Abstract
Neurons engage in causal interactions with one another and with the surrounding body and environment. Neural systems can therefore be analyzed in terms of causal networks, without assumptions about information processing, neural coding, and the like. Here, we review a series of studies analyzing causal networks in simulated neural systems using a combination of Granger causality analysis and graph theory. Analysis of a simple target-fixation model shows that causal networks provide intuitive representations of neural dynamics during behavior which can be validated by lesion experiments. Extension of the approach to a neurorobotic model of the hippocampus and surrounding areas identifies shifting causal pathways during learning of a spatial navigation task. Analysis of causal interactions at the population level in the model shows that behavioral learning is accompanied by selection of specific causal pathways—“causal cores”—from among large and variable repertoires of neuronal interactions. Finally, we argue that a causal network perspective may be useful for characterizing the complex neural dynamics underlying consciousness.
Keywords: Granger causality, Causal core, Consciousness, Neurorobotics, Hippocampus, Mesoscale
Introduction
A basic fact about neural systems is that their elements enter into causal interactions with one another, as well as with the surrounding body and environment. Because neural systems are generally composed of large numbers of elements, a useful analysis of their causal interactions must involve causal networks. Neural systems can be analyzed in terms of causal networks without assumptions about whether or not they are ‘information processing’ devices (Churchland and Sejnowski 1994), or whether or not there exist ‘neural codes’ (deCharms and Zador 2000).
In this paper, time-series analysis techniques based on Granger causality (Granger 1969) are combined with network theory in order to characterize causal networks in simulated neural systems (Seth 2005; Krichmar et al. 2005b; Seth and Edelman 2007; Seth et al. 2006). Because our aim is to elaborate a particular theoretical perspective on neural dynamics, and not to analyze existing empirical data, we illustrate causal network analysis using concrete simulation models. This approach has the advantage of providing complete knowledge of the underlying (simulated) physical system, allowing informative comparisons of structural connectivity and causal connectivity. By considering several different neural simulations we show that causal network analysis can flexibly be deployed in diverse neural modeling scenarios, yielding different insights in each case.
It is important to underline the difference between a ‘structural network’ and a ‘causal network’ as used in the present context. The former corresponds to the network defined by the synaptic connections among neurons; these connections can be unidirectional or reciprocal. The ‘causal network’ is the network of significant causal interactions among neurons. In other words, a causal network reflects an aspect of ‘functional’ and/or ‘effective’ connectivity (Friston 1994) whereas a structural network reflects physical connectivity. It also bears emphasizing that Granger causality is a statistical measure of causality, meaning that Granger-causal links do not necessarily reflect physical causal chains.
After reviewing the principles of Granger causality analysis (Granger 1969; Ding et al. 2006; Seth 2007), we illustrate the causal network approach by analyzing a simple simulation model involving sensorimotor coordination. In this model, genetic algorithms (GAs) are used to specify neural network controllers supporting target fixation behavior in a simulated head/eye system (Seth 2005; Seth and Edelman 2004), and causal networks are compared with underlying structural networks (i.e., the evolved neural network controllers). We find that causal networks provide an intuitive description of neural dynamics that reflect agent-environment interactions during behavior and we also show that they reliably predict the effects of (structural) network lesions. This study serves to validate the causal network approach and to demonstrate its potential in examining adaptation of sensorimotor networks to different environmental conditions.
In order for causal network analysis to provide useful heuristics for interpreting empirical data it is necessary to move beyond simple ‘toy’ models and to consider more detailed models incorporating aspects of neuroanatomy and neurophysiology. To address this need we extend our analysis to an embodied neural simulation—a robotic brain based device (BBD) (Krichmar and Edelman 2002)—that incorporates a detailed model of the mammalian hippocampus (Krichmar et al. 2005a, b). This BBD learns a task similar to a classical experimental paradigm in which rodents are trained to locate a ‘hidden platform’ in a pool of milky water (a Morris ‘water maze’ (Morris 1984)); a task for which an intact hippocampus is required. Causal network analysis of this BBD reveals causal pathways involving the hippocampus that mediate sensory input and motor output and that map onto known anatomical connections. The analysis also shows how these pathways are modulated as a result of learning, for example by the emergence of causal ‘short cuts’ through the hippocampus. These findings facilitate interpretation of existing empirical data and also suggest specific hypotheses that may be testable in future animal experiments.
The BBD just described has ≈90,000 neuronal units and ≈1,400,000 synaptic connections. This substantial simulated nervous system invites analysis at the mesoscale level, i.e., the level of description of neural systems that lies between small neuronal circuits and systems-level neural areas (Sporns et al. 2005; Lin et al. 2006). Due to the lack of appropriate experimental methods detailed mesoscale descriptions of biological neural systems are rare, yet theoretical approaches suggest that mesoscale dynamics are highly significant for neural function. For example, the theory of neuronal group selection (TNGS, (Edelman 1987, 1993)) proposes that functional neural circuits are selected from highly variant repertoires during development and behavior.
An important issue at the mesoscale level is the identification of causal interactions in complex neural populations that lead to specific outputs. To address this issue we introduce the concept of a causal core to refer to the set of neuronal interactions that are significant for a given output, as assessed by Granger causality (Seth and Edelman 2007). Applying this extended analysis to the neurorobotic model of the hippocampus reveals that large repertoires of neural interactions contain comparatively small causal cores and that these causal cores become smaller during learning. These results are consistent with the hypothesis, drawn from the TNGS, that behavioral learning involves selection of specific causal pathways from diverse repertoires.
Importantly, recent methodological advances promise to reveal comprehensive microscale and mesoscale details of neurobiological systems (Kelly and Strick 2004; Konkle and Bielajew 2004; Raffi and Siegel 2005). The causal core concept, and causal network analysis in general, provides both useful heuristics and practical methods for interpreting and analyzing this cutting-edge and future data. Because causality analysis makes no assumptions about information processing or neural coding, a causal network approach offers an objective framework for considering global aspects of brain function which have remained poorly understood from existing theoretical perspectives. Foremost among these global aspects are the neural mechanisms underlying consciousness.
Most attempts to understand the neural mechanisms of consciousness have proceeded by searching for the so-called ‘neural correlates of consciousness’ (Rees et al. 2002). However, correlations by themselves do not provide explanations and there is a need for theories that connect fundamental aspects of conscious phenomenology to corresponding aspects of neural dynamics. A promising approach in this regard has been to notice that all conscious scenes are both differentiated (each is composed of many parts and is therefore unique) and integrated (each is experienced as a unified whole) (Edelman 2003; Tononi and Edelman 1998). Quantitative multivariate measures that track these properties and that are applicable to neural dynamics are therefore very useful for neural theories of consciousness, and indeed several such measures have now been proposed (Edelman 2003; Tononi and Edelman 1998; Tononi 2004). Here, we adopt a causal network perspective on the neural dynamics underlying consciousness and we analyze the proposal that a particular measure applicable to causal networks, causal density, provides a useful means of quantifying the complexity of the neural dynamics relevant to consciousness (Seth et al. 2006). Causal density is argued to overcome certain limitations associated with alternative measures by (i) naturally incorporating causal interactions and (ii) providing a genuine measure of process rather than capacity.
Finally, we discuss limitations and possible extensions of the causal network approach, its relation to other methods for extracting causal representations, and future applications both in neural modeling and in the analysis of empirical data. As a whole, the research surveyed in this article indicates that a causal network perspective provides both novel concepts for understanding neural systems, and a set of practically applicable methods for illustrating, refining, and testing the usefulness of these concepts.
