Abstract
The relative toxicity and mutagenicity of Me-lex, which selectively generates 3-methyladenine (3-MeA), is dependent on the nature of the DNA repair background. Base Excision Repair (BER) defective S. cerevisiae strains mag1 and apn1apn2 were both significantly more sensitive to Me-lex toxicity, but only the latter is significantly more prone to Me-lex induced mutagenesis. To examine the contribution of translesion synthesis (TLS) DNA polymerases in the bypass of Me-lex-induced lesions, the REV3 and REV1 genes were independently deleted in the parental yeast strain and in different DNA repair deficient derivatives: the Nucleotide Excision Repair (NER) deficient rad14, and the BER deficient mag1 or apn1apn2 strains. The strains contained an integrated ADE2 reporter gene under control of the transcription factor p53. A centromeric yeast expression vector containing the wild-type p53 cDNA was treated in vitro with increasing concentrations of Me-lex and transformed into the different yeast strains. The toxicity of Me-lex induced lesions was evaluated based on the plasmid transformation efficiency compared to the untreated vector, while Me-lex mutagenicity was assessed using the p53 reporter assay. In the present study, we demonstrate that disruption of Polζ (through deletion of its catalytic subunit coded by REV3) or Rev1 (by REV1 deletion) increased Me-lex lethality and decreased Me-lex mutagenicity in both the NER defective (rad14) and BER defective (mag1; apn1apn2) strains. Therefore, Polζ and Rev1 contribute to resistance of the lethal effects of Me-lex induced lesions (3-MeA and derived AP sites) by bypassing lesions and fixing some mutations.
Keywords: Me-lex, N3-methyladenine, translesion synthesis, p53, yeast
1.INTRODUCTION
Me-lex, {1-methyl-4-[1-methyl-4-(3-methoxysulfonylpropanamido)pyrrole-2-carboxamido]-pyrrole-2-carboxamido}propane, is an alkylating agent that preferentially generates N3-methyladenine (3-MeA) adducts in A-T rich regions of double-stranded DNA due to its minor groove selectivity conferred by the lexitropsin dipeptide [1,2]. Me-lex toxicity and mutagenicity are dependent on the DNA repair background. BER defective S. cerevisiae strains that lack 3-methyladenine DNA glycosylase (mag1) or both AP endonucleases (apn1apn2) are significantly and similarly more sensitive to Me-lex toxicity than the parental strain, but only the removal of AP endonuclease activity resulted in a significant increase in mutagenicity. Furthermore, the Me-lex induced mutation spectrum determined in a yeast-based functional assay and the Me-lex induced methylation pattern determined in vitro in a human p53 cDNA showed minimal overlap [1]. This is consistent with the hypothesis that 3-MeA is a cytotoxic but weakly mutagenic lesion [1,2]. The mutation spectra induced by Me-lex were not affected by the status of Mag1 activity (MAG1 vs mag1; p = 0.10, Cariello test) nor by defects in different BER steps (mag1 vs apn1apn2; p = 0.84, Cariello test) [3]. It was also found that the mag1rad14 double mutant showed a significantly enhanced Mutation Frequency (MF), while the mag1 and rad14 single mutants had low MF similar to WT. It was suggested that the formation of abasic (AP) sites via glycosylase mediated excision and non-enzymatic hydrolysis may, in part, be repaired by different pathways, and that Rad14 plays a role in the repair of AP sites formed by hydrolysis. In total, these observations suggested that 3-MeA mutagenicity might be associated with the formation of an AP site through the enzymatic activity of Mag1 or spontaneous hydrolysis of the methylated purine or that the different lesions (3-MeA and AP site) afford the same mutation specificity, i.e., the same nucleotide is inserted with similar efficiency opposite both lesions during translesion synthesis (TLS).
The fixation of DNA damage into mutations depends on the activity of TLS DNA polymerases that allow bypass of DNA lesions that stall replication forks. In general, TLS polymerases lack 3’→5’ proofreading exonuclease activity, support the bypass of damaged DNA in vitro, and, when copying undamaged DNA, manifest both much lower fidelity and reduced processivity compared to replicative DNA polymerases. In S. cerevisiae, there are three known TLS polymerases: Polη and Rev1, which belong to the Y family of DNA polymerases, and Polζ, which is a member of the B family [4].
In S. cerevisiae, Polζ is the TLS polymerase associated with the majority of spontaneous, as well as damage induced mutagenesis [4]. Polζ is composed of two subunits, Rev3 and Rev7. Rev3 is the catalytic subunit with DNA polymerase activity. The function of Rev7 is not well understood, although it is thought to stimulate Rev3 activity. Polζ is not essential for cell viability in yeast while disruption of REV3 in mice causes embryonic lethality [4]. In vitro, Polζ acts mainly as a mispaired primer extender (frequency 10−1−10−2), although it can also incorporate nucleotides opposite a DNA lesion.
Polζ requires the Rev1 protein, which is indispensable for UV mutagenesis [4-8] and for mutagenesis resulting from TLS occurring through AP sites [9] and other damaged bases [10]. Rev1 is a member of the Y family of DNA polymerases, and specifically incorporates C opposite all template bases or AP sites [4,9].
In contrast to Polζ and Rev1, Polη suppresses UV mutagenicity due to its error free translesion synthesis activity [11]. Polη is also able to bypass bulky lesions with relatively high fidelity. Recently Zhao et al. proposed that Polη may also be involved in error prone translesion synthesis of AP sites [12]. Overall it seems that DNA lesions that severely impinge upon the minor groove block DNA synthesis by Polη [4].
