Abstract
Hedgehog signaling is an important component of cell-cell communication during bilaterian development, and abnormal Hedgehog signaling contributes to disease and birth defects. Hedgehog genes are composed of a ligand (“hedge”) domain and an autocatalytic intein (“hog”) domain. Hedgehog (hh) ligands bind to a conserved set of receptors and activate downstream signal transduction pathways terminating with Gli/Ci transcription factors. We have identified five intein-containing genes in the anthozoan cnidarian Nematostella vectensis, two of which (NvHh1 and NvHh2) contain definitive hedgehog ligand domains, suggesting that to date, cnidarians are the earliest branching metazoan phylum to possess definitive Hh orthologs. Expression analysis of NvHh1 and NvHh2, the receptor NvPatched and a downstream transcription factor NvGli (a Gli3/Ci ortholog) indicate that these genes may have conserved roles in planar and trans-epithelial signaling during gut and germline development, while the three remaining intein-containing genes (NvHint1,2,3) are expressed in a cell-type specific manner in putative neural precursors. Metazoan intein-containing genes that lack a ligand domain have previously only been identified within nematodes. However, phylogenetic analyses suggest that these nematode inteins may be derived from an ancestral nematode true hedgehog gene, and that the non-bilaterian intein-containing genes identified here may represent an ancestral state prior to the domain swapping events that resulted in the formation of true hedgehog genes in the cnidarian-bilaterian ancestor. Genomic surveys of N. vectensis suggest that most of the components of both protostome and deuterostome Hh signaling pathways are present in anthozoans and that some appear to have been lost in ecdysozoan lineages. Cnidarians possess many bilaterian cell-cell signaling pathways (Wnt, TGFß, FGF and Hh) that appear to act in concert to pattern tissues along the oral-aboral axis of the polyp. Cnidarians represent a diverse group of animals with a predominantly epithelial body plan, and perhaps selective pressures to pattern epithelia resulted in the ontogeny of the hedgehog pathway in the common ancestor of the Cnidaria and Bilateria.
Keywords: anthozoa, cnidaria, ectoderm, endoderm, evolution, genomics, gli, hedgehog, induction, Nematostella vectensis, patched, pattern formation, signal transduction
Introduction
Cell to cell signaling, or induction, governs cell fate and pattern formation during early development of essentially all metazoan embryos. In general, the principal inductive signals regulating animal embryogenesis are conveyed by members of a small number of cell signaling pathways including the Wnt, TGF-beta/Bone Morphogenetic Protein (BMP), Fibroblast Growth Factor (FGF), Notch and Hedgehog gene families (Chen et al., 2004; Cornell and Eisen, 2005; Ingham and McMahon, 2001; Kishigami and Mishina, 2005; Lai, 2004; Logan and Nusse, 2004; Tabata and Takei, 2004; Thisse and Thisse, 2005). The Hedgehog pathway in particular mediates a variety of important patterning and cell fate determination events across the Metazoa. The original hedgehog gene (hh) was discovered in Drosophila as a mutation affecting a variety of pattern formation events. Drosophila hh is first expressed in blastoderm-stage embryos where it regulates antero-posterior segment polarity by acting as a morphogen (Heemskerk and DiNardo, 1994; Mohler and Vani, 1992). Drosophila hh also governs cell fate and patterning of the primitive gut, wing disc, eye, and muscle, and in vertebrates hh paralogs display similar diverse functions in regulating cell fate and pattering of the spinal cord, somites, limb, brain, bone, smooth muscle and gut, among others (see recent reviews by (Hooper and Scott, 2005; Ingham and Placzek, 2006; McGlinn and Tabin, 2006; McMahon et al., 2003).
Hedgehogs encode proteins characterized by the presence of a soluble N-terminal ligand domain that gets released by self-proteolysis from a larger precursor through the action of a C-terminal intein domain. Mutations in genes encoding Hh-related ligands and their downstream signal transduction components cause developmental defects in Drosophila and vertebrate model organisms, human birth defects, and various diseases (McMahon et al., 2003; Mullor et al., 2002; Nieuwenhuis and Hui, 2005; Rubin and de Sauvage, 2006). Molecular aspects of signaling by hedgehog ligands, receptors and downstream components are best understood in Drosophila and vertebrates, and their respective pathways share many features yet exhibit distinct differences (Huangfu and Anderson, 2006; Ingham and Placzek, 2006). At the ligand level, just a single hh gene has been described in Drosophila, other protostomes, and most deuterostomes, but two to five hh ligands are encoded by chordate genomes, such as those of tunicates (two) and vertebrates (three – five) (Hino et al., 2003; Meyer and Schartl, 1999; Pires-daSilva and Sommer, 2003). Receptors for hh ligands in Drosophila and vertebrates are very similar and are typified by two multi-transmembrane domain receptors named Patched (Ptc) and Smoothened (Smo). One Ptc and two Smo genes have been identified in vertebrates, while one of each are known in Drosophila. Ptc and Smo regulate cytoplasmic signal transduction that ultimately affects the activity of transcription factors at the terminus of the pathway named Cubitus Interuptus (Ci) in Drosophila, and Gli proteins in vertebrates (three in all: Gli1/2/3) (Huangfu and Anderson, 2006; Wang et al., 2007). In the absence of hh ligand, Ptc inhibits signaling by Ci/Gli proteins by directing their proteolysis by an intracellular protein complex that destroys Gli1/2, or cleaves and partially destroys Ci/Gli3 to produce a stable N-terminal fragment that functions as a transcriptional repressor and blocks the activity of Hh-responsive genes. Binding of a Hh ligand to Ptc derepresses Smo and inactivates its associated Ci/Gli cleavage complex, allowing full-length Ci/Gli1/2/3 to accumulate and function as transcriptional activators of Hh-responsive genes. In vertebrates, but apparently not Drosophila, the nuclear action of Gli proteins is further regulated by Iguana, a protein essential for hh signaling (Vokes and McMahon, 2004). Another major difference between vertebrate and Drosophila hh signaling is that signaling downstream of Smo in vertebrates requires non-sensory cilia and specific intraflagellar transport proteins (Huangfu and Anderson, 2006), but in Drosophila these proteins are not essential for hh signaling (Avidor-Reiss et al., 2004; Han et al., 2003; Ray et al., 1999).
Surveys of various metazoan genomes and expressed cDNA sequences demonstrate the presence of a variety of hh gene orthologs among both deuterostomes and protostomes, with the exception of the nematode C. elegans, which lacks a clear hh ortholog but possesses at least 10 intein-containing genes (Aspock et al., 1999; Hao et al., 2006b). However, an EST (Expressed Sequence Tag) survey of the plant ectoparasitic dagger nematode, Xiphinema index, has identified a single true hedgehog gene within this more slowly evolving nematode (M. Blaxter pers. comm.). As already noted, protostomes appear to possess just a single hh gene, but deuterostomes possess several hh-related genes (three in mammals and five in zebrafish). In the genome of the choanoflagellate, Monosiga ovata, a single-celled protozoan believed to share common ancestry with the Metazoa, recent work identified a hedgehog-related gene (Hoglet) (Snell et al., 2006), composed of a C-terminal intein domain that lacks an identifiable N-terminal Hh signaling domain. A hedgehog-related, intein-only containing gene has also been identified in the mycorrhizal fungus, Glomus mosseae (Requena et al. 2002). The presence of a hedgehog-related intein gene in an outgroup to the Metazoa, a single hh gene among protostomes, and the presence of multiple vertebrate hh homologs, and the notable lack of a hh ortholog in C. elegans, raises important questions about how and when hh genes originated and diversified over the course of metazoan evolution, particularly in the Cnidaria or other non-bilaterian metazoan phyla.
To help understand the evolutionary relationships and origins of invertebrate and vertebrate Hedgehog signaling pathways, we examined the Hh pathway outside of the Bilateria, in the anthozoan cnidarian, Nematostella vectensis, the starlet sea anemone. We report the existence and expression of five N. vectensis hh-related genes, two of which encode Hh signaling ligands (NvHedgehog1 and 2), while the other three encode proteins containing an intein but no recognizable ligand domains (NvHint1, 2, and 3). The embryonic expression of both true hh genes are involved with mesentery, pharynx, and gut formation, while the three intein-only hh-related genes (NvHint1−3) are expressed in a cell-type specific manner and are likely to be involved in neural patterning. Genomic data and mRNA expression patterns suggest that downstream of the ligands, N. vectensis possesses a full suite of genes implicated in Hh signaling, from extracellular ligand modulators, through receptors, signal transduction components and Gli transcription factors, most of which appear to be orthologs of corresponding vertebrate and invertebrate signaling proteins. Furthermore, the embryonic expression of the receptor NvPatched (NvPtc) and the downstream transcriptional regulator NvGli demonstrate that Hh likely conveys both planar and trans-epithelial inductive signals within the mesendoderm. Our data suggest that the antecedents of both vertebrate and invertebrate hh ligand and signal transduction proteins arose early in metazoan evolution, with gene loss and gene duplication over the course of evolution accounting for the differences now observed between protostome and deuterostome pathways. Interestingly, particular members of the Hh signaling pathway in N. vectenis have been lost in the ecdysozoan model systems but retained in vertebrate lineages.
Materials and Methods
Isolation of genes from N. vectensis
The Joint Genome Institute (JGI) assembly of the N. vectensis genome (genome.jgipsf.org/Nemve1/Nemve1.home.html) and available ESTs (NCBI) were searched using the C-terminal intein or HINT domain of bilaterian hedgehogs utilizing TBLASTN parameters to isolate potential hedgehog genes. Gli and patched genes were identified by TBLASTN searches of the N. vectensis genome, using vertebrate orthologs. Gene-specific primers were then designed for both 5’ and 3’ RACE for each gene with annealing temperatures between 68−70C. RACE was performed using the Smart Race cDNA amplification kit (BD Biosciences Clontech). RACE products were cloned in a plasmid vector (P-Gem T easy, Promega) and sequenced at Macrogen, Inc. (South Korea). Five hedgehog-related genes were identified in this manner. Overlapping 5’ and 3’ RACE fragments were aligned and submitted to Genbank as composite transcripts (in process – we will supply prior to publication).
Phylogenetic Analyses of Hedgehog-related genes
Phylogenetic analysis of hedgehog-related genes were performed in order to determine orthology. N. vectensis genes were analyzed via BlastX searches of the GenBank database (http://www.ncbi.nlm.nih.gov/BLAST/) in order to build an alignment. Amino acid alignments of both the C-terminal signaling domain and the N-terminal HINT domain were then made using Muscle 3.6 (Edgar, 2004). and corrected by hand for obvious alignment errors. A Bayesian phylogenetic analysis was conducted using MrBayes 3.1.2 (Ronquist and Huelsenbeck, 2003) using either the mixed or “WAG + G” amino acid model option with four independent runs of 1,000,000 generations each, sampled every 100 generations with four chains. A summary “consensus tree” was produced in MrBayes, from the last 9,500 trees of each run (38,000 total trees) representing 950,000 stationary generations. Posterior probabilities were calculated from this “consensus”. Additionally, Maximum Likelihood (using PHYML; Guindon and Gascuel, 2003) with the WAG + I + G model of evolution (selected via ProtTest; Abascal et al., 2005) using 1000 bootstrap replicates) and RAxML v 2.2.3 (Stamatakis 2006) analyses were conducted. Nexus alignment files can be found in the supplemental information (SF2−4). For analyses of the hint/hog domain 800 searches were performed using RAxML v 2.2.3 (Stamatakis 2006) to find the most likely tree. Results of these analyses are available in the supplemental information (SF5−7).
Phylogenetic Analyses of downstream components
Alignments were made for both patched and Gli/Ci genes using metazoan orthologs and representative sequences from N. vectensis predicted proteins. Phylogenetic analyses were conducted as above, using MrBayes3.1.2 for 1,000,000 with four independent runs generations and a mixed model of protein evolution. For the patched phylogenetic analysis the JTT model was selected with a 99.8% posterior probability, while for the Gli/Ci analysis JTT model was selected with 100% posterior probability. Nexus alignment files can be found in the supplemental information (SF8−9). Identification of other hh pathway genes in the N. vectensis genome, presented in Table 1, was performed using vertebrate or Drosophila protein sequences for TBLASTN searches, and/or gene names to search annotated gene models on the N. vectensis genome, using search functions of the JGI Nematostella genome web server (http://genome.jgi-psf.org/Nemve1/Nemve1.home.html). Candidate Nematostella homologs were retested by BLASTP against the entire human nonredundant protein database using the NCBI server and default settings, to verify the degree of homology represented by the indicated e values in Table 1.
Table 1.
