Abstract
Polyglutamine disorders are inherited neurodegenerative diseases caused by the accumulation of expanded polyglutamine protein (polyQ). Previously, we identified a new guanosine triphosphatase, CRAG, which facilitates the degradation of polyQ aggregates through the ubiquitin–proteasome pathway in cultured cells. Because expression of CRAG decreases in the adult brain, a reduced level of CRAG could underlie the onset of polyglutamine diseases. To examine the potential of CRAG expression for treating polyglutamine diseases, we generated model mice expressing polyQ predominantly in Purkinje cells. The model mice showed poor dendritic arborization of Purkinje cells, a markedly atrophied cerebellum and severe ataxia. Lentivector-mediated expression of CRAG in Purkinje cells of model mice extensively cleared polyQ aggregates and re-activated dendritic differentiation, resulting in a striking rescue from ataxia. Our in vivo data substantiate previous cell-culture-based results and extend further the usefulness of targeted delivery of CRAG as a gene therapy for polyglutamine diseases.
Keywords: lentivirus, polyglutamine disease, gene therapy, Purkinje cell, CRAG
Introduction
Polyglutamine diseases are caused by an increase in the number of glutamine codon repeats within the open reading frame of the encoding gene (Koshy & Zoghbi, 1997). Expansion of polyglutamine repeats alters the conformation—or results in the misfolding—of the disease-associated protein, thereby conferring a toxic gain-of-function that is selectively deleterious to neurons (Sato et al, 1999; Yoshizawa et al, 2000). Recently, we identified a new guanosine triphosphatase (GTPase), CRAG, which is named after collapsin response mediator protein (CRMP)-associated molecule (CRAM[CRMP-5])-associated GTPase. CRAG contains a nuclear localization signal (NLS) sequence. In HeLa cells, CRAG interacts with expanded polyglutamine protein (polyQ) and transfers polyQ to the nucleus. Here, CRAG makes inclusions with polyQ and promyelocytic leukaemia protein body—the main component of nuclear bodies—in a GTPase-dependent manner. CRAG acts as an activator of promyelocytic leukaemia protein-associated ubiquitin ligase and leads to the degradation of polyQ through the ubiquitin–proteasome pathway (Qin et al, 2006). Because expression levels of CRAG decrease in the adult brain (Qin et al, 2006), we suggested that the reduced level of CRAG was a rate-limiting factor in the degradation of pathological forms of polyQ, and that targeted delivery of CRAG could be a potential gene therapy for polyglutamine diseases. Here, using an HIV-derived lentiviral vector, we verified this hypothesis by introducing CRAG into the cerebellar neurons of model mice overexpressing polyQ in the cerebellum.
Results
Generation of model mice with a polyglutamine disease
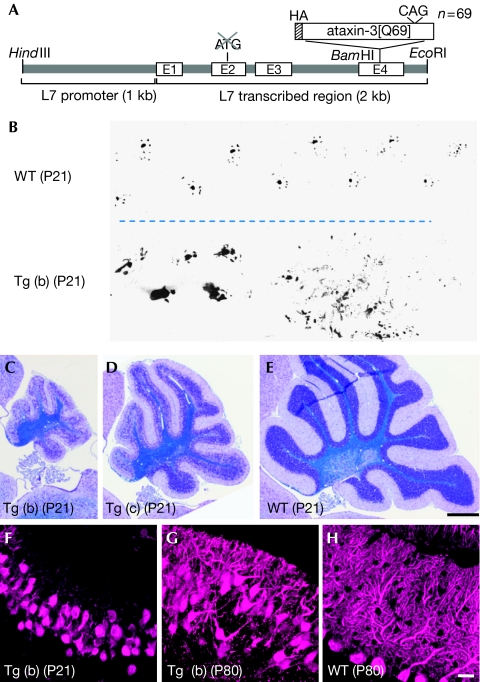
Cerebellar Purkinje cells are vulnerable to and easily degraded by various insults (Sarna & Hawkes, 2003), and a functional defect in Purkinje cells is expressed as cerebellar ataxia—that is, impaired balance and gait disturbance—which can be reliably assessed by a behavioural test. We produced transgenic mice (polyQ mice) expressing an expanded polyglutamine protein in Purkinje cells by using cDNA encoding a truncated form of human ataxin-3, the protein responsible for Machado–Joseph disease (spinocerebellar ataxia (SCA) type 3) with 69 glutamine repeats (ataxin-3[Q69]; Kawaguchi et al, 1994; Yoshizawa et al, 2000; Fig 1A). The transgene expression was driven by a Purkinje-cell-specific L7 promoter (Hirai et al, 2005). We obtained three founder lines—(a)–(c) lines—and their weight curves and rotarod performance from 3 to 14 weeks of age are shown in supplementary Fig 1A,B online. The mice of (a) and (b) lines showed severe ataxia and cerebellar atrophy from as early as 3 weeks of age, whereas the phenotypes of (c) line mice were milder (Fig 1B–E; supplementary Fig 1F and Video 1 online; data not shown). Quantitative