Abstract
Schizophrenia poses an evolutionary-genetic paradox because it exhibits strongly negative fitness effects and high heritability, yet it persists at a prevalence of approximately 1% across all human cultures. Recent theory has proposed a resolution: that genetic liability to schizophrenia has evolved as a secondary consequence of selection for human cognitive traits. This hypothesis predicts that genes increasing the risk of this disorder have been subject to positive selection in the evolutionary history of humans and other primates. We evaluated this prediction using tests for recent selective sweeps in human populations and maximum-likelihood tests for selection during primate evolution. Significant evidence for positive selection was evident using one or both methods for 28 of 76 genes demonstrated to mediate liability to schizophrenia, including DISC1, DTNBP1 and NRG1, which exhibit especially strong and well-replicated functional and genetic links to this disorder. Strong evidence of non-neutral, accelerated evolution was found for DISC1, particularly for exon 2, the only coding region within the schizophrenia-associated haplotype. Additionally, genes associated with schizophrenia exhibited a statistically significant enrichment in their signals of positive selection in HapMap and PAML analyses of evolution along the human lineage, when compared with a control set of genes involved in neuronal activities. The selective forces underlying adaptive evolution of these genes remain largely unknown, but these findings provide convergent evidence consistent with the hypothesis that schizophrenia represents, in part, a maladaptive by-product of adaptive changes during human evolution.
Keywords: adaptive evolution, schizophrenia, positive Darwinian selection, disease genetics
1. Introduction
Schizophrenia is characterized by delusions and auditory hallucinations, loss of coherence and logic in thought and language, and emotionality inappropriate to social context (Crow 1997; Horrobin 1998; Tamminga & Holcomb 2005). This disorder has a polygenic basis involving multiple genes of small effect (Riley & Kendler 2006), and its phenotypic effects represent one end of a continuum that grades into schizotypal cognition and to normality (Rossi & Daneluzzo 2002; Mata et al. 2003).
Schizophrenia has also been associated with creativity throughout recorded history (Claridge et al. 1990; Nettle 2001). Diverse evidence from psychological experiments, biographical survey studies and neurophysiology has been accumulating for many years in support of the hypothesis that schizotypal cognition is associated with creativity and divergent thinking (see reviews in Claridge et al. 1990; Nettle 2001, 2006a; Barrantes-Vidal 2004). An untested prediction of this hypothesis is that schizophrenia has evolved, and is maintained, in part as a maladaptive by-product of recent positive selection and adaptive evolution in humans (Crow 1997; Horrobin 1998).
To assess the hypothesis that genes associated with schizophrenia have been subject to positive selection, we analysed the molecular evolution of 76 genes, which have been inferred to increase schizophrenia risk. We used two complementary approaches. First, we used linkage disequilibrium-based methods to test for signatures of recent selective sweeps, which involve relatively large coding and non-coding genomic regions (extended haplotypes) inferred to have recently and rapidly increased in frequency in human ancestry, such that they have not yet been broken up by recombination (Biswas & Akey 2006; Sabeti et al. 2006; Voight et al. 2006). Second, we employed phylogeny-based maximum-likelihood methods (Yang 1997; Zhang et al. 2005) for detecting positive selection, in humans and related primates, which are based on the ratio of non-synonymous to synonymous substitution rates. We also review the literature from previous studies on the molecular evolution of schizophrenia-related genes along the human lineage.
2. Material and methods
(a) Criteria for inclusion of genes
Schizophrenia risk is mediated by many genes, each of small effect. The unambiguous identification of such genes has been difficult, because detecting small effects requires very large samples, different combinations of alleles can mediate liability, risk alleles often vary between populations and schizophrenia exhibits considerable clinical heterogeneity: it is not a simple, unitary disorder (Tamminga & Holcomb 2005; Riley & Kendler 2006). Our criterion for inclusion of schizophrenia-linked genes was genetic association with the disorder via association studies (comparing allele frequencies or genotype distributions between cases and controls), or via family-based transmission–disequilibrium studies that test for differential inheritance of alleles between affected and non-affected siblings. We excluded all genes that were linked with schizophrenia in single studies, which were subject to failed replication attempts. The list of 76 such genes used here (electronic supplementary material) is highly congruent with that of Schmidt-Kastner et al. (2006) and was fully assembled prior to our analyses.
The strength of the evidence for linkage of these genes to the development of schizophrenia varies considerably, and it is very likely that some proportion of these linkages will turn out to be false positives given the history of failed replications in this field and the high degree of clinical, genetic, gender and ethnic heterogeneity in this disorder. We appreciate this fact and, as this is the first analysis of its kind, we have chosen to initially be relatively permissive in the inclusion of genes. However, we are careful to highlight cases where evidence of selection is observed among genes for which especially strong and highly replicated association data exist.
We also note that the presence, in our analyses, of any genes that are eventually shown not to be associated with schizophrenia will make our test of enhanced positive selection on schizophrenia-associated genes (compared with control genes associated with neuronal activities) more, rather than less, conservative. Indeed, with regard to this test, the main issue at hand is whether or not the set of genes that has been statistically linked with schizophrenia in one or more studies is more likely to actually exhibit association with schizophrenia than is a random sample of control genes. We believe that this assumption is justified.
(b) Tests for positive selection using the human HapMap
Recent positive selection can usefully be inferred by the identification of selective sweeps, whereby selection for a specific allele causes a relatively large block of surrounding DNA (an extended haplotype) to increase in frequency. This method is most effective for recent selection, as recombination breaks up such extended haplotypes over time, and it can detect selection on both coding and non-coding regions.
Voight et al. (2006) have developed tests for recent selective sweeps in human populations using data from the human haplotype map. For the three genotyped populations, one European, one African and one Asian, they tested for evidence of positive selection as indicated by the tendency of recently selected alleles to sweep a set of tightly linked sites to relatively high frequency. They also provide a web interface, Haplotter (hg-wen.uchicago.edu/selection/haplotter.htm), which we used for testing the presence and significance of selective sweeps for specific genes and sets of contiguous genes, and for localizing inferences of selection to specific gene regions and SNPs. Our criterion for positive selection in these data was a probability value of 0.05 or lower for one or more of the three populations.
To test for a statistically increased signal of positive selection in schizophrenia-associated genes, we compared the frequency of positive selection in the set of 76 schizophrenia-associated genes with the frequency of positive selection in a set of 300 control genes that were derived from the Panther gene-ontology category ‘neuronal activities’ (Mi et al. 2005).
(c) PAML analyses
Analyses of historical selection on specific lineages, using the ratios of non-synonymous to synonymous amino acid substitutions for coding DNA, were carried out with the Codeml program in the PAML program package (Yang 1997; Zhang et al. 2005). This program implements maximum-likelihood methods to estimate the ratio of non-synonymous to synonymous substitution rates for each codon in a gene, across an aligned set of sequences from that gene in different species, in a phylogenetic framework (electronic supplementary material). For the analysis of each gene, we used estimates of the phylogenetic relationships of the specific taxa involved using recently published phylogenetic analyses of mammalian relationships from the literature. Only the topological relationship among the species involved was recorded for these trees. Initial branch-length estimates were determined using a preliminary Codeml run with the single ω model (M0).
We obtained orthologous mammalian gene sequences for each candidate human gene in the following manner. Orthology information was used from the Homologene database (release 50.1) at NCBI (http://www/ncbi.nih.gov/) where available. Additional mammalian orthologues were then obtained using reciprocal best BlastP hits from the GenBank protein database. Corresponding nucleotide sequences were obtained and in-frame ClustalW alignments of the coding regions were conducted. Aligned nucleotide sequences were then manually converted into PAML input files.
We used codon and branch-specific models (Zhang et al. 2005) to investigate the action of positive selection on each of the three branches in each tree: the human lineage; the human–chimpanzee lineage; and the basal-primate lineage, for each gene. In some cases, the primate lineage was the same as the human–chimpanzee lineage (i.e. no other primate sequence was available). These three lineages were chosen a priori because they represent periods when evolutionary changes underlying schizophrenia are likely to have been established: (i) at the origin of distinctive primate traits including increased encephalization and complex social behaviour, (ii) at the origin of large social-group sizes and complex communication in the lineage ancestral to chimpanzees and humans, and (iii) in the evolution of humans, the lineage expressing schizophrenia. The human lineage is most salient for analyses of positive selection in schizophrenia-associated genes, but the human–chimpanzee and primate lineage may also provide useful insights in that these lineages presumably underwent relatively strong selection for aspects of social cognition.