Identification of Nematostella vectensis counterparts of bilaterian Hedgehog signal transduction genes
|
N. vectensis Pathway Component |
Function |
Nv JGI Protein model ID (or EST ID) |
Human Homolog UniProt ID |
Drosophila homolog |
e value in BLASTp with human homolog |
|---|---|---|---|---|---|
| NvHint1 | unknown | this study | n/a | n/a | n/a |
| NvHint2 | unknown | this study | n/a | n/a | n/a |
| NvHint3 | unknown | this study | n/a | n/a | n/a |
| NvHh1 | ligand | this study | Q15465 Sonic Hh O43323 Desert Hh Q14623 Indian Hh | hedgehog | 0 |
| NvHh2 | ligand | this study | Q15465 Sonic Hh | hedgehog | 0 |
| NvSkinny Hh [rasp, sightless] | palmitoylation of Hh ligand | 217054 | Q17N53 | skinny Hh, sightless, central missing | 2 e-40 |
| NvDispatched | ligand transporter - sending cell | 199988 | Q96F81 | Q9U477 | 1 e-88 |
| NvGlypican5 | Hh movement, signaling range | 218206 | P35052 | dally-like UP:Q9GPL5 dally UP:Q966V5 | 8 e-32 |
| NvGlypican6 | Hh movement and signaling range | 218207, 247677 | Q8N158 | dally-like UP:Q9GPL5 | 7 e-92 |
| NvHIP | Hh binding membrane protein | 197846 | Q96QV1 | unknown | 1 e-40 |
| NvCdon/Boc | Hh-binding type I receptor | 185528, 105427 | Q4KMG0 (Cdon) Q9BWV1 (Boc) | iHog | Cdon 7e-30 Boc 4e-31 |
| NvGAS1 | GDNF-receptor-like Hh binding protein represses Hh activity/range | 104344 | unknown | 6 e-13 | |
| NvExostosin-1 | glycosyl transferase for heparan biosynthesis; Hh movement & reception | 173450 | Q16394 | tout-velut | 0 |
| NvExostosin-2 | glycosyl transferase for heparan biosynthesis; Hh movement & reception | 168451 | Q93063 | tout-velut | 0 |
| NvExostosin-3 | glycosyl transferase for heparan biosynthesis; Hh movement & reception | 235881 | O43909 | tout-velut | 0 |
| NvMegalin/gp330 | ligand binding and endocytosis | 196768 | Q7Z5C0 | CG12139 | 0 |
| NvPatched (Ptch) | 11 TM receptor binds Hh, represses Smo | this study | Q13635 | patched | 0 |
| NvSmoothened-1 (Smo1) | 7 TM receptor regulated by Ptch; interfaces with costal & downstream pathway | 208236 | Q99835 | smoothened | 2 e-138 |
| NvSmoothened-2 (Smo2) | 7 TM receptor regulated by Ptch; interfaces with intracellular pathway | 201646 | Q99835 | smoothened | 1 e-91 |
| Nvßarrestin1 | internalization of Smoothened | 97737 | kurtz | 3 e-111 | |
| Nvßarrestin2 | internalization of Smoothened | 149831 | kurtz | 8 e-14 | |
| NvGPRK2/ßARK-1 | G-protein coupled receptor kinase that targets Smoothened | 246204 | Q86YK6 | Gprk2, CG17998 | 0 |
| NvFKBP8 | peptidylprolyl isomerase, represses Smoothened | 91671 | Q7K3D4 | CG5482 | 7 e-34 |
| Nvrab23 | small GTPase for endocytosis & trafficking of Hh-receptor complexes | 25310 | Q9VNG6 | CG2108 | 2 e-64 |
| NvTectonic | signal transduction downstream of Rab23 | (JGI_CAGN8498) | Q2MV58 | unknown | 3 e-47 |
| NvKif3A | potential Costal 2 (Cos2) ortholog, Gli/fused scaffold interface with ciliary microtubules | 160820 | Q59EN1 | costal? | 0 |
| NvKif4A | potential Costal 2 (Cos2) ortholog, Gli/fused scaffold interface with ciliary microtubules | 233887 | O95239 | costal? | 3 e-131 |
| NvKIF27A | Costal 2 (Cos2) ortholog, Gli/fused scaffold interface with ciliary microtubules | 196177 | Q86VH2 | costal | 0 |
| NvFantom/Rpgrip1L | ciliary basal body protein required for Hh signal transduction | 181095 | Q68CZ1 | unknown | 0 |
| NvIft88/polaris/Tg737 | ciliary Hh signaling regulator | 40068 | Q13099 | nompB | 0 |
| NvIft172/wimple | ciliary Hh signaling regulator | (JGI_CAGF7669) | Q96HW4 | osm-1 | 2 e-99 |
| NvIft52/Ngd5 | ciliary Hh signaling regulator | 239245 | Q9Y366 | osm-6 | 2 e-158 |
| NvFused1 | protein kinase phosphorylates Cos2 | 219073 | Q9NRP7 | fused | 1 e-68 |
| NvFused2 | protein kinase - Gli regulation | 173313 | XP_950751* | fused | 0 |
| NvSuppressor of Fused, SuFu | binds and represses Gli | 246114 | Q9UMX1 | Su(Fu) | 6 e-120 |
| Su(Fu) | |||||
| Nvlguana | regulates Gli downstream of Smo, interacts with Sufu | 247454 | Q86YF9 | unknown | 2 e-23 |
| NvPKA | Gli phosphorylation/processing | 120559 | P22694 | Pka-C1, CG4379 | 2 e-100 |
| NvGSK3ß | Gli phosphorylation/processing | 11896 | P49841 | shaggy | 0 |
| NvCK1 | Smo and Gli phosphorylation/processing | 159193 | P48729 | gilgamesh | 4 e-152 |
| NvßTRCP1 | Fbox Ub ligase for Gli-R processing | 228088 | Q9Y297 | slimb | 0 |
| NvßTRCP2 | Fbox Ub ligase for Gli-R processing | 648 | Q9Y297 | slimb | 2 e-165 |
| NvSIL | signal transduction, function unclear | 246782 | Q15468 | unknown | 2 e-65 |
| NvGli3 | Transcriptional activator (Gli-full-length) or repressor (Gli-R) when cleaved | this study | P10071 | Cubitus interruptus (Ci) | 0 |
| NvSap18 | Gli partner: Histone deacetylase subunit:Sin3-associated polypeptide 18 | 226633 | O00422 | Bin1 | 5 e-41 |
| NvSPOP | Gli ubiquitylation and proteolysis | 177584 | O43791 | roadkill | 1 e-153 |
| NvTalpid3 | Gli regulation | (JGI_CAGG5027) | Q1G7G8** | unknown | 1 e-9** |
NCBI protein accession number
chick protein
In situ hybridization
In situ hybridizations using 1−3 kb digoxygenin labeled antisense ribonucleotide probes were performed to follow transcript distribution as previously described (Martindale et al., 2004). Probe concentrations ranged from 1.0 − 2.0 ng/μl and hybridizations were performed at 60°C for 24−48 hours. Alkaline phosphatase reaction products were visualized with NBT-BCIP. Specimens were photographed on a Zeiss Axioplan and AxioImager with a Nikon Coolpix 990 digital camera and a Zeiss HRC digital camera. Detailed protocols are available upon request (mqmartin@hawaii.edu). Lithium chloride treatments were performed as previously described (Matus et al., 2006b).
Results
Identification of Hedgehog-related genes in Nematostella vectensis
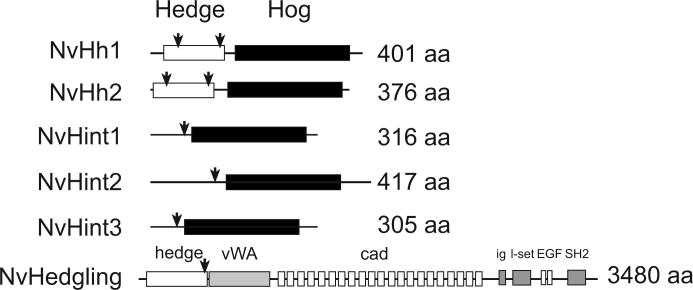
Hedgehog genes have been identified in Metazoa throughout the Bilateria (protostomes and deuterostomes), and a few homologous sequences have been identified outside of the Bilateria in both a recent Expressed Sequence Tag (EST) survey (Nicholls et al. 2006) as well as in an analysis of a preliminary annotation of the Nematostella genome (Walton et al., 2006). We performed RACE PCR on cDNA from mixed blastula through planula stage cDNA which yielded five independent hh-related cDNAs. All five clones encode predicted protein sequences with significant homology to intein domains of other metazoan Hh-related proteins (Fig. 1, SF1). These Hedgehog intein or Hint/Hog domains catalyze self-proteolytic cleavage to generate an active ligand from the N-terminus, which is typically modified by cholesterol and lipid acylation prior to secretion from the signaling cell (Guerrero and Chiang, 2007; Ingham, 2001). Additionally, the N. vectensis genome possesses one gene, NvHedgling, that has a hedge/signal domain while missing a Hint domain and is instead tethered to a large cadherin-related protein (Fig. 1, Adamska et al., 2007).
Figure 1. N. vectensis hedgehog related proteins.
The functional domains of predicted N.vectensis hedgehog-related proteins. There are two definitive hedgehog genes in the N. vectensis genome (NvHh1 and NvHh2). NvHh1 and NvHh2 encode proteins with both a “hedge” signal ligand (white box) and “hog” hint/intein (black box) domains. Three other genes encode hint/intein domains but lack hedgehog ligand domains. A sixth, hedgehog-related protein, NvHedgling lacks the hint/intein domain, but instead the hedge domain is tethered to a von-willebrand factor (vWA) domain, a series of 21 cadherin (cad) repeats, two immunoglobulin related domains (Ig and l-set), two EGF repeats, and a C-terminal SH2 domain (Adamska et al., 2007). Domain structure was identified with PFAM searches of the NCBI database (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). The two conserved intron sites with bilaterian hedgehog genes are marked with arrows. See supplemental information (SF1) for additional information.
Hedgehogs convey signals from cell to cell by an N-terminal ligand domain that gets cleaved from a precursor protein by intein-driven proteolysis. Only two of the clones we retrieved (NvHh1 and NvHh2) encode predicted ligand domains that show a high degree of amino acid identity to other metazoan Hedgehogs (Fig. 1, SF 1). As with other Hedgehogs, the signaling domains of NvHh1 and NvHh2 are located within the N-terminal half of the proteins, adjacent to an intein or Hint/Hog domain. The other three clones we retrieved, lack predicted Hh ligand domains and thus are designated NvHint1, NvHint2 and NvHint3 to reflect the presence of a Hint domain at their C-terminus. The sequences of the N-terminus of NvHint1 and NvHint3 are not homologous to other known proteins based on both Interpro Scan and BLASTP searches against the PFAM database and NCBI, while the N-terminal portion of NvHint2 shows weak blast similarity to the C-terminal portion of Pod-EPPT, a hydrozoan cnidarian, (Podocoryne carnea) gene. An Interpro Scan of NvHint2 identifies a potential domain upstream of the Hint domain: (IPR013994) a Carbohydrate-binding WSC subgroup, based on conserved cysteines.
Comparisons of the cloned and sequenced RACE PCR products to the Nematostella genome assembly allowed us to characterize intron/exon sites in the five hh-related genes. The two true hedgehog genes, NvHh1 and NvHh2 as well as the three hint genes (NvHint1−3) possess conserved intron/exon sites in common with bilaterian hedgehog genes (Fig. 1, SF 1). The first conserved site is located at the beginning of the second exon in bilaterian hedgehog genes in the signal domain and is found in both true N. vectensis hedgehog genes (NvHh1 and NvHh2). The second conserved intron/exon boundary is found in a position ∼10−11 amino acids upstream of the autocatalytic cleavage site and is found in all N. vectensis hedgehog related genes (Fig. 1, SF1).
Evolutionary relationships of Nematostella hedgehog and hedgehog-related genes
To establish potential orthology and evolutionary correspondence between Nematostella and other metazoan hedgehog and Hint-related genes, we subjected Hh-related predicted protein sequences to Bayesian and maximum likelihood phylogenetic analyses. Analyses were performed using both the N- and C-terminal domains, separately, and together. Using predicted full-length protein sequences, phylogenetic analyses support a sister group relationship between the two true Nematostella hedghehog genes (NvHh1 and NvHh2; SF6), suggesting that these genes may have arisen by gene duplication within Nematostella or within the Cnidaria. The vertebrate Hedgehog proteins cluster together in Bayesian and ML analyses (Fig. 2 and SF5−7), and suggest that sonic and indian hedgehog genes arose following a gene duplication event from a desert hedgehog-like gene, in agreement with earlier analyses (Kumar et al., 1996; Shimeld, 1999). In analyses of just the hedge or signal domain the two true Nematostella hedgehogs (NvHh1−2), show a graded relationship rather than forming a clade, but branch to the exclusion of the bilaterian hedgehog genes (SF5). In analyses of just the hint or intein domain (Fig2 and SF7), there is no support for a sister-group relationship between the two true Nematostella hedgehogs. Taken together, this suggests that either the cnidarian-bilaterian ancestor possessed two hedgehogs or that due to sequence divergence the weakly supported sister-group relationship found in full-length protein analyses is masked in analyses of individual domains. The absence of a second hedgehog gene in most bilaterians surveyed suggests that the cnidarian-bilaterian ancestor only possessed one hedgehog gene, and that the two Nematostella hedgehogs are the result of a cnidarian-specific duplication event. Analyses of just the intein domain (Fig. 2, SF7) included the five sponge hint/hog genes (Adamska et al. 2007), the two non-metazoan hint genes (G. mosseae and M. ovata), and the ten C. elegans intein-containing genes (groundhogs, warthogs, and quahog) (Aspock et al., 1999) as well as potential orthologs from related nematode ESTs identified via BlastP queries at NemBase (www.nematode.net), in addition to bilaterian true hedgehog genes. In Hint-only phylogenetic analyses, all nematode Hints form a monophyletic clade with varying levels of support, sister to the true hedgehog gene found in the slowly evolving nematode, X. index (M. Blaxter pers. comm.) (Fig. 2 and SF7). This suggests a potential single origin for nematode Hint-containing genes. The three Nematostella genes that encode intein domains, but not a conserved signaling domain, (NvHint1−3) cluster together suggesting they arose from tandem duplications either within Nematostella or during cnidarian evolution (Fig. 2 and SF7). These cnidarian hint genes fail to cluster with any of the other metazoan hint-only genes, including the five hint/hog genes from the sponge A. queenslandica (Adamska et al. 2007) and the warthog, groundhog, and quahog nematode genes (Fig. 2 and SF7). The three NvHints form a polytomy with other metazoan true hedgehogs as well as the two non-metazoan hint-containing genes, hoglet, from the choanoflagellate, Monosiga ovata, and GmGin1, from the fungi, Glomus mosseae, to the exclusion of non-hedgehog related inteins (Fig. 2 and SF7). While sequence diversity within metazoan hedgehog-related genes may mask relationships, leading to poorer statistical support, it is clear that Nematostella possesses five hedgehog-related genes, of which two possess both a hedge and a hog/hint domain that show clear orthology to other metazoan hedgehogs.
