Abstract
Micro RNAs (miRNAs) are 19-24 nucleotide long non-coding RNAs that posttranscriptionally modulate gene expression. They are found in almost all species: viruses, plants, nematodes, fly, fish, mouse, human, and are implicated in a wide array of cellular and developmental processes. Microarray-based miRNA profiling brought to the discovery of miRNAs specific to different hematopoietic lineages. Furthermore, the functional assays performed in tissue cultures to discover miRNAs involved in immune responses in combination with the reports of miRNA-transgenic or miRNA-knockout mouse models has helped elucidating the miRNA roles in the development and function of immune system. Abnormal patterns of hematopoietic-specific miRNAs have been found in different types of cancer and miRNA based gene therapy is being considered as a potential technology of choice in immunological disorders and cancer. The purpose of this review is to discuss recent findings related with the expression and function of miRNAs in hematopoietic lineages.
Keywords: Acquired immune response, Cancer, Cytokines, Drug discovery, Hematopoietic lineage, Innate immune response, TNF-α.
Introduction
Since their discovery, micro RNAs (miRNAs) have been implicated in a wide array of cellular and developmental processes 1. In particular, they are key players in the regulation of translation or degradation of target mRNAs through base pairing to partially complementary sites. Their number over-passed 500 and more miRNAs are getting cloned or in silico identified. Parallel to miRNA identification, genetic studies addressing their physiological roles in vivo in complement to different functional assays has brought a lot of information about their critical role in almost all the aspects of cell biology 2. It has now been demonstrated that miRNAs are involved in establishment, maintenance, and function of hematopoietic lineages (see below), establishment of muscle phenotype such as miR-1, -133, -206 -208, or regulation of organogenesis such as miR-1-2 or miR-133 3. They are also involved in metabolic processes and metabolic diseases such as miR-9, miR-143 or miR-122, neuronal function and neurological disorders, ageing, fragile X syndrome and hypoxia (for a thorough review on the roles of miRNAs in different biological processes and cancer refer to the review by Tili et al., 3). Expression of some of the miRNAs is mostly restricted to a single tissue or organ as e.g. miR-142 in lymphoid tissue, miR-223 in myeloid tissue or miR-1-2 in muscle 3. The majority of miRNAs are widely expressed. Abnormal patterns of miRNA expression have been found in all the examined disease states, especially cancers (for a review on abnormal expression and function of miRNAs in cancer consult the reviews by Calin & Croce 4 and Esquela-Kershner & Slack 5). Extensive genome-wide expression profiling of cells and tissues in different stages of development or differentiation, metabolic conditions, and disease models using miRNA-specific microarrays brought to the conclusion that unique miRNA profiles exist that are specific for the studied types of samples. These exciting but unexpected findings crystallized the hypothesis that genome-wide miRNA expression profiling could be used to profile tumors based on their origin and differentiation state, to help in diagnostic, prognosis, and for the use of miRNAs in therapeutic.
BIOGENESIS AND MECHANISMS OF ACTION OF miRNAs
Primary miRNA transcripts (pri-miR) are processed into precursor miRNA (pre-miRNA) by an enzymatic complex that includes the nuclear RNAse III enzyme Drosha and DGRC8 (Digeorge syndrome critical region gene 8). The resulting pre-miRNA is next transported to the cytoplasm by Exportin-5 and a RAS-like nuclear protein–guanosine triphosphate GTP, RAN. Once in cytoplasm pre-miRNA are processed into mature 22-nucleotide duplexes by another RNAse III enzyme, Dicer in association with TRBP (HIV-transactivating response RNA-binding protein). Only one of the strands of miRNA duplex is loaded onto the RNA-induced silencing complex -RISC. The mature miRNA binds then the 3' untranslated region of target mRNA transcripts and either destabilizes the target mRNA transcript, blocks its translation or both. The miRNA:mRNA recognition is mediated by complementary binding between the 5' end of the miRNA referred to as the miRNA “seed” region and the corresponding complementary sequence in the mRNA target 1, 2, 6. Although the binding of miRNAs to its target mRNA blocks the translation, a few recent reports have brought evidence that miRNAs might activate or enhance translation 7, 8. In silico bioinformatics analyses has helped a lot in predicting the potential targets of miRNAs. Elucidating miRNA-target genes and their mechanism of action has given valid clues in understanding the physiological role of miRNAs in vivo, and is paving the platform for future use of miRNAs as therapeutic tools.