For the analyses of positive selection, we used ‘model A’ as implemented in Codeml, which detects selection on specific codons along specified branches of a phylogenetic tree (Zhang et al. 2005). Model A allows four categories of selection on codon sites in a sequence: two categories (ω1 and ω0) with uniform selection across all branches (ω1=1 and 0<ω0<1; estimated from the data) and two categories for which selection pressure differs in one or a few pre-specified ‘foreground’ branches where selection is assumed to have changed. The method finds the model with the maximum likelihood, given the data. If the maximum-likelihood model includes a category of sites with an ω>1, then this provides evidence that positive selection has acted on those sites along the specific lineage analysed. To test the significance of the results, the log likelihood of the maximum-likelihood model can be compared to similar models lacking selection using a log-likelihood ratio test (LRT). We compared the maximum-likelihood model to itself with ω constrained to equal one (i.e. a category of sites under positive selection was not allowed; MAofix test in tables 2 and 3). The asymptotic null distribution for the test is a 50 : 50 mixture of point mass 0 and Χ12 (Zhang et al. 2005). Simulations have shown that this test is ‘slightly too liberal’, so we also note the results for the Χ2-test with 1 d.f., which was demonstrated to be ‘overly conservative’ (Zhang et al. 2005). Our statistical tests using PAML are expected to be prone to false-negative (rather than false-positive) results, given the relatively small number of species that were available for analyses (mean of 7, range 5–15; see electronic supplementary material). For genes showing significant evidence for selection using the LRTs, we reran the analyses with varying starting values for the parameters κ (transition: transversion ratio) and ω, to check that we had not converged on a local likelihood peak. See electronic supplementary material for further explanation of PAML methods.
Table 2.
Significant maximum-likelihood results on the human lineage for genes associated with schizophrenia.
| gene | model | ω (proportion) | λ | −2Δλ | p |
|---|---|---|---|---|---|
| NRG1a (neuregulin 1) | MA | 999 (0.050) | −2010.46 | — | — |
| MAofix | −2012.50 | 4.07 | <0.05 | ||
| PLP1 (proteolipid protein 1) | MA | 482.65 (0.004) | −1630.56 | — | — |
| MAofix | −1636.11 | 11.08 | <0.01 | ||
| TNXB (tenascin XB) | MA | 999 (0.006) | −38 734.00 | — | — |
| MAofix | −38 745.00 | 21.90 | <0.01 | ||
| TP53 (tumour protein p53) | MA | 25.38 (0.007) | −5115.85 | — | — |
| MAofix | −5118.93 | 3.78 | <0.05 |
Association with schizophrenia is especially strong and well replicated. The PAML analyses also yielded a category of sites with an ω>1, providing evidence that positive selection may have acted on those sites along the specific lineage analysed, for a set of genes that did not show significance under the likelihood ratio tests. These genes, and the putatively selected lineages, include AHI1 (human–chimpanzee), AKT1 (human), CAPON (human–chimpanzee), CCK-AR (human), CHRM5 (human–chimpanzee), CNP (human–chimpanzee), CNTF (human), DRD4 (human–chimpanzee), DRP-2 (human), GABRP (human–chimpanzee), GAD1 (primate), HOPA (human), GPR50 (human), GRIN2A (human and human–chimpanzee), GRIN2B (primate), GSTM1 (human), MLC1 (human–chimpanzee), MTHFR (primate), NOTCH4 (human–chimpanzee), NT3N (human), PCM1 (human), PDLIM5 (human), PTGS2 (human), RGS4 (human–chimpanzee), SLC6A4 (primate), SNAP29 (human) and STOP (human–chimpanzee and primate). Table 3 shows the human–chimpanzee and primate branch results.
Table 3.
Significant maximum-likelihood results in the human and chimpanzee or primate lineages for genes associated with schizophrenia.
| gene | lineage | model | ω (proportion) | λ | −2Δλ | p |
|---|---|---|---|---|---|---|
| AHI1 (Abelson helper integration site 1) | primate | MA MAofix | 14.97 (0.024) | −3512.58−3514.60 | —4.02 | —<0.05 |
| CHRNA7 (cholinergic receptor neuronal nicotinic α7) | primate | MA MAofix | 17.56 (0.002) | −4098.03−4099.89 | —3.72 | —<0.05 |
| CLDN5 (claudin-5) | human–chimpanzee | MA MAofix | 22.91 (0.061) | −1371.13−1373.85 | —5.44 | —<0.05 |
| COMT (catechol-O-methyltransferase) | primate | MA MAofix | 41.58 (0.013) | −2914.90−2918.90 | —8.00 | —<0.01 |
| DISC1a (disrupted in schizophrenia 1) | human–chimpanzee | MA MAofix | 999 (0.006) | −7704.50−7709.90 | —10.88 | —<0.01 |
| DISC1a | primate | MA | 46.90 (0.032) | −7703.50 | — | — |
| MAofix | −7710.10 | 13.2 | <0.01 | |||
| NOTCH4 | primate | MA | 294.83 (0.006) | −16 523.92 | — | — |
| MAofix | −16 526.56 | 5.26 | <0.05 | |||
| PLXNA2 (plexin A2) | human–chimpanzee | MA | 999 (0.001) | −12 045.56 | — | — |
| MAofix | −12 049.12 | 7.13 | <0.05 |
Association with schizophrenia is especially strong and well replicated.
To test for a statistically increased signal of positive selection in schizophrenia-associated genes, we compared the frequency of positive selection in these 76 genes with the frequency of positive selection in 120 control genes, randomly chosen from the total set of 300 total control genes noted above. For these genes, the taxa analysed included human, chimpanzee, mouse, rat, macaque, orangutan, dog and cow. To test for any influence of gene length on the statistical power of the PAML analysis, we used the non-parametric Kolmogorov–Smirnov test to compare the distribution of gene lengths between the schizophrenia-associated gene and control gene datasets (following the removal of outliers using Tukey's outlier filter) and found no significant difference.
3. Results
(a) HapMap analyses of positive selection in recent human evolution
HapMap-based analyses provided evidence of recent positive selection on 14 of the genes associated with schizophrenia (table 1 and electronic supplementary material). For six of these genes, the signal of selection was localized specifically to the focal gene, and for three genes, only one adjacent gene also showed significance (p<0.05). Of particular interest is the identification of a particularly strong and localized focus of selection for DTNBP1, which exhibits one of the strongest genetic and functional associations with schizophrenia of any gene analysed to date (Riley & Kendler 2006).
Table 1.
Genes with significant evidence of selection in human populations.
| gene | population | p | localized signal (gene(s)) |
|---|---|---|---|
| APC (adenomatous polyposis of colon) | Asian | 0.021 | — |
| BRD1 (bromodomain-containing protein) | European | 0.013 | — |
| CAPON (nitric oxide synthase-1 neuronal) | European | 0.036 | 2 |
| CLDN5 (claudin-5) | European | 0.042 | — |
| DGCR2 (DiGeorge syndrome region gene 2) | Asian | 0.014 | 2 |
| DTNBP1a (dystrobrevin-binding protein 1) | European | 0.003 | 1 |
| DTNBP1a | African | 0.044 | 1 |
| ENTH (epsin 1) | European | 0.027 | 1 |
| ERBB4 (erythroblastic leukaemia oncogene homolog 4) | Asian | 0.044 | 1 |
| GABRA1 (γ-aminobutyric acid receptor) | European | 0.035 | 1 |
| GABRA1 | Asian | 0.050 | 1 |
| GRM3a (glutamate receptor 3) | Asian | 0.005 | 2 |
| PCM1 (pericentriolar material 1) | European | 0.049 | 1 |
| PCM1 | African | 0.044 | 1 |
| PIK3C3 (phosphatidylinositol 3-kinase) | African | 0.037 | 1 |
| SLC6A4 (serotonin transporter) | European | 0.035 | — |
| SLC6A4 | Asian | 0.014 | — |
| SYNGR1 (synaptogyrin) | Asian | 0.031 | — |
Association with schizophrenia is especially strong and well replicated. Two genes showing significance in the PAML analyses, NOTCH4 and TNXB, were marginally non-significant in Haplotter analyses (NOTCH4: 0.094 European; TNXB: 0.06 European), and three genes showed marginal non-significance in the Haplotter analysis (HOPA: 0.060 Asian; NOGO: 0.064 African; TPH1: 0.0505 Asian).
