Abstract
Protozoa of the genus Leishmania are causative agents of leishmaniasis, an important health problem in both human and veterinary medicine. Here, we describe a new heat shock protein (HSP) in Leishmania, belonging to the small HSP (sHSP) family in kinetoplastids. The protein is highly conserved in different Leishmania species, showing instead significant divergence with sHSP's from other organisms. The humoral response elicited against this protein during Leishmania infection has been investigated in natural infected humans and dogs, and in experimentally infected hamsters. Leishmania HSP20 is a prominent antigen for canine hosts; on the contrary, the protein seems to be a poor antigen for human immune system. Time-course analysis of appearance of anti-HSP20 antibodies in golden hamsters indicated that these antibodies are produced at late stages of the infection, when clinical symptoms of disease are patent. Finally, the protective efficacy of HSP20 was assessed in mice using a DNA vaccine approach prior to challenge with Leishmania amazonensis.
1. INTRODUCTION
Leishmania spp., kinetoplastid protozoan parasites, cause a diverse collection of human diseases (known as leishmaniasis) ranging in severity from a spontaneously healing skin ulcer to overwhelming visceral disease. Leishmaniasis currently affects an estimated 12 million people in 88 countries, with approximately 500 000 new cases of visceral leishmaniasis (VL) per year and 1.5 million for cutaneous leishmaniasis [1]. The clinical manifestations of leishmaniasis depend on complex interactions between the virulence characteristics of the infecting Leishmania species and the host immune response [2, 3]. Thus, CL is caused mainly by Leishmania major and Leishmania mexicana, whereas VL is essentially caused by Leishmania donovani and Leishmania infantum (also called Leishmania chagasi in Latin America). However, a single Leishmania species can produce more than one clinical outcome; for example, Leishmania amazonensis is known to be associated with cutaneous, diffuse cutaneous, and visceral leishmaniasis in South and Central America. There are no vaccines available at present to control any form of leishmaniasis, despite considerable laboratory efforts [4]. Current chemotherapy is far from satisfactory [5] and new reagents to improve diagnosis would be desirable.
Leishmania is transmitted by the bite of infected sandflies, where the parasite lives as extracellular promastigote in the insect gut, to mammalian hosts. The promastigotes invade macrophages, where they differentiate and replicate as obligatory intracellular amastigotes. During transmission, the parasite is subjected to a drastic change of environmental temperature from the ambient temperature in the insect vector to higher temperatures in the mammalian host. The heat shock response, mediated by the induction of the heat shock proteins (HSPs), is a homeostatic mechanism that protects cells from the deleterious effects of thermal and other environmental stresses [6]. Given these circumstances, HSPs, in Leishmania, are believed to play essential roles in the host-pathogen interaction. Remarkably, many immunogenic Leishmania antigens are members of different HSP families [7]. Thus, HSP60 [8], HSP70 [9–13], Grp78 [14], HSP83/90 [11, 12, 15], and Grp94 [16] have been described as prominent antigens recognized by sera from leishmaniasis patients. The preponderance of HSPs as molecules recognized by human immune system was illustrated in a recent study [17]: from 242 protein-producing clones, which were identified by immunoscreening with sera from VL patients, 118 (49%) contained sequences coding for members of the HSP70 and HSP83/90 families. However, among the main groups of HSPs, there exists a notable absence, that is there is not any report describing members of the small heat shock protein (sHSP) family as antigens during Leishmania infections. Indeed, no descriptions of such genes have been published so far.
The sHSP family comprises the most widespread but also the most poorly conserved group of HSPs. sHSPs have been found in bacteria, archaea and eukaryotes [18]. Although proteins belonging to the sHSP family are diverse in sequence and size, they share characteristic features: (i) a conserved α-crystallin domain of approximately 90 residues; (ii) a small molecular mass of 12–43 kDa; (iii) tendency to form large oligomers; (iv) increased synthesis by stress conditions; (v) chaperone activity in suppressing protein aggregation [19].
The main aim of this work was to characterize possible sHSPs existing in Leishmania. For this purpose, we took advantage of the recently completed sequence of the L. major genome [20]. In this database, we found sequence information that helped us to identify the L. amazonensis HSP20, which represents in turn the first description of a member of the sHSP family in kinetoplastids. Furthermore, the HSP20 gene was expressed in bacteria and the purified recombinant protein was found to be antigenic during Leishmania infection. Finally, the immunoprotective properties of this protein, administered as a DNA vaccine, were analyzed in the murine model of L. amazonensis infection.
2. MATERIALS AND METHODS
2.1. Parasites
Two L. amazonensis strains were used in this work: strain IFLA/BR/67/pH-8 for gene cloning and strain MHOM/77/LTB0016 for infection of mice. The virulence of the parasite was maintained by regular passage through BALB/c mice. Parasites were cultured at 25°C in Schneider's medium supplemented with 10% fetal bovine serum (FBS). L. infantum promastigotes (M/CAN/ES/96/BCN150) were cultured at 26°C in RPMI 1640 medium supplemented with 10% FBS.