miRNAs SPECIFICALLY INVOLVED IN THE DIFFERENTIATION OF HEMATOPOIETIC LINEAGES
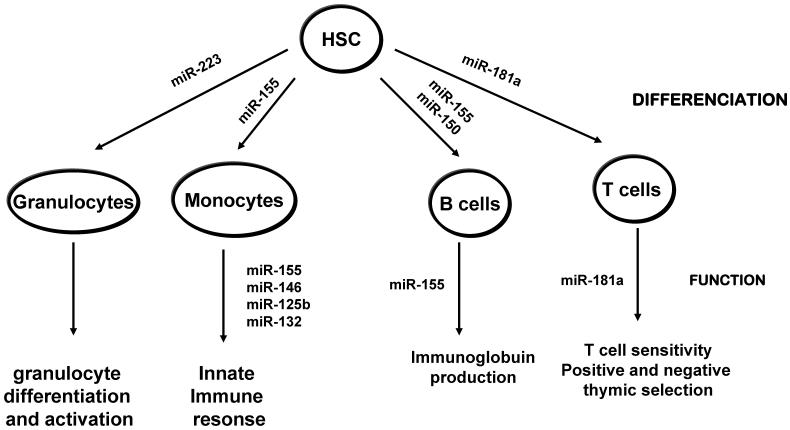
Chen et al., 9 and Monticelli et al., 10 analyzed miRNA expression profiles of different types of hematopoietic cells in murine and human respectively. They reported that miRNA expression patterns were very different not only between hematopoietic and non-hematopoietic cells but also within the hematopoietic group. Both reports are the first to show that miRNAs are implicated in the commitment of hematopoietic stem cells to a particular cell lineage. These reports together with a previous report by Calin et al., 11 stating that miRNA expression profiles can be used to characterize human tumors confirmed the use of miRNA expression profiles as tool to characterize the hematopoitic-lineage specific cells, stage specific cells or lymphoma/leukemic cells. Parallel to miRNA profiling studies, ectopic expression of miRNAs in hematopoietic stem cells substantially altered lineage differentiation giving thus strong evidence that miRNAs are not only differentially expressed in hematopoietic lineages but they also direct physiologically these processes 12. In addition to regulating hematopoietic-cell lineage differentiation, it was found that miRNAs play an important role in innate immune response and adaptive immune responses in mice (see below). Another important observation to be mentioned is the fact that miRNA levels are altered by different cytokine stimulation in immune cells. Thus, miR-125b and miR-155 levels oscillated within an hour of TNF-α stimulation in mouse Raw 264.7 cells 7. Furthermore this oscillation paralleled the rapid increase and subsequent decrease in NF-κB transcriptional activity 7. Fast increase of cellular miRNAs levels was also reported in response to INFβ 13. Whether these changes are specific to the immune cells as a way to limit the level and duration of the immune response to infections before it becomes detrimental to the health of the organism or are a common feature of other cells remains to be evaluated. Furthermore there is a dynamic change in the levels of miRNAs within the same cell type but in different activation state. Combined analyses using direct cloning of miRNAs, microarray profiling and RT-PCR, was used to identify miRNA expression profile in antigen specific naïve, effectors and memory CD8 T cells. The study brought the discovery of dynamic regulation of miRNAs during antigen-induced CD8 T cell differentiation. miR-16, -142-3p, -142-5p, -150, -15b and let-7f were downregulated in effector cells compare to naïve cells and increased back in memory T cells 14. Effector T cells originate from naïve T cells following antigen exposure and are considered cells in a high state of activity. It is interesting to emphasize the fact that the global downregulation of miRNAs in activated T cells looks similar with global downregulation of miRNA expression observed in some cancer cells 4. We will next describe a few miRNAs found to be specific to hematopoietic lineages (Figure 1) and their corresponding targets (Table 1) that seem to be crucial in development and function of the immune response. In addition we will discus their abnormal patterns of expression associated with cancer and why manipulating their levels of expression is such a promising method in the fight against cancer and immunological disorders.