Figure 2. Phylogenetic analyses of hint/intein containing genes.
Both Bayesian and Maximum Likelihood analyses were conducted utilizing MrBayes 3.1.2 (Ronquist 2003), Phyml (Guindon et al. 2003), and RAxML v.2.3.2 (Stamatakis, 2006) utilizing an amino acid alignment of hint/intein-containing genes from metazoan and non-metazoan representatives. The three Nematostella hint-only containing genes (Hint1−3) (shown in red) form a monophyletic group in Bayesian analyses. The two true Nematostella (Nv) hedgehog genes (shown in red) do not show a sister group relationship as they do in analyses of the hedge and hint domains together (see SF6). NvHh1 is weakly supported as the sister group to vertebrate hh genes (shown in green (sonic), pink (Indian) and blue (desert)) while NvHh2 forms a polytomy with other hint-containing genes. All nematode hint-only containing genes (shown in light blue) from a monophyletic group sister to the true hedgehog gene from the nematode Xiphinema index (M. Blaxter, pers. comm.). The five hint-only genes from the demosponge, Amphimedon queenslandica (shown in orange) form a monophyletic group as well. Numbers above branches indicate Bayesian posterior probabilities while numbers below indicate ML bootstrap support. Additional details of phylogenetic analyses and nexus alignments are available in the supplemental information (SF2−7).
Developmental Expression of hedgehog genes
To understand the potential role of Nematostella hedgehog genes in embryonic development, we examined the spatial and temporal expression dynamics of the hedgehog-related genes by whole mount in situ hybridization on N. vectensis embryos and early juvenile polyps. Figure 3 shows the expression patterns of the two Nematostella hh genes that encode signaling ligands, NvHedgehog1 (NvHh1) and NvHedgehog2 (NvHh2). Neither gene is detected maternally. Zygotic expression for NvHh1 begins at early gastrula stages (Fig. 3B) and in late planula stages for NvHh2 (Fig. 3I). NvHh1 expression begins at gastrulation in presumptive pharyngeal ectodermal cells during their invagination into the blastocoel (Fig. 3-E). NvHh1 expression persists in the pharyngeal ectoderm and spreads to the ectodermal components of the first two mesenteries (the directive mesenteries) throughout planula and polyp stages (Fig. 3F-H).
Figure 3. Embryonic expression of the N. vectensis Hh genes.

Transcripts for NvHh1 are not detectable until gastrulation (A). Expression begins after the onset of gastrulation in cells that will form the pharynx (pha). (B, C). Expression elevates significantly during pharynx formation (c-e). Expression persists in the ectoderm of the pharynx, and extends to the ectoderm of the primary or directive mesenteries, at mid-planula stages (F, G). In the polyp (H), expression is limited to the aboral region of the pharynx and the two directive (primary) mesenteries (d.mes). NvHh2 expression (I, J) begins during planula stages in the body-wall endodermal components forming all eight body mesenteries (I, J) (mes). As the tentacles begin to emerge, expression is strong in the underlying tentacular endoderm (J, K). Expression in the mesentery and tentacular endoderm persists into juvenile polyp stages (L, M). All embryo views are lateral, with the asterisk denoting the blastopore and future mouth, except A, which shows early cleavage and blastula stages, and B, D, G, and K, which are oral views.
In contrast to NvHh1, transcripts for NvHh2 were not detected until planula stages (Fig. 3I) where they are restricted to tentacle bud endoderm (Fig. 3J, K) and in the endoderm of the eight thickened body wall mesenteries. At polyp stages expression is more diffuse, but remains confined to endoderm, in the mesenteries and tentacle endoderm, as well as in the body wall endoderm (Fig. 3L, M).
Developmental expression of hint-only containing genes
All three hint-only genes are expressed in a cell-type specific manner during Nematostella development. NvHint1 expression commences in the early planula, peaks at late planula/early polyp, and persists into the juvenile polyp stage when tentacle buds are clearly visible (Fig. 4A-D). Throughout this timeframe, expression is punctate and primarily seen in the ectoderm, although a few endodermal cells associated with the pharynx of the polyp may also express NvHint1. There appears to be concentrated expression of NvHint1 in ectodermal cells located at the tentacle bud tips orally and at the aboral end (Fig. 4C-D), likely associated with the apical tuft of the planula, a ciliated structure with sensory neural characteristics (Matus et al., 2007; Pang et al., 2004). Expression is seen in the cell bodies, which are located towards the basal surface of the epithelium, a position characteristic of ganglionic neurons (Marlow et al., In Prep).
Figure 4. Hint/intein containing genes in N. vectensis are likely involved in neural patterning.
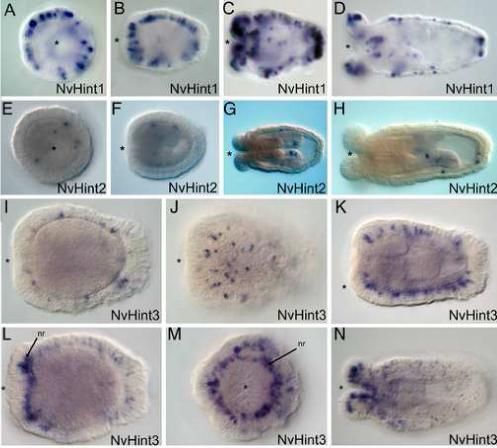
A-D. Expression of NvHint1 is punctate in the ectoderm and initiates in the early planula stage (A). Expression increases at mid-planula (B) and persists into the polyp (C,D). Expression is exclusively ectodermal, in scattered cells that are likely to be neural in origin due to their basal position. Expression is relatively uniform across the oral-aboral axis of the planula, but becomes more concentrated at both the oral and aboral ends of the polyp. E-H. Expression of NvHint2 is confined in a cell-type specific manner to body-wall endoderm and cells within the directive mesenteries. Expression is detected in a few scattered endodermal cells in the planula (E-F). In the polyp expression can be detected in both body-wall endodermal cells and within the directive mesenteries (G-H). I-N. Expression of NvHint3 is also ectodermal and begins in a punctate fashion in the planula (I-K). Expression in the early polyp becomes elevated in tentacular ectoderm as well as a ring of cells surrounding the mouth, which are likely to be components of the circumoral nerve ring (nr) (arrows, L, M). Expression persists mainly in the tentacular ectoderm with some scattered expression in the body wall ectoderm of the polyp (N). All embryo views are lateral, with the asterisk denoting the blastopore and future mouth, except A, E, M, which are oral views.
NvHint2 is expressed in a few scattered cells during planula (Fig. 4E-F) and polyp stages (Fig. 4G-H) of development in body wall endoderm and the directive mesenteries. The expression pattern of NvHint3 (Fig. 4I-N) is similar to NvHint1, with expression in a punctate pattern in the ectoderm at planula through polyp stages. The asymmetric localization of NvHint3-expressing cells share a similar distribution towards the basal surface of the epithelium but differ in their distribution along the body column (Fig. 4I-L). During tentacle-bud stages of the late planula/early polyp a greater number of cells at the oral end express NvHint3, and there is no expression in the apical sensory tuft or in the pharynx (Fig. 4L-M). The circular pattern of cells expressing NvHint3 around the blastopore/mouth corresponds to the position of the cnidarian circumoral nerve ring (Koizumi et al., 1992; Mackie, 2004; Miljkovic-Licina et al., 2004). In the polyp (Fig. 4N) NvHint3 continues to show expression throughout the ectoderm of the body column, as well as within specific ectodermal cells of the growing tentacles.
Developmental expression of patched and Gli genes
The 11 transmembrane receptor, Patched, is the ligand-binding component at the apex of the hedgehog signal reception and relay system. At the terminus of the pathway, one or more Gli/Ci transcription factors regulate Hh-responsive genes. Because these receptor and transcription factor components are critical for Hh responses, these genes would be predicted to be expressed in cells which receive hh signals, and therefore their expression patterns could be used to obtain clues about where hh signals might affect Nematostella embryos. We cloned N. vectensis embryonic cDNAs for one patched (NvPtc) and one Gli gene (NvGli), which are the only representatives of these types of genes in the N. vectensis genome (Table 1, discussed below; SF10−11). We examined their expression by in situ hybridization, and the results indicate that NvPtc and NvGli are expressed in endoderm during development. NvPtc transcripts are detected in oral bodywall endoderm and pharyngeal endoderm at mid to late gastrulation, (Fig. 5B-C). NvPtc expression is endodermal at late planula and polyp stages (Fig. 5D-H), including tentacular endoderm (Fig.5G), but its expression is noticeably higher in the endoderm of the two directive mesenteries and in pharyngeal endoderm (Fig. 5F-H). The NvGli expression pattern (Fig. 6) is almost identical to that of NvPtc, although NvGli transcripts are detected earlier than those of NvPtc. Expression commences at the onset of gastrulation in invaginating presumptive endodermal cells (Fig. 6B) and persists in the endoderm through polyp stages. NvGli expression appears restricted to the endoderm, similar to NvPtc, with a significantly higher level of expression in endodermal cells of the directive mesenteries and the endodermal layer of the pharynx that faces the gastrocoel. These expression patterns, together with those of the NvHh genes, prompt us to suggest that the embryonic endoderm, but perhaps not the ectoderm, is the principal tissue that can receive and be affected by NvHh1 and NvHh2 signals.
Figure 5. The hedgehog receptor, NvPatched is expressed in endoderm during development.
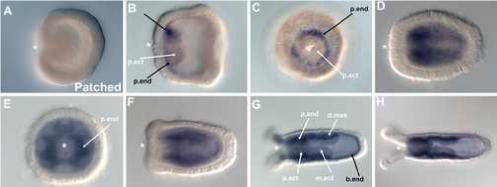
NvPtc transcripts are not detected at the onset of gastrulation (A), and are first detected in bodywall and pharyngeal endoderm (p.end) at the oral pole in late gastrula (B, C). Expression is upregulated in the planula and polyp stages (D-F), where NvPtc is expressed in endoderm of the pharynx (p.end), body wall (b.end) and directive mesenteries (d.mes). Note the absence of NvPtc transcripts in the ectodermal component of the pharynx (p.ect) and directive mesenteries (m.ect) (B-H). All embryo views are lateral, with the asterisk denoting the blastopore and future mouth, except C and E, which are oral views.
Figure 6. NvGli is expressed in the nearly the same endodermal pattern of NvPatched during development.
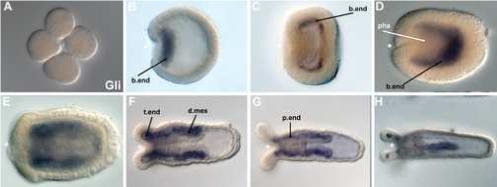
Transcripts for NvGli are not present in the blastula (A) and first detected during the onset of gastrulation in invaginating endodermal cells (B). At the end of gastrulation (C) and in the early planula (D) NvGli is expressed in bodywall endoderm (b. end) but not in the pharynx (pha). This expression pattern persists in tentacle bud stages (E). In the polyp (F-H), NvGli continues to be expressed in bodywall endoderm, as well as the endodermal component of the directive mesenteries (d.mes), in tentacular endoderm (t.end), and in the pharyngeal endoderm (p. end). All views are lateral, with the asterisk denoting the blastopore and future mouth, except A which is a cleavage stage embryo, and H, which is a dorso-ventral view.