2.2. Cloning and purification of the recombinant HSP20
The full-length open reading frame of L. amazonensis HSP20 gene was PCR amplified from genomic DNA (strain IFLA/BR/67/pH-8) with specific primers: HSP20d, 5′-CCAAGCTTATGTGGAGCCCGAGCAACAA-3′; HSP20r, 5′-CGGGATCCTTAGTCGATGGTGACTGAGT-3′ (underlined are restriction sites included in the primers for cloning purposes). The PCR product was cloned into pCR2.1 vector (Invitrogen Corp., San Diego, Calif., USA) to generate pCRHSP20La clone. The HSP20 gene in pCRHSP20La was removed by HindIII plus NotI double digestion and cloned in the corresponding restriction sites of the pET-28b prokaryotic expression plasmid (Novagen, Madison, Wis, USA) to generate pETHSP20La clone. This clone was used to express the L. amazonensis HSP20 in Escherichia coli (BL21 strain). The recombinant protein (rHSP20), which contains an N-terminal His-tag, was purified on a Ni-NTA agarose column, following the methodology provided by the supplier (Qiagen, Hilden, Germany). The tag was not removed after purification.
The L. amazonensis HSP20 coding sequence contained in the pCRHSP20La clone (see above) was obtained by HindIII-BamHI double digestion and subcloned in the corresponding sites of the eukaryotic expression plasmid pcDNA3 (Invitrogen) to produce clone pcDNA3-LaHSP20. The authenticity of the different clones and the fidelity of the PCR-amplified fragments were verified by nucleotide sequencing (Parque Científico de Madrid, UAM, Madrid). The nucleotide sequence data for L. amazonensis HSP20 coding sequence have been deposited at the EMBL, GenBank, and DDBJ Nucleotide Sequence Databases under the accession no. AM712297.
2.3. Phylogenetic analysis
The genome databases for different Leishmania species (http://www.genedb.org) were used to determine sequence homologues to members of the sHSP family. Protein sequences were scanned for the occurrence of patterns stored in PROSITE-SWISS-PROT database (http://www.expasy.org). For phylogenetic analysis, 39 sHSP sequences were used, choosing representative members for the main groups of organisms: archaea, bacteria, fungi, plant, animal, and kinetoplastids. According to previous studies [18], bacterial sHSPs can be grouped within class A or class B, and representative sequences for both groups have been included in our analysis. The alpha-crystallin domain from the sHSP sequences were aligned using the ClustalW algorithm and the data set was used to build a phylogenetic tree with the MEGA3 software [21]. The trees were made using the neighbor-joining and the minimum evolution algorithms, with Poisson-corrected amino acid distances. The reliability of clustering patterns in the tree was tested by bootstrapping (1000 replicates).
2.4. Sera and enzyme-linked immunosorbent assays (ELISA)
Sera of golden hamsters (Mesocricetus auratus) were obtained from animals experimentally infected with L. infantum (strain M/CAN/ES/96/BCN150). Blood samples were collected monthly. Further details of these sera have been published elsewhere [22]. Human sera were obtained from patients with VL living in Spain. All patients had clinical signs of leishmaniasis and the sera were positive for Leishmania by indirect immunofluorescent tests. The presence of parasites was further demonstrated in smears of bone marrow aspirates. Also, canine sera from naturally Leishmania-infected dogs from the Comunidad de Madrid (Spain) were assayed. For all animals, Leishmania infection was determined by serological and parasitological methods.
Total Leishmania antigen was prepared from L. infantum promastigotes by incubation of parasites in lysis buffer (1% Triton X-100; 150 mM NaCl; 10 mM Tris-HCl, pH 8.0; 1 mM PMSF) for 15 minutes. Afterwards, the suspension, kept on ice, was sonicated until a decrease in viscosity was observed. The insoluble material was pelleted at 10 000 g for 5 minutes and the supernatant stored at −70°C until use. ELISA assays were carried out using standard conditions. Microplate wells (Nunc A/S, Roskilde, Denmark) were coated with either 0.5 μg/ml of purified rHSP20 (see above) or 2 μg/ml of total Leishmania antigen. Primary antibodies were assayed at 1 : 200 dilution. Secondary antibodies (horseradish peroxidase immunoconjugates; Nordic Immunological Laboratories, Tilburg, The Netherlands) against human, hamster, or dog total IgGs (H + L chains) were used at 1 : 1500 dilution. Finally, ortho-phenylenediamine (Dako, Glostrup, Denmark) was used in the developing solution and the reaction product was read at 450 nm.
For each particular assay, cut-off values (Cov) were established from the reactivity values of control-negative sera, calculating the 99% confidence interval for the mean of a normal population. Thus, the Cov values were calculated with the following formula: Cov = μ + 2.576 σ (N)−1/2, where μ is the mean value, σ is the standard deviation, and N is the number of control sera. At least, ten control sera were used for each assay.
2.5. Production of mouse antisera against rHSP20
Female 6-8-week-old BALB/c mice (Harlan Interfauna Ibérica, Barcelona, Spain) were intradermically injected with a mixture of 20 μg of rHSP20 and 50 μg of oligonucleotide ODN-1826 (5′-TCCATGACGTTCCTGACGTT-3′), emulsified 1 : 1 with incomplete Freund's adjuvant (Gibco-BRL, Grand Island, NY, USA). Oligonucleotides containing CpG sequences, such as that ODN-1826, have immunostimulating properties for the mouse immune system, and they are being used in vaccination assays for inducing stronger humoral and cellular responses against coadministered antigens [23]. Injections were repeated (using only rHSP20 emulsified 1 : 1 with IFA) at 2-week intervals for a total of three immunizations. Antibody IgG-titres, evaluated by ELISA against rHSP20, were higher than 200 000.