Figure 1.
miRNAs involved in hematopoietic differenciation and function. Blood cells derive from a common hematopoietic stem cell progenitor (HSP) which differenciate into at least 8 lineages, through progenitor cells. For simplicity, the progenitor cells are not shown and only four of the main hematopoitic lineages are depicted. miRNAs that are reported to be involved in the differentiation of the shown lineages based on the mouse models are noted. Consult the text for further details.
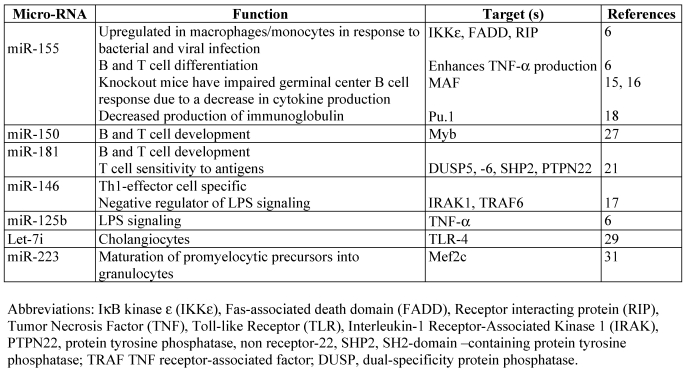
Table 1.
miRNAs implicated in the development and function of the immune system
miR-155
MiR-155 is a product of BIC (B cell integration cluster) transcript, and has been shown to be upregulated in many types of B cell lymphoma, diffuse large B‑cell lymphomas, Hodgkin lymphomas, and Burkitt lymphomas 3-5. BIC transcript was first identified as a frequent site of integration for the avian leucosis virus, and its coexpression with c-Myc have synergizing effects in lymphomagenesis 15. Costinean et al., 16 developed the first transgenic mouse that specifically overexpresses miR-155 in B cells, thus modeling the human B cell leukemia where the upregulation of miR-155 is observed. The transgenic mice developed polyclonal pre-leukaemia B-cell type followed by B-cell malignancy. Mice knockout for bic/mir‑155 gene are viable but they are unable to develop a proper immune T-, B-, or dentritic- dependent response 17, 18. Upregulation of miR-155 levels were also reported in human monocytes and mouse macrophages in response to LPS or interferon 7, 19. Detailed analyses of the function of miR-155 in B cells revealed that this miRNA plays a key role in antigen-driven B cell maturation and the persistence and/or differentiation of Ig class-switched cells and that deregulation of Pu.1 is among the factors responsible for the phenotypes observed in miR-155 deficient mice 20. Pu.1 is a member of the Ets domain-transcription factor family that plays a central role in many aspects of hematopoesis. Another exciting discovery for the function of this miRNA in the innate immune response was that it enhances TNF-α translation 7. It was among the first reports suggesting that although miRNAs negatively regulate the translation of target transcripts it is possible that they might enhance in specific instances translation. A later publication experimentally confirmed the hypothesis. 8. Thus, Vasudevan et al., 8 reported that when cultured mammalian cells are serum-starved, binding of miR-369-3 to a reporter mRNA (containing the TNF- 3′UTR) enhanced translation, whereas no stimulation was observed when this miRNA is absent. Interestingly, miR-369-3 repressed translation on actively dividing cells. It is still very early to draw any conclusion on the molecular mechanisms or cellular settings that promote the enhancement of translation by miRNAs and what are the elements that make this switch. Thai et al., 17 also demonstrated that miR‑155 might control the formation and response of germinal-centre B cells in part by controlling cytokine production. E.g. miR-155-deficient mice, showed weak production of IL‑ 2, IL‑ 4 and IFNγ by activated T cells 17 while the miR-155 transgenic mice overproduced TNF-α when challenged with LSP 7. miR-155 expression is modulated by antigenic stimulation in B cells, TNF-α in macrophages, or other Toll like receptor ligands 7, 17-20. It is still not very clear the role of NF-κB in regulation on miR-155 expression. Putative binding sites for NF-κB family members are found in human and mouse promoter regions of BIC. It is possible that this miRNA is transiently under the transcriptional control of NF-κB activity 7. In addition high levels of miR-155 were reported by Rai et al., 21 in diffuse large B-cell lymphoma characterized by constitutive activation of NF-κB signals. Interestingly, it was reported that miR-155 levels oscillate in a pattern similar to NF-κB activity 7. Whether it is the miR-155 levels that control the activity of NF-κB in this situation or is the NF-κB activity that results in the oscillatory levels of miR-155 remains to be shown. Overall knockout and transgenic mouse models developed for miR-155 proved that this miRNA plays a central role in innate and acquired immune response. Elucidating the mechanisms by which miR-155 expression is controlled and finding its targets will bring more understanding of the mechanisms of tumor induction by miR-155.
