Abstract
Background
The master regulator p53 tumor-suppressor protein through coordination of several downstream target genes and upstream transcription factors controls many pathways important for tumor suppression. While it has been reported that some of the p53's functions are microRNA-mediated, it is not known as to how many other microRNAs might contribute to the p53-mediated tumorigenesis.
Results
Here, we use bioinformatics-based integrative approach to identify and prioritize putative p53-regulated miRNAs, and unravel the miRNA-based microregulation of the p53 master regulatory network. Specifically, we identify putative microRNA regulators of a) transcription factors that are upstream or downstream to p53 and b) p53 interactants. The putative p53-miRs and their targets are prioritized using current knowledge of cancer biology and literature-reported cancer-miRNAs.
Conclusion
Our predicted p53-miRNA-gene networks strongly suggest that coordinated transcriptional and p53-miR mediated networks could be integral to tumorigenesis and the underlying processes and pathways.
Background
The p53 tumor suppressor gene has been implicated as a master regulator of genomic stability, cell cycle, apoptosis, and DNA repair [1,2]. p53 is known to act as both transcriptional activator and repressor of expression of specific genes [3]. Studies of p53-mediated repression have shown that both genes that modulate apoptotic responses and genes that promote cell cycle progression can be repressed by p53 [4].
MicroRNAs (miRNAs or miRs) are small non-coding RNAs typically of 21–25 nucleotides in length and reported to be involved in the regulation of a variety of biological processes, including development, cell death, cell proliferation, hematopoiesis and nervous systems patterning [5]. miRNAs enforce posttranscriptional silencing through the RNA interference pathway [6]. Currently more than 450 human miRNAs have been identified and deposited in the miRBase [7]. Computational prediction of miRNA targets reveal that each human miRNA can potentially target several genes [8,9], underscoring the importance of these tiny noncoding RNAs in gene regulation.
Several previous studies have identified ten positive or negative feedback loops in the p53 pathway (reviewed in [10]). All of these networks or circuits are autoregulatory in that they are either induced by p53 activity at the transcriptional level, transcriptionally repressed by p53 or are regulated by p53-induced proteins [10]. Recently, five independent studies have shown that p53 upregulates miR-34a in response to DNA damage leading to cell cycle arrest and apoptosis [11-15]. In light of these reports and based on the growing evidence that microRNAs themselves act to either promote cancer or to stop cancer spread [16-19], it is reasonable to conceive that the tumor suppressor p53 might be involved in this microRNA-related network in cancer cells. In support of this, two miRNAs (miR-372 and miR-373) were found to function as oncogenes in human testicular germ cell tumors, probably by numbing the p53 pathway and thereby allowing tumorigenesis [20]. Yang et al recently suggested that analysis of microRNA regulation in the transcriptional network of the p53 gene can reveal relationships between oncogenic microRNAs and p53 [21]. We therefore sought to explore if there are other microRNAs that are components of the p53 regulatory network. In an earlier study (Kaimal and Sinha et al., unpublished), scanning the 474 human miRNAs for putative p53 sites, we identified 143 miRNAs that have at least one p53 site (using p53MH algorithm [22]) and are predicted to target at least one known gene. We termed these 143 miRNAs as p53-miRs (Kaimal and Sinha et al., unpublished). In the current study, apart from correlating these putative p53-miRs with the upstream regulators and downstream targets of p53, we prioritize our predictions using (a) literature reported differentially expressed miRNAs in cancer; (b) miRNAs reported as induced or repressed following p53 activation [11]; and (c) miRNA targets that are functionally enriched with biological processes like apoptosis, cell cycle.
Results and discussion
Prediction of p53-miRs and the target genes
A significant fraction of human intergenic miRNAs are reported to possess TSS sites within 2 kb of the pre-miRNA [23]. Although it is accepted that intronic miRNAs are generally transcribed along with their host genes [24], TSS predictions have shown them to lie predominantly in the region from -2 kb to -6 kb [23]. Hence, we limited our search space to 10 kb flanking regions of the miRNAs. Determination of exact pri-miRNA transcript lengths however is possible only through experimental work.
We used p53MH [22], an algorithm that identifies p53-responsive genes in the human and mouse genome, to identify putative p53-binding site within miRNAs (see Methods). Out of 474 human miRNAs analyzed, 227 had at least one putative p53 site (and a total of 319 p53 sites; some miRNAs have multiple p53 sites) within their flanking regions of 10 kb (Additional File 1). Of the 319 p53 sites, 5 sites were overlapping with exonic region and we did not consider these for further analysis. The 314 p53 sites (Additional File 2) occur within 10 kb flanking sequences of 223 miRNAs. We identified two p53 binding sites in mir-34a; however neither of these corresponds to the experimental validated p53 site [12] in mir-34a. This is because in the current analysis we limit the miRNA flanking sequence search space to 10 kb only while the experimentally validated p53 site in mir-34a occurs at approximately 30 kb upstream to mir-34a [12].
To predict target genes of the 223 miRNAs, we used MAMI server [25], which has compilation of five [8,9,26-28] target prediction algorithms. A recent study showed that about 82% of unique miRNA-target pairs are predicted by only a single algorithm [19]. Therefore, a combination of predictions from all algorithms might provide a much more comprehensive list of putative miRNA-target pairs than do any single prediction. 154/223 miRNAs are predicted to target at least one gene while 69/223 have no predicted targets. The 154 miRNAs mapped to 143 miRNAs. This is because some of the miRNAs have more than one predicted hairpin precursor sequences. For example mir-135a has two predicted hairpin precursors (mir-135a-1 on chromosome 3 and mir-135a-2 on chromosome 12). The targets are however based on mir-135a only. Thus, our final set consists of 143 miRNAs; each of which has at least one p53 site and is predicted to target at least one known gene. We call these 143 miRNAs as p53-miRs. There were 12497 genes predicted as targets of these 143 p53-miRs! Assuming that most of these predicted targets are purely speculative (false positive rate of at least 4 of these algorithms was estimated to be 20–30% [29]), we sought to integrate multiple types of data resources and explore as to what number of p53-miRs could be targeting cancer-related genes.
MicroRNAs associated with p53 upstream and downstream genes
Transcriptional networks commonly contain positive- and negative-feedback loops, which provide robustness and fine-tuning to gene programs [30,31]. Recent studies have suggested a possible coordinated transcriptional and miRNA-mediated regulation as a recurrent motif to enhance the robustness of gene regulation in mammalian genomes [32-35]. It was also reported that miRNAs predominantly target positive regulatory motifs, highly connected scaffolds and most downstream network components such as signaling transcription factors, but less frequently target negative regulatory motifs, common components of basic cellular machines and most upstream network components such as ligands [32]. Using the p53 master regulatory network as a case study, we investigated the potential crosstalk between the miRNAs and the p53 transcriptional network itself involving transcription factors upstream and downstream to p53. At least four possible scenarios can be envisaged for the upstream regulators (Figures 1A–D) and downstream targets (Figures 1E–H) of p53.
Figure 1.
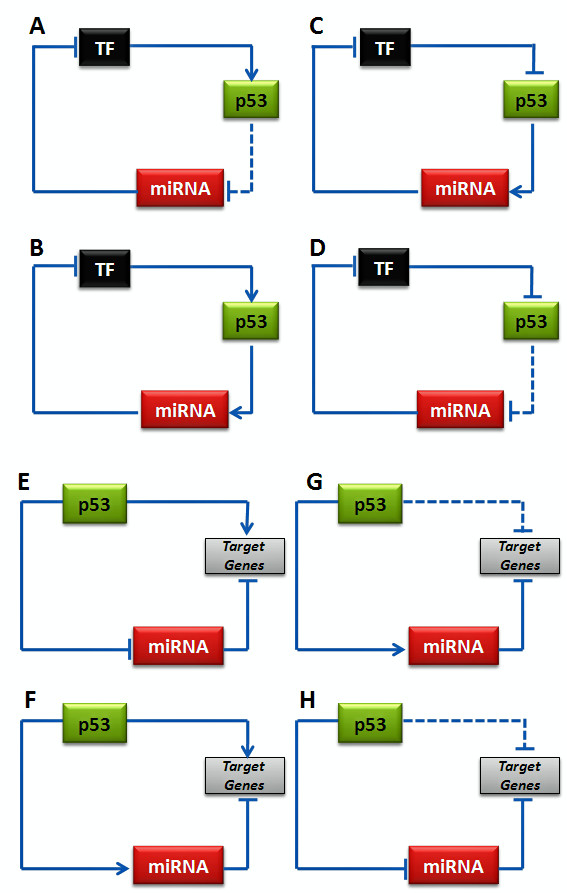
Four possible scenarios for the upstream regulators (A-D) and downstream targets (E-H) of p53. For the downstream coherent models, the p53 action on downstream target gene and miRNA is opposite (F and H). In case of downstream incoherent models (E and G), the p53 action on downstream target gene and miRNA is same. There may be a sequential gap in the activation time of the target gene and miRNA facilitating maintenance of steady states.
We downloaded the known upstream transcription factor (TF) regulators and downstream target TFs of p53 from the p53 KnowledgeBase [36]. For the 23 upstream regulators and 48 downstream TFs of p53, we then extracted the putative miRNA regulators using MAMI server [25], which has compilation of five target prediction algorithms [8,9,26-28]. Since each of these five miRNA target prediction approaches generates an unpredictable number of false positives (an estimate puts it at 20–30% [29]), we intersected the results to identify the genes commonly predicted by at least two of the five algorithms [8,9,26-28] (see Table 1; for a complete list, see Additional file 3). Although it is important to confirm that a miRNA is not a potential regulatory miRNA of a specific gene by using different bioinformatics algorithms [37], there are several experimentally validated microRNA target genes that are not predicted by more than one of the current prediction algorithms. For instance the experimentally validated mir-21-PTEN miRNA-target pair [38] is not predicted by any of the five algorithms (listed in Methods and used in MAMI compilation). To overcome this, we also checked the TarBase [39] which has experimentally validated microRNA targets irrespective of the computational predictions.