In order to examine which TLS polymerases are involved in the mutation fixation process of Me-lex induced lesions, we have begun a systematic genetic analysis by deleting REV3 and REV1 genes in the parental yeast strain and in NER defective (rad14) and BER defective (mag1; apn1apn2) derivatives. In the present study, the toxicity and mutagenicity of Me-lex induced lesions were evaluated using a p53 functional assay [13-15]. Our results are consistent with an involvement of both Polζ and Rev1 in the mutation fixation process of Me-lex induced lesions but with some subtle differences in their relative roles in protecting cells from toxicity and inducing mutations.
2. Materials and Methods
2.1 Hazardous procedures
Me-lex should be considered as a toxic compound, and was handled accordingly.
2.2 Compounds
Reagents of the highest purity were purchased from Sigma (St. Louis, MO, USA) or Aldrich (Milwaukee, WI, USA) unless otherwise stated. Me-lex was prepared as previously described [16].
2.3 Vectors, strains and media
The yeast expression vector pTS76 harbours the human wild-type p53 cDNA under the control of an ADH1 constitutive promoter and contains the TRP1 selectable marker. The haploid S. cerevisiae strain yIG397 [17], and its isogenic BER or NER deficient derivatives (Table 1), were used as recipients for pTS76. The p53-dependent reporter ADE2 gene allowed the phenotypic selection of p53 mutants as its recombinant cyc1 promoter contains three copies of the responsive element RGC [17]. Standard yeast manipulations were performed as previously described [18].
Table 1.
Yeast strains used in this study.
| Name | Genotype | Phenotype | Ref | |
|---|---|---|---|---|
| yIG397 | MATα ade2−1 leu2−3,112 trp1−1 his3−11,15 can1−100 ura3−1 URA3 3xRGC::pCYC1::ADE2 | WT | (Flaman 1995) | |
| yPM2 | same as yIG397, mag1::LEU2 | (mag1) | BER− | (Monti 2002) |
| yPC2 | same as yIG397, apn1::HIS3 apn2::KANR | (apn1apn2) | BER− | (Monti 2002) |
| yPM4 | same as yIG397, rad14:: KANR | (rad14) | NER− | (Monti 2004) |
| yPM5 | same as yIG397, rev1::HYGROR | (rev1) | TLS− | this work |
| yPM6 | same as yIG397 rev3::HYGROR | (rev3) | TLS− | this work |
| yPM7 | same as yPM2, rev1::HIGROR | (mag1rev1) | BER− TLS− | this work |
| yPM8 | same as yPM2, rev3::HIGROR | (mag1rev3) | BER− TLS− | this work |
| yPM9 | same as yPC2, rev1::HYGROR | (apn1apn2rev1) | BER− TLS− | this work |
| yPM10 | same as yPC2, rev3::HYGROR | (apn1apn2rev3) | BER− TLS− | this work |
| yPM11 | same as yPM4, rev1::HYGROR | (rad14rev1) | NER− TLS− | this work |
| yPM12 | same as yPM4, rev3::HYGROR | (rad14rev3) | NER− TLS− | this work |
2.4 DNA modification, analysis, and transformation
The experimental system is identical to that previously described [13]. The in vitro treatment of pTS76 plasmid with Me-lex was performed as previously described [14,15]. Briefly, Me-lex was dissolved in DMSO immediately before the plasmid pTS76 DNA (3.0 μg) was treated with different Me-lex concentrations (up to 12 mM) in 10 mM Tris-HCl (pH 7.4), 1 mM EDTA, 50% EtOH for 1 h at 37 °C. DNA was purified by EtOH precipitation, washed with 70% EtOH, and resuspended in sterile water. Damaged or undamaged plasmids were then transformed by the LiOAc method into the same number of yeast cells (measured by OD600), using the same growth conditions, and transformants were plated on selective synthetic medium plates specific for each strain (see below and Table 1). After 3 days of incubation at 30 °C, the colour of the colonies was evaluated. The selection for the plasmid marker (TRP1) allowed an indirect determination of the lethal effect of the damaging treatment to be calculated as the number of transformants scored in transfections with Me-lex treated plasmids relative to that obtained with undamaged vector (hereafter indicated as plasmid survival). As transformation plates contained a minimal amount of adenine, adenine auxotrophs produced small red colonies (an Ade− phenotype). Considering spontaneous mutagenesis, Ade− phenotype can be due to either inactivating mutations in the p53 cDNA or to mutations at the chromosomally located reporter gene (ADE2), whose expression is under control of 3 copies of a p53 responsive element, in the reporter strain. When induced mutagenesis is considered, since the damaging treatment is performed in vitro and is confined to the plasmid, we assume that the Ade− phenotype is almost exclusively due to plasmid targeted inactivating events (i.e. p53 cDNA targeted). This assumption is consistent with our previous work [1,2,13,19-21]. The spontaneous or induced MF (sMF or MF, respectively) was defined as the number of small red colonies with respect to the total number of transformants. For each strain and each Me-lex concentration, at least 10 experiments were performed. For MF the cumulative mutation frequencies observed in all experiments are reported. Plasmid transformation efficiency of undamaged vector was unaffected by deletion of the DNA repair genes. For each Me-lex concentration tested the effects on mutagenicity and toxicity were normalized to those observed with the undamaged plasmid in each strain.
