Abstract
The level of survivin was reported to be scarce in mouse megakaryocytes (MKs) compared with erythroid cells. Considering this finding and previously reported in vitro data showing decreased MK ploidy upon retroviral-mediated overexpression of survivin, we sought to examine whether ectopic survivin expression in the MK lineage might alter ploidy level in vivo. Here we report the generation of 2 tissue specific hematopoietic transgenic mouse models, one expressing survivin in both the erythroid and MK lineages and the other expressing survivin solely in the MK lineage. Survivin protein overexpression was confirmed in MKs and erythrocytes. Surprisingly, analysis of both transgenic mouse lines showed no detectable changes in MK number, ploidy level, and platelet and erythrocyte counts, as compared with control mice. We conclude that elevated survivin expression does not alter MK/erythroid lineage development and that elevated survivin, alone, does not interfere with MK ploidy in vivo.
Introduction
The megakaryocyte (MK) is a unique hematopoietic cell that undergoes multiple rounds of polyploidization in which the cellular DNA content can reach up to 128N, with 2N being normal diploid content. The process of polyploidization in MKs is thought to play an important role in supporting the large increase in size of mature MKs, which are capable of producing thousands of platelets upon fragmentation,1 however, much of this process remains enigmatic
Recent studies have implicated a member of the chromosome passenger complex, survivin, as being mislocalized or down-regulated at the protein or mRNA level during mitosis of murine MKs of high ploidy.2,3 In contrast, protein levels and localization of survivin was reported to be typical during mitosis of human megakaryocytes (which, in culture, are typically of low ploidy).4 Retroviral overexpression of survivin in vitro decreased murine MK ploidy level and number of MK colonies in MK colony assays in comparison to untreated MKs.3 Similarly, primary vascular smooth muscle cells, which naturally undergo polyploidization in culture, were also shown to decrease their ploidy level upon survivin overexpression.5
Lately, an increasing number of studies have identified survivin expression in normal adult cells.3,6,7 Leung et al7 recently reported on the requirement of survivin in terminal differentiation of erythroid cells and maintenance of hematopoietic stem and progenitor cells. While survivin expression has been demonstrated to be essential in various hematopoietic cell types, the effect of its ectopic expression has not yet been analyzed in vivo. We report the establishment of 2 tissue-specific hematopoietic survivin transgenic models, one overexpressing survivin in both the erythroid and megakaryocyte lineages, and the other solely in the megakaryocytic lineage.
Methods
Transgenic mouse production, MK purification, flow cytometry, and MK colony assay
Detailed procedures are outlined in Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Reverse transcriptase–polymerase chain reaction of transgenic mRNA
Total RNA was harvested from mouse bone marrow cells using TRIzol reagent, according to the manufacturer's instructions (Invitrogen, Carlsbad, CA). Transgenic mRNA in total bone marrow was detected via reverse-transcriptase polymerase chain reaction (RT-PCR) as previously described.8
β-galactosidase assay
Visualization of β-galactosidase transgene activity in MKs was performed as previously described8 using freshly isolated bone marrow (BM) from wild-type and PF4-surv mice.
Western blot analysis
Western blotting was performed as previously described.8 Primary antibodies used were anti-survivin, anti-CD41, anti-HSC70 (Santa Cruz Biotechnology, Santa Cruz, CA; sc-10811, sc-15328, and sc-7298, respectively), and anti–β-actin (Sigma-Aldrich, St Louis, MO; A5441).
Acetylcholinesterase assay
MK number was calculated based on a positive signal resulting from MK-specific acetylcholinesterase activity as previously reported.2 Slides were counterstained with 4′,6-diamidino-2-phenylindole and inspected under a light microscope.
MK ploidy analysis
Ploidy profile analyses were performed as previously reported by Nguyen et al8 and Muntean et al.9
Platelet counts and hematocrits
Blood was collected via heart puncture, and platelet counts were performed using the BD Unopette collection system (Becton Dickinson, Franklin Lakes, NJ) and a Nebauer hemacytometer. For GATA-1-surv animals and nontransgenic littermates, 50 μL of blood was collected from the tail vein and analyzed on a HemaVet 850 complete blood counter.
