Abstract
Mitogen-activated protein kinase (MAPK) cascades process myriads of stimuli, generating receptor-specific cellular outcomes. New work exploits emergent mathematics of network inference to reveal distinct feedback designs of the RAF/MEK/ERK cascade induced by two different growth factors. It shows that response specificity can arise from differential signal-induced wiring of overlapping protein networks.
The RAF/MEK/ERK cascade is activated by countless external cues that stimulate diverse receptors. How distinct receptors dictate different cellular outcomes by activating the same signalling module has long fascinated many researches. Initial clues came from observations that the duration of ERK activation is critical to cell fate decisions1. In classical experiments, PC12 cells proliferated after transient ERK activation by epidermal growth factor (EGF), but terminally differentiated after more sustained ERK activation by nerve growth factor (NGF)1. Subsequent work suggested that the duration of ERK signalling is interpreted by cells through a network of immediate-early genes2. Nevertheless, how different ERK dynamics can be robustly controlled by upstream receptors still remains unclear, although several alternative mechanisms have been proposed3,4. On page xxx of this issue, Santos et al5 offer new insight into the causes of the distinct temporal profiles of MAPK activity that determine cell fates. They show that EGF and NGF elicit different feedback architecture in the MAPK network and these distinct feedback designs precisely determine cellular decisions.
Quantitative approaches have revolutionized how we ask biological questions and carry out experiments6. The study by Santos et al5 embraces mathematical and experimental systems approaches to address key biological questions. This study has four main elements5. First, it determined the wiring of the MAPK network by exploiting new systems biology methods of unravelling a network’s connection architecture from analyzing responses to successive perturbations7. Second, it found that EGF, which stimulates EGF receptor (EGFR), elicits negative feedback from ERK, whereas NGF, which activates TrkA receptor, induces positive feedback. Third, it found positive feedback not only sustained ERK activity for long periods, but also induced switch-like, bistable dynamics of the NGF-stimulated ERK cascade. Finally, Santos et al5 could rewire the EGF-stimulated MAPK network by concomitant activation of protein kinase C (PKC). Simultaneous activation with EGF and phorbol ester transformed the negative feedback from ERK into a positive feedback and caused PC12 cells to abandon their decision to proliferate and instead to undergo differentiation as if stimulated with NGF. Conversely, PKC inhibition caused NGF-stimulated cells to proliferate rather than differentiate.
Inferring connections within complex signalling networks is a fundamental problem in cell biology. However, it is not immediately obvious how to capture interactions between individual signalling nodes, since any experimental perturbation to a particular component rapidly propagates through a network, causing widespread (global) changes. Many research groups embarked on the search for molecular interactions that cause the observed behavior of complex molecular networks8-10. Fortunately, similar questions have long been studied for metabolic networks, and a new algorithm in modular response analysis (MRA) 7 used by Bastiaens and colleagues5 is rooted in metabolic control analysis8. Fig. 1 illustrates how the topology and strength of connections between nodes is revealed by analyzing network responses to parameter perturbations at steady states.
Fig. 1. Quantification and unravelling of network connections.
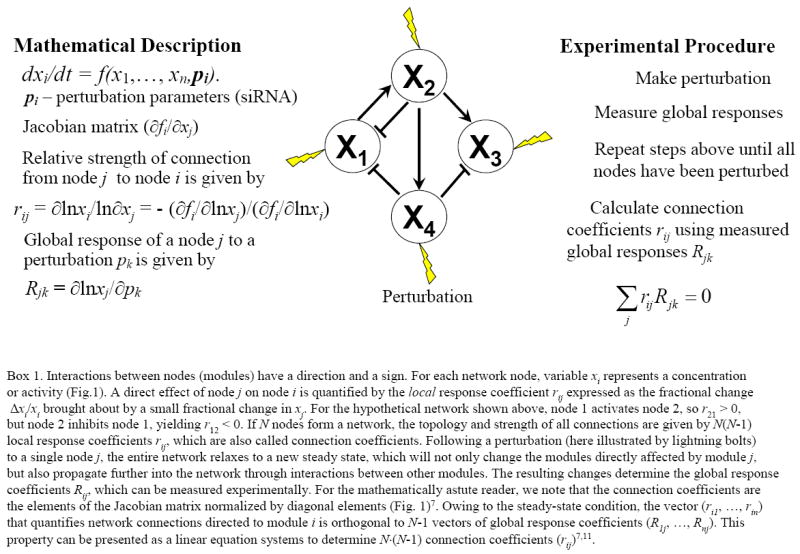
Applying four independent perturbations (shown by lightning bolts) to the network of four nodes, the global response coefficients Rkj are measured. From these data, connection coefficients rij are calculated7,11.