2.5 Construction of rev1 and rev3 yeast strains
REV1 and REV3 disruption cassettes were obtained by PCR using as a template pCOREUH plasmid [22] containing the selectable marker HYGROr (generous gift of Dr. Francesca Storici). Primers have a sequence homology to the 5’ (or 3’) end of the gene to be disrupted (underlined) and a sequence (in bold) that is complementary to the 5’ (or 3’) region of the HYGRO resistance gene present in pCOREUH. Strains were transformed with the unpurified PCR product. The deletion of the REV1 or REV3 genes was confirmed by phenotypic selection (resistance to hygromycin B) and by yeast colony PCR.
The primers used for the creation of the disruption cassettes were: REV1-hygro dw (5’-atg ggt gaa cat ggt ggt ctt gta gat tta ttg gac agc gat ttg gaa tac atc tgg gca gat gat gtc g-3’ (70 mer) REV1-up: 5’-tca aac ttc aaa gtc cat gtc aag ttt acg cac agt ctg gta agt atg ttc cgc gcg ttg gcc gat tca t-3’ (70 mer) for REV1; REV3-hygro dw: 5’-atg tcg agg gag tcg aac gac aca ata cag agc gat acg gtt aga tca tcc atc tgg gca gat gat gtc g-3’ (70 mer); REV3-up: 5’-tta cca atc att tag aga tat taa tgc ttc ttc cct ttg aac aga ttg atc cgc gcg ttg gcc gat tca t-3’ (70 mer) for REV3. The PCR conditions were 95 °C for 40 s, 55 °C for 60 s and 72 °C for 90 s repeated for 35 cycles.
The primers for yeast colony PCR were: REV1−5’: 5’-ata cct ttt ggc ata gtc t-3’; REV1−3’: 5’-gaa gac aaa tag tgt aaa aa-3’ and HYGRO-3’ : 5’-gtt ttt tta tat tgt agt tgt tc-3’ for the genomic confirmation of REV1 deletion; REV3−5’: 5’-cat ttt ttt gac gag tgc ag-3’; REV3−3’: 5’-cgt gtt tat cat ctt ttt tcc-3’; and HYGRO-3’ : 5’-gtt ttt tta tat tgt agt tgt tc-3’ for the genomic confirmation of REV3 deletion. The PCR conditions were 94 °C for 60 s, 55 °C for 60 s and 72 °C for 120 s repeated for 35 cycles; yeast colonies were heated at 94 °C for 8 min before starting.
3. RESULTS
3.1 Effects of the REV1 and REV3 deletions on spontaneous mutation frequency (sMF) in different DNA repair backgrounds
In order to determine the role of Rev1 and Rev3 (Polζ) in the biological fate of Me-lex induced lesions, the genes were deleted in the yIG397 parental strain and in NER and BER defective derivatives (Table 1; see Materials and Methods for details). All strains contain the ADE2 reporter gene under transcriptional control of wild-type human p53.
To determine sMF, untreated plasmid pTS76, containing the wild-type p53 cDNA under the control of the constitutive ADH1 promoter, was transformed into the panel of NER, BER and TLS yeast mutant strains. Transformants were selected on plates lacking tryptophan but containing sufficient adenine for adenine auxotrophs to grow and turn red. Compared to the wild type strain, the deletion of RAD14 slightly increased sMF (p<0.03, Chi square test), the deletion of MAG1 slightly decreased sMF and deletion of both APN1 and APN2 significantly increased sMFs (p < 0.05 and p < 0.0001, Chi square test, respectively) (Table 2). These results suggest that the AP sites are a major source of spontaneous mutagenesis, and that constitutive Mag1 activity contributes to the sMF. This result is consistent with the observation that Mag1 activity is associated with a weak mutator phenotype, which is even more evident when the protein is over-expressed [23,24].
Table 2.
Effects of REV1 or REV3 deletion on spontaneous p53 mutant frequencies in different DNA repair backgrounds.
| DNA repair background | p53 mutant frequency ×1000 (red colonies/total) |
relative fold decrease |
statistics |
|---|---|---|---|
| WT | 0,91 | 1 | |
| (136/149.876)*§ | |||
| rev1 | 0,3 | 3.0 | |
| (25/83.195)* | *p<0.0001 | ||
| rev3 | 0,55 | 1.6 | |
| (74/134.023)§ |
|
§ p<0.0007 |
|
| rad14 | 1,18 | 1 | WT vs rad14: p<0.03 |
| (154/130.183) *§ | |||
| rad14rev1 | 0,61 | 1.9 | |
| (32/52.813) * | * p<0.0006 | ||
| rad14rev3 | 0,73 | 1.6 | |
| (39/53.562) § |
|
§ p<0.008 |
|
| mag1 | 0,7 | 1 | WT vs mag1: p<0.05 |
| (131/186.585) *§ | |||
| mag1rev1 | 0,58 | 1.2 | |
| (52/90.070) * | *NS | ||
| mag1rev3 | 0,83 | 0.84 | |
| (33/39.799) § |
|
§NS |
|
| apn1apn2 | 3,51 | 1 | WT vs apn1,apn2: p<0.0001 |
| (344/98.052) *§ | |||
| apn1apn2rev1 | 1,52 | 2.3 | |
| (50/32.798) * | * p<0.0001 | ||
| apn1apn2rev3 | 1,92 | 1.8 | |
| (51/26.521) § | § p<0.0001 |
NS: not significant
The deletion of REV1 decreased sMF by 2−3 fold in WT, rad14 and apn1apn2 backgrounds, while the same deletion had no significant effect in the mag1 background. Similarly, the deletion of REV3 was associated with a weaker, yet still significant, reduction in sMF (1.6−1.8 fold) in all backgrounds except mag1. Only in mag1 strain there is no significant difference in sMF after deletion of REV1 or REV3.