reverse transcriptase (RT)–PCR analysis showed that the expression levels of ataxin-3 messenger RNA in polyQ mouse cerebella were much higher than that in the wild-type cerebellum (supplementary Fig 1C online). Among the three transgenic lines, (c) line mice contained the lowest amount of ataxin-3 mRNA in the cerebellum, consistent with severity of the phenotypes. With respect to CRAG, both mRNA and protein levels, examined by quantitative RT–PCR and western blots, were reduced in all three lines compared with those in wild-type littermates (supplementary Fig 1D,E online), presumably owing to overall suppression of transcription activity after severely impaired development and/or excessive accumulation polyQ in the nuclei and enhanced degradation of CRAG protein with polyQ. Immunohistochemistry showed the expression of endogenous CRAG in Purkinje cells and Bergmann glia, but not in granule cells (supplementary Fig 2A online). All three lines were fertile; however, we used mainly (b) line mice for further analysis because (c) line polyQ mice took more time to breed (polyQ mice that are mentioned in the following text are (b) line, unless otherwise stated).
Figure 1.
Severe ataxic phenotype and cerebellar defects of polyQ mice. (A) Schematic depicting the transgene that expresses mutant ataxin-3[Q69] under the control of the L7 promoter. The HA tag was fused at the amino terminus of the truncated ataxin-3. (B) Footprint of a (b) line polyQ mouse (Tg (b)) and a wild-type (WT) littermate at P21. (C–E) Sagittal sections of the cerebella of (b) and (c) line polyQ mutant and wild-type mice at P21 (C, D and E, respectively). (F–H) Purkinje cells of a (F) (b) line polyQ mouse at P21, (G) P80 and (H) a wild-type littermate at P80 that were immunolabelled for calbindin. Note the severe disarrangement of the Purkinje cell layer in the polyQ mouse as early as P21 (F). Scale bars, 500 μm (E) and 20 μm (H). HA, haemagglutinin; P, postnatal day; polyQ, expanded polyglutamine protein.
Ubiquitinated polyQ aggregates in Purkinje cells
Immunolabelling of sagittal sections of the postnatal day (P)21 and P80 polyQ mouse cerebellum for calbindin showed marked disarrangement of Purkinje cells, concomitant with substantial impairment of dendritic differentiation (Fig 1F–H). RT–PCR using various tissues from P10 polyQ mice showed that mutant ataxin-3 mRNA was present in the retina and cerebellum (supplementary Fig 3A online), consistent with the spatial expression of endogenous L7 protein (Oberdick et al, 1990). In the central nervous system, polyQ mRNA was detected in the brain stem and the cerebellum at P10, but only in the cerebellum at P80 (supplementary Fig 3B,C online). Immunostaining of the cerebellar sections for the haemagglutinin (HA) tag at the amino terminus of polyQ showed weak and diffuse accumulation of polyQ in the cytoplasm and in the nuclei of Purkinje cells at P5 (supplementary Fig 4A online); polyQ was localized to the nucleus at P10 and thereafter. Inclusion bodies started forming at P40, and the size and number were markedly increased at P80 (supplementary Fig 4B–E online). The inclusion bodies were intensely labelled with ubiquitin and expanded polyglutamine (1C2) antibodies (supplementary Fig 5A,C online); the inclusions were colocalized with CRAG (supplementary Fig 2B,C online). Immunoelectron microscopic examination for ubiquitin showed intense labelling of cytoplasmic inclusion bodies filled with abnormal filaments (supplementary Fig 5B online). In addition to the immunoreactivity in the Purkinje cell layer, small inclusion bodies with strong immunoreactivity for polyglutamine and ubiquitin were detected in the axon terminals of Purkinje cells in the deep cerebellar nuclei (supplementary Fig 5D online; data not shown). Immunolabelling for HA (mutant ataxin-3) was also observed in the superior colliculus, medial vestibular nuclei and cuneate nuclei of the P10 polyQ mouse brain, which were not detected at P80 (supplementary Fig 6 online).