A basic concept of quantifying molecular interactions between network nodes is to analyze the direct effect of a small change in one node on the activity of another node, while keeping the remaining nodes unchanged to prevent the spread of the perturbation. A node of a network can be a single protein, or a group of proteins, genes, or other cellular components, which together perform an identifiable task and are also called a module. For the plethora of differently phosphorylated protein forms and isozymes in the three-tired RAF/MEK/ERK network, MRA considers only three functional modules represented by three “communicating” nodes5,7. Interactions between nodes (modules) have a direction and a sign. For each network node, variable xi represents a concentration or activity (Fig.1). A direct effect of node j on node i is quantified by the local response coefficient rij expressed as the fractional change Δxi/xi brought about by a small fractional change in xj. For the hypothetical network shown in Fig. 1, node 1 activates node 2, so r21 > 0, but node 2 inhibits node 1, yielding r12 < 0. If N nodes form a network, the topology and strength of all connections are given by N(N-1) local response coefficients rij, which are also called connection coefficients. Following a perturbation to a single node j , the entire network relaxes to a new steady state, which will not only change the modules directly affected by module j , but also propagate further into the network through interactions between other modules. The resulting changes determine the global response coefficients Rij, which can be measured experimentally. For the mathematically astute reader, we note that the connection coefficients are the elements of the Jacobian matrix normalized by diagonal elements (Fig. 1)7. Owing to the steady-state condition, the vector (ri1, …, rin) that quantifies network connections directed to module i is orthogonal to N-1 vectors of global response coefficients (R1j, …, Rnj). This property can be presented as a linear equation systems to determine N·(N-1) connection coefficients (rij)7,11.
Santos et al5 perturbed each of the three nodes of the MAPK pathway using corresponding siRNA, and responses were measured following stimulation with EGF or NGF. These data populated the global response matrix, from which both the structure and strength of functional interactions in the MAPK network were computed. Not only were the well-known activation connections from RAF to MEK1/2 and MEK1/2 to ERK1/2 retrieved, but also were two feedback loops uncovered5. A short negative feedback from ERK1/2 to MEK1/2 was found with both EGF and NGF. This feedback was reported earlier for integrin signalling, and its presence might be related to maintaining the dynamic stability of the steady state12,13. Remarkably, feedback from ERK1/2 to RAF was negative for EGF stimulation, but strongly positive for NGF stimulation, which explained the transient MAPK activation by EGF and the sustained MAPK activation by NGF.
Although these findings are crucial for understanding the MAPK pathway dynamics, mechanistically the signalling interactions responsible for EGF-induced negative feedback and NGF-induced positive feedback await experimental scrutiny. MRA can uncover and quantify a network’s intermodular connections, but these need not be direct and can involve known or unknown proteins “external” to the network. Diverse stimuli that are transmitted through the MAPK cascade have divergent routes outside the cascade and evoke different regulatory interactions that feedback and feedforward into the MAPK cascade. Using siRNA against PKC-δ, Santos et al5 show that this isoform is involved in the NGF-induced positive feedback. Yet, there are other contributors to the different wiring of the EGF versus NGF-induced MAPK pathway, including potentially distinct feedback regulation of Raf-1 and B-Raf by ERK14 and their different levels of activation by EGFR and TrkA through signalling pathways that diverge into Ras and Rap14. Also, the distinct spatial localization of Ras isoforms on the plasma membrane and Golgi apparatus results in diverse feedback regulation15. Likewise, differences in subcellular distribution and assembly of MAPK cascades on scaffolds dramatically change the signalling output16. Remarkably, the study by Santos et al5 shows that MAPK signal specificity, which arises from the complexity of multiple signal transducers and their interactions differentially induced by NGF or EGF, can be concisely summarized in terms of the topology and strength of connections between the MAPK cascade tiers using MRA5,7.