miR-181
miR-181 is highly expressed in the thymus as well as the brain, lung, bone marrow and spleen 9, 10. This miRNA has been shown to regulate T and B cell development when ectopically expressed in hematopoietic progenitor cells 9. The first report that suggested that this miRNA might play a role in the development of T cell lineage came from the elegant work proving that the expression levels of this miRNA change dynamically during thymocyte differentiation implying also that miR-181 might be important for not only T cell development but also for T cell function 22. Indeed latter it was shown that the levels of miR-181 correspond with the sensitivity of T cells to antigens 23. In normal conditions, the successful transition of double positive thymocytes to the single positive stage depends on positive and negative selection processes that take place in the thymus. Positive and negative selection, ensure that only those thymocytes expressing a repertoire of antigen receptors that can respond to foreign antigens, but not to self, will survive. Phosphatases play a critical role in the generation of the thymocyte repertoire because they negatively control the response of T cells to antigens. Increasing levels of miR-181 resulted in higher sensitivity of T cells to antigens, while the opposite was observed when miR-181 levels were diminished 23. These results translate in a central role for miR-181 in positive and negative thymic selection, both processes depending on the strength of the signals generated by T cell receptor (TCR). miR‑181 effects on TCR signaling was found to be due to targeting by miR-181 of multiple phosphatases, such as SHP2, PTPN22, DUSP5 and DUSP6 23. By modulating T‑cell sensitivity to antigens, miR-181 therefore, plays a central role in the development and maintenance of tolerance and immune T cells. Finally, miR-181 family is considered oncogenic. Overexpression of miR-181 is observed in breast, pancreas and prostate cancers 4. This oncogenicity might be explained by targeting of Tcl1 24 or Hox 25 transcripts in addition to phosphatases. It is proposed that at the transcriptional level this miRNA is under the activity of MYCN 26.
miR-150
miR-150, is mainly expressed in the lymph nodes and spleen and is highly up-regulated during the differentiation of mature T and B cells suggesting that it may participate in B and/or T lymphopoiesis 27. When ectopically expressed in hematopoietic stem cell progenitors, miR‑150 blocked B cell development at the transition pro-B to pre-B-cell stage, impairing the maturation of B cells, while moderately enhancing T lymphopoiesis and myelopoiesis from hematopoietic stem and progenitor cells 28. As expected from the above studies, miR-150-knockout mice have a double increase in the number of B- splenocytes, but have no apparent defects in the development of other lymphocytes. Like for miR-155 the expression of miR‑150 is modulated by triggering B cells with IgM-specific antibodies, CpG-containing DNA or LPS 28. Regarding its abnormal expression in disease state miR-150 is found to be differentially expressed in certain types of cancers 29.
