Summary
In budding yeast, the four-protein CBF3 complex (Skp1p-Ctf13p-Cep3p-Ndc10p) initiates kinetochore assembly by binding to the CDEIII locus of centromeric DNA. A Cep3p dimer recruits a Ctf13p-Skp1p heterodimer and contacts two sites on CDEIII. We report here the crystal structure, determined at 2.8 Å resolution by multiple isomorphous replacement with anomalous scattering, of a truncated Cep3p [Cep3p (47−608)], comprising all but an N-terminal, Zn2Cys6-cluster, DNA-binding module. Cep3p has a well-ordered structure throughout essentially all of its polypeptide chain, unlike most yeast transcription factors, including those with Zn2Cys6 clusters like Gal4p. This difference may reflect an underlying functional distinction: while any particular transcription factor must adapt to a variety of upstream activating sites, Cep3p scaffolds kinetochore assembly on centromeres uniformly configured on all 16 yeast chromosomes. We have, using the structure of Cep3p (47−603) and the known structures of Zn2Cys6 cluster domains, modeled the interaction of Cep3p with CDEIII.
Introduction
Kinetochores are protein complexes that assemble on centromeric DNA and couple each chromosome to microtubules of the mitotic spindle. Proper assembly of a single kinetochore on each sister chromatid ensures faithful transmission of the eukaryotic genome in mitosis and meiosis. Saccharomyces cerevisiae and closely related fungi have “point” centromeres, smaller and simpler than those of higher eukaryotes, but nonetheless requiring the hierarchical assembly of at least 14 defined subcomplexes consisting of over 70 proteins, many of which are conserved in metazoans (De Wulf et al., 2003).
The sequences of point centromeres within a single organism are highly conserved, and among organisms, they are very similar. The centromeres of S. cerevisiae include three elements: two regions of conserved sequence (CDEI and CDEIII) flanking an approximately 80 bp motif with 87−98% A:T content (CDEII). CDEIII binds the CBF3 complex, which consists of the four essential proteins Skp1p, Ctf13p, Cep3p and Ndc10p (Lechner and Carbon, 1991; Sorger et al., 1995). The core CBF3 complex (one Skp1p-Ctf13p heterodimer, ∼72 kDa; one Cep3p homodimer, ∼145 kDa; and one Ndc10p homodimer, ∼220 kDa(Kaplan et al., 1997; Russell et al., 1999)) forms in the absence of DNA, and binds in vitro to a CDEIII probe of at least 56 bp (corresponding approximately to the DNAse I protected footprint(Espelin et al., 1997; Russell et al., 1999)). An extended CBF3 complex, which assembles on longer probes and contains a second Ndc10p dimer on the CDEII-distal side of CDEIII , is thought to be the form found on centromeres in vivo (Espelin et al., 1997).
Genetic interactions identify CBF3 as the cornerstone of the yeast kinetochore; association of all other known kinetochore proteins to the centromere requires functional CBF3 (De Wulf et al., 2003). None of the CBF3 proteins bind to CDEIII individually or in subcomplexes containing a subset of the subunits (although Ndc10p binds to CDEII on its own (Espelin et al., 2003)). Ndc10p, Ctf13p and Cep3p all contact the major groove of CDEIII, as detected by DNA-protein crosslinking (Espelin et al., 1997), but Cep3p is the only CBF3 subunit that contains a recognizable DNA-binding motif, an N-terminal Zn2Cys6 binuclear cluster as found in Gal4p and related fungal transcription factors (Lechner, 1994).
Most Zn2Cys6 cluster proteins bind as homodimers to pairs of CCG half-sites with specificity largely determined by the spacing and polarity of the CCG elements (MacPherson et al., 2006). CDEIII contains an absolutely conserved CCG site, which is essential for chromosome segregation(Jehn et al., 1991) and CBF3 binding(Espelin et al., 1997). The 46-residue Zn2Cys6 cluster of Cep3p is required for chromosome segregation (Lechner, 1994), and limited proteolysis of Cep3p has suggested that the Zn2Cys6 cluster is connected to the rest of the protein by a short, cleavable linker arm, as in the Gal4p family of proteins(Russell et al., 1999).
Cep3p exhibits several differences from most previously characterized Zn2Cys6 cluster proteins. Cep3p contacts CDEIII in the major groove at two non-identical sites: the conserved CCG site typical of Zn2Cys6 cluster recognition sites and at a second site (TGT) approximately one and one-quarter turns of DNA to the CDEII-proximal side (Espelin et al., 1997). The asymmetry is not critical, as neither Cep3p alone nor intact CBF3 can bind mutant CDEIII constructs in which the left half-site has been mutated to CCG or CGG (Russell et al., 1999 7). Moreover, unlike Gal4p and related transcription factors, Cep3p does not bind CDEIII DNA at detectable levels by itself, but only in the presence of the other CBF3 subunits. As a first step towards a structural understanding of the assembly and function of CBF3, we have determined the crystal structure of Cep3 (47−608), a fragment that lacks only the DNA-interacting Zn2Cys6 clusters, likely to be flexibly tethered to the rest of the protein and fixed in position only when the entire CBF3 complex binds centromeric DNA. The Zn2Cys6 clusters will then insert into the DNA major groove, and the surfaces that organize other kinetochore components will all be part of the structure described here.