The finding of self-perpetuating ERK activity, when PC12 cells are stimulated with NGF5, may have important ramifications in neurobiology. Bastiaens and colleagues5 showed that despite complete inhibition of TrkA following initial stimulation of PC12 cells with NGF, a large fraction of ERK remains active, in striking contrast with EGFR inhibition. Interestingly, this kind of biological memory brought about by NGF-induced MAPK bistability can generate a travelling wave of kinase phosphorylation17. Such phosphorylation waves can propagate along axons to the nucleus, delivering survival signals from the initial NGF-induced activation of TrkA receptors on distal axons that are located up to a meter away from neuron bodies.
Future work inferring the dynamic architecture of cellular regulatory networks from perturbation responses will likely use not only data measured at steady-states, but also time-series data11. This will enable us to uncover the temporal behaviour of the connection strengths and the dynamics of feedback loops in different cellular regimes, including cell cycle or circadian oscillations. Interestingly, monitoring time-dependent responses does not require perturbations to every network module and offers rich opportunities for understanding biological dynamics. We can expect that exciting theoretical developments will lead to additional novel biological insights, such as those provided by Bastiaens and colleagues5, generating a deeper understanding of the architecture of biological systems.
References
- 1.Marshall CJ. Cell. 1995;80:179–85. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- 2.Murphy LO, MacKeigan JP, Blenis J. Mol Cell Biol. 2004;24:144–53. doi: 10.1128/MCB.24.1.144-153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kao S, Jaiswal RK, Kolch W, Landreth GE. J Biol Chem. 2001;276:18169–77. doi: 10.1074/jbc.M008870200. [DOI] [PubMed] [Google Scholar]
- 4.Sasagawa S, Ozaki Y, Fujita K, Kuroda S. Nat Cell Biol. 2005;7:365–73. doi: 10.1038/ncb1233. [DOI] [PubMed] [Google Scholar]
- 5.Santos SDM, Verveer P, Bastiaens PIH. Nature Cell Biol. 2007 doi: 10.1038/ncb1543. [DOI] [PubMed] [Google Scholar]
- 6.Kholodenko BN. Nat Rev Mol Cell Biol. 2006;7:165–176. doi: 10.1038/nrm1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kholodenko BN, et al. Proc Natl Acad Sci U S A. 2002;99:12841–12846. doi: 10.1073/pnas.192442699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de la Fuente A, Brazhnik P, Mendes P. Trends Genet. 2002;18:395–8. doi: 10.1016/s0168-9525(02)02692-6. [DOI] [PubMed] [Google Scholar]
- 9.Vance W, Arkin A, Ross J. Proc Natl Acad Sci U S A. 2002;99:5816–21. doi: 10.1073/pnas.022049699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sachs K, Perez O, Pe’er D, Lauffenburger DA, Nolan GP. Science. 2005;308:523–9. doi: 10.1126/science.1105809. [DOI] [PubMed] [Google Scholar]
- 11.Sontag E, Kiyatkin A, Kholodenko BN. Bioinformatics. 2004;20:1877–86. doi: 10.1093/bioinformatics/bth173. [DOI] [PubMed] [Google Scholar]
- 12.Dibrov BF, Zhabotinsky AM, Kholodenko BN. J Math Biol. 1982;15:51–63. doi: 10.1007/BF00275788. [DOI] [PubMed] [Google Scholar]
- 13.Kolch W, Calder M, Gilbert D. FEBS Lett. 2005;579:1891–5. doi: 10.1016/j.febslet.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 14.Dumaz N, Marais R. Mol Cell. 2005;17:164–6. doi: 10.1016/j.molcel.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 15.Bivona TG, et al. Nature. 2003;424:694–8. doi: 10.1038/nature01806. [DOI] [PubMed] [Google Scholar]
- 16.Harding A, Tian T, Westbury E, Frische E, Hancock JF. Curr Biol. 2005;15:869–73. doi: 10.1016/j.cub.2005.04.020. [DOI] [PubMed] [Google Scholar]
- 17.Markevich NI, Tsyganov MA, Hoek JB, Kholodenko BN. Mol Syst Biol. 2006;2:61. doi: 10.1038/msb4100108. [DOI] [PMC free article] [PubMed] [Google Scholar]