Granger causality
The concept of Granger causality is based on prediction: If a signal X1 causes a signal X2, then past values of X1 should contain information that helps predict X2 above and beyond the information contained in past values of X2 alone (Granger 1969). In practice, Granger causality can be tested using multivariate regression (MVAR) modelling (Hamilton 1994). For example, suppose that the temporal dynamics of two time series, X1(t) and X2(t) (both of length T), can be described by a bivariate autoregressive model:
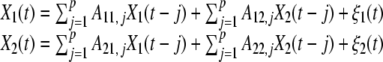 |
1 |
where p is the maximum number of lagged observations included in the model (the model order, p < T), A contains the coefficients of the model (i.e., the contributions of each lagged observation to the predicted values of X1(t) and X2(t)), and ξ1, ξ2 are the residuals (prediction errors) for each time series. If the variance of ξ1 (or ξ2) is reduced by the inclusion of the X2 (or X1) terms in the first (or second) equation, then it is said that X2 (or X1) Granger-causesX1 (or X2). In other words, X2 Granger-causes X1 if the coefficients in A12 are jointly significantly different from zero. This can be tested by performing an F-test of the null hypothesis that A12 = 0, given assumptions of covariance stationarity on X1 and X2. The magnitude of a Granger causality interaction can be estimated by the logarithm of the corresponding F-statistic (Geweke 1982). 1
As mentioned in section “Introduction”, the statistical basis of Granger causality implies that Granger-causal links need not directly reflect physical connections. In particular, variables that are not included in the regression model can induce ‘common-input’ artifacts. For example, if X1 and X2 both receive input from a third variable X3 with different time delays, then a bivariate analysis of X1 and X2 will show a causal link from one to the other even in the absence of a physical connection. Common-input problems can be avoided by extending Granger causality to the n variable case (where n > 2), by estimating an n variable autoregressive model. In this case, X2 Granger-causes X1 if knowing X2 reduces the variance in X1’s prediction error when the activities of all other variables X3 ... Xn are also taken into account. This multivariate extension is sometimes referred to as conditional Granger causality (Ding et al. 2006). As well as eliminating common-input artifacts, conditional Granger causality is useful for revealing causal interactions among sets of nodes.
Significant Granger causality interactions between variables can be represented as edges in a graph, allowing the application of graph-theoretic techniques (Seth 2005; Eichler 2005). Because Granger causality is in general not symmetric, these edges will be directed. As shown in the following, the resulting graphs, or causal networks, can provide intuitive and valuable representations of functional connectivity within a system.
Causal networks during target fixation
To illustrate causal networks in a simplified setting, we first analyze a simulation model of target fixation requiring coordination of ‘head’ and ‘eye’ movements. Full details of the model are given in Seth (2005) and Seth and Edelman (2004). Here only a minimal set of features are described.
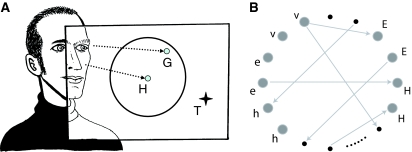
In the model, a planar environment contains a target (T) and a neural network controls a gaze direction (G) onto this xy plane by modulating head direction (H), and eye direction (E) which is relative to head direction (see Fig. 1a). Networks consisted of 32 neurons and 256 initially randomly distributed synaptic connections. Each neuron was modeled by a sigmoidal transfer function; six neurons were sensor inputs, including two visual (v-)inputs and four proprioceptive (h- and e-)inputs. V-inputs reflected displacement of G from T; h-inputs reflected displacement of H from an arbitrary origin, and e-inputs reflected displacement of H from E (see Fig. 1b). Each pair of input neurons signalled displacements in the x and y dimensions. The remaining 22 neurons were ‘interneurons’ (INs) with initially random connectivity to the remainder of the network.
Fig. 1.
Target fixation model. (a) The agent controls head-direction (H) and eye-direction (not shown) in order to move a gaze point (G) towards a target (T). (b) Neural network controller. Six input neurons are shown on the left: v-inputs reflect displacement of G from T; h-inputs and e-inputs reflect proprioceptive signals (see text). Four output neurons are shown on the right: H-outputs control head velocity and E-outputs control the velocity of E relative to H. For clarity only 4 of the remaining 22 neurons and only a subset of the 256 connections are shown.
The topology of the network (both the structural connectivity and the synaptic strengths) was evolved using a genetic algorithm (GA). The GA was permitted to explore all possible network architectures with no constraints (e.g., organisation into layers was not imposed). The fitness function F was designed to maximize target fixation while simultaneously keeping head and eye aligned. 2
To test how causal networks depended on agent-environment interactions, networks were evolved in both ‘simple’ and ‘complex’ conditions where complexity was reflected by unpredictable target movement and by variations in parameters affecting head and eye movement, including a time-delay for head movements relative to eye movements (for full details see Seth 2005; Seth and Edelman 2004). Ten evolutionary runs were carried out in each condition.
Causal networks were derived from the fittest network from each evolutionary run in each condition. From each such network, causal networks were identified by computing 10-dimensional MVAR models for time-series of the six input and four output neurons only. Before MVAR computation, each time-series was first-order differenced to ensure covariance stationarity (see section “Stationarity”), and the number of lags (p in Eq. 1) was set according to the Bayesian Information Criterion (BIC, (Schwartz 1978)) to be four. Significant Granger causality interactions were calculated between input-output neuron pairs only (P < 0.01).
Causal networks
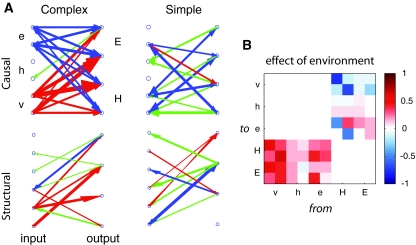
Figure 2a shows representative casual networks from each condition (top) as well as the corresponding structural networks (bottom). Networks adapted to complex conditions (C-nets) showed strong causal connectivity from visual inputs to motor outputs, suggesting that their dynamics were driven largely by visual signals. These networks also showed strong reciprocal causality involving e-inputs but little involving h-inputs, suggesting that the former may influence behavior more than the latter. By contrast, networks adapted to simple conditions (S-nets) showed significantly fewer strong driving influences and less distinction among e-inputs and h-inputs, suggesting that these networks were less functionally differentiated than C-nets.
Fig. 2.
(a) Causal connectivity in the target fixation model. Each panel shows input (v: v-inputs, e: e-inputs, h: h-inputs) and output neurons (E: E-outputs, H: H-outputs). Red arrows show input→output causality, green arrows show output→input causality, and blue arrows show reciprocal causality. Arrow width reflects magnitude of causal influence. (b) Effect of environment on causal connectivity. See text for details; color online
Causal networks overlapped with but were not identical to structural networks (Fig. 2a bottom). For C-nets the strong causal influence from v-inputs to motor outputs was reflected in network structure but the strong reciprocal causality was not structurally prominent. S-nets also showed differences between structural and causal connectivity, especially in regard to reciprocal connectivity. These differences are expected because causal networks, unlike structural networks, depend on interactions among network structure, phenotype, and environment.
It is important to note that causal network analyses are agnostic with respect to ‘feedforward’ and ‘feedback’ connections. In particular, reciprocal causal connectivity between an input and an output does not mandate the interpretation that the input→output connection is driving and that the reverse connection is modulatory. If both connections are causally significant then both should be considered to be ‘driving’, although possibly to different extents depending on magnitude (this issue is discussed further in section “Discussion”).
The effect of environment on causal connectivity is illustrated in Fig. 2b, in which warm (cool) colors indicate causal interactions that were reliably strengthened (weakened) when a C-net was tested in a complex environment as compared to a simple environment. Consistent with Fig. 2a, testing in a complex environment led to greater input→output causal connectivity and increased reciprocal causality from outputs to e-inputs. However, connections from outputs to v-inputs were weaker, suggesting that visual signals are less predictable from prior head movements in complex environments. These observations together suggest that, in this model, environmental simplification leads to a reduction in the range of causal interactions sustained by a network.
Predicting the influence of lesions
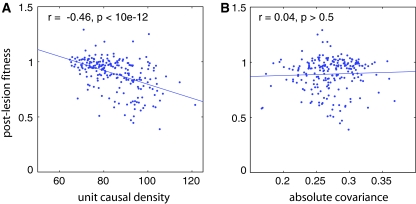
A useful way to validate causal networks derived by Granger causality is to test whether they predict the effects of network lesions. To explore this idea, we computed causal networks involving all 32 neurons of each C-net and assessed the fitness consequences of lesioning each of the 22 INs in turn. For each IN we also calculated the total strength of incoming and outgoing causal connections (‘unit causal density’), and covariance of the dynamics of each IN with the remainder of the network (calculated as the mean absolute covariance of the IN with each other neuron in the network). Figure 3 shows that post-lesion fitness has a strong negative correlation with unit causal density, but no correlation with covariance, confirming that the causal connectivity of a neuron is a useful predictor of the dynamical and behavioral consequences of network damage.