3.2 Effect of REV1 and REV3 deletion on the survival of Me-lex damaged plasmid DNA
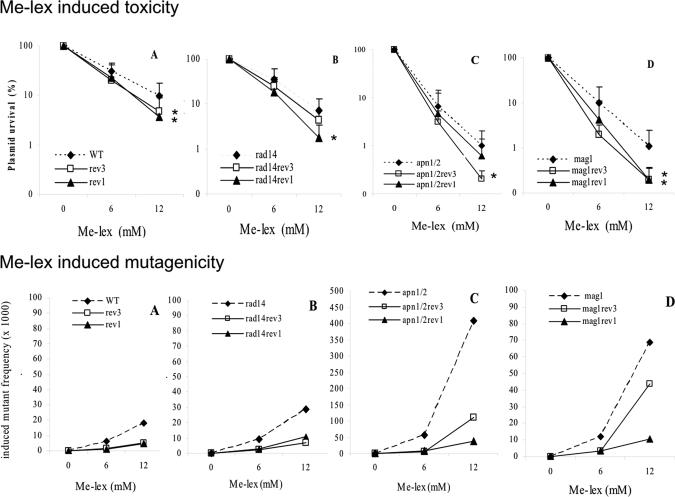
In order to evaluate the influence of Rev1 and Rev3 on the lethality of Me-lex induced lesions in relation to the different DNA repair backgrounds, plasmid pTS76 was damaged in vitro by exposure to increasing Me-lex concentrations and transformed into the appropriate yeast strains. A general Me-lex concentration-dependent decrease in plasmid survival was observed (Figure 1, upper panel, A-D). The deletion of REV1 or REV3 genes led to a reduction in plasmid survival compared to the parental strains regardless of the DNA repair background, although some quantitative differences were noted. The deletion of REV1 or REV3 in a WT background had a similar impact (mean of the reduction observed at 6 mM and 12 mM Me-lex: ∼2.0-fold reduction in plasmid survival), while in the NER deficient strain (rad14) the deletion of REV1 had a stronger impact (mean of the reduction observed at 6 mM and 12 mM Me-lex: 2.9-fold and 1.5-fold, in rev1 and rev3, respectively). The deletion of REV3 exhibited a stronger effect in the BER defective apn1 apn2 strain (mean of the reduction observed at 6 mM and 12 mM Me-lex: 3.4-fold and 1.4-fold, in rev1 and rev3, respectively). Finally, the largest difference in plasmid survival (mean of the reduction observed at 6 mM and 12 mM Me-lex : ∼5-fold) was observed for the REV1 and REV3 deletion in a mag1 background. Together these results indicate that both Rev1 and Rev3 (Polζ) are involved in the bypass of some Me-lex-induced lesions and that they provide protection against Me-lex toxicity. Interestingly, when AP sites accumulate (apn1apn2 strain) a protective effect appears to be more dependent on REV3 than on REV1, while when 3-MeA adducts accumulate (mag1 strain) there is a similar role for both TLS polymerases.
Figure 1.
Influence of deletion of REV1 or REV3 on the lethality (upper panel) and mutagenicity (lower panel) of Me-lex induced lesions in repair proficient and repair deficient yeast strains. Panel A: WT; panel B: rad14 background; panel C: apn1apn2 background; panel D: mag1 background. Upper panel: * indicates a statistically significant decrease in plasmid survival observed at 12mM Me-lex relative to the REV1, REV3 wild type strain (Student t test).
3.3 Effect of REV1 and REV3 deletion on Me-lex induced mutagenesis
We examined the impact of REV1 and REV3 deletions on the mutagenicity of Me-lex lesions induced in the p53 cDNA exploiting the ADE2 reporter gene in different yeast strains (Figure 1, lower panels, A-D). The MF increased proportionally to the Me-lex concentration. Similar to the effect on plasmid survival, deletion of REV1 or REV3 led to a significant decrease in MF, regardless of the DNA repair background (see Tables 3 A, B for raw numbers and statistical analyses). In the WT, as well as in the NER deficient rad14 strain, the deletion of REV1 or REV3 decreased MF by 3 to 5-fold. In the apn1apn2 or mag1 strains, at the highest Me-lex dose, the decrease in MF was more pronounced for the deletion of REV1 compared to that of REV3. It should be noted that the increase in mutagenesis was much greater in the apn1apn2 than in the mag1 strain (Y-axis scale is different in Figure 1 lower, panel C vs panel D).
Table 3A.
Effects of REV1 or REV3 deletion on Me-lex induced p53 mutant frequencies in different DNA repair backgrounds (6mM Me-lex).
| DNA repair background | p53 MF ×1000 (red colonies/total) | relative fold decrease | statistics |
|---|---|---|---|
| WT | 7,0 | 1 | |
| (337/47.889)*§ | |||
| rev1 | 1,4 | 5,0 | |
| (28/19.660)*^ | *p<0.0001 | ||
| rev3 | 2,1 | 3,3 | |
| |
(51/24.300)§^ |
|
§ p<0.0001; ^NS |
| rad14 | 10.3 | 1 | WT vs rad14: p<0.0001 |
| (415/40.407) *§ | |||
| rad14 rev1 | 3,7 | 2,8 | |
| (36/9.820) *^ | * p<0.0001 | ||
| rad14rev3 | 3,2 | 3,2 | |
| |
(47/14.792) §^ |
|
§ p<0.0001; ^NS |
| mag1 | 12,7 | 1 | WT vs mag1: p<0.0001 |
| (259/20.458) *§ | |||
| mag1 rev1 | 4,1 | 3,1 | |
| (18/4.399) *^ | * p<0.0001 | ||
| mag1 rev3 | 4,2 | 2,5 | |
| |
(10/2.371) §^ |
|
§ p<0.0005; ^NS |
| apn1 apn2 | 60,1 | 1 | WT vs apn1,apn2: p<0.0001 |
| (510/8.488) *§ | |||
| apn1 apn2rev1 | 10,3 | 5,8 | |
| (14/1.355) *^ | * p<0.0001 | ||
| apn1 apn2rev3 | 8,2 | 7.3 | |
| (13/1.585) §^ | § p<0.0001; ^NS |
NS: not significant
Table 3B.