2.6. Expression of Leishmania HSP20 in pcDNA3-LaHSP20-transfected cells
COS7 cells were harvested from cultures in the late-logarithmic phase of growth (85% confluence); cells were separated from plates by a short treatment (1-2 minutes at 37°C) with 0.002% EDTA and 0.25% trypsin. Afterwards, cells were washed with DMEM (supplemented with 10% FBS) and resuspended in the same medium containing 10 mM HEPES. Two hundred μl of the cell suspension (3 × 106 cells) were transferred into a 0.4-cm cuvette. After adding 5 μg of plasmid DNA, 100 μg of salmon sperm DNA and 5 μl of 1.5 mM NaCl, cells were electroporated using a capacitance of 960 μF and a voltage of 200 V. Immediately, cells were transferred to a 100-mm culture dish containing 10 ml of DMEM (with 10% FBS) and incubated for 72 hours at 37°C with atmosphere of 5% CO2. Afterwards, cells were harvested, washed two times with ice-cold PBS and immediately lysed by addition of Laemmli's load buffer. Lysates were resolved by SDS-PAGE and transferred to nitrocellulose membranes (Amersham, Little Chalfont, United Kingdom). The blots were probed with the mouse anti-HSP20 antibody diluted at 1 : 500 (see above). As secondary antibody, an antimouse IgGs (horseradish peroxidase immunoconjugate) was used at 1 : 2000 dilution. The blot was developed using the ECL system (Amersham, Little Chalfont, United Kingdom).
2.7. Vaccination and infection of mice
Endotoxin-free plasmid DNA, prepared using the EndoFree Plasmid Giga Kit (Qiagen, Hilden, Germany), was used for immunizations. BALB/c mice were obtained from the Centro Nacional para la Producción de Animales de Laboratorio (CENPALAB, La Habana, Cuba). Mice were 6–8 weeks of age when DNA immunizations were initiated. Groups of 6 mice were injected twice at 2-week interval intramuscularly in the left quadriceps with 100 μg of pcDNA3-LaHSP20 DNA plasmid. As negative controls, groups of mice were injected with either PBS or empty vector (pcDNA3, Invitrogen). Two weeks after the last injection, the mice were challenged with 105 stationary-phase L. amazonensis promastigotes that were suspended in 50 μl of PBS and injected into the left hind footpad. The development of lesions was monitored weekly using a digital calliper. The contralateral footpad of each animal represented the control value, and the swelling was calculated as follows: thickness of the left footpad—thickness of the right footpad.
3. RESULTS
3.1. Identification of Leishmania sHSPs and sequence analysis
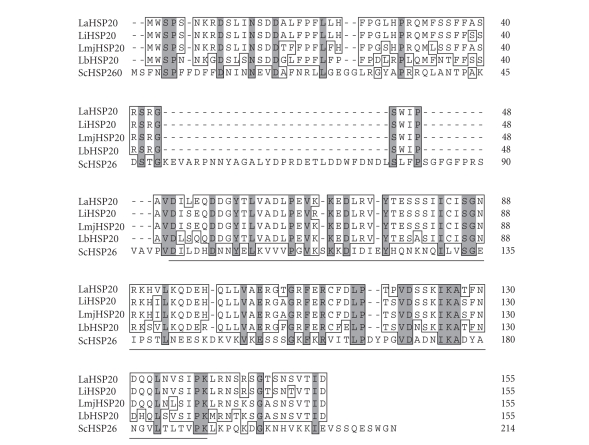
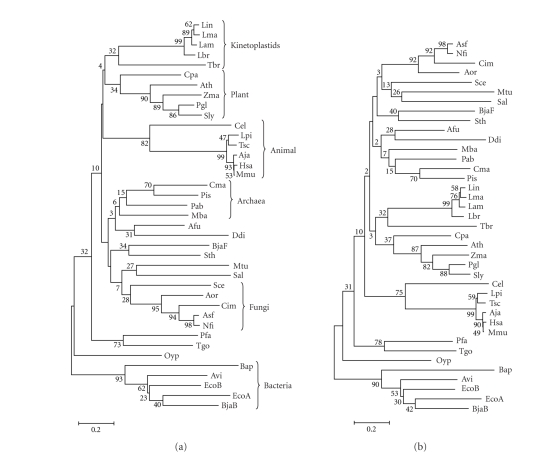
We began this study by performing a BLASTP search of the L. major, L. infantum, and L. braziliensis databases (www.genedb.org) using as query sequence the Saccharomyces cerevisiae HSP26 sequence (UniProtKB/Swiss-Prot entry P15992). Interestingly, for each Leishmania species a single entry was retrieved: LmjF29.2450(L. major), LinJ29_V3.2560 (L. infantum), and LbrM29_V2.2420(L. braziliensis). The BLAST scores were significant (114 or higher), suggesting that they may represent sHSP members in these Leishmania species. In fact, these entries in the different Leishmania databases are annotated as putative HSP20 proteins. Based on these sequences, we designed PCR primers to amplify the HSP20 gene homologue for L. amazonensis. Figure 1 shows the alignment of the various Leishmania HSP20s and the S. cerevisiae HSP26. Leishmania HSP20s share a high sequence identity (higher than 80%); instead, Leishmania HSP20s and yeast HSP26 show ≈30% sequence identity. However, all proteins present the signature motif of sHSPs, that is the HSP20/α-crystallin domain (Figure 1). In addition, we performed a phylogenetic analysis using representative sHSP sequences from the different groups of organisms (Figure 2). This analysis showed a sHSP grouping consistent with previous reports [18]. However, it should be kept in mind that phylogenetic analyses based on sHSP sequences are not adequate to establish evolutionary relationships between distant groups of organisms. Thus, the valid conclusion of our analysis was that Leishmania HSP20s are close to each other and distant from all other sequences. Although with a low bootstrap value, our data showed that Leishmania HSP20s, together Trypanosoma brucei HSP20, form a monophyletic group, separated from the other groups of organisms. This agrees with the belief that kinetoplastids evolved early in evolution from the rest of eukaryotes [24]. Furthermore, even though the bootstrap values are very low, our analyses showed that kinetoplastid sHSPs are closer to homologues from plants than to the corresponding proteins from animals. Remarkably, some plant-like traits in kinetoplastids have been reported in previous studies [25].