Fig. 3.
Post-lesion fitness following lesions to INs as a proportion of the fitness of the intact network, plotted against (a) mean unit causal density of the IN and (b) mean absolute covariance of the IN with the remainder of the network (see text). Each panel shows Pearson’s correlation coefficient (r) as well as the corresponding p-value.
Summary
The analysis of simple ‘toy’ models can be useful for validating the causal network approach and for gaining insight into the modulation of causal interactions by adaptation to different environments. In addition, simple models allow easy comparisons of structural connectivity and functional connectivity, both in normal conditions and following network damage. However, for the causal network approach to engage more directly with empirical data it is necessary to study more detailed models incorporating substantial neurobiological detail.
Causal networks in a brain-based device
Darwin X is a brain-based device (BBD), that is, a physical device which interacts with a real environment via sensors and motors, whose behavior is guided by a simulated nervous system incorporating detailed aspects of the neuroanatomy and neurophysiology of the mammalian hippocampus and surrounding areas (Fig. 4).
Fig. 4.
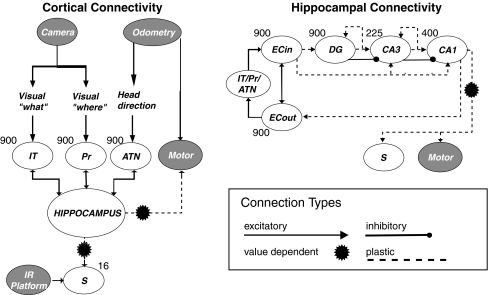
Schematic of Darwin X’s simulated nervous system. There were two visual input streams responding to the color (IT), and width (Pr), of visual landmarks on the walls, as well as one odometric input signalling Darwin X’s heading (ATN). These inputs were reciprocally connected with the hippocampus which included ‘entorhinal’ cortical areas ECin and ECout, ‘dentate gyrus’ DG, and the CA3 and CA1 hippocampal subfields. The number of simulated neuronal units in each area is indicated adjacent to each area. This figure is reprinted with permission from Seth and Edelman (2007)
The hippocampal system is particularly attractive for causal network analysis because of its distinctive functional neuroanatomy. Signals from many neocortical areas enter the hippocampus via the entorhinal cortex. Hippocampal output is also funneled via entorhinal cortex in divergent projections to neocortex. Within the hippocampus, there are connections from the entorhinal cortex to the dentate gyrus, from the dentate gyrus to CA3 (mossy fibers), from CA3 to CA1 (Schaffer collaterals), and then from CA1 back to the entorhinal cortex (Lavenex and Amaral 2000). This pathway is referred to as the trisynaptic loop. There are also perforant pathway ‘short-circuit’ projections from entorhinal cortex directly to CA3 and CA1. Functionally, an intact hippocampus is necessary for laying down episodic memories in humans as well as for learning spatial navigation behavior in rodent models. Both of these functions require integrating multimodal inputs over different timescales and it is possible that the unique looping anatomy of the hippocampal system underlies this integration. Causal network analysis of a hippocampal simulation is well placed to explore this idea.
Full details of the construction and performance of Darwin X are given in Krichmar et al. (2005a, b); for present purposes it is sufficient to mention only the following. Darwin X contained analogs of several mammalian brain areas including subareas of the hippocampus and three sensory input streams which received input from a CCD camera and from odometry (Fig. 4). Each neuronal unit in Darwin X represented a group of ≈100 real neurons and was simulated using a mean firing rate model. Synaptic plasticity was implemented using a modified version of the BCM learning rule (Bienenstock et al. 1982) in which synapses between strongly correlated neuronal units are potentiated and synapses between weakly correlated neuronal units are depressed. In some pathways, synaptic changes were further modulated by the activity of a simulated value system (area S in Fig. 4) which responded to salient events (see below). Value-dependent modulation was implemented using the temporal-difference learning rule in which learning is based on the difference between temporally successive predictions of reward (Sutton and Barto 1998). The full Darwin X model contained 50 neural areas, ≈90,000 neuronal units, and ≈1,400,000 synaptic connections.
Darwin X was trained on a ‘dry’ version of the well-known Morris water maze task (Morris 1984), in which the device learned to locate a hidden platform in a rectangular arena with diverse visual landmarks hung on the walls. Darwin X could only detect the hidden platform when it was directly overhead, by means of a downward facing infrared sensor. Each encounter with the platform stimulated the value system which modulated synaptic plasticity in the value-dependent pathways of Darwin X’s simulated nervous system. Darwin X was trained over 17 trials, each beginning from one of four initial positions. Initially, Darwin X moved randomly, but after about 10 trials, the device reliably took a comparatively direct path to the platform from any starting point (Krichmar et al. 2005b).
Causal networks in Darwin X
Causal interactions in Darwin X were analyzed by selecting ‘functional circuits’ as follows. For each neuronal unit in CA1 (the reference unit), a functional circuit of neuronal units was selected by identifying those units, from different neural areas, that covaried most strongly with the reference unit. The activity time-series corresponding to these functional circuits were then first-order differenced and circuits which were not covariance stationary were discarded. Remaining functional circuits were analyzed using a multivariate Granger causality analysis with model order p = 3 set according to the BIC.
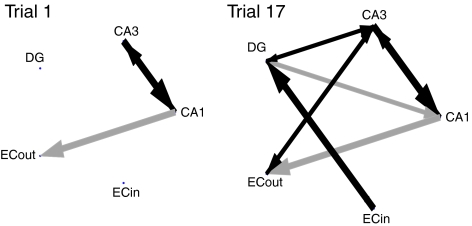
Figure 5 shows patterns of causal interactions from a representative CA1 reference unit both early in learning (left) and late in learning (right). After learning, the causal network involving the reference unit is much denser and has developed a trisynaptic loop.
Fig. 5.
Causal connectivity patterns for a representative CA1 reference unit during the first trial (left) and the last trial (right). Grey arrows show unidirectional connections and black arrows show bidirectional connections. The width of each arrow (and size of arrowhead) reflect the magnitude of the causal interaction
Analyzing all functional circuits from both trials 1 and 17 revealed an increase in the proportion of perforant pathway short-cuts in which signals from cortex causally influence the CA1 reference unit without causally involving the intermediate stages of DG and CA3, and a corresponding decrease in the proportion of causal trisynaptic pathways (Table 1). A possible explanation for this is that the trisynaptic pathway, which integrates more areas of the hippocampus, may be necessary when in unfamiliar environments, and that the comparatively direct perforant pathway may be more useful when translating sensory cues into motor actions in familiar environments.
Table 1.
Percentage of causal pathways that were either trisynaptic or (perforant) shortcut, before and after experience
| Trial 1 (n = 231) | Trial 17 (n = 162) | |
|---|---|---|
| Trisynaptic | 42.0 | 29.0 |
| Short-cut | 14.8 | 21.6 |
We also found that the causal influence exerted by neuronal units in area ATN, reflecting odometric head-direction signals, grew markedly during learning, whereas influence from the visual areas remained the same (Krichmar et al. 2005b). This suggests that, in this task, Darwin X over time relied less on integrating multisensory signals and its behavior was increasingly driven by odometric signals with modulation from visual areas. These hypotheses may be testable in future animal experiments.
Causal cores in neural populations
The causal networks just described were identified using a multivariate analysis of a small number of neuronal units from different regions of Darwin X’s simulated nervous system. We turn now to a variant of this analysis which distinguishes causal interactions within large neural populations that lead to specific outputs (Seth and Edelman 2007). This new analysis is motivated by the need to understand neural dynamics at the mesoscale, i.e., the population level that intervenes between small neuronal circuits and systems-level neural areas (Sporns et al. 2005; Lin et al. 2006). Because our analysis requires complete knowledge of neuroanatomy and dynamics of the studied system, it is also well illustrated by application to Darwin X.