Effects of REV1 or REV3 deletion on Me-lex induced p53 mutant frequencies in different DNA repair backgrounds (12mM Me-lex).
| DNA repair Background | p53 MF ×1000 (red colonies/total) | relative fold decrease | statistics |
|---|---|---|---|
| WT | 18,9 | 1 | |
| (322/17.082)*§ | |||
| rev1 | 4,8 | 3,9 | |
| (20/4.173)*^ | *p<0.0001 | ||
| rev3 | 5,8 | 3,3 | |
| |
(33/5.651)§^ |
|
§ p<0.0001; ^NS |
| rad14 | 29,8 | 1 | WT vs rad14: p<0.0001 |
| (374/12.553) *§ | |||
| rad14rev1 | 11,7 | 2,5 | |
| (16/1.363) *^ | * p<0.0002 | ||
| rad14rev3 | 7,6 | 3,9 | |
| |
(25/3.267) §^ |
|
§p<0.0001; ^NS |
| mag1 | 69,7 | 1 | WT vs mag1: p<0.0001 |
| (169/2.426) *§ | |||
| mag1rev1 | 11,4 | 6,1 | |
| (7/616) *^ | *p<0.0001 | ||
| mag1rev3 | 44,8 | 1,6 | |
| |
(6/134) §^ |
|
§NS; ^p<0.03 |
| apn1apn2 | 412,3 | 1 | WT vs apn1,apn2: p<0.0001 |
| (557/1.351) *§ | |||
| apn1apn2rev1 | 38,0 | 10,8 | |
| (7/184) *^ | *p<0.0001 | ||
| apn1apn2rev3 | 112,1 | 3,7 | |
| (24/214) §^ | §p<0.0001; ^p<0.02 |
NS: not significant
These results suggest that both Rev1 and Rev3 are involved in the bypass of Me-lex induced lesions in all DNA repair backgrounds since their absence is associated with decreased Me-lex mutagenicity.
4. DISCUSSION
Me-lex was designed to preferentially yield 3-MeA, a lesion that has been shown to be strongly cytotoxic but only weakly mutagenic in WT cells. In S. cerevisiae, 3-MeA repair is initiated by Mag1, followed by the action of AP endonucleases (Apn1, Apn2), DNA polymerase (δ, ε), and DNA ligase (III). Using a genetic approach in yeast, we previously showed that the 3-MeA lesion is critical in Me-lex induced cytotoxicity [1,15]. In contrast, its mutagenicity is only somewhat elevated in the absence of Mag1 glycosylase activity, but significantly elevated in the absence of AP endonuclease activity. Mutation hotspots observed along the p53 cDNA sequence did not always overlap with the most intense sites of methylation, but were highly associated with the A/T rich regions targeted by Me-lex [1,15]. We also demonstrated that the lethality of Me-lex induced lesions is counteracted by BER, mainly via Mag1 activity, while the role of NER is appreciable only in the absence of BER [2]. Furthermore, the mutation spectra induced by Me-lex in mag1, mag1rad14, and apn1apn2 backgrounds were indistinguishable (p = 0.74, p = 0.85) [2]. Those results were consistent with: (i) unrepaired 3-MeA and AP site in the template strand causing a similar misincorporation by DNA polymerases; and/or (ii) a single common promutagenic lesion, i.e., an AP site, formed from either the enzymatic (Mag1) or hydrolytic release of 3-MeA, being the origin of the mutation [2,21].
TLS DNA polymerases are involved in the mutagenicity of lesions produced by endogenous or exogenous DNA damaging processes [4]. These specialized DNA polymerases exhibit much lower processivity and fidelity compared to replicative DNA polymerases and can insert nucleotides opposite DNA lesions or extend from mispaired bases. A large degree of functional divergence has occurred among TLS polymerases, rendering them highly specialized for the roles they play in lesion bypass. Although some polymerases can carry out both steps of lesion bypass (nucleotide incorporation opposite the DNA lesion and the subsequent extension from the inserted nucleotide) in several cases one TLS polymerase inserts a nucleotide opposite a lesion while another one performs the extension step. This two-polymerases mechanism of lesion bypass is afforded by the high degree of structural and functional specificity of the different Y-family polymerases [4].
In yeast S. cerevisiae, three TLS polymerases are known: Polζ, Rev1 and Polη. Polζ, comprised of the Rev3 catalytic subunit and the Rev7 accessory subunit, is indispensable for UV and AP site mutagenesis [5,9,25,26]. Polζ acts primarily by extending from the mispair [27-30] and, for its role in TLS, requires the Rev1 protein. Despite the fact that Rev1 is indispensable for most Polζ-dependent TLS [6], its DNA synthetic activity is not required for many lesion bypass events [9,10,27,31,32]. This suggests that Rev1 plays a role in coordinating the assembly of Polζ at the replication fork [27]. Yeast Polη can also play a structural role (similarly to Rev1), as well as a functional role (as inserter or extender polymerase), in lesion bypass processes [12].