Figure 1.
Amino acid sequence alignment of HSP20 from Leishmania spp. and S. cerevisiae HSP26. Protein sequences of L. amazonensis (LaHSP20, this work; EMBL accession number AM712297), L. infantum (LiHSP20, GeneDB identifier LinJ29_v3.2560), L. major (LmjHSP20, GeneDB identifier LmjF29.2450), L. braziliensis (LbHSP20, GeneDB identifier LbrM29_V2.2420) sHSPs, and S. cerevisiae HSP26 (UniProtKB/Swiss-Prot entry P15992) were aligned using the default settings of ClustalW (DNAstar program). Amino acid residues conserved in all sequences are shaded; those conserved in more than 50% of the sequences are boxed. The position of the α-crystallin domain is indicated by a horizontal line. The domains were defined using the Prosite utility at the ExPASy Proteomics Server (http://www.expasy.org); high scores with the HSP20/α-crystallin family profile (accession number PS01031) were obtained.
Figure 2.
Phylogenetic analysis of sHSPs. Phylogenetic trees were constructed on the basis of α-crystallin/HSP20 domains by Neighbor Joining (a) and Minimum Evolution (b) using the MEGA 3 program as described in the Material and Methods section. Sequences (UniProtKB/TrEMBL entry): Afu, Archaeoglobus fulgidus (O28308); Aja, Artibeus jamaicensis (P02482); Aor, Aspergillus oryzae (Q2TXY8); Asf, Aspergillus fumigatus (Q4WV00); Ath, Arabidopsis thaliana (O81822); Avi, Azotobacter vinelandii (P96193); Bap, Buchnera aphidicola (P57640); BjaB, Bradyrhizobium japonicum (HspB; P70918); BjaF, Bradyrhizobium japonicum (HspF; O69243); Cel, Caenorhabditis elegans (Q7JP52); Cim, Coccidioides immitis (Q1E6R4); Cma, Caldivirga maquilingensis (A8MB44); Cpa, Carica papaya (Q69BI7); Ddi, Dictyostelium discoideum (Q54I91); EcoA, Escherichia coli (ibpA; P0C054); EcoB, Escherichia coli (ibpB; P0C058); Hsa, Homo sapiens (P02489); Lam, L. amazonensis; Lbr, L. braziliensis; Lin, L. infantum; Lma, L. major; Lpi, Lygodactylus picturatus (Q6EWI0); Mba, Methanosarcina barkeri (Q46E59); Mmu, Macaca mulatta (P02488); Mtu, Mycobacterium tuberculosis (P0A5B7); Nfi, Neosartorya fischeri (A1DEG0); Oyp, Onion yellows phytoplasma (P81958); Pab, Pyrococcus abyssi (Q9V1L0); Pfa, Plasmodium falciparum (Q8IB02); Pis, Pyrobaculum islandicum (A1RRY3); Pgl, Picea glauca (Q40852); Sal, Streptomyces albus (Q53595); Sce, S. cerevisiae (P15992); Sly, Solanum lycopersicum (O82545); Sth, Streptococcus thermophilus (P80485); Tbr, Trypanosoma brucei (Q57V53); Tgo, Toxoplasma gondii (Q6DUA8); Tsc, Trachemys scripta (Q91517); Zma, Zea mays (P24632). The scale represents mutational changes per residue.
3.2. Recognition of Leishmania HSP20 by sera from infected animals and leishmaniasis patients
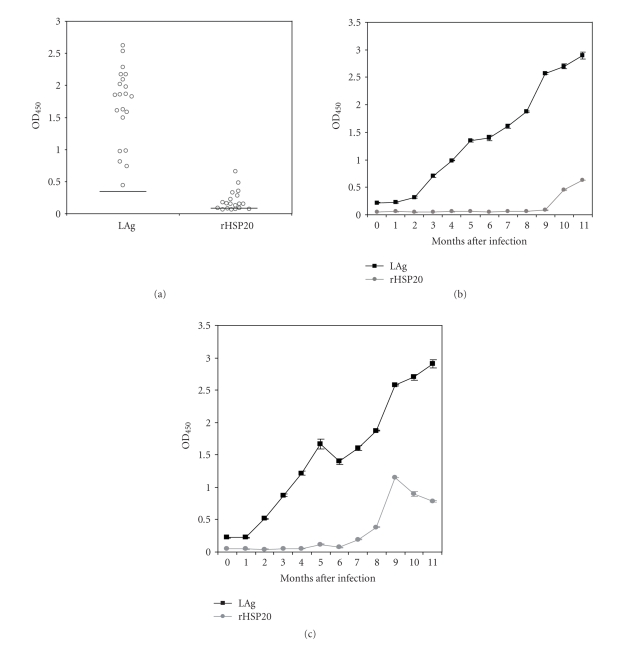
As shown in Figure 1, the HSP20 sequence is well conserved among the different Leishmania species. For instance, L. amazonensis and L. infantum HSP20s share 147 out of the 155 amino acids (95% of identity), and 4 out of the 8 amino acid changes are conservative (97.4% of similarity). With this remarkable degree of sequence conservation, it is expected that both proteins are essentially cross-reactive. Indeed, as shown below, the L. amazonensis HSP20 was recognized by all sera from L. infantum infected dogs. Thus, the L. amazonensis HSP20, expressed in E. coli as a His-tagged recombinant protein (rHSP20), was used in ELISA assays to determine whether antibodies against HSP20 are elicited in experimentally infected hamsters. Remarkably, 62% of the Leishmania infected animals developed significant humoral responses against the HSP20 (Figure 3). No correlation was observed between total antigen and HSP20 reactivity values for individual sera. Furthermore, we analyzed the time course of appearance of anti-HSP20 antibodies along the infection process. As illustrated in Figures 3(b) and 3(c), the anti-HSP20 reactivity was detected long time later than the antibodies against total Leishmania proteins.
