Abstract
The rice blast fungus, Magnaporthe grisea, generates enormous turgor pressure within a specialized cell called the appressorium to breach the surface of host plant cells. Here, we show that a mitogen-activated protein kinase, Mps1, is essential for appressorium penetration. Mps1 is 85% similar to yeast Slt2 mitogen-activated protein kinase and can rescue the thermosensitive growth of slt2 null mutants. The mps1–1Δ mutants of M. grisea have some phenotypes in common with slt2 mutants of yeast, including sensitivity to cell-wall-digesting enzymes, but display additional phenotypes, including reduced sporulation and fertility. Interestingly, mps1–1Δ mutants are completely nonpathogenic because of the inability of appressoria to penetrate plant cell surfaces, suggesting that penetration requires remodeling of the appressorium wall through an Mps1-dependent signaling pathway. Although mps1–1Δ mutants are unable to cause disease, they are able to trigger early plant-cell defense responses, including the accumulation of autofluorescent compounds and the rearrangement of the actin cytoskeleton. We conclude that MPS1 is essential for pathogen penetration; however, penetration is not required for induction of some plant defense responses.
The plant cell wall provides such an effective barrier that many microorganisms can infect plants only through wounds or natural openings in the wall (1). A variety of fungal pathogens have evolved a complex morphogenetic program to sense specific components and properties of the plant cell surface and to differentiate specialized infectious cells, called appressoria, for penetrating the plant cell wall (2). Appressoria are produced by a wide range of taxonomically diverse fungal and nonfungal species (e.g., Oomycetes), suggesting that appressorium differentiation is a widespread microbial adaptation that assists in plant cell-wall penetration.
The rice blast fungus, Magnaporthe grisea, infects most of the economically important cereal crops, particularly rice (3). Dispersed fungal spores (conidia) attach tightly to the plant surface, and under high moisture conditions, they germinate and differentiate a dome-shaped appressorium (4–6). The appressorium attaches tightly to the plant surface, and over several hours, turgor pressures within the appressorium reach 3–8 MPa, the highest measured turgor pressures for any cell (7, 8). The fact that appressoria are capable of denting and penetrating inert hydrophobic surfaces (4) suggests that turgor pressure is the major force used to drive a thin penetration hypha through the plant cell wall.
Turgor within the M. grisea appressorium is generated through a dramatic rise in intracellular glycerol levels (9). The efflux of glycerol from within the appressorium is prevented partially by a wall layer rich in polyketide-derived melanin. Mutations or chemicals that block the formation of the melanin wall layer allow the rapid efflux of glycerol, impair the maintenance of appressorium turgor, and prevent penetration (7, 9). Other cell-wall or membrane components may also be important for penetration. Ultrastructural studies reveal that several additional wall layers are laid down during appressorium formation, and these may be necessary to maintain cell-wall integrity under conditions of membrane stress and high turgor pressure (5). How turgor is directed to the highly localized site of penetration-hypha formation is not currently known. Actin is localized to the site of penetration-hypha formation, suggesting that polarization of the cytoskeleton may be involved in localized wall modification (10).
In Saccharomyces cerevisiae, the control of cell-wall growth in response to membrane stress is mediated by a protein kinase C pathway that activates the mitogen-activated protein kinase (MAPK), Slt2/Mpk1 (11–13). The Δslt2 mutants display defects in cell-wall integrity, including sensitivity to cell-wall-degrading enzymes and an inability to grow at elevated temperatures in the absence of osmotic stabilizers. The homologous MAPK in Schizosaccharomyces pombe (14, 15) also plays a role in the maintenance of cell-wall integrity; however, the function of related kinases in filamentous fungi are currently unknown. Here, we show that M. grisea has a MAPK designated Mps1 that is related functionally to the yeast Slt2 kinase. We show that MPS1 is essential for the maintenance of cell-wall integrity during specific phases of the life cycle and, most importantly, for turgor-driven penetration of plant cells. Unexpectedly, although the mps1–1Δ mutant strains are nonpathogenic and fail to penetrate the cell wall, they still activate plant defense responses.
MATERIALS AND METHODS
Strains, Growth Conditions, and Pathogenicity Assays.