Rescue from cerebellar ataxia by CRAG expression
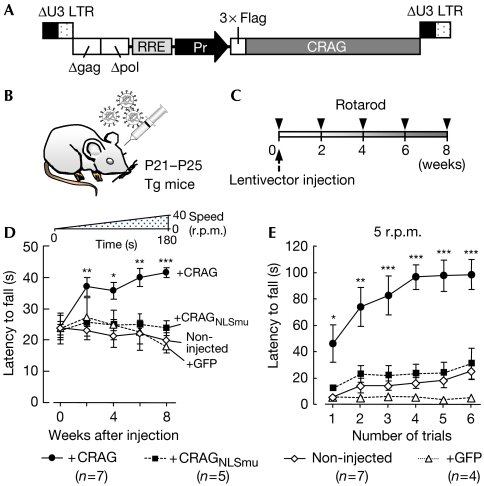
We investigated whether the delivery of recombinant CRAG in polyQ mouse Purkinje cells in vivo cleared the polyQ aggregates and, consequently, ameliorated ataxia. Lentiviral vectors expressing wild-type CRAG, mutant CRAG with a disrupted NLS sequence (CRAGNLSmu; Qin et al, 2006) or green fluorescent protein (GFP) were injected into the midline cerebellar lobules of P21–P25 mice (Fig 2A,B), as described previously (Torashima et al, 2006). The effects of protein expression on behaviour were assessed by using a rotarod test, in which mice were challenged with an accelerating rod paradigm just before the viral injection and then every 2 weeks up to 8 weeks after the viral injection (Fig 2C and inset in Fig 2D). The rotarod performance of non-injected polyQ mice and polyQ mice expressing CRAGNLSmu or GFP deteriorated slightly at 8 weeks (Fig 2D). By contrast, the performance of mice treated with wild-type CRAG improved significantly after the injection compared with the non-injected mice. The footprints of polyQ mice 8 weeks after treatment with wild-type CRAG, CRAGNLSmu or GFP are shown in supplementary Fig 7 online; the rescue was visually evident (supplementary Video 2 online).
Figure 2.
Rescue of the ataxic phenotype in polyQ mice following lentivector-mediated expression of CRAG. (A) Schematic depicting the lentivector constructs. 3 × Flag-tagged CRAG was expressed by the cytomegalovirus promoter or the murine stem-cell virus promoter (Pr). Δgag, deleted gag sequence; Δpol, deleted pol sequence; RRE, rev responsive element. (B,C) Lentivirus particles were injected into the cerebellar cortex of polyQ mice at P21–P25 (B), and the effects were examined by rotarod performance just before injection and every 2 weeks up to 8 weeks after the injection (C). (D,E) Results of the rotarod test. (D) The rod accelerated in 3 min from 0 to 40 r.p.m. as depicted above the graph. Mice treated with wild-type CRAG (closed circles), but not untreated mice (open diamonds), those treated with NLS-sequence-mutated CRAG (CRAGNLSmu, closed squares) or GFP (open triangles), showed significant improvement (n=number of individual mice in each cohort). (E) In the stable-rod-speed paradigm (5 r.p.m.) tested 8 weeks after the injection, mice treated with CRAG quickly learnt how to walk on the rod and showed better performance than non-injected, CRAGNLSmu-treated or GFP-expressing mice. *P<0.05, **P<0.01, ***P<0.001, compared with the results of non-injected mice. GFP, green fluorescent protein; LTR, long terminal repeat; NLS, nuclear localization signal; polyQ, expanded polyglutamine protein; Tg, transgenic.
