Abstract
An optimally effective AIDS vaccine would likely require the induction of both neutralizing antibody and cell-mediated immune responses, which has proven difficult to obtain in previous clinical trials. Here we report the induction of Human Immunodeficiency Virus Type-1 (HIV-1)-specific immune responses in healthy adult volunteers that received the multi-gene, polyvalent, DNA prime-protein boost HIV-1 vaccine formulation, DP6−001 in a Phase I clinical trial conducted in healthy adult volunteers of both genders. Robust cross-subtype HIV-1-specific T cell responses were detected in IFNγ ELISPOT assays. Furthermore, we detected high titer serum antibody responses that recognized a wide range of primary HIV-1 Env antigens and also neutralized pseudotyped viruses that express the primary Env antigens from multiple HIV-1 subtypes. These findings demonstrate that the DNA prime-protein boost approach is an effective immunization method to elicit both humoral and cell-mediated immune responses in humans, and that a polyvalent Env formulation could generate broad immune responses against HIV-1 viruses with diverse genetic backgrounds.
Keywords: HIV-1, DNA vaccine, recombinant protein vaccine, Phase I clinical trial, prime-boost
1. Introduction
Development of an effective HIV vaccine is critical to control the worldwide AIDS pandemic, which has caused 25 million deaths in the last 25 years and is the cause for more than 40 million people living with HIV/AIDS today [1]. Early efforts in HIV vaccine development focused on the induction of humoral responses by using recombinant Env glycoproteins [2-5]. The immunogenicity of recombinant Env protein-based vaccines was poor in humans, as shown by overall low-level binding antibodies measured by solid phase assays [6] and by the narrow spectrum of neutralizing activities mainly against T-cell line adapted (TCLA) viral isolates [7-9]. Ultimately, recombinant protein-based HIV-1 vaccines failed to show protection efficacy in Phase III clinical trials [10, 11]. In contrast, recent progress with gene-based vaccination approaches, which have used either DNA or viral vectors as delivery systems, have been effective in eliciting cell-mediated immune (CMI) responses in early phase human studies. However, these studies either did not put forth an effort to elicit protective antibody responses [12, 13] or were not effective, when used alone, in eliciting neutralizing antibodies (NAbs) against even relatively sensitive viral isolates [14, 15].
Recently, we demonstrated that a DNA prime-protein boost immunization strategy was effective in eliciting humoral and CMI responses in both small animals and non-human primates, including sterilizing immunity in a non-pathogenic SHIV model [16-18]. Our preclinical study results also indicated that this combination vaccination approach, but not recombinant protein alone, was effective in eliciting NAbs against primary HIV isolates [19], a finding that has been confirmed subsequently by other independent studies [20-26]. Furthermore, when polyvalent primary Env antigen formulations were used, the DNA prime-protein boost approach was more effective than the monovalent primary Env antigen in eliciting rabbit NAbs against a wide range of selected primary viral isolates across subtypes A, B, C, D and E [27]. In the current study, a multi-gene, polyvalent DNA prime-protein boost HIV-1 vaccine was formulated based on the above preclinical study findings, and its immunogenicity was tested in healthy adults in a Phase I clinical trial. These results demonstrate that this formulation was able to induce balanced cell-mediated and antibody immune responses against HIV-1 antigens, including low but positive neutralizing activities against selected primary HIV-1 isolates across different subtypes.
2. Materials and methods
2.1. Multi-gene, polyvalent, DNA prime-protein boost formulation DP6−001
2.1.a. DNA vaccines
The DP6−001 vaccine contains equal amounts of six individual DNA plasmid components utilizing the same vector pSW3891 [17]: five plasmids each encoding a codon-optimized gp120 gene sequence from the following primary HIV-1 Envelope proteins: subtypes A (92UG037.8), B (92US715.6 and Bal), C (96ZM651) and E (93TH976.17) and the sixth plasmid encoding a codon-optimized gag gene from subtype C (96ZM651) as previously described [28]. The cGMP plasmid DNA for this Phase I clinical trial was produced by Althea Technology (San Diego, CA). The 6 DNA plasmids used in the DP6−001 vaccine formulation was supplied in saline at a final concentration of 3 mg/ml (0.5 mg/ml/each DNA plasmid).
2.1.b. Protein Vaccines
The recombinant Env protein vaccine components included in the DP6−001 formulation contain equal amounts of five gp120 proteins matching that used in DNA prime components and were produced in CHO cell lines by Advanced BioScience Laboratories (ABL, Kensington, MD) using GMP compliance as previously described [28]. The final protein vaccine product was supplied in saline and re-formulated at the time of injection with 50μg of QS-21 adjuvant (Antigenics Inc., Woburn, MA) and 30 mg of excipient cyclodextrin (Cargill Cerestar USA Inc., Hammond, IN), to reach a final concentration of 0.375 mg/ml of five gp120 proteins (0.075 mg/ml/each protein) [28].
2.2. Plasma, sera and antibodies
The HIV human hyperimmune immunoglobulin (HIVIG), HIV positive patient plasma 91BU003 (infected with a clade C virus) and 93BR029 (infected with a clade B virus) were received from NIH AIDS Research & Reference Reagent Program. Pooled HIV-1 patient sera (infected with clade B viruses) were received from Center for AIDS Researches at UMass Medical School. Normal human sera were purchased from Sigma-Aldrich (St. Louis, MO).
2.3. Phase I clinical study design and sample collection
2.3.a. Participants
Healthy HIV-1-negative adult volunteers aged 18−50 years of both genders were screened. The individuals enrolled in this Phase I trial had no history of chronic, allergic and immunodeficient illnesses, organ transplantions or psychiatric disorder, were negative in Hepatitis B and C viral tests and a pregnancy test for all female subjects was negative. All subjects were recruited at the single clinical trial site at the University of Massachusetts Medical School (UMMS), Worcester, MA according to IRB approved study protocol.
2.3.b. Study design and immunization schedule
This open-label Phase I trial involved 2 dose levels of DNA prime and a single dose level of protein boost. The study design and numbers of volunteers included in the current analysis are provided in Table 1. Volunteers were randomly assigned to either Group A or B (1.2 mg of DNA at each immunization) at first and enrollment to Group C was started only after the safety review on Group A and B volunteers who have received the second protein boost. DNA vaccine was administrated by intradermal (ID) injection at 4 sites (0.3 mg in 0.1 ml per site) in Group A and by intramuscular (IM) injection at 2 sites (0.6 mg in 0.2 ml per site) in Group B. Group C received a 6-fold higher dose of the DNA vaccine (7.2 mg at each immunization) via IM injection at 2 sites (3.6 mg in 1.2 ml per site). Each volunteer received three priming vaccinations of DNA vaccines at Study Weeks 0, 4, and 12 and two booster immunizations of protein vaccinations via single site IM injection at Study Weeks 20 and 28 (Table 1). The adjuvant QS-21 and excipient cyclodextrin were mixed with the five gp120 proteins in a total volume of 1 ml at the time of injection.
Table 1.
Study design of DP6−001 clinical trial
| DNA Prime Phase | Protein Boost Phase | |||||||
|---|---|---|---|---|---|---|---|---|
| Group | Plasmids (6) | Dose | Route | Time (weeks) | Proteins (5) | Dose | Route | Time (weeks) |
| A (n = 10) |
gp120 (A, B, Bal, C, E) + Gag (C) | 1.2 mg | ID | 0, 4, 12 | gp120 (A, B, Bal, C, E) | 0.375 mg | IM | 20, 28 |
| B (n = 11) |
gp120 (A, B, Bal, C, E) + Gag (C) | 1.2 mg | IM | 0, 4, 12 | gp120 (A, B, Bal, C, E) | 0.375 mg | IM | 20, 28 |
| C (n = 6) |
gp120 (A, B, Bal, C, E) + Gag (C) | 7.2 mg | IM | 0, 4, 12 | gp120 (A, B, Bal, C, E) | 0.375 mg | IM | 20 |
Serum and PBMC samples were collected at Study Weeks 0, 2, 4, 6, 12, 14, 16, 20, 22, 24, 28, 30, 32, 36 and 52 to measure antibody and CMI responses. All volunteers were recruited and enrolled in the Clinical Vaccine Research Unit, UMMS. For the current immunogenicity report, samples from the following 27 volunteers are included: Group A (n=10) and Group B (n=11) volunteers who completed the entire 3 DNA and 2 protein immunizations and Group C volunteers (n=6) who received the 3 DNA and 1 protein immunization (Table 1).