miR -125b and miR-146
miR-146 is expressed at low levels in naïve T cells and is upregulated in Th1 cells, but not in Th2 cells 10. Based on this miR-146 is a considered as Th1 specific miRNA. miR-146 and miR-132 levels were reported to be upregulated in human monocytes in response to LPS 19, while the levels of miR-125b were downregulated in murine macrophages following the same stimuli 7. Upregulation of miR-146 in response to LPS was seen as a mean to negatively regulate the innate immune response, while the downregulation of miR-125b as a necessary step to allow TNF-α production due to the fact that this miRNA targets TNF-α transcripts 7, 19. Downregulated levels of miR-125b where also observed in psoriasis, a skin inflammatory disease associated with high levels of TNF-α 30.
Let-7
Let-7 was among the first miRNAs identified in C. elegance. The let-7 gene family has been evolutionarily expanded from one member in nematodes to 11 members in mice and humans. Let-7 miRNAs negatively regulate Ras, and are typical examples of miRNAs that are frequently downregulated in solid cancers such as lung, breast, gastric, colon cancers and pituitary adenomas and are considered as classical tumor suppressors genes 3-5. In addition, loss of let-7 expression can identify a less differentiated class of cancers, while, let-7a-2 low expression correlates with poor survival in lung cancer patients. At the transcriptional level MYCN activity is reported to result in the upregulation of let-7b levels in primary neuroblastoma 26. Let-7 overexpression is reported to represses cell proliferation. Interestingly, let-7f is found to promote angiogenesis 26. Chen et al., 31 reported that let-7i is involved in epithelial responses against microbial infection. Let-7i is highly expressed in human cholangiocytes (biliary epithelial cells) and regulates TLR4 expression. Infection of these cells with Cryptosporidium parvum (parasite that causes intestinal and biliary diseases) results in the reduced expression of let-7i in a MyD88/NF-kB-dependent manner.
miR-223
Due to its expression almost exclusively in bone marrow this miRNA is considered myeloid-specific. An interplay between miR-223 and two transcription factors, NFI-A and C/EBP (CCAAT/enhancer-binding protein), was reported to play an important regulatory role in granulocyte formation and miR‑223 expression is shown to be under the control of C/EBP activity, a well known transcription factor with effects on granulopoiesis 32. The knockout mice for this miRNA proved the previous reports and confirmed that miR-223 is a key player in granulocyte differentiation 33. Thus in the absence of miR-223 an increase in the production, differentiation and activation of granulocytes was observed resulting in tissue inflammation and damage. These findings have important implications for the treatment of inflammatory conditions as well as leukemia. The study suggested Mefc2 transcription factor as being a target of miR-223 33. Due to the fact that many types of leukemia have reduced levels of miR-223, finding out how lost expression of this miRNA contributes to the development of leukemias represent the next area of study.
miR-142
miR-142, is located on chromosome 17, at a site of a translocation associated with aggressive B cell leukemia 34, 35. miR-142 is among the highest expressed miRNA in almost all the hematopoietic lineages, hematopoietic stem cells, T cells, B cells, and its expression varies also within the type of cell, depending on the activation state 9, 10. There are not yet mouse models for this miRNA. However, finding out the transcription factors that control its expression and its mRNA target transcripts will bring more understanding for the oncogenic functions of this miRNA. In addition, manipulating the levels of this miRNA in aggressive B cell leukemias and monitoring its effects on proliferation, differentiation and apoptosis of these cells would be of great interest.
miR- 15, -16
miR-15 and miR-16 genes are often deleted or expressed at reduced levels in B cell chronic lymphocytic leukemias, hence the proposed function of tumor-suppressor genes 36, 37. In addition, miR-15, -16 have been found to also be downregulated or deleted in lung and colorectal cancers 4, 29. Bcl-2 is the first target described for these miRNAs 38. It was reported that when these miRNAs are deleted or downregulated the levels of Bcl-2 increase, protecting the cells from apoptosis. Furthermore, both these miRNAs were proposed to be involved in hematopoietic cell lineage differentiation 12.
miRNA-BASED GENE THERAPY?
