Abstract
The conditional expression of hairpin constructs in Drosophila melanogaster has emerged in recent years as a method of choice in functional genomic studies. To date, upstream activating site–driven RNA interference constructs have been inserted into the genome randomly using P-element–mediated transformation, which can result in false negatives due to variable expression. To avoid this problem, we have developed a transgenic RNA interference vector based on the phiC31 site-specific integration method.
Transgenic RNA interference (RNAi) has emerged as an important method for analyzing gene function in D. melanogaster and has joined the already rich arsenal of tools available for functional genomic studies in this organism1. The method relies on the Gal4–upstream activating site (UAS) system2 to control the expression of a gene fragment that is dimerized to produce a double-stranded RNA (dsRNA) hairpin structure, which then triggers a sequence-specific post-transcriptional silencing and RNAi response. Tissue- or cell-specific expression of the transgenic RNAi constructs is achieved after a cross between UAS-hairpin and Gal4 driver lines. The main advantage of the method, in addition to its relatively simple design and fast execution time, is that it allows spatial and temporal control of the knockdown construct, which is essential for characterizing genes with pleiotropic functions.
A problem with current methodology is the variability in the level of hairpin expression due to the random integration in the genome of the P-element–based UAS-hairpin constructs. For example, one recent report in which two random insertions per construct were tested showed that in 40% of cases the two behaved differently, with one insertion showing lethality and the other viability when tested with the ubiquitously expressed actin5C-Gal4 driver1. To avoid the high incidence of false negatives resulting from random integration, we decided to develop a vector for transgenic RNAi based on the phiC31 targeted integration method3. Targeting RNAi hairpin constructs to a specific region of the genome with the phiC31 integrase method offers a number of advantages over P-element–based methods, in regard to both speed of production and potency of the transgenic RNAi lines. Thus, we expect the penetrance (percentage of lines that show noteworthy phenotypes) to be higher when UAS-driven hairpins are targeted to specific attP sites that allow for consistently robust expression levels. (Our decision to use the attP2 docking site was based on observations of M.M. and N.P. (unpublished data) demonstrating optimal basal and inducible properties.) Moreover, by targeting hairpins to the same sites, we can make side-by-side comparisons between different hairpins directed against the same gene, allowing for reliable validation of the RNAi hairpin phenotypes with multiple constructs.
We built Valium (Vermilion-AttB-Loxp-Intron-UAS-MCS) (Fig. 1a), a transgenic RNAi vector based on the phiC31 site-specific integration method, and generated hairpins directed against a number of genes with known phenotypes (Supplementary Table 1 online and data not shown). To avoid potential off-target effects, which have been shown to be a significant source of false positives in cell-based D. melanogaster RNAi screens4, we designed the hairpin sequences so that they do not contain short homology stretches of ≥ 19 nucleotides (Supplementary Methods online). Further, to evaluate the differences in potency between different hairpins directed against the same genes, we generated two hairpins per gene when possible.
Figure 1.
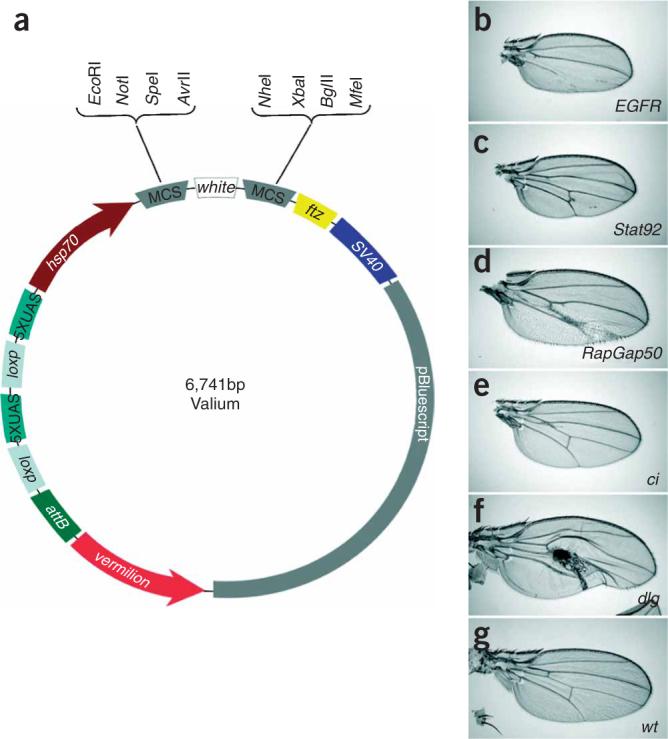