Table 1.
p53-miRs (microRNAs which have a conserved p53 binding site in their flanking 10 kb genomic sequence) which putatively target known transcription factors that are either upstream or downstream to p53 network (based on p53 knowledge base [36]) (A: activator; R: Repressor; U: uncharacterized). Only those targets are shown that are predicted as miRNA targets by 2 or more than two algorithms. miRNAs that are in bold represent miRNAs induced more than 1.5 fold (p < 0.05) after p53 activation [11]. The last column indicates whether a miRNA has been reported in literature as differentially expressed in cancer or cancer cell lines.
| miRNA | TF | TargetScanS | miRanda | microT | miRtarget | picTar | Relation to p53 (Interaction Type) [20] | Differentially expressed in cancer |
| hsa-miR-183 | EGR1 | + | + | - | + | - | Upstream (A) | Yes |
| hsa-miR-26b | HOXA5 | + | - | - | + | - | Upstream (A) | Yes |
| hsa-miR-96 | HOXA5 | + | - | - | + | - | Upstream (A) | Yes |
| hsa-miR-376a | HOXA5 | - | + | + | - | - | Upstream (A) | No |
| hsa-miR-376b | HOXA5 | - | + | + | - | - | Upstream (A) | No |
| hsa-miR-181c | FOS | + | + | - | + | - | Upstream (A/R) | Yes |
| hsa-miR-222 | FOS | + | + | - | - | - | Upstream (A/R) | Yes |
| hsa-miR-101 | FOS | + | + | - | + | - | Upstream (A/R) | No |
| hsa-miR-10a | BCL6 | + | + | - | - | - | Upstream (R) | Yes |
| hsa-miR-124a | KLF4 | + | - | - | + | - | Upstream (R) | Yes |
| hsa-miR-148a | KLF4 | + | + | + | + | - | Upstream (R) | Yes |
| hsa-miR-32 | KLF4 | + | + | - | + | - | Upstream (R) | Yes |
| hsa-miR-34a | KLF4 | + | - | - | + | - | Upstream (R) | Yes |
| hsa-miR-7 | KLF4 | + | - | - | + | - | Upstream (R) | Yes |
| hsa-miR-508 | KLF4 | - | + | - | + | - | Upstream (R) | No |
| hsa-miR-155 | CEBPB | + | + | + | - | - | Upstream (U) | Yes |
| hsa-miR-27a | BTG2 | + | + | + | - | - | Downstream (A) | Yes |
| hsa-miR-32 | BTG2 | + | + | - | - | - | Downstream (A) | Yes |
| hsa-miR-27a | CCNG1 | + | - | - | + | - | Downstream (A) | Yes |
| hsa-miR-122a | CCNG1 | + | + | - | + | - | Downstream (A) | Yes |
| hsa-miR-33 | EEF1A1 | + | + | - | - | - | Downstream (A) | No |
| hsa-miR-502 | GADD45A | - | + | - | + | - | Downstream (A) | No |
| hsa-miR-19b | IGFBP3 | + | + | - | - | - | Downstream (A) | Yes |
| hsa-miR-29a | MMP2 | - | + | - | + | - | Downstream (A) | Yes |
| hsa-miR-29b | MMP2 | - | + | - | + | - | Downstream (A) | Yes |
| hsa-miR-29c | MMP2 | - | + | - | + | - | Downstream (A) | Yes |
| hsa-miR-128b | PLK2 | + | + | - | + | + | Downstream (A) | Yes |
| hsa-miR-200b | PLK2 | + | + | - | + | - | Downstream (A) | Yes |
| hsa-miR-200c | PLK2 | + | + | - | + | - | Downstream (A) | Yes |
| hsa-miR-27a | PLK2 | + | + | - | + | + | Downstream (A) | Yes |
| hsa-miR-30b | SERPINE1 | + | + | + | + | - | Downstream (A) | Yes |
| hsa-miR-30c | SERPINE1 | + | + | + | + | - | Downstream (A) | Yes |
| hsa-miR-34a | SERPINE1 | + | - | - | + | - | Downstream (A) | Yes |
| hsa-miR-192 | RB1 | + | + | - | - | - | Downstream (A/R) | Yes |
| hsa-miR-200b | ANLN | + | + | - | - | - | Downstream (R) | Yes |
| hsa-miR-200c | ANLN | + | + | - | - | - | Downstream (R) | Yes |
| hsa-miR-142-3p | IER3 | - | + | - | + | - | Downstream (R) | Yes |
| hsa-miR-124a | SCD | + | - | - | + | - | Downstream (R) | Yes |
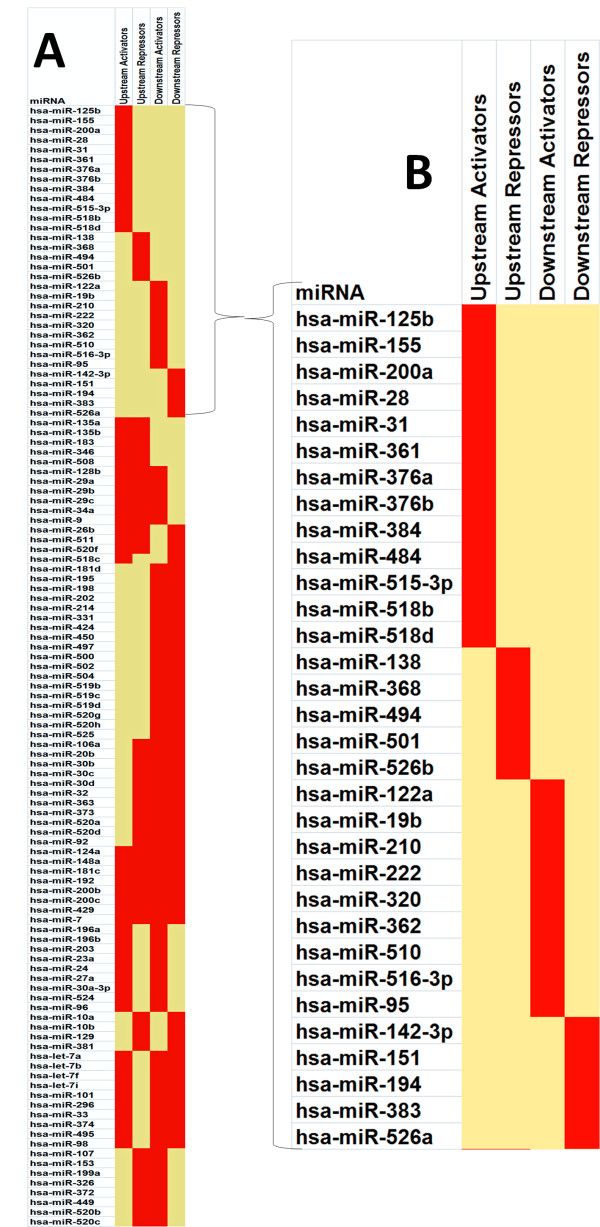
Based on MAMI predictions, a total of 242 different miRNAs suppress p53 regulators or targets. Of these, 115 (~48%) are p53-miRs (Figure 2A). Some of the p53-miRs (32/143; Figure 2B) are found to exclusively target p53 upstream activators (13/143 p53-miRs; Figure 3A) or repressors (5/143; Figure 3B) or downstream activators (9/143; Figure 3C) or repressors (5/143; Figure 3D). Interestingly, many of these miRNAs have been reported as differentially expressed in various cancerous tissues or cell lines (see last column in Table 1).
Figure 2.
Heat map representation of p53-miRs and their target genes relative to the p53 network. 115 p53-miRs are predicted to target transcription factors that are either p53 regulators (upstream) or p53 targets (downstream) (A).
Figure 3.
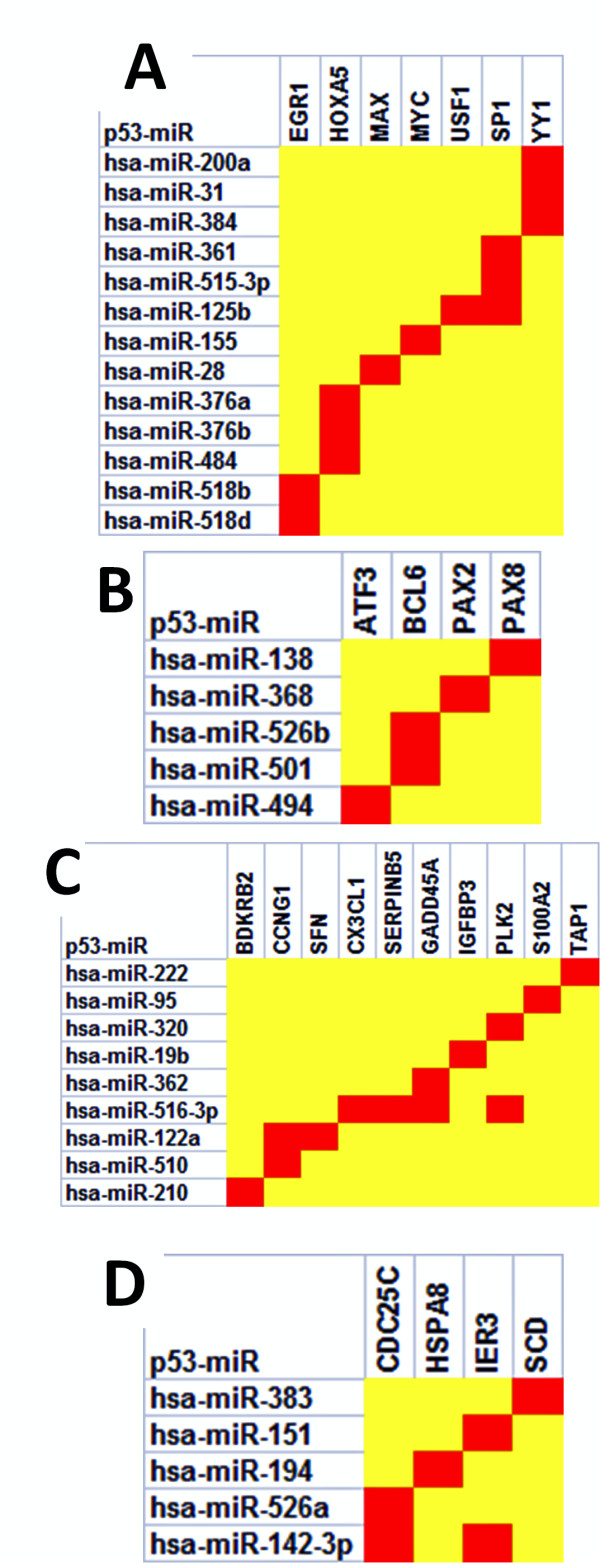
Heat map representation of p53-miRs and their target genes relative to the p53 network. Panels A, B, C and D depict p53-miRs exclusively targeting p53 upstream activators, upstream repressors, downstream activators and downstream repressors respectively along with their predicted target transcription factors.
miRNAs upstream and downstream to p53: coherent and incoherent circuits
If there is a common upstream TF that regulates both the miRNA and its target gene, ideally, the transcription of the miRNAs and their targets should be oppositely regulated by this common upstream TF. For example, p53 which is known to repress the transcription of IER3 [40] may simultaneously activate the transcription of mir-142 (predicted to target IER3) that may further inhibit IER3 posttranscriptionally. This is a coherent model for all downstream targets that are repressed by p53. Previous genome-scale studies have in fact shown that predicted target transcripts of several tissue-specific miRNAs tend to be expressed at a lower level in tissues where the miRNAs are expressed [41,42].