Figure 3.
Antigenicity of Leishmania HSP20 in experimentally infected hamsters. (a) Reactivity against total L. infantum proteins (LAg) or L. amazonensis recombinant HSP20 (rHSP20) of sera from L. infantum infected hamsters is indicated as the mean optical density read at 450 nm (OD450). Horizontal lines represent the cut-off values for the assay, which were 0.286 for LAg and 0.067 for rHSP20. (b) and (c) Time course of the humoral response against total antigens (LAg) or rHSP20 elicited in two experimentally infected hamsters.
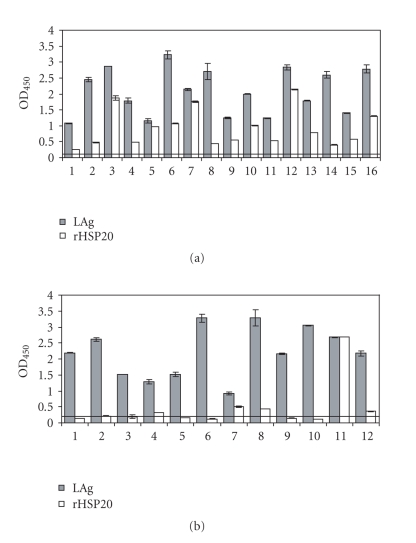
In order to know whether the Leishmania HSP20 is also immunogenic during natural Leishmania infections, the reactivity against rHSP20 of a collection of sera from canids with VL and leishmaniasis patients was determined by ELISA (Figure 4). Interestingly, all sera from dogs with VL reacted with rHSP20, showing reactivity values similar to those observed using total Leishmania proteins (Figure 4). In contrast, few sera from leishmaniasis patients showed a positive reactivity against rHSP20, suggesting that this protein is poorly antigenic for human immune system (Figure 4(b)).
Figure 4.
Antigenicity of Leishmania HSP20 in VL human patients and dogs. (a) Reactivity of 16 serum samples from VL dogs against total L. infantum proteins (LAg) or L. amazonensis recombinant HSP20 (rHSP20). (b) Reactivity of 12 sera from patients with VL against LAg or rHSP20. Horizontal lines represents the cut-off value of negative sera for the LAg antigen (0.122 for canine sera and 0.232 for human sera). All sera were tested in duplicate at a 1 : 200 dilution.
3.3. Immunoprotective potential of Leishmania HSP20 as a DNA vaccine
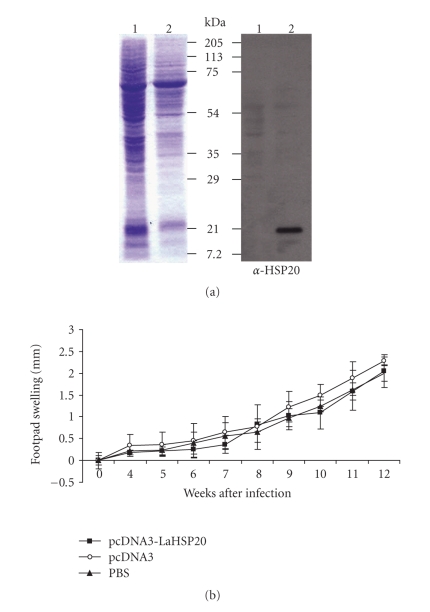
The potential of parasite HSP20 to induce an immunoprotective response was assessed in DNA vaccination experiments using the infection model of L. amazonensis in mice. For that purpose, the L. amazonensis HSP20 gene was cloned into the eukaryotic expression vector pcDNA3 and its appropriate expression was assessed in COS7 cells transfected with the construct (Figure 5). Groups of BALB/c mice were immunized with either pcDNA3-LaHSP20 DNA, empty plasmid DNA or PBS (see materials and methods for further details). A very low, but detectable, reactivity against rHSP20 protein was observed in the sera of LaHSP20-DNA vaccinated mice (data not shown). After challenge, L. amazonensis infection was evaluated by measuring lesion development in the infected footpads (Figure 5). Lesions progressed similarly in the three groups with no statistical differences. Thus, it was concluded that no significant protection or reduction in lesion development was induced in the animals immunized with the HSP20 DNA vaccine in the experimental conditions used in this assay.
Figure 5.