Results and Discussion
Structure of Cep3p (47−608)
We expressed and crystallized a truncated Cep3p lacking the N-terminal Zn2Cys6 domain [Cep3p (47−608)]. We chose this strategy, because the Zn2Cys6 clusters of the well-studied transcription factors are flexibly attached to the rest of the protein, and crystals of these proteins have been obtained only as DNA complexes. The Cep3p crystals diffracted to a minimum Bragg spacing of 2.8Å, and we determined the structure using multiple isomorphous replacement with anomalous scattering (Table 1). The model contains 517 of the 563 residues in the construct; residues 47−53, 322−340, and 564−587 are not visible in the electron density map.
TABLE 1.
Diffraction, Phasing and Refinement of Cep3p (47−608)
| |
Native |
Hg Peak |
Hg Inflection |
Pt Peak |
Pt Inflection |
|---|---|---|---|---|---|
|
DIFFRACTION |
|
|
|
|
|
| Space Group | P43212 | P43212 | P43212 | P43212 | P43212 |
| Unit Cell (Å) | a=b=84.63 | a=b=85.77 | a=b=85.77 | a=b=85.77 | a=b=85.77 |
| c=230.95 | c=230.54 | c=230.54 | c=231.03 | c=231.03 | |
| Wavelength (Å) | 1.07182 | 1.00794 | 1.00912 | 1.07179 | 1.07206 |
| Resolution (Å) | 45.0 − 2.8 | 50.0−3.0 | 50.0−3.0 | 50.0−3.5 | 50.0−3.5 |
| (2.9 − 2.8) | (3.11−3.0) | (3.11−3.0) | (3.63−3.5) | (3.63−3.5) | |
| Completeness (%) | 94.4 (67.5) | 84.4 (33.6) | 86.4 (32.9) | 98.5 (91.5) | 98.7 (94.4) |
| Redundancy | 6.8 (4.8) | 8.9 (6.4) | 15.2 (9.5) | 14.7 (6.0) | 13.6 (5.2) |
| Average I/σ(I) | 25.4 (1.86) | 45.8 (2.9) | 53.9 (2.9) | 54.2 (2.0) | 53.5 (1.3) |
| RSym (%) | 0.072 (0.307) | 0.076 (0.351) | 0.086 (0.395) | 0.086 (0.442) | 0.078 (0.611) |
|
PHASING |
|
|
|
|
|
| # Sites | 4 | 4 | 2 | 2 | |
| Phasing Power (iso/ano) | 5.30/1.51 | 6.37/1.00 | 0.58/0.56 | 0.61/0.69 | |
| Mean FOM (acentric/centric) | 0.39/0.37 | ||||
|
REFINEMENT |
|
|
|
|
|
| Resolution Range | (Å) | 45.0 − 2.8 | |||
| Reflections | Theoretical Number | 21542 | |||
| Total Observed | 20325 (94.4%) | ||||
| Working Set | 18327 (85.1%) | ||||
| Test Set | 1998 (9.3%) | ||||
| Rfree | 0.285 | ||||
| Rworking | 0.228 | ||||
| No. of atoms: | Protein | 4292 | |||
| Water | 15 | ||||
| Ramachandran | Core | 85.9% | |||
| Plot: | Allowed | 13.7% | |||
| Generously Allowed | 0.4% | ||||
| Disallowed | 0.0% | ||||
| RMS Deviations | Bond Lengths | 0.008 | |||
| Bond Angles | 1.336 | ||||
| Dihedral Angles | 19.801 | ||||
| Improper Angles | 0.778 | ||||
| Average B-Factors | Protein | 98.97 | |||
| Water | 87.46 |
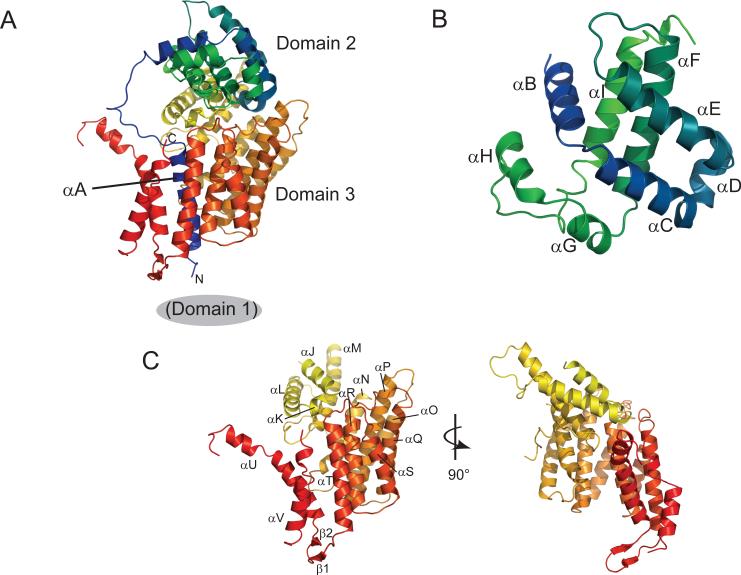
Cep3p has a largely α-helical fold, with three domains (Fig. 1A): the N-terminal Zn2Cys6 cluster, absent in this truncated form and designated here “domain 1” (residues 1−46); domain 2 (residues 78−229), consisting of 8 relatively short helices connected by 3−4 residue turns; and domain 3 (residues 230−608), a set of helical zig-zags. A long helical connector (αA) between domains 1 and 2, residues 54−77, runs through domain 3, which effectively surrounds the connector with an irregular helical cage.