Of the 14 genes inferred as selected from the HapMap data, four displayed significant evidence for recent selection in more than one human population, while the rest were found to be significant in only one population. This is suggestive of extremely recent selection acting on these genes since the time of human demographic expansion out of Africa, and it is also generally consistent with the presence of variation among human populations in the genetic basis of schizophrenia (e.g. Bulayeva et al. 2007).
The proportion of schizophrenia genes inferred as selected in Haplotter (18.4%, 14 of 76) was significantly higher than the proportion of neuronal activities control genes inferred as selected (9.0%, 27 of 300, Χ2=5.52, p=0.019). Similar results were obtained when we restricted the analyses to the subset of schizophrenia genes (N=44) for which differences in gene expression levels or gene-associated neuroanatomy have been documented between schizophrenics and controls (electronic supplementary material): 9 (20%) of 44 positively selected compared with 27 (9%) of 300 for neuronal activities controls (Χ2=5.37, p=0.02).
These statistical comparisons with control genes indicate that schizophrenia-associated genes may show an enriched signal of positive selection in recent human evolution, and that the evidence of selection is often heterogeneous across human populations. Additionally, 12 genes, AHI1, CHL1, GRIA4, GRIN2A, GRIN2B, NOS-1, PDLIM5, PLA2G4A, RELN, SYN2, TNXB and TPH1, exhibited non-significance in gene-wide analyses using Haplotter, but had one or more single nucleotide polymorphisms that were inferred to have been subject to positive selection.
(b) PAML analyses of positive selection in primate evolution
PAML analyses yielded significant evidence of positive selection on four genes on the human lineage (table 2 and electronic supplementary material). Of particular interest is the identification of NRG1 (neuregulin 1) that shows notably strong genetic association with schizophrenia (Hall et al. 2006; Riley & Kendler 2006; Thomson et al. 2007). This gene shows an absence of evidence for selective sweeps in recent human evolution (Gardner et al. 2007), which suggests that the positive selection that we have inferred may have taken place earlier in the human lineage, such that selected haplotypes have been broken up by recombination. An additional seven genes displayed evidence of positive selection either in the human–chimpanzee lineage, in the ancestral primate lineage leading to humans, or in both lineages (table 3).
Overall, 11 (15%) of the 76 schizophrenia-associated genes showed evidence of positive selection in the PAML analyses (tables 2 and 3), compared with 10 (8.3%) of the 120 control genes analysed (Χ2=1.83, p=0.176). For the human lineage, a higher proportion, 4 (5.6%) of 76 of schizophrenia-associated genes, showed evidence of positive selection than did the control genes (none of 120; Fisher's exact test, p=0.022). However, there was no such difference for the human–chimpanzee lineage (with three positively selected control genes versus three positively selected schizophrenia-associated genes; Fisher's exact test, p>0.50), or for the primate-origin lineage (seven positively selected control genes versus five positively selected schizophrenia-associated genes; Fisher's exact test, p>0.50). If the overly conservative (Zhang et al. 2005) test using Χ2-test with 1 d.f. is used, then one of the schizophrenia-associated genes, TP53, shows marginally non-significant evidence of positive selection in the human lineage, such that three genes show positive selection in this lineage (Fisher's exact test, p=0.057).
(c) Positive selection of DISC1
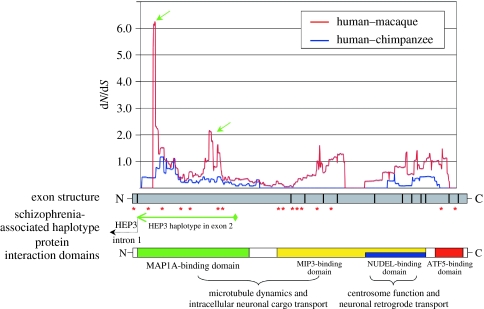
DISC1 (disrupted in schizophrenia), a gene with strong, well-replicated genetic ties to this disorder (Mackie et al. 2007), was inferred as positively selected in both the human–chimpanzee and primate-origin lineages. Remarkably, this gene showed 16 amino acid sites as having evolved under the influence of positive selection, of which six are within the HEP3 haplotype that segregates with schizophrenia (see figure 1 and the following; Porteous et al. 2006). To further investigate the DISC1 coding region in the HEP3 haplotype, we conducted a sliding-window analysis and found a localized peak of accelerated evolution in this region (figure 1). DISC1 thus evolved extremely rapidly during primate evolution, with significant evidence for acceleration in the only coding region of the gene showing reproducible evidence of a genetic association with schizophrenia.
Figure 1.
Sliding-window analysis of DISC1 (disrupted in schizophrenia 1). A sliding-window analysis of DISC1 between humans and other primates identified several localized peaks of highly accelerated evolution. The most pronounced peaks (green arrows) co-localize to the HEP3 haplotype region, a region reproducibly shown to associate with schizophrenia. The locations of positively selected amino acid residues are indicated by red asterisks. Of the 16 selected residues, six fall within the HEP3 haplotype. Schematic of protein interaction domains within DISC1 are provided (Morris et al. 2003). A window size of 180 bp and a 3 bp sliding increment were used in the analysis.
4. Discussion
Our results provide evidence for a significantly stronger signal of positive selection specific to the human lineage for schizophrenia-associated genes than for a control set of neuronal activities genes, for both the HapMap-based analyses of recent selective sweeps and the PAML analyses of positive selection. Furthermore, significant evidence of positive selection was found for several genes that exhibit some of the best-replicated and best-understood genetic and functional associations with this disorder, including DISC1, DTNBP1 and NRG1.
The primary, non-exclusive implications of these results are that schizophrenia may represent, in part, a maladaptive by-product of adaptive changes in human and primate evolution, and that some alleles increasing liability to the cognitive impairments associated with schizophrenia may have been selected against. These hypotheses can be evaluated by testing the same sets of polymorphisms for association with schizophrenia and for signatures of positive selection or other evolutionary-genetic processes (Mutsuddi et al. 2006). Such integrated evolutionary–genomic analyses will also provide a novel, powerful perspective into the functions of genes, alleles and neurological substrates that underlie the aetiology of schizophrenia and schizotypal cognition, since positively selected variants are expected to exhibit functional significance. Genetic and phenotypic changes along the human lineage are most directly salient to the evolutionary basis of schizophrenia, but changes earlier in primate evolution, as apparently exemplified by DISC1, may have generated some of the basic social-cognitive substrates that are dysregulated in this disorder.
Additional convergent evidence that selection has impacted the evolutionary underpinnings of schizophrenia comes from two sources. First, neurological studies have shown that brain areas differentially dysregulated in schizophrenia include the regions most-notably subject to differential evolutionary change along the human lineage (Randall 1998; Brüne 2004; Burns 2006). Second, Wayland et al. (2006) found that genes exhibiting positive selection for differential expression between humans and chimpanzees are differentially dysregulated in dorsolateral prefrontal and orbitofrontal cortices of individuals with schizophrenia (Wayland et al. 2006). These findings link recent evolutionary changes in human neuroanatomy and gene expression with alterations of these phenotypes in schizophrenia and demonstrate that the phenotype substrates of this disorder, as well as its genetic basis, show evidence of effects from positive selection and adaptive evolution.
(a) Evidence for positive selection from previous studies
Our maximum-likelihood protein-coding and haplotype-based analyses probe only for a subset of the possible signatures of selection on genes increasing liability to schizophrenia. Moreover, these methods detect signals over different time scales, and for different types of positively selected variants, such that the general absence of concordance of genes identified as selected between the two types of method (HapMap and PAML) is not unexpected. For example, the great majority of positively selected alleles in humans and other mammals are non-coding (Williamson et al. 2007; Resch et al. 2007), such that recent selective sweeps are expected to involve mainly non-coding regions that will be detected only by analyses based on HapMap data.