Most of the miRNAs that are reported to be necessary for the differentiation and function of immune cells are found to be abnormally expressed in both solid and liquid tumors. Thus the first miRNA microarray profiling reported by Calin et al., 11 showed significant differences between B cell chronic lymphocytic leukemia and normal CD5+ B cells. Furthermore, analyses of hematopoietic tissue specific miRNAs such as miR-142, 155, 181 and 223 in malignant hematopoietic cell lines showed that although they have similar pattern of expression compared to normal cell lineages, the levels of expression were significantly altered, suggesting for the important roles of these miRNAs in hematopoietic diseases specially leukemias/lymphomas 39.
miRNA expression profiling studies suggested that miRNAs might be used as diagnostic, prognostic and therapeutic tools. Promising results have been obtained from studies of miRNAs on mammalian cell culture systems and animal in vivo models. miRNAs are natural antisense interactors that modulate the expression of multiple genes. The use of anti-miRNA oligonucleotides (antisense to the miRNA impairing the interaction between miRNAs and target mRNAs) in different cancer cell lines seems to be very promising techniques for modulating miRNA action. The same stands for the use of mimic miRNAs in order to reconstitute the expression of lost miRNAs in tumors. The development and in vivo use of "antagomirs", a novel class of chemically engineered oligonucleotides, showed that antagomirs are specific and effective silencers of miRNA expression in mice 40. Antagomir technology can therefore be promising as therapeutic tools in many diseases, from metabolic disorders, to cancer and immune related diseases.
CONCLUSION
Discovered 15 years ago in C.elegans 41, miRNAs play important roles in all the cellular processes studied so far and their deregulated expression is associated with different diseases including cancer or immunological disorders. Both increased and decreased expression of miRNAs is observed in neoplasias, and therefore the terms onco-miRNAs and tumour-suppressor-miRNAs are used 4, 5. The mouse models developed so far for miRNAs primarily expressed in the hematopoietic lineages showed that miRNAs contribute in the development, differentiation and function of immune cells. These models showed to be important not only for analyzing miRNA function in vivo but also for future drug development. Overall we can conclude that miRNAs are lineage-specific and important components of hematopoitic lineage differentiation. When their expression is modified the harboring cells become prone to cancer or other diseases. miRNA-based gene therapy targeting deregulated miRNAs will be the future tool for gene therapy.
References
- 1.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Tili E, Michaille JJ, Gandhi V et al. miRNAs and their potential for use against cancer and other diseases. Future Oncol. 2007;3:521–537. doi: 10.2217/14796694.3.5.521. [DOI] [PubMed] [Google Scholar]
- 4.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 5.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 6.Cullen BR. Transcription and processing of human microRNA precursors. Mol Cell. 2004;16:861–865. doi: 10.1016/j.molcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 7.Tili E, Michaille JJ, Cimino A et al. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol. 2007;179(8):5082–5089. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- 8.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318(5858):1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 9.Chen CZ, Li L, Lodish HF et al. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 10.Monticelli S, Ansel KM, Xiao CM et al. MicroRNA profiling of the murine hematopoietic system. Genome Biol. 2005;6:R71. doi: 10.1186/gb-2005-6-8-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calin GA, Liu CG, Sevignani C et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci U S A. 2004;101(32):11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Georgantas RW 3rd, Hildreth R, Morisot S et al. CD34+ hematopoietic stem-progenitor cell microRNA expression and function: a circuit diagram of differentiation control. Proc Natl Acad Sci U S A. 2007;104(8):2750–2755. doi: 10.1073/pnas.0610983104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pedersen IM, Cheng G, Wieland S et al. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449(7164):919–922. doi: 10.1038/nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu H, Neilson JR, Kumar P et al. miRNA Profiling of Naïve, Effector and Memory CD8 T Cells. PLoS ONE. 2007;2(10):e1020. doi: 10.1371/journal.pone.0001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tam W, Dahlberg JE. miR-155/BIC as an oncogenic microRNA. Genes Chromosomes Cancer. 2006;45:211–212. doi: 10.1002/gcc.20282. [DOI] [PubMed] [Google Scholar]