Figure 1. Structure of Cep3p.
A. Ribbon diagram of Cep3p, colored from blue at the N-terminus (54) to red at the C-terminus (608). Domain 1 (the N-terminal Zn2Cys6 cluster), absent in this construct, is located immediately before helix αA. B. Domain 2 (residues 78−229) has a structure similar to the globin fold. C. Domain 3 (residues 230−608) forms a left-handed solenoid composed of seven helical zig-zags which encircles αA. Disordered loops absent from electron density link αM to αN and αU to αV.
Domain 2 contains helices αB-αI (Fig. 1B). Its polypeptide chain emerges from αA through an extended segment (residues 78−94), which participates in the dimer contact. The three-dimensional arrangement of helices in domain 2 is a sandwich of two antiparallel helical hairpins (αC-αE and αF-αI), with segmented helical connectors (αD at the tip of the first hairpin and αG-αH at the tip of the second). A rough twofold axis relates the two halves of the domain. The fold is topologically related to the globin fold. Together with its dimer partner, domain 2 forms the DNA-distal apex of Cep3p.
Domain 3 contains helices αJ-αV (Fig. 1C). Successive helices extend in antiparallel directions, except for those joined by the two disordered loops between αN and αO and between αU and αV. We imagine that parts of these loops might become ordered, perhaps as helices, upon incorporation into CBF3. In addition to about 10 residues that might form a short helix, the UV loop contains an extended segment of acidic residues as well as a potential Ipl1p phosphorylation site at Ser577(Westermann et al, 2003). If we include the loops in our description, then the entire domain is a set of seven helical zig-zags plus a final helix. The zig-zags form a tight, left-handed solenoid, rather like certain HEAT-repeat domains. The αA helix runs through the core of the solenoid. A type II′ β-turn between αT and αU projects like a paddle from the solenoid.
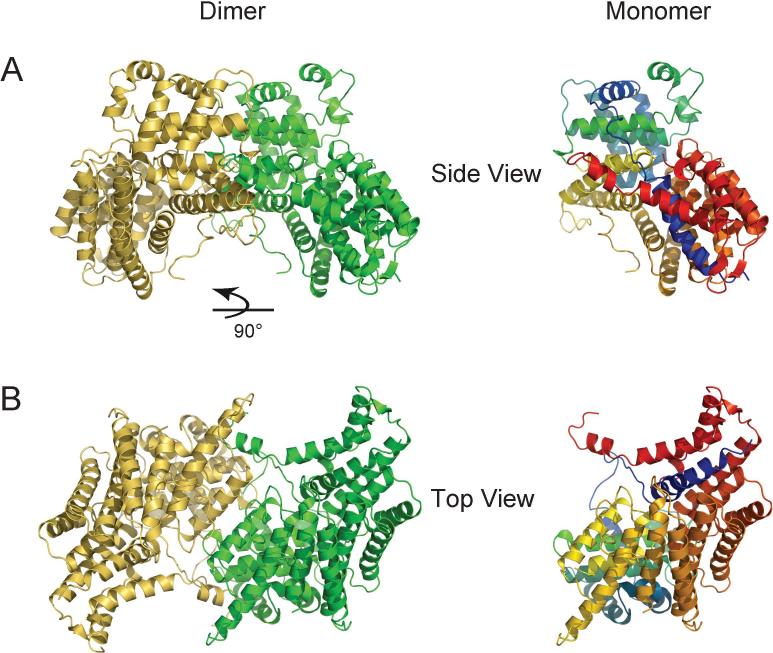
The dimer interface of Cep3p buries 3540 Å2 extending over both domains 2 and 3 (Fig. 2). It establishes so robust a contact that distortions or changes upon association with Skp1p-Ctf13p and with DNA appear unlikely. This substantial and intimately associated surface includes main-chain hydrogen bonds between the αA-αB linker of one subunit and the αE-αF loop of the other, as well as a large variety of side-chain contacts. Helix αV is kinked at residue 552 and protrudes into the dimer partner, where it makes hydrophobic contacts with sidechains from αL and αM and terminates in an Arg-rich pocket where Arg139 and Arg232 of the dimer partner are hydrogen bonded to the main chain carbonyls of Gly561, Leu562 and Gly563 and the hydroxyl of Ser564. The kink is induced by side chain-main chain hydrogen bonds from Arg307 to the carbonyl of Asn552 and from Gln82 to the carbonyl of Asp553. The kink is further stabilized by hydrogen bonds between the Asn552 sidechain and the sidechains of Thr80 and Arg307.
Figure 2. The Cep3p dimer.
A. Ribbon diagram of the Cep3p dimer viewed perpendicular to the dyad (side view). A ribbon diagram of a monomer in the same orientation, colored as in Figure 1, is provided for reference. B. The Cep3p dimer viewed along the dyad, with domain 3 closest to the viewer (bottom view).