To investigate the effect of CRAG expression on motor learning, mice were evaluated by using a rotarod test with a different paradigm, in which the rod speed was fixed at 5 r.p.m. and the trial was repeated six times with a 5-min interval. Non-injected polyQ mice and those treated with CRAGNLSmu or GFP showed almost no improvement in the performance, even on the sixth trial (Fig 2E). By contrast, mice treated with CRAG quickly learnt how to stay on the rod, indicating the rescue of motor learning ability.
Clearance of polyQ aggregates by CRAG expression
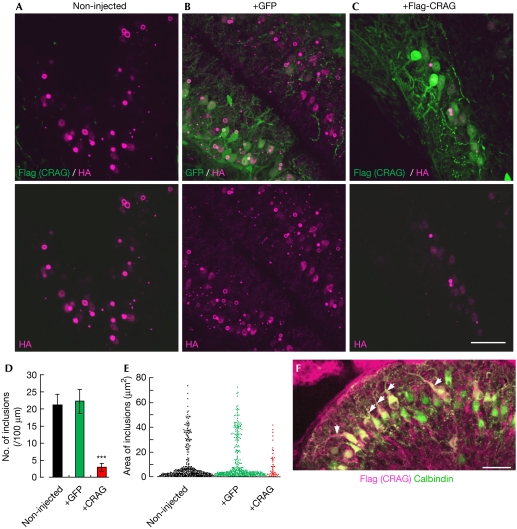
Next, we immunohistochemically examined the cerebellar sections from untreated mice or mice treated with the lentiviral vectors. Slices were double immunolabelled for HA (tagged with ataxin-3[Q69]) and Flag (tagged with CRAG). HA immunoreactivity in the Purkinje cell layer was markedly decreased 8 weeks after treatment with CRAG, but not with GFP, compared with that in non-injected mice (Fig 3A–C). Supplementary Fig 8 online shows an example obtained from a CRAG-treated polyQ mouse in which ataxia was markedly rescued 8 weeks after the viral injection (supplementary Fig 8A online): CRAG was expressed in lobule 10 (vestibulocerebellum; supplementary Fig 8B online, arrow), and immunoreactivity for polyQ was substantially lower at the corresponding area (supplementary Fig 8C,D online).
Figure 3.
Marked reduction in polyQ and the extension of dendrites in mutant Purkinje cells that express CRAG. PolyQ mice 8 weeks after viral injection (∼P80) were used for the analysis. (A–C) The cerebellar slices from (A) non-injected, (B) GFP-expressing and (C) CRAG-expressing polyQ mice that were double immunolabelled for Flag (tagged with CRAG; green) and HA (tagged with ataxin-3[Q69]; magenta). The lower panels show only immunoreactivity for HA (ataxin-3[Q69]). (D,E) Graphs showing (D) the number and (E) area of polyQ inclusions. Numbers of inclusions per 100 μm of Purkinje cell layer are presented in (D), whereas the areas of all inclusions and aggregates detected by imaging software are plotted in (E). (F) Dendritic extension of Purkinje cells that expressed recombinant CRAG. A cerebellar section of a polyQ mouse that received an injection of the lentivector expressing CRAG was double immunolabelled for CRAG (magenta) and calbindin (green). Note that only transduced (CRAG-expressing) Purkinje cells extend their dendrites (arrows). Scale bars, 50 μm (C,F). ***P<0.001, compared with the results of non-injected mice. GFP, green fluorescent protein; HA, haemagglutinin; polyQ, expanded polyglutamine protein.
