcel.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 73.Matsuda T, Bebenek K, Masutani C, Hanaoka F, Kunkel TA. Low fidelity DNA synthesis by human DNA polymerase-η. Nature. 2000;404:1011–1013. doi: 10.1038/35010014. [DOI] [PubMed] [Google Scholar]
- 74.Kunkel TA, Hamatake RK, Motto-Fox J, Fitzgerald MP, Sugino A. Fidelity of DNA polymerase I and the DNA polymerase I-DNA primase complex from Saccharomyces cerevisiae. Mol. Cell. Biol. 1989;9:4447–4458. doi: 10.1128/mcb.9.10.4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fortune JM, Pavlov YI, Welch CM, Johansson E, Burgers PM, Kunkel TA. Saccharomyces cerevisiae DNA polymerase δ: high fidelity for base substitutions but lower fidelity for single- and multi-base deletions. J. Biol. Chem. 2005;280:29980–29987. doi: 10.1074/jbc.M505236200. [DOI] [PubMed] [Google Scholar]
- 76.Shcherbakova PV, Pavlov YI, Chilkova O, Rogozin IB, Johansson E, Kunkel TA. Unique error signature of the four-subunit yeast DNA polymerase ε. J. Biol. Chem. 2003;278:43770–43780. doi: 10.1074/jbc.M306893200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Spectra of forward assay mutations