Growth, mating assays, transformation, and nucleic-acid extraction from M. grisea were performed as described (16–18). Infection assays on rice, epidermal-cell penetration assays on onions, germination assays, and appressorium-formation assays were performed as described (18, 19). S. cerevisiae strains were cultured on 1% yeast extract/1% peptone, supplemented with either 1% galactose/1% raffinose or 1% dextrose.
Cloning of MPS1.
Procedures for amplifying and cloning MAPK-related genes from M. grisea are described in ref. 18. Amplified DNA fragments were used to screen cDNA and genomic libraries. Automated fluorescent DNA sequencing was performed on an ALF-Express DNA sequencer (Pharmacia). DNA-protein sequence database searches were performed with the blast programs (20). Amino-acid-sequence comparisons and alignments were made with the bestfit, pileup, and boxshade programs from the Genetics Computer Group’s software package (Madison, WI).
Yeast Complementation.
The MPS1 ORF was amplified with primers M3E5 (GGAATTCTAATGTCGGATCTCCAGGGCC) and M3S3 (GCCGAGCTCTCACCTCCTCTGATCCAAA), and subcloned between EcoRI and SacI sites in the yeast expression vector pYES2 (Invitrogen). This construct was then transformed into yeast strain DL456 (MATa Δslt2 ura3-52). Ura+ transformants were tested for the ability to grow at 37°C.
MPS1 Gene Replacement.
A 1.4-kb HindIII fragment extending upstream of MPS1 was end-filled with DNA polymerase I and cloned into the SmaI site on pBluescript II KS(+) as pM3L7. The 2.4-kb XhoI–HindIII fragment (downstream of MPS1) from pM3R24 was cloned into the XhoI–HindIII sites on pM3L7. The hygromycin phosphotransferase gene was then cloned into the EcoRI site to create the mps1–1Δ gene-replacement vector pM3H22.
Microscopy.
Onion epidermal peels were isolated, and conidia were allowed to germinate on the adaxial surface (19). After 48 h in the dark, epidermal peels were fixed for 1 h with 500 μM 3-maleimidobenzoic acid N-hydroxysuccinimide ester (Sigma) prepared in PBS, 0.1% Triton X-100, and 0.5% dimethyl sulfoxide. Peels were rinsed of fixative by three 10-min washes with PBS, and then filamentous-actin stained for 45 min with 0.33 μM rhodamine-phalloidin (Molecular Probes) in PBS plus 5% methanol. After two 10-min PBS washes, the peels were mounted in SlowFade reagent (Molecular Probes) and visualized by fluorescence microscopy.
Cell-Wall Integrity Assays.
M. grisea cells at various stages of germination and appressorium formation were incubated in 1 M sorbitol with 5 mg/ml Novozyme 234 (InterSpex Products, Foster City, CA). Spheroplast release and cell-wall degradation were examined with differential-interference-contrast microscopy after incubation for various periods at 30°C.
RESULTS
Mps1 Is Functionally Related to the Yeast Slt2 MAPK.
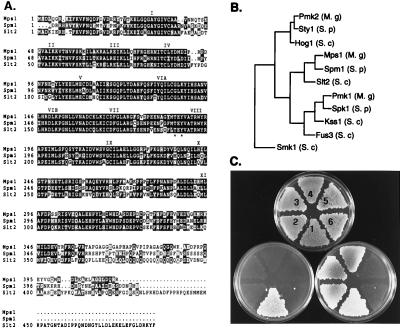
We identified sequences from the rice blast fungus that are related to yeast MAPKs (ref. 18; see Materials and Methods). The cDNA and genomic sequence of MPS1 identified an ORF encoding a 415-aa peptide (Fig. 1A) that is 80% similar and 64% identical to S. cerevisiae Slt2/Mpk1 and 77% similar and 65% identical to S. pombe MAPK Spm1/Pmk1 (14, 15). Phylogenetic analysis indicated that Mps1 clustered with kinases related to Slt2/Mpk1 and was less related to S. cerevisiae Hog1 or Fus3/Kss1 MAPKs (Fig. 1B). To test whether MPS1 could function in place of SLT2, the MPS1 ORF was cloned into a galactose-inducible yeast expression vector and transformed into yeast strain DL456 (MATa Δslt2 ura3-52). Ura+ transformants were tested for the ability to grow at 37°C with either glucose or galactose as a carbon source (Fig. 1C). MPS1 complemented the Δslt2 autolysis defect at 37°C (Fig. 1C). This complementation was specific for MPS1, as another MAPK gene from M. grisea, PMK1, failed to complement the Δslt2 defect (data not shown).