As a validation for its structural role in TLS, Rev1 has recently been shown to be part of a tight physical complex with Polζ in two independent studies [33,34]. It is likely that Rev1 interacts with the intact Polζ complex, as the association of Rev1 with either Rev3 or Rev7 is disrupted when either REV7 or REV3 is deleted. However, Acharya et al. [33] suggest that Rev1 interacts separately with either Rev7 or Rev3, that Rev1-Rev7 and Rev3-Rev7 complexes do not physically interact, and that the two complexes represent separate functional entities. In Polζ, Rev7 is needed for Rev3 to express its DNA polymerase activity, whereas in the Rev1-Rev7 complex Rev7 has no significant effect upon Rev1 DNA synthetic activity. This raises the possibility that in the Rev1- Rev7 complex, the role of Rev7 is to modulate protein-protein interactions at the replication fork.
In the present study, we demonstrate that the disruption of Polζ (through deletion of its catalytic subunit coded by REV3) or Rev1 caused an increase in lethality and a decrease in mutagenicity induced by Me-lex. Thus, Polζ and Rev1 contribute to overcoming some of the lethal effects of Me-lex induced lesions at the cost of creating mutations. The somewhat surprising differences between the rev1 and rev3 mutants on Me-lex mutagenicity and lethality are pointing towards independent functions in mutagenesis and/or survival in response to 3-MeA. This is a subject of future investigations.
The relative contribution of specialized TLS DNA polymerases to the toxicity and mutagenicity after 12 mM Me-lex treatment, where the effects were exacerbated, is summarized in Table 4. Relative to the parental strain, the decrease in plasmid survival (increase in Me-lex toxicity) caused by BER defects is strongly enhanced by the combined presence of TLS defects. Either rev1 or rev3 deficiency are synergistic with mag1 deficiency at increasing Me-lex toxicity (∼50-fold), while, in an apn1apn2 background, rev1 deficiency is additive and rev3 deficiency is synergistic. The anti-mutator effect of TLS deletions is retained in BER deficient backgrounds with the exception of rev3mag1 background. In apn1apn2 cells, REV1 deletion has an even stronger anti-mutator effect (10-fold reduction). The combined deletion of MAG1 and REV1 results in a strong increase in Me-lex toxicity and in a small reduction in MF. In total, these results suggest that the best therapeutic window (i.e. maximum killing with minimal mutagenesis) for Me-lex, would be accomplished by inactivation of the MAG1 and REV1 gene products.
Table 4.
Overall impact of NER, BER, TLS defects on Me-lex toxicity and mutagenicity, based only on 12mM Me-lex treatment, relative to the parental (wild type) strain.
| Decrease (↓) in S (a) | Decrease (↓) or increase (↑) in MF(b) | |||||
|---|---|---|---|---|---|---|
| wt | rev1 | rev3 | wt | rev1 | rev3 | |
| Wild type | 1 | ↓ 2.7 | ↓ 2.1 | 1.0 | ↓3.9 | ↓3.3 |
| rad14 | ↓ 1.2 | ↓ 4.8 | ↓ 1.8 | ↑ 1.7 | ↓1.5 | ↓2.3 |
| apn1 apn2 | ↓ 9.1 | ↓ 12.7 | ↓ 50.1 | ↑ 21.7 | ↑ 2.0 | ↑ 5.0 |
| mag1 | ↓ 7.7 | ↓ 50.1 | ↓ 50.1 | ↑ 3.3 | ↓1.6 | ↑ 2.5 |
fold decrease in plasmid survival due to NER, BER, or TLS defects.
fold decrease (↓) or increase (↑) in MF due NER, BER, or TLS defects.
Can we understand which Me-lex induced lesion(s) is (are) bypassed by Polζ or Rev1? The two candidates are 3-MeA, which is directly formed, and AP sites, which are derived from glycosylase excision or non-enzymatic hydrolysis of the labile 3-MeA base. The former is highly cytotoxic and poorly mutagenic, while the latter is both cytotoxic and highly mutagenic. In the absence of their repair (apn1apn2 background), AP sites are expected to accumulate. In this situation deletion of REV1 has a more dramatic impact on the reduction of mutagenicity than on Me-lex toxicity. On the other hand, the deletion of REV3 has a dramatic impact on Me-lex toxicity. In the presence of 3-MeA accumulation (mag1 background), the deletion of REV3 or REV1 has similar effect on 3-MeA toxicity, while mutagenicity decreases substantially only when REV1 is deleted. The stronger dependency of AP site induced mutagenesis on Rev1 rather than Polζ (Rev3) is consistent with the results reported by Auerbach et al., [35] who found a similar phenomenon due to inactivation of Polζ via REV7 rather than REV3 deletion. These results suggest that both lesions are substrates for TLS bypass, an hypothesis that could be tested in vivo or in vitro assays using single lesions introduced at specific locations (work in progress). It is important to note, however, that AP sites can produce secondary lesions (such as 3'-blocked single strand breaks with 3'-dRP termini), spontaneously (via hydrolysis) or through the action of different enzymes, whose contribution to the end point analyzed in the present work is unknown. Thus, we cannot exclude that the difference between the mag1 and apn1apn2 may reflect differences in the composition of lesions present at the replication fork.
In summary, by combining a chemical and genetic approach we have shown that in yeast both Rev1 and Rev3 play an active role in mediating the biological effects (toxicity and mutagenicity) of Me-lex induced lesions (3-MeA or AP site intermediate).