Analysis of protective effect of DNA vaccination with L. amazonensis HSP20 gene. (a) Expression of Leishmania HSP20 in pcDNA3-LaHSP20 transfected COS7 cells. COS7 cells were transiently transfected by electroporation with either pcDNA3 DNA (lane 1) or pcDNA3-LaHSP20 DNA (lane 2). Protein extracts were analyzed by 12% SDS-PAGE and stained with Coomassie blue (left panel). The expression of Leishmania HSP20 was assayed by Western blotting using a mouse anti-HSP20 polyclonal antibody (right panel). (b) BALB/c mice (six per group) were immunized twice at 2-week interval with PBS, empty vector or pcDNA3-LaHSP20. After challenge with L. amazonensis, lesion development (footpad swelling) was monitored weekly. Each point represents the average and standard deviations for the group.
4. DISCUSSION
HSPs represent dominant antigens in many infections and autoimmune diseases, inducing strong humoral and cellular immune responses. Among the main HSP families, HSP60, HSP70, and HSP90 have been described as major antigens in a large number of infectious diseases caused by nematodes, protozoa, fungi, or bacteria. Furthermore, in various infectious disease models, vaccination strategies using HSPs have induced significant protection [26]. In addition, the unique and potent immunostimulatory properties of some HSPs have been applied to the development of new vaccines in which HSPs act as immunomodulatory/carrier agents [27]. However, there are few reports describing members of the sHSP family as antigens during infectious diseases [28, 29]. Unlike large HSPs, the sHSPs are highly divergent in both size and primary sequence; this feature has impaired the characterization of the coding genes and, in consequence, the analysis of the possible antigenicity of this class of proteins.
In Leishmania, several families of HSPs have been described as prominent antigens [7, 17], but no description of sHSPs as antigens exists. Thus, as first step, we analyzed whether sHSPs are encoded in the genome of Leishmania. For this purpose, we took advantage of the recent completion of the genome sequences for L. major [20], L. infantum, and L. braziliensis [30]. A bioinformatics analysis of the L. major database allowed us to identify a protein entry (LmjF29.2450) containing the α-crystallin motif, which characterizes sHSPs. The predicted protein (named HSP20) has 155 amino acids, accounting for a molecular mass of 17.5 kDa. This seems to be the sole sHSP family member in Leishmania, since no new protein entries were recovered from the L. major database when the LmjF29.2450 sequence was used as query in further bioinformatics studies. Homologous sequences were identified in the sequence databases for L. infantum (LinJ29_V3.2560) and L. braziliensis (LbrM29_V2.2420). Based on these sequences, we designed oligonucleotides that allowed us to PCR amplify the corresponding gene in L. amazonensis. Sequence alignments (Figure 1) and phylogenetic studies (Figure 2) revealed that HSP20 is well conserved among Leishmania species but it is highly divergent when compared with sHSPs from other groups of organisms.
In order to analyze the antigenic properties of Leishmania HSP20, the L. amazonensis protein was expressed in E. coli and used to determine the presence of specific IgG in the sera of Leishmania-infected animals and human patients with VL. Remarkably, the protein was recognized by 100% of the sera from dogs with VL, suggesting that this protein would be useful for serodiagnosis of canine leishmaniasis. However, the protein has a limited antigenicity for the human immune system, since only about 30% of the assayed sera showed a positive reactivity. Also, we assayed the recognition of the Leishmania HSP20 by sera from experimentally infected hamsters. In this infection model, the protein was recognized by sera from about 62% of infected animals. In experimentally infected hamsters, analysis of the time-course appearance of anti-Leishmania humoral response indicated that anti-HSP20 antibodies seem to be produced late during infection (Figure 3) coinciding with the onset of disease symptoms. These findings would indicate that HSP20 behaves as a pathoantigen [31], and consequently, anti-HSP20 antibodies could be involved in the pathological processes leading to disease progression. In addition, these findings would point to a usefulness of this protein for serodiagnosis of active leishmaniasis disease.
Finally, we examined whether Leishmania HSP20 could elicit protective immunity. Recently, it has been reported that vaccination with Toxoplasma gondii HSP30 gene, a member of the sHSP family, induced protection in mice against a challenge with the parasite [32]. We chose to use a DNA-based vaccination because this approach has been demonstrated to be adequate for inducing protection against diseases that require cell-mediated immunity such as intracellular protozoan parasites [33]. DNA vaccination has proven to be a successful approach for development immunoprotective responses in different Leishmania-infection models, particularly for protection against L. major [4]. However, our results indicate that immunization of susceptible BALB/c mice with the DNA encoding Leishmania HSP20 provided negligible protection against L. amazonensis infection. Thus, it appears that HSP20 does not represent a feasible vaccine candidate against L. amazonensis infection. Nevertheless, this finding does not exclude that HSP20 would be able to elicit an immunoprotective response against infection with other Leishmania species as occurs for other antigens [34]. In particular, given the strong humoral response elicited by HSP20 in naturally infected dogs, it would be of high-value testing in dogs the immunoprotective potential of this antigen against development of visceral leishmaniasis. On the other hand, the results showed here do not discard a possible use of the Leishmania HSP20 within a multivalent vaccine. For example, a vaccine combination, including DNA encoding P4 and HSP70, induced a significant protection in mice against L. amazonensis, but no protection was observed after administration of these genes separately [35]. Recently, it has been published that the L. major HSP70 is not protective in murine models of cutaneous leishmaniasis using a prime-boost strategy [36]. Overall, these findings could be indicating that Leishmania HSPs alone, using conventional vaccination strategies, are unsuitable for inducing immunoprotection against Leishmania infections; however, these studies do not preclude their use as either immunomodulators or, alternatively, vaccine candidates if different vaccination approaches are followed.