Figure 1.
Structure, function, and relatedness of MPS1. (A) Sequence alignment of Mps1 with S. cerevisiae Slt2 and S. pombe Spm1. Identical residues are shown in black boxes and similar residues are shaded. The 11 protein-kinase subdomains are labeled with roman numerals. (B) Relatedness of Mpk1 to yeast MAPKs. GenBank accession nos. used for yeast MAPKs are StyI (X89262), Hog1 (L06279), Spm1 (U65405), Slt2 (X59262), Spk1 (X57334), Kss1 (M26398), Fus3 (M31132), and Smk1 (L35047). Accession nos. for M. grisea MAPKs are Mps1 (AF020316) and Pmk1 (U70134). A third MAPK from M. grisea, Pmk2, is most closely related to yeast Hog1 and StyI kinases (J.-R.X. and N. Talbot, unpublished work). The phylogram was prepared with the growtree program from the Genetics Computer Group’s software package (Madison, WI). (C) Complementation of a yeast slt2 mutant. S. cerevisiae strains numbered 2–6 are DL456 (slt2Δ, ura3-52); strain 1 is CG219 (ura3–52 SLT2); strains numbered 2, 3, and 4 contain the plasmid pNX9 (MPS1 controlled by the GAL1 promoter in pYES2); strains 5 and 6 contain the vector pYES2. The upper plate (1% yeast extract/1% peptone/1% dextrose) was incubated at 30°C and replicated to 1% yeast extract/1% peptone/1% galactose/1% raffinose (lower right) or 1% yeast extract/1% peptone/1% dextrose (lower left) plates that were incubated at 37°C before being photographed.
MPS1 Is Required for Efficient Sporulation, Cell-Wall Integrity, and Female Fertility.
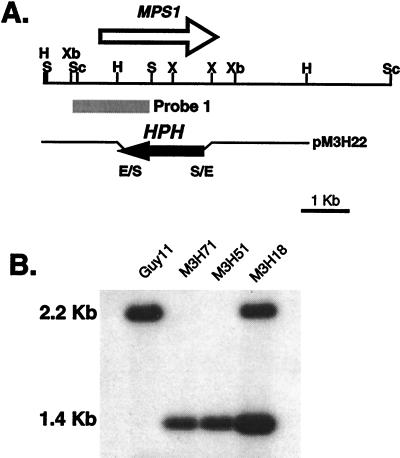
The MPS1 gene-replacement vector pM3H22 (Fig. 2A) was constructed by replacing the HindIII–XhoI fragment, containing the majority of the MPS1 coding region, with the hygromycin phosphotransferase gene, HPH. This construct was used to transform wild-type strain Guy11. Strains containing the MPS1 deletion (mps1–1Δ) were characterized by Southern blot analysis (Fig. 2B). Guy11 and transformant M3H18 contained the MPS1 2.2-kb SalI fragment. M3H18 also contained a 1.4-kb SalI fragment from an ectopic insertion event. Transformants M3H51 and M3H71 do not contain the 2.2-kb SalI fragment, but have the 1.4-kb fragment indicative of the gene-replacement event (Fig. 2B). The mps1–1Δ mutants were crossed with wild-type strain 2359 (MAT1-1; ref. 21). Progeny that segregated for the mps1–1Δ allele (n = 34: 15 mps1–1Δ, hygromycin-resistant; 19 MPS1, hygromycin-sensitive) were isolated and used to confirm cosegregation of all mps1–1Δ phenotypes (described below).
Figure 2.
MPS1 gene-replacement vector and transformants. (A) Physical map of the MPS1 genomic region and the gene-replacement vector pM3H22. The restriction enzymes are E, EcoRI; H, HindIII; S, SalI; Sc, SacI; X, XhoI; Xb, XbaI. (B) Southern blot of wild-type strain Guy11, ectopic-integration transformant M3H18, and MPS1 gene-replacement transformants M3H51 and M3H71. All DNA samples were digested with SalI. The blot was probed with the 1.6-kb SacI–SalI fragment. Guy11 and transformant M3H18 contained the MPS1 2.2-kb SalI fragment. M3H18 also contained a 1.4-kb SalI fragment, indicative of an ectopic insertion event. Transformants M3H51 and M3H71 do not contain the 2.2-kb SalI fragment but have the 1.4-kb SalI fragment, indicative of a gene replacement.