2.4. ELISPOT
The interferon (IFN)-γ ELISPOT assay for HIV-1 peptide-specific T cells used the human IFN-γ ELISPOT kit from Mabtech (Cat # 3420−2) (Cincinnati, OH) with Millipore (Billerica, MA) ELISPOT plates, MSIPS4W10. The assay was performed according to manufacturers' directions with the minor modifications that monoclonal antibody, 1-D1K, was diluted to 5μg/ml and biotinylated monoclonal antibody, 7-B6−1, was diluted to 2ug/ml. The ELISPOT assay was performed using pools of peptides covering gp120 of HIV-1 92UG037.8 (Env-A), 92US715.6 (Env-B), 96ZM651 (Env-C) and 93TH976.17 (Env-E) or Gag of 96ZM651 that are present in the vaccine formulation. These peptides, 20-mer in length and overlapping by 10 amino acids, were produced in the UMMS Peptide Core Facility. Each peptide pool consisted of 5 to 7 peptides with a final concentration of 2 ug/ml per peptide. Plates were developed using the Vector Nova Red Kit (SK-4800, Vector labs, Burlingame, CA).
Cryopreserved PBMCs were thawed, washed and diluted to 2×105 / well in RPMI 10 (10% fetal bovine serum, 2 mM L-glutamine, 50ug/ml streptomycin and 50 U/ml penicillin). Wells containing media alone served as a negative control and wells containing 1 μg/ml phytohemagglutinin (PHA) served as a positive control. A CEF peptide pool containing defined epitopes of cytomegalovirus, Epstein-Barr virus and influenza A virus was used as a peptide positive control [30]. PBMC from HIV-1-infected donors and healthy control donors (with a known positive response to the CEF peptide pool) were included in each assay to assess reproducibility. Selected samples were repeated to test inter-assay variation. The final IFNγ ELISPOT responses against Env or Gag peptides of each individual antigen were calculated as the cumulative response across non-overlapping peptide pools. Spots were counted and analyzed on a CTL immunospot 3 reader and recorded as the mean spot-forming cells (SFC) per million PBMCs of replicate or triplicate wells. The final numbers of peptide-specific SFC were obtained by subtracting the background spots in medium control wells. The cut-off for positive responses was determined as at least 10 peptide-specific SFC per million PBMCs.
2.5. ELISA
Enzyme-linked immunosorbent assay (ELISA) was used to detect the gp120-specific IgG responses. The gp120 antigens used in the ELISA assays were 5 proportionally pre-mixed gp120 antigens included in the DP6−001 formulation produced in CHO cells with a purity of >99% based on size exclusion HPLC. Microtiter plates were coated with the 5 mixed gp120 antigens at 100ng/well (20ng/antigen) in PBS (100μL) at 4°C for 1 hour; plates were then washed and blocked in 200μL blocking buffer (PBS, 0.5% Tween-20, 5% NGS, 5% non-fat dry milk) overnight at 4°C. On the following day, serum dilutions were prepared in Dilsim II (BioMerieux, Durham, NC) and incubated on the plates (100μL/well) for 1 hour at room temperature. Biotinylated goat anti-human IgG (Vector Laboratories, CA) was diluted to 1:5000 in Dilsim II and then incubated on the plates (100μL/well) for 1 hour at room temperature. Horseradish peroxidase-streptavidin (Vector Laboratories, CA) diluted to 1:10000 in Dilsim II was added (100μL/well) to the plates and incubated for 1 hour at room temperature. Between steps, the plates were washed 5 times with 1× PBS-0.1% Triton X-100. The assays were developed with 3, 3’, 5, 5’-tetramethylbenzidine (TMB) substrate (Sigma, St. Louis, MO) and stopped with sulfuric acid after 3 minutes. Assays were read immediately at 450 and 630 nanometers using the Opsys MR Microplate Reader (Dynex Technologies, Chantilly, VA). The gp120-specific antibody titer was determined as the highest serum dilution which achieved 2 fold higher OD value than the pre-bleed control for each subject.
2.6. Western blot
The individual gp120 antigens used in the Western blot analysis were obtained from different sources: A, B, Bal, C, E and 92BR025.9 from ABL as the same reagents used for ELISA; JR-FL, ADA, SF162, CN54 and CM235 from NIH AIDS Reagent Program; CA1 and UG21−9 from NIH/NIAID; 92UG021, 93BR020.17 and 92UG975.10 produced from 293T cells at UMMS. Individual gp120 antigens (100ng/lane) were subjected to SDS-PAGE and blotted onto PVDF membrane, as previous described [27]. Blocking of PVDF membrane was done with 0.1% I-Block (Tropix, Bedford, MA). The membranes were incubated with sera at 1:100 dilution for 45 min and subsequently reacted with AP-conjugated goat anti-human IgG (Tropix, Bedford, MA) at 1:5000 dilution for 30 min. Membranes were washed with blocking buffer after each step. Western-light substrate was then applied to the membranes for 5 min. Once the membranes were dried, X-ray films were exposed to the membrane and developed by a Kodak processor. Individual HIV+ patient sera or purified HIV+ immunoglobulin (HIV-IG) were used as controls.
2.7. Neutralizing antibody assays
One virus neutralization assay was based on reductions in luciferase (Luc) reporter gene expression after a single round of virus infection with pseudotyped HIV-1 viruses in TZM-bl cells, as previously described [31]. Neutralizing antibody levels in the human sera from DP6−001 vaccines and normal control donors were measured against 2 panels of pseudotyped HIV-1 viruses expressing primary Envs antigens. There were 6 pseudotyped HIV-1 viruses in the first panel (Tier-1) including MN and five homologous to the Env immunogens included in the DP6−001 formulation. There were 12 viruses in the second panel (Tier 2), 4 each from subtypes B (QH0692042, SC422661.8; PVO.4 and AC10.0.29), A (Q23.17, Q168.A2, Q461.E2 and Q769.D22), and C (Du123.6, Du151.2, Du156.12 and Du172.17) [32, 33]. In this assay, 200 TCID50 of virus was incubated with diluted serum samples in triplicate in a total volume of 150μl for 1 h at 37 °C in 96-well flat-bottom culture plates. Freshly trypsinized cells (10,000 cells in 100μl of growth medium containing 75μg/ml DEAE dextram) were added to each well. One set of control wells received cells plus virus (virus control) and another set received cells only (background control). After 48 h incubation, 100μl of cells was transferred to a 96-well black solid plate (Costar) for measurements of luminescence using Bright Glo substrate solution, as described by the supplier (Promega). The percent neutralization was calculated by comparing experimental wells to virus control wells. Neutralization titer was the dilution at which RLUs were reduced by 50% compared to virus control wells after subtraction of background RLUs using prebleed sera.
The second neutralization assay (PhenoSense Assay) used recombinant viruses pseudotyped with the virus envelope proteins and a firefly luciferase indicator gene [34]. The pseudoviruses were incubated for 18 h at 37 °C with serial 3-fold dilutions of heat-inactivated human sera. U87 cells that express CD4 plus the CCR5 and CXCR4 co-receptors were inoculated with virus dilutions in the absence of added cations. Virus infectivity was determined 3 days later by measuring the amount of luciferase activity expressed in infected cells. Neutralizing activity was calculated as the percent inhibition of viral replication (luciferase activity) at each antibody dilution compared with an antibody-negative control: % inhibition = {1 – [luciferase + Ab/luciferase – Ab]} × 100. Titers were presented as the reciprocal of the plasma dilution conferring 50% inhibition (IC50) [34]. The specificity control was composed of a virus pseudotyped with an aMuLV envelope. An HIV-serum combination was considered to have positive neutralization if the inhibition of HIV was at least 50% and >3X higher IC50 than the same plasmas tested with aMuLV while the prebleed was not scored positive. The starting sera dilution used in the neutralization assays was 1:20.
2.8. Statistical analysis
Wilcoxon rank sum test was used to analyze the differences of HIV-1 antigen-specific T cell ELISPOT and antibody responses between vaccination groups. Student T test was used to analyze the difference of T cell ELISPOT results between low and high dose DNA prime (Groups A/B vs. Group C).
3. RESULTS
3.1. Design of the Phase I clinical trial in healthy adult volunteers
The multi-gene, polyvalent primary Env DNA prime-protein boost HIV vaccine, DP6−001, included 6 DNA plasmids (one expressing a subtype C full length Gag antigen and the other five each expressing one of the 5 primary gp120 antigens from subtypes A, B, C or E) as the prime and 5 recombinant gp120 proteins matching the Env DNA prime as the boost (Table 1). The study was a 3-group trial that tested two dosing levels of DNA administered either intradermally (ID) or intramuscularly (IM) and one standard dose of protein with adjuvant QS-21 administered IM (Table 1). Groups A and B received three DNA immunizations, either ID or IM, respectively, with 1.2 mg total DNA at each immunization, equally divided among one Gag and five gp120 DNA plasmids. For Group C, a higher dose of DNA prime with 7.2mg total DNA was administered IM at each immunization. Protein boosts contained a fixed dose of five recombinant gp120 proteins delivered twice with adjuvant QS-21.