ACKNOWLEDGMENTS
This work was supported by grants from the Ministerio de Ciencia y Tecnología (BFU2006-08346), Fondo de Investigaciones Sanitarias (ISCIII-RETIC RD06/0021/0008-FEDER and ISCIII-RETIC RD06/0021/0009-FEDER), and Centro de Estudios de América Latina (UAM-SCH-2005). Also, an institutional grant from Fundación Ramón Areces is acknowledged.
References
- 1.Desjeux P. Leishmaniasis: current situation and new perspectives. Comparative Immunology, Microbiology and Infectious Diseases. 2004;27(5):305–318. doi: 10.1016/j.cimid.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 2.Pearson RD, Sousa AQ. Clinical spectrum of Leishmaniasis. Clinical Infectious Diseases. 1996;22(1):1–13. doi: 10.1093/clinids/22.1.1. [DOI] [PubMed] [Google Scholar]
- 3.Murray HW, Berman JD, Davies CR, Saravia NG. Advances in leishmaniasis. The Lancet. 2005;366(9496):1561–1577. doi: 10.1016/S0140-6736(05)67629-5. [DOI] [PubMed] [Google Scholar]
- 4.Requena JM, Iborra S, Carrión J, Alonso C, Soto M. Recent advances in vaccines for leishmaniasis. Expert Opinion on Biological Therapy. 2004;4(9):1505–1517. doi: 10.1517/14712598.4.9.1505. [DOI] [PubMed] [Google Scholar]
- 5.Alvar J, Croft S, Olliaro P. Chemotherapy in the treatment and control of Leishmaniasis. Advances in Parasitology. 2006;61:223–274. doi: 10.1016/S0065-308X(05)61006-8. [DOI] [PubMed] [Google Scholar]
- 6.Lindquist S. The heat-shock response. Annual Review of Biochemistry. 1986;55:1151–1191. doi: 10.1146/annurev.bi.55.070186.005443. [DOI] [PubMed] [Google Scholar]
- 7.Requena JM, Alonso C, Soto M. Evolutionarily conserved proteins as prominent immunogens during Leishmania infections. Parasitology Today. 2000;16(6):246–250. doi: 10.1016/s0169-4758(00)01651-3. [DOI] [PubMed] [Google Scholar]
- 8.Rey-Ladino JA, Joshi PB, Singh B, Gupta R, Reiner NE. Leishmania major: molecular cloning, sequencing, and expression of the heat shock protein 60 gene reveals unique carboxy terminal peptide sequences. Experimental Parasitology. 1997;85(3):249–263. doi: 10.1006/expr.1996.4137. [DOI] [PubMed] [Google Scholar]
- 9.Amorim AG, Carrington M, Miles MA, Barker DC, de Almeida MLC. Identification of the C-terminal region of 70 kDa heat shock protein from Leishmania (Viannia) braziliensis as a target for the humoral immune response. Cell Stress Chaperones. 1996;1(3):177–187. doi: 10.1379/1466-1268(1996)001<0177:iotctr>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.MacFarlane J, Blaxter ML, Bishop RP, Miles MA, Kelly JM. Identification and characterisation of a Leishmania donovani antigen belonging to the 70-kDa heat-shock protein family. European Journal of Biochemistry. 1990;190(2):377–384. doi: 10.1111/j.1432-1033.1990.tb15586.x. [DOI] [PubMed] [Google Scholar]
- 11.de Andrade CR, Kirchhoff LV, Donelson JE, Otsu K. Recombinant Leishmania Hsp90 and Hsp70 are recognized by sera from visceral leishmaniasis patients but not Chagas' disease patients. Journal of Clinical Microbiology. 1992;30(2):330–335. doi: 10.1128/jcm.30.2.330-335.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skeiky YAW, Benson DR, Guderian JA, et al. Immune responses of leishmaniasis patients to heat shock proteins of Leishmania species and humans. Infection and Immunity. 1995;63(10):4105–4114. doi: 10.1128/iai.63.10.4105-4114.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quijada L, Requena JM, Soto M, Alonso C. Analysis of the antigenic properties of the L. infantum Hsp70: design of synthetic peptides for specific serodiagnosis of human leishmaniasis. Immunology Letters. 1998;63(3):169–174. doi: 10.1016/s0165-2478(98)00071-6. [DOI] [PubMed] [Google Scholar]
- 14.Jensen AT, Curtis J, Montgomery J, Handman E, Theander TG. Molecular and immunological characterisation of the glucose regulated protein 78 of Leishmania donovani. Biochimica et Biophysica Acta. 2001;1549(1):73–87. doi: 10.1016/s0167-4838(01)00240-0. [DOI] [PubMed] [Google Scholar]
- 15.Celeste BJ, Angel SO, Castro LGM, Gidlund M, Goto H. Leishmania infantum heat shock protein 83 for the serodiagnosis of tegumentary leishmaniasis. Brazilian Journal of Medical and Biological Research. 2004;37(11):1591–1593. doi: 10.1590/s0100-879x2004001100001. [DOI] [PubMed] [Google Scholar]
- 16.Larreta R, Guzman F, Patarroyo ME, Alonso C, Requena JM. Antigenic properties of the Leishmania infantum GRP94 and mapping of linear B-cell epitopes. Immunology Letters. 2002;80(3):199–205. doi: 10.1016/s0165-2478(01)00331-5. [DOI] [PubMed] [Google Scholar]
- 17.Martins DRA, Jeronimo SMB, Donelson JE, Wilson ME. Leishmania chagasi T-cell antigens identified through a double library screen. Infection and Immunity. 2006;74(12):6940–6948. doi: 10.1128/IAI.02032-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu X, Jiao W, Chang Z. Phylogenetic and biochemical studies reveal a potential evolutionary origin of small heat shock proteins of animals from bacterial class A. Journal of Molecular Evolution. 2006;62(3):257–266. doi: 10.1007/s00239-005-0076-5. [DOI] [PubMed] [Google Scholar]