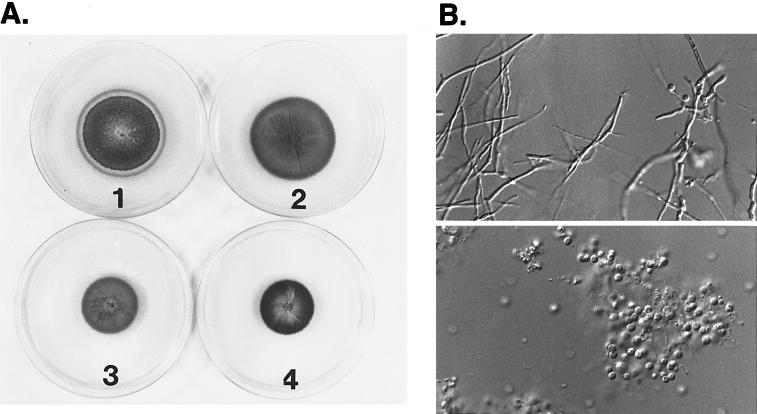
The mps1–1Δ mutants have radial growth rates on plate media similar to those of wild-type strains (Fig. 3A). Mycelial dry-weight determinations of mps1–1Δ mutant and wild-type strains grown in liquid complete media also show no difference (data not shown). However, mps1–1Δ mutants were reduced dramatically in aerial hyphal growth and conidiation (Fig. 3A). Wild-type strains produced an average of 107 conidia per plate, whereas mps1–1Δ mutants produced <4,000 conidia per plate. The reduction in conidiation was caused by the limited ability of mps1–1Δ strains to produce aerial hyphae. Conidia produced by mps1–1Δ strains have normal morphology and germination rates (data not shown).
Figure 3.
mps1-1Δ mutants have defects in cell-wall integrity. (A) Growth of mps1–1Δ strains on complete media without 1 M sorbitol (plate 1); growth of MPS1 strains on complete media without 1 M sorbitol (plate 2); growth of mps1–1Δ strains on complete media with 1 M sorbitol (plate 3); and growth of MPS1 strains on complete media with 1 M sorbitol (plate 4). mps1–1Δ mutants sporulate poorly and undergo progressive autolysis in the absence of osmotic stabilization. Radial growth rates are identical to those of wild-type strains. (B) Sensitivity of mps1–1Δ strains to cell-wall-digesting enzymes. Wild-type (Upper) or mps1–1Δ (Lower) strains were photographed 15 min after treatment with Novozyme 234. The mps1–1Δ mutants are reduced to spheroplasts with very short enzyme treatment. (×230.)
mps1–1Δ colonies underwent progressive autolysis beginning at the central part of the colony and radiating outwards (Fig. 3A). This novel growth-related phenotype was suppressed by the addition of 1 M sorbitol. mps1–1Δ mutants are also hypersensitive to the fungal cell-wall-digesting enzyme Novozyme 234 (Fig. 3B). Enzyme digestion of mycelia from mps1–1Δ strains released spheroplasts within 5 min and resulted in the dissolution of all intact hyphae after 30 min at 30°C (Fig. 3B). These findings suggest that MPS1 has a role similar to that of the SLT2 gene in S. cerevisiae and is required to maintain cell-wall integrity in M. grisea. However, unlike Δslt2 mutants, mps1–1Δ mutants are not temperature-sensitive for growth, and unlike S. pombe spm1−/pmk1− mutants (15), they are not sensitive to growth in medium containing either 1 M sorbitol, 1 M NaCl, 1 M MgCl2, or 1.4 M KCl.
A novel phenotype was observed during genetic analysis of strains carrying the mps1–1Δ allele. The mps1–1Δ strains and segregants mated only as males. As female sterility is encountered commonly in cultures of M. grisea, the 1.7-kb SacI fragment containing the MPS1 gene was reintroduced at an ectopic site in the mps1–1Δ strain M3H51. All cotransformed strains (n = 20) were rescued for the mps1–1Δ phenotypes and were restored for female fertility.
