Chromatin insulators have evolved to regulate transcription by using regulatory elements that are often distant from each other on the linear genome. This feature is position-dependent—that is, a functional insulator must be positioned between the enhancer and its target promoter (Wallace & Felsenfeld, 2007). The molecular basis of insulation seems to be tightly linked to, and might depend on, several long-range physical interactions between different portions of the chromatin fibre (Wallace & Felsenfeld, 2007). This is exemplified by the H19 imprinting control region, which not only regulates long-range interactions but also the epigenetic states of a crucial cis element (Kurukuti et al, 2006). Independent suggestions have promoted the idea that components of the cohesin complex might participate in the formation of such complexes and contribute to the insulator function (Hagstrom & Meyer, 2003). This proposal has been partly borne out in three seminal reports published in recent issues of Nature (Wendt et al, 2008), Cell (Parelho et al, 2008) and The EMBO Journal (Stedman et al, 2008). Although the full implications of these observations remain to be elucidated, they have shed new light on how the enigmatic chromatin insulators might operate.
How do the cohesins work and how are they organized? The cohesin complex comprises four components termed Smc1, Smc3, Scc3 and Scc1 (Smc for structural maintenance of chromosomes; Scc for Subunit of the cohesin complex), also known as Rad21. It has been proposed that Smc1 and Smc3 form a heterodimer together with Scc1, and that Scc3 organizes a ring structure. In addition, Scc2 and Scc4 are required for the loading of the cohesin complex onto the chromatin fibre. Functionally, the cohesin complex holds sister chromatids together after DNA replication until their sequential separation during G2/M phases. This function is essential for genomic DNA stability and repair (for reviews, see Hagstrom & Meyer, 2003; Uhlmann, 2008).
All three of the new reports have used so-called ChIP-on-chip analysis (ChIP for chromatin immunoprecipitation) to score for patterns of cohesin epitopes along the chromatin fibre. The outcomes of these studies are similar—that cohesin subunits are distributed both at transcription units and within intergenic regions in mouse and human cells. This is in contrast to the situation reported in yeast in which cohesins have been proposed to be pushed to intergenic regions by the transcriptional machinery (Glynn et al, 2004; Lengronne et al, 2004). It is of note that cohesin distribution on the chromatin fibre in Drosophila shows yet another pattern: most of the cohesin distributes on transcriptionally active regions and co-localizes with RNA polymerase II (Misulovin et al, 2008). Therefore, it seems that a conserved set of genes have acquired mechanistically different functions during evolution.
It is of substantial interest that all three reports document that many sequences that interact with Smc3 and Scc1 (Parelho et al, 2008), Smc3, Scc1 and Scc2 (Wendt et al, 2008) and Smc1, Smc3, as well as Scc1 (Stedman et al, 2008), also interact with CCCTC-binding factor (CTCF)-binding sites. Stedman and colleagues examined viral sequences, as well as a limited set of genomic sequences, whereas the other two studies included a larger set of genomic sequences. In particular, Wendt and colleagues revealed extensive overlap in the binding patterns of SCC1 and CTCF for the whole human genome. This association is exciting as CTCF, which has been linked with not only chromatin insulation but also transcriptional activation and silencing (Ohlsson et al, 2001), is emerging as a crucial player in the relationship between gene-expression domains and long-range interactions between chromatin fibres (Kurukuti et al, 2006; Splinter et al, 2006; Zhao et al, 2006).
Although it is clear that cohesins recognize CTCF, directly or indirectly, for their positioning throughout the genome, two of the new studies argue that cohesins can also interact with the chromatin fibre in a CTCF-independent manner. This conclusion was drawn from experiments using deletion mutants and RNA interference (RNAi) to suppress the expression of Smc3, Scc1, Scc2 and CTCF. The available data suggest that the bulk of chromatin-bound cohesin is formed independently of CTCF-binding sites. This deduction was reinforced by the demonstrations that downregulation of CTCF did not impede mitosis and hence sister chromatid cohesion (Parelho et al, 2008; Wendt et al, 2008). Therefore, the emerging message is that CTCF might not have a significant role in sister chromatid cohesion but that cohesins might determine the function of CTCF, at least at some specific sites.
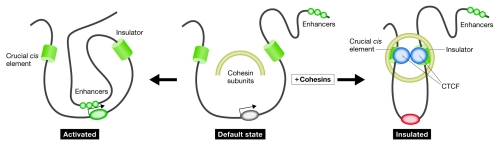
On the basis of both direct and indirect evidence, all three reports suggest that cohesins regulate the insulator function of CTCF (Fig 1). These data were obtained using mouse cells transfected with either a maternal or paternal human chromosome 11, or cells transfected with plasmid constructs designed to determine insulator function. Although the data are largely convincing, the effects of cohesin on endogenous imprinted genes were not examined. More importantly, none of the reports addressed the scenario that cohesin subunits might organize higher-order chromatin structures perceived to be crucial for the insulator function at endogenous loci. It is essential to perform 3C (chromosome conformation capture) analysis of cells, manipulated to express different levels of CTCF or cohesin subunits, before more firm conclusions can be drawn about the role of cohesin in insulation.
Figure 1.
Hypothetical model of how the cohesin complexes might cooperate with CCCTC-binding factor to establish a chromatin insulator. The model is based on data obtained largely with G2/M cells. It is as yet unknown whether this structure can also exist during the remainder of the cell cycle. The figure also proposes that the CTCF–cohesin complexes might establish and stabilize long-range interactions. The proposal that this interaction involves CTCF–CTCF interaction is entirely hypothetical. See text for further details. CTCF, CCCTC-binding factor.