Conservation and potential contact surfaces for other CBF3 components
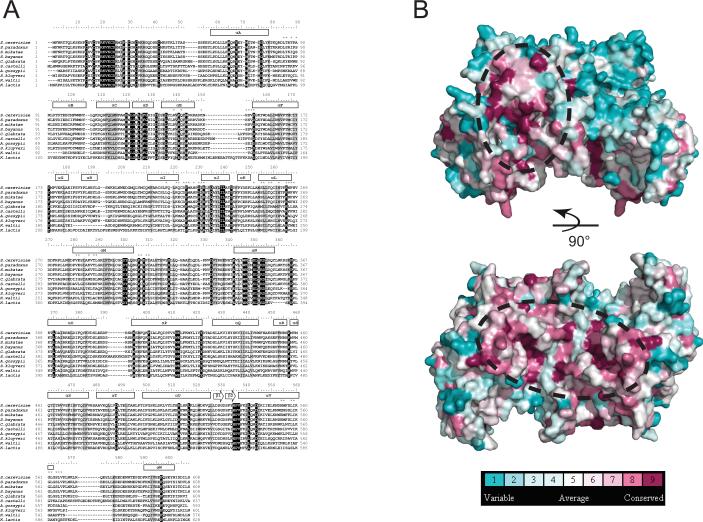
In addition to binding to DNA, Cep3p contacts Ctf13p and possibly other kinetochore or checkpoint components whose centromere association is CBF3-dependent. We would expect regions of protein-protein contact on Cep3p to be conserved among the budding yeasts with point centromeres (De Wulf et al., 2003; McAinsh et al., 2003; Meraldi et al., 2006; Stoyan and Carbon, 2004). Multiple sequence alignment of Cep3p orthologs from ten such species (Fig. 3A) reveals two highly conserved surface patches (Fig. 3B) with potentially important functions, one composed of residues from αC,αD, and αE in domain 2 and αN and αO in domain 3, the other, of residues from αL, αM, and αN (all in domain 3). Mutation of these sites should help uncover the identities of relevant partners.
Figure 3. Evolutionary conservation of Cep3p.
A. Multiple sequence alignment of Cep3p orthologs from ten point centromere-containing fungi. The alignment is numbered according to the S. cerevisiae Cep3p sequence. Secondary structural elements are indicated above the alignment. Residues participating in dimer contacts are indicated by asterisks above the alignment. Shaded boxes indicate conserved (gray) and invariant (black) residues. B. Side view and bottom view of the Cep3p dimer molecular surface colored by degree of conservation. The dotted ovals indicate two regions of high conservation that may represent Ctf13p interaction surfaces. Residues on the apex are highly variable, and they are presumably not involved in conserved protein-protein contacts.
Model for Cep3p-DNA interactions
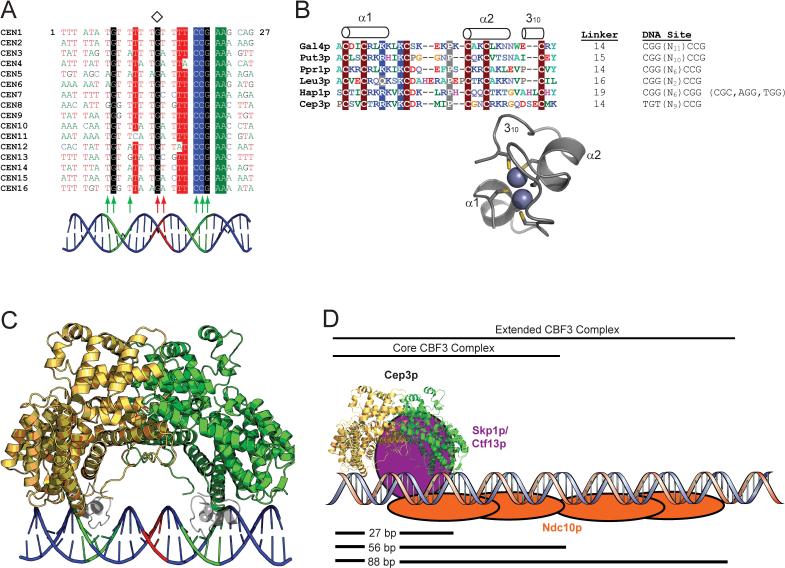
Intact CBF3, an asymmetric protein complex, binds CDEIII, a DNA site whose asymmetry is essential for function. Inspection of the centromere sequences of all 16 S. cerevisiae chromosomes, as well as those of other fungal species with identified CDEIII loci, suggests a pseudo-palindrome at the left-hand end of CDEIII coincident with the region contacted by Ctf13p and Cep3p, with a center of symmetry at the absolutely conserved G at position 14 (Fig. 4A) (Note that this position is displaced from the palindrome center suggested originally on the basis of conservation immediately flanking the conserved CCG(Jehn et al., 1991).) Thus, at the core of the CBF3 complex is the twofold symmetric Cep3p bound to an asymmetric but pseudo-palindromic site. All of the base pairs in CDEIII that are both conserved and required for in vivo kinetochore function and for in vitro CBF3 binding are in within this 27 base pair pseudo-palindrome, as are all of the major-groove contacts of Cep3p and Ctf13p detected by DNA-protein crosslinking: Cep3p and Ctf13p have major-groove contacts with CDEIII between base pairs 8 and 21 (numbering from the left, or CDEII-proximal, end of the 56-bp, DNAse I footprint; Fig. 4A). A single dimer of Ndc10p is then believed to make multiple contacts with CDEIII over nearly the entire 56 bp segment, and an extended complex containing a second Ndc10p dimer binds between base pairs 56 and 88 (Espelin et al., 1997). We have therefore modeled the complex of Cep3p with CBF3 as follows.
Figure 4. A model of CBF3-DNA assembly.