Given that selection involving only one or few amino acid sites, selective sweeps older than HapMap data can distinguish, or balancing selection that maintains human polymorphisms, may have influenced the genetic architecture of schizophrenia, we also reviewed the literature for evidence of selection or rapid evolution of genes associated with schizophrenia risk, from previous studies. Sixteen genes that exhibit genomic and functional links with schizophrenia, ADCYAP1, AHI1, APOE, CCR5, DRD4, DTNBP1, FOXP2, GABRB2, GRM3, HOPA, LRRTM1, MAOA, MCPH1, OLIG2, PDLIM5 and SLC6A4, also show evidence of positive or balancing selection in the human lineage from previous published studies, and five genes, CHRM5, GRIK4, ENTH, HLA-DRB and NPAS3, show evidence of rapid evolution (table 4 and electronic supplementary material). Furthermore, Voight et al. (2006) noted an unusual preponderance of phosphatidylinositol 3-kinase signal-transduction pathway genes subject to recent selective sweeps, and evidence from functional analyses, physiology, pharmacology and genetic-association studies indicates that dysregulated activation of this pathway (via effects of genes such as AKT1, DTNBP1, ENTH, ERBB4, NRG1, PIK3C3 and RELN) represents an important risk factor for schizophrenia (Kalkman 2006). Finally, the schizophrenia-related dopamine receptor genes DRD2, DRD3 and DRD4 exhibit heterosis in some of their effects on human physiology and behaviour (Comings & MacMurray 2000). These findings provide additional evidence for selection and rapid evolution of genes underlying schizophrenia, which is generally concordant with the results from PAML and HapMap analyses and localizes signatures of selection to the human lineage.
Table 4.
Evidence for selection and rapid evolution of genes associated with schizophrenia from previous studies.
| gene | evidence of selection | references |
|---|---|---|
| ADCYAP1 (adenylate cyclase-activating polypeptide 1) | positive selection | Bustamante et al. (2005); Wang et al. (2005); Hashimoto et al. (in press) |
| AHI1 (Abelson helper integration site 1) | rapid evolution; positive selection | Ferland et al. (2004); Amann-Zalcenstein et al. (2006) |
| APOE (apolipoprotein E) | positive and balancing selection | Fullerton et al. (2000); Finch & Stanford (2004); Xu et al. (2006) |
| CCR5 (chemokine, C–C motif, receptor 5) | balancing selection | Bamshad et al. (2002); Wooding et al. (2005); Geroldi et al. (2006); Rasmussen et al. (2006) |
| CHRM5 (cholinergic receptor muscarinic 5) | rapid evolution in primate compared with rodents | Dorus et al. (2004) |
| DRD4 (dopamine receptor D4) | selective sweep (approx. 40 000 yrs ago) | Ding et al. (2002); Lung et al. (2002); Jönsson et al. (2003a); Wang et al. (2004) |
| DTNBP1a (dystrobrevin-binding protein 1) | haplotype structure suggests positive selection | van den Oord et al. (2003); Williams et al. (2005); Riley & Kendler (2006); Tochigi et al. (2006) |
| ENTH (epsin 1) | nearest gene to human-accelerated genomic region | Pollard et al. (2006); Tang et al. (2006); Gurling et al. (2007) |
| FOXP2 (forkhead box P2) | positive selection | Enard et al. (2002); Zhang et al. (2002); Sanjuán et al. (2006) |
| GABRB2 (γ-aminobutyric acid receptor β2) | positive selection | Lo et al. (2007a,b) |
| GRIK4 (glutamate receptor ionotropic kainate 4) | rapid evolution in primate compared to rodents | Dorus et al. (2004); Pickard et al. (2006a) |
| GRM3 (glutamate receptor metabotropic 3) | selective sweep | Diller et al. (2002); Egan et al. (2004); Chen et al. (2005) |
| HLA-DRB (major histocompatibility complex, class II, DR β-5) | rapid evolution | Bergström et al. (1998); Wright et al. (2001); Hohjoh et al. (2003) |
| HOPA (mediator of RNA polymerase II transcription) | selective sweep; positive selection | DeLisi et al. (2000); Philibert et al. (2001, 2007); Spinks et al. (2004); Bustamante et al. (2005) |
| LRRTM1 (leucine-rich repeat transmembrane protein 1) | selective sweep | Voight et al. (2006); Laurén (2007); Francks et al. (in press) |
| MAOA (monoamine oxidase A) | selective sweep | Balciuniene et al. (2001); Gilad et al. (2002); Jönsson et al. (2003b); Andrés et al. (2004) |
| MCPH1 (microcephalin 1) | positive selection | Evans et al. (2004, 2005); Wang & Su (2004); Lencz et al. (2007) |
| NPAS3 (neuronal PAS domain protein 3) | nearest gene to human-accelerated genomic region | Pickard et al. (2006b); Pollard et al. (2006) |
| OLIG2 (oligodendrocyte transcription factor 2) | selective sweep | Wright et al. (2001); Diller et al. (2002); Georgieva et al. (2006) |
| PDLIM5 (PDZ and LIM domain 5) | positive selection | Bustamante et al. (2005); Kato et al. (2005); Horiuchi et al. (2006) |
| SLC6A4 (solute carrier family 6; serotonin transporter) | positive selection | Bustamante et al. (2005) |
Association with schizophrenia is especially strong and well replicated. Detailed information on evidence for selection, linkages to schizophrenia and cognitive effects of each gene is provided in the electronic supplementary material. Prabhakar et al. (2006) also reported evidence for accelerated evolution of CHL1, ERBB4, GRIK4, GRM8, LRRTM1 and QKI genes in the human lineage, involving human-specific substitutions in non-coding regions directly adjacent to these genes.
(b) Potential mechanisms mediating the evolution of genes that underlie schizophrenia
Positive selection of genes implicated in schizophrenia may be mediated by an array of neurodevelopmental, neurophysiological and psychological mechanisms. Our analyses provide complementary types of evidence for positive selection of genes underlying this disorder, but do not provide any direct information concerning how selective pressures at the phenotypic level may have impacted the evolution of these genes.
Evidence from previous work salient to the hypothesis that the adaptive evolution of genes underlying schizophrenia has involved phenotypic selection comes from studies of genetic linkages between schizophrenia-risk genes and aspects of creativity. Allelic variants of three genes associated with schizophrenia, SLC6A4, TPH1 and DRD2, have recently been associated with measures of creativity and imagination in normal population (Bachner-Melman et al. 2005; Reuter et al. 2006). Notably, the STin2.12 allele of an intronic polymorphism in the SLC6A4 gene and the A allele of the A779C polymorphism in the TPH1 gene are each associated with both increased creativity and increased risk of schizophrenia (Bachner-Melman et al. 2005; Reuter et al. 2006). Alleles of several genes associated with schizophrenia also influence measures of schizotypal cognition and ‘openness to experience’ in non-clinical populations, including COMT and HTR2A (Ott et al. 2005), MAOA (Samochowiec et al. 2004) and SLC6A4 (Golimbet et al. 2003). Description of the linkages of creativity and cognition with schizotypy and schizophrenia is provided for the four genes, SLC6A4, TPH1, COMT and DRD2, with sufficient integrative information available for detailed analysis (electronic supplementary material). We also describe here the neurological and psychological evidence that links schizotypy with aspects of creativity and divergent thinking, and epidemiological data for variation in measures of fitness between schizophrenics, first-order relatives and controls.
The mechanisms connecting schizotypal cognition and creativity with postulated fitness benefits are unclear, but may include sexual selection, creative and artistic skills, or general benefits from insight problem solving (Shaner et al. 2004; Nettle 2006a; Karimi et al. 2007). Such processes could potentially help to explain the paradoxical high heritability and persistence of schizophrenia, owing to some combination of multilocus balancing selection, antagonistic pleiotropy affecting fitness and mutation–selection balance.
We stress that enhanced creativity is not expected to be a function of schizophrenia itself (which involves profound cognitive impairments), but rather is inferred to be a function of schizotypal cognition (Claridge et al. 1990; Nettle 2001, 2006a,b; Barrantes-Vidal 2004). Indeed, alleles underlying the deleterious aspects of schizophrenia or schizotypal cognition are expected to be selected against (Keller & Miller 2006; Nettle 2006b). We also note that most studies of the cognitive correlates of genetic liability to schizophrenia have focused on documenting deficits in clinical populations, rather than testing for potentially positive aspects of cognition in healthy relatives or other individuals who carry schizophrenia-associated alleles or genotypes.