- 19.Haslbeck M, Franzmann T, Weinfurtner D, Buchner J. Some like it hot: the structure and function of small heat-shock proteins. Nature Structural and Molecular Biology. 2005;12(10):842–846. doi: 10.1038/nsmb993. [DOI] [PubMed] [Google Scholar]
- 20.Ivens AC, Peacock CS, Worthey EA, et al. The genome of the kinetoplastid parasite, Leishmania major. Science. 2005;309(5733):436–442. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar S, Tamura K, Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Briefings in Bioinformatics. 2004;5(2):150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 22.Requena JM, Soto M, Doria MD, Alonso C. Immune and clinical parameters associated with Leishmania infantum infection in the golden hamster model. Veterinary Immunology and Immunopathology. 2000;76(3-4):269–281. doi: 10.1016/s0165-2427(00)00221-x. [DOI] [PubMed] [Google Scholar]
- 23.Tewary P, Sukumaran B, Saxena S, Madhubala R. Immunostimulatory oligodeoxynucleotides are potent enhancers of protective immunity in mice immunized with recombinant ORFF leishmanial antigen. Vaccine. 2004;22(23-24):3053–3060. doi: 10.1016/j.vaccine.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 24.Moreira D, López-García P, Vickerman K. An updated view of kinetoplastid phylogeny using environmental sequences and a closer outgroup: proposal for a new classification of the class Kinetoplastea. International Journal of Systematic and Evolutionary Microbiology. 2004;54(5):1861–1875. doi: 10.1099/ijs.0.63081-0. [DOI] [PubMed] [Google Scholar]
- 25.Hannaert V, Bringaud F, Opperdoes FR, Michels PAM. Evolution of energy metabolism and its compartmentation in Kinetoplastida. Kinetoplastid Biology and Disease. 2003;2, article 11:1–30. doi: 10.1186/1475-9292-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zügel U, Kaufmann SHE. Role of heat shock proteins in protection from and pathogenesis of infectious diseases. Clinical Microbiology Reviews. 1999;12(1):19–39. doi: 10.1128/cmr.12.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mizzen L. Immune responses to stress proteins: applications to infectious disease and cancer. Biotherapy. 1998;10(3):173–189. doi: 10.1007/BF02678295. [DOI] [PubMed] [Google Scholar]
- 28.Norimine J, Mosqueda J, Palmer GH, Lewin HA, Brown WC. Conservation of Babesia bovis small heat shock protein (Hsp20) among strains and definition of T helper cell epitopes recognized by cattle with diverse major histocompatibility complex class II haplotypes. Infection and Immunity. 2004;72(2):1096–1106. doi: 10.1128/IAI.72.2.1096-1106.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferrer E, González LM, Foster-Cuevas M, et al. Taenia solium: characterization of a small heat shock protein (Tsol-sHSP35.6) and its possible relevance to the diagnosis and pathogenesis of neurocysticercosis. Experimental Parasitology. 2005;110(1):1–11. doi: 10.1016/j.exppara.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 30.Peacock CS, Seeger K, Harris D, et al. Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nature Genetics. 2007;39(7):839–847. doi: 10.1038/ng2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang K-P, Reed SG, McGwire BS, Soong L. Leishmania model for microbial virulence: the relevance of parasite multiplication and pathoantigenicity. Acta Tropica. 2003;85(3):375–390. doi: 10.1016/s0001-706x(02)00238-3. [DOI] [PubMed] [Google Scholar]
- 32.Mohamed RM, Aosai F, Chen M, et al. Induction of protective immunity by DNA vaccination with Toxoplasma gondii HSP70, HSP30 and SAG1 genes. Vaccine. 2003;21(21-22):2852–2861. doi: 10.1016/s0264-410x(03)00157-9. [DOI] [PubMed] [Google Scholar]
- 33.Gurunathan S, Klinman DM, Seder RA. DNA vaccines: immunology, application, and optimization. Annual Review of Immunology. 2000;18:927–974. doi: 10.1146/annurev.immunol.18.1.927. [DOI] [PubMed] [Google Scholar]
- 34.Coelho EAF, Tavares CAP, Carvalho FAA, et al. Immune responses induced by the Leishmania (Leishmania) donovani A2 antigen, but not by the LACK antigen, are protective against experimental Leishmania (Leishmania) amazonensis infection. Infection and Immunity. 2003;71(7):3988–3994. doi: 10.1128/IAI.71.7.3988-3994.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Campbell K, Diao H, Ji J, Soong L. DNA immunization with the gene encoding P4 nuclease of Leishmania amazonensis protects mice against cutaneous Leishmaniasis. Infection and Immunity. 2003;71(11):6270–6278. doi: 10.1128/IAI.71.11.6270-6278.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rafati S, Gholami E, Hassani N, et al. Leishmania major heat shock protein 70 (HSP70) is not protective in murine models of cutaneous leishmaniasis and stimulates strong humoral responses in cutaneous and visceral leishmaniasis patients. Vaccine. 2007;25(21):4159–4169. doi: 10.1016/j.vaccine.2007.03.006. [DOI] [PubMed] [Google Scholar]