A. Multiple sequence alignment of the first 27 bases of the 56-bp DNAse I footprint of CDEIII from the 16 S. cerevisiae centromeres. The center of pseudosymmetry at the conserved base 14G is denoted by a diamond. Highly conserved positions are shaded. The positions of Cep3p (green) and Ctf13p (red) protein-DNA crosslinks (Espelin et al., 1997) are indicated by arrows. A B-DNA model of CDEIII is shown in cartoon form, with the putative Cep3p half-sites in green and the putative Ctf13p major groove binding site in red. B. The Zn2Cys6 cluster domain. Multiple sequence alignment of Zn2Cys6 cluster domains from six S. cerevisiae proteins, with conserved residues shaded and secondary structural elements indicated above the alignment. The length of the linker connecting the Zn2Cys6 cluster with αA (in Cep3p) or with the coiled-coil dimerization element (in the other proteins), is indicated, along with the DNA sites recognized by each protein. The Zn2Cys6 cluster from Hap1p (PDB ID 1WHT) is presented as a ribbon diagram with the Zn atoms and Cys sidechains shown (King et al., 1999). The two-residue insertion between Cys5 and Cys6 that forms the 310 helix in Hap1p is also present in Cep3p. C. A model for Cep3p-CDEIII binding. Zn2Cys6 clusters from the Hap1p structure have been docked into the two Cep3p half-sites on CDEIII. The twofold axis of Cep3p and the pseudo twofold axis of CDEIII have been superimposed. The polarity of the left-half site is ambiguous; this model approximates what the Cep3p-CDEIII complex could look like if the left half-site forms an inverted repeat (as in Gal4p) retaining overall two-fold symmetry. D. Model of CBF3-CDEIII Assembly. Ctf13p must bind asymmetrically to the Cep3p dimer and contact CDEIII in the major groove halfway between the Cep3p half-sites, as well as link Cep3p to Ndc10p. The Skp1p-Ctf13p heterodimer (purple) has been placed in an arbitrary position that fulfills those constraints. Ndc10p (orange) is modeled to reflect DNA crosslinking and electrophoretic mobility shift data, which suggest that one Ndc10p dimer binds the first 56 bases of CDEIII (as part of the core CBF3 complex), and a second Ndc10p dimer binds the region between 56 and 88 bp (to form the extended CBF3 complex) (Espelin et al., 1997).
Beginning with a 27 bp double helix of B-DNA representing CDEIII (Fig. 4A), we model the Zn2Cys6 clusters based on previously determined crystal structures of Gal4p family transcription factors (Fig. 4B) (Fitzgerald et al., 2006; King et al., 1999; Marmorstein et al., 1992; Marmorstein and Harrison, 1994; Swaminathan et al., 1997). We assume that the conserved and required CCG triplet of the right Cep3p half-site binds one of the Zn2Cys6 clusters just as in the DNA complexes of these proteins. Most of these transcription factors are homodimers (although in some cases asymmetric ones) and recognize identical half-sites. The half-sites can form inverted repeats (CCG...CGG), such as those that bind Gal4p, but they can also have symmetrically opposite orientations (“everted” repeats, CGG...CCG) and tandem orientations (CGG...CGG, as seen in the Hap1p:DNA complex). We have chosen to model the second Zn2Cys6 cluster bound to the left (TGT) triplet in an orientation twofold related to the one on the conserved triplet, but a tandem orientation is also possible. The latter would probably disturb alignment of the Cep3p and DNA twofold axes (as assumed in the next step of the model building; this alignment is not formally required by available data).
Each half of the Cep3p dimer has a “notch” formed by the N-terminus of αA, the αN-αO loop, and the β-turn, and the spacing of the dyad-related notches corresponds to the spacing of the Zn2Cys6 clusters positioned over their DNA binding sites. We can therefore dock the Cep3p model onto the Zn2Cys6 cluster:DNA model by aligning the twofold axis of Cep3p with the approximate dyad of the DNA model and placing the C-terminus of each cluster in the notch, close to the position into which the polypeptide chain must connect to αA (Fig. 4C). This docking also positions the N-terminus of αO so that the positive end of its helix dipole points towards a phosphate (5′ to base 7 on the top strand and to base 20 on the bottom strand). The phosphate could then receive hydrogen bonds from the amides of residues 367 and/or 368, a position held by a water in the Cep3p structure; the sidechains of Lys364 and Lys368 are also available to interact with the phosphate backbone. In this manner, the interaction of Cep3p with the 27-base pair pseudo-palindrome of CDEIII may retain overall two-fold symmetry, broken by the binding of Ctf13 across the dyad (Fig. 4D).
The Zn2Cys6 cluster of Cep3p deviates from most of the transcription factor clusters in having eight, rather than six, residues between the last two cysteines (Fig 4B). The Hap1p Zn2Cys6 cluster resembles Cep3p in this respect: the extra residues in Hap1p give rise to a turn of 310 helix, which helps place the two Zn2Cys6 clusters in the dimer in their asymmetric, tandem orientation by interacting with residues in the dimerization region(King et al., 1999). The modeling of Cep3p summarized by the diagrams in Fig. 4 strongly suggests that the Zn2Cys6 clusters of Cep3p will contact domain 3. The two-residue insertion may provide additional contact surface, as in Hap1p.
We also note that the Zn2Cys6 cluster of Hap1p appears to recognize degenerate CGG sites more readily than many others, by forming a “looser” major-groove interface with fewer protein-DNA contacts(King et al., 1999), and Cep3p's Zn2Cys6 cluster probably binds the left half-site in a similarly “loose” way. The central G:C base pair is invariant in the Hap1p sites, as it is on the CDEII-proximal half-site of CDEIII.