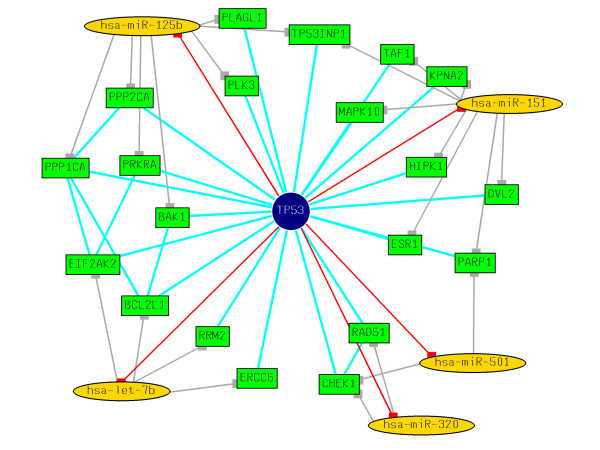
Alternately, a common TF (p53 in this case) can activate a downstream target gene and also a miRNA which then suppresses the downstream target gene. A well known example is the experimentally confirmed microregulatory network comprising miR-17-5p and E2F1 – both of which are transcriptionally activated by c-Myc in human cells [43]. We report a similar example of mir-122a, a putative p53-miR suppressing CCNG1, a downstream target of p53. CCNG1 is a known transcriptional target (activator) of the p53 tumor suppressor protein [44] and it was recently reported that mir-122a (predicted as p53-miR in our analysis) has an inverse correlation with cyclin G1 expression in primary liver carcinomas [45]. In other words, p53 activates CCNG1 and also mir-122a which in turn suppresses CCNG1. This is an incoherent model. Adding to this complexity further, it is also reported that CCNG1 negatively regulates the stabilization of p53 in a possible negative feedback loop [46] (Figure 4A). Thus, the regulatory network wherein p53 activates a downstream target and a miR (which target the downstream target) simultaneously appears "inefficient". However, feedforward loops have the potential to provide temporal control, because expression of the ultimate target may depend on the accumulation of adequate levels of master regulator and the secondary regulators [47]. Therefore, if we consider a sequential gap in the activation time of the target genes and the miRNA, then this downstream p53-gene and p53-miR regulation appears coherent. Feedforward loops may provide a form of multistep ultrasensitivity [48], as any deviation from the p53's steady state would drive the downstream targets and miRNA (p53-miR) away from their steady states in the same direction. p53-miRs could therefore tune the production rate of the downstream p53 target gene opposite to the direction of p53's fluctuation. Such noise buffering probably helps to maintain target protein homeostasis and ensures more uniform expression [49] of the p53 downstream target genes within a cell population. Additionally, since the level of p53-miR defines the p53 downstream target's translation rate, their coexpression may allow p53-miR to fine-tune the downstream target's steady state. Thus, p53-miRs acting on p53 downstream targets could significantly shorten the response delay, leading to more effective noise buffering, as well as precise definition and maintenance of steady states.
Figure 4.
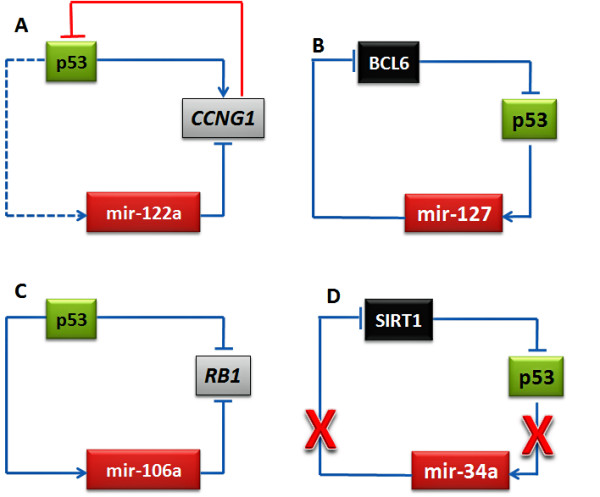
Schematic representation of p53-miR-mediated models downstream (A and C) and upstream (B and D) to p53.
We found two entries in TarBase (database of experimentally validated microRNA target genes) [39] which are relevant and could be interesting to p53 network (a) mir-127 targeting BCL6 [50], an upstream repressor of p53 [51] (Figure 4B) and (b) mir-106a reported to suppress RB1 [52]. A previous study has shown that mir-106a is induced more than 1.5 fold (p < 0.05) after p53 activation [11]. p53 is known to suppress RB1 transcription through inhibition of the basal promoter activity [53]. Additionally, posttranscriptionally, p53 might suppress RB1 further by inducing mir-106a, a known suppressor of RB1 (Figure 4C).
As a master transcriptional regulator, p53 is also known to trans-repress genes including those affecting signaling pathways such as cell proliferation, apoptosis and cytoskeleton organization [54,55]. Although, the mechanism(s) of repression is often unclear and may not be dependent on site-specific DNA-binding activity [54-57], the responsiveness of some genes to p53 can be indirect in that target genes (transactivated or transrepressed) might be other transcription factors [55,58] or microRNAs, which then modulate downstream responses to p53 signaling. We, therefore, hypothesize that in case of p53 downstream targets, the foremost activated positive feedback in a coupled feedback circuit can rapidly induce the "on" state transition of the signaling system (p53 activating downstream target genes), and that then another delayed positive feedback (p53 suppressing miRNAs that target p53 downstream target genes) robustly maintains this "on" state. Finally, the most delayed negative feedback reinstates the system in the original "off " state, preventing any further excessive response specific to the applied stimulus.
Loss of p53 expression in some tumors has also been shown to occur through inhibition of transcription of the p53 gene itself [51,59,60]. For example, mir-31, one of the p53-miRs predicted to suppress the upstream activator (YY1) of p53, is reported to be upregulated in colorectal cancer [61]. Shalgi et al recently reported that the extent of miR mediated regulation varies extensively among different genes, some of which, especially those who serve as regulators themselves, are subject to enhanced miRNA-based silencing [34].
p53-miR target genes – functional over-representation analysis
Functional over-representation analysis was performed to objectively identify biological processes potentially affected by p53-miR target genes. Specifically, the percentage of p53-miR target genes with a given gene ontology (GO) annotation was compared with the percentage of whole miRNA-target genes genome-wide with the same annotation. A significant p value (p < 0.05) indicates that the observed percentage of p53-miR target genes with a given annotation could not likely occur by chance given the frequency of whole miRNA-target genes genome-wide with the same annotation. MAMI predicted target genes for the 143 p53-miRs were 12497. We used DAVID [62], and our in-house developed ToppGene server [63] for the functional enrichment analysis. Since apoptosis and growth arrest are common known consequences of p53 activation, we also tested whether p53-miRs tend to target apoptotic and cell proliferation related genes. Interestingly, the enrichment analysis revealed that while cell cycle regulation (p = 8.41E-4) was among the top in the p53-miR targets functional enrichment (p < 0.05), apoptosis was not significant. The other significantly enriched GO terms include transcription, vesicle-mediated transport, intracellular signaling cascade and cellular metabolism (Additional File 4). Interestingly, vesicle-mediated transport (p = 3.9 E-6) and endocytosis (p = 5.1 E-4) were specifically enriched in cancer-associated p53-miR target genes. Abnormal expression or mutation of endocytosis proteins has been reported in human cancers [64-66]. The possibility of harnessing the endocytic machinery to cancer through miRNAs could offer viable and promising leads in cancer therapy.
We extended p53-miR target gene functional enrichment analysis further to identify specific p53-miRs that significantly target apoptotic and cell proliferation genes (see Tables 2 and 3 for a list of top 25 p53-miRs and their target apoptotic and cell proliferation genes respectively). There was an overlap of 10 p53-miRs (mir-181c, mir-200a, mir-200b, mir-19b, mir-101, mir-106a, mir-142-3p, mir-195, mir-124a and mir-24) between these two groups. Earlier studies have shown that mir-23 is indeed critical for cell proliferation [67]. A recent study also implicated mir-23a, mir-24 and mir-195 in cardiac hypertrophy and reported that these miRNAs were regulated in response to stress signaling in the heart [68]. The Additional File 4 enlists the other functional categories significantly enriched within the putative p53-miR targets. It is to be noted that 18/25 (72%) apoptotic p53-miRs and 22/25 (88%) cell proliferation p53-miRs have been reported in literature as differentially regulated in various cancerous tissues or cancer cell lines (see last columns in Tables 2 and 3).
Table 2.
Top 25 most significant p53-miRs that target known apoptotic genes (n = 685). 18/25 apoptotic p53-miRs have been reported in literature as differentially regulated in various cancerous tissues or cancer cell lines (see last column). The p-values represent the probability of a p53-miR targeting an apoptotic gene purely by chance.