Additional Hedgehog signaling pathway components in Nematostella
In addition to directly cloning and examining the expression of Hh, Ptc and Gli genes in N. vectensis, we surveyed the N. vectensis genome for the presence of sequences predicted to correspond to genes for other components of Hh signaling pathways known from studies of bilaterian animals. We used protein sequences and nomenclature for Drosophila and vertebrate hh signaling components to search the N. vectensis genome, EST sequences, and predicted protein using BLAST and name queries at the N. vectensis genome website, http://genome.jgi-psf.org/Nemve1/Nemve1.home.html. Table 1 summarizes N. vectensis gene sequences we identified that encode predicted proteins with significant homology to known bilaterian hh pathway components. Table 1 reveals that N. vectensis appears to contain a near full scope of hh pathway components identified in protostome and deuterostome model systems. Genomic sequence for a few of these (Ptc, Smo, Disp, and Gli) were also identified in a recent study (Walton et al. 2006). In bilaterians, Hh ligand activity can be affected by many extracellular modifiers, and the N. vectensis genome contains sequences predicted to encode many of these genes, including skinny hedgehog/rasp/sightless, which encodes an acetyltransferase that regulates hh activity by palmitoylation; (Lee and Treisman, 2001), Dispatched, which encodes a membrane protein for hh ligand transmission from the source cell; (Burke et al., 1999), two glypicans which govern extracellular hh ligand movement (Desbordes and Sanson, 2003; Han et al., 2004); and orthologs of HIP and GAS1 which bind extracellular hh ligands and regulate their activity and range of movement (Cabrera et al., 2006; Chuang and McMahon, 1999; Lee et al., 2001). We also found likely orthologs of vertebrate exostosins and Drosophila tout-velut, which encode glycosyl transferases that affect heparan biosynthesis, which in turn regulates hh intercellular movement and reception by target cells (Bellaiche et al., 1998; Siekmann and Brand, 2005).
At the receptor level and downstream we identified two homologs of Smoothened (NvSmo1, 2), the partner receptor that is regulated by Patched, (Fig. 5). We also identified a N. vectensis ortholog of another type of Hh receptor, megalin, which encodes a LDL-related protein that binds and can endocytose extracellular Hh ligand (McCarthy et al., 2002; Morales et al., 2006). Predicted regulators of intracellular Hh receptor trafficking (Eggenschwiler et al., 2006; Kalderon, 2005; Meloni et al., 2006) are also present in the genome, including two forms of β-arrestin, GRK2/βARK-1 and the small GTPase, Rab23. We further identified a N. vectensis homolog of tectonic, which functions downstream of Smo by an unknown mechanism that can enhance or repress Hh signal transduction (Reiter and Skarnes, 2006).
At the signal transduction level, in addition to a Gli3/Ci transcription factor ortholog NvGli described above, N. vectensis possesses predicted genes encoding core components of the Gli cleavage complex (reviewed by (Huangfu and Anderson, 2006; Osterlund and Kogerman, 2006), consisting of costal2, fused, suppressor of fused (SuFu), and βTRCP/slimb, as well as associated Gli kinases (PKA, GSK3β, Casein Kinase 1). We identified other Gli regulators as well, including a homolog of Drosophila roadkill and its vertebrate ortholog SPOP, which are substrate adaptors for cullin-type E3 ubiquitin ligases that affect Ci/Gli proteasomal processing and degradation (Kent et al., 2006; Zhang et al., 2006). We also found a potential homolog of a vertebrate Gli1/2/3 transcriptional partner protein sap18 (Cheng and Bishop, 2002) in the N. vectensis genome. Furthermore, Hh signal transduction in vertebrates (but not Drosophila) requires intact cilia and associated intraflagellar transport proteins (Ifts), particularly Ift88/polaris, Ift172/wimple, and Ift52/Ngd5 (Huangfu et al., 2003; Liu et al., 2005), as well as Fantom, an apparently vertebrate-specific, ciliary basal body protein required for Hh signal transduction (Vierkotten et al., 2007). Homologs of each of these are represented in the N. vectensis genome (Table 1). Additionally, other genes implicated in vertebrate but not Drosophila hh signaling (with unknown mechanisms) are present in the N. vectensis genome: sil, a novel gene encoding an essential intracellular signaling regulator (Izraeli et al., 2001), iguana (Sekimizu et al., 2004), and talpid3 (Davey et al., 2006), the latter two being novel regulators of Gli activity. Perhaps orthologs of these genes were lost during the evolution of Drosophila and other ecdysozoan lineages. Conversely, a few bilaterian Hh regulators appear to be absent in the N. vectensis genome, including homologs of Drosophila interference Hedgehog (iHog) and its vertebrate ortholog Cdon, which bind Hh ligands (Okada et al., 2006; Yao et al., 2006), and WIF (wnt inhibitory factor), which can bind Hh as well as Wnt ligands (Glise et al., 2005; Gorfinkiel et al., 2005). This suggests these genes evolved after the divergence of cnidarians and bilaterians. The sum of our cloning, expression and genomic survey results indicate that N. vectensis possesses essentially the full repertoire of Hh signaling pathway components found among bilaterian model organisms. Thus, the Hh signaling pathway appears to have evolved its high degree of complexity in an ancient, common ancestor of the Cnidaria and Bilateria.
Discussion
The function of cell signaling pathways during embryogenesis is well documented in representative protostome ecdysozoans (e.g. nematode and insect) and deuterostomes, but the extent to which such signaling systems function in the embryogenesis of non-bilaterians such as sponges, ctenophores and cnidarians has only recently begun to be investigated (Adell et al., 2003; Nichols et al., 2006; Suga et al., 1999; Wiens et al., 2007). Recent exploration of some of these gene families in cnidarian embryos have been particularly revealing, and have demonstrated the existence and embryonic deployment of diverse Wnt (Kusserow et al., 2005), TGF-beta and BMP (Finnerty et al., 2004; Hayward et al., 2002; Hobmayer et al., 2001; Matus et al., 2006a; Matus et al., 2006b; Reber-Muller et al., 2006; Rentzsch et al., 2006; Samuel et al., 2001), and FGF-related genes (Matus et al., 2007). Hedgehog signaling plays a critical role in pattern formation and cell fate decisions in bilaterians (Fuccillo et al., 2006; Huangfu and Anderson, 2006; McMahon et al., 2003; Wang et al., 2007). Hedgehog-related genes have been described in protostomes, within ecdysozoan lineages (Tashiro et al., 1993), as well as in several lophotrochozoans (Kang et al., 2003; Nederbragt et al., 2002; Seaver and Kaneshige, 2006), and in non-vertebrate deuterostomes including sea urchins (Walton et al., 2006), urochordates (Hino et al., 2003) and cephalochordates (Shimeld, 1999).
To understand the evolutionary origins of hedgehog signaling we investigated whether hedgehog genes are present in the genome of N. vectensis, an anthozoan cnidarian. From genomic searches using the Hint/Hog domain, we found that N. vectensis possesses five hedgehog-related genes, two of which are predicted to encode ligands and are thus “true” hedgehog genes, while three others encode proteins missing the N-terminal signaling domain, possessing only the C-terminal intein or Hint/Hog domain, a key feature of Hedgehogs, but also found outside of the Metazoa in protists (e.g., choanoflagellates (Snell et al. 2006) and the fungus, Glomus messeae (Requena et al. 2002). We also identified an ortholog of the poriferan hedgling gene from A. queenslandica in N. vectensis (Adamska et al. 2007), that appears to have been lost in bilaterians.
Ancient origins and evolution of metazoan Hedgehog-related genes
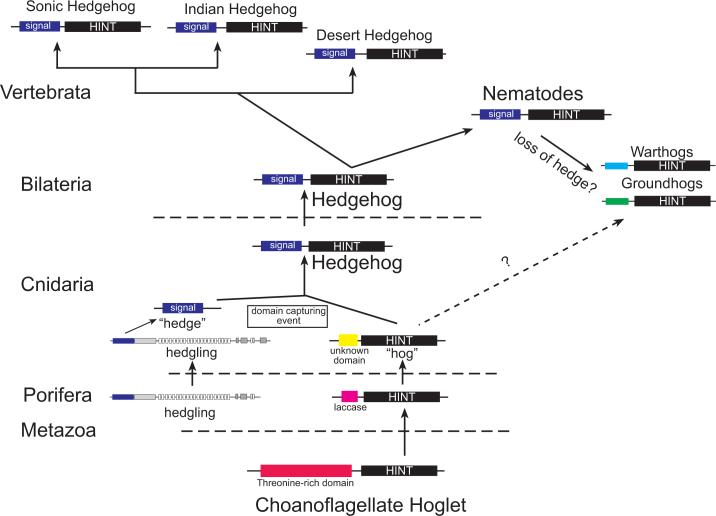
The presence of Hint genes outside of the Metazoa, such as the hoglet gene (Snell et al. 2006) identified in the freshwater choanoflagellate, Monosiga ovata, and GmGin1 (Requena et al. 2002) in the mycorhrhizal fungi, Glomus mosseae, suggests that evolutionary precursors of hedgehog signaling existed prior to the metazoan radiation. Choanoflagellagtes are unicellular protozoa considered to be the likely sister group to the Metazoa (King and Carroll, 2001; Snell et al., 2001). M. ovata hoglet encodes an N-terminal, threonine-rich domain of unknown function linked to a C-terminal Hint domain (Snell et al. 2006), while GmGin1 possesses a C-terminal Hedgehog-related intein domain linked to an N-terminal domain that shares sequence similarity with a novel family of GTP binding proteins (Requena et al. 2002). Phylogenetic analysis of Hint/Hogs in N. vectensis and other species, suggest that the Hint/Hogs of Nematostella and those of the sponge, Amphimedon, likely arose from independent gene duplication events within each lineage (Fig. 2). The lack of true hedgehogs in the Amphimedon genome (Adamska et al. 2007) suggests that the origins of the metazoan hedgehog ligand may have occurred following the divergence of sponges with Eumetazoa (Adamska et al. 2007). The identification of both Hint/Hog genes and hedgehog genes in Nematostella argues that the evolution of a hedgehog gene in the cnidarian-bilaterian ancestor occurred by a domain-capturing event of a N-terminal signaling (or “hedge”) domain and a Hint/Hog domain-containing gene (“hog”) (Fig. 7).
Figure 7. A model for Hedgehog evolution.
Data from the genomes of the anthozoan cnidarian Nematostella vectensis and the demosponge, Amphimedon queenslandica suggest that the origin of the Hedgehog signaling molecule arose in the eumetazoan ancestor (Adamska et al. 2007). The choanoflagellate, Monosiga ovata possesses a hint/intein “hog” containing gene (Snell et al., 2006), but no gene possessing a signal or “hedge” domain has been found. The demosponge, A. queenslandica possesses both hint/intein genes as well as a gene possessing a “hedge” domain tethered to a fat-like cadherin (Adamska et al. 2007). An ortholog of this gene has been found in the N. vectensis genome. However, N. vectensis also possesses two true hedgehogs as well as three hint/intein genes. While Hint/intein containing genes have not been identified in most bilaterians surveyed, C. elegans, which lacks true hedgehog genes (Aspock et al., 1999; Hao et al., 2006a), possesses two families of Hint/intein containing genes, with novel N-terminal domains (warthog and groundhog). However, a true hedgehog gene has been identified in the slowly evolving nematode, Xiphinema index (M. Blaxter pers. comm.). Based on phylogenetic analyses (Fig. 2) it appears likely that nematode groundhog, warthog, and quahog genes are derived from an ancestral nematode true hedgehog gene (solid arrow), however additional data may be necessary to rule out the possibility that these genes could also be derived from non-bilaterian hint-only genes (dashed arrow) The hedgehog ligands diversified within the Vertebrata.
With the addition of two lophotrochozoan genomes (the polychaete annelid, Capitella sp. I, and the gastropod mollusc, Lottia scutum in draft assembly (Seaver and Rohksar pers. comm.), the only other metazoan group to definitively possess both hint-only genes and true hedgehogs appears to be the nematodes. Previously, it has been suggested that nematodes possess as many as ten different hedgehog-related genes (e.g., warthogs, groundhogs, and quahog). We have identified orthologs of these genes within publicly available ESTs available at NemBase (www.nematode.net) along with a single EST cluster from the slowly evolving nematode, Xiphinema index, that appears to be a definitive hedgehog gene possessing both a signaling or hedge domain as well as a hint/hog domain (M. Blaxter pers. comm. and Fig. 2). The identification of a definitive nematode hedgehog gene supports earlier hypotheses that the C. elegans hh-related genes are derived from an ancestral hedgehog (Aspock et al., 1999; Hao et al., 2006a). The conservation of intron/exon boundaries in all of the Nematostella hedgehog-related genes with bilaterian hedgehogs also supports a monophyletic origin of the hedgehog in the cnidarian-bilaterian ancestor. Hedgehog genes later underwent gene duplication events (Fig. 7), likely correlated with the genome duplication events within the vertebrate lineages, to give rise to the hedgehog paralogs (sonic, indian, and desert) found in extant vertebrates.