Centromeric evolution
Kinetochore proteins and centromeric DNA have undergone rapid evolution, but there is considerable evidence that many architectural features are conserved among all eukaryotes (Kitagawa and Hieter, 2001) (Meraldi et al., 2006). Cep3p has no direct homolog in higher eukaryotes, however, and it belongs to a subset of budding yeast kinetochore proteins that are specific to point centromeres. All centromeric chromatin contains specialized nucleosomes in which histone H3 has been replaced by the centromere-specific variant CenH3 (Cse4p in yeast and CENP-A in humans), which is thought to provide an epigenetic mark defining sites for kinetochore assembly (Mellone and Allshire, 2003) (Choo, 2001). In the case of fungi with point centromeres, deposition of Cse4p-containing nucleosomes, mediated by the conserved protein Scm3p, requires prior binding of CBF3 to CDEIII(Camahort et al., 2007; Mizuguchi et al., 2007; Stoler et al., 2007); in this sense, it is CBF3 that defines the positions of point centromeres . The regional centromeres of higher eukaryotes lack a structure analogous to the sequence specific CBF3-CDEIII complex. In those organisms, establishment of CENP-A-containing nucleosomes at sites not previously marked as centromeres (“neocentromeres”) requires binding of a protein known as CENP-B at specific, 17 bp sites within the α-satellite repeats of centromeric DNA(Ohzeki et al., 2002). The role, if any, of an Scm3p homolog has not yet been defined, but Scm3p homologs are indeed present in organisms with regional centromeres.
The structure of the N-terminal, 129-residue DNA-binding domain of CENP-B (a dimer of 80 kDa polypeptide chains and hence comparable in size to Cep3p) has been determined, in complex with its specific target site (Tanaka et al., 2001). It has two, small helix-turn-helix domains, positioned over the major groove about one DNA turn apart. The C-terminal CENP-B dimerization region consists of a pair of antiparallel α-helices (Tawaramoto et al., 2003). Association with the dimer partner is such that the two N-termini are at opposite ends of the dimer module, and the two DNA-binding domains may therefore associate with distant sites, separated by a large loop. Much of the amino-acid sequence between the N-terminal and C-terminal domains of CENP-B is of low complexity. Its architecture outside of the DNA-binding domain itself thus resembles that of a typical UAS or enhancer-binding protein more closely than does the fully ordered structure of Cep3p. This difference suggests that the mechanism by which CBF3 specifies the deposition of Cse4p-containing nucleosomes is structurally and evolutionarily distinct from the mechanism used to establish CENP-A-containing nucleosomes.
Conclusion
Point centromeres and the proteins that bind them appear to be relatively recent evolutionary innovations (Meraldi et al., 2006). The CDEI binding protein Cbf1p is a transcription factor as well as a kinetochore component. Skp1p is part of the ubiquitous SCF ubiquitin ligase complex, and Ctf13p appears to derive from one its F-box containing components. As discussed above, Cep3p has an N-terminal Zn2Cys6 cluster DNA-binding domain, which is exclusively found in fungal transcription factors, but like Ctf13p, it is an anomaly among its family members. Thus, many kinetochore proteins specific to point centromeres have apparently been “borrowed” from other cellular functions.
The structure of the Cep3p (47−608) dimer and the DNA-binding model derived from it are the beginnings of a three-dimensional representation of how CBF3 nucleates kinetochore assembly. The centromere resembles an enhancer/promoter in certain respects: it is composed of specifically spaced DNA sites that bind relatively dissimilar proteins, and kinetochore assembly (or transcriptional activation) requires reorganization of local chromatin structure. Unlike transcription factors, which generally serve at a variety of promoters with diverse organization of sub-sites, kinetochore components have apparently evolved to fit a fixed geometry, which is undoubtedly critical for ensuring the high selectivity of kinetochore assembly. Thus, Cep3p (47−608) has a well-ordered structure throughout nearly all of its roughly 560 amino-acid residues, while many transcription factors have large, unstructured regions and flexible hinges, so that they can adapt to multiple contexts. It should therefore be possible to “hang” additional kinetochore components, experimentally and conceptually, onto the Cep3p scaffold described here..
Experimental Procedures
Previously reported limited proteolysis experiments defined a stable fragment of Cep3p beginning at residue 47, which is 10 residues beyond the predicted end of the Zn2Cys6 cluster (Russell et al., 1999). The coding sequence for Cep3p (47−608) was cloned into pET3aTr and expressed in Rosetta(DE3)pLysS bacterial cells. The protein was purified by ammonium sulfate precipitation, followed by anion exchange chromatography and gel filtration. Cep3p (47−608) migrates as a dimer under gel filtration.
Crystals were grown by hanging drop vapor diffusion. Drops containing 0.5 μL 12 mg/mL Cep3 (47−608) in 50 mM HEPES pH 7.0, 300 mM KCl and 10 mM DTT and 0.5 μL reservoir solution were equilibrated against a reservoir of 0.7 mL 100 mM HEPES pH 7.0, 5% PEG 4000 and 500 mM NaCl. Crystals grew rapidly and reached maximum dimensions of 0.5 × 0.2 × 0.2 um in less than 48 hours. Crystals were transferred to a stabilization solution containing 100 mM HEPES pH 7.0 and 150 mM Na2SO4, then to cryoprotectant solution (stabilization solution containing 25%(v/v) glycerol) and flash-frozen in liquid nitrogen. Heavy atom derivatives were obtained from overnight soaks in stabilization solution plus 0.1 mM Na2PtCl4 and 0.1 mM ethyl mercuric thiosalicylate. Crystals soaked in Na2PtCl4 were backsoaked in stabilization solution lacking Na2PtCl4 before freezing.