Overall the DP6−001 vaccine was well tolerated and the most frequent adverse events were skin reactions. A higher reactogenicity, in the form of transient low grade fever and one case of lower extremity leukocytoclastic vasculitis (LCV), a skin form vasculitis with unknown etiology, which was resolved shortly without specific treatment, was observed in the high dose DNA prime group (Group C) after receiving a protein boost. Due to such reactogenicity, the clinical trial was terminated early and only 6 subjects in Group C received one protein boost. Detailed information on the safety and reactogenicity of this clinical trial is summarized in a separate report [29]. The current report focuses on the immunogenicity results in volunteers received DP6−001.
3.2. Cross-subtype HIV-1-specific cell-mediated immune (CMI) responses
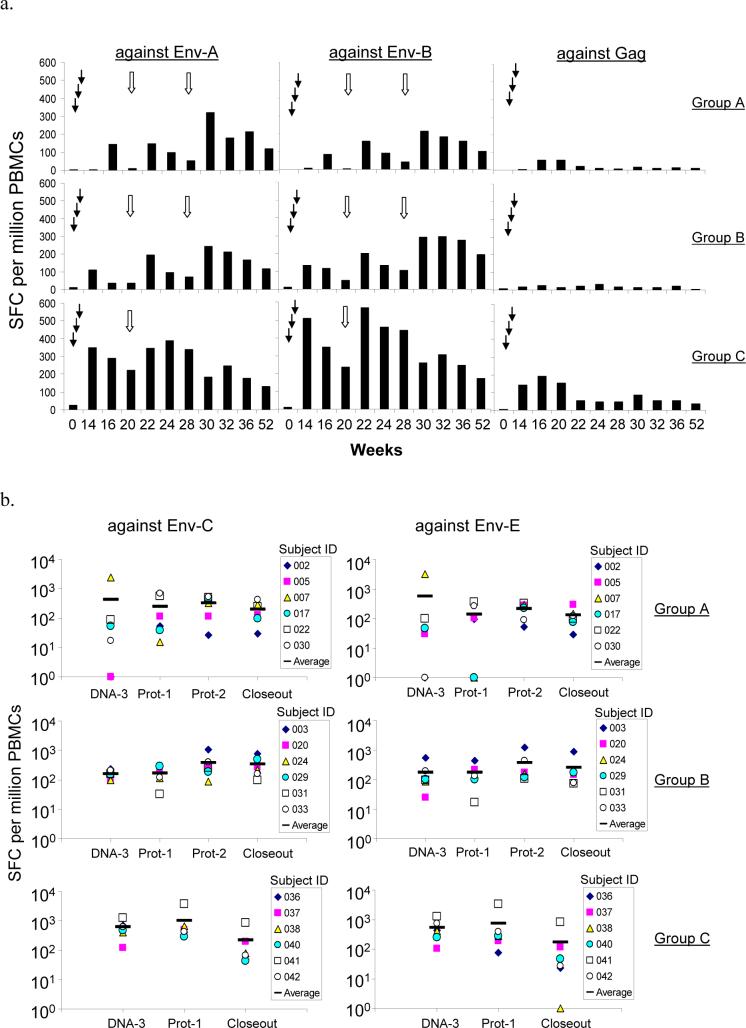
CMI responses to two primary Env (Env-A and Env-B) antigens and one Gag antigen were first analyzed by an IFNγ ELISPOT assay. Positive CMI responses against pooled Env peptides were observed at the end of the three DNA prime immunizations for both low-dose DNA priming groups (Groups A and B) but the levels of CMI responses were low (Fig. 1-A). The high-dose DNA priming group (Group C) was able to induce a significantly higher CMI response at the end of DNA prime when compared to Groups A and B (p<0.01 for CMI responses against both Env-A and Env-B antigens).
Figure 1.
DP6−001 formulation induced HIV-1 specific cell-mediated immune responses in volunteers' PBMC. (a) Group averages of Env- and Gag-specific IFNγ ELISPOT responses at different time points following DNA or protein immunizations (see Table 2 for standard error and percentage of responders). The solid arrows indicate DNA immunizations and the open arrows indicate protein immunizations. Pools of overlapping peptides from either gp120 antigens of subtype A isolate 92UG037.8 (Env-A) and subtype B isolate 92US715.6 (Env-B) or Gag antigen of subtype C isolate 96ZM651 (Gag) were used for the assay. (b) HIV-1-specific IFNγ ELISPOT responses were detected at 2 weeks after either the 3rd DNA immunization (DNA-3), the 1st or 2nd protein boosts (Prot-1 and Prot-2), or at Week 52 (Closeout) against pools overlapping peptides from gp120 antigens of subtype C isolate 96ZM651 (Env-C) and subtype E isolate 93TH976.17 (Env-E). Results are shown as responses from each of six randomly selected volunteers in each group as well as the group average (short horizontal bars).
Levels of Env-specific CMI were further boosted by each of the two subsequent protein immunizations and maintained at relatively high levels even at the end of trial (24 weeks from the last protein immunization). Higher CMI responses to Env-A and Env-B antigens were observed in Group C volunteers when compared to Groups A (p<0.05) and B (p<0.05), showing the response of a 6-fold higher doses of the DNA vaccines during priming immunizations (Fig. 1-A).
Also, while CMI responses to Gag peptides were poor in Groups A and B, these responses increased substantially in Group C (p<0.05 compared with either Groups A or B) (Fig. 1-A and Table 2). The Gag-specific CMI response declined more than 5-fold from its peak level at the end of DNA prime immunization to barely positive responses by 52 weeks while the Env-specific CMI declined only 2- to 3-fold from peak levels (Table 2). It is not clear whether such a difference was due to the fact that only an Env protein was used as the boost.
Table 2.
Summary of HIV-1 antigen-specific cell mediated and gp120-specific antibody responses in DP6−001 vaccinees
| Samples |
Peptide specific IFN-□ELISpot (SFC/million PBMCs)* |
gp120-specific IgG titers** |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| against Env-A |
against Env-B |
against Gag |
|||||||
| Average ± SE | % responders | Average ± SE | % responders | Average ± SE | % responders | Median | Range | % responders | |
| Group A (n = 10) | |||||||||
| DNA-3 | 149.5 ± 112.3 | 70 | 83 ± 60.9 | 50 | 56 ± 42.3 | 30 | <50 | <50 − 50 | 20 |
| Protein-1 | 147.5 ± 70 | 70 | 185.1 ± 81.7 | 80.0 | NA | NA | 25,600 | 200 − 1,638,400 | 100 |
| Protein-2 | 322.9 ± 82.3 | 100 | 214.1 ± 68.1 | 90.0 | NA | NA | 204,800 | 25,600 − 819,200 | 100 |
| Closeout | 119.5 ± 25 | 80 | 102.2 ± 33.9 | 80.0 | 10.1 ± 7.7 | 20 | 12,800 | <50 − 51,200 | 90 |
| Group B (n = 11) | |||||||||
| DNA-3 | 108.6 ± 27.2 | 100 | 131.7 ± 35.1 | 81.8 | 25 ± 13.6 | 36.4 | <50 | <50 | 0 |
| Protein-1 | 195.8 ± 94.3 | 81.8 | 201.4 ± 97.7 | 100 | NA | NA | 25,600 | 200 − 819,200 | 100 |
| Protein-2 | 242.4 ± 100.4 | 100 | 291.5 ± 132.7 | 100 | NA | NA | 204,800 | 1600 − 819,200 | 100 |
| Closeout | 151.3 ± 57.1 | 81.8 | 192.7 ± 70.3 | 90 | 4.3 ± 2.8 | 18.2 | 8,000 | 200 − 102,400 | 100 |
| Group C (n = 6) | |||||||||
| DNA-3 | 348.1 ± 122.9 | 100 | 507.4 ± 161.4 | 100 | 191.5 ± 86.1 | 100 | 1,600 | <50 − 409,600 | 66.7 |
| Protein-1 | 347.5 ± 203.1 | 100 | 569.2 ± 394.2 | 100 | NA | NA | 51,200 | 51,200 − 819,200 | 100 |
| Protein-2 | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Closeout | 131.4 ± 97.2 | 83.3 | 171.0 ± 114.2 | 83.3 | 35.6 ± 16.8 | 50 | 8,000 | 800 − 25,600 | 100 |
SFC: spot forming cells were measured against Env-A, Env-B and Gag peptided at 2 weeks after the 3rd DNA (DNA-3), the 1st and 2nd protein (Protein-1 and Protein-2) immunizations, and at Week 52 (Closeout) respectively.
gp120 specific IgG titers were measured against 5-mixed gp120 proteins (A, B, Bal, C and E) at 2 weeks after the 3rd DNA (DNA-3), the 1st and 2nd protein (Protein-1 and Protein-2) immunizations, and at Week 52 (Closeout) respectively.
NA: not applicable.