MPS1 Is Required for Pathogenicity.
Infection assays performed on rice seedlings showed that mps1–1Δ mutants were completely nonpathogenic (Fig. 4). However, when mps1–1Δ conidia are sprayed onto an abraded leaf surface (not shown) or injected into the leaf sheath, they produce small lesions (Fig. 4). The mps1–1Δ mutants could be recovered from these lesions (data not shown). These results show that MPS1 is essential for plant penetration but not necessarily for invasive growth in rice plants.
Figure 4.
Infection assays. (Upper) Typical infected leaf areas from 14-day-old rice seedlings spray-inoculated with either wild-type (WT) or mps1–1Δ (mps1) strains. (Lower) Typical infected areas from rice leaves inoculated by leaf sheath injection of either wild-type (WT) or mps1–1Δ (mps1) strains. A wounded control (CON) inoculated with water is also shown. Small areas of fungal growth occur near the injection site in mps1–1Δ strains.
The kinetics of conidial germination, germ tube differentiation, and appressorium formation in mps1–1Δ mutants were indistinguishable from those of isogenic wild-type strains (data not shown). In addition, appressorium formation could be stimulated by 10 mM cAMP (data not shown; ref. 22). Calcofluor (cell wall) and Hoechst 33258 (nuclei) staining showed that septation, cell-wall deposition, nuclear division, and migration were also indistinguishable from those of wild type (data not shown). Finally, mps1–1Δ mutant appressoria underwent melanization and were resistant to plasmolysis by 1 M KCl (23) suggesting that they were not osmotically sensitive (data not shown).
mps1 Mutants Elicit Plant Defense Responses.
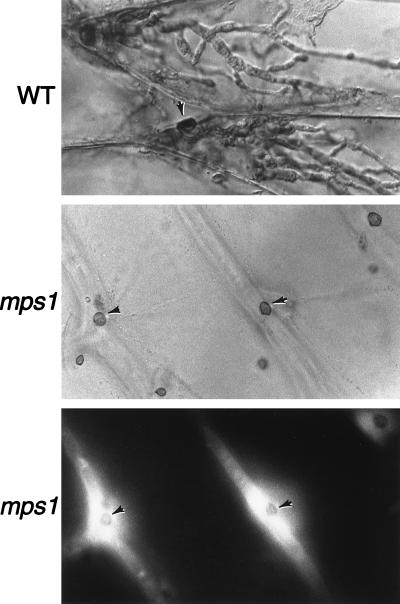
To define more precisely the pathogenicity defects of mps1–1Δ, we examined infection-related morphogenesis with a sensitive penetration assay of onion epidermal cells (19). In this assay, wild-type strains form appressoria, penetrate into epidermal cells, and produce invasive hyphae (Fig. 5 Top). mps1–1Δ mutant strains also formed appressoria efficiently in this assay, but the mutant strains were unable to produce intracellular infectious hyphae and were not observed to be able to penetrate plant cells (n = 200). Because mps1–1Δ mutants can infect plant cells through wounds, we conclude that the primary pathogenicity defect in mps1–1Δ mutants is their failure to penetrate plant cells.
Figure 5.
Fungal penetration assays on plant epidermal cells. (Top) Wild-type strains (WT) form appressoria (arrow) that penetrate and produce abundant infectious hyphae. (Middle and Bottom) Strains containing the mps1–1Δ allele (mps1) produce numerous appressoria (arrows) but do not penetrate or form infectious hyphae. Lower shows fluorescence (465 nm) illumination of the same area depicted in Middle. Plant-cell-derived autofluorescence surrounds nonpenetrating appressoria. (×465.)
Interestingly, although there was no penetration of onion epidermal cells by mps1–1Δ mutants, we noticed that the plant cells that underlie appressoria seemed to undergo cytoplasmic alterations. Fluorescence microscopy showed the accumulation of intense plant-cell-derived autofluorescence directly around mps1–1Δ mutant appressoria, consistent with the activation of early plant defense responses (Fig. 5 Lower). Similar levels of plant-cell autofluorescence and cytoplasmic alterations were visible at early stages of infection by wild-type appressoria (data not shown).