Valium is an effective vector for transgenic RNAi. (a) Structure of Valium: Valium contains vermilion as the selectable marker9; an attB sequence to allow for phiC31 targeted integration at genomic attP landing sites3; two pentamers of UAS, one of which can be removed using the Cre-loxP system10; the hsp70 TATA promoter; a multiple cloning site (MCS) that allows a single PCR product to be cloned in both orientations to generate the hairpin construct; the white intron, located between the two halves of the inverted DNA repeat, which has been shown to reduce toxicity in bacteria; and the ftz intron, followed by the SV40 poly(A) tail, to facilitate hairpin RNA expression, processing and export from the nucleus. We chose vermillion rather than mini-white because the exact gene dosage of white has been found to be important in behavioral studies11. (b–g) Examples of hairpin-induced wing phenotypes in en-Gal4; UAS-hairpin flies. Note that the various phenotypes generated have no common features, indicating that there are no nonspecific patterning defects generated as a result of general overexpression of dsRNAs. Further, control hairpins do not show any unusual wing phenotypes (data not shown).
We chose a set of 11 genes encoding proteins well known for their functions in wing development and crossed them to the engrailed-Gal4 (en-Gal4) driver to allow us to detect phenotypes in the posterior wing compartment. The phenotypes generated were specific and expected (Fig. 1 and Supplementary Table 1). Knockdown of the epidermal growth factor receptor (EGFR) gene led to defects in growth and vein formation (Fig. 1b). A similar phenotype was observed with RNAi lines against son of sevenless (sos), which encodes a downstream transducer of the EGFR. RNAi directed against the sprouty (sty) gene was consistent with the protein's wild-type role as a negative regulator of EGFR in wing vein patterning. Knockdown of decapentaplegic (dpp) removed all veins, as expected (data not shown). Notably, knockdowns of the genes dome, hop and STAT92E, which encode three components of the JAK/STAT pathway (the receptor domeless, the JAK kinase hopscotch and the transcription factor STAT92E, respectively), all produced the same ectopic vein phenotype previously reported for classical mutants of these genes (Fig. 1c). The vein phenotype associated with knockdown of cubitus interruptus (ci) (Fig. 1e) as well as the polarity defects caused by knockdown of armadillo (arm) were consistent with the reported functions of these proteins in vein and wing margin formation, respectively. Knockdown of discs-large (dlg1) caused a crumbled and necrotic wing phenotype that is most likely due to a loss of apical-basal polarity and overproliferation (Fig. 1f). Finally, knockdown of RacGap50C (also known as tum) generated a range of defects, including increased width and fusion of veins and enlarged cells (Fig. 1d). A description of the wing-mutant phenotypes of the genes tested here can be found at FlyBase. Notably, in the seven cases in which two independent hairpin lines were generated (Supplementary Table 1), their phenotypes were very similar if not identical, suggesting that the specific phenotypes obtained with these hairpins are due to knockdown of the intended gene and not to off-target effects. Further, expression of hairpin constructs directed against control dsRNAs did not generate patterning defects1 (data not shown).
Notably, we consistently observed a stronger effect in males than in females, most likely reflecting a difference in the level of hairpin expression. The origin of this effect is unclear and could be due to a difference in developmental timing between males and females. Hairpin constructs may produce a higher abundance of dsRNAs in males because they develop slightly more slowly than females. Alternatively, this phenomenon may be due to dosage compensation, as the transgenes contain X-linked sequences of the white intron and the vermilion gene as well as being inserted in an attP site flanked by the yellow gene that originates from the X chromosome.