The crystal structure of Cep3p (47−608) was determined using multiple isomorphous replacement with anomalous scattering. Table 1 contains data collection, phasing and refinement statistics. X-ray diffraction data on native crystals as well as Pt and Hg derivatives were collected at beamline ID-24 at the Advanced Photon Source. HKL2000 (Otwinowski et al., 1997) was used to index, integrate and scale the diffraction data, SHARP (Bricogne et al., 2003), to locate and refine the heavy atom positions and calculate phases, and Solomon (Abrahams and Leslie, 1996), for density modification by solvent flipping.
The initial model was built into a 3.0 Å density-modified experimental map using O (Kleywegt et al., 1997). CNS 1.2(Brunger et al., 1998) was used to refine the model against the 2.8 Å native dataset using maximum likelihood energy minimization, torsion angle simulated annealing, and unrestrained grouped B-factor refinement. All available data were used in all stages of refinement - the full resolution range of reflections (45−2.8 Å) and the experimental phase probabilities. Phi/psi restraints were placed on helical residues during refinement. Iterative cycles of refinement and rebuilding were carried out until the Rfree converged to 28.5%. Coordinates and structure factors have been deposited in the Protein Data Bank with accession number 2QUQ.
CDEIII was modeled as a 27-mer of B-DNA using 3DNA (Lu and Olson, 2003). Zn2Cys6 cluster coordinates were taken from the Hap1p crystal structure (PDBID 1WHT, (King et al., 1999)) and docked into the CDEIII model using lsqman (Kleywegt, 1996). Ribbon and surface figures were rendered using Pymol (DeLano, 2002). The Consurf server (Landau et al., 2005) was used to calculate conservation scores for coloring the molecular surface in Figure 3B.
Acknowledgments
The authors thank Kim Simons and JJ Miranda for helpful discussions. J.J.B. is the recipient of a Leukemia and Lymphoma Society Fellows Award. S.C.H. is an investigator of the Howard Hughes Medical Institute. Diffraction data were recorded at the Northeastern Collaborative Access Team beamlines of the Advanced Photon Source, supported by award RR-15301 from the National Center for Research Resources at the National Institute of Health. Use of the Advanced Photon Source is supported by the U.S. Department of Energy, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abrahams JP, Leslie AG. Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr D Biol Crystallogr. 1996;52:30–42. doi: 10.1107/S0907444995008754. [DOI] [PubMed] [Google Scholar]
- Bricogne G, Vonrhein C, Flensburg C, Schiltz M, Paciorek W. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr D Biol Crystallogr. 2003;59:2023–2030. doi: 10.1107/s0907444903017694. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Camahort R, Li B, Florens L, Swanson SK, Washburn MP, Gerton JL. Scm3 is essential to recruit the histone h3 variant cse4 to centromeres and to maintain a functional kinetochore. Mol Cell. 2007;26:853–865. doi: 10.1016/j.molcel.2007.05.013. [DOI] [PubMed] [Google Scholar]
- Choo KH. Domain organization at the centromere and neocentromere. Dev Cell. 2001;1:165–177. doi: 10.1016/s1534-5807(01)00028-4. [DOI] [PubMed] [Google Scholar]
- De Wulf P, McAinsh AD, Sorger PK. Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes Dev. 2003;17:2902–2921. doi: 10.1101/gad.1144403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. Delano Scientific; Palo Alto, CA: 2002. [Google Scholar]
- Espelin CW, Kaplan KB, Sorger PK. Probing the architecture of a simple kinetochore using DNA-protein crosslinking. J Cell Biol. 1997;139:1383–1396. doi: 10.1083/jcb.139.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espelin CW, Simons KT, Harrison SC, Sorger PK. Binding of the essential S. cerevisiae kinetochore protein Ndc10p to CDEII. Mol Biol Cell. 2003 doi: 10.1091/mbc.E02-08-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald MX, Rojas JR, Kim JM, Kohlhaw GB, Marmorstein R. Structure of a Leu3-DNA complex: recognition of everted CGG half-sites by a Zn2Cys6 binuclear cluster protein. Structure. 2006;14:725–735. doi: 10.1016/j.str.2005.11.025. [DOI] [PubMed] [Google Scholar]
- Jehn B, Niedenthal R, Hegemann JH. In vivo analysis of the Saccharomyces cerevisiae centromere CDEIII sequence: requirements for mitotic chromosome segregation. Mol Cell Biol. 1991;11:5212–5221. doi: 10.1128/mcb.11.10.5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan KB, Hyman AA, Sorger PK. Regulating the yeast kinetochore by ubiquitin-dependent degradation and Skp1p-mediated phosphorylation. Cell. 1997;91:491–500. doi: 10.1016/s0092-8674(00)80435-3. [DOI] [PubMed] [Google Scholar]
- King DA, Zhang L, Guarente L, Marmorstein R. Structure of a HAP1-DNA complex reveals dramatically asymmetric DNA binding by a homodimeric protein. Nat Struct Biol. 1999;6:64–71. doi: 10.1038/4940. [DOI] [PubMed] [Google Scholar]