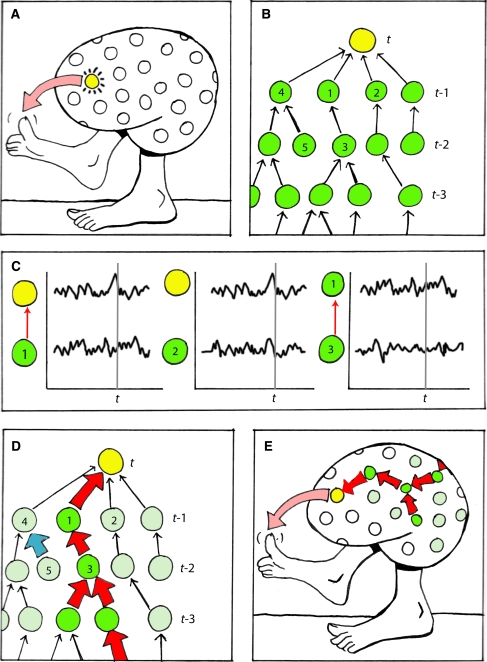
The general framework for this mesoscale analysis is illustrated in Fig. 6. First, a neural reference (NR) is selected from among many possible neuronal events, in this example by virtue of its relation to a specific behavioral output (Fig. 6a). In the terminology introduced in the foregoing, a NR refers to the activity of a reference unit at a particular time. Second, a context network is identified by recursively examining the activity of all neurons that led to each NR, a procedure referred to as a backtrace (Krichmar et al. 2005a) (Fig. 6b). The first iteration of a backtrace identifies those neurons that were both anatomically connected to the NR neuron and active (above a threshold) at the previous time-step. This procedure can be iterated as allowed by computational tractability, but in general a low iteration depth ensures the identification of the most salient neural interactions for a particular NR while avoiding a combinatorial explosion. Third, a Granger causality analysis is applied to assess the causal significance of each connection in the context network (Fig. 6c). In order to ensure robust statistical inferences, each connection is assessed over a time period considerably longer than that used to identify the context network.3 Also, each connection is assessed separately; i.e., using a repeated bivariate design with correction for multiple comparisons. The resulting networks of significant Granger causality interactions are referred to as Granger networks. Last, the causal core of each NR is identified by extracting the subset of the corresponding Granger network consisting of all causally significant connections leading, via other causally significant connections, to the NR (Fig. 6d–e).
Fig. 6.
Distinguishing causal interactions in neuronal populations. (a) Select a neural reference (NR), i.e., the activity of a particular neuron (yellow) at a particular time (t). (b) The context network of the NR corresponds to the network of all coactive and connected precursors, assessed over a short time period. (c) Assess the Granger causality significance of each interaction in the context network, based on extended time-series of the activities of the corresponding neurons. Red arrows indicate causally significant interactions. (d, e) The causal core of the NR (red arrows) is defined as that subset of the context network that is causally significant for the activity of the corresponding neuron (i.e., excluding both non-causal interactions (black arrows) and ‘dead-end’ causal interactions such as 5→4, indicated in blue). Color online
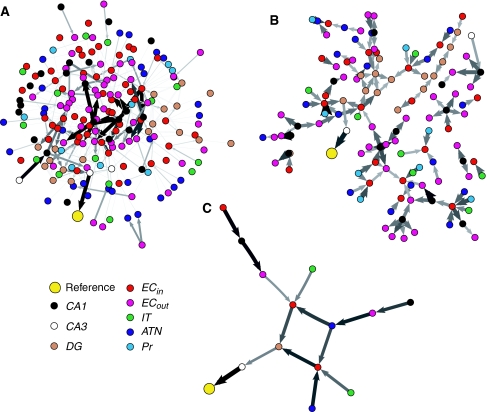
As reported in detail in Seth and Edelman (2007), application of the above analysis to Darwin X involved selecting 93 NRs corresponding to bursts of activity in CA1 neuronal units at different time points spanning the learning process. Context networks for each NR were identified by iterating the backtrace algorithm for six time steps. Figure 7 shows the context network, Granger network, and causal core for a representative NR. The causal core is strikingly small as compared to the context and Granger networks, and is largely free from the entorhinal interactions which dominate these other networks. These observations generalized to the remaining 92 NRs (Table 2), suggesting that (i) even in large neural populations, only comparatively small subsets may be causally recruited at a given time for a given function, and (ii) trisynaptic and perforant pathways had greater causal influence on the selected NRs than did entorhinal interactions. Importantly, as shown in Seth and Edelman (2007), causal cores could not in general be identified on the basis of synaptic strengths alone.
Fig. 7.
(a) The context network for a representative NR in Darwin X. The thickness of each line (and size of each arrowhead) is determined by the product of synaptic strength and the activity of the presynaptic neuron. (b) The corresponding Granger network. Line thickness here reflects magnitude of the corresponding causal interaction. (c) The corresponding causal core. Networks were visualized using the Pajek program (http://vlado.fmf.uni-lj.si/pub/networks/pajek/), which implements the Kamada-Kawai energy minimization algorithm. Color online
Table 2.
Mean composition and size of context networks, Granger networks, and causal cores
| % entorhinal | % cortical input | % hippocampal | Size | |
|---|---|---|---|---|
| Context | 71 | 20 | 9 | 723 |
| Granger | 26 | 52 | 22 | 223 |
| Causal core | 7 | 48 | 45 | 29 |
Causal core refinement during learning
The functional circuit analysis described in section “Causal networks in brain-based device” showed that learning in Darwin X was accompanied by a shift in the balance of causal pathways from trisynaptic loops to perforant ‘short-cuts’. Here, we explore how population causal interactions are modulated during learning, which raises the general issue of how synaptic plasticity and behavioral learning are related. It is often assumed that learned behavior depends on the cumulative effects of long-term potentiation (LTP) and depression (LTD) of synapses. However, synaptic plasticity and behavioral learning operate over vastly different temporal and spatial scales (Drew and Abbott 2006) and although empirical evidence indicates that learning can induce LTP and LTD in certain cases (Malenka and Bear 2004; Dityatev and Bolshakov 2005), the precise functional contributions of synaptic plasticity to learned behavior have so far remained unclear.
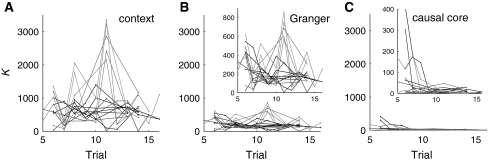
Figure 8 shows that causal cores reliably diminish in size as learning progresses; we have called this reduction in size refinement (Seth and Edelman 2007). Furthermore, causal core size appeared to reach an asymptote after about 10 trials, which corresponds to the number of trials required for Darwin X to learn the task. Because neither context networks nor Granger networks showed similar refinement during learning, and because causal cores were not composed solely of the strongest synaptic interactions, causal core refinement in Darwin X can be understood as arising from the selection of particular causal pathways from a diverse and dynamic repertoire of neural interactions. This is consistent with the notion that synaptic plasticity underlies behavioral learning via modulation of causal networks at the population level, and not by strengthening or weakening associative links between neural representations of objects and actions. In this view, the comparatively large size of causal cores early in learning reflects a situation where selection has not yet acted to consolidate a causal pathway within a large space of possible pathways.
Fig. 8.
(a) Size of context networks as a function of trial number during learning, in terms of number of edges (K), for 15 different CA1 neuronal units. (b) Sizes of the corresponding Granger networks. (c) Sizes of the corresponding causal cores. Insets of panels (b) and (c) show the same data on a magnified scale. Reprinted with permission from Seth and Edelman (2007)
Summary
The analyses described above demonstrate the usefulness of a causal network perspective for probing the functional connectivity of neural circuits and for analyzing the principles governing causal interactions in large neural populations. By performing these analyses in detailed neurorobotic models we have generated both heuristics and hypotheses engaging with empirical data. Indeed it is at this level that interaction with empirical data is most plausible, whether via existing methods (e.g., by using multielectrode recordings to explore the increasing causal dominance of head-direction information seen in Darwin X during learning) or by leveraging cutting-edge and future techniques to look for causal cores (see section “Relation to neurobiological data”).