Assigning the origin of the first true hedgehog gene within the Cnidaria is somewhat contentious due to conflicting data between two sponges, Oscarella carmella (Nicholls et al. 2006) and A. queenslandica (Adamska et al. 2007). A recent Expressed Sequence Tag (EST) survey in the demosponge, O. carmella. (Nicholls et al. 2006) used blast searches of ∼11,000 ESTs to identify both cell adhesion and signal transduction genes that had previously never been identified in sponges. Included in this survey were a putative hedgehog ligand, three clones with Blast identity to the hh receptor patched, and one membrane-modifier, dispatched. While this suggests that components of the hedgehog pathway predated the cnidarian-bilaterian ancestor, this data is preliminary. Their putative hedgehog sequence is missing a definitive Hint domain, and the three non-overlapping patched sequence fragments are of insufficient length to establish definitive orthology beyond that of simple Blast identity. Contrary to the Oscarella data, the recently sequenced genome of the demosponge, A. queenslandica, lacks a true hedgehog ortholog, but does possess several Hint-only containing genes, as well as one hedge containing gene, hedgling, that is tethered to a series of cadherin domains (Adamska et al. 2007). Additionally, the A. queenslandica genome is missing a definitive Ptc ortholog (B. Degnan pers. comm.). Together, this suggests that either there are large differences in the genomic content between these two sponge species, or that the identified EST sequences in O. carmella need to be verified by either genomic southerns or in situ hybridizations. However, given the lack of true hedghog genes in the genome of the sponge, A. queenslandica, and the presence of a hedge-only containing ortholog, hedgling, in both A. queenslandica and N. vectensis, this would suggest that the origin of the first hedgehog ligand occurred after the divergence of sponges from the Eumetazoa (Adamska et al., 2007). With the recent sequencing of a placozoan genome, and more extensive sampling within the ctenophores, the exact timing of the origin and diversification of the hedgehog pathway within the Metazoa will soon be determined.
Coordination between the Wnt, TGF-ß, FGF, and hedgehog signaling pathways
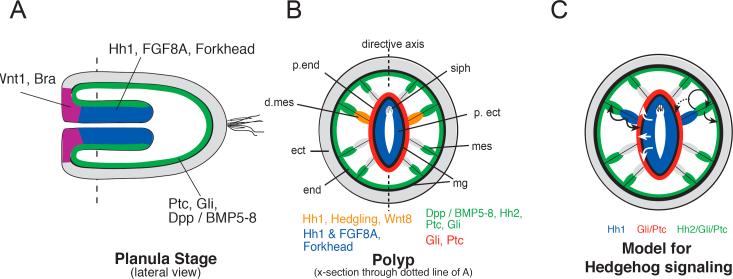
Signal transduction pathways are often functionally coordinated during ontogeny. Interactions between the Wnt, TGFß, FGF, and Hh pathways in particular, have been described in a variety of contexts, including Drosophila body segmentation (DiNardo et al., 1994; Sanson, 2001) and retinal development (Silver and Rebay, 2005), mammalian jaw and tooth development (Cobourne and Sharpe, 2003; Tompkins, 2006), insect and vertebrate limb patterning (Angelini and Kaufman, 2005; Stopper and Wagner, 2005), gut development (Fukuda and Yasugi, 2002; Grapin-Botton, 2005; Tam et al., 2003), vertebrate somitogenesis (Aulehla and Herrmann, 2004; Baker et al., 2006; Pourquie, 2003) and neural tube development (Cayuso and Marti, 2005; Roussa and Krieglstein, 2004; Wilson and Maden, 2005). Recent evidence has suggested a role for Wnt (Kusserow et al., 2005; Wikramanayake et al., 2003), TGFß/BMP (Matus et al., 2006a; Matus et al., 2006b), and FGF signaling (Matus et al., 2007) in gastrulation and endoderm development in Nematostella. Various components of these signal transduction pathways along with the hh pathway are deployed at the onset of and during gastrulation in invaginating mesendoderm or later in mesendodermally associated structures such as the pharynx (Fig. 8A, B) or mesenteries (Fig. 8B).
Figure 8. Summary of Hedgehog, Wnt, TGFß, and FGF signaling in Nematostella vectensis.
Signal transduction pathway ligands of the Wnt, TGFß, FGF, and Hedgehog pathways appear to be involved in patterning the oral/aboral axis during N. vectensis development. (A) Wnt ligands (e.g., NvWnt1 (Kusserow et al. 2005), and the T-box transcription factor NvBra (Scholz and Technau, 2003), are expressed in oral ectoderm. Ligands for both Hedgehog (NvHh1) and FGF (NvFGF8A) (Matus et al., 2007), are expressed along with the transcription factor, forkhead (Martindale et al., 2004), in the ectodermal component of the pharynx, indicating a conserved role in gut formation between cnidarians and bilaterians. TGFß ligands NvDpp and NvBMP5−8 (Finnerty et al. 2004, Matus et al. 2006) are expressed in bodywall endoderm along with downstream components of the hedgehog pathway, including a receptor, NvPtc, and transcription factor, NvGli. (B) During polyp stages, these same signal transduction pathways appear to be coordinately involved in patterning the mesenteries. Both TGFß (NvDpp and NvBMP5−8) (Finnerty et al., 2004; Matus et al., 2006), Wnt ligands (e.g., NvWnt8) (Kusserow et al., 2005) along with an FGF ligand (NvFGF8A) (Matus et al., 2007) are all expressed in endodermally derived structures. Hedgehog ligands are expressed in either the pharyngeal ectodermal component of the directive mesenteries (NvHh1) or the bodywall endodermal component of the eight mesenteries (NvHh2). (C) Downstream components of the hedgehog pathway, NvPtc and NvGli are expressed exclusively in bodywall endoderm and in the endodermal components of the eight mesenteries, suggesting that the Hedeghog ligands may be acting in both a trans-epithelial (signaling from NvHh1 positive cells in pharyngeal ectoderm to NvPtc / NvGli positive cells in the bodywall endoderm and mesenteries) and a planar fashion (NvHh2 in the same endodermal cells as NvPtc / NvGli) within the endoderm.
Specifically, NvHh1 shows co-expression with FGF8A (Matus et al. 2007) and the transcription factor NvForkhead (Martindale et al. 2004) in the developing pharynx (Fig. 8A). The Wnt1 (Wg) ortholog, NvWnt1 is expressed along with several other Wnt ligands (Kusserow et al. 2005) and the T-box transcription factor NvBra (Scholz and Technau, 2003) in oral ectoderm in a domain abutting NvHh1 and NvFGF8A expression (Fig. 8A). During mesentery patterning, two TGFß ligands (NvDpp and NvBMP5−8 (Matus et al., 2006b) are co-expressed in the bodywall endodermal components of the eight mesenteries along with NvHh2 (Fig. 8B). The close coordination of the expression of these signal transduction pathways during gastrulation and in structures such as the pharynx and mesenteries may represent a conserved developmental cassette of cell-cell signaling components, and may provide clues to the evolutionarily conserved roles of these signal transduction pathways during development. In support of this, treatment with lithium chloride, a known activator of the canonical Wnt signaling pathway via GSK3ß inhibition and nuclear stabilization of ß-catenin (Klein and Melton, 1996) appears to downregulate the expression of hedgehog pathway components during Nematostella development (SF11). An ectopic increase in Wnt signaling has also been shown to have a repressive affect on Hh signaling in chick Sonic hedgehog neural plate explants, both via exposure to Wnt1 and Wnt3 as well as the addition of lithium chloride (Robertson et al., 2004) suggesting that the relationship between these pathways is an ancient one.
A conserved role for Hh and TGFß signaling in germ cell development
TGFß and Hh signaling have been implicated in germ cell development in Drosophila, where TGFß signaling via dpp and glass bottom boat (gbb) are essential for promoting the self-renewal and proliferation of germline stem cells (Kirilly et al. 2005, Chen and McKearin 2003), while the hedgehog pathway may be functioning in a cell-autonomous fashion for germ cell migration (Deshpande et al. 2001). In vertebrates, the ligand, desert hedeghog regulates spermatogenesis in mice via Sertoli cell signaling (Bitgood et al. 1996). Hedgehog signaling has been suggested to play an ancient conserved role in mediating somatic and germ cell interactions (Bitgood et al. 1996). Data from N. vectensis supports this claim, as both TGFß ligands (NvDpp and NvBMP5−8, an ortholog of Drosophila gbb) (Finnerty et al. 2004, Matus et al. 2006) and both of the Hh ligands are expressed in the mesenteries during development, with NvHh1 expressed in the pharyngeal component of the directive mesenteries and NvHh2 expressed in the endodermal components of all eight mesenteries (Fig. 8B). Previous work has shown that germ cells in Nematostella first appear late in development, associated with compartments within the mesenteries, and that a vasa gene, Nvvas1, and a nanos ortholog, Nvnos2, are expressed in putative germ cells within the developing mesenteries (Extavour et al. 2005). While functional experiments are needed, based upon observed expression patterns and data from bilaterians, it is likely that the involvement of Hh and TGFß signaling in germline development pre-dates the cnidarian-bilaterian divergence. If the two Nematostella hedgehogs share a common origin, based on both amino acid identity and phylogenetic analyses (SF6), it is likely that they split their ancestral function (duplication and subfunctionalization) in the Nematostella-specific or cnidarian-line of descent.
A conserved role for hedgehog signaling in gut development
Similar to most signal transduction pathways, Hedgehog signaling is utilized in a variety of different contexts during bilaterian development. A conserved role for the Hedgehog pathway in gut development has been suggested on the basis of hedgehog gene expression in deuterostomes (e.g. sea urchin, Walton et al. 2006; zebrafish and medaka, (Kobayashi et al., 2006; Strahle et al., 1996); frog, (Ekker et al., 1995); chick, (Roberts et al., 1995; Sukegawa et al., 2000), mouse, (Bitgood and McMahon, 1995), and protostomes (e.g., the arthropods Drosophila (Hoch and Pankratz, 1996), Gryllus (Inoue et al., 2002), Euscorpius and Aretemia (Simonnet et al., 2004), and recently in the annelids Helobdella, Capitella and Hydroides (Kang et al., 2003; Seaver and Kaneshige, 2006). Indeed, gain and loss-of-function studies in many of these animals demonstrates that abnormal hedgehog signaling causes gut malformation and disease (Diiorio et al., 2007; Hatini et al., 2005; Inoue et al., 2002; Iwaki et al., 2001; Mullor et al., 2002; Takashima and Murakami, 2001; Wang et al., 2002; Warburton et al., 2000; Zhang et al., 2001) with a recent review by (Lees et al., 2005). The endodermal and pharyngeal expression of hedgehog genes in N. vectensis (Figs. 3, 8) suggests that the role of hh in endoderm specification and gut development may also have predated the origin of the Bilateria, and was likely present in the cnidarian-bilaterian ancestor.
In bilaterians, hedgehog ligands are often found expressed in endodermal epithelia during gut and lung formation (Narita et al., 1998; Sukegawa et al., 2000), and have been shown to signal to downstream components, such as patched and gli/Ci which are found in neighboring mesenchyme (Ramalho-Santos et al., 2000; Rubin, 2007; Shannon and Hyatt, 2004; Zhang et al., 2001). This scenario has recently been described in the sea urchin, Lytechinus variegatus, where hedgehog is expressed at the vegetal pole at the onset of gastrulation, in the invaginating endodermal vegetal plate, and is expressed in the endoderm through prism and larval pluteus stages, while patched and smoothened are expressed in neighboring secondary mesenchyme cells (Walton et al., 2006). A similar situation in observed to Nematostella, as NvHh1 is expressed in neighboring tissue to Ptc and gli positive cells (Fig. 8). It has been suggested that the cnidarian endoderm is either a remnant of the endomesoderm/mesendoderm of a triploblastic ancestor or the evolutionary precursor to triploblasty (Martindale et al. 2004). The Nematostella gastrodermis is a bi-functional (contractive and absorptive) endoderm, and has been previously been shown to express many of the same transcription factors and signaling molecules associated with patterning endomesoderm in bilaterians (Wikramanyake et al., 2003; Martindale et al., 2004; Finnerty et al., 2004; Matus et al., 2006a; Matus et al., 2006b; Matus et al., 2007; Rentzsch et al., 2006; Scholz and Technau, 2003). The expression of hedgehog pathway components described here provides additional support for homology between the endoderm of cnidarians and the endomesoderm of bilaterians.
Spatial dynamics of Hh signaling in Nematostella
The potential function of these hedgehog genes is difficult to predict, as the ligands produced by each of the genes would have the potential to either signal within the plane of the endodermal (i.e., NvHh1 and NvHh2) and ectodermal (NvHint1−3) epithelia, or between these germ layers. Hedgehog signaling typically occurs within the plane of an epithelial tissue, often through the formation of a gradient of Hh ligand (Vincent and Dubois, 2002), such as observed in Drosophila in the cellular blastoderm of the early embryo, or the wing or leg imaginal discs in the larvae. Hedgehog ligands are particularly “equipped” for planar signaling within epithelia, as Hh ligands can either move from a source cell to receiving cells through the extracellular space (where cholesterol modification can affect their diffusion) or through the epithelial cells via endocytic vesicles called argosomes (Cadigan, 2002). Based on the expression data presented here, it appears likely that both trans-epithelial and planar signaling is occurring: NvHh1 expressed in pharyngeal and directive mesentery ectoderm signaling in trans to adjacent endoderm, and NvHh2 in bodywall and mesentery endoderm signaling in a planar fashion within the endoderm. This prediction is supported by the expression of both NvPtc and NvGli in the same, endodermal epithelial layer as the Hh ligands, overlapping with NvHh2 expression in the bodywall endoderm and eight mesenteries, and adjacent to the NvHh1 expressing cells of the pharyngeal ectoderm (Fig. 8C).