Compared to P-element–based transgenic RNAi, the targeted insertion RNAi method has several practical advantages. The frequency at which transformants are recovered using the integrase method is almost fivefold higher than that obtained using conventional P-element transformation3,5 (unpublished observations). No mapping of the transformants to a specific chromosome is needed, and insertions into the attP2 landing site are homozygous viable. Of the 7,623 lines generated by R. Ueda et al. by P-element transformation, 54% are associated with lethality or semilethality when homozygous (numbers extracted from the information available at http://www.shigen.nig.ac.jp/fly/nigfly/). Similarly, among the 22,270 lines established by G. Dietzl et al. by the same mechanism, 5.4% were associated with sterility and 17.5% with lethality when homozygous (numbers extracted from Supplementary Table 2 of ref. 1). The inability to create homozygotes of P-element–integrated UAS-hairpin lines is a potential drawback to the transgenic hairpin technology, given that a twofold increase in gene expression can have a strong effect on phenotype (as discussed below). Finally, the high rate of false negatives, resulting from random integration into poorly expressed loci, is automatically eliminated by inserting RNAi constructs into an optimal site such as attP2.
To ask how temperature, the amount of Gal4 and the number of UASs affect the severity of hairpin-induced phenotypes, we carried out a number of tests using a sensitive wing assay whereby an RNAi construct against Notch is expressed under the control of the enhancer trap C96-Gal4 driver6. C96-Gal4 is expressed in the region of the wing imaginal disc that gives rise to the dorsal-ventral margin7, and a reduction of Notch activity at the margin generates dose-dependent phenotypes ranging from mild loss of margin bristles to strong wing notching (Fig. 2a). We generated two independent hairpins against Notch and examined their phenotypes when driven by C96-Gal4 at both 25 °C and 29 °C. As expected from our previous results, both Notch hairpin constructs behaved similarly (Supplementary Fig. 1 online). However, we observed marked differences in the expressivity of the phenotypes between males and females, as has been seen with other hairpin constructs (as discussed above). Further, the severity of the phenotypes was dependent on temperature, most likely reflecting the temperature sensitivity of the Gal4-UAS system8. Control hairpins did not produce any phenotype of note.
Figure 2.
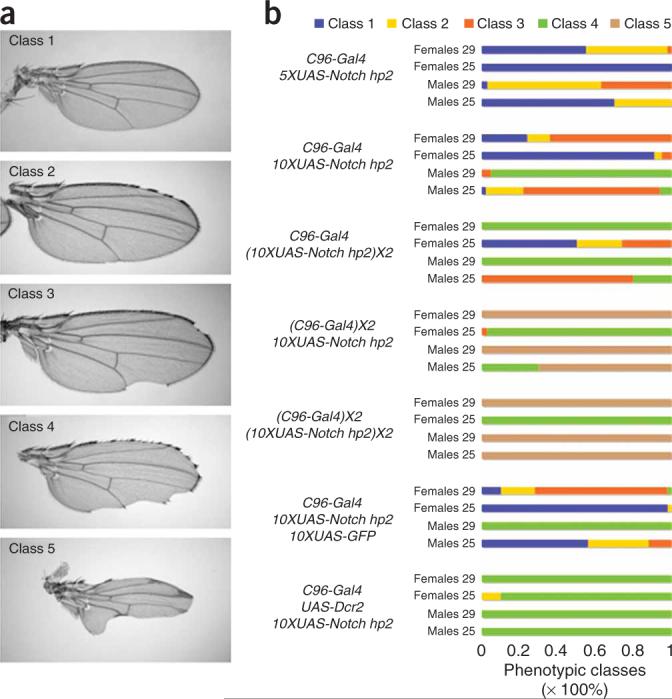
Parameters of in vivo RNAi using Valium. (a) Five classes of wing phenotypes were distinguished when a Notch hairpin was expressed using the wing margin C96-Gal4 driver. Class 1, wild type or a few bristles missing; class 2, margin bristles missing but no notches; class 3, moderate wing notching; class 4, extensive wing notching; class 5, most of the wing margin missing. (b) Phenotypes of C96-Gal4, Notch-hp2 flies. The phenotypes are stronger at 29 °C than at 25 °C and in males than in females; the two hairpins have similar expressivity. The effect of varying the number of UASs, the amounts of Gal4 and Dcr2 are shown using UAS-Notch hp2. More than 50 wings were scored in each experiment. Details of the construction of the various genotypes can be found in the Supplementary Note online.