Overall, 90−100% of the volunteers had positive Env-specific IFNγ ELISPOT responses against both Env-A and Env-B peptides after two protein boosts in low-dose Groups A and B, and 100% had positive responses by the end of DNA priming in high-dose Group C (Table 2). At Week 52, greater than 80% of the volunteers still had positive Env-specific CMI responses. In contrast, although 100% of volunteers in Group C had positive Gag-specific IFNγ ELISPOT with DNA prime alone, the percentage of responders decreased to about 50% by Week 52 (Table 2).
Additional IFNγ ELISPOT analyses using peptides from the subtypes C and E Env antigens included in DP6−001 were conducted with samples from 6 randomly selected volunteers from each group. Env-C- and Env-E-specific CMI responses were detected in all three groups but higher peak levels of positive CMI responses at the end of DNA prime were observed in Group C when compared to Groups A or B (Fig. 1-B). Flow cytometry assays further demonstrated that the HIV-1-specific IFNγ responses observed in this study were mediated predominantly by CD4+ T cells. HIV-1-specific CD8+ T cell responses were also detected but at a lower frequency. Detailed flow cytometry results will be summarized in a separate report [35].
3.3. Broadly reactive antibody responses against a wide range of primary HIV-1 Env antigens
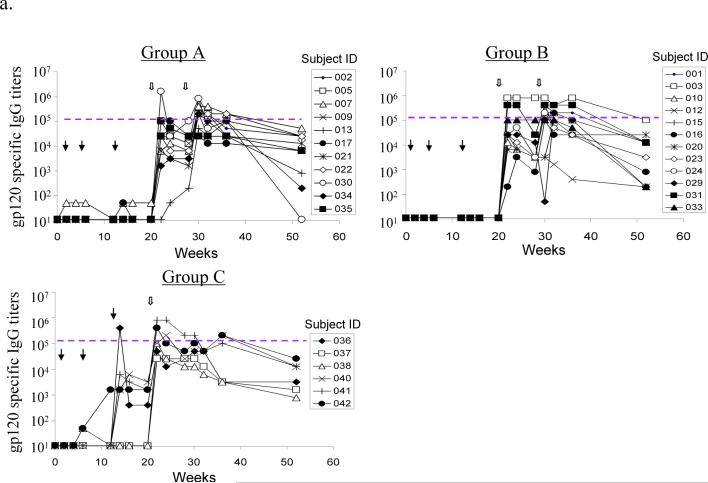
High titer HIV-1 Env-specific antibody responses were generated with the DNA prime-protein boost DP6−001 formulation (Fig. 2-A). For Groups A and B, most volunteers did not have detectable Env-specific antibody responses at the end of three DNA prime immunizations, as expected at such a low dose of DNA [14, 36]. However, the antibody titers rose quickly after just one protein boost. The anti-gp120 IgG titers reached 1:105 or higher with 1 or 2 protein boosts, levels that are comparable to those observed in chronically infected HIV patients (Fig. 2-A). The majority of volunteers maintained significant serum anti-gp120 IgG titers at the end of trial. Four out of six volunteers (66.7%) in Group C showed detectable Env-specific IgG responses even before protein boosting (Fig. 2-A and Table 2). The gp120 antigens used in the ELISA assay were the same GMP lot proteins produced from CHO cells used for DP6−001 formulation. The specificity of antibody responses were further confirmed by another ELISA against Env antigens produced from a different cell line 293T cells (data not shown) and Western blot analysis (see below).
Figure 2.
DP6−001 formulation induced HIV-1 gp120-specific antibody responses in volunteers' sera. (a) Titers of serum gp120-specific IgG were measured by ELISA in individual volunteers of different study groups. Each curve represents one volunteer. The solid arrows indicate DNA immunizations and the open arrows indicate protein immunizations. The dotted line denotes the average titer of gp120-specific IgG in sera of three patients chronically infected with HIV-1 (titer range: 1:102,400 to 1:204,800). (b) Reactivity of two DP6−001 immune sera (#022 of Group A and #023 of Group B at 1:100 dilution) at different time points of immunization against five autologous primary HIV-1 gp120 glycoproteins as measured by Western blot analysis. Normal human sera and HIV-1 positive patient sera, both at 1:100 dilution, or HIV-1 positive immunoglobulin (HIV-IG) (at 0.5 mg/ml total human IgG) were included as controls. (c) Cross reactivity of sera from one representative DP6−001 immune serum #003 (Group B) against a panel of heterologous primary HIV-1 gp120 glycoproteins by Western blot analysis. Two individual HIV+ patient sera (91BU003 and 93BR029) were included as the controls. Serum dilution of 1:100 was used for both DP6−001 and HIV+ patient sera.
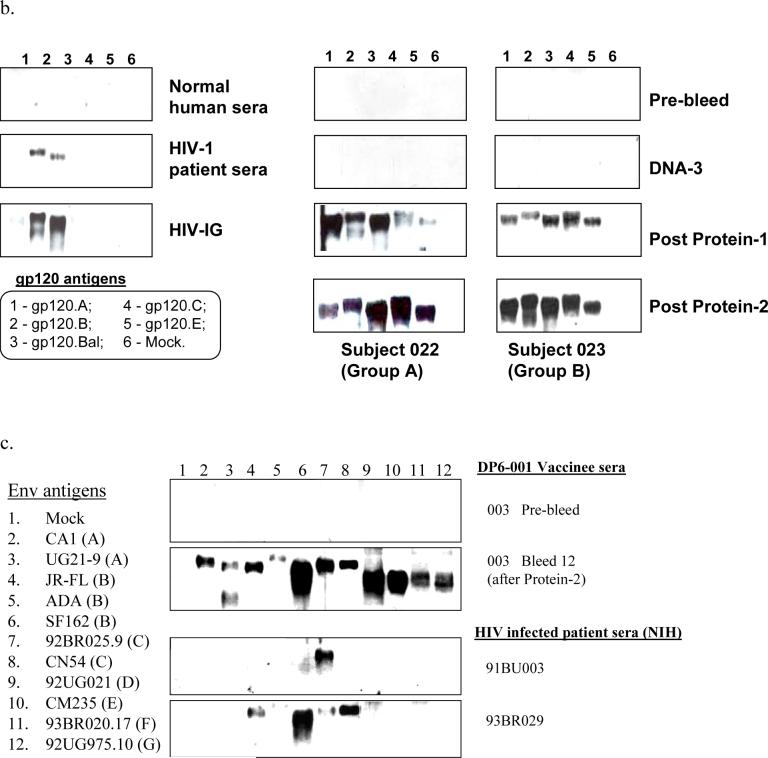
Western blot analysis indicated that antibodies elicited by the polyvalent Env formulation DP6−001 were broadly cross-reactive against diverse primary HIV-1 Env antigens (Fig. 2-B). Sera from all 27 volunteers included in this report recognized all of the five homologous primary gp120 antigens included in DP6−001 after 1 or 2 protein boosts, as shown by two representative volunteer sera from Groups A (#022) and B (#023). Neither randomly selected individual HIV+ patient serum nor pooled HIV-1 positive human immunoglobulin (HIV-IG) from patients infected with subtype B viruses could recognize more than 2 or 3 primary gp120 antigens (Fig. 2-B). Additional Western blot analysis confirmed reactivity of immunized volunteer sera against a wide range of 11 heterologous primary HIV-1 gp120 antigens from subtypes A to G. One representative sample blot with volunteer #013 sera is shown in Fig. 2-C. Two control individual HIV+ patient sera could only recognize a small fraction of these primary gp120 antigens.