(c) Limitations and implications
Interpretation of our results is limited by the observation that few of the genes associated with increased schizophrenia risk are specific to this disorder: some such genes are also known to affect liability to bipolar disorder, and most of them influence a diverse range of neurodevelopmental and physiological processes, and exert only a small effect on schizophrenia risk that may vary among populations (Kendler 2005; Riley & Kendler 2006). Bipolar disorder and schizophrenia in particular exhibit substantial overlap in cognitive symptoms as well as their genetic basis, and links of bipolar disorder with creativity and imagination have also been repeatedly described (Claridge et al. 1990; Nettle 2001).
A second limitation of our study is that there remains considerable controversy about what constitutes adequate statistical demonstration of association between allelic variants of a specific gene and schizophrenia. Taking the most conservative approach on this question would result in too few genes for any meaningful analyses, whereas including all genes that have ever been linked with schizophrenia in a single study or population would be relatively liberal, although it would not be expected to bias analytic results towards a finding of an enhanced signal of positive selection, relative to control genes. We have taken a middle ground and used genes that have not been subject to failed replication attempts. However, it is important to note that significant evidence for selection was found in multiple genes showing the strongest, best-replicated associations with schizophrenia to date, including DISC1, DTNBP1 and NRG1. We have also repeated the HapMap analyses using only the 44 genes for which expression or gene-associated neuroanatomy differences have been recorded between schizophrenics and controls, and found the same results.
An important implication of our findings is that evolutionary–genomic analyses can provide novel insights into the nature and functions of the genes that underlie the aetiologies of schizophrenia. More generally, analyses that probe for signatures of selection on genes implicated in schizophrenia may localize functional sites affecting gene expression, enzyme activity, neurocognitive phenotypes and disease risk, and thus highlight potential allelic variants salient to the cognitive changes that have driven the origin of modern humans (Crow 1997; Horrobin 1998).
Acknowledgments
We thank F. Breden and B. Voight for advice and comments and four anonymous reviewers for their insightful suggestions. This work was funded by grants from NSERC and the Canada Council for the Arts to B.C., an ECU College Research Award to K.S. and an NIH Ruth L. Kirschstein National Research Service Award to S.D.
Supplementary Material
Table of genes analyzed and recent citations for association of these genes with schizophrenia
Detailed information for genes in Table 3 of main text, including information on gene function, evidence of selection or rapid evolution in the human lineage, associations with schizophrenia, and links with cognitive functions
Explanation of PAML methods, and complete results from PAML and Haplotter analyses, including control-gene results from PAML analyses
Sets of taxa analyzed for each gene in PAML analyses
Detailed information on linkages of schizotypy and schizophrenia with aspects of creativity, for the genes COMT, DRD2, SLC6A4 and TPH1
Neurological and physiological evidence relating schizophrenia and schizotypy to cognition
Epidemiological evidence on variation in correlates of fitness, between schizophrenics, first-order relatives, and controls
References
- Amann-Zalcenstein D, et al. AHI1, a pivotal neurodevelopmental gene, and C6orf217 are associated with susceptibility to schizophrenia. Eur. J. Hum. Genet. 2006;14:1111–1119. doi: 10.1038/sj.ejhg.5201675. doi:10.1038/sj.ejhg.5201675 [DOI] [PubMed] [Google Scholar]
- Andrés A.M, Soldevila M, Navarro A, Kidd K.K, Oliva B, Bertranpetit J. Positive selection in MAOA gene is human exclusive: determination of the putative amino acid change selected in the human lineage. Hum. Genet. 2004;115:377–386. doi: 10.1007/s00439-004-1179-6. doi:10.1007/s00439-004-1179-6 [DOI] [PubMed] [Google Scholar]
- Bachner-Melman R, et al. AVPR1a and SLC6A4 gene polymorphisms are associated with creative dance performance. PLoS Genet. 2005;1:e42. doi: 10.1371/journal.pgen.0010042. doi:10.1371/journal.pgen.0010042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balciuniene J, Syvanen A.C, McLeod H.L, Pettersson U, Jazin E.E. The geographic distribution of monoamine oxidase haplotypes supports a bottleneck during the dispersion of modern humans from Africa. J. Mol. Evol. 2001;52:157–163. doi: 10.1007/s002390010144. [DOI] [PubMed] [Google Scholar]
- Bamshad M.J, et al. A strong signature of balancing selection in the 5’ cis-regulatory region of CCR5. Proc. Natl Acad. Sci. USA. 2002;99:10 539–10 544. doi: 10.1073/pnas.162046399. doi:10.1073/pnas.162046399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrantes-Vidal N. Creativity & madness revisited from current psychological perspectives. J. Conscious. Stud. 2004;11:58–78. [Google Scholar]
- Bergström T.F, Josefsson A, Erlich H.A, Gyllensten U. Recent origin of HLA-DRB1 alleles and implications for human evolution. Nat. Genet. 1998;18:237–242. doi: 10.1038/ng0398-237. doi:10.1038/ng0398-237 [DOI] [PubMed] [Google Scholar]
- Biswas S, Akey J.M. Genomic insights into positive selection. Trends Genet. 2006;22:437–446. doi: 10.1016/j.tig.2006.06.005. doi:10.1016/j.tig.2006.06.005 [DOI] [PubMed] [Google Scholar]
- Brüne M. Schizophrenia—an evolutionary enigma? Neurosci. Biobehav. Rev. 2004;28:41–53. doi: 10.1016/j.neubiorev.2003.10.002. doi:10.1016/j.neubiorev.2003.10.002 [DOI] [PubMed] [Google Scholar]
- Bulayeva K.B, Glatt S.J, Bulayev O.A, Pavlova T.A, Tsuang M.T. Genome-wide linkage scan of schizophrenia: a cross-isolate study. Genomics. 2007;89:167–177. doi: 10.1016/j.ygeno.2006.10.001. doi:10.1016/j.ygeno.2006.10.001 [DOI] [PubMed] [Google Scholar]
- Burns J.K. Psychosis: a costly by-product of social brain evolution in Homo sapiens. Prog. Neuropsychopharmacol. Biol. Psych. 2006;30:797–814. doi: 10.1016/j.pnpbp.2006.01.006. doi:10.1016/j.pnpbp.2006.01.006 [DOI] [PubMed] [Google Scholar]
- Bustamante C.D, et al. Natural selection on protein-coding genes in the human genome. Nature. 2005;437:1153–1157. doi: 10.1038/nature04240. doi:10.1038/nature04240 [DOI] [PubMed] [Google Scholar]
- Chen Q, He G, Chen Q, Wu S, Xu Y, Feng G, Li Y, Wang L, He L. A case-control study of the relationship between the metabotropic glutamate receptor 3 gene and schizophrenia in the Chinese population. Schizophr. Res. 2005;73:21–26. doi: 10.1016/j.schres.2004.07.002. doi:10.1016/j.schres.2004.07.002 [DOI] [PubMed] [Google Scholar]
- Claridge G, Pryor R, Watkins G. The Macmollan Press Ltd; London, UK: 1990. Sounds from the bell jar: ten psychotic authors. [Google Scholar]
- Comings D.E, MacMurray J.P. Molecular heterosis: a review. Mol. Genet. Metab. 2000;71:19–31. doi: 10.1006/mgme.2000.3015. doi:10.1006/mgme.2000.3015 [DOI] [PubMed] [Google Scholar]
- Crow T.J. Is schizophrenia the price that Homo sapiens pays for language? Schizophr. Res. 1997;28:127–141. doi: 10.1016/s0920-9964(97)00110-2. doi:10.1016/S0920-9964(97)00110-2 [DOI] [PubMed] [Google Scholar]
- DeLisi L.E, Smith A.B, Razi K, Stewart J, Wang Z, Sandhu H.K, Philibert R.A. Investigation of a candidate gene for schizophrenia on Xq13 previously associated with mental retardation and hypothyroidism. Am. J. Med. Genet. 2000;96:398–403. doi: 10.1002/1096-8628(20000612)96:3<398::aid-ajmg30>3.0.co;2-z. doi:10.1002/1096-8628(20000612)96:3<398::AID-AJMG30>3.0.CO;2-Z [DOI] [PubMed] [Google Scholar]