| miRNA | Total Number of Predicted Targets | Apoptosis Genes (n = 685) | Genes | p-value | Cancer miRNA |
| hsa-miR-30b | 1054 | 64 | ABL1, ACTN1, ACVR1, ARHGEF6, ATG5, BCL2, BCL2L11, BCL6, BDNF, BECN1, BIRC6, BNIP3L, CASP3, CLCF1, CUL2, EBAG9, ELMO1, FOXO3A, FXR1, GAS2, GCLC, HSPA5, IFI16, IFNG, IHPK3, IL1A, JAG2, MAGI3, MAL, MAP3K5, MNT, MYBL2, NOTCH1, PAK1, PAK7, PAWR, PAX3, PDCD10, PDCD5, PIK3R2, PLAGL2, PPP2R1B, PRLR, PSEN2, RARG, RASA1, RHOB, RNF7, SGK3, SIAH2, SIRT1, SMNDC1, SNCA, SOCS3, SON, SOX9, TESK2, TIA1, TIMP3, TNFSF9, TP53INP1, UNC5C, UNC5D, VCP | 1.50E-12 | Yes |
| hsa-miR-30c | 1040 | 61 | ABL1, ACTN1, ACVR1, ARHGEF6, ATG5, BCL2, BCL2L11, BCL6, BDNF, BECN1, BNIP3L, CASP3, CLCF1, CUL2, EBAG9, ELMO1, FOXO3A, FXR1, GAS2, GCLC, HSPA5, IFI16, IHPK3, IL1A, JAG2, MAGI3, MAL, MAP3K5, MNT, MYBL2, NOTCH1, PAK1, PAK7, PAWR, PAX3, PDCD10, PDCD5, PIK3R2, PLAGL2, PPP2R1B, PRLR, PSEN2, RARG, RASA1, RHOB, RNF7, SGK3, SIAH2, SIRT1, SMNDC1, SNCA, SOCS3, SON, SOX9, TIA1, TIMP3, TNFSF9, TP53INP1, UNC5C, UNC5D, VCP | 2.16E-11 | Yes |
| hsa-miR-30d | 1034 | 57 | ABL1, ACTN1, ACVR1, ARHGEF6, ATG5, BCL2, BCL2L11, BCL6, BDNF, BECN1, BNIP3L, CARD8, CASP3, CLCF1, CUL2, EBAG9, ELMO1, FOXO3A, FXR1, GAS2, HSPA5, IFI16, IHPK3, IL1A, IL31RA, JAG2, MAL, MAP3K5, MNT, MYBL2, NOTCH1, PAK1, PAK7, PAWR, PAX3, PDCD10, PIK3R2, PLAGL2, PPP2R1B, PRLR, PSEN2, RARG, RASA1, RHOB, SGK3, SIAH2, SIRT1, SMNDC1, SOCS3, SON, SOX9, TIMP3, TNFSF9, TP53INP1, UNC5C, UNC5D, VCP | 1.03E-09 | Yes |
| hsa-miR-495 | 389 | 31 | ACTN2, AKT1, ATOH1, AVEN, BAG4, CARD6, CASP2, CTNNAL1, DDX41, DIDO1, EEF1E1, EIF2AK2, FOXO3A, IAPP, IGFBP3, MTL5, NOTCH2, OPA1, PAWR, PDCD10, PHLPP, PIM2, RTN4, SGK, SMNDC1, SOX9, STK17A, TGFB2, TMEM23, TNFRSF9, TXNDC5 | 2.17E-09 | No |
| hsa-miR-106a | 751 | 44 | ACIN1, ACVR1B, APBB2, APP, BCL2L11, BCL2L2, BCL6, BIRC4, BNIP2, BTG1, CASP6, CASP8, CDKN1A, CFLAR, COL4A3, DAPK2, DEDD, DNASE2, DNM2, E2F1, EGLN3, EP300, FASTK, FOXL2, HIF1A, INHBA, LALBA, MAP3K5, PAK7, PIK3R1, PLAGL2, PPARD, PPP2CA, PTEN, PURB, SQSTM1, STK17B, TAOK2, TAX1BP1, TIMP3, TMEM23, TNFRSF21, TOPORS, TP53INP1 | 1.54E-08 | Yes |
| hsa-miR-124a | 1333 | 62 | ACTN4, AHR, AKT1S1, ANXA5, APBB2, APPBP1, ARHGDIA, AXIN1, BAG3, BCL2L11, BCL6, BECN1, CCL2, CDKN1A, CUL5, DHCR24, DNM2, DPF2, ERN1, FXR1, GAS2, GLO1, HIP1, HTATIP2, IHPK2, JAG2, LITAF, MITF, NR4A1, PDCD10, PDCD6, PEA15, PEG3, PIK3CA, PIM1, PLAGL2, PPM1F, PPP1R13L, PROC, PURB, RAF1, RARG, RASSF5, RFFL, RHOT1, RIPK2, RP6-213H19.1, SEMA6A, SFRP5, SGK, SGPL1, SGPP1, SH3KBP1, SIRT1, SLK, SOX9, SPHK1, STK4, TMEM23, TP53INP1, TRIB3, ZDHHC16 | 1.06E-07 | Yes |
| hsa-miR-20b | 751 | 42 | ACIN1, ACVR1B, APBB2, APP, BCL2L11, BCL2L2, BCL6, BIRC4, BNIP2, BTG1, CASP6, CASP8, CASP8AP2, CDKN1A, COL4A3, DAPK2, DEDD, DNASE2, DNM2, E2F1, EGLN3, EP300, FASTK, FOXL2, HIF1A, IKIP, LALBA, MAP3K5, PAK7, PIK3R1, PLAGL2, PPP1R13L, PPP2CA, PTEN, PURB, SQSTM1, TAOK2, TIMP3, TMEM23, TNFRSF21, TOPORS, TP53INP1, | 1.18E-07 | No |
| hsa-miR-24 | 445 | 30 | ACVR1B, ADAMTSL4, ADORA1, BCL2L11, BCL2L12, BCL2L2, CARD10, CD28, CDK5, CIDEA, CLCF1, CSF2, FASLG, IHPK2, LALBA, MNT, PAK7, PCSK9, PEA15, PIM1, PIM2, PURB, RARG, RASA1, SPHK1, TMEM23, TNFRSF19, TNFRSF1B, TP53INP1, TRAF7, | 1.66E-07 | Yes |
| hsa-miR-27a | 579 | 32 | ANXA5, AXUD1, BAG2, BIRC4, BNIP3L, BTG1, CDK5R1, CFDP1, EI24, FAIM, FOXO1A, HIP1, IFNG, IGF1, IL10, JMY, MITF, MLH1, NEK6, NGFR, NGFRAP1, PHB, PLAGL2, PRKRA, RASSF5, RYBP, SAP30BP, SEMA6A, SERINC3, SFRP1, TMEM23, ZNF443 | 4.86E-06 | Yes |
| hsa-miR-195 | 766 | 38 | ALOX12, APP, ARHGDIA, BAG5, BCAP29, BCL2, BCL2L2, BDNF, BFAR, CARD10, CBX4, CD28, CD40, CDK5R1, CUL2, DEDD, MNT, PAK7, PCBP4, PDCD4, PIM1, PLAGL1, PPP2R1A, PTH, PURB, RAD9A, RAF1, RASSF5, RIPK2, RYBP, SEMA6A, SGK, SIAH1, SNCA, SNRK, SON, UBE4B, ZDHHC16, | 8.00E-06 | Yes |
| hsa-miR-182 | 485 | 28 | ACVR1, ARHGDIA, BAG4, BCL2, BCL2L12, BDNF, CFL1, CUL5, ELMO1, EP300, FOXO3A, FXR1, MITF, NCKAP1, NPM1, PIK3R1, PRKCE, RARG, RASA1, RNF130, RRAGA, RTN4, SERPINB2, TMEM23, TOPORS, TP53INP1, UNC5D, ZAK | 8.50E-06 | No |
| hsa-miR-153 | 464 | 27 | ACTN4, ACVR1B, APBB2, APP, ATF5, ATG5, BCL2, CIB1, FOXO1A, GRIK2, IGF1R, LCK, MAP3K7, MITF, POU4F1, PRKAA1, RASA1, RTN4, SGK3, SH3KBP1, SIRT1, SNCA, SOCS2, SPHK2, TESK2, TIAL1, TP53INP1 | 1.06E-05 | Yes |
| hsa-miR-200a | 634 | 33 | APBB2, ATF5, BAG2, CROP, CUL3, FOXO3A, HGF, IL1A, MAL, MAP3K7, MSH2, NME1, PAX3, PDCD4, PERP, PHLPP, PLAGL2, POU4F1, PPP2CA, PRKCE, SEMA6A, SIAH1, SIRT1, SON, STAT5A, STAT5B, TESK2, TGFB2, TIAL1, TP53INP1, TXNL1, UBE4B, UNC5C | 1.20E-05 | Yes |
| hsa-miR-141 | 413 | 24 | ACVR1B, ALB, APBB2, BAG2, BCL10, DCC, EBAG9, FOXO3A, HMGB1, IL1A, NME1, PAK1, PDCD4, PERP, PHLPP, PPP2CA, PTK2B, SIAH1, SMNDC1, SNCA, SON, STAT5A, TGFB2, TP53INP1 | 3.32E-05 | Yes |
| hsa-miR-19b | 715 | 34 | ACTN1, BAG5, BCL3, CASP8, CSE1L, CUL5, DLX1, FASTK, GULP1, HIF1A, HIP1, IGF1, IGFBP3, MAEA, MAP3K1, NGFRAP1, PIK3CA, PRKAA1, PTK2B, RAF1, RHOB, RYBP, SGK, SGPP1, SLC25A6, SOCS3, SPHK2, SULF1, TESK2, TIA1, TIMP3, TNFRSF12A, TP53INP1, TP73L | 5.53E-05 | Yes |
| hsa-miR-518e | 312 | 19 | ACTN4, ALS2CR2, ANXA5, BNIP3, CD5, CDC2L2, DFFB, FGFR1, FXR1, IFNG, MAPK1, PIM1, PML, PPP1R13B, RRAGC, SON, TRAIP, TRIAP1, VCP | 0.000116642 | No |
| hsa-miR-101 | 627 | 30 | APP, BAG1, BUB1B, CD28, CDK5R1, CUL3, EBAG9, EDAR, EIF2AK3, FOXO1A, ING4, MAL, NCR1, NGFRAP1, PAX3, PLAGL1, PPP1R13B, PRKAA1, PRKCE, PURA, PURB, SEMA6A, SFRP1, SGK, SGPL1, SOCS2, SOX9, STAMBP, STK4, TP73L | 0.000134035 | No |
| hsa-miR-183 | 428 | 23 | AXUD1, BCL6, BNIP3L, BTG1, CD3E, CDK5R1, CTSB, DAP, EI24, FOXO1A, LCK, NOTCH1, PDCD11, PDCD4, PLAGL2, PPP2CA, PRKCA, PSEN2, PURA, RHOB, TP53BP2, UNC13B, VDAC1 | 1.54E-04 | Yes |
| hsa-miR-519c | 428 | 23 | ACVR1, ANXA4, AVEN, BAG3, BECN1, BIRC6, CAMK1D, DEDD, FASLG, FASTK, GLO1, IKIP, INHBA, NFKBIA, PTH, TAX1BP1, TEGT, TMEM23, TNFRSF10D, TNFSF18, TOPORS, TPT1, VCP | 0.000154374 | No |
| hsa-miR-181c | 824 | 36 | AHR, APIP, ASAH2, ATG5, BCL2, BCL2L11, BCL6, BCLAF1, CBX4, CCAR1, CEBPG, CECR2, CUL3, DNAJA3, GRIK2, HMGB1, HSPA5, IL1A, MAEA, MAP3K10, MGMT, NOTCH2, PAK7, PAWR, PDCD4, PHLDA1, PRKCE, PURB, RAD21, RNF34, SIRT1, SPP1, TIMP3, TNFRSF11B, TUBB, UNC5B | 1.82E-04 | Yes |
| hsa-miR-107 | 436 | 23 | AATF, ACVR1, APP, BCL2L2, BDNF, CFL1, CIAPIN1, ICEBERG, IHPK3, MAP3K7, NME5, NOTCH2, PDCD10, PIK3R1, PRKCE, PTH, PURB, RAD21, RASSF5, RYBP, SNRK, THY1, VCP | 2.02E-04 | Yes |
| hsa-miR-200b | 737 | 33 | ALS2CR2, APBB2, ARHGDIA, BAG2, BCAP29, BIRC4, CBX4, CROP, EP300, ERN1, FOXO3A, HGF, IL1A, MLH1, MYC, NPM1, NTF3, PAK7, PDCD10, PERP, PPM1F, PPP2CA, PRKCA, PROK2, RHOT1, SIAH1, SLK, STAT5A, SULF1, TIAL1, TNFRSF11B, TUBB, TUBB2C | 2.15E-04 | Yes |
| hsa-miR-29a | 530 | 26 | ANGPTL4, BAK1, BCL2L7P1, CD40, DAXX, DPF1, DUSP22, ELMO2, FEM1B, IFNG, MCL1, MSH6, MYBL2, POLB, PPP1R13B, PPP2CA, PRKRA, PTEN, SGK, TNFRSF1A, TNFRSF9, TRAF4, UACA, UBE4B, UNC13B, ZNF346 | 2.51E-04 | Yes |
| hsa-miR-142-3P | 364 | 20 | AKT1S1, BCL2L1, BIRC3, BUB1B, CRTAM, FOXO1A, HDAC1, HMGB1, IER3, IL6, PPT1, PRKDC, PRLR, PURB, RARG, RNF7, SGK, TESK2, TMEM23, TNFSF18 | 3.05E-04 | Yes |
| hsa-miR-520h | 238 | 15 | ATF5, BRE, CASP6, CDKN2A, CFLAR, DNASE2, FASTK, HBXIP, HIF1A, IFT57, LALBA, PDIA3, STK17A, TAX1BP1, VHL | 0.000412436 | No |
Table 3.