3.4. Neutralizing antibody activities of DP6−001 vaccinated human sera
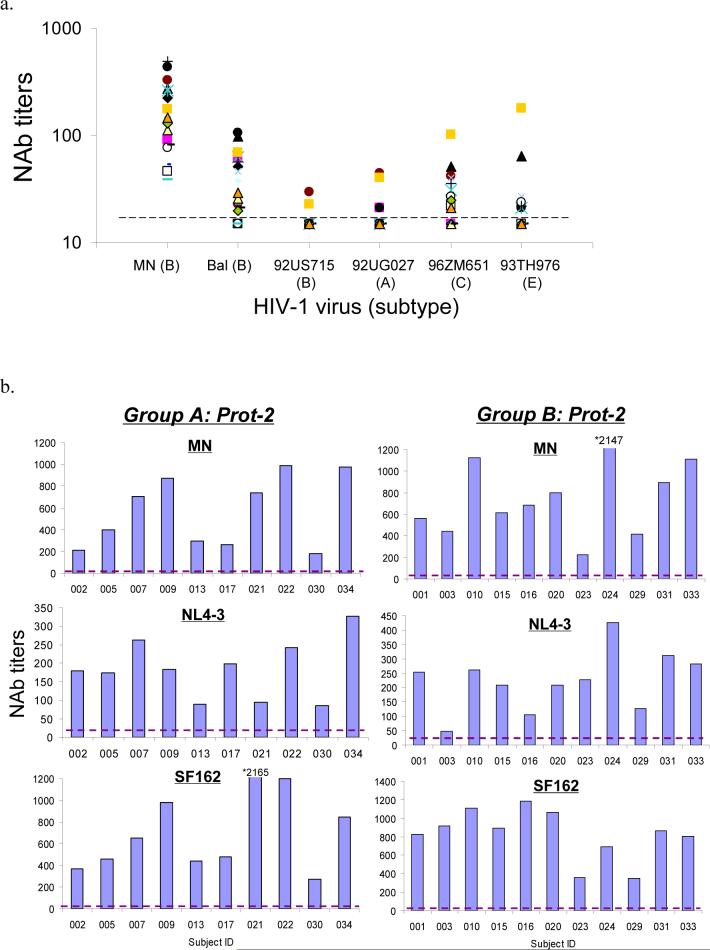
Three studies were organized to assess the neutralizing antibody (NAb) activities in DP6−001 vaccinee sera. The first study analyzed the ability of these sera to neutralize the Tier 1 viruses: a TCLA HIV-1 virus (MN) and five pseudotyped viruses each expressing one of the five homologous primary Env antigens included in DP6−001 (Table 3 and Fig. 3-A). Positive neutralizing activities against MN were seen in 100% of Groups A and B vaccinee sera at the peak antibody level (after the 2nd protein boost). The percentage of positive NAb responses against different homologous pseudotyped viruses varied: 71% neutralized Bal (subtype B), 62% neutralized 96ZM652 (subtype C), 38% neutralized 93TH976 (subtype E), 28% neutralized 92UG037 (subtype A), and less than 9% neutralized 92US715 (subtype B). None of the 7 control human serum samples had positive neutralizing activity against the above viruses (Table 3). More than 50% of the vaccinees had NAb titers greater than 1:100 against MN. The titers of NAbs against other homologous pseudotyped viruses were between 1:20 to 1:100 for vaccinee sera with positive NAb activities (Fig. 3-A). Group C showed a higher percentage of individual positive NAb sera after only one protein boost when compared to corresponding sera from Groups A and B. One vaccinee (16.67%) in Group C even had positive NAb activities against MN, Bal and 96ZM651 following only the DNA immunizations (Table 3).
Table 3.
Percent of responders with neutralizing antibody (NAb) titers greater than 1: 20
| Serum samples |
% with positive NAb against autologous pseudotyped HIV-1 viruses (subtype) |
|||||
|---|---|---|---|---|---|---|
| MN (B) | Bal (B) | 92US715 (B) | 92UG037 (A) | 96ZM652 (C) | 93TH976 (E) | |
| Groups A and B (n = 21) | ||||||
| DNA-3 | 0.00 | 4.76 | 0.00 | 4.76 | 0.00 | 0.00 |
| Protein-1 | 28.57 | 28.57 | 0.00 | 4.76 | 14.29 | 4.76 |
| Protein-2 | 100.00 | 71.43 | 9.52 | 28.57 | 61.90 | 38.10 |
| Group C (n = 6) | ||||||
| DNA-3 | 16.67 | 16.67 | 0.00 | 0.00 | 16.67 | 0.00 |
| Protein-1 | 66.67 | 50.00 | 0.00 | 0.00 | 33.33 | 0.00 |
| Protein-2 | N/A | N/A | N/A | N/A | N/A | N/A |
| Negative control (n = 7) | ||||||
| 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
Figure 3.
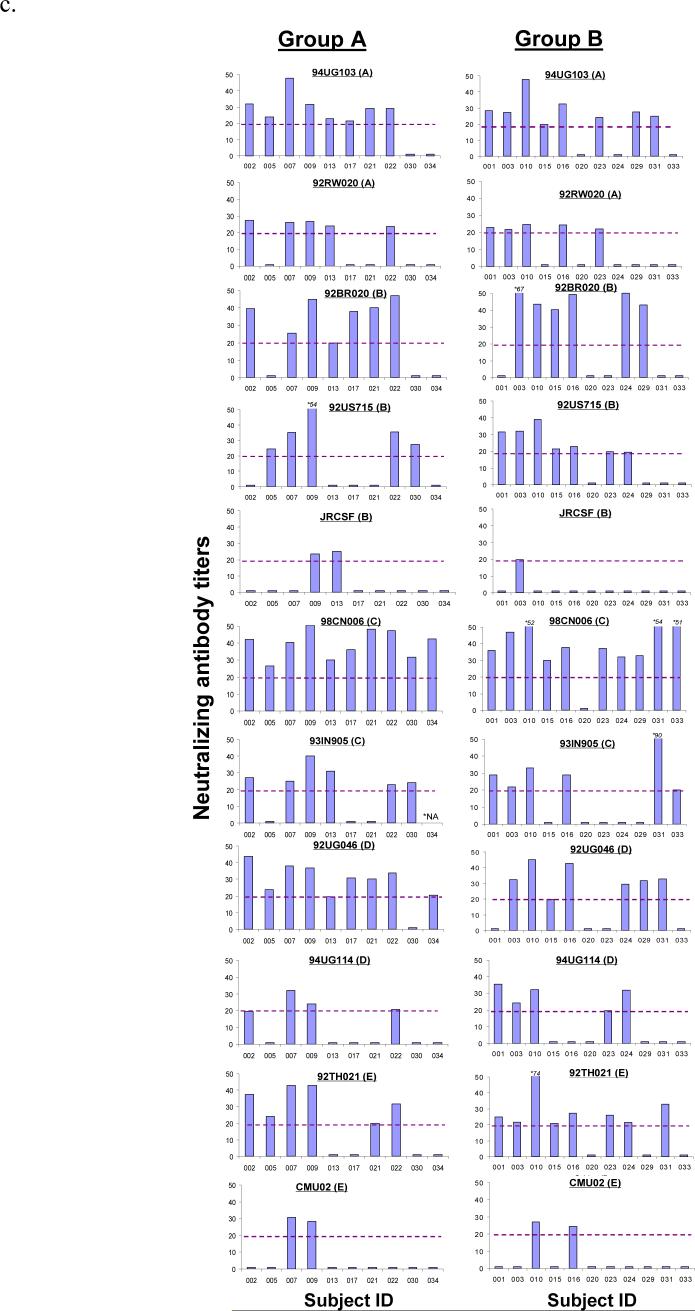
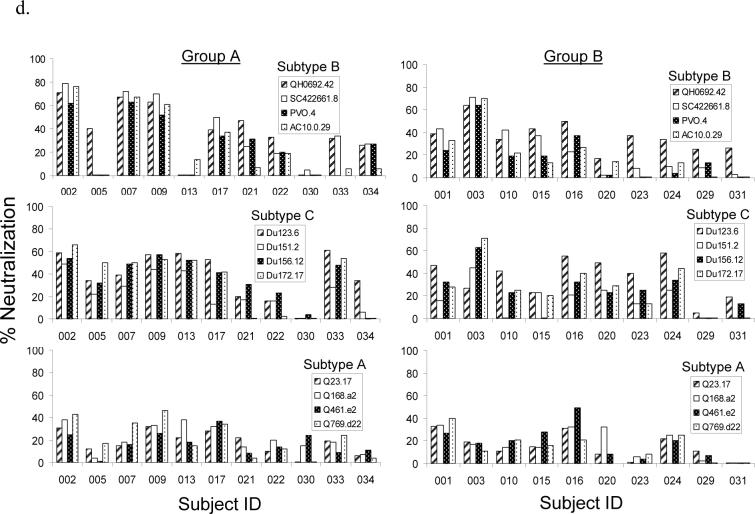
Neutralizing antibody (NAb) responses against different panels of pseudotyped viruses. (a) NAb titers against Tier 1/autologous viruses including a T-cell line adapted (TCLA) HIV-1 virus (MN) and five pseudotyped viruses each expressing one of the five autologous primary Env antigens included in the DP6−001 formulation. The neutralization assays were done in TZM-bl cells and the NAb titers were measured using individual volunteer sera from Groups A and B (N = 21) at 2 weeks after the 2nd protein boost. (b)-(c) NAb titers determined by PhenoSense™ assay in U87 cells with sera from Groups A and B at two weeks after the 2nd protein boost against pseudotyped viruses expressing either (b) three relatively sensitive to neutralization Env antigens of subtype B (MN, NL4−3 and SF162) or (c) eleven additional primary Env antigens of subtypes A, B, C, D and E. A serum dilution of 1:20 was used as the cut-off to score the positive NAb (shown as the broken line in each graph). (d) NAb responses against Tier 2 pseudotyped viruses expressing primary Env from subtypes A, B and C (4 viruses for each subtype). Individual subject sera from Groups A and B were collected at two weeks post the 2nd protein boost. The percent neutralization was measured at 1:10 serum dilution. The neutralization assays were done in TZM-bl cells.