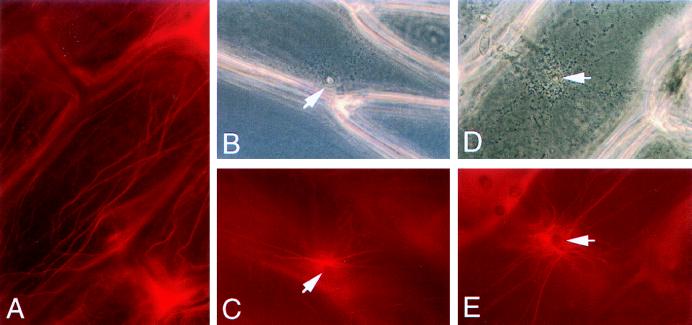
Because previous studies suggested that plant cells respond to pathogen attack with rearrangement of cytoskeletal elements (24, 25), onion epidermal peels were inoculated with either wild-type or mps1–1Δ mutant strains and stained with rhodamine-phalloidin to visualize the plant actin cytoskeleton. Uninfected onion epidermal cells contain prominent longitudinal bundles and a fine cortical meshwork of actin microfilaments (Fig. 6A). After the formation of an appressorium by a wild-type strain on the epidermal cell surface, the longitudinal actin cables accumulated and seemed to radiate from the penetration site (Fig. 6 B and C). Moreover, the fine cortical network and longitudinal actin cables were reduced substantially elsewhere in the plant-cell cytoplasm. Although they fail to penetrate, the mps1–1Δ mutant appressoria elicited an identical cytoplasmic-actin response in the onion epidermal cells (Fig. 6 D and E).
Figure 6.
Actin rearrangements in response to mps1–1Δ mutants. Onion epidermal cells were left untreated (A), inoculated with wild-type conidia (B and C), or inoculated with mps1–1Δ mutants (D and E). Plant cells were visualized with phase-contrast microscopy (B and D) and with epifluorescence (C and E) after staining with rhodamine-phalloidin. Arrows indicate appressoria (B and D) and underlying actin rearrangements (C and E). (×280.)
DISCUSSION
Several of the most economically devastating fungal crop pathogens use appressoria to penetrate the plant cell wall. Elegant experiments with the rice blast fungus on synthetic surfaces showed that penetration is driven by the generation of enormous turgor pressure (5, 6). Here, we provide evidence that a signaling pathway related to the yeast protein kinase C/Slt2/Mpk1 cell-integrity pathway is required to allow turgor-driven penetration of plant cells by fungal appressoria. In the rice blast fungus, this pathway is also required for sporulation and the maintenance of hyphal cell-wall integrity. Components of this signaling pathway could provide targets for the development of antifungals that would block both penetration by appressoria and sporulation from developing lesions.
The compromised cell integrity of mps1–1Δ mutants of M. grisea is reminiscent of Δslt2/Δmpk1 mutants of yeast but with several important differences. S. cerevisiae mutants undergo lysis at high temperature, and analogous mutants in S. pombe are salt-sensitive as well as temperature-sensitive. These phenotypes were not observed with M. grisea mutants. Rather, cells became osmotically sensitive as a result of cell aging. In addition, specific defects were observed in certain developmental processes, such as aerial-hypha formation, conidiation, female fertility, and appressorium-driven penetration of plant cells. These findings suggest that the Mps1 signaling pathway in M. grisea is required for cell-wall remodeling in response to specific developmental cues. It has been shown that yeast Mpk1 activates a MADS-box transcription factor, Rlm1, that may be required to activate cell-wall biosynthetic genes (26). Although we do not know which gene products could be activated by this pathway in M. grisea, it is known that, for emergent growth and sporulation, filamentous fungi secrete a group of fungal cell-wall proteins, called hydrophobins, that are arrayed on the outer cell-wall surface (27). Interestingly, the hydrophobin Mpg1p from M. grisea has been shown to be necessary for sporulation and pathogenicity (16, 17). It will be interesting to see whether developmental expression of Mpg1p depends on Mps1 signaling.