Results and discussion
Generation of 2 survivin transgenic mouse models
To explore survivin's role in MK polyploidization in vivo, we created a transgenic mouse using the tetracycline/doxycycline Tet-Off system. We used the previously established transactivator mouse line (PF4-tTA), which expresses the Tet transactivator element (tTA-VP16) under the control of the MK-specific platelet factor 4 promoter (PF4).8 To use this system, we generated a responder line (termed TRE-surv) with a bi-directional Tet-responsive minimal cytomegalovirus promoter (TRE-bidirec-mCMV) driving a mouse survivin cDNA, and the prokaryotic β-galactosidase gene as a cellular reporter (Figure S1A). Establishment of the transgenic lines was confirmed by Southern blot and PCR genotyping (Figure S1C-E). Double transgenic mice, containing the PF4-tTA and the TRE-surv transgenes, were used for analysis and are termed PF4-surv mice.
Independently, to examine the effects of survivin overexpression on erythroid progenitor cells and polyploidizing MKs, a separate survivin transgenic mouse model was created using the GATA-1 promoter to drive expression of a human survivin cDNA (Figure S1B).2,3 This promoter has been previously employed to highly express transgenes specifically in these 2 cell lineages.10,11 Transgenic founders harboring the survivin transgene were genotyped as determined by PCR of tail DNA and are referred as GATA-1-surv mice (Figure S1B,F).
Expression of transgenes
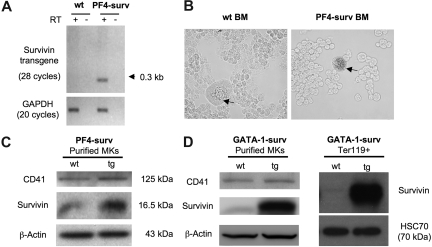
Expression of the PF4-surv transgenic transcript was confirmed by RT-PCR using total RNA isolated from femoral BM. No transgenic transcript mRNA was detected in wild-type mice (Figure 1A). Lineage specificity of expression of transgenes was confirmed by 2 experiments. First, β-galactosidase activity in freshly isolated transgenic and wild-type BM revealed nominal blue β-galactosidase staining in wild-type MKs, while transgenic MKs showed dark staining (Figure 1B). Secondly, overexpression of survivin protein in purified BM MKs in both mouse models is depicted via Western blot in Figure 1C and D. In addition, a Western blot of Ter119 + erythrocytes displayed highly expressed survivin in erythrocytes of GATA-1-surv transgenic animals (Figure 1D).
Figure 1.
Transgene expression in PF4-surv and GATA-1-surv MKs and GATA-1-surv erythrocytes. (A) RT-PCR of the survivin transgene on RNA isolated from fresh BM of PF4-surv mice confirms survivin transgene expression. A single line of transgenic mice produced an abundant spliced transcript PCR product of about 300 bp in comparison to an unspliced size of 1.125 kb. Survivin transgene primers: sense 5′-cagccaccactttctgatag-3′ and antisense 5′-aaactggacagacagagagccaag-3′; GAPDH loading control primers: sense 5′-tcaccatcttccaggag-3′ and antisense 5′-gcttcaccaccttcttg-3′. (B) β-galactosidase transgene activity (blue) was highly detected in MKs from PF4-surv BM but not in other BM cells nor in control mouse BM, demonstrating lineage specificity of expression. Staining was performed in suspension in fixed cells before cells were cytospun and viewed via light microscopy. (C) Western blotting of magnetic bead-purified PF4-surv and wild-type (wt) MKs. MK marker, CD41, and β-actin were used as loading controls. (D) Western blotting of magnetic bead–purified GATA-1-surv MKs and MoFlo-sorted Ter119 + erythrocytes. β-actin and HSC70 were used as loading controls. Band quantitation using ImageJ 1.34s software (National Institutes of Health, Bethesda, MD) indicated an increase in transgenic survivin level over control in the range of 2- to 3-fold in PF4-suv MKs and 5- to 6-fold in GATA-1-surv MKs.