The second neutralization study was done using a high-throughput, pseudotyped virus assay system. Sera collected 2 weeks after the second protein boost (peak of anti-gp120 antibody responses) were assayed against a panel of 11 heterologous primary viruses covering subtypes A to E in addition to three sensitive viruses (MN, NL4−3 and SF162). Because Group C volunteers did not complete two protein boost immunizations, only sera from Groups A and B volunteers are included in this study. High-titer NAb responses (up to 1:2147) were identified in all human immune sera against the three sensitive viruses (Fig. 3-B). Using a NAb titer of 1:20 as the cut-off, more than 50% of vaccinees also showed positive neutralizing activities against 8 pseudotyped viruses (92UG103 and 92RW020 of subtype A, 92BR020 and 92US715 of subtype B, 98CN006 and 93IN905 of subtype C, 92UG046 of subtype D and 92TH021 of subtype E), while the three remaining pseudotyped viruses (JR-CSF of subtype B, 94UG114 of subtype D and CMU02 of subtype E) were more resistant to neutralization (Fig 3-C). The overall patterns of NAb responses were similar between Group A and Group B. Two vaccinees from Group A (#007 and #009) and two from Group B (#010 and #016) had NAb activities against a majority of the pseudotyped viruses included in the analysis.
The third neutralization study was conducted to test the ability of DP6−001 vaccinee sera to neutralize standardized panels of Tier 2 viruses. Viruses included in these panels are more recently isolated (the “contemporary viruses”) from acute and early infections [32]. Although neutralizing activity was clearly higher than the negative control sera, most volunteer sera displayed viral inhibition of less than 50% at a serum dilution of 1:10 (Fig. 3-D).
4. DISCUSSION
Induction of anti-Env antibody responses in small animals was one of the first pieces of evidence that established DNA immunization as a novel approach for vaccination [37]. Although significant progress has been made using DNA immunization to elicit HIV-1-specific CMI in small animals, non-human primates and humans over the past 15 years [12-16, 18, 38-41], there has been no report of using Env DNA immunization to elicit broadly cross-reactive antibodies in humans. Levels of anti-Env antibodies elicited in previous DNA vaccine clinical trials were either low or undetectable [42, 43], nor was there a clear induction of NAbs against even sensitive viruses [14]. In the current study, one Env protein immunization following DNA prime was able to elicit human anti-Env antibody responses to a level that has proven difficult to achieve in previous studies that employed multiple injections of recombinant HIV-1 Env proteins [2, 3, 6]. Therefore, data from this study provides evidence that DNA vaccination can effectively prime the induction of high-level anti-Env antibody responses in humans.
In prior recombinant Env protein-based HIV vaccine studies, up to 2 Env antigens were included in the vaccine formulation and as many as seven Env protein immunizations had to be administered in order to achieve high antibody responses [44]. While the overall levels of Env-specific antibodies were low and not broadly cross-reactive, their binding specificity was measured against selected peptides of Env proteins rather than the primary Env antigens themselves [4, 45]. Our results clearly demonstrate the benefit of delivering polyvalent Env antigens through the DNA prime-protein boost approach. Not only did we elicit Env-specific antibody responses comparable to those seen in persons chronically infected with HIV-1, but the immune sera were able to react with primary Env antigens from every HIV sub-type included in the study. Furthermore, the antibody responses appeared to be long lasting, with only a moderate decrease in titers at the end of the 52-week study period.
Given the importance of NAb responses in a prophylactic HIV vaccine and the complexity of measuring such responses, three neutralization studies were conducted, each with a slightly different approach and each using different panels of HIV viruses, to provide a more complete picture of the spectrum of neutralizing activities contained in the immune sera. The DP6−001 formulation elicited neutralizing activities against the sensitive viruses (TCLA and SF-162) that were comparable to or better than those elicited by recombinant gp120 alone [31], and clearly much better than a recently reported DNA vaccine alone approach which did not show neutralizing antibody activities [14]. In current studies, neutralization activity, against pseudoviruses expressing the homologous or randomly selected primary Env antigens, was detected in most of the post-immunization sera against approximately half of the viruses tested, independent of subtype. Not all homologous Env viruses showed high sensitivity to neutralization by post-immunization sera, which was not surprising given previous reports showing that infected HIV patient sera did not always have a high neutralizing activity against autologous viruses [46, 47].
The neutralizing activities against the standard panel of Tier 2 HIV-1 viruses were low in the current study, even at low serum dilutions. While it is unclear whether this panel represents more resistant viruses than the viruses included in the other two neutralization assays, it is possible that the difference reflects the inclusion of more contemporary viral isolates in the Tier 2 panel while Env antigens included in the DP6−001 formulation were cloned from patient samples collected around or before the early 1990s. While the current DP6−001 formulation, as a proof of concept study, may not have the optimal profile to move to more advanced clinical trials, the results included in this report clearly indicate that the use of a polyvalent, DNA prime-protein boost approach is feasible in order to elicit human neutralizing activities against a wide range of HIV-1 isolates. Future studies should test whether modified formulations including contemporary Env antigens can improve neutralizing activities against Tier 2 viruses.
Recently published studies with optimized DNA or other gene-based HIV vaccines elicited robust CMI responses [14, 15]. One key question asked in the current trial was whether robust CMI responses would be maintained when DNA immunization was used in conjunction with a protein boost component. The levels of Env-specific CMI responses elicited after three DNA immunizations in this study were similar to those previously reported [12, 14, 15]. One new finding from the current study is that the Env protein boost further increased the magnitude of Env-specific CMI responses when compared to the Gag-specific CMI, for which there was no protein boost. The Env-specific CMI responses were cross-reactive against at least 4 different primary Env antigens that were included in this study. In further support for the presence of HIV-1-specific CMI responses in DP6−001 vaccinees, a subset of volunteers developed DTH-like skin reactions at the sites of DNA immunization after receiving an Env protein boost at a distant inoculation [29].
We also detected a robust Gag-specific CMI response in Group C volunteers, which was higher than observed by others [14, 15]. Given the 1:5 ratio of Gag to Env DNA vaccines in DP6−001, the actual dose of Gag DNA vaccine was 1.2 mg in Group C. It suggested that the minimum immunogenic dose for naked DNA vaccines would be at least 1 mg with the current design of DNA vaccines. There is a clear dose response relationship for both Env and Gag DNA vaccines. Due to the small sample size in the current study, future trials should study the boost effect of proteins on CMI responses. Furthermore, this study also shows that the ID route of administration was not more effective than IM in eliciting an immune response by DNA vaccines.
In summary, results from this first report of a human study with DNA prime-protein boost HIV-1 vaccine confirms the immunogenicity of this novel approach in eliciting balanced humoral and cell-mediated immune responses in a healthy adult population. These results are significant in that they confirm initial observations on the immunogenicity of DNA vaccines in animal models [37, 48-50]. The DNA prime-protein boost approach will not only accelerate the testing of more candidate HIV vaccines that aim to achieve improved neutralizing antibody responses, but will also provide a new platform for the development of future vaccines against a wide range of existing or emerging pathogens. Future studies should include in-depth analysis on the structural basis for both antibody and CMI cross reactivities observed in the current report. The composition of Env antigens should be further optimized to identify a polyvalent formulation that may expand the breadth of neutralizing activities against viruses that were resistant to immune sera elicited by DP6−001. The immunization schedule including the use of adjuvant should be also optimized to reduce the reactogenicity of DP6−001 before moving this DNA prime and protein boost approach to more advanced human studies.
Acknowledgements
This work was supported in part by the HIV Vaccine Design and Development Teams contract N01AI05394, and grants R29AI40337, R21AI46294 and R01AI65250 from the National Institute of Allergy and Infectious Diseases. The PhenoSense neutralization assay was conducted at the Mongram, Inc. and funded by the International AIDS Vaccine Initiative. The project also used core facility resources at the University of Massachusetts Medical School supported by NIH grant 5P30DK32520 from the NIDDK. The authors thank Drs. Beatrice Hahn and Feng Gao for providing the DNA plasmids coding for primary HIV-1 envelope antigens, Drs. Wayne Koff and Emily Carrow for their support to this project, Dr. Terri Wrin for her neutralization assay and Drs. Jill M. Grimes-Serrano and Anthony Cristillo for their critical reading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.HIV/AIDS JUNPo [cited 2007 14 February];Report on the global AIDS epidemic 2006. 2006 Availablefrom: http://data.unaids.org/pub/GlobalReport/2006/2006_GR_CH02_en.pdf.