- Diller K.C, Gilbert W.A, Kocher T.D. Selective sweeps in the human genome: a starting point for identifying genetic differences between modern humans and chimpanzees. Mol. Biol. Evol. 2002;19:2342–2345. doi: 10.1093/oxfordjournals.molbev.a004061. [DOI] [PubMed] [Google Scholar]
- Ding Y, et al. Evidence of positive selection acting at the human dopamine receptor D4 gene locus. Proc. Natl Acad. Sci. USA. 2002;99:309–314. doi: 10.1073/pnas.012464099. doi:10.1073/pnas.012464099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorus S, Vallender E, Evans P, Anderson J, Gilbert S, Mahowald M, Wyckoff G, Malcom C, Lahn B. Accelerated evolution of the nervous system genes in the origin of Homo sapiens. Cell. 2004;119:1027–1040. doi: 10.1016/j.cell.2004.11.040. doi:10.1016/j.cell.2004.11.040 [DOI] [PubMed] [Google Scholar]
- Egan M.F, et al. Variation in GRM3 affects cognition, prefrontal glutamate, and risk for schizophrenia. Proc. Natl Acad. Sci. USA. 2004;101:12 604–12 609. doi: 10.1073/pnas.0405077101. doi:10.1073/pnas.0405077101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enard W, Przeworski M, Fisher S.E, Lai C.S.L, Wiebe V, Kitano T, Monaco A.P, Pääbo S. Molecular evolution of FOXP2, a gene involved in speech and language. Nature. 2002;418:869–872. doi: 10.1038/nature01025. doi:10.1038/nature01025 [DOI] [PubMed] [Google Scholar]
- Evans P.D, Anderson J.R, Vallender E.J, Choi S.S, Lahn B.T. Reconstructing the evolutionary history of microcephalin, a gene controlling human brain size. Hum. Mol. Genet. 2004;13:1139–1145. doi: 10.1093/hmg/ddh126. doi:10.1093/hmg/ddh126 [DOI] [PubMed] [Google Scholar]
- Evans P.D, Gilbert S.L, Mekel-Bobrov N, Vallender E.J, Anderson J.R, Vaez-Azizi L.M, Tishkoff S.A, Hudson R.R, Lahn B.T. Microcephalin, a gene regulating brain size, continues to evolve adaptively in humans. Science. 2005;309:1717–1720. doi: 10.1126/science.1113722. doi:10.1126/science.1113722 [DOI] [PubMed] [Google Scholar]
- Ferland R.J, et al. Abnormal cerebellar development and axonal decussation due to mutations in AHI1 in Joubert syndrome. Nat. Genet. 2004;36:1008–1013. doi: 10.1038/ng1419. doi:10.1038/ng1419 [DOI] [PubMed] [Google Scholar]
- Finch C.E, Stanford C.B. Meat-adaptive genes and the evolution of slower aging in humans. Q. Rev. Biol. 2004;79:3–50. doi: 10.1086/381662. doi:10.1086/381662 [DOI] [PubMed] [Google Scholar]
- Francks, C. et al In press. LRRTM1 on chromosome 2p12 is a maternally suppressed gene that is associated paternally with handedness and schizophrenia. Mol. Psych. [DOI] [PMC free article] [PubMed]
- Fullerton S.M, et al. Apolipoprotein E variation at the sequence haplotype level: implications for the origin and maintenance of a major human polymorphism. Am. J. Hum. Genet. 2000;67:881–900. doi: 10.1086/303070. doi:10.1086/303070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner M, Williamson S, Casals F, Bosch E, Navarro A, Calafell F, Bertranpetit J, Comas D. Extreme individual marker F(ST) values do not imply population-specific selection in humans: the NRG1 example. Hum. Genet. 2007;121:759–762. doi: 10.1007/s00439-007-0364-9. doi:10.1007/s00439-007-0364-9 [DOI] [PubMed] [Google Scholar]
- Georgieva L, et al. Convergent evidence that oligodendrocyte lineage transcription factor 2 (OLIG2) and interacting genes influence susceptibility to schizophrenia. Proc. Natl Acad. Sci. USA. 2006;103:12 469–12 474. doi: 10.1073/pnas.0603029103. doi:10.1073/pnas.0603029103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geroldi D, Emanuele E, Martinelli V, Pesenti S. Schizophrenia and cancer risk: can the CCR5-delta 32 chemokine receptor gene mutation play a role? Med. Hypotheses. 2006;67:427–428. doi: 10.1016/j.mehy.2006.03.008. doi:10.1016/j.mehy.2006.03.008 [DOI] [PubMed] [Google Scholar]
- Gilad Y, Rosenberg S, Przeworski M, Lancet D, Skorecki K. Evidence for positive selection and population structure at the human MAO-A gene. Proc. Natl Acad. Sci. USA. 2002;99:862–867. doi: 10.1073/pnas.022614799. doi:10.1073/pnas.022614799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golimbet V.E, Alfimova M.V, Shcherbatikh T, Kaleda V.G, Abramova L.I, Rogaev E.I. Serotonin transporter gene polymorphism and schizoid personality traits in the patients with psychosis and psychiatrically well subjects. World J. Biol. Psych. 2003;4:25–29. doi: 10.3109/15622970309167907. [DOI] [PubMed] [Google Scholar]
- Gurling H, Pimm J, McQuillin A. Replication of genetic association studies between markers at the epsin 4 gene locus and schizophrenia in two Han Chinese samples. Schizophr. Res. 2007;89:357–359. doi: 10.1016/j.schres.2006.08.024. doi:10.1016/j.schres.2006.08.024 [DOI] [PubMed] [Google Scholar]
- Hall J, et al. A neuregulin 1 variant associated with abnormal cortical function and psychotic symptoms. Nat. Neurosci. 2006;9:1477–1478. doi: 10.1038/nn1795. doi:10.1038/nn1795 [DOI] [PubMed] [Google Scholar]
- Hashimoto, R. et al In press. Pituitary adenylate cyclase-activating polypeptide is associated with schizophrenia. Mol. Psych. [DOI] [PubMed]
- Hohjoh H, Ohashi J, Takasu M, Nishioka T, Ishida T, Tokunaga K. Recent divergence of the HLA-DRB1*04 allelic lineage from the DRB1*0701 lineage after the separation of the human and chimpanzee species. Immunogenetics. 2003;54:856–861. doi: 10.1007/s00251-003-0539-z. [DOI] [PubMed] [Google Scholar]
- Horiuchi Y, et al. A polymorphism in the PDLIM5 gene associated with gene expression and schizophrenia. Biol. Psych. 2006;59:434–439. doi: 10.1016/j.biopsych.2005.07.041. doi:10.1016/j.biopsych.2005.07.041 [DOI] [PubMed] [Google Scholar]
- Horrobin D.F. Schizophrenia: the illness that made us human. Med. Hypotheses. 1998;50:269–288. doi: 10.1016/s0306-9877(98)90000-7. doi:10.1016/S0306-9877(98)90000-7 [DOI] [PubMed] [Google Scholar]
- Jönsson E.G, Sedvall G.C, Nöthen M.M, Cichon S. Dopamine D4 receptor gene (DRD4) variants and schizophrenia: Meta-analyses. Schizophr. Res. 2003a;61:111–119. doi: 10.1016/s0920-9964(02)00287-6. doi:10.1016/S0920-9964(02)00287-6 [DOI] [PubMed] [Google Scholar]
- Jönsson E.G, Norton N, Forslund K, Mattila-Evenden M, Rylander G, Asberg M, Owen M.J, Sedvall G.C. Association between a promoter variant in the monoamine oxidase A gene and schizophrenia. Schizophr. Res. 2003b;61:31–37. doi: 10.1016/s0920-9964(02)00224-4. doi:10.1016/S0920-9964(02)00224-4 [DOI] [PubMed] [Google Scholar]
- Kalkman H.O. The role of the phosphatidylinositide 3-kinase-protein kinase B pathway in schizophrenia. Pharmacol. Therapy. 2006;110:117–134. doi: 10.1016/j.pharmthera.2005.10.014. doi:10.1016/j.pharmthera.2005.10.014 [DOI] [PubMed] [Google Scholar]
- Karimi Z, Windmann S, Gunturkun O, Abraham A. Insight problem solving in individuals with high versus low schizotypy. J. Res. Pers. 2007;41:473–480. doi:10.1016/j.jrp.2006.03.008 [Google Scholar]
- Kato T, et al. Gene expression and association analyses of LIM (PDLIM5) in bipolar disorder and schizophrenia. Mol. Psych. 2005;10:1045–1055. doi: 10.1038/sj.mp.4001719. doi:10.1038/sj.mp.4001719 [DOI] [PubMed] [Google Scholar]
- Keller M.C, Miller G. Resolving the paradox of common, harmful, heritable mental disorders: which evolutionary genetic models work best? Behav. Brain Sci. 2006;29:385–404. doi: 10.1017/S0140525X06009095. doi:10.1017/S0140525X06009095 [DOI] [PubMed] [Google Scholar]
- Kendler K.S. “A gene for…”: the nature of gene action in psychiatric disorders. Am. J. Psych. 2005;162:1243–1252. doi: 10.1176/appi.ajp.162.7.1243. doi:10.1176/appi.ajp.162.7.1243 [DOI] [PubMed] [Google Scholar]
- Laurén, J. 2007 Characterization of LRRTM and NGR gene families: expression and functions. PhD dissertation, University of Helsinki. See https://oa.doria.fi/bitstream/handle/10024/5795/characte.pdf?sequence=1.