Top 25 most significant p53-miRs that target known cell proliferation genes (n = 712). 22/25 of these have been reported in literature as differentially regulated in various cancerous tissues or cancer cell lines (see last column). The p-values represent the probability of a p53-miR targeting a cell proliferation gene purely by chance.
| miRNA | Total Number of Predicted Targets | Cell Proliferation Genes (n = 712) | Genes | p-value | Cancer miRNA |
| hsa-miR-23a | 656 | 44 | ALDH1A2, BCL11B, CBFA2T2, CBFA2T3, CDC6, CDK5R1, CFDP1, CSE1L, CTNNBIP1, CUL3, CXCL5, ELF4, ELF5, EPS15, GPC4, HDGFRP3, HOXA3, IHH, IL11, IL6R, IRF2, KITLG, LRP5, MAPRE1, MDK, MET, MLLT7, NAP1L1, NOTCH2, NPM1, NR6A1, ODZ1, OPRM1, PRKRIR, PTK2B, RAP1B, SMAD3, SPOCK1, STAT5B, TGFA, TOB2, TRIB1, TUSC2, ZAK | 8.42E-10 | Yes |
| hsa-miR-92 | 614 | 41 | ADM, AGGF1, APRIN, BMPR2, BRCA1, BTG2, CBFA2T3, CDC27, CDK6, CDKN1C, CTNNBIP1, CUL3, CXCL5, DERL2, DLG5, EDG1, EPS8, GDF11, HHIP, IGF1R, INSIG1, IRS2, KLF4, LBX1, MITF, NCK2, NF1, NOTCH1, NOX4, PAFAH1B1, PAX3, PCAF, RAP1B, RBM9, SPHK2, TBX3, TCFL5, TOB1, TOB2, UTP20, ZAK | 3.64E-09 | Yes |
| hsa-miR-32 | 630 | 39 | ADM, AGGF1, APRIN, BMPR2, BTG2, BTLA, CBFA2T3, CDK6, CDKN1C, CHRM5, CTNNBIP1, CUL3, CXCL5, EDD1, EDG1, EPS8, EVI5, GDF11, HHIP, INSIG1, IRS2, KLF4, LBX1, MITF, NCK2, NOTCH1, NOX4, PAFAH1B1, PAX3, PCAF, RAP1B, RBM9, SPHK2, TBX3, TCFL5, TOB1, TOB2, TTK, ZAK | 6.67E-08 | Yes |
| hsa-miR-200b | 737 | 42 | APRIN, BAP1, BCL11B, BHLHB3, CD274, CDC25B, CNOT8, CNTFR, CTBP2, EDNRA, EPS8, ETS1, EVI5, GAB1, GLI3, HDAC4, HELLS, HGF, IL1A, KHDRBS1, KLF10, KLF4, LAMC1, LRP1, LRPAP1, MAFG, MAPRE1, MXD4, MYC, NCK2, NDN, NPM1, PCAF, POU3F2, PROK2, PTHLH, RAP1B, SESN1, SHC1, STAT5A, SUZ12, TOB1 | 2.02E-07 | Yes |
| hsa-miR-200c | 751 | 42 | APRIN, BAP1, BCL11B, BHLHB3, BIN1, CD274, CNOT8, CNTFR, CTBP2, EDNRA, EPS8, ETS1, EVI5, GAB1, GLI3, GNAT1, HDAC4, HELLS, HGF, KHDRBS1, KLF10, KLF4, LAMC1, LRP1, LRPAP1, MAFG, MAPRE1, MXD4, MYC, NCK2, NDN, NPM1, PCAF, POU3F2, PROK2, PTHLH, RAP1B, SCG2, SESN1, SHC1, STAT5A, TOB1 | 3.33E-07 | Yes |
| hsa-miR-195 | 766 | 42 | ALOX12, AXIN2, BAI1, BDNF, BHLHB3, BIN1, BTG2, CBFA2T3, CD164, CD28, CD40, CDC25A, CDC27, CDK5R1, CHEK1, CUL2, DNAJA2, EFNB1, FGF2, FGF7, FOSL1, HDGF, HOXA3, LAMC1, MAPRE1, MNT, MTCP1, NF2, PAFAH1B1, PDAP1, PIM1, POU3F2, PPAP2A, PPM1D, PRDM4, PRKCD, PTHLH, PURB, RAF1, RARRES1, SESN1, SPEG | 5.60E-07 | Yes |
| hsa-miR-10a | 230 | 20 | BCAR1, BCL6, BDNF, CDK4, CTNNBIP1, FLT1, HDAC4, HOXA3, ID4, IFNAR2, JARID2, KLF11, KLF4, MAPRE1, PAFAH1B1, PURB, SERTAD1, SHC1, TRAIP, ZMYND11 | 7.09E-07 | Yes |
| hsa-miR-10b | 237 | 20 | BCAR1, BCL6, BDNF, CDK4, CNOT8, CTNNBIP1, FLT1, HDAC4, HOXA3, ID4, IFNAR2, JARID2, KLF11, KLF4, MAPRE1, PAFAH1B1, PURB, SHC1, TRAIP, ZMYND11 | 1.14E-06 | Yes |
| hsa-miR-24 | 445 | 29 | ADM, CD164, CD28, CDK5, CDKN1B, CDV3, CNTFR, CSF2, CSK, DCTN2, EDG1, FLT1, GPC4, ING5, INSIG1, MAFG, MNT, MXI1, NUDC, PDGFRA, PIM1, PIM2, PURB, RAP1B, SERTAD1, SESN1, SPHK1, TOB2, ZNF259 | 1.16E-06 | Yes |
| hsa-miR-203 | 505 | 31 | ALDH1A2, BCL11B, CASP3, CNTFR, DLG5, EDD1, EDN1, EDNRA, FGG, GLI3, HBEGF, ID4, INSIG1, IRS2, KHDRBS1, MAPRE1, NUMBL, PDAP1, PDGFRA, PPM1D, PRKRIR, PURB, RERG, SMAD1, SMAD3, TBX3, TGFB2, TNFRSF8, TUSC2, USP8, ZMYND11 | 1.77E-06 | Yes |
| hsa-miR-124a | 1333 | 60 | ANXA7, AR, BCL11B, BCL6, BTG2, CAPN1, CAV1, CD164, CD276, CDK6, CDKN1A, CUL5, DLG5, DLG7, EFNB1, EMP2, EPS8, ERF, ETS1, EVI1, EVI5, FGFR1OP, GAS1, GNAI2, GPC4, HDAC4, IL9R, ING1, JAG1, JAG2, KLF4, LAMC1, MAP3K11, MAPRE1, MDK, MITF, MXD4, NCK2, NRP1, OVOL2, PAFAH1B1, PBEF1, PDZK1, PGF, PIM1, PRKD1, PTHLH, PURB, QSCN6, RAF1, RAP1B, RARRES1, RHOG, SHC1, SOX9, SPHK1, SPOCK1, TAL1, TOB2, UHRF1 | 1.82E-06 | Yes |
| hsa-miR-106a | 751 | 40 | BCL11B, BCL6, BHLHB3, BMPR2, BTG1, BTG2, BTG3, CDKN1A, COL4A3, CSF1, DERL2, E2F1, EBI3, EDD1, EDG1, EFNB1, EREG, FLT1, FZD3, GAB1, HDAC4, KLF11, LIF, MAP3K11, MAPRE1, PAFAH1B1, PCAF, PDGFRA, PPARD, PTEN, PTHLH, PURB, RB1, RBBP7, TAL1, TBX3, TGFB1, TOPORS, TSG101, TUSC2, | 2.13E-06 | Yes |
| hsa-miR-142-3P | 364 | 25 | BTG4, BUB1B, CDC14A, CDC25C, CRTAM, EDG3, EHF, ERG, FGFR1OP, FNTB, FOXO1A, FRK, GRN, IL6, IL6ST, KHDRBS1, KLF10, MAP3K11, MLLT7, NUMBL, PCAF, PURB, SKP2, TNFRSF13C, UTP20, | 2.50E-06 | Yes |
| hsa-miR-33 | 312 | 22 | APRIN, BCL11B, BMPR2, CD40, CDK6, CKS2, DOCK2, FGA, FGFBP1, HIPK2, HOXC10, IRS2, LGI1, LMO1, NPM1, NPY, PDGFRA, PIM1, PURB, RB1, TSG101, UBE2V2, | 6.53E-06 | No |
| hsa-miR-372 | 378 | 24 | BCL6, BTG1, CDCA7, CDK2, CXCR4, DERL2, DNAJA2, E2F1, ERBB4, FGF9, FRK, IRF2, MAP3K11, MNT, PCAF, PRDM4, PURB, RBBP7, TAL1, TOB2, TUSC2, UCHL1, ZFP36L2, ZMYND11, | 1.47E-05 | Yes |
| hsa-miR-101 | 627 | 33 | BUB1B, CALCRL, CBFA2T2, CD28, CDC14A, CDC27, CDH5, CDK5R1, CDK6, CSRP2, CUL3, DLG5, ELN, EMP1, FGA, FOXO1A, GLI3, GNB1, ING4, PAX3, PBEF1, PPBP, PPP1R8, PURA, PURB, RAP1B, RBBP7, SOX9, STAMBP, TAL1, TCF19, TGFBR1, TRIB1, | 2.09E-05 | No |
| hsa-miR-19b | 715 | 36 | ANXA7, BMPR2, CD164, CDC2L5, CHERP, CLK1, CNTFR, CSE1L, CUL5, DLG5, DNAJA2, EDG1, EPS15, ERBB4, EREG, FOSL1, GCG, GRN, HDAC4, HHIP, HPRT1, ID4, IGF1, IMPDH1, KLF10, LIF, PCAF, POU3F2, PTK2B, RAF1, RAP1B, SMARCA2, SPHK2, ST8SIA1, SUZ12, ZMYND11 | 2.29E-05 | Yes |
| hsa-miR-196a | 243 | 18 | ACSL6, ANXA1, ATP6AP1, CASP3, CBFA2T3, CCL23, CDKN1B, ELF4, ERG, IGFBP7, ING5, MYC, NFKBIA, PA2G4, PDGFRA, SSR1, UHRF2, ZMYND11 | 2.30E-05 | Yes |
| hsa-miR-26b | 633 | 33 | ADM, BCL6, BTG1, CDC27, CDC2L5, CDK6, CKS2, DLG5, DNAJA2, ENPEP, EPS15, ERBB4, HGF, HIPK2, HOXD13, IGF1, IL6, JAG1, KLF10, KLF4, LIF, MITF, MXI1, PAWR, PBEF1, PDGFRA, PIM1, PRKCD, PRKCQ, PTEN, SMAD1, SMAD4, TOB1, | 2.52E-05 | Yes |
| hsa-miR-196b | 245 | 18 | ACSL6, ANXA1, ATP6AP1, CBFA2T3, CCL23, CDKN1B, ELF4, ERG, FOXN1, IGFBP7, ING5, MYC, NF2, NFKBIA, PA2G4, PDGFRA, SSR1, ZMYND11 | 2.57E-05 | Yes |
| hsa-miR-9 | 737 | 36 | ARHGEF2, BCL6, BIN1, BTG2, CBFA2T2, CNTFR, COL18A1, CUL4A, CXCR4, EDD1, ERG, ETS1, FGF5, FOXO1A, HDAC4, HES1, ID4, INSIG1, KITLG, LEPRE1, LIFR, NAP1L1, NOTCH2, NOX4, NRP1, ODZ1, PDGFC, PMP22, POU3F2, PPARD, RAP1B, RBM9, SHC1, TBC1D8, TGFBI, UHRF1 | 4.29E-05 | Yes |
| hsa-miR-29b | 541 | 29 | BAX, BCL11B, CAV2, CD40, CDC7, CREG1, DUSP22, EPS15, FRAT2, FSCN1, GAB1, HDAC4, INSIG1, JARID2, LAMC1, MAFG, NASP, PDGFB, PDGFC, PINX1, PMP22, PPM1D, PRKRA, PTEN, SETDB1, SMARCA2, SPEG, TNFRSF9, UHRF2 | 4.71E-05 | Yes |
| hsa-miR-200a | 634 | 32 | APRIN, ATP8A2, BAP1, BIN1, CD274, CDC14A, CDC25A, CDC25B, CDC2L5, CDK6, CSF3, CTBP2, CTNNB1, CUL3, FLT1, GAB1, HDAC4, HGF, IL1A, IRS2, JAG1, NME1, NRP1, PAX3, PBEF1, POU3F2, PRKD1, SCG2, STAT4, STAT5A, STAT5B, TGFB2 | 6.15E-05 | Yes |
| hsa-miR-34a | 666 | 33 | AREG, ARX, BCL11B, BCL6, CBFA2T3, CD28, CD3E, CDC25A, CSF1R, EFNB1, ENPP7, FOSL1, GFER, GNAI2, IL6R, JAG1, KITLG, KLF4, LGI1, MET, MOV10L1, NOTCH1, NOTCH2, NUMBL, PDGFRA, PGF, PRKD1, PURB, SPEG, TGFBI, TOB2, TOPORS, UHRF2 | 6.76E-05 | Yes |
| hsa-miR-181c | 824 | 38 | ABI1, ADAMTS1, ADM, ARHGEF2, BCL6, BMPR2, CALCRL, CBFA2T2, CBFA2T3, CKS1B, CUL3, EDG1, EPS15, ERF, ERG, ETS1, FLT1, IL1A, IRS2, LIF, LMO1, NOTCH2, NR6A1, PAFAH1B1, PAWR, PCAF, PDAP1, PDGFRA, PRDM4, PRKCD, PURB, RAP1B, RBBP7, TGFBI, TGFBR1, TGFBR2, USP8, VIP | 8.89E-05 | Yes |