Acknowledgements
This work was supported by the National Institute of Health Grant RO1 CA 29088 (to B.G.), and partially by the Associazione Italiana per la Ricerca sul Cancro, by Alleanza contro il Cancro. Y.C. and D.R. were on a PhD programme.
Abbreviations1
- BER
Base Excision Repair
- 3-MeA
N3-methyladenine
- Me-lex
{1-methyl-4-[1-methyl-4-(3-methoxysulfonylpropanamido)pyrrole-2-carboxamido]-pyrrole-2-carboxamido}propane
- MF
mutation frequency
- BER
Base Excision Repair
- NER
Nucleotide Excision Repair
- TLS
translesion synthesis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kelly JD, Inga A, Chen FX, Dande P, Shah D, Monti P, Aprile A, Burns PA, Scott G, Abbondandolo A, Gold B, Fronza G. Relationship between DNA methylation and mutational patterns induced by a sequence selective minor groove methylating agent. J Biol Chem. 1999;274:18327–18334. doi: 10.1074/jbc.274.26.18327. [DOI] [PubMed] [Google Scholar]
- 2.Monti P, Iannone R, Campomenosi P, Ciribilli Y, Varadarajan S, Shah D, Menichini P, Gold B, Fronza G. Nucleotide excision repair defect influences lethality and mutagenicity induced by Me-lex, a sequence-selective N3-adenine methylating agent in the absence of base excision repair. Biochemistry. 2004;43:5592–5599. doi: 10.1021/bi035968x. [DOI] [PubMed] [Google Scholar]
- 3.Cariello NF, Piegorsch WW, Adams WT, Skopek TR. Computer program for the analysis of mutational spectra: application to p53 mutations. Carcinogenesis. 1994;15:2281–2285. doi: 10.1093/carcin/15.10.2281. [DOI] [PubMed] [Google Scholar]
- 4.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu Rev Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 5.Lawrence CW, Christensen RB. Ultraviolet-induced reversion of cyc1 alleles in radiation-sensitive strains of yeast. III. rev3 mutant strains. Genetics. 1979;92:397–408. doi: 10.1093/genetics/92.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lawrence CW, Christensen RB. Ultraviolet-induced reversion of cyc1 alleles in radiation-sensitive strains of yeast. I. rev1 Mutant strains. J Mol Biol. 1978;122:1–21. doi: 10.1016/0022-2836(78)90104-3. [DOI] [PubMed] [Google Scholar]
- 7.Lawrence CW, Nisson PE, Christensen RB. UV and chemical mutagenesis in rev7 mutants of yeast. Mol Gen Genet. 1985;200:86–91. doi: 10.1007/BF00383317. [DOI] [PubMed] [Google Scholar]
- 8.Lawrence CW, O'Brien T, Bond J. UV-induced reversion of his4 frameshift mutations in rad6, rev1, and rev3 mutants of yeast. Mol Gen Genet. 1984;195:487–490. doi: 10.1007/BF00341451. [DOI] [PubMed] [Google Scholar]
- 9.Johnson RE, Torres-Ramos CA, Izumi T, Mitra S, Prakash S, Prakash L. Identification of APN2, the Saccharomyces cerevisiae homolog of the major human AP endonuclease HAP1, and its role in the repair of abasic sites. Genes Dev. 1998;12:3137–3143. doi: 10.1101/gad.12.19.3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baynton K, Bresson-Roy A, Fuchs RP. Distinct roles for Rev1p and Rev7p during translesion synthesis in Saccharomyces cerevisiae. Mol Microbiol. 1999;34:124–133. doi: 10.1046/j.1365-2958.1999.01583.x. [DOI] [PubMed] [Google Scholar]
- 11.Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Poleta. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 12.Zhao B, Xie Z, Shen H, Wang Z. Role of DNA polymerase eta in the bypass of abasic sites in yeast cells. Nucleic Acids Res. 2004;32:3984–3994. doi: 10.1093/nar/gkh710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inga A, Iannone R, Monti P, Molina F, Bolognesi M, Abbondandolo A, Iggo R, Fronza G. Determining mutational fingerprints at the human p53 locus with a yeast functional assay: a new tool for molecular epidemiology. Oncogene. 1997;14:1307–1313. doi: 10.1038/sj.onc.1200952. [DOI] [PubMed] [Google Scholar]
- 14.Kelly JD, Inga A, Chen FX, Dande P, Shah D, Monti P, Aprile A, Burns PA, Scott G, Abbondandolo A, Gold B, Fronza G. Relationship between DNA methylation and mutational patterns induced by a sequence selective minor groove methylating agent [In Process Citation] J Biol Chem. 1999;274:18327–18334. doi: 10.1074/jbc.274.26.18327. [DOI] [PubMed] [Google Scholar]
- 15.Monti P, Campomenosi P, Ciribilli Y, Iannone R, Inga A, Shah D, Scott G, Burns PA, Menichini P, Abbondandolo A, Gold B, Fronza G. Influences of base excision repair defects on the lethality and mutagenicity induced by Me-lex, a sequence-selective N3-adenine methylating agent. J Biol Chem. 2002;277:28663–28668. doi: 10.1074/jbc.M203384200. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y, Chen FX, Mehta P, Gold B. Groove- and sequence-selective alkylation of DNA by sulfonate esters tethered to lexitropsins. Biochemistry. 1993;32:7954–7965. doi: 10.1021/bi00082a017. [DOI] [PubMed] [Google Scholar]