- Kitagawa K, Hieter P. Evolutionary conservation between budding yeast and human kinetochores. Nat Rev Mol Cell Biol. 2001;2:678–687. doi: 10.1038/35089568. [DOI] [PubMed] [Google Scholar]
- Kleywegt GJ. Use of non-crystallographic symmetry in protein structure refinement. Acta Crystallogr D Biol Crystallogr. 1996;52:842–857. doi: 10.1107/S0907444995016477. [DOI] [PubMed] [Google Scholar]
- Kleywegt GJ, Alwyn Jones T, Charles W. C. J. a. R. M. S. Methods in Enzymology. Academic Press; 1997. Model building and refinement practice; pp. 208–230. [DOI] [PubMed] [Google Scholar]
- Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-Tal N. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner J. A zinc finger protein, essential for chromosome segregation, constitutes a putative DNA binding subunit of the Saccharomyces cerevisiae kinetochore complex, Cbf3. Embo J. 1994;13:5203–5211. doi: 10.1002/j.1460-2075.1994.tb06851.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner J, Carbon J. A 240 kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell. 1991;64:717–725. doi: 10.1016/0092-8674(91)90501-o. [DOI] [PubMed] [Google Scholar]
- Lu X-J, Olson WK. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacPherson S, Larochelle M, Turcotte B. A fungal family of transcriptional regulators: the zinc cluster proteins. Microbiol Mol Biol Rev. 2006;70:583–604. doi: 10.1128/MMBR.00015-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmorstein R, Carey M, Ptashne M, Harrison SC. DNA recognition by GAL4: structure of a protein-DNA complex. Nature. 1992;356:408–414. doi: 10.1038/356408a0. [DOI] [PubMed] [Google Scholar]
- Marmorstein R, Harrison SC. Crystal structure of a PPR1-DNA complex: DNA recognition by proteins containing a Zn2Cys6 binuclear cluster. Genes Dev. 1994;8:2504–2512. doi: 10.1101/gad.8.20.2504. [DOI] [PubMed] [Google Scholar]
- McAinsh AD, Tytell JD, Sorger PK. Structure, Function and Regulation of Budding Yeast Kinetochores. Annu Rev Cell Dev Biol. 2003;19:519–539. doi: 10.1146/annurev.cellbio.19.111301.155607. [DOI] [PubMed] [Google Scholar]
- Mellone BG, Allshire RC. Stretching it: putting the CEN(P-A) in centromere. Curr Opin Genet Dev. 2003;13:191–198. doi: 10.1016/s0959-437x(03)00019-4. [DOI] [PubMed] [Google Scholar]
- Meraldi P, McAinsh AD, Rheinbay E, Sorger PK. Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol. 2006;7:R23. doi: 10.1186/gb-2006-7-3-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Nonhistone Scm3 and Histones CenH3-H4 Assemble the Core of Centromere-Specific Nucleosomes. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- Ohzeki J, Nakano M, Okada T, Masumoto H. CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA. J Cell Biol. 2002;159:765–775. doi: 10.1083/jcb.200207112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W, Carter Charles W., Jr. Methods in Enzymology. Academic Press; 1997. [20] Processing of X-ray diffraction data collected in oscillation mode; pp. 307–326. [DOI] [PubMed] [Google Scholar]
- Russell ID, Grancell AS, Sorger PK. The unstable F-box protein p58-Ctf13 forms the structural core of the CBF3 kinetochore complex. J Cell Biol. 1999;145:933–950. doi: 10.1083/jcb.145.5.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger PK, Doheny KF, Hieter P, Kopski KM, Huffaker TC, Hyman AA. Two genes required for the binding of an essential Saccharomyces cerevisiae kinetochore complex to DNA. Proc Natl Acad Sci U S A. 1995;92:12026–12030. doi: 10.1073/pnas.92.26.12026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoler S, Rogers K, Weitze S, Morey L, Fitzgerald-Hayes M, Baker RE. Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proc Natl Acad Sci U S A. 2007;104:10571–10576. doi: 10.1073/pnas.0703178104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoyan T, Carbon J. Inner kinetochore of the pathogenic yeast Candida glabrata. Eukaryot Cell. 2004;3:1154–1163. doi: 10.1128/EC.3.5.1154-1163.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan K, Flynn P, Reece RJ, Marmorstein R. Crystal structure of a PUT3-DNA complex reveals a novel mechanism for DNA recognition by a protein containing a Zn2Cys6 binuclear cluster. Nat Struct Biol. 1997;4:751–759. doi: 10.1038/nsb0997-751. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Nureki O, Kurumizaka H, Fukai S, Kawaguchi S, Ikuta M, Iwahara J, Okazaki T, Yokoyama S. Crystal structure of the CENP-B protein-DNA complex: the DNA-binding domains of CENP-B induce kinks in the CENP-B box DNA. Embo J. 2001;20:6612–6618. doi: 10.1093/emboj/20.23.6612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawaramoto MS, Park SY, Tanaka Y, Nureki O, Kurumizaka H, Yokoyama S. Crystal structure of the human centromere protein B (CENP-B) dimerization domain at 1.65-A resolution. J Biol Chem. 2003;278:51454–51461. doi: 10.1074/jbc.M310388200. [DOI] [PubMed] [Google Scholar]