Another interesting model is SIRT1-mir-34a-p53; SIRT1 is known to inhibit p53-mediated apoptosis by attenuation of the transcriptional activation potential of p53 primarily through deacetylation of p53 [69]. Several recent studies have shown that p53 activates mir-34a. A speculative model (Figure 4D) therefore is, in normal state mir-34a, a known p53-miR, suppresses SIRT1 (which is known to suppress p53) balancing the SIRT1-mediated transcriptional repression of p53. However, during tumorigenesis, increased levels of SIRT1 will suppress p53 triggering further suppression of p53 downstream mir-34a in a feedback loop. Examples of such models may have potential implications for cancer therapy. For example, combining DNA damage drugs (since p53 is strongly activated in response to DNA damage), SIRT1-mediated deacetylase inhibitors, and mir-34a activation (with "agomiR") may have synergistic effects in cancer therapy for maximally activating p53.
p53-miR and p53 protein interaction networks
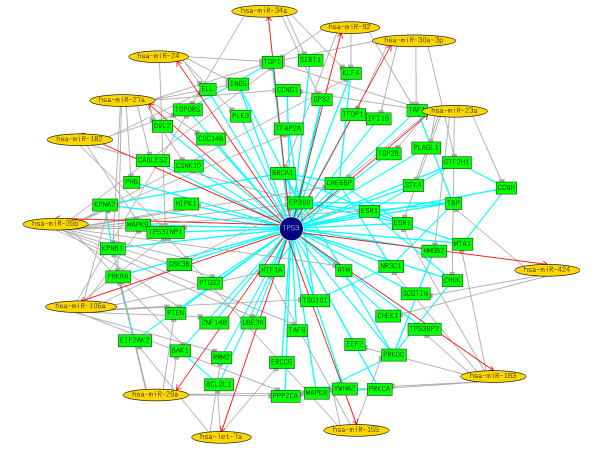
Since the functional state of a protein-protein interaction network depends on gene expression, a fundamental question is what relationships exist between protein interaction network and gene regulation [70]. Liang and Li in their recent study of evidence for global correlation between microRNA repression and protein-protein interactions reported that interacting proteins tend to share more microRNA target-site types than random pairs [70]. In the current study, using the known p53-interactants, we calculated the probability of p53-miRs regulating the p53 interactants. We mined the BioGRID database [71], and currently there are 141 known interactants of p53. Surprisingly, 114 of these 141 p53-interactants were predicted to be regulated by p53-miRs and were shown to be statistically significant (p = 1.41e-21) (see Additional File 5 for details). Using the known data of miRNAs induced or repressed following p53 activation [11], we further filtered the p53-miRs that could be associated with the p53 interactome. Figures 5 and 6 show the p53 interactome along with the putative p53-miR regulators that are shown to be induced or repressed [11] respectively following the activation of p53.
Figure 5.
p53 interactome along with the putative miR regulators that are shown to be induced following the activation of p53 [11]. miRNAs are represented by yellow ellipses, target genes are represented by green boxes. p53 is represented in the center as blue circle. Induction of the miRNAs by p53 is represented by directed red lines. Negative regulation of the target genes by miRNAs are represented by dark grey lines. The protein interactions are represented by undirected light blue lines. Networks (using force-directed layouts) were generated using aiSee [76] network visualization software.
Figure 6.
p53 interactome along with the putative miR regulators that are shown to be repressed following the activation of p53 [11]. miRNAs are represented by yellow ellipses, target genes are represented by green boxes. p53 is represented in the center as blue circle. Induction of the miRNAs by p53 is represented by directed red lines. Negative regulation of the target genes by miRNAs are represented by dark grey lines. The protein interactions are represented by undirected light blue lines. Networks (using force-directed layouts) were generated using aiSee [76] network visualization software.
Conclusion
In conclusion, our bioinformatic-based analysis yielded several putative regulatory circuits that involve p53 and microRNAs along with their target genes. Although these predicted networks need to be experimentally validated to ascertain a bona fide regulatory relationship, it is still exciting to speculate that these p53-miRs may be critical to tumorigenesis through their regulation of the p53 master regulatory network. We strongly believe that our results substantially expand the hitherto little known repertoire of miRNAs in p53 regulatory network. It can be anticipated that further characterization of our predicted p53-miRs and their target genes will improve our understanding of the underlying processes of cancer, tumorigenesis, and ultimately impact response to therapy.
Methods
Human miRNA genomic analysis
The human miRNAs analyzed in this study (474 in total) were derived from the miRNA registry release 9.2 (May 2007; Genome Assembly NCBI-36 [72]). The genomic sequences, including 10 kb flanking sequences 5' and 3' for each of the miRNAs, were downloaded using Table Browser from the UCSC Golden Path [73].
Out of a total 474 human miRNAs analyzed, 100 microRNAs were part of a cluster (2 or more miRNAs occurring together on the genome). In all, there were 100 miRNAs organized into 37 clusters (see Additional Files 6 and 7 for details). The cluster definitions were based on those defined by Yu et al [74]. The nearest neighboring gene information (gene name, distance from the nearest miRNA, orientation with respect to the host or neighboring gene) for each of the miRNA was extracted from the public database RefFlat provided by the UCSC Golden Path [73].
Screening miRNA genomic sequences for p53 binding sites
The genomic sequences (10 kb flanking region, 5' and 3') of the 474 human miRNAs were scanned for potential p53-binding motifs using the p53MH algorithm [22]. Briefly, p53-binds to a specific sequence comprised of two decamers (RRRCWWGYYY) separated by 0–13 bp [75]. Nearly all established human response element sequences (REs) have at least one mismatch in this degenerate sequence [55]. The computer algorithm p53MH identifies potential p53-binding sites by a scoring system based on the percentage of similarity to a consensus sequence. We set the parameter conditions to include the following restrictions: (i) a p53MH score ≥ 85%, and (ii) the spacer between the 10-bp motifs is ≤ 4 bp. Although p53 is known to bind to sites with a spacer as high as 13 bp [75], for the current analysis we considered 4 bp spacer as the maximum limit because majority of the validated p53 binding sites are reported to have a spacer less than 4 bp (see Additional Files 1 and 2 for the putative p53-miR distribution).
p53 upstream and downstream activators and repressors
The group of TFs that act upstream to p53 either as activators or repressors and the p53 target genes involved in specific p53 downstream effects (activator or repressor) were downloaded from the compiled and curated dataset of the p53 knowledge-base [36]. We also mined the BioGRID database [71] for all known p53 interactants.
p53-miR targets
We used MAMI [25] to download the predicted targets for the p53-miRs. MAMI has a compilation of target prediction tools, DIANA-microT (version 5/05) [26], miRanda (version 4/06) [8], TargetScanS (version 4/05) [9], miRtarget (version 3/06) [28] and PicTar (version 5/05) [27]. The miRNAs are based on miRs registry version 8.1 (5/06).