- Lencz T, Burdick K.E, DeRosse P, Morgan T.V, Kane J.M, Kucherlapati R, Malhotra A.K. MCPH1 is associated with risk for illness and cognitive ability in patients with schizophrenia. Biol. Psych. 2007;61(Suppl. 190):609. [Google Scholar]
- Lo W.S, et al. Positive selection within the schizophrenia-associated GABA(A) receptor beta(2) gene. PLoS. 2007a;2:e462. doi: 10.1371/journal.pone.0000462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo W.S, et al. GABRB2 association with schizophrenia: commonalities and differences between ethnic groups and clinical subtypes. Biol. Psych. 2007b;61:653–660. doi: 10.1016/j.biopsych.2006.05.003. doi:10.1016/j.biopsych.2006.05.003 [DOI] [PubMed] [Google Scholar]
- Lung F, Tzeng D, Shu B. Ethnic heterogeneity in allele variation in the DRD4 gene in schizophrenia. Schizophr. Res. 2002;57:239–245. doi: 10.1016/s0920-9964(01)00313-9. doi:10.1016/S0920-9964(01)00313-9 [DOI] [PubMed] [Google Scholar]
- Mackie S, Millar J.K, Porteous D.J. Role of DISC1 in neural development and schizophrenia. Curr. Opin. Neurobiol. 2007;17:95–102. doi: 10.1016/j.conb.2007.01.007. doi:10.1016/j.conb.2007.01.007 [DOI] [PubMed] [Google Scholar]
- Mata I, Gilvarry C.M, Jones P.B, Lewis S.W, Murray R.M, Sham P.C. Schizotypal personality traits in nonpsychotic relatives are associated with positive symptoms in psychotic probands. Schizophr. Bull. 2003;29:273–283. doi: 10.1093/oxfordjournals.schbul.a007004. [DOI] [PubMed] [Google Scholar]
- Mi H, et al. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res. 2005;33:D284–D288. doi: 10.1093/nar/gki078. doi:10.1093/nar/gki078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J.A, Kandpal G, Ma L, Austin C.P. DISC1 is a centrosome-associated protein that interacts with MAP1A, MIPT3, ATF4/5 and NUDEL: regulation and loss of interaction with mutation. Hum. Mol. Genet. 2003;12:1591–1608. doi: 10.1093/hmg/ddg162. doi:10.1093/hmg/ddg162 [DOI] [PubMed] [Google Scholar]
- Mutsuddi M, Morris D.W, Waggoner S.G, Daly M.J, Scolnick E.M, Sklar P. Analysis of high-resolution HapMap of DTNBP1 (dysbindin) suggests no consistency between reported common variant associations and schizophrenia. Am. J. Hum. Genet. 2006;79:903–909. doi: 10.1086/508942. doi:10.1086/508942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nettle D. Oxford University Press; Oxford, UK: 2001. Strong imagination: madness, creativity and human nature. [Google Scholar]
- Nettle D. Schizotypy and mental health amongst poets, visual artists, and mathematicians. J. Res. Pers. 2006a;40:876–890. doi:10.1016/j.jrp.2005.09.004 [Google Scholar]
- Nettle D. Reconciling the mutation–selection balance model with the schizotypy–creativity connection. Behav. Brain Sci. 2006b;29:418. doi:10.1017/S0140525X06359092 [Google Scholar]
- Ott U, Reuter M, Hennig J, Vaitl D. Evidence for a common biological basis of the absorption trait, hallucinogen effects, and positive symptoms: epistasis between 5-HT2a and COMT polymorphisms. Am. J. Med. Genet. B Neuropsych. Genet. 2005;137:29–32. doi: 10.1002/ajmg.b.30197. doi:10.1002/ajmg.b.30197 [DOI] [PubMed] [Google Scholar]
- Philibert R.A, Sandhu H.K, Hutton A.M, Wang Z, Arndt S, Andreasen N.C, Crowe R, Wassink T.H. Population-based association analyses of the HOPA12bp polymorphism for schizophrenia and hypothyroidism. Am. J. Med. Genet. 2001;105:130–134. doi:10.1002/1096-8628(20010108)105:1<130::AID-AJMG1076>3.0.CO;2-P [PubMed] [Google Scholar]
- Philibert R.A, Bohle P, Secrest D, Deaderick J, Sandhu H, Crowe R, Black D.W. The association of the HOPA (12bp) polymorphism with schizophrenia in the NIMH genetics initiative for schizophrenia sample. Am. J. Med. Genet. B Neuropsych. Genet. 2007;144:743–747. doi: 10.1002/ajmg.b.30489. doi:10.1002/ajmg.b.30489 [DOI] [PubMed] [Google Scholar]
- Pickard B.S, et al. Cytogenetic and genetic evidence supports a role for the kainate-type glutamate receptor gene, GRIK4, in schizophrenia and bipolar disorder. Mol. Psych. 2006a;11:847–857. doi: 10.1038/sj.mp.4001867. doi:10.1038/sj.mp.4001867 [DOI] [PubMed] [Google Scholar]
- Pickard B.S, Pieper A.A, Porteous D.J, Blackwood D.H, Muir W.J. The NPAS3 gene—emerging evidence for a role in psychiatric illness. Ann. Med. 2006b;38:439–448. doi: 10.1080/07853890600946500. doi:10.1080/07853890600946500 [DOI] [PubMed] [Google Scholar]
- Pollard K.S, et al. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443:167–172. doi: 10.1038/nature05113. doi:10.1038/nature05113 [DOI] [PubMed] [Google Scholar]
- Porteous D.J, Thomson P, Brandon N.J, Millar J.K. The genetics and biology of DISC1—an emerging role in psychosis and cognition. Biol. Psych. 2006;60:123–131. doi: 10.1016/j.biopsych.2006.04.008. doi:10.1016/j.biopsych.2006.04.008 [DOI] [PubMed] [Google Scholar]
- Prabhakar S, Noonan J.P, Paabo S, Rubin E.M. Accelerated evolution of conserved noncoding sequences in humans. Science. 2006;314:786. doi: 10.1126/science.1130738. doi:10.1126/science.1130738 [DOI] [PubMed] [Google Scholar]
- Randall P.L. Schizophrenia as a consequence of brain evolution. Schizophr. Res. 1998;30:143–148. doi: 10.1016/s0920-9964(97)00143-6. doi:10.1016/S0920-9964(97)00143-6 [DOI] [PubMed] [Google Scholar]
- Rasmussen H.B, Timm S, Wang A.G, Søeby K, Lublin H, Fenger M, Hemmingsen R, Werge T. Association between the CCR5 32-bp deletion allele and late onset of schizophrenia. Am. J. Psych. 2006;163:507–511. doi: 10.1176/appi.ajp.163.3.507. doi:10.1176/appi.ajp.163.3.507 [DOI] [PubMed] [Google Scholar]
- Resch A.M, Carmel L, Mariño-Ramírez L, Ogurtsov A.Y, Shabalina S.A, Rogozin I.B, Koonin E.V. Widespread positive selection in synonymous sites of mammalian genes. Mol. Biol. Evol. 2007;24:1821–1831. doi: 10.1093/molbev/msm100. doi:10.1093/molbev/msm100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter M, Roth S, Holve K, Hennig J. Identification of first candidate genes for creativity: a pilot study. Brain Res. 2006;1069:190–197. doi: 10.1016/j.brainres.2005.11.046. doi:10.1016/j.brainres.2005.11.046 [DOI] [PubMed] [Google Scholar]
- Riley B, Kendler K.S. Molecular genetic studies of schizophrenia. Eur. J. Hum. Genet. 2006;14:669–680. doi: 10.1038/sj.ejhg.5201571. doi:10.1038/sj.ejhg.5201571 [DOI] [PubMed] [Google Scholar]
- Rossi A, Daneluzzo E. Schizotypal dimensions in normals and schizophrenic patients: a comparison with other clinical samples. Schizophr. Res. 2002;54:67–75. doi: 10.1016/s0920-9964(01)00353-x. doi:10.1016/S0920-9964(01)00353-X [DOI] [PubMed] [Google Scholar]
- Sabeti P.C, et al. Positive natural selection in the human lineage. Science. 2006;312:1614–1620. doi: 10.1126/science.1124309. doi:10.1126/science.1124309 [DOI] [PubMed] [Google Scholar]