Unexpected complexity in the N. vectensis Hedgehog signaling pathway
In addition to directly cloning cDNAs for Hh-related ligands, receptors and Gli proteins, we further surveyed the N. vectensis genome and EST databases for the presence of sequences that encode additional components of Hh signaling regulation. The most striking feature revealed by this survey is that homologs for nearly every protein implicated in hedgehog signaling in Bilateria can be found in N. vectensis (Table 1). These findings indicate that a complex Hh signaling pathway had already evolved in the cnidarian-bilaterian ancestor. From the diversity of ligand and signal transduction components we have uncovered, the putative N. vectensis hedgehog pathway more closely resembles the vertebrate pathway than that of Drosophila, although some gene duplication has occurred in vertebrates (e.g. the Gli's). The Drosophila hh pathway appears to be a somewhat simplified (derived) version of a more complex pathway likely present in the cnidarian-bilaterian ancestor. C. elegans presents an even more extreme case of apparent gene loss in an ecdysozoan lineage. Although numerous Hint genes and patched homologs are present, C. elegans lacks virtually all other components of Hh signaling (Hao et al., 2006a), although more slowly evolving nematodes (e.g., Xiphinema index) appear to possess true hedgehog ligands (Fig. 2 and M. Blaxter pers. comm.). Such findings correlate with studies showing extensive gene loss within the model ecdysozoans, Drosophila and C. elegans (Copley et al., 2004; Miller et al., 2005; Wolf et al., 2004);Kusserow et al., 2005; Putnam et al., 2007; Technau et al., 2005) To fully understand the evolutionary picture of Hedgehog signaling we need to tap emerging genomes of non-model bilaterians, particularly within the Lophotrochozoa and Ecdysozoa, as well as in nonbilaterians such as sponges, placozoans, and ctenophores, to obtain a complete picture of the evolution of this important signaling pathway.
Acknowledgements
We thank Bernie Degnan and members of the Martindale lab for useful discussions and Mark Blaxter for alerting us to slow evolving nematode taxa. MQM is supported by grants from NSF and NASA, and GHT and CM are supported by grants from the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary Material
Literature Cited
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–5. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Adamska M, Matus DQ, Adamski M, Green K, Rokhsar DS, Martindale MQ, Degnan BM. Domain shuffling and the evolutionary origin of hedgehogs. Curr. Biol. 2007 doi: 10.1016/j.cub.2007.08.010. in press. [DOI] [PubMed] [Google Scholar]
- Adell T, Nefkens I, Muller WE. Polarity factor ‘Frizzled’ in the demosponge Suberites domuncula: identification, expression and localization of the receptor in the epithelium/pinacoderm(1). FEBS Lett. 2003;554:363–8. doi: 10.1016/s0014-5793(03)01190-6. [DOI] [PubMed] [Google Scholar]
- Angelini DR, Kaufman TC. Insect appendages and comparative ontogenetics. Dev Biol. 2005;286:57–77. doi: 10.1016/j.ydbio.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Aspock G, Kagoshima H, Niklaus G, Burglin TR. Caenorhabditis elegans has scores of hedgehog-related genes: sequence and expression analysis. Genome Res. 1999;9:909–23. doi: 10.1101/gr.9.10.909. [DOI] [PubMed] [Google Scholar]
- Aulehla A, Herrmann BG. Segmentation in vertebrates: clock and gradient finally joined. Genes Dev. 2004;18:2060–7. doi: 10.1101/gad.1217404. [DOI] [PubMed] [Google Scholar]
- Avidor-Reiss T, Maer AM, Koundakjian E, Polyanovsky A, Keil T, Subramaniam S, Zuker CS. Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell. 2004;117:527–39. doi: 10.1016/s0092-8674(04)00412-x. [DOI] [PubMed] [Google Scholar]
- Baker RE, Schnell S, Maini PK. A clock and wavefront mechanism for somite formation. Dev Biol. 2006;293:116–26. doi: 10.1016/j.ydbio.2006.01.018. [DOI] [PubMed] [Google Scholar]
- Bellaiche Y, The I, Perrimon N. Tout-velu is a Drosophila homologue of the putative tumour suppressor EXT-1 and is needed for Hh diffusion. Nature. 1998;394:85–8. doi: 10.1038/27932. [DOI] [PubMed] [Google Scholar]
- Bitgood MJ, McMahon AP. Hedgehog and Bmp genes are coexpressed at many diverse sites of cell-cell interaction in the mouse embryo. Dev Biol. 1995;172:126–38. doi: 10.1006/dbio.1995.0010. [DOI] [PubMed] [Google Scholar]
- Burke R, Nellen D, Bellotto M, Hafen E, Senti KA, Dickson BJ, Basler K. Dispatched, a novel sterol-sensing domain protein dedicated to the release of cholesterol-modified hedgehog from signaling cells. Cell. 1999;99:803–15. doi: 10.1016/s0092-8674(00)81677-3. [DOI] [PubMed] [Google Scholar]
- Cabrera JR, Sanchez-Pulido L, Rojas AM, Valencia A, Manes S, Naranjo JR, Mellstrom B. Gas1 is related to the glial cell-derived neurotrophic factor family receptors alpha and regulates Ret signaling. J Biol Chem. 2006;281:14330–9. doi: 10.1074/jbc.M509572200. [DOI] [PubMed] [Google Scholar]
- Cadigan KM. Regulating morphogen gradients in the Drosophila wing. Semin Cell Dev Biol. 2002;13:83–90. doi: 10.1016/s1084-9521(02)00014-9. [DOI] [PubMed] [Google Scholar]
- Cayuso J, Marti E. Morphogens in motion: growth control of the neural tube. J Neurobiol. 2005;64:376–87. doi: 10.1002/neu.20169. [DOI] [PubMed] [Google Scholar]
- Chen D, Zhao M, Mundy GR. Bone morphogenetic proteins. Growth Factors. 2004;22:233–41. doi: 10.1080/08977190412331279890. [DOI] [PubMed] [Google Scholar]
- Cheng SY, Bishop JM. Suppressor of Fused represses Gli-mediated transcription by recruiting the SAP18-mSin3 corepressor complex. Proc Natl Acad Sci U S A. 2002;99:5442–7. doi: 10.1073/pnas.082096999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang PT, McMahon AP. Vertebrate Hedgehog signalling modulated by induction of a Hedgehog-binding protein. Nature. 1999;397:617–21. doi: 10.1038/17611. [DOI] [PubMed] [Google Scholar]
- Cobourne MT, Sharpe PT. Tooth and jaw: molecular mechanisms of patterning in the first branchial arch. Arch Oral Biol. 2003;48:1–14. doi: 10.1016/s0003-9969(02)00208-x. [DOI] [PubMed] [Google Scholar]
- Copley RR, Aloy P, Russell RB, Telford MJ. Systematic searches for molecular synapomorphies in model metazoan genomes give some support for Ecdysozoa after accounting for the idiosyncrasies of Caenorhabditis elegans. Evol Dev. 2004;6:164–9. doi: 10.1111/j.1525-142X.2004.04021.x. [DOI] [PubMed] [Google Scholar]
- Cornell RA, Eisen JS. Notch in the pathway: the roles of Notch signaling in neural crest development. Semin Cell Dev Biol. 2005;16:663–72. doi: 10.1016/j.semcdb.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Davey MG, Paton IR, Yin Y, Schmidt M, Bangs FK, Morrice DR, Smith TG, Buxton P, Stamataki D, Tanaka M, Munsterberg AE, Briscoe J, Tickle C, Burt DW. The chicken talpid3 gene encodes a novel protein essential for Hedgehog signaling. Genes Dev. 2006;20:1365–77. doi: 10.1101/gad.369106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbordes SC, Sanson B. The glypican Dally-like is required for Hedgehog signalling in the embryonic epidermis of Drosophila. Development. 2003;130:6245–55. doi: 10.1242/dev.00874. [DOI] [PubMed] [Google Scholar]
- Diiorio P, Alexa K, Choe SK, Etheridge L, Sagerstrom CG. TALE-Family homeodomain proteins regulate endodermal sonic hedgehog expression and pattern the anterior endoderm. Dev Biol. 2007;304:221–31. doi: 10.1016/j.ydbio.2006.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNardo S, Heemskerk J, Dougan S, O'Farrell PH. The making of a maggot: patterning the Drosophila embryonic epidermis. Curr Opin Genet Dev. 1994;4:529–34. doi: 10.1016/0959-437x(94)90068-e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggenschwiler JT, Bulgakov OV, Qin J, Li T, Anderson KV. Mouse Rab23 regulates hedgehog signaling from smoothened to Gli proteins. Dev Biol. 2006;290:1–12. doi: 10.1016/j.ydbio.2005.09.022. [DOI] [PubMed] [Google Scholar]
- Ekker SC, McGrew LL, Lai CJ, Lee JJ, von Kessler DP, Moon RT, Beachy PA. Distinct expression and shared activities of members of the hedgehog gene family of Xenopus laevis. Development. 1995;121:2337–47. doi: 10.1242/dev.121.8.2337. [DOI] [PubMed] [Google Scholar]
- Finnerty JR, Pang K, Burton P, Paulson D, Martindale MQ. Origins of bilateral symmetry: Hox and dpp expression in a sea anemone. Science. 2004;304:1335–7. doi: 10.1126/science.1091946. [DOI] [PubMed] [Google Scholar]
- Fuccillo M, Joyner AL, Fishell G. Morphogen to mitogen: the multiple roles of hedgehog signalling in vertebrate neural development. Nat Rev Neurosci. 2006;7:772–83. doi: 10.1038/nrn1990. [DOI] [PubMed] [Google Scholar]
- Fukuda K, Yasugi S. Versatile roles for sonic hedgehog in gut development. J Gastroenterol. 2002;37:239–46. doi: 10.1007/s005350200030. [DOI] [PubMed] [Google Scholar]
- Glise B, Miller CA, Crozatier M, Halbisen MA, Wise S, Olson DJ, Vincent A, Blair SS. Shifted, the Drosophila ortholog of Wnt inhibitory factor-1, controls the distribution and movement of Hedgehog. Dev Cell. 2005;8:255–66. doi: 10.1016/j.devcel.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Gorfinkiel N, Sierra J, Callejo A, Ibanez C, Guerrero I. The Drosophila ortholog of the human Wnt inhibitor factor Shifted controls the diffusion of lipid-modified Hedgehog. Dev Cell. 2005;8:241–53. doi: 10.1016/j.devcel.2004.12.018. [DOI] [PubMed] [Google Scholar]
- Grapin-Botton A. Antero-posterior patterning of the vertebrate digestive tract: 40 years after Nicole Le Douarin's PhD thesis. Int J Dev Biol. 2005;49:335–47. doi: 10.1387/ijdb.041946ag. [DOI] [PubMed] [Google Scholar]
- Guerrero I, Chiang C. A conserved mechanism of Hedgehog gradient formation by lipid modifications. Trends Cell Biol. 2007;17:1–5. doi: 10.1016/j.tcb.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Han C, Belenkaya TY, Wang B, Lin X. Drosophila glypicans control the cell-to-cell movement of Hedgehog by a dynamin-independent process. Development. 2004;131:601–11. doi: 10.1242/dev.00958. [DOI] [PubMed] [Google Scholar]
- Han YG, Kwok BH, Kernan MJ. Intraflagellar transport is required in Drosophila to differentiate sensory cilia but not sperm. Curr Biol. 2003;13:1679–86. doi: 10.1016/j.cub.2003.08.034. [DOI] [PubMed] [Google Scholar]
- Hao L, Johnsen R, Lauter G, Baillie D, Burglin TR. Comprehensive analysis of gene expression patterns of hedgehog-related genes. BMC Genomics. 2006a;7:280. doi: 10.1186/1471-2164-7-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao L, Mukherjee K, Liegeois S, Baillie D, Labouesse M, Burglin TR. The hedgehog-related gene qua-1 is required for molting in Caenorhabditis elegans. Dev Dyn. 2006b;235:1469–81. doi: 10.1002/dvdy.20721. [DOI] [PubMed] [Google Scholar]
- Hatini V, Green RB, Lengyel JA, Bray SJ, Dinardo S. The Drumstick/Lines/Bowl regulatory pathway links antagonistic Hedgehog and Wingless signaling inputs to epidermal cell differentiation. Genes Dev. 2005;19:709–18. doi: 10.1101/gad.1268005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward DC, Samuel G, Pontynen PC, Catmull J, Saint R, Miller DJ, Ball EE. Localized expression of a dpp/BMP2/4 ortholog in a coral embryo. Proc Natl Acad Sci U S A. 2002;99:8106–11. doi: 10.1073/pnas.112021499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heemskerk J, DiNardo S. Drosophila hedgehog acts as a morphogen in cellular patterning. Cell. 1994;76:449–60. doi: 10.1016/0092-8674(94)90110-4. [DOI] [PubMed] [Google Scholar]