However, causal networks also offer new opportunities for developing theoretical perspectives on global aspects of brain function. In the following section we turn to the utility of a causal network perspective for characterizing the possible neural bases of consciousness.
Causal density and consciousness
A prominent theory of global brain function that has been applied to consciousness, the TNGS, proposes that consciousness is entailed by complex interactions among neural populations in the thalamocortical system, the so-called dynamic core (Edelman 2003; Edelman and Tononi 2000; Tononi and Edelman 1998; Seth and Baars 2005). This proposal raises the question: How can these complex interactions best be characterized quantitatively?
A rewarding approach is to consider phenomenology. A fundamental property of conscious scenes is that they are both differentiated (reflecting the discriminatory capability of consciousness; every conscious scene is one among a vast repertoire of different possible scenes) and integrated (reflecting the unity of conscious experience; every conscious scene is experienced “all of a piece”) (Edelman 2003; Edelman and Tononi 2000; Tononi and Edelman 1998). Therefore, a useful measure of complex neural interactions relevant to consciousness should reflect a balance between integration and differentiation in neural dynamics; this balance can be referred to as the relevant complexity of the system (Seth et al. 2006). Searching for neural dynamics of high relevant complexity marks an important departure from standard approaches seeking to isolate anatomical correlates of conscious experience (Rees et al. 2002). Unlike anatomical correlates, relevant complexity provides an explanatory link between specific aspects of conscious phenomenology and corresponding aspects of neural dynamics (Tononi and Edelman 1998).
As we have argued previously (Seth et al. 2006), a useful quantitative measure of relevant complexity should also reflect the fact that consciousness is a dynamic process (James 1904), and not a thing or a capacity; it should also take account of causal interactions within a neural system and between a neural system and its surroundings, i.e., bodies and environments. To be practically applicable, a useful measure should also be calculable for systems composed of large numbers of interacting elements.
Several measures of relevant complexity have now been proposed, including ‘neural complexity’ (CN) (Tononi et al. 1994), ‘information integration’ (Φ) (Tononi 2004), and, most recently, causal density (cd) (Seth 2005; Seth et al. 2006). All of these measures reflect in some way the balance between differentiation and integration within multivariate neural dynamics. In the present, we focus on causal density; a detailed comparative analysis of all three measures is provided in Seth et al. (2006).
Causal density
The causal density (cd) of a network’s dynamics measures the fraction of interactions among nodes that are causally significant (Seth 2005; Seth et al. 2006). cd is calculated as α/(n(n − 1)), where α is the total number of significant causal links observed, according to a multivariate Granger causality analysis, and n is the number of elements in the network; calculated this way, the value of cd will be bounded in the range [0,1]. It is also possible to calculate a ‘weighted’ version of cd which takes into account the varying contributions of each causally significant interaction; this version of cd is unbounded.
In terms of the criteria just described, cd naturally reflects a process because it is based on ongoing dynamics and it reflects causal interactions because it is based on an explicit statistical measure of causality. However, it is presently difficult to calculate for large systems because multivariate autoregressive models become difficult to estimate as the number of variables increases. Extended approaches based on Bayesian methods (Zellner 1971) may overcome this practical limitation (see section “Limitations and extensions”).
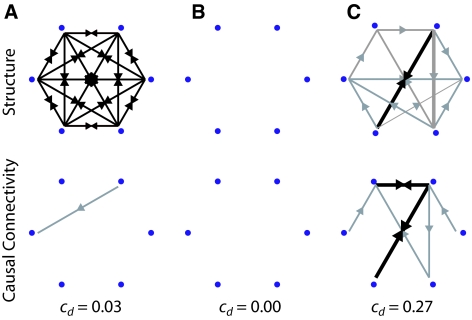
Does causal density capture aspects of relevant complexity? Figure 9 shows a comparison of structural connectivity and causal connectivity for three example networks, along with the corresponding values of cd. The dynamics for each network were generated using a mean-firing-rate neuronal model in which each node received an independent Gaussian noise input. While both a fully connected network (having near-identical dynamics at each node) and a fully disconnected network (having independent dynamics at each node) have low cd, a randomly connected network has a much higher value. These results support the notion that high values of cd indicate that elements in a system are both globally coordinated in their activity (in order to be useful for predicting each other’s activity) and at the same time dynamically distinct (reflecting the fact that different elements contribute in different ways to these predictions). High causal density is therefore consistent with high relevant complexity in that it reflects a dynamical balance between differentiation and integration.
Fig. 9.
Example simple networks (top row) and corresponding causal connectivity patterns (bottom row). (a) Fully connected network. (b) Fully disconnected network. (c) Randomly connected network. Grey arrows show unidirectional connections and black arrows show bidirectional connections. The width of each arrow (and size of arrowhead) reflect the magnitude of the causal interaction. Corresponding values of causal density (cd) are also given
It is important to recognize the limited role that a quantitative measure of neural dynamics can play within a scientific theory of consciousness. For example, at present it is not possible to measure the cd (or CN or Φ) exhibited by a human brain. Quantitative measures are therefore offered for their theoretical and conceptual utility which can be further elucidated by additional modeling and by application to simplified experimental preparations.
In addition, the relevant complexity of neural dynamics is likely to be multidimensional, involving spatial, temporal, and recursive aspects (Seth et al. 2006). Because phenomenal states appear to exhibit a balance between differentiation and integration along each of these dimensions, a satisfying measure of relevant complexity must be sensitive to these aspects of complexity in neural dynamics; indeed, the simultaneous application of multiple measures may be required. 4 Like other measures of relevant complexity (e.g., CN (Tononi et al. 1994) and Φ (Tononi 2004)), cd is best suited to measuring complexity in the spatial domain, although its basis in autoregressive modelling implies some integration over time.
Finally, some aspects of consciousness are likely to resist quantification altogether. Conscious scenes have many diverse features, several of which do not appear to be readily quantifiable (Edelman 2003; Seth et al. 2005, 2006). These features include subjectivity, the attribution of conscious experience to a self, and intentionality, which reflects the observation that consciousness is largely about events and objects. It is therefore clear that the quantitative characterization of relevant complexity can only constitute one aspect of a scientific theory of consciousness.
Summary
The concept of causal density cd provides a measure of dynamical complexity that reflects a balance between integration and differentiation in neural dynamics. This balance has previously been proposed to provide an explanatory link to conscious phenomenology (Edelman 2003; Tononi and Edelman 1998; Tononi 2004). cd overcomes some of the limitations of alternative measures described in the papers just cited: Unlike CN, cd incorporates causal interactions, and unlike Φ it provides a measure of process rather than capacity.
Discussion
The experiments reviewed in this paper have illustrated a variety of applications of causal network analysis. Section “Causal networks during target fixation” showed that causal networks can trace functional pathways connecting sensory input to motor output in a simple target fixation model. This model further showed how causal interactions are modulated by the network environment and verified that causal connectivity is a useful predictor of the behavioral consequences of network damage. Section “Causal networks in a brain-based device” extended the causal network approach to the analysis of a large-scale embodied model of the hippocampus and surrounding cortical areas, showing how causal networks were modified during learning of a spatial navigation task. This analysis found an increasing dominance of perforant pathway signals over trisynaptic signals, and of head-direction signals over those from other sensory modalities; these observations constitute hypotheses which can be tested in animal experiments. Section “Causal cores in neural populations” leveraged the power of large scale simulation modelling to develop the concept of a causal core which provides a mesoscale description of causal interactions. The causal core ‘refinement’ observed in Darwin X suggests an intriguing mesoscale connection between synaptic plasticity and behavioral learning, one which may be testable in future animal experiments using cutting-edge technologies (see section “Relation to neurobiological data”). Finally, section “Causal density and consciousness” broadened the perspective to speculate on possible causal interactions underlying conscious experience, arguing that a measure of causal connectivity—causal density—can connect global features of neural dynamics to corresponding features of conscious scenes.