- 2.Gorse GJ, Corey L, Patel GB, Mandava M, Hsieh RH, Matthews TJ, et al. HIV-1MN recombinant glycoprotein 160 vaccine-induced cellular and humoral immunity boosted by HIV-1MN recombinant glycoprotein 120 vaccine. National Institute of Allergy and Infectious Diseases AIDS Vaccine Evaluation Group. AIDS Res Hum Retroviruses. 1999 Jan 20;15(2):115–32. doi: 10.1089/088922299311547. [DOI] [PubMed] [Google Scholar]
- 3.Kim JH, Pitisuttithum P, Kamboonruang C, Chuenchitra T, Mascola J, Frankel SS, et al. Specific antibody responses to vaccination with bivalent CM235/SF2 gp120: detection of homologous and heterologous neutralizing antibody to subtype E (CRF01.AE) HIV type 1. AIDS Res Hum Retroviruses. 2003 Sep;19(9):807–16. doi: 10.1089/088922203769232601. [DOI] [PubMed] [Google Scholar]
- 4.Lee SA, Orque R, Escarpe PA, Peterson ML, Good JW, Zaharias EM, et al. Vaccine-induced antibodies to the native, oligomeric envelope glycoproteins of primary HIV-1 isolates. Vaccine. 2001 Nov 12;20(3−4):563–76. doi: 10.1016/s0264-410x(01)00334-6. [DOI] [PubMed] [Google Scholar]
- 5.Nitayaphan S, Khamboonruang C, Sirisophana N, Morgan P, Chiu J, Duliege AM, et al. A phase I/II trial of HIV SF2 gp120/MF59 vaccine in seronegative thais.AFRIMS-RIHES Vaccine Evaluation Group. Armed Forces Research Institute of Medical Sciences and the Research Institute for Health Sciences. Vaccine. 2000 Feb 14;18(15):1448–55. doi: 10.1016/s0264-410x(99)00421-1. [DOI] [PubMed] [Google Scholar]
- 6.Ackers ML, Parekh B, Evans TG, Berman P, Phillips S, Allen M, et al. Human immunodeficiency virus (HIV) seropositivity among uninfected HIV vaccine recipients. J Infect Dis. 2003 Mar 15;187(6):879–86. doi: 10.1086/368169. [DOI] [PubMed] [Google Scholar]
- 7.Beddows S, Lister S, Cheingsong R, Bruck C, Weber J. Comparison of the antibody repertoire generated in healthy volunteers following immunization with a monomeric recombinant gp120 construct derived from a CCR5/CXCR4-using human immunodeficiency virus type 1 isolate with sera from naturally infected individuals. J Virol. 1999 Feb;73(2):1740–5. doi: 10.1128/jvi.73.2.1740-1745.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mascola JR, Snyder SW, Weislow OS, Belay SM, Belshe RB, Schwartz DH, et al. Immunization with envelope subunit vaccine products elicits neutralizing antibodies against laboratory-adapted but not primary isolates of human immunodeficiency virus type 1. The National Institute of Allergy and Infectious Diseases AIDS Vaccine Evaluation Group. J Infect Dis. 1996 Feb;173(2):340–8. doi: 10.1093/infdis/173.2.340. [DOI] [PubMed] [Google Scholar]
- 9.Bures R, Gaitan A, Zhu T, Graziosi C, McGrath KM, Tartaglia J, et al. Immunization with recombinant canarypox vectors expressing membrane-anchored glycoprotein 120 followed by glycoprotein 160 boosting fails to generate antibodies that neutralize R5 primary isolates of human immunodeficiency virus type 1. AIDS Res Hum Retroviruses. 2000 Dec 10;16(18):2019–35. doi: 10.1089/088922200750054756. [DOI] [PubMed] [Google Scholar]
- 10.Gilbert PB, Peterson ML, Follmann D, Hudgens MG, Francis DP, Gurwith M, et al. Correlation between immunologic responses to a recombinant glycoprotein 120 vaccine and incidence of HIV-1 infection in a phase 3 HIV-1 preventive vaccine trial. J Infect Dis. 2005 Mar 1;191(5):666–77. doi: 10.1086/428405. [DOI] [PubMed] [Google Scholar]
- 11.Flynn NM, Forthal DN, Harro CD, Judson FN, Mayer KH, Para MF. Placebo-controlled phase 3 trial of a recombinant glycoprotein 120 vaccine to prevent HIV-1 infection. J Infect Dis. 2005 Mar 1;191(5):654–65. doi: 10.1086/428404. [DOI] [PubMed] [Google Scholar]
- 12.Mwau M, Cebere I, Sutton J, Chikoti P, Winstone N, Wee EG, et al. A human immunodeficiency virus 1 (HIV-1) clade A vaccine in clinical trials: stimulation of HIV-specific T-cell responses by DNA and recombinant modified vaccinia virus Ankara (MVA) vaccines in humans. J Gen Virol. 2004 Apr;85(Pt 4):911–9. doi: 10.1099/vir.0.19701-0. [DOI] [PubMed] [Google Scholar]
- 13.Goonetilleke N, Moore S, Dally L, Winstone N, Cebere I, Mahmoud A, et al. Induction of multifunctional human immunodeficiency virus type 1 (HIV-1)-specific T cells capable of proliferation in healthy subjects by using a prime-boost regimen of DNA- and modified vaccinia virus Ankara-vectored vaccines expressing HIV-1 Gag coupled to CD8+ T-cell epitopes. J Virol. 2006 May;80(10):4717–28. doi: 10.1128/JVI.80.10.4717-4728.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graham BS, Koup RA, Roederer M, Bailer RT, Enama ME, Moodie Z, et al. Phase 1 safety and immunogenicity evaluation of a multiclade HIV-1 DNA candidate vaccine. J Infect Dis. 2006 Dec 15;194(12):1650–60. doi: 10.1086/509259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Catanzaro AT, Koup RA, Roederer M, Bailer RT, Enama ME, Moodie Z, et al. Phase 1 safety and immunogenicity evaluation of a multiclade HIV-1 candidate vaccine delivered by a replication-defective recombinant adenovirus vector. J Infect Dis. 2006 Dec 15;194(12):1638–49. doi: 10.1086/509258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pal R, Kalyanaraman VS, Nair BC, Whitney S, Keen T, Hocker L, et al. Immunization of rhesus macaques with a polyvalent DNA prime/protein boost human immunodeficiency virus type 1 vaccine elicits protective antibody response against simian human immunodeficiency virus of R5 phenotype. Virology. 2006 May 10;348(2):341–53. doi: 10.1016/j.virol.2005.12.029. [DOI] [PubMed] [Google Scholar]
- 17.Pal R, Wang S, Kalyanaraman VS, Nair BC, Whitney S, Keen T, et al. Polyvalent DNA prime and envelope protein boost HIV-1 vaccine elicits humoral and cellular responses and controls plasma viremia in rhesus macaques following rectal challenge with an R5 SHIV isolate. J Med Primatol. 2005 Oct;34(5−6):226–36. doi: 10.1111/j.1600-0684.2005.00120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cristillo AD, Wang S, Caskey MS, Unangst T, Hocker L, He L, et al. Preclinical evaluation of cellular immune responses elicited by a polyvalent DNA prime/protein boost HIV-1 vaccine. Virology. 2006 Mar 1;346(1):151–68. doi: 10.1016/j.virol.2005.10.038. [DOI] [PubMed] [Google Scholar]
- 19.Wang S, Arthos J, Lawrence JM, Van Ryk D, Mboudjeka I, Shen S, et al. Enhanced immunogenicity of gp120 protein when combined with recombinant DNA priming to generate antibodies that neutralize the JR-FL primary isolate of human immunodeficiency virus type 1. J Virol. 2005 Jun;79(12):7933–7. doi: 10.1128/JVI.79.12.7933-7937.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beddows S, Schulke N, Kirschner M, Barnes K, Franti M, Michael E, et al. Evaluating the immunogenicity of a disulfide-stabilized, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. Journal of virology. 2005 Jul;79(14):8812–27. doi: 10.1128/JVI.79.14.8812-8827.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Srivastava IK, Ulmer JB, Barnett SW. Role of neutralizing antibodies in protective immunity against HIV. Hum Vaccin. 2005 Mar-Apr;1(2):45–60. doi: 10.4161/hv.1.2.1764. [DOI] [PubMed] [Google Scholar]
- 22.Lian Y, Srivastava I, Gomez-Roman VR, Zur Megede J, Sun Y, Kan E, et al. Evaluation of envelope vaccines derived from the South African subtype C human immunodeficiency virus type 1 TV1 strain. J Virol. 2005 Nov;79(21):13338–49. doi: 10.1128/JVI.79.21.13338-13349.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vajdy M, Singh M, Kazzaz J, Soenawan E, Ugozzoli M, Zhou F, et al. Mucosal and systemic anti-HIV responses in rhesus macaques following combinations of intranasal and parenteral immunizations. AIDS Res Hum Retroviruses. 2004 Nov;20(11):1269–81. doi: 10.1089/aid.2004.20.1269. [DOI] [PubMed] [Google Scholar]
- 24.Leung L, Srivastava IK, Kan E, Legg H, Sun Y, Greer C, et al. Immunogenicity of HIV-1 Env and Gag in baboons using a DNA prime/protein boost regimen. Aids. 2004 Apr 30;18(7):991–1001. doi: 10.1097/00002030-200404300-00006. [DOI] [PubMed] [Google Scholar]
- 25.Shu Y, Winfrey S, Yang ZY, Xu L, Rao SS, Srivastava I, et al. Efficient protein boosting after plasmid DNA or recombinant adenovirus immunization with HIV-1 vaccine constructs. Vaccine. 2007 Feb 9;25(8):1398–408. doi: 10.1016/j.vaccine.2006.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Law M, Cardoso RM, Wilson IA, Burton DR. Antigenic and Immunogenic Study of Membrane-Proximal External Region-Grafted gp120 Antigens by a DNA Prime-Protein Boost Immunization Strategy. J Virol. 2007 Apr;81(8):4272–85. doi: 10.1128/JVI.02536-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang S, Pal R, Mascola JR, Chou TH, Mboudjeka I, Shen S, et al. Polyvalent HIV-1 Env vaccine formulations delivered by the DNA priming plus protein boosting approach are effective in generating neutralizing antibodies against primary human immunodeficiency virus type 1 isolates from subtypes A, B, C, D and E. Virology. 2006 Jun 20;350(1):34–47. doi: 10.1016/j.virol.2006.02.032. [DOI] [PubMed] [Google Scholar]
- 28.Pal R, Yu Q, Wang S, Kalyanaraman VS, Nair BC, Hudacik L, et al. Definitive toxicology and biodistribution study of a polyvalent DNA prime/protein boost human immunodeficiency virus type 1 (HIV-1) vaccine in rabbits. Vaccine. 2006 Feb 20;24(8):1225–34. doi: 10.1016/j.vaccine.2005.07.112. [DOI] [PubMed] [Google Scholar]
- 29.Kennedy JS, Co M, Green S, Longtine K, Longtine J, O'Neill MA, Adams JP, Rothman AL, Yu. Q, Johnson-Leva R, Pal R, Wang S, Lu S, Markham P. Phase 1 clinical trial evaluating the safety, tolerability, and antibody sero-conversion following administration of an HIV-1 DNA Prime-Protein Boost Vaccine (DP6−001) in healthy adult volunteers. Submitted.