- Hino K, Satou Y, Yagi K, Satoh N. A genomewide survey of developmentally relevant genes in Ciona intestinalis. VI. Genes for Wnt, TGFbeta, Hedgehog and JAK/STAT signaling pathways. Dev Genes Evol. 2003;213:264–72. doi: 10.1007/s00427-003-0318-8. [DOI] [PubMed] [Google Scholar]
- Hobmayer B, Rentzsch F, Holstein TW. Identification and expression of HySmad1, a member of the R-Smad family of TGFbeta signal transducers, in the diploblastic metazoan Hydra. Dev Genes Evol. 2001;211:597–602. doi: 10.1007/s00427-001-0198-8. [DOI] [PubMed] [Google Scholar]
- Hoch M, Pankratz MJ. Control of gut development by fork head and cell signaling molecules in Drosophila. Mech Dev. 1996;58:3–14. doi: 10.1016/s0925-4773(96)00541-2. [DOI] [PubMed] [Google Scholar]
- Hooper JE, Scott MP. Communicating with Hedgehogs. Nat Rev Mol Cell Biol. 2005;6:306–17. doi: 10.1038/nrm1622. [DOI] [PubMed] [Google Scholar]
- Huangfu D, Anderson KV. Signaling from Smo to Ci/Gli: conservation and divergence of Hedgehog pathways from Drosophila to vertebrates. Development. 2006;133:3–14. doi: 10.1242/dev.02169. [DOI] [PubMed] [Google Scholar]
- Huangfu D, Liu A, Rakeman AS, Murcia NS, Niswander L, Anderson KV. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature. 2003;426:83–7. doi: 10.1038/nature02061. [DOI] [PubMed] [Google Scholar]
- Ingham PW. Hedgehog signaling: a tale of two lipids. Science. 2001;294:1879–81. doi: 10.1126/science.1064115. [DOI] [PubMed] [Google Scholar]
- Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 2001;15:3059–87. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- Ingham PW, Placzek M. Orchestrating ontogenesis: variations on a theme by sonic hedgehog. Nat Rev Genet. 2006;7:841–50. doi: 10.1038/nrg1969. [DOI] [PubMed] [Google Scholar]
- Inoue Y, Niwa N, Mito T, Ohuchi H, Yoshioka H, Noji S. Expression patterns of hedgehog, wingless, and decapentaplegic during gut formation of Gryllus bimaculatus (cricket). Mech Dev. 2002;110:245–8. doi: 10.1016/s0925-4773(01)00584-6. [DOI] [PubMed] [Google Scholar]
- Iwaki DD, Johansen KA, Singer JB, Lengyel JA. drumstick, bowl, and lines are required for patterning and cell rearrangement in the Drosophila embryonic hindgut. Dev Biol. 2001;240:611–26. doi: 10.1006/dbio.2001.0483. [DOI] [PubMed] [Google Scholar]
- Izraeli S, Lowe LA, Bertness VL, Campaner S, Hahn H, Kirsch IR, Kuehn MR. Genetic evidence that Sil is required for the Sonic Hedgehog response pathway. Genesis. 2001;31:72–7. doi: 10.1002/gene.10004. [DOI] [PubMed] [Google Scholar]
- Kalderon D. Hedgehog signaling: an Arrestin connection? Curr Biol. 2005;15:R175–8. doi: 10.1016/j.cub.2005.02.046. [DOI] [PubMed] [Google Scholar]
- Kang D, Huang F, Li D, Shankland M, Gaffield W, Weisblat DA. A hedgehog homolog regulates gut formation in leech (Helobdella). Development. 2003;130:1645–57. doi: 10.1242/dev.00395. [DOI] [PubMed] [Google Scholar]
- Kent D, Bush EW, Hooper JE. Roadkill attenuates Hedgehog responses through degradation of Cubitus interruptus. Development. 2006;133:2001–10. doi: 10.1242/dev.02370. [DOI] [PubMed] [Google Scholar]
- King N, Carroll SB. A receptor tyrosine kinase from choanoflagellates: molecular insights into early animal evolution. Proc Natl Acad Sci U S A. 2001;98:15032–7. doi: 10.1073/pnas.261477698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishigami S, Mishina Y. BMP signaling and early embryonic patterning. Cytokine Growth Factor Rev. 2005;16:265–78. doi: 10.1016/j.cytogfr.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Klein PS, Melton DA. A molecular mechanism for the effect of lithium on development. Proc Natl Acad Sci U S A. 1996;93:8455–9. doi: 10.1073/pnas.93.16.8455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi D, Jindo T, Naruse K, Takeda H. Development of the endoderm and gut in medaka, Oryzias latipes. Dev Growth Differ. 2006;48:283–95. doi: 10.1111/j.1440-169X.2006.00870.x. [DOI] [PubMed] [Google Scholar]
- Koizumi O, Itazawa M, Mizumoto H, Minobe S, Javois LC, Grimmelikhuijzen CJ, Bode HR. Nerve ring of the hypostome in hydra. I. Its structure, development, and maintenance. J Comp Neurol. 1992;326:7–21. doi: 10.1002/cne.903260103. [DOI] [PubMed] [Google Scholar]
- Kumar S, Balczarek KA, Lai ZC. Evolution of the hedgehog gene family. Genetics. 1996;142:965–72. doi: 10.1093/genetics/142.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusserow A, Pang K, Sturm C, Hrouda M, Lentfer J, Schmidt HA, Technau U, von Haeseler A, Hobmayer B, Martindale MQ, Holstein TW. Unexpected complexity of the Wnt gene family in a sea anemone. Nature. 2005;433:156–60. doi: 10.1038/nature03158. [DOI] [PubMed] [Google Scholar]
- Lai EC. Notch signaling: control of cell communication and cell fate. Development. 2004;131:965–73. doi: 10.1242/dev.01074. [DOI] [PubMed] [Google Scholar]
- Lee CS, Buttitta L, Fan CM. Evidence that the WNT-inducible growth arrest-specific gene 1 encodes an antagonist of sonic hedgehog signaling in the somite. Proc Natl Acad Sci U S A. 2001;98:11347–52. doi: 10.1073/pnas.201418298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JD, Treisman JE. Sightless has homology to transmembrane acyltransferases and is required to generate active Hedgehog protein. Curr Biol. 2001;11:1147–52. doi: 10.1016/s0960-9822(01)00323-2. [DOI] [PubMed] [Google Scholar]
- Lees C, Howie S, Sartor RB, Satsangi J. The hedgehog signalling pathway in the gastrointestinal tract: implications for development, homeostasis, and disease. Gastroenterology. 2005;129:1696–710. doi: 10.1053/j.gastro.2005.05.010. [DOI] [PubMed] [Google Scholar]
- Liu A, Wang B, Niswander LA. Mouse intraflagellar transport proteins regulate both the activator and repressor functions of Gli transcription factors. Development. 2005;132:3103–11. doi: 10.1242/dev.01894. [DOI] [PubMed] [Google Scholar]
- Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- Mackie GO. Central neural circuitry in the jellyfish Aglantha: a model ‘simple nervous system’. Neurosignals. 2004;13:5–19. doi: 10.1159/000076155. [DOI] [PubMed] [Google Scholar]
- Martindale MQ, Pang K, Finnerty JR. Investigating the origins of triploblasty: ‘mesodermal’ gene expression in a diploblastic animal, the sea anemone Nematostella vectensis (phylum, Cnidaria; class, Anthozoa). Development. 2004;131:2463–74. doi: 10.1242/dev.01119. [DOI] [PubMed] [Google Scholar]
- Matus DQ, Pang K, Marlow H, Dunn CW, Thomsen GH, Martindale MQ. Molecular evidence for deep evolutionary roots of bilaterality in animal development. Proc Natl Acad Sci U S A. 2006a;103:11195–200. doi: 10.1073/pnas.0601257103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus DQ, Thomsen GH, Martindale MQ. Dorso/ventral genes are asymmetrically expressed and involved in germ-layer demarcation during cnidarian gastrulation. Curr Biol. 2006b;16:499–505. doi: 10.1016/j.cub.2006.01.052. [DOI] [PubMed] [Google Scholar]
- Matus DQ, Thomsen GH, Martindale MQ. FGF signaling in gastrulation and neural development in Nematostella vectensis, an anthozoan cnidarian. Dev Genes Evol. 2007;217:137–148. doi: 10.1007/s00427-006-0122-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy RA, Barth JL, Chintalapudi MR, Knaak C, Argraves WS. Megalin functions as an endocytic sonic hedgehog receptor. J Biol Chem. 2002;277:25660–7. doi: 10.1074/jbc.M201933200. [DOI] [PubMed] [Google Scholar]
- McGlinn E, Tabin CJ. Mechanistic insight into how Shh patterns the vertebrate limb. Curr Opin Genet Dev. 2006;16:426–32. doi: 10.1016/j.gde.2006.06.013. [DOI] [PubMed] [Google Scholar]
- McMahon AP, Ingham PW, Tabin CJ. Developmental roles and clinical significance of hedgehog signaling. Curr Top Dev Biol. 2003;53:1–114. doi: 10.1016/s0070-2153(03)53002-2. [DOI] [PubMed] [Google Scholar]
- Meloni AR, Fralish GB, Kelly P, Salahpour A, Chen JK, Wechsler-Reya RJ, Lefkowitz RJ, Caron MG. Smoothened signal transduction is promoted by G protein-coupled receptor kinase 2. Mol Cell Biol. 2006;26:7550–60. doi: 10.1128/MCB.00546-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer A, Schartl M. Gene and genome duplications in vertebrates: the one-to-four (-to-eight in fish) rule and the evolution of novel gene functions. Curr Opin Cell Biol. 1999;11:699–704. doi: 10.1016/s0955-0674(99)00039-3. [DOI] [PubMed] [Google Scholar]
- Miljkovic-Licina M, Gauchat D, Galliot B. Neuronal evolution: analysis of regulatory genes in a first-evolved nervous system, the hydra nervous system. Biosystems. 2004;76:75–87. doi: 10.1016/j.biosystems.2004.05.030. [DOI] [PubMed] [Google Scholar]
- Miller DJ, Ball EE, Technau U. Cnidarians and ancestral genetic complexity in the animal kingdom. Trends Genet. 2005;21:536–9. doi: 10.1016/j.tig.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Mohler J, Vani K. Molecular organization and embryonic expression of the hedgehog gene involved in cell-cell communication in segmental patterning of Drosophila. Development. 1992;115:957–71. doi: 10.1242/dev.115.4.957. [DOI] [PubMed] [Google Scholar]
- Morales CR, Zeng J, El Alfy M, Barth JL, Chintalapudi MR, McCarthy RA, Incardona JP, Argraves WS. Epithelial trafficking of Sonic hedgehog by megalin. J Histochem Cytochem. 2006;54:1115–27. doi: 10.1369/jhc.5A6899.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullor JL, Sanchez P, Altaba AR. Pathways and consequences: Hedgehog signaling in human disease. Trends Cell Biol. 2002;12:562–9. doi: 10.1016/s0962-8924(02)02405-4. [DOI] [PubMed] [Google Scholar]
- Narita T, Ishii Y, Nohno T, Noji S, Yasugi S. Sonic hedgehog expression in developing chicken digestive organs is regulated by epithelial-mesenchymal interactions. Dev Growth Differ. 1998;40:67–74. doi: 10.1046/j.1440-169x.1998.t01-5-00008.x. [DOI] [PubMed] [Google Scholar]
- Nederbragt AJ, van Loon AE, Dictus WJ. Evolutionary biology: hedgehog crosses the snail's midline. Nature. 2002;417:811–2. doi: 10.1038/417811b. [DOI] [PubMed] [Google Scholar]
- Nichols SA, Dirks W, Pearse JS, King N. Early evolution of animal cell signaling and adhesion genes. Proc Natl Acad Sci U S A. 2006;103:12451–6. doi: 10.1073/pnas.0604065103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwenhuis E, Hui CC. Hedgehog signaling and congenital malformations. Clin Genet. 2005;67:193–208. doi: 10.1111/j.1399-0004.2004.00360.x. [DOI] [PubMed] [Google Scholar]
- Okada A, Charron F, Morin S, Shin DS, Wong K, Fabre PJ, Tessier-Lavigne M, McConnell SK. Boc is a receptor for sonic hedgehog in the guidance of commissural axons. Nature. 2006;444:369–73. doi: 10.1038/nature05246. [DOI] [PubMed] [Google Scholar]
- Osterlund T, Kogerman P. Hedgehog signalling: how to get from Smo to Ci and Gli. Trends Cell Biol. 2006;16:176–80. doi: 10.1016/j.tcb.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Pang K, Matus DQ, Martindale MQ. The ancestral role of COE genes may have been in chemoreception: evidence from the development of the sea anemone, Nematostella vectensis (Phylum Cnidaria; Class Anthozoa). Dev Genes Evol. 2004;214:134–8. doi: 10.1007/s00427-004-0383-7. [DOI] [PubMed] [Google Scholar]
- Pires-daSilva A, Sommer RJ. The evolution of signalling pathways in animal development. Nat Rev Genet. 2003;4:39–49. doi: 10.1038/nrg977. [DOI] [PubMed] [Google Scholar]
- Pourquie O. Vertebrate somitogenesis: a novel paradigm for animal segmentation? Int J Dev Biol. 2003;47:597–603. [PubMed] [Google Scholar]
- Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, Terry A, Shapiro H, Lindquist E, Kapitonov VV, Jurka J, Genikhovich G, Grigoriev IV, Lucas SM, Steele RE, Finnerty JR, Technau U, Martindale MQ, Rokhsar DS. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- Ramalho-Santos M, Melton DA, McMahon AP. Hedgehog signals regulate multiple aspects of gastrointestinal development. Development. 2000;127:2763–72. doi: 10.1242/dev.127.12.2763. [DOI] [PubMed] [Google Scholar]
- Ray K, Perez SE, Yang Z, Xu J, Ritchings BW, Steller H, Goldstein LS. Kinesin-II is required for axonal transport of choline acetyltransferase in Drosophila. J Cell Biol. 1999;147:507–18. doi: 10.1083/jcb.147.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reber-Muller S, Streitwolf-Engel R, Yanze N, Schmid V, Stierwald M, Erb M, Seipel K. BMP2/4 and BMP5−8 in jellyfish development and transdifferentiation. Int J Dev Biol. 2006;50:377–84. doi: 10.1387/ijdb.052085sr. [DOI] [PubMed] [Google Scholar]
- Reiter JF, Skarnes WC. Tectonic, a novel regulator of the Hedgehog pathway required for both activation and inhibition. Genes Dev. 2006;20:22–7. doi: 10.1101/gad.1363606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentzsch F, Anton R, Saina M, Hammerschmidt M, Holstein TW, Technau U. Asymmetric expression of the BMP antagonists chordin and gremlin in the sea anemone Nematostella vectensis: implications for the evolution of axial patterning. Dev Biol. 2006;296:375–87. doi: 10.1016/j.ydbio.2006.06.003. [DOI] [PubMed] [Google Scholar]
- Requena N, Mann P, Hampp R, Franken P. Early developmentally regulated genes in the arbuscular mycorrhizal fungus Glomus mosseae: identification of GmGIN1, a novel gene with homology to the C-terminus of metazoan hedgehog proteins. Plant and Soil. 2002;244:129–139. [Google Scholar]
- Roberts DJ, Johnson RL, Burke AC, Nelson CE, Morgan BA, Tabin C. Sonic hedgehog is an endodermal signal inducing Bmp-4 and Hox genes during induction and regionalization of the chick hindgut. Development. 1995;121:3163–74. doi: 10.1242/dev.121.10.3163. [DOI] [PubMed] [Google Scholar]
- Robertson CP, Braun MM, Roelink H. Sonic hedgehog patterning in chick neural plate is antagonized by a Wnt3-like signal. Dev Dyn. 2004;229:510–9. doi: 10.1002/dvdy.10501. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–4. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Roussa E, Krieglstein K. Induction and specification of midbrain dopaminergic cells: focus on SHH, FGF8, and TGF-beta. Cell Tissue Res. 2004;318:23–33. doi: 10.1007/s00441-004-0916-4. [DOI] [PubMed] [Google Scholar]
- Rubin DC. Intestinal morphogenesis. Curr Opin Gastroenterol. 2007;23:111–4. doi: 10.1097/MOG.0b013e3280145082. [DOI] [PubMed] [Google Scholar]
- Rubin LL, de Sauvage FJ. Targeting the Hedgehog pathway in cancer. Nat Rev Drug Discov. 2006;5:1026–33. doi: 10.1038/nrd2086. [DOI] [PubMed] [Google Scholar]
- Samuel G, Miller D, Saint R. Conservation of a DPP/BMP signaling pathway in the nonbilateral cnidarian Acropora millepora. Evol Dev. 2001;3:241–50. doi: 10.1046/j.1525-142x.2001.003004241.x. [DOI] [PubMed] [Google Scholar]
- Sanson B. Generating patterns from fields of cells. Examples from Drosophila segmentation. EMBO Rep. 2001;2:1083–8. doi: 10.1093/embo-reports/kve255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz CB, Technau U. The ancestral role of Brachyury: expression of NemBra1 in the basal cnidarian Nematostella vectensis (Anthozoa). Dev Genes Evol. 2003;212:563–70. doi: 10.1007/s00427-002-0272-x. [DOI] [PubMed] [Google Scholar]
- Seaver EC, Kaneshige LM. Expression of ‘segmentation’ genes during larval and juvenile development in the polychaetes Capitella sp. I and H. elegans. Dev Biol. 2006;289:179–94. doi: 10.1016/j.ydbio.2005.10.025. [DOI] [PubMed] [Google Scholar]
- Sekimizu K, Nishioka N, Sasaki H, Takeda H, Karlstrom RO, Kawakami A. The zebrafish iguana locus encodes Dzip1, a novel zinc-finger protein required for proper regulation of Hedgehog signaling. Development. 2004;131:2521–32. doi: 10.1242/dev.01059. [DOI] [PubMed] [Google Scholar]
- Shannon JM, Hyatt BA. Epithelial-mesenchymal interactions in the developing lung. Annu Rev Physiol. 2004;66:625–45. doi: 10.1146/annurev.physiol.66.032102.135749. [DOI] [PubMed] [Google Scholar]
- Shimeld SM. The evolution of the hedgehog gene family in chordates: insights from amphioxus hedgehog. Dev Genes Evol. 1999;209:40–7. doi: 10.1007/s004270050225. [DOI] [PubMed] [Google Scholar]
- Siekmann AF, Brand M. Distinct tissue-specificity of three zebrafish ext1 genes encoding proteoglycan modifying enzymes and their relationship to somitic Sonic hedgehog signaling. Dev Dyn. 2005;232:498–505. doi: 10.1002/dvdy.20248. [DOI] [PubMed] [Google Scholar]
- Silver SJ, Rebay I. Signaling circuitries in development: insights from the retinal determination gene network. Development. 2005;132:3–13. doi: 10.1242/dev.01539. [DOI] [PubMed] [Google Scholar]
- Simonnet F, Deutsch J, Queinnec E. hedgehog is a segment polarity gene in a crustacean and a chelicerate. Dev Genes Evol. 2004;214:537–45. doi: 10.1007/s00427-004-0435-z. [DOI] [PubMed] [Google Scholar]
- Snell EA, Brooke NM, Taylor WR, Casane D, Philippe H, Holland PW. An unusual choanoflagellate protein released by Hedgehog autocatalytic processing. Proc Biol Sci. 2006;273:401–7. doi: 10.1098/rspb.2005.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell EA, Furlong RF, Holland PW. Hsp70 sequences indicate that choanoflagellates are closely related to animals. Curr Biol. 2001;11:967–70. doi: 10.1016/s0960-9822(01)00275-5. [DOI] [PubMed] [Google Scholar]
- Stopper GF, Wagner GP. Of chicken wings and frog legs: a smorgasbord of evolutionary variation in mechanisms of tetrapod limb development. Dev Biol. 2005;288:21–39. doi: 10.1016/j.ydbio.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Strahle U, Blader P, Ingham PW. Expression of axial and sonic hedgehog in wildtype and midline defective zebrafish embryos. Int J Dev Biol. 1996;40:929–40. [PubMed] [Google Scholar]
- Suga H, Ono K, Miyata T. Multiple TGF-beta receptor related genes in sponge and ancient gene duplications before the parazoan-eumetazoan split. FEBS Lett. 1999;453:346–50. doi: 10.1016/s0014-5793(99)00749-8. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–90. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Sukegawa A, Narita T, Kameda T, Saitoh K, Nohno T, Iba H, Yasugi S, Fukuda K. The concentric structure of the developing gut is regulated by Sonic hedgehog derived from endodermal epithelium. Development. 2000;127:1971–80. doi: 10.1242/dev.127.9.1971. [DOI] [PubMed] [Google Scholar]
- Tabata T, Takei Y. Morphogens, their identification and regulation. Development. 2004;131:703–12. doi: 10.1242/dev.01043. [DOI] [PubMed] [Google Scholar]
- Takashima S, Murakami R. Regulation of pattern formation in the Drosophila hindgut by wg, hh, dpp, and en. Mech Dev. 2001;101:79–90. doi: 10.1016/s0925-4773(00)00555-4. [DOI] [PubMed] [Google Scholar]
- Tam PP, Kanai-Azuma M, Kanai Y. Early endoderm development in vertebrates: lineage differentiation and morphogenetic function. Curr Opin Genet Dev. 2003;13:393–400. doi: 10.1016/s0959-437x(03)00085-6. [DOI] [PubMed] [Google Scholar]
- Tashiro S, Michiue T, Higashijima S, Zenno S, Ishimaru S, Takahashi F, Orihara M, Kojima T, Saigo K. Structure and expression of hedgehog, a Drosophila segment-polarity gene required for cell-cell communication. Gene. 1993;124:183–9. doi: 10.1016/0378-1119(93)90392-g. [DOI] [PubMed] [Google Scholar]
- Technau U, Rudd S, Maxwell P, Gordon PM, Saina M, Grasso LC, Hayward DC, Sensen CW, Saint R, Holstein TW, Ball EE, Miller DJ. Maintenance of ancestral complexity and non-metazoan genes in two basal cnidarians. Trends Genet. 2005;21:633–9. doi: 10.1016/j.tig.2005.09.007. [DOI] [PubMed] [Google Scholar]
- Thisse B, Thisse C. Functions and regulations of fibroblast growth factor signaling during embryonic development. Dev Biol. 2005;287:390–402. doi: 10.1016/j.ydbio.2005.09.011. [DOI] [PubMed] [Google Scholar]
- Tompkins K. Molecular mechanisms of cytodifferentiation in mammalian tooth development. Connect Tissue Res. 2006;47:111–8. doi: 10.1080/03008200600727756. [DOI] [PubMed] [Google Scholar]
- Vierkotten J, Dildrop R, Peters T, Wang B, Ruther U. Ftm is a novel basal body protein of cilia involved in Shh signalling. Development. 2007;134:2569–77. doi: 10.1242/dev.003715. [DOI] [PubMed] [Google Scholar]
- Vincent JP, Dubois L. Morphogen transport along epithelia, an integrated trafficking problem. Dev Cell. 2002;3:615–23. doi: 10.1016/s1534-5807(02)00323-4. [DOI] [PubMed] [Google Scholar]
- Vokes SA, McMahon AP. Hedgehog signaling: iguana debuts as a nuclear gatekeeper. Curr Biol. 2004;14:R668–70. doi: 10.1016/j.cub.2004.08.015. [DOI] [PubMed] [Google Scholar]
- Walton KD, Croce JC, Glenn TD, Wu SY, McClay DR. Genomics and expression profiles of the Hedgehog and Notch signaling pathways in sea urchin development. Dev Biol. 2006;300:153–64. doi: 10.1016/j.ydbio.2006.08.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LC, Nassir F, Liu ZY, Ling L, Kuo F, Crowell T, Olson D, Davidson NO, Burkly LC. Disruption of hedgehog signaling reveals a novel role in intestinal morphogenesis and intestinal-specific lipid metabolism in mice. Gastroenterology. 2002;122:469–82. doi: 10.1053/gast.2002.31102. [DOI] [PubMed] [Google Scholar]
- Wang Y, McMahon AP, Allen BL. Shifting paradigms in Hedgehog signaling. Curr Opin Cell Biol. 2007;19:159–65. doi: 10.1016/j.ceb.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Warburton D, Schwarz M, Tefft D, Flores-Delgado G, Anderson KD, Cardoso WV. The molecular basis of lung morphogenesis. Mech Dev. 2000;92:55–81. doi: 10.1016/s0925-4773(99)00325-1. [DOI] [PubMed] [Google Scholar]
- Wiens M, Belikov SI, Kaluzhnaya OV, Adell T, Schroder HC, Perovic-Ottstadt S, Kaandorp JA, Muller WE. Regional and modular expression of morphogenetic factors in the demosponge Lubomirskia baicalensis. Micron. 2007 doi: 10.1016/j.micron.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Wikramanayake AH, Hong M, Lee PN, Pang K, Byrum CA, Bince JM, Xu R, Martindale MQ. An ancient role for nuclear beta-catenin in the evolution of axial polarity and germ layer segregation. Nature. 2003;426:446–50. doi: 10.1038/nature02113. [DOI] [PubMed] [Google Scholar]
- Wilson L, Maden M. The mechanisms of dorsoventral patterning in the vertebrate neural tube. Dev Biol. 2005;282:1–13. doi: 10.1016/j.ydbio.2005.02.027. [DOI] [PubMed] [Google Scholar]
- Wolf YI, Rogozin IB, Koonin EV. Coelomata and not Ecdysozoa: evidence from genome-wide phylogenetic analysis. Genome Res. 2004;14:29–36. doi: 10.1101/gr.1347404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao S, Lum L, Beachy P. The ihog cell-surface proteins bind Hedgehog and mediate pathway activation. Cell. 2006;125:343–57. doi: 10.1016/j.cell.2006.02.040. [DOI] [PubMed] [Google Scholar]
- Zhang J, Rosenthal A, de Sauvage FJ, Shivdasani RA. Downregulation of Hedgehog signaling is required for organogenesis of the small intestine in Xenopus. Dev Biol. 2001;229:188–202. doi: 10.1006/dbio.2000.9953. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Zhang L, Wang B, Ou CY, Chien CT, Jiang J. A hedgehog-induced BTB protein modulates hedgehog signaling by degrading Ci/Gli transcription factor. Dev Cell. 2006;10:719–29. doi: 10.1016/j.devcel.2006.05.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.