To quantify the effect of CRAG expression on polyQ inclusions, we measured the number and area of polyQ inclusions. A total of 1,792, 1,763 and 1,700 μm along the Purkinje cell layer in CRAG-treated (n=6 mice), GFP-treated (n=4 mice) and non-injected (n=4 mice) mouse cerebella, respectively, were assessed. In non-injected mice, the same area that was measured for CRAG-treated slices was used for the analysis. Non-injected and GFP-expressing polyQ mice had 368 and 382 inclusions (21.6 and 21.7 inclusions/100 μm Purkinje cell layer), respectively, whereas the number markedly decreased in CRAG-treated mice to 47 inclusions (2.6 inclusions/100 μm Purkinje cell layer; P<0.001; Fig 3D). Subsequently, we measured the area of those inclusions. CRAG, but not GFP, treatment shifted the area of distribution in nontreated polyQ mice downwards: large inclusions almost disappeared with CRAG expression (Fig 3E). The Purkinje cell layer in a CRAG-treated polyQ mouse showed a monolayer arrangement (supplementary Fig 9A online, arrows), in contrast to the severely disarranged Purkinje cell layers in non-injected (Fig 1G) and GFP-treated (supplementary Fig 9B online) polyQ mice. Furthermore, immunolabelling of Purkinje cells for calbindin and Flag (CRAG) showed that only those cells that expressed CRAG extended their dendrites (Fig 3F, arrows).
As shown in supplementary Fig 10A–F online, recombinant CRAG was also colocalized with polyQ inclusions (arrows) and aggregates (arrowheads). Next, we examined the expression levels of recombinant CRAG in the infected lobules compared with the endogenous levels in non-infected lobules. To detect cerebellar lobules expressing CRAG without immunostaining, CRAG was fused with GFP directly (GFP-CRAG) or indirectly (GFP-P2A-CRAG) using the Picornavirus ‘self-cleaving' 2A peptide sequence (P2A). By using lentiviral vectors, one of the GFP-CRAG fusion proteins was expressed in the polyQ mouse cerebellum. Eight weeks after the injection, cerebellar slices were taken and CRAG-expressing lobules were identified by GFP fluorescence. Total mRNA was isolated from lobules with and without CRAG expression, followed by quantitative real-time PCR analysis after reverse transcription of CRAG mRNA. The results showed that the expression levels of CRAG mRNA in transduced lobules were about seven times higher than that of non-injected lobules (supplementary Fig 10G online).
We conducted further quantitative RT–PCR to confirm that the effect of CRAG was due to the enhancement of polyQ degradation and not due to the suppression of transgene expression. The results showed that the amount of ataxin-3[Q69] mRNA in transduced (recombinant CRAG-expressing) lobules of the polyQ mouse cerebellum was 92% of that in non-transduced lobules of the same mouse, indicating that the CRAG-induced removal of polyQ inclusions was not due to the reduction in polyQ transgene expression.
Involvement of the ubiquitin–proteasome pathway
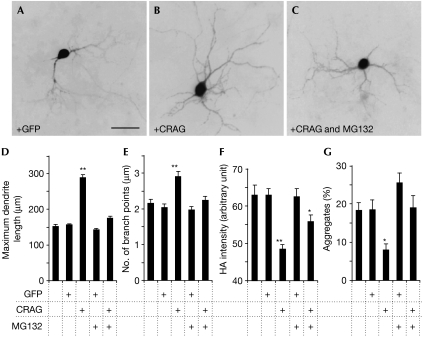
Our previous study using HeLa cells showed that CRAG-mediated degradation of polyQ is by means of the ubiquitin–proteasome pathway (Qin et al, 2006). Therefore, we examined whether this is true for polyQ mouse Purkinje cells. GFP-CRAG was expressed in the cerebella of polyQ mice, and CRAG-expressing lobules were separated from non-expressing lobules by using GFP fluorescence. Western blot analysis showed enhanced ubiquitination in lobules expressing CRAG (supplementary Fig 11A online). Furthermore, we examined ubiquitination of polyQ by immunoprecipitation assay: polyQ (HA-tagged ataxin-3[Q69]) was immunoprecipitated, using expanded polyQ antibody (1C2), from lobules with or without GFP-CRAG and analysed by immunoblot with ubiquitin and HA antibodies. As shown in supplementary Fig 11B online, ubiquitinated polyQ was clearly increased by GFP-CRAG expression. Involvement of the ubiquitin–proteasome pathway in CRAG-mediated degradation of polyQ was verified by using a primary culture of Purkinje cells. Cerebellar cells from P0 polyQ mouse pups were infected at day 0 in vitro with lentiviral vectors expressing CRAG or GFP under the control of the Purkinje-cell-specific L7 promoter so as to express CRAG and GFP only in Purkinje cells. Purkinje cells or polyQ aggregates were visualized by immunolabelling for calbindin or HA (tagged with polyQ). The length and number of branch points in Purkinje cell dendrites significantly increased with CRAG expression but not with GFP expression (Fig 4A,B,D,E). The effect of CRAG on dendritic differentiation was reversed by the presence of MG132—a proteasome inhibitor—in the culture medium (Fig 4C–E). Similarly, both HA intensity and the percentage ratio of Purkinje cells with inclusions were significantly decreased by CRAG expression, but not with GFP expression, and this effect was reversed by the addition of MG132 to the medium (Fig 4F,G). These results indicate that, as shown previously in HeLa cells, the effects of CRAG on polyQ degradation and dendritic differentiation in Purkinje cells were closely associated with the ubiquitin–proteasome pathway.