Mps1 signaling is required for appressorium penetration of plant cells. Mps1 may be required to bring about modifications to the appressorium cell wall in response to increasing turgor. These modifications are not immediately obvious; mps1–1Δ mutant appressoria undergo melanization normally, and Calcofluor staining did not reveal anomalies in appressorium formation. In addition, mps1–1Δ appressoria did not undergo autolysis during prolonged incubations on artificial surfaces, a condition that leads to dramatic increases in intracellular turgor (7). The Pkc1-Mpk1 signaling pathway of S. cerevisiae is also required for bud emergence and polarization of the actin cytoskeleton (28). Thus we conclude, by analogy to yeast, that Mps1 signaling may also be necessary for polarization of the actin cytoskeleton to facilitate the formation of a penetration hypha. This role must be rather stage-specific, because Mps1 seems to be dispensable for actin-dependent polarized growth of fungal hyphae.
Mps1 is one of two MAPKs found to be essential for the infection process in M. grisea. Pmk1, functionally homologous to the yeast MAPKs Fus3/Kss1, is required specifically for the differentiation of appressoria and plays no role in mating processes (18). Studies of S. cerevisiae have shown that outputs from the pheromone signaling pathway are required for activation of the Mpk1-cell-integrity pathway (29). An analysis of the phenotypes of Δpmk1 strains (failure to form appressoria) and mps1–1Δ strains (appressoria that fail to penetrate plant cells) also would suggest that Mps1 function is subsequent to Pmk1 function during the infection process. It is tempting to speculate that although the output of MAPK-signaling pathways between yeast and the rice blast fungus are very different, remarkably, their integration into a common cell-morphogenesis pathway may be conserved (30).
A surprising finding is that although mps1–1Δ mutants are nonpathogenic and fail to penetrate plant cell walls, mps1–1Δ mutants still elicit certain physiological behaviors characteristic of the plant defense response. These include accumulation of autofluorescent compounds, changes in the pattern of cytoplasmic streaming, and dramatic rearrangement of the actin cytoskeleton. Reorganization of the plant cytoskeleton has been observed usually as a consequence of pathogen penetration (24), particularly in the case of barley–Erysiphe graminis interactions (25). Experiments with actin-microfilament inhibitors suggest that this response is part of a general defense mechanism to prevent fungal penetration (31). Here, the surprising finding is that these dramatic cytological events can be triggered in the complete absence of fungal penetration. Although we have not investigated whether these effects also occur in rice plants, careful examination of rice leaves inoculated with mps1–1Δ mutants did not reveal any hypersensitive plant cell death and indicates that mps1–1Δ mutants most likely failed to penetrate rice cells. Together, these findings suggest that certain aspects of the rice-plant defense response may be activated before fungal penetration and thus may be elicited at the plant cell surface. It remains to be determined whether this plant response is triggered by mechanical pressure exerted by appressoria attempting (but failing) to penetrate or by soluble factors released from the invading fungus. The array of M. grisea mutants defective in various aspects of appressorium formation and penetration should allow a more detailed investigation of this process.
Acknowledgments
We thank Drs. David Levin and Takashi Toda for yeast strains, Jane Einstein for assistance with DNA sequencing, and members of the Hamer and Staiger laboratories for helpful discussions. Research in the laboratory of C.J.S. is supported by the U.S. Department of Agriculture’s National Research Initiative Competitive Grants Program. This work was also supported in part by a National Science Foundation Grant and a Presidential Faculty Fellow Award to J.E.H.
ABBREVIATION
- MAPK
mitogen-activated protein kinase
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF020316).