Functional annotation and prioritization of p53-miRs
To annotate the p53-miRs and to identify/prioritize p53-miRs that might have potential implications in tumorigenesis we performed functional enrichment analysis and also intersected our predictions with cancer-associated miRNAs. Specifically, we used (a) literature reported differentially expressed miRNAs in cancer; (b) miRNAs reported as induced or repressed following p53 activation; and (c) miRNA targets that were functionally enriched with biological processes like apoptosis, cell cycle, etc.
Known tumor suppressor and oncogenic microRNAs
We manually compiled the list of microRNAs that were reported in literature as differentially expressed (up- or down-regulated) in various cancers in human (Additional File 8).
miRNAs induced or repressed following p53 activation
We intersected our predicted p53-miRs with the data from Tarasov et al [11] to identify potential p53 induced/repressed miRNAs. Following a genome-wide screen for microRNAs regulated by the transcription factor encoded by the p53 tumor suppressor gene, the authors reported that after p53-activation the abundance of thirty-four miRNAs was significantly increased, whereas sixteen miRNAs were suppressed [11].
p53-miR target genes – Functional enrichment
Functional over-representation analysis was performed to objectively identify biological processes potentially affected by p53-miR target genes. Specifically, the percentage of p53-miR target genes with a given gene ontology (GO) or pathway annotation was compared with the percentage of all miRNA-target genes genome-wide with the same annotation. A significant p value (p < 0.05) indicates that the observed percentage of p53-miR target genes with a given annotation could not likely occur by chance given the frequency of miRNA-target genes genome-wide with the same annotation. For functional enrichment analysis, we used DAVID [62] and ToppGene [63].
p53-miR and p53-interactant gene networks
p53-miR and p53-interactant gene networks were generated using aiSee [76] with force-directed layouts. The GDL (Graphics Description Language) files, generated by a locally-implemented JAVA GDL library, were used as input to aiSee to create SVG (Scalable Vector Graphics) network graphs. The p53-related interactions were downloaded from BioGRID [71].
Statistical methods
Fisher's exact test was used to calculate a p-value estimating the probability that a particular category of miRNAs is associated with a particular pattern or cluster of miRs more than would be expected by chance. Fisher's exact test was performed online at the MATFORSK [77]. For testing the functional enrichment of p53-miR target genes, DAVID [62] and ToppGene server [63] were used. For statistical analysis, the programming language and statistical environment R [78] were used.
Authors' contributions
AS, VK and AJ conceived the study design which was coordinated by AJ. AS and VK designed and implemented the p53-miR discovery and prioritization approaches and along with AJ participated in the analysis and interpretation of results. JC carried out the p53 interactome data analysis. AJ drafted the manuscript. All the authors have read and approved the final manuscript
Supplementary Material
Systematic break-down of the putative p53-regulated miRNAs (p53-miRs). Schematic classification of putative p53 sites and p53-miRs based on the miRNA types (intragenic or intergenic).
Genomic coordinates of putative p53 sites within 10 kb flanking regions of human miRNAs. Chromosome cooridnates (based on Human Mar. 2006 – hg18 – assembly) of putative p53 sites in the flanking 10 kb regions of human microRNAs.
Putative miRNAs regulating known 23 upstream regulators and 48 downstream TFs of p53. For the known 23 upstream regulators and 48 downstream TFs of p53 (based on p53 Knowledgebase; http://p53.bii.a-star.edu.sg), putative miRs regulating them were extracted using MAMI server and the database http://mami.med.harvard.edu/.
Functional enrichment (based on GO and pathways) in target genes of 143 p53-miRs. Results of functional over-representation analysis, performed to objectively identify biological processes potentially affected by p53-miR target genes.
Details of Fisher's exact test used to test whether p53-miRs tend to target p53-interactants. Fisher's exact test was used to calculate the probability of p53-miRs regulating the p53 interactants (downloaded from the BioGRID database).
Systematic break-down of the 474 human miRNAs based on their genomic location. Classification of the known 474 human miRNAs based on their genomic location (intergenic or intragenic/intronic).
Details of the 474 human miRNAs analyzed. Additional details of genomic locations of the 474 human miRNAs analyzed (whether part of a cluster, i.e., 2 or more miRNAs occurring together on the genome; or located in the intergenic or intragenic/intronic regions of known genes).
Manually compiled list of miRNAs that are reported in the literature as either up or down regulated in various human cancers or cancer cell lines. This file has a list of miRNAs that are reported in the literature as either up or down regulated in various human cancers or cancer cell lines. These "cancer-associated" miRNAs were used to prioritize putative p53-miR targets.
Acknowledgments
Acknowledgements
This study was supported by the State of Ohio Computational Medicine Center (ODD TECH 04–042) and NCI UO1 CA84291-07 (Mouse Models of Human Cancer Consortium). The authors also acknowledge the support of Drs. Bruce Aronow (Division of Biomedical Informatics, Cincinnati Children's Hospital Medical Center) and Raj Bhatnagar (Department of Computer Science, University of Cincinnati).
Contributor Information
Amit U Sinha, Email: sinhaam@ececs.uc.edu.
Vivek Kaimal, Email: Vivek.Kaimal@cchmc.org.
Jing Chen, Email: Jing.Chen@cchmc.org.
Anil G Jegga, Email: Anil.Jegga@cchmc.org.
References
- Heinrichs S, Deppert W. Apoptosis or growth arrest: modulation of the cellular response to p53 by proliferative signals. Oncogene. 2003;22:555–571. doi: 10.1038/sj.onc.1206138. [DOI] [PubMed] [Google Scholar]
- Sionov RV, Haupt Y. The cellular response to p53: the decision between life and death. Oncogene. 1999;18:6145–6157. doi: 10.1038/sj.onc.1203130. [DOI] [PubMed] [Google Scholar]
- Dickins RA, Hemann MT, Zilfou JT, Simpson DR, Ibarra I, Hannon GJ, Lowe SW. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat Genet. 2005;37:1289–1295. doi: 10.1038/ng1651. [DOI] [PubMed] [Google Scholar]
- Spurgers KB, Gold DL, Coombes KR, Bohnenstiehl NL, Mullins B, Meyn RE, Logothetis CJ, McDonnell TJ. Identification of cell cycle regulatory genes as principal targets of p53-mediated transcriptional repression. J Biol Chem. 2006;281:25134–25142. doi: 10.1074/jbc.M513901200. [DOI] [PubMed] [Google Scholar]
- Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113:673–676. doi: 10.1016/S0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S. miRBase: the microRNA sequence database. Methods Mol Biol. 2006;342:129–138. doi: 10.1385/1-59745-123-1:129. [DOI] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/S0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Harris SL, Levine AJ. The p53 pathway: positive and negative feedback loops. Oncogene. 2005;24:2899–2908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- Tarasov V, Jung P, Verdoodt B, Lodygin D, Epanchintsev A, Menssen A, Meister G, Hermeking H. Differential Regulation of microRNAs by p53 Revealed by Massively Parallel Sequencing: miR-34a is a p53 Target That Induces Apoptosis and G(1)-arrest. Cell Cycle. 2007;6 doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, Arking DE, Beer MA, Maitra A, Mendell JT. Transactivation of miR-34a by p53 Broadly Influences Gene Expression and Promotes Apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, Bentwich Z, Oren M. Transcriptional Activation of miR-34a Contributes to p53-Mediated Apoptosis. Mol Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ. A microRNA component of the p53 tumour suppressor network. Nature. 2007. [DOI] [PMC free article] [PubMed]
- Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, Zhai Y, Giordano TJ, Qin ZS, Moore BB, Macdougald OA, Cho KR, Fearon ER. p53-Mediated Activation of miRNA34 Candidate Tumor-Suppressor Genes. Curr Biol. 2007 doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM. MicroRNAs as tumor suppressors. Nat Genet. 2007;39:582–583. doi: 10.1038/ng0507-582. [DOI] [PubMed] [Google Scholar]
- Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- Wu W, Sun M, Zou GM, Chen J. MicroRNA and cancer: Current status and prospective. Int J Cancer. 2007;120:953–960. doi: 10.1002/ijc.22454. [DOI] [PubMed] [Google Scholar]
- Voorhoeve PM, le Sage C, Schrier M, Gillis AJ, Stoop H, Nagel R, Liu YP, van Duijse J, Drost J, Griekspoor A, Zlotorynski E, Yabuta N, De Vita G, Nojima H, Looijenga LH, Agami R. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124:1169–1181. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]
- Yang Y, Tantoso E, Chua GH, Yeo ZX, Ng FS, Wong ST, Chung CW, Li KB. In silico analysis of p53 using the P53 Knowledgebase: mutations, polymorphisms, microRNAs and pathways. In Silico Biol. 2007;7:61–75. [PubMed] [Google Scholar]
- Hoh J, Jin S, Parrado T, Edington J, Levine AJ, Ott J. The p53MH algorithm and its application in detecting p53-responsive genes. Proc Natl Acad Sci U S A. 2002;99:8467–8472. doi: 10.1073/pnas.132268899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini HK, Griffiths-Jones S, Enright AJ. Genomic analysis of human microRNA transcripts. Proc Natl Acad Sci U S A. 2007;104:17719–17724. doi: 10.1073/pnas.0703890104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAMI http://mami.med.harvard.edu/
- Kiriakidou M, Nelson PT, Kouranov A, Fitziev P, Bouyioukos C, Mourelatos Z, Hatzigeorgiou A. A combined computational-experimental approach predicts human microRNA targets. Genes Dev. 2004;18:1165–1178. doi: 10.1101/gad.1184704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Wang X, Wang X. Systematic identification of microRNA functions by combining target prediction and expression profiling. Nucleic Acids Res. 2006;34:1646–1652. doi: 10.1093/nar/gkl068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentwich I. Prediction and validation of microRNAs and their targets. FEBS Lett. 2005;579:5904–5910. doi: 10.1016/j.febslet.2005.09.040. [DOI] [PubMed] [Google Scholar]
- Arias AM, Hayward P. Filtering transcriptional noise during development: concepts and mechanisms. Nat Rev Genet. 2006;7:34–44. doi: 10.1038/nrg1750. [DOI] [PubMed] [Google Scholar]