The indirect character of the RNAi strategy calls for some caution in the interpretation of these data. Parelho and colleagues point out that as the levels of cohesins decrease beyond a crucial point in RNAi-treated cells, a G2/M arrest is an expected outcome. Will such cells faithfully record functionality in the insulator studies? This issue is compounded by the general appearance of the ring structure of the cohesin complex only during S phase (Hagstrom & Meyer, 2003). One possibility is that a non-canonical (possibly ring-like) structure is formed between cohesin components from two different chromosomal regions to bring together and stabilize pivotal cis elements that collaborate to manifest the insulator function in the G1 phase (Fig 1). This scenario has a bearing on the observation that a chromodomain factor, CHD8, also associates with CTCF and is essential for its insulator function (Ishihara et al, 2006). As CHD8 also has sucrose non-fermenter 2 (SNF2)-like helicase activity, it might release torsional stress at DNA strands engaged in long-range interactions. However, if a cohesin-like ring structure is formed between insulators on different chromosomes, cells might be arrested at the decatenation checkpoint, unless insulator–cohesin complexes are disassociated at this point of the cell cycle. Therefore, we cannot rule out that any higher-order chromatin structure organized by the CTCF-cohesin link applies primarily in cis.
It goes without saying that even three seminal reports cannot cover all the ground of this exciting new perspective of cohesin functions. The repeat elements stand out amongst the missing pieces in all three reports as they are not scored for in the screening strategies. Cohesins are known to be loaded onto extensive repeat elements within constitutive heterochromatin such as centromeres. Do CTCF and cohesins also cooperate in such instances? Alternatively, do cohesin complexes form through different pathways with only a subfraction associated to CTCF (Parelho et al, 2008)? RNAi experiments alone might not be sufficient to resolve this question as many juxtaposed CTCF-binding sites within such heterochromatic blocks might cooperate to achieve full occupancy even at low levels of CTCF. The other side of this issue is whether cohesin complexes would actually be able to function as insulators in instances when their association with the chromatin fibre is CTCF-independent? On a more practical level, will the CTCF–cohesin link explain human diseases from new perspectives? Mutations influencing the composition of the cohesin complex lead to the development of the Cornelia de Lange syndrome. Will such patients also display epimutations, such as loss of Igf2 imprinting? Notably, loss of Igf2 imprinting—a common epimutation in human cancer and an expected outcome with loss of cohesin functions—has been associated with a predisposition to colon cancer (Sakatani et al, 2005). Stedman and colleagues also linked CTCF and cohesin to the life cycle of DNA viruses (Stedman et al, 2008), which might open new strategies for the intervention of pathological DNA viruses. Clearly, the discovery of the cohesin–CTCF link has not only provided a new perspective on the partitioning of expression domains, but also opened a Pandora's box of new questions and opportunities.


Acknowledgments
We acknowledge valuable comments from M. Merkenschlager. The authors are supported by grants from the European Union (HEROIC), Swedish Cancer Research Foundation and Swedish Research Council.
References
- Glynn EF, Megee PC, Yu HG, Mistrot C, Unal E, Koshland DE, DeRisi JL, Gerton JL (2004) Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol 2: E259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagstrom KA, Meyer BJ (2003) Condensin and cohesin: more than chromosome compactor and glue. Nat Rev Genet 4: 520–534 [DOI] [PubMed] [Google Scholar]
- Ishihara K, Oshimura M, Nakao M (2006) CTCF-dependent chromatin insulator is linked to epigenetic remodeling. Mol Cell 23: 733–742 [DOI] [PubMed] [Google Scholar]
- Kurukuti S, Tiwari VK, Tavoosidana G, Pugacheva E, Murrell A, Zhao Z, Lobanenkov V, Reik W, Ohlsson R (2006) CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc Natl Acad Sci USA 103: 10684–10689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengronne A, Katou Y, Mori S, Yokobayashi S, Kelly GP, Itoh T, Watanabe Y, Shirahige K, Uhlmann F (2004) Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature 430: 573–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misulovin Z et al. (2008) Association of cohesin and nipped-B with transcriptionally active regions of the Drosophila melanogaster genome. Chromosoma 117: 89–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsson R, Renkawitz R, Lobanenkov V (2001) CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet 17: 520–527 [DOI] [PubMed] [Google Scholar]
- Parelho V et al. (2008) Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 132: 422–433 [DOI] [PubMed] [Google Scholar]
- Sakatani T, Kaneda A, Lacobuzio-Donahue CA, Carter MG, de Boom Witzel S, Okano H, Ko MS, Ohlsson R, Longo DL, Feinberg AP (2005) Loss of imprinting of Igf2 alters intestinal maturation and tumorigenesis in mice. Science 307: 1976–1978 [DOI] [PubMed] [Google Scholar]
- Splinter E, Heath H, Kooren J, Palstra RJ, Klous P, Grosveld F, Galjart N, de Laat W (2006) CTCF mediates long-range chromatin looping and local histone modification in the β-globin locus. Genes Dev 20: 2349–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedman W, Kang H, Lin S, Kissil JL, Bartolomei MS, Lieberman PM (2008) Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J 27: 654–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F (2008) Molecular biology: cohesin branches out. Nature 451: 777–778 [DOI] [PubMed] [Google Scholar]
- Wallace JA, Felsenfeld G (2007) We gather together: insulators and genome organization. Curr Opin Genet Dev 17: 400–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt KS et al. (2008) Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451: 796–801 [DOI] [PubMed] [Google Scholar]
- Zhao Z et al. (2006) Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet 38: 1341–1347 [DOI] [PubMed] [Google Scholar]