References
- 1. Walton J D. Plant Physiol. 1994;104:1113–1118. doi: 10.1104/pp.104.4.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Emmett R W, Parbery D G. Annu Rev Phytopathol. 1975;13:147–167. [Google Scholar]
- 3.Ou S H. Rice Diseases. Surrey, U.K.: Commonwealth Mycological Institute; 1985. [Google Scholar]
- 4.Howard R J, Bourett T M, Ferrari M A. In: Infection by Magnaporthe grisea: An in Vitro Analysis. Mendgen K, Lesemann D E, editors. Berlin: Springer; 1991. pp. 251–264. [Google Scholar]
- 5.Howard R J. In: Cell Biology of Pathogenesis. Zeigler R S, Leong S A, Teng P S, editors. Wallingford, U.K.: CAB International; 1994. pp. 3–22. [Google Scholar]
- 6.Howard R J, Valent B. Annu Rev Microbiol. 1996;50:491–512. doi: 10.1146/annurev.micro.50.1.491. [DOI] [PubMed] [Google Scholar]
- 7.Howard R J, Ferrari M A, Roach D H, Money N P. Proc Natl Acad Sci USA. 1991;88:11281–11284. doi: 10.1073/pnas.88.24.11281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Money N P, Howard R J. Fungal Genet Biol. 1996;20:217–227. [Google Scholar]
- 9.Dejong J C, McCormack B J, Smirnoff N, Talbot N J. Nature (London) 1997;389:244–245. [Google Scholar]
- 10.Bourett T M, Howard R J. Protoplasma. 1992;168:20–26. [Google Scholar]
- 11.Costigan C, Gehrung S, Snyder M. Mol Cell Biol. 1992;12:1162–1178. doi: 10.1128/mcb.12.3.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee K S, Irie K, Gotoh Y, Watanabe Y, Araki H, Nishida E, Matsumoto K, Levin D E. Mol Cell Biol. 1993;13:3067–3075. doi: 10.1128/mcb.13.5.3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davenport K R, Sohaskey M, Kamada Y, Levin D E, Gustin M C. J Biol Chem. 1995;270:30157–30161. doi: 10.1074/jbc.270.50.30157. [DOI] [PubMed] [Google Scholar]
- 14.Zaitseveskaya-Carter T, Cooper J A. EMBO J. 1997;16:1318–1331. doi: 10.1093/emboj/16.6.1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toda T, Dhut S, Superti-Furga G, Gotoh Y, Nishida E, Sugiura R, Kuno T. Mol Cell Biol. 1996;16:6752–6764. doi: 10.1128/mcb.16.12.6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Talbot N J, Ebbole D J, Hamer J E. Plant Cell. 1993;5:1575–1590. doi: 10.1105/tpc.5.11.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Talbot N J, Kershaw M J, Wakley G E, de Vries O M H, Wessels J G H, Hamer J E. Plant Cell. 1996;8:985–999. doi: 10.1105/tpc.8.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu J-R, Hamer J E. Genes Dev. 1996;10:2696–2706. doi: 10.1101/gad.10.21.2696. [DOI] [PubMed] [Google Scholar]
- 19.Xu J-R, Urban M, Sweigard J A, Hamer J E. Mol Plant–Microbe Interact. 1997;10:187–194. [Google Scholar]
- 20.Altschul S F, Gish W, Miller W, Myers C W, Lipman D L. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 21.Leung H, Borromeo E S, Bernardo M A, Notteghem J L. Phytopathology. 1988;78:1227–1233. [Google Scholar]
- 22.Lee Y-H, Dean R A. Plant Cell. 1993;5:693–700. doi: 10.1105/tpc.5.6.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Howard R J, Ferrari M A. Exp Mycol. 1989;13:403–418. [Google Scholar]
- 24.Gross P, Julius C, Schmelzer E, Hahlbrock K. EMBO J. 1993;12:1735–1744. doi: 10.1002/j.1460-2075.1993.tb05821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kobayashi Y, Yamada M, Kobayashi I, Kunoh H. Plant Cell Physiol. 1997;38:723–733. [Google Scholar]
- 26.Watanabe Y, Takaesu G, Hagiwara M, Irie K, Matsumoto K. Mol Cell Biol. 1997;17:2615–2623. doi: 10.1128/mcb.17.5.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wessels J G H. Trends Plant Sci. 1996;1:9–15. [Google Scholar]
- 28.Zarzov P, Mazzoni C, Mann C. EMBO J. 1996;15:83–91. [PMC free article] [PubMed] [Google Scholar]
- 29.Buehrer B M, Errede B. Mol Cell Biol. 1997;17:6517–6525. doi: 10.1128/mcb.17.11.6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Banuett F. Microbiol Mol Biol Rev. 1998;62:249–274. doi: 10.1128/mmbr.62.2.249-274.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kobayashi Y, Kobayashi I, Funaki Y, Fujimoto S, Takemoto T, Kunoh H. Plant J. 1997;11:525–537. [Google Scholar]