Causal networks
The identification of causal networks in neural systems involves no assumptions about whether these systems ‘process information’ or operate according to ‘neural codes’. For example, causal cores are not information-bearing representations nor do they require explicit encoding and decoding. Instead, they are dynamic causally effective processes that give rise to specific outputs. This perspective can lead to different interpretations of commonly observed phenomena. For example, variability in neural activity (Knoblauch and Palm 2005) is often treated as an inconvenience and is minimized using averaging. From a population coding perspective, attempts have been made to identify whether variation is unavoidable neural noise, or is part of a signal transmitted via a neural code (Stein et al. 2005). According to the present view, however, such variability is expected and indicates the existence of diverse and dynamic repertoires of neuronal interactions underlying selection.
Causal networks can be interpreted in a variety of ways. One attractive view is that causal networks distinguish those aspects of neural dynamics that are driving (i.e., causal) from those aspects that are modulatory (i.e., non-causal). The distinction between driving and modulatory connections appears frequently in descriptions of neural activity (see, for example, Sherman and Guillery 2002; Friston 2005) where it is usually based on anatomical considerations (e.g., bottom-up versus top-down connections, input-to-output versus output-to-input) or on the differential involvement of particular neurotransmitters (e.g., NMDA-independent versus NMDA-dependent (Grossberg 1999)). Causal networks provide an alternative means of distinguishing driving from modulatory connections that does not depend on these assumptions, which is based instead on principled inference directly from dynamics.
It is important to emphasize that non-causal connections may still be significant for neural system operation. For example, one cannot expect Darwin X to behave normally after removing all parts of its simulated nervous system apart from a few causal cores. These ‘non-causal’ parts may not only be causal with respect to other NRs, they may play significant modulatory roles as well. Furthermore, causal network analysis may be able to differentiate a scale of possible influences ranging from ‘pure driving’ to ‘pure modulatory’ according to the relative magnitudes of causal influence.
Another interpretation of causal networks is that they are useful inasmuch as they allow inference of underlying unknown structural connectivity from neural dynamics (Makarov et al. 2005; Horwitz et al. 2005). The contrasting view taken here is that causal networks characterize dynamical interactions that result from interactions among network structure, embodiment, and behavior. Causal networks are rich representations of brain-body-environment interactions that should not be expected to recapitulate structural descriptions.
Relation to neurobiological data
Granger causality analysis has been used for several years to identify causal relations in neurobiological data. Many types of neurobiological data are suitable for Granger causality analysis, including local field potentials (Bernasconi and Konig 1999; Liang et al. 2000; Brovelli et al. 2004), spike trains (Kaminski et al. 2001), 5 electroencephalographic (EEG) and magnetoencephalographic (MEG) recordings (Hesse et al. 2003; Seth 2007), and functional magnetic resonance imaging (fMRI) data (Roebroeck et al. 2005; Valdes-Sosa et al. 2005). Even though these data sources provide incomplete representations of the underlying neurophysiology, informative causal networks can nevertheless be constructed.
Having said this, of the analyses described in this paper the causal core analysis imposes stricter requirements: it requires extensive knowledge of both anatomical and functional connectivity during behavior. At present, this data is fully available only in simulation and as such the causal core analysis is best considered as providing heuristics for interpreting neural dynamics. However, recent methodological advances suggest that investigators soon may be able to characterize both anatomical and functional architectures in biological systems at the required microscopic and mesoscopic scales. These advances include retrograde transneuronal transport of virus particles (Kelly and Strick 2004), metabolic markers for neuronal activity (Konkle and Bielajew 2004), and high-resolution optical imaging (Raffi and Siegel 2005). Causal networks provide both a general perspective and specific methods suited to making sense of the data generated by these new techniques. In particular, the causal core analysis and its future extensions will allow exploration of mesoscale dynamics, a level of description which has so far been neglected as compared to both the single neuron level and the systems level (Sporns et al. 2005; Lin et al. 2006).
Limitations and extensions
Linearity
Because it is based on linear regression Granger causality can only give information about linear features of signals. Extensions to nonlinear cases now exist: In the approach of (Friewald et al. 1999) the globally nonlinear data is divided into a locally linear neighborhoods (see also Chen et al. 2004), whereas (Ancona et al. 2004) use a radial basis function method to perform a global nonlinear regression. However, these extensions are more difficult to use in practice and their statistical properties are less well understood. In the analysis of neurophysiological signals simple, linear methods can be tried first before moving on to more complicated alternatives.
Stationarity
Application of Granger causality assumes that the analyzed signals are covariance stationary, i.e., that the mean and variance of each signal does not change over time. Non-stationary data are commonly treated, as in this paper, by first-order differencing. Higher-order differencing can be applied if first-order differenced series remain non-covariance-stationary, but interpretation of the resulting causal networks becomes more difficult when the data are repeatedly transformed.
Alternative methods for ensuring covariance stationarity include windowing across relatively short data segments (Hesse et al. 2003), assuming that sufficiently short windows of a non-stationary signal are locally stationary. A related approach takes advantage of the trial-by-trial nature of many neurophysiological experiments (Ding et al. 2000). In this approach, time series from different trials are treated as separate realizations of a non-stationary stochastic process with locally stationary segments.
Dependence on observed variables
All dynamical methods for inferring causal networks depend on the appropriate selection of variables. In the case of Granger causality, causal factors that are not incorporated into the regression model (latent variables) obviously cannot be represented in the output. Furthermore, omission of variables can lead to misidentification of causal links between variables that are included in the analysis leading to ‘common-input’ artifacts (see section “Granger causality”). These considerations emphasize the statistical nature of Granger causality: Granger causality networks should not be interpreted as directly reflecting physical causal chains. A useful discussion of the important problem of latent variables is provided in Eichler (2005) and a new approach based on adapting methods from partial coherence may also help ameliorate this problem.6
Scaling to large networks
Multivariate Granger causality analyses of large systems face the problem that the number of parameters estimated grows as n2 where n is the number of variables in the system (Eq. 1). If the amount of data available also grows exponentially with the number of variables then this poses a severe computational bottleneck but the resulting coefficient matrices, and the subsequent causal networks, will still be reliable. Normally, however, the amount of data grows only linearly with the number of variables, in which case the regression models can be underdetermined by the data and the resulting causal networks can be unreliable. One approach to overcoming this problem is to use Bayesian methods to place limits (priors) on certain coefficients, based on existing knowledge of the system (Zellner 1971). A Bayesian approach provides a middle way between a purely data-driven approach in which no assumptions are made about possible causal relations, and a model-driven approach in which a priori hypotheses about causal networks are tested for their fit to the data.
Relation to other causal methods
The Granger causality method for identifying causal networks can be contrasted with alternative techniques which divide into two general classes. In the first, causal networks are inferred from data using techniques similar to Granger causality but which have different properties and which make different assumptions about the data. In the second, causal networks are inferred by repeatedly perturbing or lesioning the studied system.
There are several alternatives to Granger causality which can be used to identify causal networks which lie on a scale from data-driven to model-driven. Among data-driven methods, transfer entropy (Schreiber 2000) is an asymmetric version of mutual information which has the advantage of responding to both linear and nonlinear interactions, but which has the disadvantage of applying to multivariate situations only with difficulty. Sporns and Lungarella (2006) have used transfer entropy to show how information flow in sensorimotor networks is affected by behavior, learning, embodiment, and body morphology, in a variety of simulation and robotic experiments. For a useful comparative analysis of transfer entropy, Granger causality, and other data-driven methods see Lungarella et al. (2007).
Among model-driven methods, structural equation modeling (SEM) has gained prominence within neuroscience (McIntosh and Gonzalez-Lima 1994). In SEM, connections between brain areas are based on known neuroanatomy, and the interregional activity covariances are used to calculate magnitudes of the influence of each directional path. A recent extension of SEM, dynamic causal modeling, is targeted towards fMRI data in particular by employing a plausible generative model of the fMRI signal (Friston et al. 2003). Lastly, the ‘dynamic Bayesian network’ method (Smith et al. 2006) provides a compromise between model-driven and data-driven methods by including an optimization procedure in which the tested model is progressively refined according to its fit to the data.