Figure 4.
Involvement of the ubiquitin–proteasome pathway in CRAG-mediated dendritic extension and degradation of polyQ aggregates in cultured Purkinje cells. (A–C) Fluorescence images of polyQ mouse Purkinje cells expressing (A) GFP or (B,C) CRAG. Dissociated cerebellar neurons were infected with lentivectors at day 0 in vitro and cultured for 14 days in the (A,B) absence or (C) presence of the proteasome inhibitor MG132 (0.1 nM). Then, the cells were immunostained for Flag (tagged with CRAG) and calbindin or HA (tagged with polyQ). (D–G) The maximum dendrite length (D), number of branch points per longest dendrite (E), fluorescence intensity of the polyQ aggregates (F), and ratio (%) of Purkinje cells with polyQ aggregates (G). The results were obtained from four independent cultures using two batches of lentiviral vectors. A total of 40–90 Purkinje cells were analysed to obtain each result. Note that in all cases, the presence of MG132 in the culture medium blocked the effects of CRAG. *P<0.05, **P<0.01, compared with the results of non-infected Purkinje cells. Scale bar, 50 μm. GFP, green fluorescent protein; HA, haemagglutinin; polyQ, expanded polyglutamine protein.
Discussion
Previously, SCA3 model mice, which express a truncated form of ataxin-3—with an expanded polyglutamine stretch—under the control of the L7 promoter, were generated by Ikeda et al (1996). Although the mice showed distinct ataxia and marked cerebellar atrophy, mutant ataxin-3 was hardly detected by immunohistochemistry. Furthermore, there were almost no Purkinje cells present in the cerebellum. Thus, these model mice cannot be used to examine the potential of CRAG for polyQ degradation. Recently, two SCA3 model mice that express full-length ataxin-3 with expanded polyglutamine repeats by the mouse prion promoter were independently reported (Goti et al, 2004; Bichelmeier et al, 2007), whereas Cemal et al (2002) generated model mice using a yeast artificial chromosome construct spanning the human SCA3 locus containing up to 84 CAG repeats in the SCA3 gene under the control of its endogenous promoter. These three model mice showed a phenotype similar to that observed in Machado–Joseph disease pathology, including neurodegeneration with intranuclear inclusions in different regions such as the cortex, hippocampus, pons and cerebellar nuclei. Therefore, they are superior in the study of SCA3 pathogenesis to our polyQ mice, which are affected only in the cerebellum. However, by taking advantage of the confined lesions in our model mice, we could effectively evaluate a causal relationship between lentivector-mediated expression of CRAG, degradation of polyQ aggregates and recovery from ataxia. Thus, our polyQ mice will be useful, as shown here, in viral-vector-based screening of genes for therapeutic molecules that have the potential to rescue cerebellar neurons and, consequently, motor control ability.