- Milo R, Shen-Orr S, Itzkovitz S, Kashtan N, Chklovskii D, Alon U. Network motifs: simple building blocks of complex networks. Science. 2002;298:824–827. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- Cui Q, Yu Z, Purisima EO, Wang E. Principles of microRNA regulation of a human cellular signaling network. Mol Syst Biol. 2006;2:46. doi: 10.1038/msb4100089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Q, Yu Z, Pan Y, Purisima EO, Wang E. MicroRNAs preferentially target the genes with high transcriptional regulation complexity. Biochem Biophys Res Commun. 2007;352:733–738. doi: 10.1016/j.bbrc.2006.11.080. [DOI] [PubMed] [Google Scholar]
- Shalgi R, Lieber D, Oren M, Pilpel Y. Global and Local Architecture of the Mammalian microRNA-Transcription Factor Regulatory Network. PLoS Comput Biol. 2007;3:e131. doi: 10.1371/journal.pcbi.0030131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang J, Zhu J, van Oudenaarden A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol Cell. 2007;26:753–767. doi: 10.1016/j.molcel.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim YP, Lim TT, Chan YL, Song AC, Yeo BH, Vojtesek B, Coomber D, Rajagopal G, Lane D. The p53 knowledgebase: an integrated information resource for p53 research. Oncogene. 2007;26:1517–1521. doi: 10.1038/sj.onc.1209952. [DOI] [PubMed] [Google Scholar]
- Hua Z, Lv Q, Ye W, Wong CK, Cai G, Gu D, Ji Y, Zhao C, Wang J, Yang BB, Zhang Y. MiRNA-Directed Regulation of VEGF and Other Angiogenic Factors under Hypoxia. PLoS ONE. 2006;1:e116. doi: 10.1371/journal.pone.0000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethupathy P, Corda B, Hatzigeorgiou AG. TarBase: A comprehensive database of experimentally supported animal microRNA targets. Rna. 2006;12:192–197. doi: 10.1261/rna.2239606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im HJ, Pittelkow MR, Kumar R. Divergent regulation of the growth-promoting gene IEX-1 by the p53 tumor suppressor and Sp1. J Biol Chem. 2002;277:14612–14621. doi: 10.1074/jbc.M109414200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- Sood P, Krek A, Zavolan M, Macino G, Rajewsky N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc Natl Acad Sci U S A. 2006;103:2746–2751. doi: 10.1073/pnas.0511045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Beach D. Cyclin G is a transcriptional target of the p53 tumor suppressor protein. Embo J. 1994;13:4816–4822. doi: 10.1002/j.1460-2075.1994.tb06807.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramantieri L, Ferracin M, Fornari F, Veronese A, Sabbioni S, Liu CG, Calin GA, Giovannini C, Ferrazzi E, Grazi GL, Croce CM, Bolondi L, Negrini M. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 2007;67:6092–6099. doi: 10.1158/0008-5472.CAN-06-4607. [DOI] [PubMed] [Google Scholar]
- Ohtsuka T, Ryu H, Minamishima YA, Ryo A, Lee SW. Modulation of p53 and p73 levels by cyclin G: implication of a negative feedback regulation. Oncogene. 2003;22:1678–1687. doi: 10.1038/sj.onc.1206306. [DOI] [PubMed] [Google Scholar]
- Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, Zeitlinger J, Jennings EG, Murray HL, Gordon DB, Ren B, Wyrick JJ, Tagne JB, Volkert TL, Fraenkel E, Gifford DK, Young RA. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- Goldbeter A, Koshland DE., Jr. Ultrasensitivity in biochemical systems controlled by covalent modification. Interplay between zero-order and multistep effects. J Biol Chem. 1984;259:14441–14447. [PubMed] [Google Scholar]
- Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, Jones PA. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- Phan RT, Dalla-Favera R. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature. 2004;432:635–639. doi: 10.1038/nature03147. [DOI] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiio Y, Yamamoto T, Yamaguchi N. Negative regulation of Rb expression by the p53 gene product. Proc Natl Acad Sci U S A. 1992;89:5206–5210. doi: 10.1073/pnas.89.12.5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho J, Benchimol S. Transcriptional repression mediated by the p53 tumour suppressor. Cell Death Differ. 2003;10:404–408. doi: 10.1038/sj.cdd.4401191. [DOI] [PubMed] [Google Scholar]
- Menendez D, Inga A, Jordan JJ, Resnick MA. Changing the p53 master regulatory network: ELEMENTary, my dear Mr Watson. Oncogene. 2007;26:2191–2201. doi: 10.1038/sj.onc.1210277. [DOI] [PubMed] [Google Scholar]
- Ho JS, Ma W, Mao DY, Benchimol S. p53-Dependent transcriptional repression of c-myc is required for G1 cell cycle arrest. Mol Cell Biol. 2005;25:7423–7431. doi: 10.1128/MCB.25.17.7423-7431.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai W, Comai L. Repression of RNA polymerase I transcription by the tumor suppressor p53. Mol Cell Biol. 2000;20:5930–5938. doi: 10.1128/MCB.20.16.5930-5938.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, El-Gewely MR. Differentially expressed downstream genes in cells with normal or mutated p53. Oncol Res. 2003;13:429–436. doi: 10.3727/096504003108748456. [DOI] [PubMed] [Google Scholar]
- Kang JH, Kim SJ, Noh DY, Park IA, Choe KJ, Yoo OJ, Kang HS. Methylation in the p53 promoter is a supplementary route to breast carcinogenesis: correlation between CpG methylation in the p53 promoter and the mutation of the p53 gene in the progression from ductal carcinoma in situ to invasive ductal carcinoma. Lab Invest. 2001;81:573–579. doi: 10.1038/labinvest.3780266. [DOI] [PubMed] [Google Scholar]
- Raman V, Martensen SA, Reisman D, Evron E, Odenwald WF, Jaffee E, Marks J, Sukumar S. Compromised HOXA5 function can limit p53 expression in human breast tumours. Nature. 2000;405:974–978. doi: 10.1038/35016125. [DOI] [PubMed] [Google Scholar]
- Bandres E, Cubedo E, Agirre X, Malumbres R, Zarate R, Ramirez N, Abajo A, Navarro A, Moreno I, Monzo M, Garcia-Foncillas J. Identification by Real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol Cancer. 2006;5:29. doi: 10.1186/1476-4598-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman BT, Huang DW, Tan Q, Guo Y, Bour S, Liu D, Stephens R, Baseler MW, Lane HC, Lempicki RA. DAVID Knowledgebase: a gene-centered database integrating heterogeneous gene annotation resources to facilitate high-throughput gene functional analysis. BMC Bioinformatics. 2007;8:426. doi: 10.1186/1471-2105-8-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Xu H, Aronow BJ, Jegga AG. Improved human disease candidate gene prioritization using mouse phenotype. BMC Bioinformatics. 2007;8:392. doi: 10.1186/1471-2105-8-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaccorsi L, Nosi D, Muratori M, Formigli L, Forti G, Baldi E. Altered endocytosis of epidermal growth factor receptor in androgen receptor positive prostate cancer cell lines. J Mol Endocrinol. 2007;38:51–66. doi: 10.1677/jme.1.02155. [DOI] [PubMed] [Google Scholar]
- Floyd S, De Camilli P. Endocytosis proteins and cancer: a potential link? Trends Cell Biol. 1998;8:299–301. doi: 10.1016/S0962-8924(98)01316-6. [DOI] [PubMed] [Google Scholar]
- Polo S, Pece S, Di Fiore PP. Endocytosis and cancer. Curr Opin Cell Biol. 2004;16:156–161. doi: 10.1016/j.ceb.2004.02.003. [DOI] [PubMed] [Google Scholar]
- Kawasaki H, Taira K. Functional analysis of microRNAs during the retinoic acid-induced neuronal differentiation of human NT2 cells. Nucleic Acids Res Suppl. 2003:243–244. doi: 10.1093/nass/3.1.243. [DOI] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, Richardson JA, Olson EN. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci U S A. 2006;103:18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/S0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- Liang H, Li WH. MicroRNA regulation of human protein protein interaction network. Rna. 2007;13:1402–1408. doi: 10.1261/rna.634607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–9. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human miRNA registry ftp://ftp.sanger.ac.uk/pub/mirbase/sequences/CURRENT/genomes/hsa.gff
- Kuhn RM, Karolchik D, Zweig AS, Trumbower H, Thomas DJ, Thakkapallayil A, Sugnet CW, Stanke M, Smith KE, Siepel A, Rosenbloom KR, Rhead B, Raney BJ, Pohl A, Pedersen JS, Hsu F, Hinrichs AS, Harte RA, Diekhans M, Clawson H, Bejerano G, Barber GP, Baertsch R, Haussler D, Kent WJ. The UCSC genome browser database: update 2007. Nucleic Acids Res. 2007;35:D668–73. doi: 10.1093/nar/gkl928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Wang F, Yang GH, Wang FL, Ma YN, Du ZW, Zhang JW. Human microRNA clusters: genomic organization and expression profile in leukemia cell lines. Biochem Biophys Res Commun. 2006;349:59–68. doi: 10.1016/j.bbrc.2006.07.207. [DOI] [PubMed] [Google Scholar]
- el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nat Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- aiSee http://www.aisee.com
- MATFORSK http://www.langsrud.com/fisher.htm
- R programming language and statistical environment http://www.r-project.org
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Systematic break-down of the putative p53-regulated miRNAs (p53-miRs). Schematic classification of putative p53 sites and p53-miRs based on the miRNA types (intragenic or intergenic).
Genomic coordinates of putative p53 sites within 10 kb flanking regions of human miRNAs. Chromosome cooridnates (based on Human Mar. 2006 – hg18 – assembly) of putative p53 sites in the flanking 10 kb regions of human microRNAs.
Putative miRNAs regulating known 23 upstream regulators and 48 downstream TFs of p53. For the known 23 upstream regulators and 48 downstream TFs of p53 (based on p53 Knowledgebase; http://p53.bii.a-star.edu.sg), putative miRs regulating them were extracted using MAMI server and the database http://mami.med.harvard.edu/.
Functional enrichment (based on GO and pathways) in target genes of 143 p53-miRs. Results of functional over-representation analysis, performed to objectively identify biological processes potentially affected by p53-miR target genes.
Details of Fisher's exact test used to test whether p53-miRs tend to target p53-interactants. Fisher's exact test was used to calculate the probability of p53-miRs regulating the p53 interactants (downloaded from the BioGRID database).
Systematic break-down of the 474 human miRNAs based on their genomic location. Classification of the known 474 human miRNAs based on their genomic location (intergenic or intragenic/intronic).
Details of the 474 human miRNAs analyzed. Additional details of genomic locations of the 474 human miRNAs analyzed (whether part of a cluster, i.e., 2 or more miRNAs occurring together on the genome; or located in the intergenic or intragenic/intronic regions of known genes).
Manually compiled list of miRNAs that are reported in the literature as either up or down regulated in various human cancers or cancer cell lines. This file has a list of miRNAs that are reported in the literature as either up or down regulated in various human cancers or cancer cell lines. These "cancer-associated" miRNAs were used to prioritize putative p53-miR targets.