Effect of survivin overexpression on MK and erythroid cell development
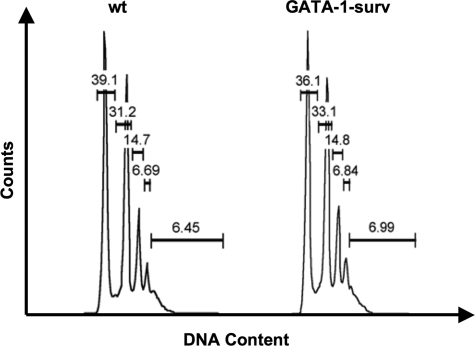
Intriguingly, although we previously reported that survivin mRNA and protein is much lower in murine MK maturation in comparison to erythroid development,2,3 here we found that in vivo survivin overexpression in MKs does not impact MK number or polyploidy level in either transgenic mouse model (Figures 2,S2A,B, Tables 1,2). Blood platelet counts confirmed normal platelet levels in control mice and PF4-surv and GATA-1-surv transgenic animals (Table 3). Further analysis revealed that ectopic survivin expression did not influence MK, GR1/Mac myeloid, erythroid, and c-kit–positive progenitor cell populations isolated from GATA-1-surv and wild-type mouse BM and spleens (Figure S2A,B). Moreover, analysis of the erythroid cells by CD71/Ter119 staining (Figure S2A,B) demonstrates that increased survivin expression does not alter erythrocyte development nor hematocrit levels (Table 4). This finding agrees with our previous observation that overexpression of survivin did not affect erythroid burst-forming unit (BFU-E) colony formation.3
Figure 2.
Analysis of MK ploidy status of GATA-1-surv mice. Representative MK ploidy profiles of wild-type (wt) and GATA-1-surv mice. Average MK ploidy displayed no significant change between wild-type and transgenic mice (n = 5 mice per group). Differences in ploidy profiles between GATA-1-surv MKs and PF4-surv mice (Table 2) could be due to the different mouse strains (FVB and CD-1; see Document S1).
Table 1.
Change in MK number in PF4-surv transgenic mice
| Genotype | MK number as percent of wt | P |
|---|---|---|
| wt | 4100 | — |
| tg | 98.90 | .915 |
Wild-type (wt) and transgenic (tg) PF4-surv MKs in fresh bone marrow cytospins were quantified via positive acetylcholinesterase staining. 375 wild-type MKs and 436 tg MKs were counted and normalized to cell density as calculated using Image-Pro software (Media Cybernetics, Silver Spring, MD) and quantitation of the 4′,6-diamidino-2-phenylindole counterstain signal. The number of MKs in the tg animals is presented as a percentage of the wild-type control. A 2-tailed Student t test was used to generate a P value, indicating no difference between the 2 groups (n = 3 mice per group). — indicates not applicable.
Table 2.
Summary of ploidy status of fresh PF4-surv MKs
| Ploidy | Percent of wt MKs (± SD) | Percent of tg MKs (± SD) |
|---|---|---|
| 2n | 9.975 ± 1.83 | 8.935 ± 2.88 |
| 4n | 8.035 ± 1.96 | 6.91 ± 1.70 |
| 8n | 23.07 ± 0.95 | 21.21 ± 1.74 |
| 16n | 47.025 ± 0.47 | 51.13 ± 3.86 |
| 32n | 6.67 ± 2.38 | 7.835 ± 2.71 |
MK ploidy profiles from wild-type (wt) and transgenic (tg) PF4-surv mice. Fresh BM was stained with CD41-FITC antibody followed by propidium iodide staining for MK ploidy analysis via FACS. Means (± SD) represent analysis from 3 independent experiments (n = 5 mice per group).
Table 3.