- 17.Flaman JM, Frebourg T, Moreau V, Charbonnier F, Martin C, Chappuis P, Sappino AP, Limacher IM, Bron L, Benhattar J, et al. A simple p53 functional assay for screening cell lines, blood, and tumors. Proc Natl Acad Sci U S A. 1995;92:3963–3967. doi: 10.1073/pnas.92.9.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guthrie C, Fink GR. Academic press; San Diego: 1991. [Google Scholar]
- 19.Monti P, Inga A, Scott G, Aprile A, Campomenosi P, Menichini P, Ottaggio L, Viaggi S, Abbondandolo A, Burns PA, Fronza G. 5-methylcytosine at HpaII sites in p53 is not hypermutable after UVC irradiation. Mutat Res. 1999;431:93–103. doi: 10.1016/s0027-5107(99)00187-6. [DOI] [PubMed] [Google Scholar]
- 20.Fronza G, Inga A, Monti P, Scott G, Campomenosi P, Menichini P, Ottaggio L, Viaggi S, Burns PA, Gold B, Abbondandolo A. The yeast p53 functional assay: a new tool for molecular epidemiology. Hopes and facts. Mutat Res. 2000;462:293–301. doi: 10.1016/s1383-5742(00)00011-9. [DOI] [PubMed] [Google Scholar]
- 21.Monti P, Campomenosi P, Ciribilli Y, Iannone R, Inga A, Shah D, Scott G, Burns PA, Menichini P, Abbondandolo A, Gold B, Fronza G. Influences of base excision repair defects on the lethality and mutagenicity induced by Me-lex, a sequence-selective N3-adenine methylating agent. J Biol Chem. 2002;277:28663–28668. doi: 10.1074/jbc.M203384200. [DOI] [PubMed] [Google Scholar]
- 22.Storici F, Resnick MA. Delitto perfetto targeted mutagenesis in yeast with oligonucleotides. Genet Eng (N Y) 2003;25:189–207. [PubMed] [Google Scholar]
- 23.Glassner BJ, Rasmussen LJ, Najarian MT, Posnick LM, Samson LD. Generation of a strong mutator phenotype in yeast by imbalanced base excision repair. Proc Natl Acad Sci U S A. 1998;95:9997–10002. doi: 10.1073/pnas.95.17.9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Posnick LM, Samson LD. Imbalanced base excision repair increases spontaneous mutation and alkylation sensitivity in Escherichia coli. J Bacteriol. 1999;181:6763–6771. doi: 10.1128/jb.181.21.6763-6771.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nelson JR, Lawrence CW, Hinkle DC. Thymine-thymine dimer bypass by yeast DNA polymerase zeta. Science. 1996;272:1646–1649. doi: 10.1126/science.272.5268.1646. [DOI] [PubMed] [Google Scholar]
- 26.Lawrence CW, Das G, Christensen RB. REV7, a new gene concerned with UV mutagenesis in yeast. Mol Gen Genet. 1985;200:80–85. doi: 10.1007/BF00383316. [DOI] [PubMed] [Google Scholar]
- 27.Haracska L, Unk I, Johnson RE, Johansson E, Burgers PM, Prakash S, Prakash L. Roles of yeast DNA polymerases delta and zeta and of Rev1 in the bypass of abasic sites. Genes Dev. 2001;15:945–954. doi: 10.1101/gad.882301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson RE, Washington MT, Haracska L, Prakash S, Prakash L. Eukaryotic polymerases iota and zeta act sequentially to bypass DNA lesions. Nature. 2000;406:1015–1019. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- 29.Johnson RE, Yu SL, Prakash S, Prakash L. Yeast DNA polymerase zeta (zeta) is essential for error-free replication past thymine glycol. Genes Dev. 2003;17:77–87. doi: 10.1101/gad.1048303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prakash S, Prakash L. Translesion DNA synthesis in eukaryotes: a one- or two-polymerase affair. Genes Dev. 2002;16:1872–1883. doi: 10.1101/gad.1009802. [DOI] [PubMed] [Google Scholar]
- 31.Gibbs PE, Borden A, Lawrence CW. The T-T pyrimidine (6−4) pyrimidinone UV photoproduct is much less mutagenic in yeast than in Escherichia coli. Nucleic Acids Res. 1995;23:1919–1922. doi: 10.1093/nar/23.11.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gibbs PE, Kilbey BJ, Banerjee SK, Lawrence CW. The frequency and accuracy of replication past a thymine-thymine cyclobutane dimer are very different in Saccharomyces cerevisiae and Escherichia coli. J Bacteriol. 1993;175:2607–2612. doi: 10.1128/jb.175.9.2607-2612.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Acharya N, Haracska L, Johnson RE, Unk I, Prakash S, Prakash L. Complex formation of yeast Rev1 and Rev7 proteins: a novel role for the polymerase-associated domain. Mol Cell Biol. 2005;25:9734–9740. doi: 10.1128/MCB.25.21.9734-9740.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hirano Y, Sugimoto K. ATR homolog Mec1 controls association of DNA polymerase zeta-Rev1 complex with regions near a double-strand break. Curr Biol. 2006;16:586–590. doi: 10.1016/j.cub.2006.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Auerbach P, Bennett RA, Bailey EA, Krokan HE, Demple B. Mutagenic specificity of endogenously generated abasic sites in Saccharomyces cerevisiae chromosomal DNA. Proc Natl Acad Sci U S A. 2005;102:17711–17716. doi: 10.1073/pnas.0504643102. [DOI] [PMC free article] [PubMed] [Google Scholar]