- 16.Costinean S, Zanesi N, Pekarsky Y et al. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci USA. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thai TH, Calado DP, Casola S et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 18.Rodriguez A, Vigorito E, Clare S et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taganov KD, Boldin MP, Chang KJ et al. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103(33):12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vigorito E, Perks KL, Abreu-Goodger C et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27(6):847–859. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rai D, Karanti S, Jung I et al. Coordinated expression of microRNA-155 and predicted target genes in diffuse large B-cell lymphoma. Cancer Genet Cytogenet. 2008;181(1):8–15. doi: 10.1016/j.cancergencyto.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neilson JR, Zheng GX, Burge CB et al. Dynamic regulation of miRNA expression in ordered stages of cellular development. Genes Dev. 2007;21:578–589. doi: 10.1101/gad.1522907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li QJ, Chau J, Ebert PJ et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147–161. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 24.Pekarsky Y, Santanam U, Cimmino A et al. Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Res. 2006;66(24):11590–11593. doi: 10.1158/0008-5472.CAN-06-3613. [DOI] [PubMed] [Google Scholar]
- 25.Naguibneva I, Ameyar-Zazoua M, Polesskaya A et al. The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nat Cell Biol. 2006;8:278–284. doi: 10.1038/ncb1373. [DOI] [PubMed] [Google Scholar]
- 26.Nicoloso MS, Calin GA. MicroRNA Involvement in Brain Tumors: From Bench to Bedside. Brain Pathol. 2008;18(1):122–129. doi: 10.1111/j.1750-3639.2007.00119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou B, Wang S, Mayr C et al. miR-150, a microRNA expressed in mature B and T cells, blocks early B cell development when expressed prematurely. Proc Natl Acad Sci U S A. 2007;104(17):7080–7085. doi: 10.1073/pnas.0702409104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao C, Calado DP, Galler G et al. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131(1):146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 29.Volinia S, Calin GA, Liu CG et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103(7):2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sonkoly E, Wei T, Janson PC et al. MicroRNAs: novel regulators involved in the pathogenesis of Psoriasis? PLoS ONE. 2007;2(7):e610. doi: 10.1371/journal.pone.0000610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen XM, Splinter PL, O'Hara SP et al. A cellular micro-RNA, let-7i, regulates Toll-like receptor 4 expression and contributes to cholangiocyte immune responses against Cryptosporidium parvum infection. J Biol Chem. 2007;282(39):28929–28938. doi: 10.1074/jbc.M702633200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fazi F, Racanicchi S, Zardo G et al. Epigenetic silencing of the myelopoiesis regulator microRNA-223 by the AML1/ETO oncoprotein. Cancer Cell. 2007;12(5):457–466. doi: 10.1016/j.ccr.2007.09.020. [DOI] [PubMed] [Google Scholar]
- 33.Johnnidis JB, Harris MH, Wheeler RT et al. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451(7182):1125–9. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- 34.Michael MZ, O' Connor SM, van Holst Pellekaan NG et al. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1:882–891. [PubMed] [Google Scholar]
- 35.Kasashima K, Nakamura Y, Kozu T. Altered expression profiles of microRNAs during TPA-induced differentiation of HL-60 cells. Biochem Biophys Res Commun. 2004;322:403–410. doi: 10.1016/j.bbrc.2004.07.130. [DOI] [PubMed] [Google Scholar]
- 36.Calin GA, Dumitru CD, Shimizu M et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Calin GA, Ferracin M, Cimmino A et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353(17):1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 38.Cimmino A, Calin GA, Fabbri M et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102(39):13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramkissoon SH, Mainwaring LA, Ogasawara Y et al. Hematopoietic- specific microRNA expression in human cells. Leuk Res. 2006;30(5):643–647. doi: 10.1016/j.leukres.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 40.Krützfeldt J, Rajewsky N, Braich R et al. Silencing of microRNAs in vivo with 'antagomirs'. Nature. 2005;438(7068):685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 41.Lee RC Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]