Platelet counts in wild-type and survivin transgenic mice
| Genotype | Number (n) | Platelets, 1000/μL (± SD) |
|---|---|---|
| wt | 6 | 1483.3 ± 147.6 |
| PF4-surv | 5 | 1503.6 ± 138.9 |
| wt | 7 | 1028.5 ± 186.1 |
| GATA-1-surv | 7 | 1085.7 ± 152.4 |
Platelet counts in wild-type (wt) and survivin transgenic models. Platelet counts are in thousands per microliter (± SD). Note differing platelet counts between the 2 models may also reflect strain differences.
Table 4.
Summary of hematocrits (%) for GATA-1-surv mice
| Age, mo |
|||||||
|---|---|---|---|---|---|---|---|
| 2 | 3 | 4 | 6 | 8 | 10 | 12 | |
| wt | 48 ± 3.3 | 49.5 ± 3.8 | 42.6 ± 4.7 | 44.3 ± 2.6 | 40.9 ± 9.6 | 44.1 ± 2.7 | 42.4 ± 3.0 |
| tg | 47.2 ± 2.4 | 45.1 ± 3.7 | 44.3 ± 3.2 | 44.6 ± 2.6 | 42.4 ± 4.5 | 42.2 ± 2.5 | 43.2 ± 3.4 |
| P | 0.59 | 0.05 | 0.47 | 0.88 | 0.73 | 0.21 | 0.65 |
Summary of mean hematocrit (%; ± SD) for GATA-1-surv transgenic (tg) and wild-type (wt) mice over the course of 1 year (n = 7 mice per group). No statistical differences were found between the 2 groups except at the 3-month time point, which displayed a statistical significance of P = .0485 (unpaired, 2-tailed Student t test). Overall, we interpreted these results to demonstrate that survivin overexpression in GATA-1-surv mice had no effect on hematocrit levels.
Our results show that despite the requirement of survivin by hematopoietic progenitor cells for maintenance and differentiation,3,7 increased survivin expression in erythrocytes and MKs is not sufficient to alter lineage commitment nor ploidy level of MKs in vivo. Interestingly, results from MK colony assays with GATA-1-surv BM replicated the in vitro study,3 demonstrating a significant decrease in MK number and colony size in comparison to wild-type BM (Figure S3). This finding highlights survivin's surprising ability to exert this effect on MK development only when cultured ex vivo and accentuates the importance of its in vivo study. Additionally, although we cannot exclude the possibility that our in vivo expression models do not express survivin as highly as observed with in vitro retroviral expression, the MK colony assay validates the high level of survivin expression achieved in the mouse model. We argue that the observed survivin expression levels in the 2 different transgenic models (a range of 2- to 6-fold over control) are in line with those achieved in other cell-targeted survivin transgenic mouse models, in which effects were observed on melanocytes and keratinocytes.12,13 Our results exemplify the complexity of regulation in vivo and suggests that elevated survivin expression alone may not be sufficient to influence detectable changes in MK and erythroid development in vivo. These findings further clarify the role of survivin in MK/erythroid development with respect to its overexpression in vivo.
Supplementary Material
Acknowledgments
We thank Greg Martin and Robin MacDonald at the Boston University School of Medicine Transgenic Facility.
This work was supported by National Heart, Lung, and Blood Institute grant HL80442 (K.R.) and National Institute of Diabetes and Digestive and Kidney Diseases grant DK074693 (J.D.C.). J.D.C. is a Scholar of the Leukemia and Lymphoma Society. K.R. is an Established Investigator with the American Heart Association.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Author contribution: D.J.M. generated the PF4-surv mice and carried out all the related experiments, participated in analysis of expression in GATA-1-surv transgenic mice, and participated in writing the paper along with K.R. and J.D.C.; T.Y. generated and analyzed the GATA-1-surv mice; H.G.N. generated the PF4-tTA transgenic line and participated in generation and initial analysis of the TRE-surv founder mice; N.P. participated in analysis of transgene expression and cell preparation; H.L. and Q.W. analyzed the hematopoietic profile in GATA-1-surv mice; J.D.C. designed and supervised the GATA-1-surv model and oversaw its analysis; and K.R. designed and directed the generation and analysis the PF4-surv model.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Katya Ravid, Department of Biochemistry, 15 Stoughton St, K225, Boston University School of Medicine, Boston, MA 02118; e-mail: ravid@biochem.bumc.bu.edu; or John D. Crispino, Department of Medicine, Feinberg School of Medicine, 303 East Superior St, Lurie 5-113, Northwestern University, Chicago, IL 60611; e-mail: j-crispino@northwestern.edu.