The second class of causal inference methods involves perturbation or lesioning the studied system (Pearl 1999). For example, Tononi and Sporns (2003) stimulate selected subsets of a network with Gaussian noise and interpret the resulting mutual information between the subset and the rest of the network as a measure of causal influence. Similarly, Keinan et al. (2004) assess causal influence by measuring network performance following selective lesions to subsets of elements. These interventionist approaches are in principle robust to artifacts induced by common input and thus allow identification of physical causal chains (Timme 2007). However, their practical use is limited to situations in which networks can be repeatedly and reversibly perturbed, which for biological neural systems is rarely the case. Furthermore, the interpretation of causal inference based on lesions of perturbations is complicated by the fact that the studied system is either no longer intact (for lesions) or may display different behavior (for perturbations).
Conclusions
Current theories of embodied cognition stress that adaptive behavior results from the continuous interplay of causal influences among brain, body, and environment (Clark 1997). Quantitative development of these theories requires a framework that restores balance among these factors. One way to achieve this balance is to treat neural systems not as information processing devices, nor as devices that encode and decode stimulus representations, but as causal networks that are embedded in larger networks of causal factors reflecting morphological and environmental constraints. This framework can be articulated quantitatively using a combination of Granger causality and graph theory. The present results have demonstrated how such a quantitative framework can shed new light on many areas of embodied cognitive science including (i) the emergence of adaptive responses in sensorimotor networks, (ii) how causal pathways shift during learning in a neurobiologically plausible architecture, (iii) the relations among synaptic plasticity, mesoscale causal interactions, and behavioral learning, and (iv) possible dynamical correlates of conscious experience. In addition, a causal network perspective could provide useful design heuristics to guide the development of artificially intelligent and perhaps even artificially conscious devices.
Acknowledgment
The research described in this paper was largely carried out while the author was at The Neurosciences Institute, La Jolla, California, with support from the Neurosciences Research Foundation. Many thanks to my colleagues there and in particular to Jeffrey L. Krichmar and Gerald M. Edelman who were instrumental in enabling this work. I am also grateful for the comments of two anonymous reviewers.
Footnotes
Many applications in neurophysiology make use of a frequency-domain version of Granger causality (Geweke 1982; Kaminski et al. 2001). However, because in this paper we analyze simulation models without oscillatory dynamics, we remain in the (simpler) time domain.
The fitness function was F = tfix + 0.25(35 − d̄), where tfix denotes the proportion of time for which the target was fixated and d̄ the mean offset between H and G (the environment was a toroidal square plane with side length 100).
The analyzed time series varied in length from 450 to 4,994 time-steps. Robustness to different lengths was assessed by reanalyzing causal interactions after dividing each time series into two parts; results were qualitatively identical (see (Seth and Edelman 2007) for details).
Recursive complexity refers to the balance between differentiation and integration across different levels of description. The phenomenal structure of consciousness appears to be recursive inasmuch as individual features of conscious scenes are themselves Gestalts which share organizational properties with the conscious scene as a whole.
Because Granger causality is based on linear regression it assumes a continuous signal, but neural systems at the level of spikes are discontinuous. A straightforward adaptation of the technique is to convolve spikes with a continuous function (e.g., a half-Gaussian) in order to generate a continuous signal. A more principled but more complex alternative is to substitute linear regression modelling with a point-process prediction algorithm (Okatan et al. 2005; Nykamp 2007).
J. Feng, personal communication.
References
- Ancona N, Marinazzo D, Stramaglia S (2004) Radial basis function approaches to nonlinear granger causality of time series. Phys Rev E 70:056221 [DOI] [PubMed]
- Bernasconi C, Konig P (1999) On the directionality of cortical interactions studied by structural analysis of electrophysiological recordings. Biol Cybern 81:199–210 [DOI] [PubMed]
- Bienenstock EL, Cooper LN, Munro PW (1982) Theory for the development of neuron selectivity: orientation specificit and binocular interaction in the visual cortex. J Neurosci 2(1):32–48 [DOI] [PMC free article] [PubMed]
- Brovelli A, Ding M, Ledberg A, Chen Y, Nakamura R, Bressler S (2004) Beta oscillations in a large-scale sensorimotor cortical network: directional influences revealed by Granger causality. Proc Natl Acad Sci USA 101(26):9849–9854 [DOI] [PMC free article] [PubMed]
- Chen Y, Rangarajan G, Feng J, Ding M (2004) Analyzing multiple nonlinear time series with extended Granger causality. Phys Lett A 324:26–35 [DOI]
- Churchland P, Sejnowski T (1994) The computational brain. MIT Press, Cambridge, MA
- Clark A (1997) Being there: putting brain, body, and world together again. MIT Press, Cambridge, MA
- deCharms RC, Zador A (2000) Neural representation and the cortical code. Annu Rev Neurosci 23:613–647 [DOI] [PubMed]
- Ding M, Bressler S, Yang W, Liang H (2000) Short-window spectral analysis of cortical event-related potentials by adaptive multivariate autoregressive modeling: data prepocessing, model validation, and variability assessment. Biol Cybern 83:35–45 [DOI] [PubMed]
- Ding M, Chen Y, Bressler S (2006) Granger causality: basic theory and application to neuroscience. In: Schelter S, Winterhalder M, Timmer J (eds) Handbook of time series analysis. Wiley, Wienheim, pp 438–460
- Dityatev AE, Bolshakov VY (2005) Amygdala, long-term potentiation, and fear conditioning. Neuroscientist 11:75–88 [DOI] [PubMed]
- Drew PJ, Abbott LF (2006) Extending the effects of spike-timing-dependent plasticity to behavioral timescales. Proc Natl Acad Sci USA 103(23):8876–8881 [DOI] [PMC free article] [PubMed]
- Edelman GM (1987) Neural Darwinism. Basic Books, New York
- Edelman GM (1993) Selection and reentrant signaling in higher brain function. Neuron 10:115–125 [DOI] [PubMed]
- Edelman GM (2003) Naturalizing consciousness: a theoretical framework. Proc Natl Acad Sci USA 100(9):5520–5524 [DOI] [PMC free article] [PubMed]
- Edelman GM, Tononi G (2000) A universe of consciousness: how matter becomes imagination. Basic Books, New York
- Eichler M (2005) A graphical approach for evaluating effective connectivity in neural systems. Philos Trans R Soc B 360:953–967 [DOI] [PMC free article] [PubMed]
- Friewald WA, Valdes P, Bosch J, Biscay R, Jimenez JC, Rodriguez LM, Rodriguez V, Kreiter AK, Singer W (1999) The use of time-variant EEG Granger causality for inspecting directed interdependencies of neural assemblies. J Neurosci Methods 94:105–119 [DOI] [PubMed]
- Friston K (1994) Functional and effective connectivity in neuroimaging: a synthesis. Hum Brain Mapp 2:56–78 [DOI]
- Friston K (2005) A theory of cortical responses. Philos Trans R Soc Lond B Biol Sci 360:815–836 [DOI] [PMC free article] [PubMed]
- Friston K, Harrison L, Penny W (2003) Dynamic causal modeling. Neuroimage 19(4):1273–1302 [DOI] [PubMed]