- Samochowiec J, Syrek S, Michal P, Ryzewska-Wódecka A, Samochowiec A, Horodnicki J, Zakrzewska M, Kucharska-Mazur J. Polymorphisms in the serotonin transporter and monoamine oxidase A genes and their relationship to personality traits measured by the temperament and character inventory and NEO five-factor inventory in healthy volunteers. Neuropsychobiology. 2004;50:174–181. doi: 10.1159/000079111. doi:10.1159/000079111 [DOI] [PubMed] [Google Scholar]
- Sanjuán J, Tolosa A, González J.C, Aguilar E.J, Pérez-Tur J, Nájera C, Moltó M.D, de Frutos R. Association between FOXP2 polymorphisms and schizophrenia with auditory hallucinations. Psych. Genet. 2006;16:67–72. doi: 10.1097/01.ypg.0000185029.35558.bb. doi:10.1097/01.ypg.0000185029.35558.bb [DOI] [PubMed] [Google Scholar]
- Schmidt-Kastner R, van Os J.W.M, Steinbusch H, Schmitz C. Gene regulation by hypoxia and the neurodevelopmental origin of schizophrenia. Schizophr. Res. 2006;84:253–271. doi: 10.1016/j.schres.2006.02.022. doi:10.1016/j.schres.2006.02.022 [DOI] [PubMed] [Google Scholar]
- Shaner A, Miller G, Mintz J. Schizophrenia as one extreme of a sexually selected fitness indicator. Schizophr. Res. 2004;70:101–109. doi: 10.1016/j.schres.2003.09.014. doi:10.1016/j.schres.2003.09.014 [DOI] [PubMed] [Google Scholar]
- Spinks R, Sandhu H.K, Andreasen N.C, Philibert R.A. Association of the HOPA12bp allele with a large X-chromosome haplotype and positive symptom schizophrenia. Am. J. Med. Genet. B Neuropsych. Genet. 2004;127:20–27. doi: 10.1002/ajmg.b.20175. doi:10.1002/ajmg.b.20175 [DOI] [PubMed] [Google Scholar]
- Tamminga C.A, Holcomb H.H. Phenotype of schizophrenia: A review and formulation. Mol. Psych. 2005;10:27–39. doi: 10.1038/sj.mp.4001563. doi:10.1038/sj.mp.4001563 [DOI] [PubMed] [Google Scholar]
- Tang R.Q, et al. Family-based association study of epsin 4 and schizophrenia. Mol. Psych. 2006;11:395–399. doi: 10.1038/sj.mp.4001780. doi:10.1038/sj.mp.4001780 [DOI] [PubMed] [Google Scholar]
- Thomson P.A, et al. Association of neuregulin 1 with schizophrenia and bipolar disorder in a second cohort from the Scottish population. Mol. Psych. 2007;12:94–104. doi: 10.1038/sj.mp.4001889. doi:10.1038/sj.mp.4001889 [DOI] [PubMed] [Google Scholar]
- Tochigi M, et al. Association study of the dysbindin (DTNBP1) gene in schizophrenia from the Japanese population. Neurosci. Res. 2006;56:154–158. doi: 10.1016/j.neures.2006.06.009. doi:10.1016/j.neures.2006.06.009 [DOI] [PubMed] [Google Scholar]
- van den Oord E.J.C.G, Sullivan P.F, Jiang Y, Walsh D, O'Neill F.A, Kendler K.S, Riley B.P. Identification of a high-risk haplotype for the dystrobrevin binding protein 1 (DTNBP1) gene in the Irish study of high-density schizophrenia families. Mol. Psych. 2003;8:499–510. doi: 10.1038/sj.mp.4001263. doi:10.1038/sj.mp.4001263 [DOI] [PubMed] [Google Scholar]
- Voight B.F, Kudaravalli S, Wen X, Pritchard J.K. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. doi:10.1371/journal.pbio.0040072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.Q, Su B. Molecular evolution of microcephalin, a gene determining human brain size. Hum. Mol. Genet. 2004;13:1131–1137. doi: 10.1093/hmg/ddh127. doi:10.1093/hmg/ddh127 [DOI] [PubMed] [Google Scholar]
- Wang E, et al. The genetic architecture of selection at the human dopamine receptor D4 (DRD4) gene locus. Am. J. Hum. Genet. 2004;74:931–944. doi: 10.1086/420854. doi:10.1086/420854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.Q, et al. Accelerated evolution of the pituitary adenylate cyclase-activating polypeptide precursor gene during human origin. Genetics. 2005;170:801–806. doi: 10.1534/genetics.105.040527. doi:10.1534/genetics.105.040527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayland M, Khaitovich P, Ryan M, Paabo S, Bahn S. The molecular basis of cognitive impairment on schizophrenia: evidence from comparative transcriptomics. Schizophr. Res. 2006;81:s17. doi:10.1016/j.schres.2005.09.016 [Google Scholar]
- Williams N.M, O'Donovan M.C, Owen M.J. Is the dysbindin gene (DTNBP1) a susceptibility gene for schizophrenia? Schizophr. Bull. 2005;31:800–805. doi: 10.1093/schbul/sbi061. doi:10.1093/schbul/sbi061 [DOI] [PubMed] [Google Scholar]
- Williamson S.H, Hubisz M.J, Clark A.G, Payseur B.A, Bustamante C.D, Nielsen R. Localizing recent adaptive evolution in the human genome. PLoS Genet. 2007;3:e90. doi: 10.1371/journal.pgen.0030090. doi:10.1371/journal.pgen.0030090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooding S, Stone A.C, Dunn D.M, Mummidi S, Jorde L.B, Weiss R.K, Ahuja S, Bamshad M.J. Contrasting effects of natural selection on human and chimpanzee CC chemokine receptor 5. Am. J. Hum. Genet. 2005;76:f291–f301. doi: 10.1086/427927. doi:10.1086/427927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright P, Nimgaonkar V.L, Donaldson P.T, Murray R.M. Schizophrenia and HLA: a review. Schizophr. Res. 2001;47:1–12. doi: 10.1016/s0920-9964(00)00022-0. doi:10.1016/S0920-9964(00)00022-0 [DOI] [PubMed] [Google Scholar]
- Xu M.Q, St Clair D, He L. Meta-analysis of association between ApoE epsilon4 allele and schizophrenia. Schizophr. Res. 2006;84:228–235. doi: 10.1016/j.schres.2006.02.015. doi:10.1016/j.schres.2006.02.015 [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Zhang J, Webb D.M, Podlaha O. Accelerated protein evolution and origins of human-specific features: Foxp2 as an example. Genetics. 2002;162:1825–1835. doi: 10.1093/genetics/162.4.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.Z, Nielsen R, Yang Z.H. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol. Biol. Evol. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. doi:10.1093/molbev/msi237 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table of genes analyzed and recent citations for association of these genes with schizophrenia
Detailed information for genes in Table 3 of main text, including information on gene function, evidence of selection or rapid evolution in the human lineage, associations with schizophrenia, and links with cognitive functions
Explanation of PAML methods, and complete results from PAML and Haplotter analyses, including control-gene results from PAML analyses
Sets of taxa analyzed for each gene in PAML analyses
Detailed information on linkages of schizotypy and schizophrenia with aspects of creativity, for the genes COMT, DRD2, SLC6A4 and TPH1
Neurological and physiological evidence relating schizophrenia and schizotypy to cognition
Epidemiological evidence on variation in correlates of fitness, between schizophrenics, first-order relatives, and controls