- 30.Kennedy JS, Frey SE, Yan L, Rothman AL, Cruz J, Newman FK, et al. Induction of human T cell-mediated immune responses after primary and secondary smallpox vaccination. J Infect Dis. 2004 Oct 1;190(7):1286–94. doi: 10.1086/423848. [DOI] [PubMed] [Google Scholar]
- 31.Montefiori DC. Evaluating neutralizing antibodies against HIV, SIV and SHIV in luciferase reporter gene assays. In: Coligan JE, Kruisbeek AM, Margulies DH, Shevach EM, Strober W, Coico R, editors. Current Protocols in Immunology. John Wiley & Sons; 2004. pp. 12.1.1–.1.5. [DOI] [PubMed] [Google Scholar]
- 32.Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, et al. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol. 2005 Aug;79(16):10108–25. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li M, Salazar-Gonzalez JF, Derdeyn CA, Morris L, Williamson C, Robinson JE, et al. Genetic and neutralization properties of subtype C human immunodeficiency virus type 1 molecular env clones from acute and early heterosexually acquired infections in Southern Africa. J Virol. 2006 Dec;80(23):11776–90. doi: 10.1128/JVI.01730-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richman DD, Wrin T, Little SJ, Petropoulos CJ. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc Natl Acad Sci U S A. 2003 Apr 1;100(7):4144–9. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bansal A, Jackson B, West K, Wang S, Kennedy JS, Lu S, Goepdert Polyfunctional HIV-1 gp120 DNA vaccine-induced responses are enhanced with intra-muscular compared to intra-dermal administration. In preparation.
- 36.Mulligan MJ, Russell ND, Celum C, Kahn J, Noonan E, Montefiori DC, et al. Excellent safety and tolerability of the human immunodeficiency virus type 1 pGA2/JS2 plasmid DNA priming vector vaccine in HIV type 1 uninfected adults. AIDS Res Hum Retroviruses. 2006 Jul;22(7):678–83. doi: 10.1089/aid.2006.22.678. [DOI] [PubMed] [Google Scholar]
- 37.Wang B, Ugen KE, Srikantan V, Agadjanyan MG, Dang K, Refaeli Y, et al. Gene inoculation generates immune responses against human immunodeficiency virus type 1. Proc Natl Acad Sci U S A. 1993 May 1;90(9):4156–60. doi: 10.1073/pnas.90.9.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Amara RR, Villinger F, Altman JD, Lydy SL, O'Neil SP, Staprans SI, et al. Control of a mucosal challenge and prevention of AIDS by a multiprotein DNA/MVA vaccine. Science. 2001 Apr 6;292(5514):69–74. doi: 10.1126/science.1058915. [DOI] [PubMed] [Google Scholar]
- 39.Barouch DH, Santra S, Schmitz JE, Kuroda MJ, Fu TM, Wagner W, et al. Control of viremia and prevention of clinical AIDS in rhesus monkeys by cytokine-augmented DNA vaccination. Science. 2000 Oct 20;290(5491):486–92. doi: 10.1126/science.290.5491.486. [DOI] [PubMed] [Google Scholar]
- 40.Shiver JW, Fu TM, Chen L, Casimiro DR, Davies ME, Evans RK, et al. Replication-incompetent adenoviral vaccine vector elicits effective anti-immunodeficiency-virus immunity. Nature. 2002 Jan 17;415(6869):331–5. doi: 10.1038/415331a. [DOI] [PubMed] [Google Scholar]
- 41.Yasutomi Y, Robinson HL, Lu S, Mustafa F, Lekutis C, Arthos J, et al. Simian immunodeficiency virus-specific cytotoxic T-lymphocyte induction through DNA vaccination of rhesus monkeys. J Virol. 1996 Jan;70(1):678–81. doi: 10.1128/jvi.70.1.678-681.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.MacGregor RR, Boyer JD, Ugen KE, Lacy KE, Gluckman SJ, Bagarazzi ML, et al. First human trial of a DNA-based vaccine for treatment of human immunodeficiency virus type 1 infection: safety and host response. J Infect Dis. 1998 Jul;178(1):92–100. doi: 10.1086/515613. [DOI] [PubMed] [Google Scholar]
- 43.Ugen KE, Nyland SB, Boyer JD, Vidal C, Lera L, Rasheid S, et al. DNA vaccination with HIV-1 expressing constructs elicits immune responses in humans. Vaccine. 1998 Nov;16(19):1818–21. doi: 10.1016/s0264-410x(98)00180-7. [DOI] [PubMed] [Google Scholar]
- 44.Pitisuttithum P. HIV-1 prophylactic vaccine trials in Thailand. Curr HIV Res. 2005 Jan;3(1):17–30. doi: 10.2174/1570162052772933. [DOI] [PubMed] [Google Scholar]
- 45.Berman PW, Huang W, Riddle L, Gray AM, Wrin T, Vennari J, et al. Development of bivalent (B/E) vaccines able to neutralize CCR5-dependent viruses from the United States and Thailand. Virology. 1999 Dec 5;265(1):1–9. doi: 10.1006/viro.1999.0031. [DOI] [PubMed] [Google Scholar]
- 46.Bures R, Morris L, Williamson C, Ramjee G, Deers M, Fiscus SA, et al. Regional clustering of shared neutralization determinants on primary isolates of clade C human immunodeficiency virus type 1 from South Africa. J Virol. 2002 Mar;76(5):2233–44. doi: 10.1128/jvi.76.5.2233-2244.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deeks SG, Schweighardt B, Wrin T, Galovich J, Hoh R, Sinclair E, et al. Neutralizing antibody responses against autologous and heterologous viruses in acute versus chronic human immunodeficiency virus (HIV) infection: evidence for a constraint on the ability of HIV to completely evade neutralizing antibody responses. J Virol. 2006 Jun;80(12):6155–64. doi: 10.1128/JVI.00093-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang DC, DeVit M, Johnston SA. Genetic immunization is a simple method for eliciting an immune response. Nature. 1992 Mar 12;356(6365):152–4. doi: 10.1038/356152a0. [DOI] [PubMed] [Google Scholar]
- 49.Ulmer JB, Donnelly JJ, Parker SE, Rhodes GH, Felgner PL, Dwarki VJ, et al. Heterologous protection against influenza by injection of DNA encoding a viral protein. Science. 1993 Mar 19;259(5102):1745–9. doi: 10.1126/science.8456302. [DOI] [PubMed] [Google Scholar]
- 50.Robinson HL, Hunt LA, Webster RG. Protection against a lethal influenza virus challenge by immunization with a haemagglutinin-expressing plasmid DNA. Vaccine. 1993;11(9):957–60. doi: 10.1016/0264-410x(93)90385-b. [DOI] [PubMed] [Google Scholar]