References
- 1.Ravid K, Lu J, Zimmet JM, Jones MR. Roads to polyploidy: the megakaryocyte example. J Cell Physiol. 2002;190:7–20. doi: 10.1002/jcp.10035. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y, Nagata Y, Yu G, et al. Aberrant quantity and localization of Aurora-B/AIM-1 and survivin during megakaryocyte polyploidization and the consequences of Aurora-B/AIM-1-deregulated expression. Blood. 2004;103:3717–3726. doi: 10.1182/blood-2003-09-3365. [DOI] [PubMed] [Google Scholar]
- 3.Gurbuxani S, Xu Y, Keerthivasan G, Wickrema A, Crispino JD. Differential requirements for survivin in hematopoietic cell development. Proc Natl Acad Sci U S A. 2005;102:11480–11485. doi: 10.1073/pnas.0500303102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geddis AE, Kaushansky K. Megakaryocytes express functional Aurora-B kinase in endomitosis. Blood. 2004;104:1017–1024. doi: 10.1182/blood-2004-02-0419. [DOI] [PubMed] [Google Scholar]
- 5.Nagata Y, Jones MR, Nguyen HG, et al. Vascular smooth muscle cell polyploidization involves changes in chromosome passenger proteins and an endomitotic cell cycle. Exp Cell Res. 2005;305:277–291. doi: 10.1016/j.yexcr.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 6.Xu YG, Zhou SH, Li YG, et al. The mechanism underlying vascular smooth muscle cell apoptosis induced by atorvastatin may be mainly associated with down-regulation of survivin expression. Cardiovasc Drugs Ther. 2007;21:145–153. doi: 10.1007/s10557-007-6018-2. [DOI] [PubMed] [Google Scholar]
- 7.Leung CG, Xu Y, Mularski B, Liu H, Gurbuxani S, Crispino JD. Requirements for survivin in terminal differentiation of erythroid cells and maintenance of hematopoietic stem and progenitor cells. J Exp Med. 2007;204:1603–1611. doi: 10.1084/jem.20062395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nguyen HG, Yu G, Makitalo M, et al. Conditional overexpression of transgenes in megakaryocytes and platelets in vivo. Blood. 2005;106:1559–1564. doi: 10.1182/blood-2005-02-0638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muntean AG, Pang L, Poncz M, Dowdy SF, Blobel GA, Crispino JD. Cyclin D-Cdk4 is regulated by GATA-1 and required for megakaryocyte growth and polyploidization. Blood. 2007;109:5199–5207. doi: 10.1182/blood-2006-11-059378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Visvader JE, Fujiwara Y, Orkin SH. Unsuspected role for the T-cell leukemia protein SCL/tal-1 in vascular development. Genes Dev. 1998;12:473–479. doi: 10.1101/gad.12.4.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hock H, Meade E, Medeiros S, et al. Tel/Etv6 is an essential and selective regulator of adult hematopoietic stem cell survival. Genes Dev. 2004;18:2336–2341. doi: 10.1101/gad.1239604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomas J, Liu T, Cotter MA, et al. Melanocyte expression of survivin promotes development and metastasis of UV-induced melanoma in HGF-transgenic mice. Cancer Res. 2007;67:5172–5178. doi: 10.1158/0008-5472.CAN-06-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grossman D, Kim PJ, Blanc-Brude OP, et al. Transgenic expression of survivin in keratinocytes counteracts UVB-induced apoptosis and cooperates with loss of p53. J Clin Invest. 2001;108:991–999. doi: 10.1172/JCI13345. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.