Although the accelerating rotarod performance of CRAG-treated polyQ mice (40–45 s; Fig 2D) was still poorer than that of wild-type mice (∼150 s; supplementary Fig 1B online), it should be noted that the cerebella of our transgenic mice at around P25 are very small, with gross morphological defects in Purkinje cells (Fig 1C–H). Nevertheless, clearance of polyQ aggregates from Purkinje cells markedly improved the ataxic phenotype of these polyQ mice. Thus, excessive accumulation of polyQ aggregates in Purkinje cells is thought to cause profound but reversible defects in these cells, and a limited number of rescued Purkinje cells could substantially compensate the cerebellar function. As a next step, the potential of CRAG delivery in fully matured animals should be verified, as P25 mutant cerebella could be still in the process of development.
The misarrangement and disorientation of the soma of Purkinje cells in the polyQ mice were ameliorated as the polyQ aggregates were cleared (supplementary Fig 9 online). This finding suggests that normal function of Purkinje cells is required to maintain the correct dendrite/axon orientation and monolayer arrangement of Purkinje cell bodies.
Previously, using adeno-associated viral vectors, Davidson and co-workers (Xia et al, 2004) expressed RNA interference in Purkinje cells of SCA1 model mice to suppress the expression of a mutant SCA1 gene, which significantly slowed down disease progression. Our approach, of enhancing the ubiquitin–proteasome pathway, provides an alternative method to protect neurons from toxic polyQ accumulation. This approach might be promising as gene therapy for not only polyQ diseases, but also disorders that are caused by the accumulation of toxic substances in neuronal and/or glial cells such as Alzheimer disease, Parkinson disease and prion diseases.
Methods
Transgenic mice. The mice used express N-terminal truncated ataxin-3 with Q69 together with an N-terminal HA epitope in cerebellar Purkinje cells. The ataxin-3 in these mice lacks the 286 N-terminal amino-acid residues (see the supplementary information online for details; Yoshizawa et al, 2000). All procedures relating to the care and treatment of animals were carried out according to National Institutes of Health guidelines, and the experimental protocol was approved by the Animal Resource Committees of Gunma University.
RT–PCR analysis. Total RNA (400 ng) was reverse transcribed using a RETRO script (Ambion, Austin, TX, USA). Quantitative RT–PCR was run on a Thermal Cycler Dice TP800 system (Takara Bio, Otsu, Japan) using SYBR Premix Ex Taq II (Takara Bio) with cycles of 95°C for 5 s and 60°C for 30 s (see the supplementary information online for details).
Histological methods. Brain sections were immunostained with rabbit polyclonal calbindin D28K (Sigma, St Louis, MO, USA), mouse monoclonal HA (InvivoGen, San Diego, CA, USA), rabbit polyclonal ubiquitin (Dako, Carpinteria, CA, USA) or mouse monoclonal expanded polyglutamine (1C2; Chemicon, CA, USA) antibodies. We also carried out Klüver–Barrera staining and immunoelectron microscopy (see the supplementary information online for details including western blotting).
Virus preparation and cerebellar injection. Vesicular stomatitis virus-G protein-pseudotyped lentiviral vectors provided by St Jude Children's Research Hospital were used in this study. Four microlitres of virus solution was injected into the molecular layer of the cerebellar vermis (see the supplementary information online for details).
Acquisition and analysis of fluorescent images. Fluorescent images were obtained by using a confocal laser-scanning microscope (LSM 5 PASCAL; Zeiss, Oberkochen, Germany) or a cooled CCD camera attached to a fluorescence microscope (DMI6000 B; Leica, Nusslock, Germany). In the confocal microscopic analysis, cerebellar slices were scanned at 1 μm intervals, and 10 sections were projected (see the supplementary information online for details).
Cerebellar neuronal culture. Dissociated cerebellar neuronal cultures were prepared from P0 mouse pups (see the supplementary information online for details).
Statistics. Statistical differences were analysed by Dunnett's post hoc test after one-way analysis of variance, two-tailed t-test or Mann–Whitney U-test. Data are expressed as the mean±s.e.m.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Video 1
Supplementary Video 2
Supplementary Video 3
Supplementary Video 4
Supplementary Video 5
Supplementary Information
Acknowledgments
This work was supported by the SORST programme of the Japan Science and Technology Agency, a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science and grants from the Mitani Foundation (to H.H.).