Next, we compared the effect of 5×, 10× and 20× UAS-Notch hp2 in the presence of one copy of C96-Gal4. The 5× UAS line was generated as a derivative of the 10×UAS-Notch hp2 line after excision of the Cre-loxP cassette (Supplementary Methods). A recombinant chromosome was generated between C96-Gal4 and 10×UAS-Notch hp2 to produce the various genotypes shown in Figure 2b. Analyses of the various combinations revealed a continuous phenotypic series from the 5× to the 20× UAS hairpin dose, demonstrating that expressivity can be manipulated by altering the number of UAS sites in addition to the hairpin copy number. To examine the effect of varying the amount of Gal4, we compared the effects of one copy versus two copies of C96-Gal4 in the presence of one copy and two copies of 10×UAS-Notch hp2. In all situations in which the dose of C96-Gal4 was increased, a significant enhancement of the phenotype was observed, as we detected a large fraction of flies with the most extreme (Class 5) wing-notching phenotypes. Notably, little enhancement was observed between one copy and two copies of 10×UAS-Notch hp2 when two copies of C96-Gal4 were present. Further, doubling the amount of Gal4 had a more profound effect than doubling the number of UAS hairpins, indicating that the amount of Gal4 in these experiments is more limiting. This is consistent with the relatively weaker phenotype observed in C96-Gal4, 10×UAS-Notch hp2/10×-UASGFP compared to C96-Gal4, 10×UAS-Notch hp2/+, where presumably the amount of Gal4 available for activation of the 10× UAS is titrated by the 10× UASGFP (Fig. 2b). Note, however, that the effect is only clearly detected at 25 °C, which is consistent with the temperature sensitivity of the Gal4-UAS system.
In conclusion, in our experiments with C96-Gal4, the amount of Gal4 was more critical than the number of UASs for generating strong hairpin-induced phenotypes. It is important to note that because the C96-Gal4 line is a relatively weak driver (data not shown), we expect that, in conditions where the driver is stronger, the number of UASs may be the most important parameter to optimize in order to improve the severity of hairpin-based phenotypes.
Recently, Dietzl et al. reported that increasing the level of Dicer 2 (Dcr 2) increases the potency of the RNAi hairpin phenotype, most likely by increasing the processing of long dsRNAs1. Thus, we compared the phenotype of C96-Gal4, 10×UAS-Notch hp2/+ and UAS-Dcr2/+; C96-Gal4, 10×UAS-Notch hp2/+ flies. Consistent with these findings, overexpressing Dcr2 along with the hairpin construct increased the severity of the phenotype, but not as much as doubling the amount of Gal4 (Fig. 2b).
ACKNOWLEDGMENTS
We thank G. Rubin, C. Zuker, K. Moses and B. Mathey-Prevot for critical input on the project, B. Dickson (IMP, Vienna) for the gift of UAS-Dcr2 and F. Karch (University of Geneva) for nanos-integrase. M.M. is a fellow of the Jane Coffin Childs Memorial Fund. M.B. is supported by R01 GM067761 from the US National Institute of General Medical Sciences. This work was supported in part by the Janelia Farm Visitor program. N.P. is an investigator of the Howard Hughes Medical Institute.
Supplementary Material
References
- 1.Dietzl G, et al. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 2.Brand AH, Perrimon N. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 3.Groth AC, Fish M, Nusse R, Calos MP. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perrimon N, Mathey-Prevot B. Genetics. 2007;175:7–16. doi: 10.1534/genetics.106.069963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bischof J, Maeda RK, Hediger M, Karch F, Basler K. Proc. Natl. Acad. Sci. USA. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Presente A, Shaw S, Nye JS, Andres AJ. Genesis. 2002;34:165–169. doi: 10.1002/gene.10149. [DOI] [PubMed] [Google Scholar]
- 7.Gustafson K, Boulianne GL. Genome. 1996;39:174–182. doi: 10.1139/g96-023. [DOI] [PubMed] [Google Scholar]
- 8.Mondal K, et al. J. Mol. Biol. 2007;370:939–950. doi: 10.1016/j.jmb.2007.05.035. [DOI] [PubMed] [Google Scholar]
- 9.Fridell YW, Searles LL. Nucleic Acids Res. 1991;19:5082. doi: 10.1093/nar/19.18.5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siegal ML, Hartl DL. Genetics. 1996;144:715–726. doi: 10.1093/genetics/144.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.An X, Armstrong JD, Kaiser K, O'Dell KM. J. Neurogenet. 2000;14:227–243. doi: 10.3109/01677060009084500. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