- Geweke J (1982) Measurement of linear dependence and feedback between multiple time series. J Am Stat Assoc 77:304–313 [DOI]
- Granger CWJ (1969) Investigating causal relations by econometric models and cross-spectral methods. Econometrica 37:424–438 [DOI]
- Grossberg S (1999) The link between brain learning, attention, and consciousness. Conscious Cogn 8:1–44 [DOI] [PubMed]
- Hamilton JD (1994) Time series analysis. Princeton University Press, Princeton, NJ
- Hesse W, Möller E, Arnold M, Schack B (2003) The use of time-variant EEG Granger causality for inspecting directed interdependencies of neural assemblies. J Neurosc Methods 124:27–44 [DOI] [PubMed]
- Horwitz B, Warner B, Fitzer J, Tagamets M, Husain F, Long T (2005) Investigating the neural basis for functional and effective connectivity. Application to fmri. Philos Trans R Soc Lond B Biol Sci 360:1093–1108 [DOI] [PMC free article] [PubMed]
- James W (1904) Does consciousness exist? J Philos Pyschol Sci Methods 1:477–491 [DOI]
- Kaminski M, Ding M, Truccolo WA, Bressler SL (2001) Evaluating causal relations in neural systems: Granger causality, directed transfer function and statistical assessment of significance. Biol Cybern 85:145–157 [DOI] [PubMed]
- Keinan A, Sandbank B, Hilgetag CC, Meilijson I, Ruppin E (2004) Fair attribution of functional contribution in artificial and biological networks. Neural Comput 16:1887–1915 [DOI] [PubMed]
- Kelly RM, Strick PL (2004) Macro-architecture of basal ganglia loops with the cerebral cortex: use of rabies virus to reveal multisynaptic circuits. Prog Brain Res 143:449–459 [DOI] [PubMed]
- Knoblauch A, Palm G (2005) What is signal and what is noise in the brain? Biosystems 79(1–3):83–90 [DOI] [PubMed]
- Konkle AT, Bielajew C (2004) Tracing the neuroanatomical profiles of reward pathways with markers of neuronal activation. Rev Neurosci 15(6):383–414 [DOI] [PubMed]
- Krichmar JL, Edelman GM (2002) Machine psychology: autonomous behavior, perceptual categorization and conditioning in a brain-based device. Cereb Cortex 12(8):818–830 [DOI] [PubMed]
- Krichmar JL, Nitz DA, Gally JA, Edelman GM (2005a) Characterizing functional hippocampal pathways in a brain-based device as it solves a spatial memory task. Proc Natl Acad Sci USA 102(6):2111–2116 [DOI] [PMC free article] [PubMed]
- Krichmar JL, Seth AK, Nitz DA, Fleischer JG, Edelman GM (2005b) Spatial navigation and causal analysis in a brain-based device modeling cortical-hippocampal interactions. Neuroinformatics 3(3):197–222 [DOI] [PubMed]
- Lavenex P, Amaral D (2000) Hippocampal-neocortical interaction: a hierarchy of associativity. Hippocampus 10:420–430 [DOI] [PubMed]
- Liang H, Ding M, Nakamura R, Bressler SL (2000) Causal influences in primate cerebral cortex during visual pattern discrimination. Neuroreport 11(13):2875–2880 [DOI] [PubMed]
- Lin L, Osan R, Tsien J (2006) Organizing principles of real-time memory encoding: neural clique assemblies and universal neural codes. Trends Neurosci 29(1):48–57 [DOI] [PubMed]
- Lungarella M, Ishiguro K, Kuniyoshi Y, Otsu N (2007) Methods for quantifying the causal structure of bivariate time series. Int J Bifurcat Chaos 17(3):903–921 [DOI]
- Makarov V, Panetsos F, de Feo O (2005) A method for determining neural connectivity and inferring the underlying neural dynamics using extracellular spike recordings. J Neurosci Methods 144:265–279 [DOI] [PubMed]
- Malenka RC, Bear M (2004) LTP and LTD: an embarrasment of riches. Neuron 44:5–21 [DOI] [PubMed]
- McIntosh AR, Gonzalez-Lima F (1994) Structural equation modeling and its application to network analysis in functional brain imaging. Hum Brain Mapp 2:2–22 [DOI]
- Morris RGM (1984) Developments of a water-maze procedure for studying spatial learning in the rat. J Neurosci Methods 11:47–60 [DOI] [PubMed]
- Nykamp D (2007) A mathematical framework for inferring connectivity in probabilistic neuronal networks. Math Biosci 205(2):204–251 [DOI] [PubMed]
- Okatan M, Wilson MA, Brown EN (2005) Analyzing functional connectivity using a network likelihood model of ensemble neural spiking activity. Neural Comput 17(9):1927–1961 [DOI] [PubMed]
- Pearl J (1999) Causality: models, reasoning, and inference. Cambridge University Press, Cambridge, UK
- Raffi M, Siegel RM (2005) Functional architecture of spatial attention in the parietal cortex of the behaving monkey. J Neurosci 25:5171–5186 [DOI] [PMC free article] [PubMed]
- Rees G, Kreiman G, Koch C (2002) Neural correlates of consciousness in humans. Nat Rev Neurosci 3(4):261–270 [DOI] [PubMed]
- Roebroeck A, Formisano E, Goebel R (2005) Mapping directed influence over the brain using granger causality and fmri. Neuroimage 25(1):230–242 [DOI] [PubMed]
- Schreiber T (2000) Measuring information transfer. Phys Rev Lett 85(2):461–464 [DOI] [PubMed]
- Schwartz G (1978) Estimating the dimension of a model. Ann Stat 5(2):461–464 [DOI]
- Seth AK (2005) Causal connectivity of evolved neural networks during behavior. Network Comput Neural Syst 16:35–54 [DOI] [PubMed]
- Seth AK (2007) Granger causality. Scholarpedia, page 15501
- Seth AK (2007) Granger causality analysis of MEG signals during a working memory task. Abst Soc Neurosci
- Seth AK, Baars BJ (2005) Neural Darwinism and consciousness. Conscious Cogn 14:140–168 [DOI] [PubMed]
- Seth AK, Baars BJ, Edelman DB (2005) Criteria for consciousness in humans and other mammals. Conscious Cogn 14(1):119–139 [DOI] [PubMed]
- Seth AK, Edelman GM (2004) Environment and behavior influence the complexity of evolved neural networks. Adapt Behav 12:5–21 [DOI]
- Seth AK, Edelman GM (2007) Distinguishing causal interactions in neural populations. Neural Comput 19:910–933 [DOI] [PubMed]
- Seth AK, Izhikevich E, Reeke GN, Edelman GM (2006) Theories and measures of consciousness: an extended framework. Proc Natl Acad Sci USA 103(28):10799–10804 [DOI] [PMC free article] [PubMed]
- Sherman M, Guillery R (2002) The role of the thalamus in the flow of information to the cortex. Philos Trans R Soc B Biol Sci 357:1695–1708 [DOI] [PMC free article] [PubMed]
- Smith V, Yu J, Smulders T, Hartemink A, Jarvis E (2006) Computational inference of neural information flow networks. PLoS Comput Biol 2(11):e161 [DOI] [PMC free article] [PubMed]
- Sporns O, Lungarella M (2006) Information flow in sensorimotor networks. PLoS Comput Biol 2(10):e144 [DOI] [PMC free article] [PubMed]
- Sporns O, Tononi G, Kotter R (2005) The human connectome: a structural description of the human brain. PLoS Comput Biol 1(4):e42 [DOI] [PMC free article] [PubMed]
- Stein RB, Gossen ER, Jones KE (2005) Neuronal variability: noise or part of the signal? Nat Rev Neurosci 6(5):389–397 [DOI] [PubMed]
- Sutton R, Barto A (1998) Reinforcement learning. MIT Press, Cambridge, MA
- Timme M (2007) Revealing network connectivity from response dynamics. Phys Rev Lett 98:224101 [DOI] [PubMed]
- Tononi G (2004) An information integration theory of consciousness. BMC Neurosci 5(1):42 [DOI] [PMC free article] [PubMed]
- Tononi G, Edelman GM (1998) Consciousness and complexity. Science 282:1846–1851 [DOI] [PubMed]
- Tononi G, Sporns O (2003) Measuring information integration. BMC Neurosci 4(1):31 [DOI] [PMC free article] [PubMed]
- Tononi G, Sporns O, Edelman GM (1994) A measure for brain complexity: relating functional segregation and integration in the nervous system. Proc Natl Acad Sci USA 91:5033–5037 [DOI] [PMC free article] [PubMed]
- Valdes-Sosa P, Sanchez-Bornot J, Lage-Castellanos A, Vega-Hernandez M, Bosch-Bayard J, Melie-Garcia L, Canales-Rodriguez E (2005) Estimating brain functional connectivity with sparse multivariate autoregression. Philos Trans R Soc Lond B Biol Sci 360:969–981 [DOI] [PMC free article] [PubMed]
- Zellner A (1971) An introduction to Bayesian inference in econometrics. Wiley, New York