Footnotes
The authors declare that they have no conflict of interest.
References
- Bichelmeier U et al. (2007) Nuclear localization of ataxin-3 is required for the manifestation of symptoms in SCA3: in vivo evidence. J Neurosci 27: 7418–7428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cemal CK, Carroll CJ, Lawrence L, Lowrie MB, Ruddle P, Al-Mahdawi S, King RHM, Pook MA, Huxley C, Chamberlain S (2002) YAC transgenic mice carrying pathological alleles of the MJD1 locus exhibit a mild and slowly progressive cerebellar deficit. Hum Mol Genet 11: 1075–1094 [DOI] [PubMed] [Google Scholar]
- Goti D et al. (2004) A mutant ataxin-3 putative-cleavage fragment in brains of Machado–Joseph disease patients and transgenic mice is cytotoxic above a critical concentration. J Neurosci 24: 10266–10279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai H, Miyazaki T, Kakegawa W, Matsuda S, Mishina M, Watanabe M, Yuzaki M (2005) Rescue of abnormal phenotypes of the δ2 glutamate receptor-null mice by mutant δ2 transgenes. EMBO Rep 6: 90–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda H, Yamaguchi M, Sugai S, Aze Y, Narumiya S, Kakizuka A (1996) Expanded polyglutamine in the Machado–Joseph disease protein induces cell death in vitro and in vivo. Nat Genet 13: 196–202 [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y et al. (1994) CAG expansions in a novel gene for Machado–Joseph disease at chromosome 14q32.1. Nat Genet 8: 221–228 [DOI] [PubMed] [Google Scholar]
- Koshy BT, Zoghbi HY (1997) The CAG/polyglutamine tract diseases: gene products and molecular pathogenesis. Brain Pathol 7: 927–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberdick J, Smeyne RJ, Mann JR, Zackson S, Morgan JI (1990) A promoter that drives transgene expression in cerebellar Purkinje and retinal bipolar neurons. Science 248: 223–226 [DOI] [PubMed] [Google Scholar]
- Qin Q, Inatome R, Hotta A, Kojima M, Yamamura H, Hirai H, Yoshizawa T, Tanaka H, Fukami K, Yanagi S (2006) A novel GTPase, CRAG, mediates promyelocytic leukemia protein-associated nuclear body formation and degradation of expanded polyglutamine protein. J Cell Biol 172: 497–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarna JR, Hawkes R (2003) Patterned Purkinje cell death in the cerebellum. Prog Neurobiol 70: 473–507 [DOI] [PubMed] [Google Scholar]
- Sato A et al. (1999) Adenovirus-mediated expression of mutant DRPLA proteins with expanded polyglutamine stretches in neuronally differentiated PC12 cells. Preferential intranuclear aggregate formation and apoptosis. Hum Mol Genet 8: 997–1006 [DOI] [PubMed] [Google Scholar]
- Torashima T, Okoyama S, Nishizaki T, Hirai H (2006) In vivo transduction of murine cerebellar Purkinje cells by HIV-derived lentiviral vectors. Brain Res 1082: 11–22 [DOI] [PubMed] [Google Scholar]
- Xia H, Mao Q, Eliason SL, Harper SQ, Martins IH, Orr HT, Paulson HL, Yang L, Kotin RM, Davidson BL (2004) RNAi suppresses polyglutamine-induced neurodegeneration in a model of spinocerebellar ataxia. Nat Med 10: 816–820 [DOI] [PubMed] [Google Scholar]
- Yoshizawa T, Yamagishi Y, Koseki N, Goto J, Yoshida H, Shibasaki F, Shoji S, Kanazawa I (2000) Cell cycle arrest enhances the in vitro cellular toxicity of the truncated Machado–Joseph disease gene product with an expanded polyglutamine stretch. Hum Mol Genet 9: 69–78 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Video 1
Supplementary Video 2
Supplementary Video 3
Supplementary Video 4
Supplementary Video 5
Supplementary Information